Abstract
Recent crystal structures of G protein-coupled receptors (GPCRs) show the remarkable structural diversity of extracellular loop 2 (ECL2), implying its potential role in ligand binding and ligand-induced receptor conformational selectivity. Here we have applied molecular modeling and mutagenesis studies to the TM4/ECL2 junction (residues Pro174(4.59)–Met180(4.66)) of the human gonadotropin-releasing hormone (GnRH) receptor, which uniquely has one functional type of receptor but two endogenous ligands in humans. We suggest that the above residues assume an α-helical extension of TM4 in which the side chains of Gln174(4.60) and Phe178(4.64) face toward the central ligand binding pocket to make H-bond and aromatic contacts with pGlu1 and Trp3 of both GnRH I and GnRH II, respectively. The interaction between the side chains of Phe178(4.64) of the receptor and Trp3 of the GnRHs was supported by reciprocal mutations of the interacting residues. Interestingly, alanine mutations of Leu175(4.61), Ile177(4.63), and Met180(4.66) decreased mutant receptor affinity for GnRH I but, in contrast, increased affinity for GnRH II. This suggests that these residues make intramolecular or intermolecular contacts with residues of transmembrane (TM) domain 3, TM5, or the phospholipid bilayer, which couple the ligand structure to specific receptor conformational switches. The marked decrease in signaling efficacy of I177A and F178A also indicates that IIe177(4.63) and Phe178(4.64) are important in stabilizing receptor-active conformations. These findings suggest that the TM4/ECL2 junction is crucial for peptide ligand binding and, consequently, for ligand-induced receptor conformational selection.
Keywords: 7-transmembrane Receptor, G Protein-coupled Receptors (GPCR), G Proteins, Peptide Hormones, Receptor Structure-Function, Signal Transduction
Introduction
G protein-coupled receptors (GPCRs),2 which comprise the largest family of cell surface receptors, are characterized by seven-transmembrane (7-TM) domains joined by three extracellular loops (ECLs) and three intracellular loops. It is of fundamental importance to understand how these receptors, which share a common architecture, are activated by a variety of structurally diverse ligands. In this way, therapeutics that are better targeted to mimic or block these molecules can be designed. The crystal structure of bovine rhodopsin provided the first structural insight into this family of receptors (1) and aided much of our understanding of the molecular mechanisms of GPCR activation, in combination with a number of biochemical and biophysical studies.
The gonadotropin-releasing hormone (GnRH) receptor is a member of the rhodopsin-like subfamily of GPCRs. GnRH I is released in pulses from the hypothalamus into the portal blood system where it is transported to the anterior pituitary gland. Here it binds to GnRH receptors expressed on gonadotrope cells, which predominantly causes Gq/11 activation. The resultant signaling pathway brings about the release of luteinizing hormone (LH) and follicle-stimulating hormone (FSH) to regulate steroidogenesis and gametogenesis. As such, GnRH analogues are used extensively to treat many hormone-dependent diseases, including infertility, endometriosis, benign prostatic hyperplasia, and breast cancer (2). However, it has become evident that GnRH also acts on many cells outside of the pituitary gland. There are two endogenous ligands, hypothalamic GnRH I and extrahypothalamic GnRH II, despite there being only one functional receptor subtype expressed in humans. Both ligands exert their effects through the same receptor subtype, although they exhibit distinct pharmacological and signaling profiles (3). Different physiological outcomes are proposed to be mediated by different receptor coupling to Gs (4) or Gi/o (5) in addition to the classical Gq/11 pathway, but this remains controversial (6). However, we have shown that the human GnRH receptor can activate other Gq/11-independent signaling pathways such as G12/13 (7). Hence, the divergent signaling is believed to be mediated by different receptor-active conformations induced by differential ligand-receptor interactions. Binding of GnRH I and II to the GnRH receptor may cause different intramolecular interactions to be broken, allowing the receptor to adopt varied conformations and thus enabling divergent signaling pathways. We have termed this ligand-induced selective signaling (3, 8, 9).
Investigations into the structure of the GnRH receptor and identification of ligand binding contacts between GnRH and its receptor will enable refinement of molecular models for structure-based drug design. Previous site-directed mutagenesis studies and modeling have identified multiple contact sites of the GnRH receptor with its ligands (2, 3, 10–12). As these peptide ligands are larger than many biogenic amines, the binding pocket for GnRH is located toward the extracellular side of the receptor rather than buried further down the TM domains. It is likely to involve interactions with the extracellular loops as well as the TM domains. Interestingly, the molecular model of GnRH I docking to the human GnRH receptor suggests a potential interaction between Trp3 of the ligand with a region at the extracellular end of TM4 leading into ECL2 (12).
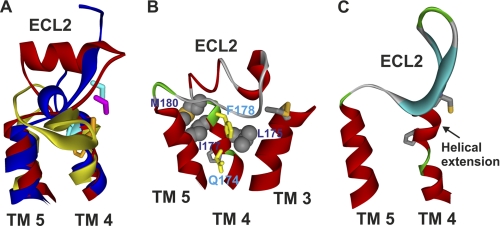
From the recently obtained crystal structures of the β2-adrenergic receptor (β2-AR) (13), β1-AR (14), A2A adenosine receptor (15), D3 dopamine receptor (16), and CXCR4 receptor (17), it has been shown that the ECL2 region flanked by TM4 and TM5 is highly variable compared with the better known structure of rhodopsin (1). In rhodopsin the ECL2 has two short anti-parallel β-sheets that form a stable cap over the covalently bound ligand 11-cis-retinal. Conversely, the ECL2 regions in β1-AR and β2-AR form an α-helix that is constrained by two disulfide bonds and is more solvent-exposed to allow diffusion of ligands into the binding pocket (Fig. 1A). In contrast, the ECL2 region in the A2A adenosine receptor lacks any prominent secondary structure, with random coils and low electron density in the middle portion of the loop indicating a more mobile structure. Functionally, ECL2 is implicated in ligand binding (18, 19), regulating whether ligands are agonists/antagonists (20, 21), acting as a binding site gate-keeper (22, 23), and constraining the receptor in the inactive conformation by acting as an activation dampener (24, 25). Therefore, alanine-scanning mutagenesis of residues Pro173(4.59)–Met180(4.66) (receptor residues are identified by the amino acid sequence number followed in parentheses by Ballesteros and Weinstein numbering, where the position of the most conserved amino acid in the TM domain, X, is designated X.50) was employed to better understand the role of this TM4/ECL2 junction of the GnRH receptor in terms of structure, ligand binding, and function.
FIGURE 1.
Molecular model of ECL2 of the human GnRH receptor. A, structural diversity of ECL2 revealed by crystal structures of bovine rhodopsin (yellow), β2-AR (red), and A2A adenosine receptor (blue). B, modeled structure of ECL2 of the human GnRH receptor. The residues Pro173(4.59)–Met180(4.66) assume an α-helical extension of TM4, which positions Glu174(4.60) and Phe178(4.64) (yellow) toward the central ligand binding pocket. Residues Leu175(4.61), Ile177(4.63), and Met180(4.80) face toward TM3, TM5, or the phospholipid bilayer to make intramolecular or intermolecular interactions. The side chain of the highly conserved Pro4.59 is also shown. C, crystal structure of CXCR4 (PDB code 3OE6) showing an α-helix extension of TM4 toward ECL2.
Our studies have demonstrated that Phe178(4.64) is a contact site in the human GnRH receptor for GnRH I and GnRH II binding, presumably through aromatic pi interactions with Trp3 of GnRH. Molecular modeling indicates that the residues surrounding Phe178(4.64) form an extended α-helical structure from the extracellular end of TM4 into ECL2, with mutagenesis studies suggesting that certain residues play a role in ligand-dependent receptor conformational selection. This may lead to different ligand-induced selective signaling as suggested in previous studies (8, 9).
EXPERIMENTAL PROCEDURES
Materials
GnRH I (pGlu1-His2-Trp3-Ser4-Tyr5-Gly6-Leu7-Arg8-Pro9-Gly10-NH2) and GnRH II ([His5,Trp7,Tyr8]GnRH) were purchased from Sigma and Bachem (Bubendorf, Switzerland). Teverelix (Ac-d-Nal1-d-Cpa2-d-Pal3-Ser4-Tyr5-d-Hci6-Leu7-Lys-(iPr)8-Pro9-d-Ala10-NH2), [Phe3]GnRH I, [Ala3]GnRH I, and [His3]GnRH I were synthesized as described previously (11). DeepVent polymerase was from New England Biolabs (Hertfordshire, UK). EcoRI, BsrGI, and XhoI restriction endonucleases and T4 ligase were from Promega (Madison, WI). d-[myo-3H]Inositol was from GE Healthcare. NBI-42902 (1-(2,6-difluorobenzyl)-3-[(2R)-amino-2-phenethyl]-5-(2-fluoro-3-methoxyphenyl)-6-methyluracil) (26) was obtained from Neurocrine Biosciences Inc. (San Diego). The MultiScreen system 96-well plates with fitted filters (Durapore membrane, pore size 0.22 μm) and the vacuum manifold for the multiwell plates were obtained from Millipore (Bedford, MA).
GnRH Docking and Molecular Dynamics Simulations
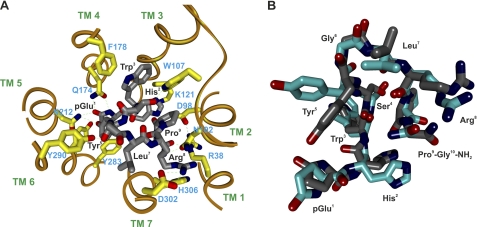
Homology models of human GnRH receptor in the inactive and active states were constructed based on the crystal structures of β2-AR (PDB code 2RH1 (13)) and opsin in its G protein-interacting conformation (PDB code 3DQB (27)) using MODELLER implemented in Discovery Studio (version 2.5; Accelrys, San Diego) as described previously (3, 8). The models with the best scores were selected for GnRH docking and molecular dynamics (MD) simulations. A βII′-type turn conformation of GnRH I (derived from an NMR structure (PDB code 1YY1)) was docked into the active state model according to the previously experimentally identified contact points between GnRH and its receptor (pGlu1 with Asn212(5.39), His2 with Asp98(2.61)/Lys121(3.32), Tyr5/His5 with Tyr290(6.58), and Gly10NH2 with Arg38(1.35)/Asn102(2.65) (2, 3, 10, 12)). The above prepared molecules were inserted into the membrane bilayer. The membrane thickness, centered at Z = 0, was set to 30 Å. The N terminus and loops were built ab initio using MODELLER and/or LOOPER (a molecular mechanics-based algorithm (28)). The MD simulations were performed in implicit membrane using the generalized Born with simple switching (GBSW) method implemented using CHARMM (29). The empty and GnRH I-occupied GnRH receptor structures were first energy-minimized and then subjected to 1-ns MD simulations using a setting similar to that described previously (8) at a temperature of 300 K with time steps of 0.002 ps and SHAKE constraints for all bond lengths involving hydrogen atoms. Harmonic restraints of 5 kcal/mol/Å2 on the receptor backbone atoms of the 7-TM domains were applied to allow small conformational changes but preserve the helical structure of the TM domains.
Site-directed Mutagenesis and Receptor Expression
The human GnRH receptor was cloned previously into the pcDNA1 expression vector. Mutant sequences were constructed using a polymerase chain reaction method (30). Wild-type and mutant receptors were transiently expressed in COS-7 cells by transfection using the Bio-Rad Gene Pulser (Bio-Rad Laboratories) at 230 V, 960 microfarads, with 15 μg of DNA/0.4 cm cuvette (1.5 × 107 cells; 0.7 ml). After transfection, cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum, 1% penicillin (10, 000 units/ml)/streptomycin (10, 000 μg/ml), and 2 mm glutamine (complete DMEM) in the absence or presence of 1 μm NBI-42902 (a membrane-permeant, nonpeptide GnRH receptor antagonist (26)) at 37 °C in a humidified 5% CO2 atmosphere for 48 h to allow receptor expression prior to binding or functional assays. Cells were washed four times, with each wash lasting for 30 min, with 2% Me2SO and 0.1% BSA/HEPES/DMEM at 37 °C after 28 h of incubation. The cells were further incubated with complete DMEM overnight (∼16 h) and then washed again as above before assays were performed (9).
Ligand Binding Assays
Radioligand binding assays (31) were performed on intact cells 48 h after transfection. After washing the transfected cells in 12-well culture plates (as described above) they were incubated with [125I]Teverelix (32) at 100, 000 cpm/well and various concentrations of unlabeled GnRH ligands in 0.1% BSA/HEPES/DMEM for 4 h at 4 °C. After incubation, free radioligand was removed from the cells by two rapid washes with ice-cold phosphate-buffered saline, pH 7.4, and the cells were solubilized in 0.5 ml of 0.1 m NaOH. Radioactivity was counted by γ-spectrometry. All experiments were performed in triplicate and repeated at least three times.
Inositol Phosphate (IP) Accumulation Assays
Assays for ligand stimulation of IP production were carried out as described previously (31, 33) using multiwell filtration plates (34). Briefly, transfected cells were seeded onto 24-well plates in the absence or presence of 1 μm NBI-42902. After 28 h, cells were washed as described above and labeled overnight with 1 μCi/ml d-[myo-3H]inositol in inositol-free DMEM containing 1% dialyzed fetal calf serum. Before the IP assay was conducted, the medium was removed, and cells were washed again as above. Cells were then preincubated with 0.5 ml of buffer (140 mm NaCl, 20 mm HEPES, 8 mm glucose, 4 mm KCl, 1 mm MgCl2, 1 mm CaCl2, and 1 mg/ml BSA containing 10 mm LiCl) at 37 °C for 30 min followed by the addition of GnRH peptides for an additional 60 min. This was shown to be within the linear period of the assay. The stimulation was terminated by the removal of the medium and the addition of 200 μl of 10 mm formic acid. The 3H-labeled IPs were isolated from the formic acid extracts using 300 μl of Dowex AG 1-X8 ion exchange resin in 96-well MultiScreen filtration plates, collected with 1 m ammonium formate/0.1 m formic acid, and quantified by liquid scintillation counting.
Data Analysis
Binding curves were fitted to a one-site model of binding using Prism 5.0 (GraphPad Software Inc., San Diego), yielding an IC50 value. The mutant receptor expression levels (Rexp) were expressed relative to a wild-type control included in each transfection. IP dose-response curves were fitted to a sigmoidal dose-response model, yielding a basal activity, a maximum response (Emax), and an EC50 value.
RESULTS
Comparative Modeling of the Human GnRH Receptor and MD Simulations
Initial models of the human GnRH receptor in the inactive and active conformations were built based on the crystal structures of β2-AR in its inverse agonist binding conformation and opsin in its G protein-interacting conformation. The crystal structures of bovine rhodopsin, the β-ARs, and A2A adenosine receptor reveal the conformational diversity of ECLs, especially ECL2 (35), but all of the above GPCR structures predict ECL2 as part of the ligand binding pocket (36, 37). A βII′-turn conformation of GnRH I (derived from a recent NMR structure) was docked into the active state model according to previous experimentally determined interactions (2, 10, 12). In our current GnRH receptor models, the 7-TM domains were constructed based on β2-AR and opsin, whereas the ECLs and intracellular loops were modeled ab initio by means of a molecular mechanics-based algorithm implemented in LOOPER (28), which often finds near native loop conformations. Because of the length limitation (maximum 25 amino acids) of the LOOPER program and the fact that ECL2 is pinned to the extracellular end of TM3 by a highly conserved disulfide bond (Cys114–Cys196), the ECL2 of GnRH receptor was built as N (Gly172–Cys196, ECL2a)- and C-terminal segments (Cys196–Ala209, ECL2b) by LOOPER. The models were then subjected to energy minimization and MD simulations in the membrane environments. Our molecular modeling shows that the residues at the N terminus of ECL2 (Gly172(4.58)–Met180(4.66)) of the human GnRH receptor form a helical extension from TM4 (Fig. 1B), which leaves ECL2 with a trajectory similar to that of β-ARs. It is noteworthy that Phe178 faces toward the binding pocket in the current model, but it pointed away from the binding pocket in the rhodopsin-based homology models prior to refinements. To validate our modeling predictions, Ala-scanning mutagenesis studies were applied to the residues from Pro173(4.59) to Met180(4.66) of the human GnRH receptor.
Expression of Human GnRH Receptors in COS-7 Cells
When 125I-[His5,d-Tyr6]GnRH I, a traditional, labeled peptide agonist in the laboratory, was used for receptor binding assays, we found that Ala mutation of Phe178(4.64) displayed little binding. Because mutations of the GnRH receptor often have less effect on the mutant receptor binding affinity for peptide antagonists (10, 12), 125I-Teverelix, a water-soluble peptide antagonist, was applied to validate the mutant receptor binding. Surprisingly, the mutant F178A displayed a markedly increased receptor cell surface expression level as compared with the wild type (see below). Hence, we used 125I-Teverelix as a labeled ligand for all receptor binding assays. This preliminary result indicated that Ala mutation of Phe178(4.64) led to a markedly decreased receptor binding affinity for peptide agonist but not a decreased receptor expression. In view of the fact that the Ala mutation of Phe178(4.64) caused a large effect on [His5,d-Tyr6]GnRH I binding affinity, we further mutated this residue to Trp and Leu to examine the role of the aromatic ring in GnRH binding.
Homologous competition binding experiments of 125I-Teverelix on intact cells transiently transfected with wild-type and mutant receptors demonstrated little or only a marginal shift in affinity for any of the mutants compared with the pIC50 of the wild-type receptor, 8.34 ± 0.11 (4.6 nm, Table 1). The mutants P173A, L175A, Y176A, and R179A gave 3–6-fold increases in the IC50 values. Therefore, in most cases, the B0 values of the specific binding measured in the absence of competitive ligands reflect the relative expression levels (Rexp) of receptors on the cell surface. Overall there were reductions in cell surface expression of the mutant receptors except for the F178A mutant, which displayed approximately a 3-fold increase in cell surface expression (Table 1). The cell surface expression levels of the mutants with greatly reduced Rexp (less than one-fourth of the wild type: L175A, F178L, and R179A mutant receptors) were rescued by a non-peptide antagonist, NBI-42902 (26). We propose that this cell-permeant small molecule may also act as a pharmacological chaperone to stabilize the receptor structure, increasing trafficking through the ER, preventing degradation of misfolded receptors in a fashion similar to that of IN3 (8, 9) and other non-peptide GnRH antagonists (38). After rescue, the cell surface expression levels of the mutant receptors were about 3-fold higher relative to their non-rescued mutants (Table 1). Interestingly, cell surface expression of wild-type receptors was also enhanced 164% by NBI-42902 pretreatment, whereas their affinity for Teverelix was unaffected (Table 1).
TABLE 1.
Binding of GnRH ligands to wild-type and mutant human GnRH receptors
Ligand binding affinity (pIC50) was measured on intact COS-7 cells at 48 h after transfection with wild-type and mutant receptors by competition binding assays using 125I-Teverelix as a radioligand with increasing concentrations of unlabeled GnRH ligands. Relative expression levels of mutants (Rexp) were compared with wild-type controls in each transfection. Values are mean ± S.E. of three or more independent experiments converted to nm (in parentheses).
| Mutant | Rexp | Binding |
||
|---|---|---|---|---|
| Teverelix | GnRH I | GnRH II | ||
| % WT | pIC50 (nm) | |||
| WT | 100 | 8.34 ± 0.11 (4.6) | 8.47 ± 0.09 (3.4) | 7.28 ± 0.09 (52.5) |
| WT + NBI-42902a | 164 ± 24 | 8.38 ± 0.18 (4.2) | 8.57 ± 0.05 (2.7) | 7.28 ± 0.09 (52.1) |
| P173A | 41 ± 6 | 7.57 ± 0.12 (26.8) | 7.77 ± 0.13 (17.1) | 6.40 ± 0.22 (401) |
| Q174A | 51 ± 10 | 8.31 ± 0.18 (4.9) | 7.52 ± 0.11 (30.3) | 6.39 ± 0.29 (404) |
| L175A | 12 ± 1 | UD | UD | UD |
| L175A + NBI-42902a | 45 ± 6 | 7.82 ± 0.20 (15.1) | 8.29 ± 0.26 (5.1) | 7.47 ± 0.12 (33.7) |
| Y176A | 26 ± 5 | 7.90 ± 0.20 (12.5) | 8.16 ± 0.03 (7.0) | 6.66 ± 0.32 (221) |
| I177A | 77 ± 24 | 8.37 ± 0.09 (4.2) | 8.04 ± 0.03 (9.1) | 7.79 ± 0.04 (16.1) |
| F178A | 324 ± 40 | 8.23 ± 0.11 (5.9) | 5.03 ± 0.02 (9310) | 4.58 ± 0.10 (26600) |
| F178L | 23 ± 3 | UD | UD | UD |
| F178L + NBI-42902a | 78 ± 1 | 8.22 ± 0.19 (6.0) | 6.71 ± 0.39 (195) | 5.34 ± 0.08 (4530) |
| F178W | 55 ± 8 | 8.16 ± 0.13 (7.0) | 7.68 ± 0.03 (21.0) | 6.90 ± 0.05 (125) |
| R179A | 8 ± 6 | UD | UD | UD |
| R179A + NBI-42902a | 31 ± 9 | 7.79 ± 0.47 (16.3) | 7.96 ± 0.39 (11.1) | 7.10 ± 0.31 (80.3) |
| M180A | 45 ± 8 | 8.00 ± 0.03 (10.1) | 7.48 ± 0.06 (33.1) | 7.57 ± 0.01 (26.6) |
a Pretreated with NBI-42902. UD, undetectable.
Effect of Mutations of the Human GnRH Receptor TM4/ECL2 Junction on GnRH Affinity
Competition binding studies of receptors with Ala mutations of residues from Pro173(4.59) to Met180(4.66) were performed to investigate any effects on the binding affinity of GnRH I and GnRH II (Table 1). Consistent with previous studies (8–10, 12), GnRH I binds to wild-type receptors expressed in COS-7 cells with high affinity (3.4 nm), which is unaffected by pretreatment with NBI-42902 (Table 1). Ala mutations of residues Leu175(4.61), Tyr176(4.62), Ile177(4.63), and Arg179(4.65) of the human GnRH receptor caused a minor (2–3 fold) reduction, whereas mutation of Phe178(4.64) to Ala led to a 2738-fold reduction in receptor affinity for GnRH I. Ala mutation of Pro173(4.59), Gln174(4.60), and Met180(4.66) led to a moderate (5–10 fold) reduction in mutant receptor binding affinity for GnRH I.
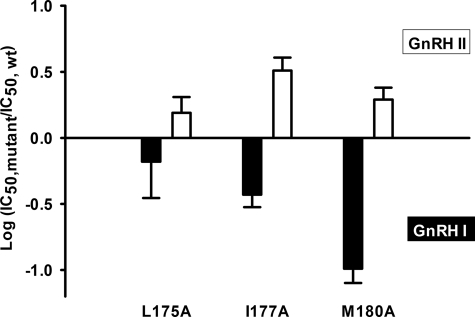
As described previously (8–10, 12), GnRH II has lower affinity for the human GnRH receptor than GnRH I (52.5 compared with 3.4 nm). The affinity of GnRH II for wild-type receptors was also unaffected by pretreatment with NBI-42902 (Table 1). In contrast to GnRH I, Ala mutations of the residues from Pro173(4.59) to Met180(4.66) gave differential effects on the mutant receptor binding affinity for GnRH II. Ala mutations of Pro173(4.59), Gln174(4.60), Tyr176(4.62), Phe178(4.64), and Arg179(4.65) gave a phenotypic effect similar to that of GnRH I on the receptor affinity for GnRH II. However, mutations of Leu175(4.61), Ile177(4.63), and Met180(4.66) led to 2–3 fold increases in affinity for GnRH II but 2–10 fold reductions for GnRH I (Table 1 and Fig. 2). The above results suggest differential functions of these residues in ligand binding and ligand-induced receptor conformational selections.
FIGURE 2.
Differential effects of mutations on the receptor binding affinity for GnRH I and GnRH II. Ala mutations of Leu175(4.61), Ile177(4.63), and Met180(4.80) had differential effects on the affinities for GnRH I and GnRH II, with decreased affinities for GnRH I but increased affinities for GnRH II. Closed bars represent GnRH I and open bars GnRH II. At least three independent pIC50 values were obtained from competition binding experiments (Table 1). Results are expressed as the log of the ratio of the IC50 values of GnRH I and GnRH II for the mutant to the IC50 values for the wild-type receptor ± S.E.
Mutation of Phe178(4.64) of the Human GnRH Receptor Decreases Affinity for GnRH I and II
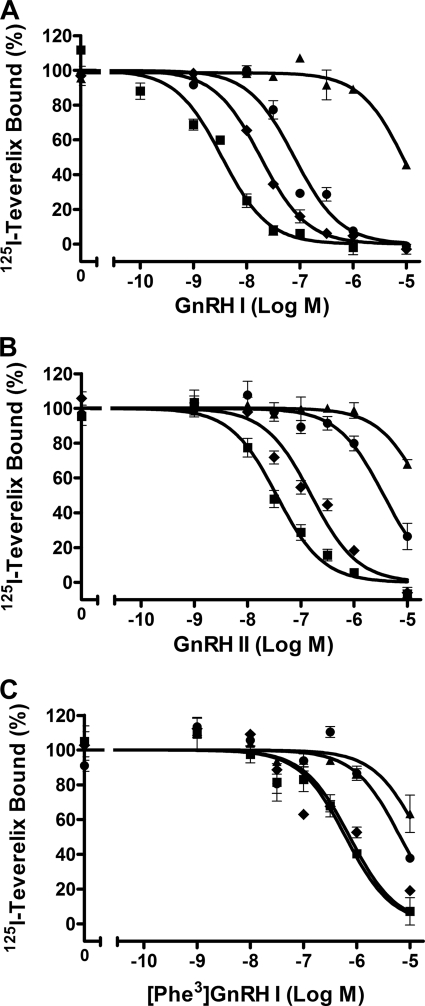
The affinities of GnRH I and GnRH II for the wild-type and Phe178(4.64) mutant receptors were determined by competition binding experiments (Fig. 3, A and B). Mutation of Phe178(4.64) to Ala, which deletes the side chain beyond the β-carbon, brought about a 2738-fold reduction in affinity for GnRH I. Mutation of Phe178(4.64) to Leu, which is also lipophilic but lacks the aromatic group, was a more tolerated mutation with only a 57-fold reduction in affinity for GnRH I, whereas mutation of Phe178(4.64) to the bulkier aromatic residue Trp gave only a 6-fold decrease in affinity, indicating that mutations of Phe178(4.64) to Leu and Trp retain part of the original receptor-ligand interactions (Fig. 3A and Table 1). Similar patterns were seen with GnRH II, whereby mutations of Phe178(4.64) to Ala, Leu, or Trp increased the IC50 values by 506-, 86-, or 2-fold, respectively (Fig. 3B and Table 1). These results suggest that the aromatic ring of Phe178(4.64) is important for GnRH I and II binding to the receptor.
FIGURE 3.
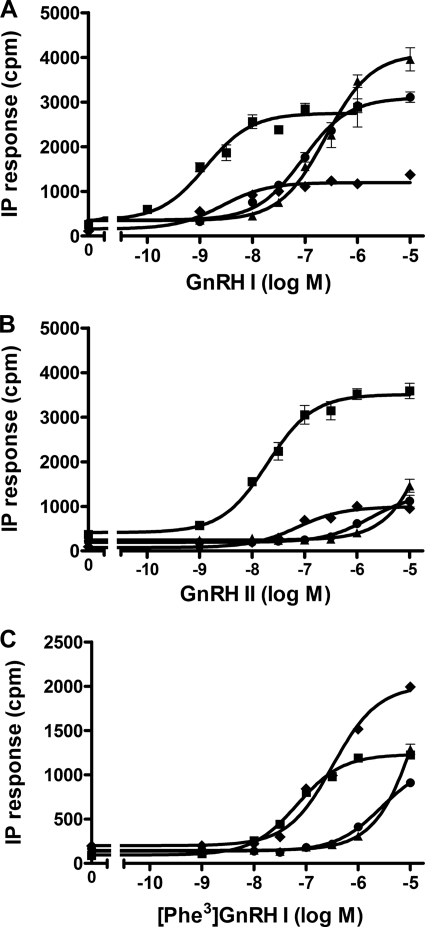
Competition binding of GnRH peptides to wild-type and Phe178(4.64) mutant receptors. COS-7 cells transfected with wild-type or mutant receptors were preincubated with (F178L) or without (F178A and F178W) 1 μm NBI-42902 for 48 h and then washed prior to the binding assay. Details are given under “Experimental Procedures.” Normalized competitive binding of GnRH I (A), GnRH II (B), and [Phe3]GnRH I (C) at the wild-type ■, F174A ▴, F178L ●, and F178W ⧫ receptors. Results are representative experiments repeated at least three times with essentially the same results. Points are mean ± S.E. of triplicate measurements.
Importance of Trp3 in GnRH for Receptor Binding
Previous molecular modeling and docking of GnRH to the receptor (12) suggests a potential interaction between Trp3 of GnRH and the extracellular end of TM4/ECL2. To determine the role of Trp3 of GnRH in binding with the receptor, Trp3-substituted analogues of GnRH I were examined in competition binding experiments. [His3]GnRH I and [Ala3]GnRH I were unable to displace 125I-Teverelix (data not shown), suggesting that they are low affinity ligands. However, [Phe3]GnRH I could bind to the wild-type receptor but had a 146-fold reduced affinity compared with GnRH I (with IC50 values of 499 versus 3.4 nm; Fig. 3C and Table 2). Pretreatment with NBI-42902 had little effect on the affinity of [Phe3]GnRH I (Table 2). To determine whether Phe178(4.64) of the receptor interacts with Trp3 of GnRH I, as suggested by the molecular model (Fig. 4A), the affinity of [Phe3]GnRH I for the Phe178(4.64) mutant receptors was determined (Table 2). Similar to the results with GnRH I, mutation of Phe178(4.64) to Ala caused the largest shift in affinity for [Phe3]GnRH I with a 61-fold increase in the IC50 value to 30500 nm, whereas mutation of Phe178(4.64) to Leu led to a 9-fold reduction in the affinity for [Phe3]GnRH I. Most interestingly, mutation of Phe178(4.64) to Trp produced effectively no shift in affinity for [Phe3]GnRH I, with an IC50 of 672 nm, compared with the IC50 of 499 nm for the wild-type receptor. This demonstrates a partial rescue of binding with reciprocal mutations between Phe and Trp in the receptor and ligand.
TABLE 2.
Ligand binding and [Phe3]GnRH I-elicited IP responses at the wild-type and Phe178(4.64) mutant human GnRH receptors
Ligand binding affinity (pIC50) was measured on intact COS-7 cells at 48 h after transfection with wild-type and mutant receptors by competition binding assays using 125I-Teverelix as a radioligand with increasing concentrations of unlabeled [Phe3]GnRH I. IP responses (pEC50) were determined in COS-7 cells transfected with wild-type and mutant receptors. Maximum IP responses (Emax) were expressed relative to wild-type controls in each transfection. Values are mean ± S.E. of three or more independent experiments converted to nm (in parentheses).
| Mutant | Binding |
IP response |
|
|---|---|---|---|
| pIC50 | pEC50 | Emax | |
| (nm) | (nm) | % WT | |
| WT | 6.30 ± 0.08 (499) | 7.12 ± 0.17 (76.6) | 100 |
| WT + NBI-42902a | 6.45 ± 0.31 (354) | 7.68 ± 0.23 (20.8) | 98 ± 9 |
| F178A | 4.52 ± 0.08 (30500) | 4.94 ± 0.15 (11400) | 338 ± 174 |
| F178L + NBI-42902a | 5.33 ± 0.08 (4680) | 5.39 ± 0.06 (4060) | 178 ± 61 |
| F178W | 6.17 ± 0.08 (672) | 6.21 ± 0.17 (621) | 209 ± 62 |
a Pretreated with NBI-42902.
FIGURE 4.
Molecular model of the human GnRH receptor-GnRH I complex. A, A βII′-turn conformation of GnRH I derived from the NMR structure (PDB code 1YY1) was docked into the receptor model in the active conformation derived from the active state of bovine rhodopsin (3DQB) according to experimentally identified intermolecular interactions between GnRH I (gray) and the receptor contact sites (yellow). pGlu1 of GnRH I interacts with Asn212(5.39); His2 interacts with Asp98(2.61) and Lys121(3.32); Tyr5 interacts with Tyr290(6.58); Arg8 interacts with Asp302(7.32); and Pro9-Gly10NH2 interacts with Arg38(1.35) and Asn102(2.65). The H-bonds are indicated by dashed lines. The model suggests that the side chain of Gln174(4.60) makes H-bond contacts with pGlu1 of GnRH, whereas the aromatic ring of Phe178(4.64) interacts with the aromatic ring of Trp3 in GnRHs through displaced pi-stacking connections. B, NMR (cyan) and docked (gray) structures of GnRH I showing that the docked GnRH I has a trajectory similar to that of the NMR structure.
Effects of Mutations of the Human GnRH Receptor TM4/ECL2 Junction on GnRH-elicited IP Responses
GnRH I and GnRH II elicited robust IP responses from COS-7 cells expressing wild-type GnRH receptors with EC50 values of 3.4 and 20.1 nm (Table 3). The maximal IP responses (Emax) for all experiments with wild-type receptors were typically eight times the basal activity. The effects of alanine mutations of residues Pro173(4.59)–Met180(4.66) on the IP responses are summarized in Table 3. The changes in potency of the mutant receptors in mediating IP responses for both GnRH I and GnRH II are closely related to the changes of the mutant receptor binding affinities for the ligands (Fig. 5A). In parallel with the decreased mutant receptor binding affinities, all of the mutants gave decreased potency in GnRH I-stimulated IP responses (Tables 1 and 3). A similar change in affinity and potency of GnRH II was observed for most of the mutants, except for L175A, I177A, and M180A, which displayed 2–3-fold increases in receptor binding affinity for GnRH II, but gave little change (L175A), or a 2-fold (M180A) and 5-fold (I177A) decrease in GnRH II potency (Table 3).
TABLE 3.
GnRH-elicited IP responses at wild-type and mutant human GnRH receptors
IP responses (pEC50) were determined in COS-7 cells transfected with wild-type and mutant receptors. Maximum IP responses (Emax) were expressed relative to wild-type controls in each transfection. Values are mean ± S.E. of three or more independent experiments, converted to nm (in parentheses).
| Mutant | GnRH I |
GnRH II |
||
|---|---|---|---|---|
| pEC50 | Emax | pEC50 | Emax | |
| (nm) | % WT | (nm) | % WT | |
| WT | 8.77 ± 0.11 (1.7) | 100 | 7.70 ± 0.07 (20.1) | 100 |
| WT + NBI-42902a | 8.74 ± 0.08 (1.8) | 86 ± 10 | 7.69 ± 0.04 (20.4) | 92 ± 7 |
| P173A | 7.39 ± 0.17 (41.2) | 79 ± 8 | 6.10 ± 0.12 (787) | 82 ± 10 |
| Q174A | 7.56 ± 0.18 (27.7) | 81 ± 4 | 6.33 ± 0.12 (464) | 90 ± 1 |
| L175A + NBI-42902a | 8.41 ± 0.06 (3.9) | 60 ± 8 | 7.66 ± 0.04 (22.1) | 59 ± 7 |
| Y176A | 7.82 ± 0.11 (15.0) | 37 ± 3 | 6.81 ± 0.01 (153) | 31 ± 7 |
| I177A | 8.50 ± 0.34 (3.1) | 17 ± 4 | 7.02 ± 0.12 (94.6) | 17 ± 8 |
| F178A | 6.41 ± 0.10 (386) | 105 ± 23 | 4.71 ± 0.24 (19400) | 103 ± 79 |
| F178L + NBI-42902a | 6.86 ± 0.11 (139) | 82 ± 18 | 5.44 ± 0.16 (3640) | 46 ± 15 |
| F178W | 8.30 ± 0.26 (5.1) | 46 ± 8 | 6.86 ± 0.25 (138) | 39 ± 15 |
| R179A + NBI-42902a | 8.53 ± 0.15 (3.0) | 59 ± 4 | 7.05 ± 0.17 (90.2) | 45 ± 14 |
| M180A | 7.79 ± 0.08 (16.4) | 82 ± 2 | 7.35 ± 0.14 (45.1) | 70 ± 21 |
a Pretreated with NBI-42902.
FIGURE 5.
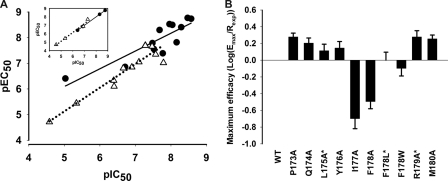
Effects of mutations on the functional responses. A, effects of mutational changes in receptor binding affinity (pIC50 values) on the functional potency (pEC50 values). Inset, mutants of Phe178(4.64) only: GnRH I, ●; GnRH II, △. B, effects of mutations on the maximum signaling efficacy. *, denotes mutants pretreated with NBI-42902. For detailed results see Tables 1 and 3.
It has been shown that there is a near linear relationship between the GnRH receptor binding sites and the maximum IP responses (9, 39). Hence, the relative maximum signaling efficacy of GnRH can be calculated by normalization of the maximum IP responses to the cell surface receptor binding sites as the receptor population is saturated, and the affinity is not an issue (9, 40). Most mutants, except for I177A and F178A, had little or only a marginal effect on the signaling efficacy of GnRH I (Fig. 5B) and GnRH II. The above results indicate that conformational changes of this region are involved in GnRH-induced receptor activation. The mutant, I177A, was expressed at the cell surface to 77% of wild-type level but gave reduced Emax to 17% that of the wild-type for both GnRH I and GnRH II. The differential effects of Ala mutation of Ile177(4.63) on GnRH affinity and potency, combined with the reduced efficacy of this mutant, indicate that Ile177(4.63) is important for the stabilization of the GnRH-induced receptor-active conformational states and may also act as a determinant of ligand-induced receptor conformational selection.
Effects of Mutations of Phe178(4.64) on GnRH-elicited IP Responses
In parallel with the decreased receptor binding affinity, the mutations of Phe178(4.66) also gave decreased potency for both GnRH I and GnRH II (Fig. 5A, insert). The effects of the mutation of Phe178 to Ala, Leu, and Trp on the IP responses are shown in Fig. 6. Similar to the outcome on affinity, mutation of Phe178 to Ala had the largest effect on potency, with 228- and 962-fold increases in EC50 in response to GnRH I (Fig. 6A) and GnRH II (Fig. 6B). Mutation of Phe178 to Leu gave intermediate decreases of potency with 82- and 181-fold increases in the EC50, whereas mutation of Phe178 to Trp had only a small effect on potency with 3- and 7-fold increases in the EC50 values. Despite the mutant F178A having an increased expression of 324% compared with the wild-type receptors, the maximum responses of GnRH I at this mutant were similar to the wild-type receptors with Emax values of 105%, implying a potential decrease in signaling efficacy (Fig. 5B). However, mutations of Phe178(4.64) to Leu and Trp had little or only a marginal effect on the GnRH I-induced signaling efficacy (Fig. 5B). A similar phenotype was also observed for GnRH II, derived from the calculated Emax, which could not be measured accurately because of the solubility limit of the maximum GnRH II concentration (10−5 m) to be used for F178A and F178L (Fig. 6B and Table 3).
FIGURE 6.
IP responses of wild-type and Phe178(4.64) mutant receptors. GnRH I- (A), GnRH II- (B), and [Phe3]GnRH I (C)-stimulated IP responses of Phe178(4.64) receptors mutated to Ala ▴, Leu ●, or Trp ⧫ compared with wild-type receptors ■. COS-7 cells were transiently transfected with the wild-type or mutant receptors, and IP accumulation was measured after stimulation with GnRH ligands for 60 min. Results are representative experiments repeated at least three times with essentially the same results. Points are mean ± S.E. of triplicate measurements.
[Phe3]GnRH I-elicited IP responses at the Phe178(4.64) mutant receptors followed a similar pattern to that of GnRH I (Fig. 5C and Table 2). However, although the affinity of [Phe3]GnRH I was similar at both the wild-type and F178W mutant receptors, the EC50 value of [Phe3]GnRH I at the F178W mutant was 8-fold higher than the wild-type receptor. Interestingly, [Phe3]GnRH I-stimulated IP responses at F178W mutant receptors (reciprocal mutation between Trp3 of GnRH I and Phe178(4.66) of the receptor) gave higher maximal IP responses than that of the wild-type receptor (Fig. 6C), suggesting the reciprocal mutations between the ligand and the receptor can rescue the maximum functional response. Consistent with the previous report (41), [Phe3] GnRH I is a partial agonist at wild-type receptors with decreased Emax, but the reciprocal mutations of Phe and Trp between the ligand and the receptor converts this partial agonist to a full or nearly full agonist.
DISCUSSION
The recently available crystal structures of GPCRs show significant structural diversity of ECL2, which connects TM4 and TM5 and is pinned onto the extracellular end of TM3 (1, 13, 15–17, 42). The roles of the extracellular residues for GPCRs remain poorly defined compared with that of residues in the TMs. Our previous docking models of GnRH I suggest an interaction of Trp3 of GnRH I with the extracellular end of TM4 bordering ECL2 (12). The aim of this study was to apply molecular modeling and mutagenesis to the eight residues, Pro173(4.59)–Met180(4.66), of the N-terminal region of ECL2 of the human GnRH receptor to define the functional role of the targeted residues within the receptor structure. Our molecular modeling suggests that the above residues assume an α-helical extension of TM4, which positions the side chains of Gln174(4.60) and Phe178(4.64) toward the central ligand binding pocket while leaving the subsequent ECL2 away from the receptor TM core (Fig. 1B). A similar helical extension was also observed in the crystal structure of the CXCR4 receptor (Fig. 1C) (17).
In the rhodopsin-like family of GPCRs (43), TM4 appears as an outlier of the helical bundle, making extensive contacts with the phospholipid bilayer. Two mutations, L175A and R179A, had a destabilized effect on the receptor structure, reflected by the strongly decreased receptor cell surface expression level to less than 80% of the wild-type level that could be rescued by NBI-42902. In the models, the side chain of Leu175((4.61) interacted with the side chains of Tyr119(3.30) and Leu122(3.33) of TM3. However, Arg179(4.65), located one helix above Leu175(4.61), appears to face toward the phospholipid bilayer and may interact with the negatively charged phospholipid head group. In addition, Arg179(4.65) interacts intramolecularly with the negatively charged Asp185 of ECL2, in which the Ala mutation also gave a reduction in the mutant receptor cell surface expression level similar to that of Arg179(4.65).3 Interestingly, Ala mutation of Phe178(4.64) createed a stabilized mutant receptor reflected by a 3-fold increase in cell surface expression level relative to the wild type, whereas mutation of Phe178(4.64) to Leu destabilized the receptor, giving a 4-fold reduction in cell surface expression. This suggests that the side chain of Phe178(4.64) may also play a role in the configuration of the receptor structure.
One of the main findings of this study was the identification of a residue, Phe178(4.64), that is crucial for binding of both GnRH I and GnRH II. Mutation of Phe178(6.64) to Ala led to a 2737- and 511-fold reduction in the affinity for GnRH I and GnRH II, respectively. Mutation of Phe178(4.64) to a non-aromatic Leu also reduced receptor binding affinity for GnRH I and GnRH II by 57–86-fold, whereas mutation to Trp gave only a minor reduction in affinities (2–6 fold; Table 1). The much smaller effect of mutation to Leu rather than Ala, which deletes the side chain beyond the β-carbon, indicates that the non-polar side chain of Leu retains some interactions with GnRH peptides, whereas substitution of Phe with aromatic Trp is well tolerated. The minor reduction of F178W in affinity for GnRH suggests that substitution of Phe178(4.64) with a bulkier Trp may have steric detrimental effect on agonist-induced receptor conformational changes. This is more obvious from the functional assay results whereby F178W gave a reduced Emax for both GnRH I and GnRH II to 39–46% that of the wild type (Table 3 and Fig. 6, A and B). These results indicate the importance of Phe178(4.64) of the GnRH receptor in high affinity ligand binding and receptor activation.
Our new molecular modeling studies revealed that residues Pro173(4.59)–Met180(4.66) assume an α-helical extension of TM4 and that Phe178(4.64) of the receptor may interact with Trp3 of GnRH, consistent with our previous prediction (12). The involvement of Trp3 of GnRH in binding the receptor was confirmed using Trp3-substituted peptides. [Ala3]GnRH I and [His3]GnRH I were unable to bind to the receptor, whereas [Phe3]GnRH I exhibited a 146-fold reduction in affinity compared with the parent peptide. This is consistent with the finding from a previous structure-activity report that substitution of Trp3 of GnRH with non-aromatic amino acids gives rise to very low activity, whereas some activity is present with Phe substitution and the activity can be substantially increased in [pentamethyl-Phe3]GnRH I (41). This indicates that Trp3 of GnRH makes a crucial aromatic contact with the receptor, which facilitates other intermolecular interactions between the ligand and receptor.
Trp3 of GnRH analogues were proposed to interact with Trp280(6.48) of the human GnRH receptor or the equivalent residue of the receptors from different species (44, 45). However, the above proposed interaction not only contradicts the NMR structure of GnRH I (PDB code 1YY1), which was docked into our GnRH receptor model (Fig. 4), but also is inconsistent with our mutational results of Trp280(6.48), as none of the mutations has significant effect on either ligand binding or receptor activation (46). Here, we have shown that Trp3 of GnRH makes an aromatic contact with Phe178(4.64). The reduction in affinities for GnRH I seen when Phe178(4.64) was mutated to Ala or Leu was also observed with [Phe3]GnRH I but to lesser extent, with affinity reductions of only 61- and 9-fold for [Phe3]GnRH I compared with 2737- and 57-fold with GnRH I. We propose that the side chain of Phe178(4.64) makes extensive contacts with Trp3 of GnRH but less so for [Phe3]GnRH I, which possesses a smaller aromatic residue at position 3, reflected by a 146-fold decreased affinity of [Phe3]GnRH I compared with the wild-type receptor. The much smaller effect of the mutations of Phe178(4.64) to Ala and Leu on mutant receptor binding affinity for [Phe3]GnRH I supports a direct contact between Trp3 of GnRH and Phe178(4.64). A similar phenotype was observed in the mutation of other ligand-receptor-interacting residues (10, 12). When the mutations in the receptor and ligand were reciprocal (Phe178(4.64)Trp in the receptor and [Phe3]GnRH I), their affinity was similar to [Phe3]GnRH I binding to the wild-type receptor. More interestingly, the functional response was partially rescued by the reciprocal mutations between the receptor and ligand, as the maximum IP responses of [Phe3]GnRH I in the F178W mutant exceeded that of [Phe3]GnRH I in the wild-type receptor (Fig. 6C). Taken together, these results support our molecular modeling prediction that the side chain of Phe178(4.64) makes a direct contact with Trp3 of GnRH peptide agonists. This hydrophobic interaction is also important for GnRH-induced receptor activation, as F178A significantly reduced the maximum efficacy of GnRH I (Fig. 5B). However, this residue is not important for the binding of peptide antagonists, as mutations of Phe178(4.64) had little effect on the mutant receptor binding affinity of Teverelix, which was used as the radiolabeled ligand in our competition binding assays. Interestingly, a cation-pi interaction between Trp186(4.64) of the sphingosine 1-phosphate receptor and the ammonium head group of sphingosine 1-phosphate was proposed (47).
The extracellular portion of TM4 from 4.59 to 4.80 exhibits high structural divergence, implying its importance in receptor functional selectivity. Consistent with other GPCRs (48), mutation of Pro173(4.59) affected ligand affinity for both peptide agonists and antagonist by 5–8-fold, although this residue faces away from the central ligand binding pocket toward the phospholipid bilayer, close to the side of TM5. We suggest that Pro173(4.59) may be involved as a ligand-induced receptor conformational switch (see below). Ala mutation of Gln174(4.60), next to Pro173(4.59), specifically decreased the affinity for peptide agonists GnRH I and GnRH II by 8–9-fold but not for the antagonist Teverelix. Our molecular modeling studies suggest that the side chain of Gln174(4.60) makes an H-bond contact with pGlu1 of GnRH I and GnRH II (Fig. 4A).
Our other important finding is that, in addition to playing a role in ligand binding, the TM4/ECL2 junction of the GnRH receptor is likely to be involved in ligand binding selectivity via ligand-induced receptor conformational selectivity, a concept we proposed previously (8, 9). Our Ala scanning mutagenesis supported our modeling prediction that the residues Pro173(4.59)–Met180(4.66) form an extended α-helix of TM4. Interestingly, Ala mutations of Ile177(4.63) and Met180(4.80), located on the same face as Pro173(5.59) in the boundary of TM5, and of Leu175(4.61), facing toward TM3, led to 2–10 fold reductions in affinity for GnRH I but 2–3 fold increases in affinity for GnRH II (Fig. 2). The mutation-induced differential effects on the affinities of GnRH I and GnRH II have led us to propose that the binding of GnRH I and GnRH II stabilizes different receptor-active conformations (8, 9). In our GnRH receptor models, Leu175(4.61) and Ile177(4.63) may make intramolecular contacts (Leu175(4.61) with Tyr119(3.30) and Leu122(3.33) of TM3 and Ile177(4.63) with Phe210(5.37) and Tyr211(5.38) of TM5), whereas Met180(4.80) may make contacts with the phospholipid bilayer. The latter can also regulate receptor conformations (49). Ala mutation of Ile177(4.63) markedly reduced the signaling efficacy of GnRH I (Fig. 5B). The GnRH II-induced IP responses at the I177A mutant receptor also demonstrated a 5-fold decrease in potency compared with wild-type receptors, yet there was a 3-fold increase in affinity in addition to the decreased Emax (Table 3). This indicates that Ile177(4.63) is important in stabilizing receptor-active conformational states. Binding of GnRH I and GnRH II appears to be involved in the differential rearrangements of the above contacts, bringing on distinct receptor-active conformations. Mutation of these key residues to Ala allows the receptor to adopt new conformations that selectively affect (increase or decrease) receptor binding affinity for GnRH I and GnRH II. We therefore propose that the TM4/ECL2 junction of the GnRH receptor may act as a ligand-dependent receptor conformational selector, which is coupled to helix movements and/or ligand-specific conformational switches (37, 50, 51). The discovery of structural elements for ligand and receptor conformational selection could have implications for the development of novel ligands that selectively activate one signaling pathway and bypass others, allowing improved pharmacological specificity and profiles.
In summary, we have identified that the side chain of Phe178(4.64) of the GnRH receptor is important for high affinity binding of both GnRH I and GnRH II. Site-directed mutagenesis of the receptor and substitution of GnRH at position 3 indicates that the aromatic ring of Phe178(4.64) of the GnRH receptor interacts directly with Trp3 of GnRH. The molecular docking also reveals an H-bond interaction between Gln174(4.60) of the receptor and pGlu1 of GnRH peptides, confirmed by the mutagenesis results. We propose that residues Pro173(4.59)–Met180(4.66) of the GnRH receptor form an extended α-helix. The side chains of Leu175(4.61), Ile177(4.63), and Met180(4.66) make contact with other TM domains or the phospholipid bilayer, which couple ligand structure to specific receptor conformational switches in the human GnRH receptor, thereby accounting for the ligand-induced selective signaling of GnRH analogues described previously (3).
Acknowledgments
We greatly appreciate the gift of NBI-42902, provided by Neurocrine Biosciences. We thank Nicola Millar and Adele Whay for technical assistance and Robin Sellar for the preparation of 125I-labeled ligands.
This work was supported by Programme Grant U1276.00.005.00001.01 from the Medical Research Council, United Kingdom.
R. Forfar and Z. L. Lu, unpublished observation.
- GPCR
- G protein-coupled receptor
- TM
- transmembrane domain
- ECL
- extracellular loop
- GnRH
- gonadotropin-releasing hormone
- IP
- inositol phosphate
- AR
- adrenergic receptor
- PDB
- Protein Data Bank
- MD
- molecular dynamics.
REFERENCES
- 1. Palczewski K., Kumasaka T., Hori T., Behnke C. A., Motoshima H., Fox B. A., Le Trong I., Teller D. C., Okada T., Stenkamp R. E., Yamamoto M., Miyano M. (2000) Science 289, 739–745 [DOI] [PubMed] [Google Scholar]
- 2. Millar R. P., Lu Z. L., Pawson A. J., Flanagan C. A., Morgan K., Maudsley S. R. (2004) Endocr. Rev. 25, 235–275 [DOI] [PubMed] [Google Scholar]
- 3. Millar R. P., Pawson A. J., Morgan K., Rissman E. F., Lu Z. L. (2008) Front. Neuroendocrinol. 29, 17–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ulloa-Aguirre A., Stanislaus D., Arora V., Väänänen J., Brothers S., Janovick J. A., Conn P. M. (1998) Endocrinology 139, 2472–2478 [DOI] [PubMed] [Google Scholar]
- 5. Gründker C., Völker P., Emons G. (2001) Endocrinology 142, 2369–2380 [DOI] [PubMed] [Google Scholar]
- 6. White C. D., Coetsee M., Morgan K., Flanagan C. A., Millar R. P., Lu Z. L. (2008) Mol. Endocrinol. 22, 2520–2530 [DOI] [PubMed] [Google Scholar]
- 7. Lu Z. L. (2010) 92nd Annual Meeting of the Endocrine Society, San Diego, June 19–22, 2010, pp. 3–217, The Endocrine Society, Chevy Chase, MD [Google Scholar]
- 8. Lu Z. L., Coetsee M., White C. D., Millar R. P. (2007) J. Biol. Chem. 282, 17921–17929 [DOI] [PubMed] [Google Scholar]
- 9. Lu Z. L., Gallagher R., Sellar R., Coetsee M., Millar R. P. (2005) J. Biol. Chem. 280, 29796–29803 [DOI] [PubMed] [Google Scholar]
- 10. Coetsee M., Millar R. P., Flanagan C. A., Lu Z. L. (2008) Biochemistry 47, 10305–10313 [DOI] [PubMed] [Google Scholar]
- 11. Mamputha S., Lu Z. L., Roeske R. W., Millar R. P., Katz A. A., Flanagan C. A. (2007) Mol. Endocrinol. 21, 281–292 [DOI] [PubMed] [Google Scholar]
- 12. Stewart A. J., Sellar R., Wilson D. J., Millar R. P., Lu Z. L. (2008) Mol. Pharmacol. 73, 75–81 [DOI] [PubMed] [Google Scholar]
- 13. Cherezov V., Rosenbaum D. M., Hanson M. A., Rasmussen S. G., Thian F. S., Kobilka T. S., Choi H. J., Kuhn P., Weis W. I., Kobilka B. K., Stevens R. C. (2007) Science 318, 1258–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Warne T., Serrano-Vega M. J., Baker J. G., Moukhametzianov R., Edwards P. C., Henderson R., Leslie A. G., Tate C. G., Schertler G. F. (2008) Nature 454, 486–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jaakola V. P., Griffith M. T., Hanson M. A., Cherezov V., Chien E. Y., Lane J. R., Ijzerman A. P., Stevens R. C. (2008) Science 322, 1211–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chien E. Y., Liu W., Zhao Q., Katritch V., Han G. W., Hanson M. A., Shi L., Newman A. H., Javitch J. A., Cherezov V., Stevens R. C. (2010) Science 330, 1091–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wu B., Chien E. Y., Mol C. D., Fenalti G., Liu W., Katritch V., Abagyan R., Brooun A., Wells P., Bi F. C., Hamel D. J., Kuhn P., Handel T. M., Cherezov V., Stevens R. C. (2010) Science 330, 1066–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shi L., Javitch J. A. (2002) Annu. Rev. Pharmacol. Toxicol. 42, 437–467 [DOI] [PubMed] [Google Scholar]
- 19. Conner M., Hawtin S. R., Simms J., Wootten D., Lawson Z., Conner A. C., Parslow R. A., Wheatley M. (2007) J. Biol. Chem. 282, 17405–17412 [DOI] [PubMed] [Google Scholar]
- 20. Ott T. R., Troskie B. E., Roeske R. W., Illing N., Flanagan C. A., Millar R. P. (2002) Mol. Endocrinol. 16, 1079–1088 [DOI] [PubMed] [Google Scholar]
- 21. Herold C. L., Qi A. D., Harden T. K., Nicholas R. A. (2004) J. Biol. Chem. 279, 11456–11464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Avlani V. A., Gregory K. J., Morton C. J., Parker M. W., Sexton P. M., Christopoulos A. (2007) J. Biol. Chem. 282, 25677–25686 [DOI] [PubMed] [Google Scholar]
- 23. Goodwin J. A., Hulme E. C., Langmead C. J., Tehan B. G. (2007) Mol. Pharmacol. 72, 1484–1496 [DOI] [PubMed] [Google Scholar]
- 24. Klco J. M., Wiegand C. B., Narzinski K., Baranski T. J. (2005) Nat. Struct. Mol. Biol. 12, 320–326 [DOI] [PubMed] [Google Scholar]
- 25. Storjohann L., Holst B., Schwartz T. W. (2008) Biochemistry 47, 9198–9207 [DOI] [PubMed] [Google Scholar]
- 26. Tucci F. C., Zhu Y. F., Struthers R. S., Guo Z., Gross T. D., Rowbottom M. W., Acevedo O., Gao Y., Saunders J., Xie Q., Reinhart G. J., Liu X. J., Ling N., Bonneville A. K., Chen T., Bozigian H., Chen C. (2005) J. Med. Chem. 48, 1169–1178 [DOI] [PubMed] [Google Scholar]
- 27. Scheerer P., Park J. H., Hildebrand P. W., Kim Y. J., Krauss N., Choe H. W., Hofmann K. P., Ernst O. P. (2008) Nature 455, 497–502 [DOI] [PubMed] [Google Scholar]
- 28. Spassov V. Z., Flook P. K., Yan L. (2008) Protein Eng. Des. Sel. 21, 91–100 [DOI] [PubMed] [Google Scholar]
- 29. Brooks B. R., Bruccoleri R. E., Olason B. D., States D. J., Swaminathan S., Karplus M. (1983) J. Comp. Chem. 4, 187–217 [Google Scholar]
- 30. Lu Z. L., Curtis C. A., Jones P. G., Pavia J., Hulme E. C. (1997) Mol. Pharmacol. 51, 234–241 [DOI] [PubMed] [Google Scholar]
- 31. Millar R. P., Davidson J., Flanagan C., Wakefield I. (1995) Methods in Neurosciences: Receptor Molecular Biology, (Sealfon S. C. ed) pp. 145–162, Academic Press, San Diego, CA [Google Scholar]
- 32. Deghenghi R., Boutignon F., Wüthrich P., Lenaerts V. (1993) Biomed. Pharmacother. 47, 107–110 [DOI] [PubMed] [Google Scholar]
- 33. Imai A., Gershengorn M. C. (1987) Methods Enzymol. 141, 100–101 [DOI] [PubMed] [Google Scholar]
- 34. Chengalvala M., Kostek B., Frail D. E. (1999) J. Biochem. Biophys. Methods 38, 163–170 [DOI] [PubMed] [Google Scholar]
- 35. Hanson M. A., Stevens R. C. (2009) Structure 17, 8–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nygaard R., Frimurer T. M., Holst B., Rosenkilde M. M., Schwartz T. W. (2009) Trends Pharmacol. Sci. 30, 249–259 [DOI] [PubMed] [Google Scholar]
- 37. Unal H., Jagannathan R., Bhat M. B., Karnik S. S. (2010) J. Biol. Chem. 285, 16341–16350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Janovick J. A., Brothers S. P., Cornea A., Bush E., Goulet M. T., Ashton W. T., Sauer D. R., Haviv F., Greer J., Conn P. M. (2007) Mol. Cell. Endocrinol. 272, 77–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Arora K. K., Cheng Z., Catt K. J. (1997) Mol. Endocrinol. 11, 1203–1212 [DOI] [PubMed] [Google Scholar]
- 40. Kenakin T. (2002) Annu. Rev. Pharmacol. Toxicol. 42, 349–379 [DOI] [PubMed] [Google Scholar]
- 41. Sealfon S. C., Weinstein H., Millar R. P. (1997) Endocr. Rev. 18, 180–205 [DOI] [PubMed] [Google Scholar]
- 42. Warne T., Moukhametzianov R., Baker J. G., Nehmé R., Edwards P. C., Leslie A. G., Schertler G. F., Tate C. G. (2011) Nature 469, 241–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lu Z. L., Saldanha J. W., Hulme E. C. (2002) Trends Pharmacol. Sci. 23, 140–146 [DOI] [PubMed] [Google Scholar]
- 44. Chauvin S., Bérault A., Lerrant Y., Hibert M., Counis R. (2000) Mol. Pharmacol. 57, 625–633 [DOI] [PubMed] [Google Scholar]
- 45. Hövelmann S., Hoffmann S. H., Kühne R., ter Laak T., Reiländer H., Beckers T. (2002) Biochemistry 41, 1129–1136 [DOI] [PubMed] [Google Scholar]
- 46. Coetsee M., Gallagher R., Millar R. P., Flanagan C. A., Lu Z. L. (2006) BioScience 2006, Glasgow, United Kingdom, July 23–27, 2006, Abstract 0518, The Biochemical Society, London [Google Scholar]
- 47. Inagaki Y., Pham T. T., Fujiwara Y., Kohno T., Osborne D. A., Igarashi Y., Tigyi G., Parrill A. L. (2005) Biochem. J. 389, 187–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Javitch J. A., Shi L., Simpson M. M., Chen J., Chiappa V., Visiers I., Weinstein H., Ballesteros J. A. (2000) Biochemistry 39, 12190–12199 [DOI] [PubMed] [Google Scholar]
- 49. Fantini J., Barrantes F. J. (2009) Biochim. Biophys. Acta 1788, 2345–2361 [DOI] [PubMed] [Google Scholar]
- 50. Ahuja S., Hornak V., Yan E. C., Syrett N., Goncalves J. A., Hirshfeld A., Ziliox M., Sakmar T. P., Sheves M., Reeves P. J., Smith S. O., Eilers M. (2009) Nat. Struct. Mol. Biol. 16, 168–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bokoch M. P., Zou Y., Rasmussen S. G., Liu C. W., Nygaard R., Rosenbaum D. M., Fung J. J., Choi H. J., Thian F. S., Kobilka T. S., Puglisi J. D., Weis W. I., Pardo L., Prosser R. S., Mueller L., Kobilka B. K. (2010) Nature 463, 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]