Background: Phosphorylation at Ser-776 of the polyglutamine disease-associated protein Ataxin-1 modulates its function.
Results: 14-3-3 binding stabilizes Ataxin-1 by blocking dephosphorylation of pS776 and impedes Ataxin-1 transport to the nucleus.
Conclusion: 14-3-3 must disassociate from Ataxin-1 for its transport to the nucleus.
Significance: 14-3-3 regulates Ataxin-1 function by protecting phosphorylation of Ser-776 and Ataxin-1 entry into the nucleus.
Keywords: Ataxia, Neurodegeneration, Nuclear Translocation, Phosphatase, PP2A, 14-3-3
Abstract
Spinocerebellar ataxia type 1 (SCA1) is a lethal neurodegenerative disorder caused by expansion of a polyglutamine tract in ATXN1. A prominent site of pathology in SCA1 is cerebellar Purkinje neurons where mutant ATXN1 must enter the nucleus to cause disease. In SCA1, phosphorylation of ATXN1 at Ser-776 modulates disease. Interestingly, Ser-776 is located within a region of ATXN1 that harbors several functional motifs including binding sites for 14-3-3, and splicing factors RBM17 and U2AF65. The interaction of ATXN1 with these proteins is thought to be regulated by the phosphorylation status of Ser-776. In addition, Ser-776 is adjacent to the NLS in ATXN1. Although pS776-ATXN1 is enriched in nuclear extracts of cerebellar cells, the vast majority of 14-3-3 is in the cytoplasmic fraction. We found that dephosphorylation of cytoplasmic pS776-ATXN1 is blocked by virtue of it being in a complex with 14-3-3. In addition, data suggest that binding of 14-3-3 to cytoplasmic ATXN1 impeded its transport to the nucleus, suggesting that 14-3-3 must disassociate from ATXN1 for transport of ATXN1 to the nucleus. Consistent with this hypothesis is the observation that once in the nucleus pS776 is able to be dephosphorylated. Evidence is presented that PP2A is the pS776-ATXN1 phosphatase in the mammalian cerebellum. In the nucleus, we propose that dephosphorylation of pS776-ATXN1 by PP2A regulates the interaction of ATXN1 with the splicing factors RBM17 and U2AF65.
Introduction
Expansion of a CAG trinucleotide repeat that encodes a polyglutamine tract within the protein ATXN13 causes SCA1, a fatal progressive neurodegenerative disease (1). Other neurodegenerative disorders that are also caused by expansions of polyglutamine tracts include Huntington disease, spinobulbar muscular atrophy (Kennedy disease), dentatorubral-pallidoluysian atrophy, and SCAs 2, 3 (Machado-Joseph disease), 6, 7, and 17 (2). CAG repeats in mutant SCA1 alleles vary from 39 to 82, with age of onset (ranging from 4 to 74 years of age) and severity of disease being inversely correlated with length of the repeat (1). A prominent site of neurodegeneration in SCA1 is Purkinje cells of the cerebellar cortex. In addition, subsets of neurons in the brainstem are affected as well (3).
SCA1 is caused by an expansion of the glutamine tract in ATXN1. Yet, considerable data indicate that ATXN1 residues outside of the glutamine tract have a substantial impact on severity of disease (4–11). Two highly conserved regions outside of the polyglutamine tract in ATXN1 that have such a role are the 120 residue ATXN1/HBP1 (AXH) domain, amino acids 570–689 (12), and a short stretch of amino acids, residues 771–778, at the C terminus (13, 14). While several transcription factors as well as RNA interact with ATXN1 via the AXH domain (6, 9, 15), the C-terminal region is of particular interest since the interaction of certain proteins with this region is impacted by length of the polyglutamine and/or phosphorylation of Ser-776 (11, 14). Moreover, data indicate that the phosphorylation status of Ser-776 has a critical role in regulating SCA1 pathogenesis (5, 16, 17). Transgenic mice expressing ATXN1[82Q] containing a phosphorylation resistant Ser to Ala (S776A) substitution fail to develop neurodegeneration (5). In contrast, the potentially phospho-mimicking amino acid substitution S776D enhances pathogenesis induced by ATXN1[82Q] and converts wild type ATXN1[30Q] to a pathogenic protein (17).
Ser-776 is positioned within or immediately adjacent to three functional motifs in the C terminus of ATXN1; the NLS, amino acids 771–774 (4), the 14-3-3 binding site, amino acids 774–776 (14, 16), and the U2AF-homology ligand motif (ULM), amino acids 771–776 (14). The ULM in ATXN1 binds to the U2AF-homology motifs (UHM) in RBM17 and U2AF65 (11, 14). Moreover, as seen for other ULM/UHM interactions (18), phosphorylation of Ser-776 in the ULM of ATXN1 seems to regulate its interaction with RBM17 (11) and U2AF65 (14). Thus, understanding how phosphorylation of Ser-776 is regulated is critical for understanding the biology of ATXN1.
The phosphorylation state of a protein is a dynamic process dictated by both protein kinases and phosphatases. Ser-776 of ATXN1 lies within strong consensus phosphorylation sites for the kinases AKR mouse thymoma (Akt) and cyclic AMP-dependent protein kinase (PKA). While initial data using tissue culture cells and a Drosophila model of SCA1 indicated that Akt can phosphorylate Ser-776 (16), more recent data suggests that PKA is the S776-ATXN1 kinase in the mammalian cerebellum (19). In this study, we utilized a mouse cerebellar extract-based dephosphorylation assay to investigate the dephosphorylation of pS776-ATXN1. We present data indicating that the protein phosphatase 2A (PP2A) dephosphorylation of pS776-ATXN1 is restricted to the nucleus of cerebellar cells. In addition, data indicate that binding of 14-3-3 to pS776-ATXN1 is restricted to the cytoplasm and impedes the dephosphorylation of pS776 as well as entry of ATXN1 into nuclei of cerebellar cells. These data, along with pervious data (19), support a model in which phosphorylation and dephosphorylation of ATXN1 at Ser-776 take place in separate subcellular compartments in the cerebellum. The binding of 14-3-3 to pS776-ATXN1 protects against dephosphorylation in the cytoplasm. Moreover, the pS776-ATXN1/14-3-3 complex must be disassociated in order for ATXN1 to be transported to the nucleus, implying that this disassociation is regulated.
MATERIALS AND METHODS
Reagents
Reagent sources were: okadaic acid (Calbiochem); antibodies to IKB-α (1:2500) and histone-3 (1:5000) (Cell Signaling), 14-3-3 ϵ (1:500), β (1:10,000), ζ (1:10,000) (Santa Cruz Biotechnology), PP2A (1:2000) (Millipore), PP1 (1:4000) (Santa Cruz Biotechnology), PP5 (1:4000) (Bethyl Laboratory), PP4 (1:2000) (R&D Systems), α-Tubulin (1:20000) (Sigma), Lamin b (1:5000) (Millipore); purified PP2A A/C (14-111) (Millipore); purified PP1 (14-110) (Millipore); protein G-Sepharose beads (GE Healthcare); glutathione-agarose resin (Invitrogen); control siRNA, PP2A siRNA, 14-3-3 ϵ, ζ siRNA (Dharmacon); ATXN1 antibody 11750 (1:5000) (20) and phospho-S776 specific ATXN1 antibody PN1248 (1:1000) were as described previously (5). Glutathione S-transferase (GST) and GST-ATXN1 were purified as described (10).
Cerebellar/DAOY Cytoplasmic and Nuclear Fractionation
DAOY nuclear and cytoplasmic extracts were prepared using the NE-PER kit (Pierce) according to the manufacturer's protocol. Cerebellar nuclear and cytoplasmic extracts were also prepared using the NE-PER kit according to the manufacturer's protocol with slight modifications. Briefly, cerebella were homogenized in Cytoplasmic Extraction Reagent I, vortexed, incubated on ice for 10 min, Cytoplasmic Extraction Reagent II added, vortexed, incubated on ice for 1 min and votexed again. Extracts were centrifuged for 5 min at 16,000 × g and supernatant was saved as the cytoplasmic extract. Pellets were washed once with 200 μl of ice-cold PBS, suspended in ice-cold Nuclear Extraction Reagent, vortexed extensively and centrifuged for 10 min (16,000 × g). Supernatants were collected and saved as nuclear extract. IKB-α and histone-3 were used as cerebellar cytoplasmic and nuclear markers, respectively. α-Tubulin and Lamin b were used as DAOY cytoplasmic and nuclear markers, respectively.
Dephosphorylation Assays
For the cerebellar cytoplasmic and nuclear dephosphorylation assays, aliquots of cytoplasmic and nuclear extracts prepared in absence of phosphatase inhibitors were incubated at 37 °C. Dephosphorylation was terminated by addition of sample buffer and heating in boiling water for 5 min. Aliquots of cytoplasmic and nuclear extracts were electrophoresed using Bis-Tris polyacrylamide gels (Invitrogen). Western blots were generated as described (19) and probed for pS776-ATXN1 with the pS776 specific antibody PN1248, stripped, and probed with the ATXN1 antibody 11750.
Disassociation of ATXN1/14-3-3 complexes in cytoplasmic extracts was accomplished by adding the high affinity 14-3-3 binding 20-mer peptide R18 (AnaSpec), a potent general inhibitor of 14-3-3/ligand interactions (21). R18 was added to each aliquot of cytoplasmic extract to a final concentration of 25 μm.
Co-immunoprecipitation and Immunodepletion
Mouse cerebella were homogenized in a buffer containing 20 mm Tris, pH 7.5, 150 mm NaCl, 1% Nonidet P-40, protease inhibitors (Complete® EDTA free; Roche Diagnostics), and phosphatase inhibitors (Cocktails 1 and 2, Sigma), and extracted for 40 min on ice. After centrifugation, the supernatant was recovered, and protein concentration was measured by the Bradford assay. For co-immunoprecipitation assays, 0.5 mg of protein was diluted with lysis buffer to 500 μl. Lysate was precleared for 1 h with 50 μl of protein G-Sepharose bead slurry. After centrifugation, the precleared supernatant was rocked overnight at 4 °C with the PP2A catalytic subunit antibody. The next day, 50 μl of bovine serum albumin (BSA)-blocked protein G-Sepharose beads were added to co-immunoprecipitate the antibody-antigen complex. After centrifugation, the pellet was washed four times with lysis buffer, suspended in sample buffer, boiled and supernatant analyzed by PAGE. Negative controls for co-immunoprecipitations were lysates prepared under the same conditions without addition of antibodies.
For the immunodepletion of PP2A catalytic subunit from the nuclear extract, 0.05 mg of protein was diluted to 150 μl with the lysis buffer. The lysate was precleared with protein G-Sepharose beads and spun. PP2A catalytic subunit antibody was added to the supernatant to immunodeplete PP2A. After 3 h with rocking at 4 °C BSA-blocked protein G-Sepharose beads were added to immunoprecipitate the antibody-antigen complex. After centrifugation the post-immunodepletion supernatant (post-ID supernatant) was kept on ice. Aliquots of post-ID supernatant were incubated at 37 °C for varying times. In the immunodepletion assay, nuclear extracts contained a high concentration of salts (400 mm) and were vortexed four times at high-speed to disrupt PP2A/ATXN1 complexes.
GST Pull-downs
For pull-down analyses, recombinant GST-ATXN1 immobilized on glutathione-agarose was incubated with cerebellar lysate for 3 h at 4 °C with constant mixing in co-immunoprecipitation buffer as described above. After centrifugation, protein complexes bound to the agarose were washed three times with the lysis buffer. Bound proteins were resolved by Bis-Tris protein PAGE and immunoblotted with phosphatase-specific antibodies.
siRNA Knockdown Assay
Hela and DAOY cells were maintained in Dulbecco's modified Eagle's medium (GIBCO) containing 10% fetal bovine serum (Invitrogen), 1% penicillin/streptomycin (Invitrogen) at 37 °C in 5% CO2. Hela cells were transfected with control siRNA or siRNA targeted to PP2A catalytic subunit using Qiagen HiPerFect transfection Reagent. 24 h post-transfection, cells were transfected with ATXN1 plasmid using Invitrogen Lipofectamine and Plus reagent. 48 h after ATXN1 plasmid transfection, cells were lysed, and the lysate loaded onto a gel for Western blotting.
DAOY cells were transfected with control siRNA or combined 14-3-3 ϵ and ζ siRNA. 72 h post-transfection, aliquots of cytoplasmic and nuclear extracts were electrophoresed using Bis-Tris polyacrylamide gels. Western blotting was performed as described above. Protection of pS776-ATXN1 from PP2A by 14-3-3 in the cytoplasm was tested by knockdown of 14-3-3 and incubating the cytoplasmic extracts prepared in absence of phosphatase inhibitors at 37 °C for varying time.
In Vitro Phosphatase Assay
Purified GST-ATXN1[30Q] was phosphorylated by purified PKA in kinase buffer at 30 °C for 1 h (19). Phosphorylated GST-ATXN1 was collected by GST-pull-down with glutathione-agarose resin. The resin containing phosphorylated GST-ATXN1 was incubated with 0.15 unit purified PP2A core enzyme or PP1 enzyme for varying time at 30 °C in a reaction mixture consisting of 20 mm Tris, pH 7.5, 1 mm DTT, and protease inhibitors. The reaction was stopped by adding sample buffer and heating in boiling water for 5 min. Samples were subject to SDS-PAGE and Western blot analysis using the pS776-specific antibody PN1248 and ATXN1 antibody 11750.
Statistical Analysis
Data are expressed as the means ± S.E. Statistical comparisons were made using a Student's t test.
RESULTS
pS776-Atxn1 Phosphatase Activity Is Restricted to Nuclei of Cerebellar Cells
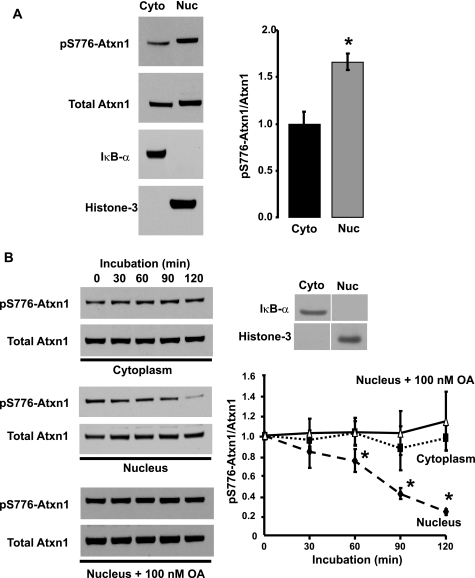
We previously showed that the ability to phosphorylate Atxn1 at Ser-776 localized to a cerebellar cytoplasmic extract and was not present in a nuclear extract (19). To explore further the subcellular distribution of the components of the pS776-Atxn1 pathway, we determined the subcellular distribution of endogenous pS776-Atxn1 in the mouse cerebellum. Fig. 1A shows that, relative to the subcellular distribution of total Atxn1, pS776-Atxn1 was enriched in a cerebellar nuclear fraction. Next the subcellular distribution of pS776 dephosphorylation activity was assessed. To accomplish this, we first determined if pS776-Atxn1 could be dephosphorylated by a mouse cerebellar lysate. After a 1-h incubation at 37 °C, the amount of endogenous pS776-Atxn1 detected was substantially reduced (supplemental Fig. S1). Importantly, adding phosphatase inhibitors at the onset of the incubation blocked the reduction in amount of pS776-Atxn1 detected with incubation (supplemental Fig. S1). These results indicated a pS776 phosphatase was present in cerebellar lysates.
FIGURE 1.
Subcellular distribution of endogenous pS776-Atxn1 and pS776 dephosphorylation activity in the cerebellum. A, subcellular distribution of pS776-Atxn1 in the cerebellum was determined relative to that of total Atxn1 by Western blot analyses of cytoplasmic and nuclear fractions. Purity of the cytoplasmic and nuclear fractions were assessed by probing for IκB-α (a cytoplasmic-specific marker) and histone-3 (a nuclear-specific marker). Quantitative analysis of the data is depicted to the right, n = 3. *, p < 0.05; nuclear versus cytoplasmic levels of pS776-Atxn1. B, cerebellar cytoplasmic and nuclear fractions were assessed for their ability to dephosphorylate endogenous pS776-Atxn1. Dephosphorylation of pS776-Atxn1 occurred only in the nuclear extract, which was inhibited by 100 nm OA. Assessment of cytoplasmic and nuclear fraction purity (IκB-α, a cytoplasmic-specific marker and histone-3, a nuclear-specific marker) and quantitative analysis of the data are depicted to the right, n = 3. *, p < 0.05; nuclear versus cytoplasmic pS776-Atxn1 dephosphorylation.
To determine the subcellular site of pS776 dephosphorylation activity, mouse cerebellar cytoplasmic and nuclear extracts were incubated in the absence of phosphatase inhibitors at 37 °C. As shown in Fig. 1B, endogenous pS776-Atxn1 levels in the cytoplasmic fraction remained constant over the 2-h incubation period, indicating that dephosphorylation did not occur in the cytoplasmic fraction. In contrast, the amount of pS776-Atxn1 decreased in the nuclear fraction with incubation at 37 °C. Importantly, the reduction in nuclear pS776-Atxn1 was blocked by 100 nm okadaic acid (OA), a serine/threonine phosphatase inhibitor (22) (Fig. 1B). From these results we concluded that the endogenous pS776-Atxn1 phosphatase activity is restricted to nuclei of cerebellar cells.
Association of 14-3-3 Blocks Dephosphorylation of Cytoplasmic pS776-Atxn1
Data presented above raises the question as to why pS776-Atxn1 was unable to be dephosphorylated in the cytoplasm of cerebellar cells? Earlier work showed that pS776 is a binding site for 14-3-3 (13, 16). It is known that 14-3-3 binds to phosphoserine as well as phosphothreonine motifs in a variety of cellular proteins, thus inhibiting their dephosphorylation (23). Therefore, we next determined the cerebellar subcellular distribution of the three isoforms of 14-3-3 previously shown to bind pS776-Atxn1, 14-3-3ϵ, 14-3-3β, and 14-3-3ζ (16). The vast majority of all three of these isoforms of 14-3-3 localized to the cytoplasmic fraction of cerebellar cells (Fig. 2A). Thus, the subcellular distribution of these isoforms of 14-3-3 inversely correlated with the subcellular distribution of pS776-Atxn1 phosphatase activity, raising the possibility that the high levels of 14-3-3 in the cytoplasm underlie the inability of cytoplasmic pS776-Atxn1 to be dephosphorylated.
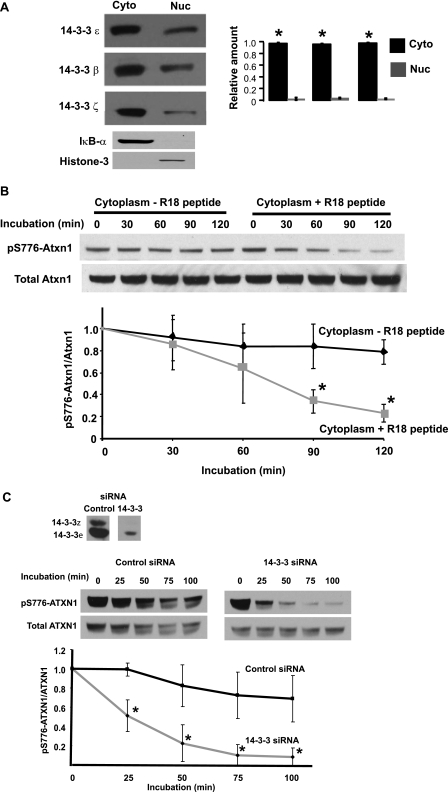
FIGURE 2.
14-3-3 binding inhibits the dephosphorylation of pS776-Atxn1 in the cytoplasm of cerebellar cells. A, Western blot analysis of the subcellular distribution of the 14-3-3ϵ, β, and ζ isoforms in the cerebellum, showing that each is highly enriched in the cytoplasmic fraction. B, dephosphorylating activity of pS776-Atxn1 in cerebellar cytoplasmic extracts in which the Atxn1/14-3-3 complexes were intact compared with extracts in which the complexes were disrupted by the addition of peptide R18, a general inhibitor of 14-3-3/ligand interaction. Cytoplasmic extracts prepared and used to assess dephosphorylating activity with incubation at 37 °C. Levels of pS776-Atxn1 and total Atxn1 were determined using the PN1248 and 11750 antibodies, respectively. The quantitative analysis of the data is depicted below. The error bar indicates ± S.E., n = 3, *, p < 0.05. C, dephosphorylating activity of pS776-ATXN1 in DAOY cells in which endogenous 14-3-3 levels were reduced using a siRNA. DAOY cells were transfected with a control or 14-3-3 siRNA that reduce endogenous levels of 14-3-3ϵ and 14-3-3ζ. Cytoplasmic extracts prepared and used to assess dephosphorylating activity with incubation at 37 °C. Levels of pS776-ATXN1 and total ATXN1 were determined using the PN1248 and 11750 antibodies, respectively. Quantitative analysis of the Western blot data is depicted below. The error bar indicates ± S.E., n = 3, *, p < 0.05.
To examine whether association of 14-3-3 with Atxn1 interferes with dephosphorylation of pS776, we utilized the R18 peptide to disrupt the Atxn1/14-3-3 complex in a cerebellar cytoplasmic extract. R18 is a 20-mer peptide that specifically binds to all isoforms of 14-3-3 and competitively interferes with the 14-3-3/ligand interactions (21). Upon addition of R18 to a cerebellar cytoplasmic extract, endogenous pS776-Atxn1 was dephosphorylated with incubation at 37 °C (Fig. 2B).
Next we used siRNA to examine whether depleting 14-3-3 from human DAOY cells altered dephosphorylation of endogenous pS776-ATXN1. As shown in Fig. 2C, transfecting DAOY cells with siRNAs targeting both 14-3-3ϵ and ζ reduced 14-3-3 protein levels with a corresponding increase in the ability of cytoplasmic pS776-ATXN1 to be dephosphorylated. Together, these results indicate that 14-3-3 bound to pS776-Atxn1/ATXN1 in the cytoplasm inhibited dephosphorylation of S776. The relatively low amount of 14-3-3 in the nuclei of cerebellar cells (Fig. 2A), we suggest, is insufficient to affect dephosphorylation of pS776-Atxn1/ATXN1 in this compartment.
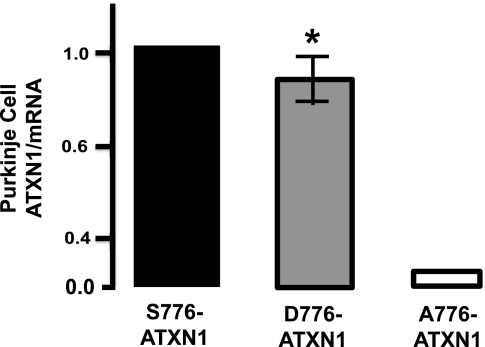
In a previous study, it was suggested that 14-3-3 binding to ATXN1 stabilized ATXN1 and slowed its degradation (16). Consistent with this is the observation that in vivo A776-ATXN1, which is resistant to phosphorylation and thus does not bind to 14-3-3 (16), is considerably less stable than S776-ATXN1 (19), which upon phosphorylation is able to bind 14-3-3 (16). These results raise the question whether phosphorylation is in itself sufficient to stabilize ATXN1 or must pS776-ATXN1 also bind 14-3-3 to be stabilized. To approach this question, the stability of phospho-mimicking D776-ATXN1 in Purkinje cells in vivo was examined. To accomplish this, transgenic mice with Purkinje cell-specific expression of S776- and D776-ATXN1 generated using the Pcp2/L7 regulatory region (17) were crossed to Sca1−/− mice (24) to eliminate contamination of measurements by endogenous Atxn1. The ratio of protein to mRNA in cerebellar extracts was used as a measure of the relative stabilities of S776-ATXN1 and D776-ATXN1. As shown in Fig. 3, the ratio of ATXN1 protein to RNA in D776-ATXN1 mice was comparable to that in S776-ATXN1 mice indicating the two proteins have similar stabilities. This is in marked contrast to A776-ATXN1 mice where the protein to RNA is dramatically reduced indicative of a considerably less stable protein (Fig. 3 and Ref. 19). Because D776-ATXN1 does not bind to 14-3-3 (13), these results suggest that a phospho-mimicking residue at position 776 is sufficient to generate a stable form of ATXN1.
FIGURE 3.
Effect of the amino acid at position 776 on stability of ATXN1 in vivo. The ratio of ATXN1 protein to ATXN1 RNA levels in mice expressing S776-ATXN1 and D776-ATXN1. RNA and protein levels were determined by Western and qPCR, respectively. Data for A776-ATXN1 is from Ref. (19). Error bars indicate ± S.E., n = 3, *, p < 0.05.
PP2A and Dephosphorylation of pS776-Atxn1
Toward identifying the pS776 phosphatase in the cerebellum, we generated a dose response curve for cerebellar nuclear extract dephosphorylation of pS776-Atxn1 (Fig. 4A). This analysis indicated an IC50 between 1 and 10 nm for the cerebellar nuclear pS776-Atxn1 phosphatase.
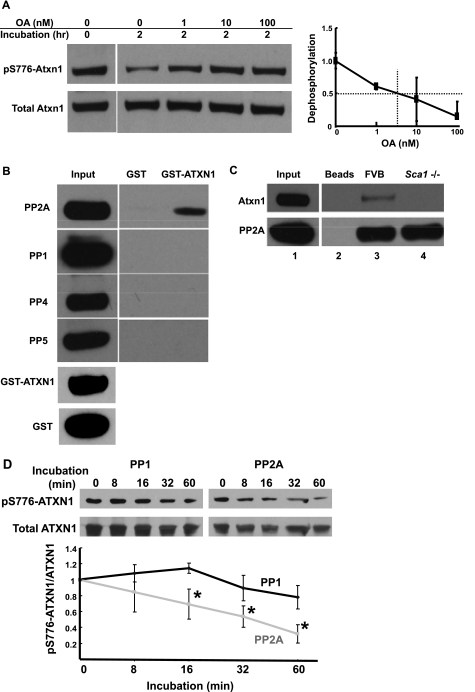
FIGURE 4.
Evidence supporting PP2A as the pS776-ATXN1/Atxn1 phosphatase. A, inhibition of endogenous nuclear pS776-Atxn1 dephosphorylation with varying concentrations of OA (0–100 nm). The resulting dose response curve for OA inhibition of nuclear pS776-Atxn1 dephosphorylation is at the right. Error bars indicate ± S.E., n = 3, *, p < 0.05. B, Western blots showing that PP2A but not PP1 or PP4 or PP5 in a cerebellar lysate bound to exogenous GST-ATXN1. GST-ATXN1 brought down 33% of input PP2A. C, co-immunoprecipitation of endogenous Atxn1 from a wild type FVB mouse cerebellar lysate using an antibody directed against PP2A catalytic subunit (lane 3). Lane 1 depicts the input amount of Atxn1. Control immunoprecipitations included beads without PP2A antibody (lane 2) and a PP2A immunoprecipitation using extract from a Sca1−/− mouse (lane 4). D, pS776-ATXN1 is dephosphorylated by exogenous PP2A but not by PP1. pS776-ATXN1 was detected using antibody PN1248 and total ATXN1 by immunoblotting with antibody 11750. Quantitative analysis of Western blots data is depicted below. Error bars indicate ± S.E., n = 3, *, p < 0.05.
Several phosphatases are reported to have IC50 values for OA in the nm range (22). Because protein phosphatases often form relatively stable complexes with their substrates (24), GST-ATXN1 pull-downs were used to test whether a specific cerebellar phosphatase can complex with ATXN1. Cerebellar phosphatases brought down by GST-ATXN1 were identified by probing Western blots with antibodies against each of the phosphatases having an IC50 for OA in the nm range and expressed highly in the cerebellum (25–28). The results indicated that while PP2A catalytic subunit was brought down by GST-ATXN1, other phosphatases expressed highly in the cerebellum, PP1, PP4, and PP5, did not form detectable complexes with GST-ATXN1 (Fig. 4B). Quantitation of the ratio of PP2A interacting with GST-ATXN1 to total PP2A indicated that interacting fraction was 0.33 of total PP2A, indicating that a substantive proportion of endogenous PP2A was in a complex with ATXN1. Moreover, endogenous Atxn1 could be co-immunoprecipitated along with endogenous PP2A from a wild type mouse cerebellar lysate (Fig. 4C). The capability of PP2A to dephosphorylate pS776 was tested directly by incubating pS776-ATXN1 with the PP2A core enzyme. As a control, the ability of PP2A to dephosphorylate pS776-ATXN1 was compared with PP1, a phosphatase for which the evidence indicated did not interact with ATXN1 (Fig. 4B). Within 16 min at 30 °C, PP2A decreased the level of pS776-ATXN1 while PP1 did not (Fig. 4D).
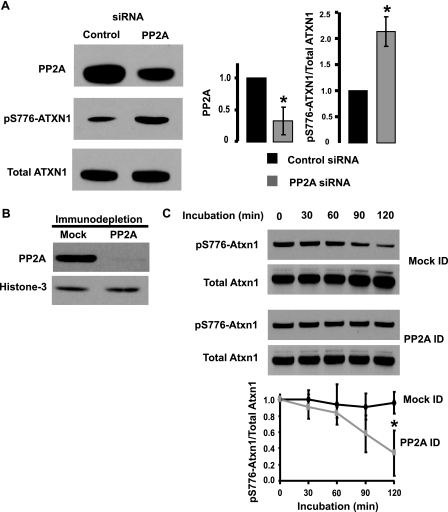
To obtain further evidence in support of PP2A as the pS776-ATXN1 phosphatase, a siRNA specific to the mRNA encoding the catalytic subunit of PP2A was used to deplete endogenous PP2A levels in Hela cells co-transfected with a plasmid encoding ATXN1. Fig. 5A shows that the PP2A siRNA effectively reduced the amount of PP2A in HeLa cells to a degree sufficient to significantly increase the amount of pS776-ATXN1 obtained from transfected HeLa cells.
FIGURE 5.
Depletion of PP2A decreases endogenous pS776-ATXN1 phosphatase activity. A, siRNA-mediated depletion of PP2A increases the amount of transfected pS776-ATXN1 in HeLa cells. Western blotting was used to assess PP2A and pS776-ATXN1 in HeLa cells transfected with control and PP2A siRNAs. ATXN1 amounts were used as a loading control. Quantitative analysis of Western blots data is presented to the right. Error bars indicate ± S.E., n = 3, *, p < 0.05. B, immunodepletion of PP2A from a cerebellar nuclear fraction. Western blot analysis of PP2A in PP2A-immunodepleted and mock depleted cerebellar nuclear extracts with histone-3 levels as loading controls. C, immunodepletion of PP2A from a cerebellar nuclear fraction decreases the dephosphorylation of endogenous pS776-Atxn1. Dephosphorylation of pS776-Atxn1 in cerebellar nuclear extracts either mock or immunodepleted for PP2A assessed by Western blotting. Levels of pS776-Atxn1 and total Atxn1 were determined using the PN1248 and 11750 antibodies, respectively. Quantitative analysis of the relative amount of pS776-Atxn1 is depicted below. Error bars indicate ± S.E., n = 3, *, p < 0.05.
As a means of assessing more directly whether PP2A is the phosphatase that dephosphorylates pS776-Atxn1 in the cerebellum, we utilized an immunodepletion strategy. In contrast to the PP2A/Atxn1 co-immunoprecipitations (Fig. 5C), we selected buffer and cell-lysing conditions that disrupted PP2A/ATXN1 complexes (see “Materials and Methods” for details). The catalytic subunit of PP2A was immunodepleted from a cerebellar nuclear extract (Fig. 5B) and the ability of the PP2A-depleted extract to dephosphorylate endogenous pS776-Atxn1 was evaluated. As presented in Fig. 5C immunodepletion of PP2A substantially reduced the ability of a cerebellar nuclear extract to dephosphorylate endogenous pS776-Atxn1. In contrast, the mock-treated nucleus retained the ability to dephosphorylate pS776-Atxn1, demonstrating that the buffer conditions used to weaken the interaction between PP2A and Atxn1 did not ablate the dephosphorylation. These data provided direct evidence supporting that PP2A as the cerebellar phosphatase for pS776-Atxn1.
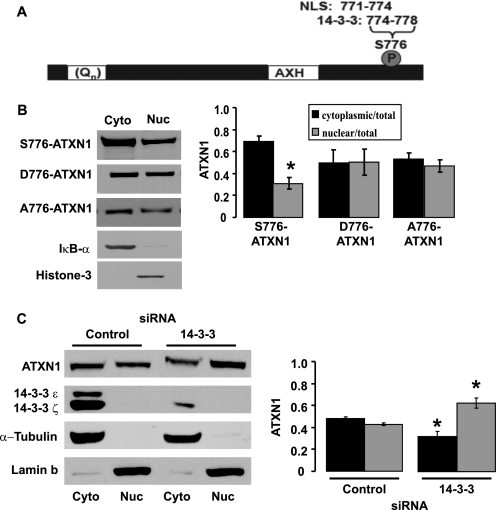
14-3-3: A Regulator of ATXN1 Transport to the Nucleus
As depicted in Fig. 6A, the 14-3-3 binding site involving pS776, is immediately adjacent to the NLS in ATXN1. This relationship of a 14-3-3 binding site in proximity to the NLS in ATXN1 is similar to that seen in Cdc25 where 14-3-3 binding to Cdc25 masks the nearby NLS and suppresses nuclear entry of Cdc25 (29). To investigate whether 14-3-3 association with pS776-ATXN1 affects localization of ATXN1 to the nucleus, we examined the cerebellar cytoplasmic and nuclear distribution of ATXN1 in mice expressing forms of ATXN1 with varied ability to bind 14-3-3. These included mice with Purkinje cell expression of S776-ATXN1 that binds to 14-3-3 (16) and mice with Purkinje cell expression of two forms of ATXN1 that do not bind to 14-3-3, A776-ATXN1(16) and D776-ATXN1 (13). In all cases mice expressing these forms of transgenic ATXN1 were bred to Sca1−/− mice to eliminate complications due to endogenous murine Atxn1. In mice with Purkinje cells expressing a form of ATXN1, S776-ATXN1, which binds 14-3-3 the amount of ATXN1 in the cytoplasmic fraction was twice that in the nuclear fraction (Fig. 6B). In contrast, when a form of ATXN1 that does not bind 14-3-3, A776-ATXN1 and D776-ATXN1, was expressed in Purkinje cells there was an even distribution of ATXN1 in cytoplasmic and nuclear fractions (Fig. 6B). Thus, the ability of 14-3-3 to bind ATXN1 provides a cytoplasmic anchor controlling subcellular distribution.
FIGURE 6.
Association of 14-3-3 with ATXN1 impedes its entry into the nucleus. A, diagram of ATXN1 indicating the polyglutamine tract (Qn), the AXH domain and Ser-776. In addition, the location of two functional motifs, the nuclear localization signal (NLS) and the 14-3-3 binding motif, are depicted. B, inability to bind 14-3-3 increases the amount of ATXN1 in the nucleus of Purkinje cells. Representative Western blots showing the subcellular distribution of S776-ATXN1 (able to bind 14-3-3), D776-ATXN1 (does not bind 14-3-3), and A776-ATXN1 (does not bind 14-3-3) in the cerebellum of transgenic mice are depicted. In each case the Pcp2-ATXN1 transgene was crossed onto Sca−/− background. This approach enabled analyses of the subcellular distribution of the ATXN1 transgenic protein specifically in Purkinje cells to be examined. The quantitative analysis of the subcellular distribution of ATXN1 is depicted to the right. Error bars indicate ± S.E., n = 3, *, p < 0.05. C, siRNA-mediated depletion of endogenous 14-3-3 in DAOY cells increases presence of endogenous ATXN1 in the nucleus. Representative Western blots showing the subcellular fractionation of ATXN1 upon 14-3-3 depletion by siRNA. Purity of cytoplasmic and nuclear fractions was assessed by blotting for α-tubulin and lamin b, respectively. Quantitative analysis of the subcellular distribution of ATXN1 is depicted to the right. Error bars indicate ± S.E., n = 3, *, p < 0.05.
Next, we utilized a siRNA strategy to examine whether depletion of endogenous 14-3-3 affects the subcellular distribution of ATXN1 in DAOY cells, a human medulloblastoma cell line that expresses ATXN1. Medulloblastoma cells are derived from pluripotent, neuroepithelial stem cells of the cerebellum, a region affected in SCA1. Transfection of siRNAs to 14-3-3ϵ and 14-3-3ζ, two isoforms of 14-3-3 expressed by DAOY cells and shown to bind to pS776-ATXN1 (16), decreased 14-3-3 protein and significantly decreased the fraction of ATXN1 in the cytoplasm and correspondingly increased the proportion of endogenous ATXN1 localized to the nucleus (Fig. 6C). Together these data from the SCA1 transgenic mice and DAOY cells suggest that 14-3-3 binding to ATXN1 impedes its entry into the nucleus. An implication of these data is that for ATXN1 to be transported to the nucleus it must first dissociate from 14-3-3.
DISCUSSION
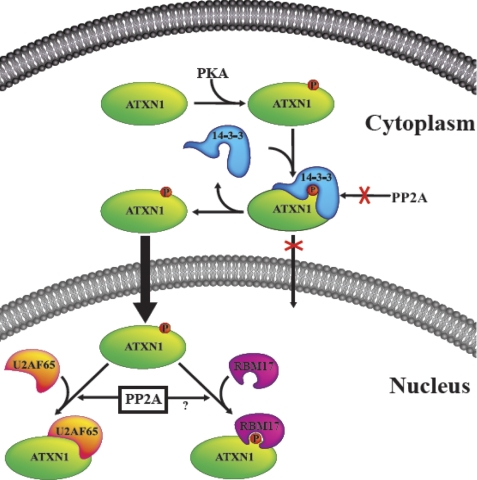
ATXN1, the protein affected in the polyglutamine neurodegenerative disease SCA1, is phosphorylated at Ser-776 (5, 31). Phosphorylation of Ser-776 in ATXN1 occurs in the cytoplasm (19), creating a binding site for members of a family of small acidic proteins collectively designated as the 14-3-3 proteins (13, 16). The functional consequence(s) of 14-3-3 binding to ATXN1 was previously unclear. Earlier work indicated that binding of 14-3-3 to pS776-ATXN1 stabilized ATXN1 (16) with a subsequent study suggesting that 14-3-3 binding also regulates the interaction of ATXN1 with the splicing factors RBM17 and U2AF65 in the nucleus (13). Here we provide evidence for two functional consequences of 14-3-3 binding to pS776-ATXN1 that are likely to be restricted to the cytoplasm of cerebellar cells (Fig. 7). First, binding of 14-3-3 to cytoplasmic ATXN1 blocks dephosphorylation of pS776 by PP2A. Because unphosphorylated ATXN1 is more rapidly degraded than pS776-ATXN1, we propose that binding of 14-3-3 indirectly stabilizes ATXN1 by blocking pS776 dephosphorylation. In addition, binding of 14-3-3 to pS776-ATXN1 reduces the proportion of ATXN1 localized to the nucleus, presumably by masking the NLS in ATXN1 adjacent to Ser-776.
FIGURE 7.
A scheme depicting the subcellular compartmentalization of ATXN1 phosphorylation. ATXN1 is phosphorylated in the cytoplasm upon which 14-3-3 associates with ATXN1-pS776 and protects it from dephosphorylation in the cytoplasm. Association of 14-3-3 with ATXN1 also interferes with its transport to the nucleus. Thus, in order for ATXN1-pS776 to move to the nucleus, the ATXN1/14-3-3 complex is dissociated. In the nucleus, ATXN1 interacts with other nuclear proteins such as splicing factors RBM17 and U2AF65. It is suggested that these interactions in the nucleus are regulated by PP2A.
A previous study revealed that 14-3-3 bound to both wild type and expanded mutant ATXN1 in transfected cells, promoting accumulation of nuclear ATXN1 in the absence of detectable 14-3-3 co-localizing with nuclear ATXN1 (16). The evidence presented here suggests that any increase in nuclear ATXN1 is an indirect effect of increasing the amount of cytoplasmic ATXN1 by 14-3-3. Even though pS776-ATXN1 was enriched in the nucleus, data indicate that binding of 14-3-3 to endogenous wild type ATXN1 in the cerebellum was largely restricted to the cytoplasm. First, the vast majority of 14-3-3 in the cerebellum localized to the cytoplasmic fraction. Because protection of a target protein from dephosphorylation is a previously described function of 14-3-3 (32), the inability of cytoplasmic ATXN1 to be dephosphorylated in an in vitro assay is consistent with the majority of it being in a complex with 14-3-3 protecting it from dephosphorylation. Furthermore, either disrupting the cytoplasmic 14-3-3/ATXN1 complex by adding the competing R18 peptide or reducing the level of 14-3-3 by siRNA promoted pS776-ATXN1 dephosphorylation.
Earlier work shows that compared with S776-ATXN1 phospho-resistant A776-ATXN1 is relatively unstable in transfected cells (16) and in the cerebellum of transgenic mice (19). Here we showed that the stability of D776-ATXN1 in vivo, as assessed by relative ratio of protein to RNA, in the cerebellum of transgenic mice was comparable to the stability of S776-ATXN1. Because the majority of S776-ATXN1 is phosphorylated in the cerebellum (data not shown), these data argue that at least in terms of promoting the stability of ATXN1 D776 mimics pS776. Intriguingly, D776-ATXN1 is as stable as S776-ATXN1 even though it is unable to bind to 14-3-3 (13), suggesting that phosphorylation of Ser-776 is sufficient to stabilize ATXN1 and that 14-3-3 binding, by blocking dephosphorylation of pS776, protects ATXN1 from subsequent proteolysis in the cytoplasm.
In contrast to the cytoplasm of cerebellar cells, we found that nuclear 14-3-3 levels were dramatically lower and that nuclear ATXN1 was readily dephosphorylated. Both of these results are consistent with little to no ATXN1 in a stable complex with 14-3-3 in the nucleus. Moreover, we found that in transgenic mice D776- and A776-ATXN1, forms of ATXN1 not able to bind to 14-3-3 (13, 16), had a higher proportion of ATXN1 in the nuclear fraction than did mice expressing S776-ATXN1 that binds 14-3-3. In addition, siRNA-mediated knockdown of 14-3-3 in DAOY cells decreased the portion of endogenous ATXN1 in the cytoplasm and increased the fraction of ATXN1 in the nucleus. One should be cautious in concluding that the results obtained in DAOY cells accurately reflect the situation in Purkinje cells. DAOY cells are mitotic cells whereas the cerebellar neuronal cells are post-mitotic. Regardless, these data indicate that upon binding to pS776-ATXN1 14-3-3 masks the adjacent NLS and blocks transport of ATXN1 to the nucleus.
The ability to affect subcellular location and function of a protein by concealing the NLS is known for several 14-3-3 targets (23, 32, 33). In order for these target proteins to be transported into the nucleus 14-3-3 must disassociate. For example, in the case of the FoxO transcription factors 14-3-3 dissociation is mediated by dephosphorylation of the target protein (31). Our observation that cytoplasmic ATXN1 was not dephosphorylated unless its interaction with 14-3-3 was first disrupted argues against pS776 dephosphorylation as a means by which the ATXN1/14-3-3 complex is dissociated. The mitotic activator Cdc25 is suppressed by phosphorylation at Ser-287 and docking with 14-3-3. In this case, phosphorylation of Cdc25 at a second site, Thr-138, reduces its affinity for 14-3-3 and is required for release of 14-3-3 (33, 34). We speculate that like Cdc25, a second post-translational modification of ATXN1 is required for its discharge from 14-3-3. Besides enabling ATXN1 to enter the nucleus, a potential advantage of a second post-translational mark on ATXN1 is that once in the nucleus its low affinity for 14-3-3 would presumably persist. de Chiara et al. (13) recently reported, using Iso-Thermal Calorimetry, that the Kd of a pS776-ATXN1 peptide for 14-3-3 (0.4 nm) was two orders of magnitude less than for U2AF65 and RBM17 (35.8 and 40.0 μm, respectively). Thus, it is likely to be critical for ATXN1's affinity for 14-3-3 to remain reduced for it to effectively interact with splicing factors RBM17 and U2AF65 in the nucleus, even in the presence of small amounts of 14-3-3.
Last, several lines of evidence presented here support PP2A as the phosphatase that dephosphorylates pS776-ATXN1. In vitro PP2A was able to readily dephosphorylate pS776-ATXN1 and selective siRNA knockdown of PP2A increased the amount of pS776-ATXN1 in transfected HeLa cells. Notably, immunodepletion of PP2A decreased the ability of a cerebellar nuclear extract to dephosphorylate endogenous pS776-ATXN1. PP2A is a major brain serine/threonine phosphatase that participates in a variety of signaling pathways that modulate neuronal function and activity (23). Of note is the reported role of PP2A in the second step of pre-RNA splicing (36) given that ATXN1 interacts with RBM17, which is also implicated in regulating the second step of splicing (37). An important feature of PP2A activity, as well as that of PPPs in general, is the critical role regulatory subunits have in determining substrate specificity and cellular pattern of activity (23). In the brain, the subcellular localization and cellular expression of the variable B regulatory subunits of PP2A is known to vary (38). Of note, while Purkinje cells express both Bα and Bβ isoforms, cerebellar stellate and basket cells also express Bα with Bβ expression being restricted to Purkinje cells. In addition, Bα is found in the soma and nuclei of Purkinje cells while Bβ is not detected by immunohistochemistry in Purkinje cell nuclei.
In summary, we found that 14-3-3 binding to cytoplasmic pS776-ATXN1 regulates the dephosphorylation and proteolytic degradation of ATXN1 as well as nuclear entry of pS776-ATXN1, thus, providing mechanistic insight into how 14-3-3 modulates the function of ATXN1.
Supplementary Material
This research was supported, in whole or in part, by National Institutes of Health/NINDS Grant NS045667.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- ATXN1/Atxn1
- human Ataxin-1/mouse Ataxin-1
- SCA1
- spinocerebellar ataxia type 1
- NLS
- nuclear localization signal
- OA
- okadaic acid.
REFERENCES
- 1. Orr H. T., Chung M. Y., Banfi S., Kwiatkowski T. J., Jr., Servadio A., Beaudet A. L., McCall A. E., Duvick L. A., Ranum L. P., Zoghbi H. Y. (1993) Nat. Genet. 4, 221–226 [DOI] [PubMed] [Google Scholar]
- 2. Orr H. T., Zoghbi H. Y. (2007) Annu. Rev. Neurosci. 30, 575–621 [DOI] [PubMed] [Google Scholar]
- 3. Robitaille Y., Schut L., Kish S. J. (1995) Acta Neuropathol. 90, 572–581 [DOI] [PubMed] [Google Scholar]
- 4. Klement I. A., Skinner P. J., Kaytor M. D., Yi H., Hersch S. M., Clark H. B., Zoghbi H. Y., Orr H. T. (1998) Cell 95, 41–53 [DOI] [PubMed] [Google Scholar]
- 5. Emamian E. S., Kaytor M. D., Duvick L. A., Zu T., Tousey S. K., Zoghbi H. Y., Clark H. B., Orr H. T. (2003) Neuron 38, 375–387 [DOI] [PubMed] [Google Scholar]
- 6. Tsuda H., Jafar-Nejad H., Patel A. J., Sun Y., Chen H. K., Rose M. F., Venken K. J., Botas J., Orr H. T., Bellen H. J., Zoghbi H. Y. (2005) Cell 122, 633–644 [DOI] [PubMed] [Google Scholar]
- 7. Okazawa H., Rich T., Chang A., Lin X., Waragai M., Kajikawa M., Enokido Y., Komuro A., Kato S., Shibata M., Hatanaka H., Mouradian M. M., Sudol M., Kanazawa I. (2002) Neuron 34, 701–713 [DOI] [PubMed] [Google Scholar]
- 8. Tsai C. C., Kao H. Y., Mitzutani A., Banayo E., Rajan H., McKeown M., Evans R. M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 4047–4052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lam Y. C., Bowman A. B., Jafar-Nejad P., Lim J., Richman R., Fryer J. D., Hyun E. D., Duvick L. A., Orr H. T., Botas J., Zoghbi H. Y. (2006) Cell 127, 1335–1347 [DOI] [PubMed] [Google Scholar]
- 10. Serra H. G., Duvick L., Zu T., Carlson K., Stevens S., Jorgensen N., Lysholm A., Burright E., Zoghbi H. Y., Clark H. B., Andresen J. M., Orr H. T. (2006) Cell 127, 697–708 [DOI] [PubMed] [Google Scholar]
- 11. Lim J., Crespo-Barreto J., Jafar-Nejad P., Bowman A. B., Richman R., Hill D. E., Orr H. T., Zoghbi H. Y. (2008) Nature 452, 713–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. de Chiara C., Giannini C., Adinolfi S., de Boer J., Guida S., Ramos A., Jodice C., Kioussis D., Pastore A. (2003) FEBS Lett. 551, 107–112 [DOI] [PubMed] [Google Scholar]
- 13. de Chiara C., Menon R. P., Strom M., Gibson T. J., Pastore A. (2009) PLoS One 4, e8372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Carlson K. M., Melcher L., Lai S., Zoghbi H. Y., Clark H. B., Orr H. T. (2009) J. Neurogenetics 23, 313–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yue S., Serra H. G., Zoghbi H. Y., Orr H. T. (2001) Hum. Mol. Genet. 10, 25–30 [DOI] [PubMed] [Google Scholar]
- 16. Chen H. K., Fernandez-Funez P., Acevedo S. F., Lam Y. C., Kaytor M. D., Fernandez M. H., Aitken A., Skoulakis E. M., Orr H. T., Botas J., Zoghbi H. Y. (2003) Cell 113, 457–468 [DOI] [PubMed] [Google Scholar]
- 17. Duvick L., Barnes J., Ebner B., Agrawal S., Andresen M., Lim J., Giesler G. J., Zoghbi H. Y., Orr H. T. (2010) Neuron 67, 929–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Selenko P., Gregorovic G., Sprangers R., Stier G., Rhani Z., Krämer A., Sattler M. (2003) Mol. Cell 11, 965–976 [DOI] [PubMed] [Google Scholar]
- 19. Jorgensen N. D., Andresen J. M., Lagalwar S., Armstrong B., Stevens S., Byam C. E., Duvick L. A., Lai S., Jafar-Nejad P., Zoghbi H. Y., Clark H. B., Orr H. T. (2009) J. Neurochem. 110, 675–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Servadio A., Koshy B., Armstrong D., Antalffy B., Orr H. T., Zoghbi H. Y. (1995) Nat. Genet. 10, 94–98 [DOI] [PubMed] [Google Scholar]
- 21. Wang B., Yang H., Liu Y. C., Jelinek T., Zhang L., Ruoslahti E., Fu H. (1999) Biochemistry 38, 12499–12504 [DOI] [PubMed] [Google Scholar]
- 22. Swingle M., Ni L., Honkanen R. E. (2007) Meth. Mol. Biol. 365, 23–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fu H., Subramanian R. R., Masters S. C. (2000) Annu. Rev. Pharmacol. Toxicol. 40, 617–647 [DOI] [PubMed] [Google Scholar]
- 24. Matilla A., Roberson E. D., Banfi S., Morales J., Armstrong D. L., Burright E. N., Orr H. T., Sweatt J. D., Zoghbi H. Y., Matzuk M. M. (1998) J. Neurosci. 18, 5508–5516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Virshup D. M., Shenolikar S. (2009) Mol. Cell 33, 537–545 [DOI] [PubMed] [Google Scholar]
- 26. Hashikawa T., Nakazawa K., Mikawa S., Shima H., Nagao M. (1995) Neurosci. Res. 22, 133–136 [DOI] [PubMed] [Google Scholar]
- 27. Rossie S., Jayachandran H., Meisel R. L. (2006) Brain Res. 21, 1–11 [DOI] [PubMed] [Google Scholar]
- 28. Tanimuko H., Grundke-Iqbal I., Iqbal (2004) Mol. Brain Res. 126, 146–156 [DOI] [PubMed] [Google Scholar]
- 29. Deleted in proof.
- 30. Yang J., Winkler K., Yoshida M., Kornbluth S. (1999) EMBO J. 18, 2174–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huttin E. L., Jedrychowski M. P., Elias J. E., Goswami T., Rad R., Ceausoleil S. A., Villén J., Haas W., Sowa M. E., Gygi S. P. (2010) Cell 143, 1174–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Morrison D. K. (2008) Trends Cell Bio. 19, 16–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tzivion G., Dobson M., Ramakrishnan G. (2011) Biochim. Biophys. Acta, in press [DOI] [PubMed] [Google Scholar]
- 34. Margolis S. S., Perry J. A., Forester C. M., Nutt L. K., Guo Y., Jardim M. J., Thomenius M. J., Freel C. D., Darbandi R., Ahn J. H., Arroyo J. D., Wang X. F., Shenolikar S., Nairn A. C., Dunphy W. G., Hahn W. C., Virshup D. M., Kornbluth S. (2006) Cell 127, 759–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Deleted in proof.
- 36. Shi Y., Reddy B., Manley J. L. (2006) Mol. Cell 23, 819–829 [DOI] [PubMed] [Google Scholar]
- 37. Lallena M. J., Chalmers K. J., Llamazares S., Lamond A. I., Valcárcel J. (2002) Cell 109, 285–296 [DOI] [PubMed] [Google Scholar]
- 38. Strack S., Zaucha J. A., Ebner F. F., Colbran R. J., Wadzinski B. E. (1998) J. Comp. Neurol. 392, 515–527 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.