Abstract
Interactions between polymorphonuclear neutrophils (PMNs) and tumor cells have been reported to facilitate the adhesion and subsequent extravasation of tumor cells through the endothelium under blood flow, both of which are mediated by binding β2-integrin to intercellular adhesion molecule 1 (ICAM-1). Here the adhesions between human WM9 metastatic melanoma cells, PMNs, and human pulmonary microvascular endothelial cells (HPMECs) were quantified by a gas-driven micropipette aspiration technique (GDMAT). Our data indicated that the cellular binding affinity of PMN-WM9 pair was 3.9-fold higher than that of the PMN-HPMEC pair. However, the effective binding affinities per molecular pair were comparable between the two cell pairs no matter whether WM9 cells or HPMECs were quiescent or cytokine-activated, indicating that the stronger adhesion between PMN-WM9 pair is mainly attributed to the high expression of ICAM-1 on WM9 cells. These results proposed an alternative mechanism, where WM9 melanoma cells adhere first with PMNs near vessel-wall regions and then bind to endothelial cells via PMNs under blood flow. In contrast, the adhesions between human MDA-MB-231 metastatic breast carcinoma cells and PMNs showed a comparable cellular binding affinity to PMN-HPMEC pair because the ICAM-1 expressions on MDA-MB-231 cells and HPMECs are similar. Furthermore, differences were observed in the intrinsic forward and reverse rates of the β2-integrin-ICAM-1 bond between PMN-TC and PMN-EC pairs. This GDMAT assay enables us to quantify the binding kinetics of cell adhesion molecules physiologically expressed on nucleated cells. The findings also further the understanding of leukocyte-facilitated tumor cell adhesion from the viewpoint of molecular binding kinetics.
Keywords: Cell Adhesion, Integrin, Kinetics, Neutrophil, Tumor Metastases, Micropipette Aspiration Frequency Assay
Introduction
Tumor metastasis requires the detachment of malignant cells from primary tumor, invasion through blood/lymph vessels, transmigration across the endothelium, and finally, adhesion to the host cells (1). The adhesion and subsequent extravasation of tumor cells through the vascular endothelium are critical steps in this complex cascade (2). Malignant melanoma is the most deadly skin cancer with high metastatic potential (3). Although the binding of such adhesion molecules as P-selectin (4, 5), Lu-ECAM-1 (6), or integrin α4β1 (VLA-4) (7) to their ligands has been found to support the adhesion of melanoma tumor cells (TCs)3 with endothelial cells (ECs), the expression of the molecules seems insufficient to support the direct adhesion between TCs and ECs in blood flow (8). Rather, the presence of polymorphonuclear neutrophils (PMNs) potentially enhances TCs adhesion to the ECs and induces the subsequent extravasation under flow conditions (9–11). A “two-step adhesion” hypothesis was introduced that involves initial PMN tethering on ECs followed by subsequent TCs being captured by tethered PMNs (10). Another line of in vitro evidence has indicated independently that TCs are able to aggregate with PMNs in a shear flow (12), which proposes an alternative hypothesis that TCs interact first with PMNs to form aggregates in a near-wall region of vascular endothelium prior to binding to EC through PMNs. Thus, it is physiologically important to elucidate how TCs, PMNs, and/or ECs interact with each other and what their underlying molecular mechanisms are.
Regulations of PMN-EC or PMN-TC adhesion have been extensively studied at the cellular and molecular levels. Tethering and rolling of leukocytes on the endothelium are initiated by selectins and stabilized by β2-integrins, which is followed by PECAM-1-induced transmigration through the extracellular matrix (13, 14). Tumor cells are found to interact with PMNs under shear flow in a cone-plate viscometer (12, 15–17). Evidently, the interactions between β2-integrin (LFA-1 or Mac-1) expressed on PMNs and intercellular adhesion molecule 1 (ICAM-1) expressed on TCs or ECs dominate both PMN-EC and PMN-TC adhesions (9, 10, 12). Binding kinetics between the receptors and ligands anchored on two opposing surfaces (so-called two-dimensional binding) is different from that for the interacting molecules in solution and governs how likely they are to bind and how long they will remain bound, which is the essential determinant of multistep adhesions among the three types of cells. For example, the readout on binding kinetics of LFA-1 and ICAM-1 interactions indicated that the cell adhesion was initiated by the formation of a single bond with a dissociation rate ∼0.3 s−1 (18). The TC fraction in PMN-TC heterotypic aggregates measured was used to estimate the kinetic rates and binding affinities of β2-integrin-ICAM-1 interactions (12). It was also found that PMNs tethering on ECs was regulated by both shear rate and shear stress (10, 19), whereas TCs adhesion to PMNs was predominantly regulated by shear rate (9, 10, 17). Thus, it is critical from the viewpoint of two-dimensional kinetics of β2-integrin-ICAM-1 interactions to compare the adhesions between PMN-EC and PMN-TC pairs and to further the understanding of leukocyte-facilitated tumor cell adhesion.
Two-dimensional binding kinetics of surface-bound receptors and ligands has been extensively studied using various techniques such as a parallel-plate flow chamber, cone-plate viscometer, optical tweezers, atomic force microscopy, micropipette aspiration technique (MAT), and biomembrane force probe (20). Whereas the other techniques are frequently applied to quantify the cell rolling/tethering or aggregation dynamics and the forced rupture of receptor-ligand bond, the adhesion frequency assay with MAT or biomembrane force probe technique is widely used to determine the kinetic rates and binding affinity of surface-bound molecules (21–26). In this regard, a red blood cell (RBC) bearing the purified receptors or ligands serves as a force transducer to visualize the occurrence of an adhesive event by taking the advantage of deflection of RBC membrane via a precise micromanipulation (21). This conventional MAT or biomembrane force probe technique is no longer applicable when both receptors and ligands are constitutively expressed on two nucleated cells, because no membrane deflection is measurable to determine the adhesion event. In the current work, a gas-driven micropipette aspiration technique (GDMAT), first introduced for visualizing the membrane tether formation (27) and microvillus extension (28) and later used for quantifying the binding of E-selectin to its antibody coated on latex beads (29), was modified to determine the two-dimensional kinetics of surface-bound β2-integrin and ICAM-1 on TCs, PMNs, or ECs using a well established kinetic model (21). Kinetic rates and binding affinity of interacting molecules were compared between PMN-TC and PMN-EC pairs, and the regulation of ICAM-1 expression on binding kinetics by cytokine TNF-α was quantified.
EXPERIMENTAL PROCEDURES
Reagents and Cell Culture
Mouse IgG anti-human monoclonal blocking antibodies (mAbs) against αL chain (CD11a, clone MEM-25) and β2 chain (CD18, clone CLB-LFA-1/1) were purchased from Invitrogen. Mouse IgG anti-human blocking mAb against αM chain (CD11b, clone 44) was from Chemicon International (Temecula, CA). Mouse IgG anti-human blocking mAbs against β2 chain (CD18, clone 212701), ICAM-1 (CD54, clone BBIG-I1(11C81)), and E-selectin (CD62E, clone BBIG-E4(5D11)) were from R & D System (Minneapolis, MN). FITC-conjugated goat anti-mouse IgG (5.7 equivalent of FITC per molecule) was from Sigma.
Human WM9 metastatic melanoma cells (kindly provided by Dr. M. Herlyn, Wistar Institute, Philadelphia, PA) were grown in RPMI 1640 medium supplemented with 1 mm l-glutamine, 100 units/ml penicillin, 10 mg/ml streptomycin, and 10% FBS. Human metastatic breast adenocarcinoma cell line MDA-MB-231 (purchased from the Cell Culture Center of Union Medical University, Beijing, China) was cultured in L-15 medium supplemented with 1 mm l-glutamine, 100 units/ml penicillin, 10 mg/ml streptomycin, and 10% FBS. Human pulmonary microvascular endothelial cell line HPMEC ST1.6R (a kind gift from Dr. C. J. Kirkpatrick, Institute of Pathology, Johannes-Gutenberg University, Mainz, Germany) was cultured as previously described (30). For adhesion frequency measurement, confluent cells were trypsinized and washed twice with fresh medium. Collected cells were then resuspended in the medium and allowed to recover for 1 h while being rocked at 8 rpm at 37 °C. To up-regulate the expression of ICAM-1, WM9 or HPMEC cells were incubated with TNF-α (R & D System) at respective 110 and 300 units/ml for 24 h before use. For blocking measurements, the cells were preincubated with blocking mAbs at a concentration of 10 μg/ml for 45 min on ice.
Human PMNs were obtained from whole blood samples collected by venipuncture and then isolated using a Ficoll-Hypaque density gradient (Histopaque-1077 and Histopaque-1119 from Sigma). Collected cell mixture with ∼20–50% of PMNs were kept at 4 °C in Dulbecco's PBS containing 0.1% human serum albumin for up to 4 h (12). Individual PMNs were directly used for adhesion frequency assays without further lysing RBCs from cell mixture to minimize the activation of PMNs.
To conduct the adhesion frequency measurements, the cells were suspended in HBSS with Ca2+ and Mg2+ containing 10 mm HEPES and 4% FBS. In some cases, the cells were suspended in HBSS without Ca2+ and Mg2+ containing 10 mm HEPES, 4% FBS, and 5 mm EDTA.
Site Density Determination
Site density of adhesion molecules expressed on cell surface was determined by flow cytometry. Three aliquots of PMNs were respectively incubated with anti-αL (clone MEM-25), anti-αM (clone 44), and anti-β2 (clone 212701) mAbs, and one aliquot of WM9 cells, MDA-MB-231 cells, or HPMECs was incubated with anti-ICAM-1 mAbs (clone BBIG-I1(11C81)) at a concentration of 10 μg/ml for 45 min on ice. The cells were then incubated with FITC-conjugated goat anti-mouse secondary mAbs for 45 min on ice. Washed cells were analyzed by flow cytometer (BD Biosciences, San Jose, CA). The expression of β2-integrin on PMNs was isolated from mixed RBCs by gating them in FSC-SSC dot plot. The fluorescent intensities of the cells were then compared with standard calibration beads (Bangs, Fishers, IN) to determine the site densities (21), where the calibration curves for PMN or WM9/HPMEC/MDA-MB-231 were obtained by running standard beads labeled with specific molecules of equivalent soluble fluorochromes in the same instrument setting. Site densities of β2-integrin and ICAM-1 molecules were calculated using mr or ml = (S × GMFI)/[4πr2 × (F/P)], where S denotes the slope of calibration curve, GMFI is the geometric mean fluorescence intensity, r is the cell radius, and F/P is the equivalent of FITC/molecule.
Gas-driven Micropipette Aspiration Technique
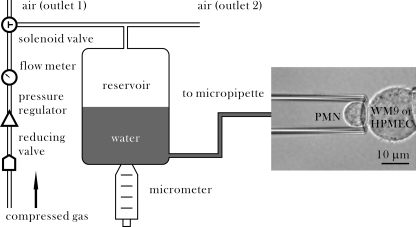
The GDMAT, first introduced to quantify the membrane tether formation (27) and microvillus extension (28), was modified to measure the adhesion of two nucleated cell bearing respective receptors and ligands. The system was set up by adding a gas-driven pressure unit in a conventional MAT system (22, 23). Although a tumor or endothelial cell was held steadily in the right pipette, a neutrophil was sucked into the left pipette and driven by the pressure unit to approach to, contact with, and withdraw from the steady cell on the right (Fig. 1). To manipulate the neutrophil movement, a negative pressure was applied via adjusting the height of reservoir gauged by a micrometer, and a positive pressure was exerted via controlling gas flow rate gauged by a pressure regulator. The switch between negative and positive pressures was implemented via a solenoid valve combined with a time relay to force the cell to move backwards or forwards and to repeat automatically the approach-contact-retraction cycle at given contact duration. The images of cell movement were recorded in a compressed MPEG-2 format (720 × 576 pixel, 25 frames/second) using a charge-coupled device camera (WAT-902H, Watec, Japan) and then decoded using a video encoder board (MP-400, Gotron, China). The moving distance of the cell inside the pipette was calibrated at a resolution of ∼74.6 nm/pixel using a standard microruler, and the time course of cell movement was analyzed off-line frame-by-frame.
FIGURE 1.
Schematic of GDMAT. A PMN was sucked into the left pipette, and a WM9 cell or a HPMEC was held steadily by the right pipette. Compressed gas stored in a high pressure container was forced to flow through a reducing valve to generate a very low gas flow, which was controlled by a pressure regulator and gauged by a flow meter. A three-way solenoid valve was used to shunt the gas flow into two ways (outlet 1 or 2). A water reservoir, scaled by a micrometer at a resolution of 0.01 mm, was moved up and down to generate a negative pressure and to drive the PMN moving backward. A positive pressure was generated by the gas flow onto water surface to force the cell moving forward. A solenoid valve combined with a time relay was bridged to switch between negative and positive pressures acting on the PMN. This process was repeated automatically to implement the test cycles of PMN approach to, contact with, and withdraw from the WM9 or HPMEC cell until the adhesion probability was obtained statistically.
Mechanically, the calibration between gas flow rate and suction pressure was performed in the following procedure. First, a PMN was sucked into the tip of the pipette and allowed to move freely. Next, the pressure was zeroed when the PMN stayed still inside the pipette. Finally, once a suction pressure was applied via adjusting the micrometer to withdraw the cell, the gas flow was set on and adjusted to stop its movement. Six systematically varied suction pressures at 0.05–0.5 mmH2O were employed, and the corresponding gas flow rates were measured to obtain the calibration curve. Meanwhile, the impinging or pulling force F imposed on a static PMN with a suction pressure (Δp) can be calculated as F = ΔpπRp2(1 − 4ϵ/3Rp) (28, 31), where Rp is the radius of the pipette, and ϵ is the gap width between the cell and the pipette wall. When a tether is pulled out from a cell membrane, the force will be modified as F = ΔpπRp2(1 − 4ϵ/3Rp)(1 − Ut/Uf) (31), where Ut is the velocity of the cell during tethering, and Uf is the free moving velocity of the cell under the same pressure Δp. In the current study, the suction pressure was set to 0.3 mmH2O, and the gas flow rate was given to 200 ml/min that was calibrated to a positive pressure of 0.5 mmH2O. The impinging force applied on PMN was estimated to be 99 pN, and the pulling force was to be 149 pN.
Data Analysis
Binding kinetics between β2-integrin and ICAM-1 interactions was determined from the measured time dependence of adhesion probability between two nucleated cells. Here the adhesion probability (or frequency) is defined as the fraction of adhesive events in total tests, which serves as a measure of cell adhesion. The resulted binding curve, Pa(t), was fitted using a small system probabilistic kinetic model (21),
to estimate a pair of kinetic parameters: the zero force reverse rate, kr0, and the cellular binding affinity, AcmrmlKa0, where Ac is the contact area, and mr and ml are site densities of β2-integrin receptors and ICAM-1 ligands, respectively. The effective binding affinity, AcKa0, and intrinsic forward rate, kf, per molecular pair were obtained by calculating from AcKa0 = (AcmrmlKa0)/(mr × ml) and kf = (AcKa0) × kr0/Ac. Note that AcmrmlKa0 and AcKa0 serve as the respective measures to estimate the binding capability at cellular and molecular levels.
The site densities of β2-integrin on PMNs (mr) and ICAM-1 on WM9, MDA-MB-231, and HPMEC cells (ml), as well as the cell-cell contact areas (Ac) were presented as the means ± S.D. The best fit of measured data to Equation 1 and the 95% confidence intervals of the best fit curves were performed by SigmaPlot (Systat Software, San Jose, CA), and the best fit parameters were presented as the means ± S.E. The statistical significance was assessed using a paired t test for measured values and an unpaired t test for best fit parameters. The entire binding curves of PMNs adhesion to quiescent cells or TNF-α-stimulated cells were compared by an F test.
RESULTS
Adhesion between Nucleated Cells Is Identified
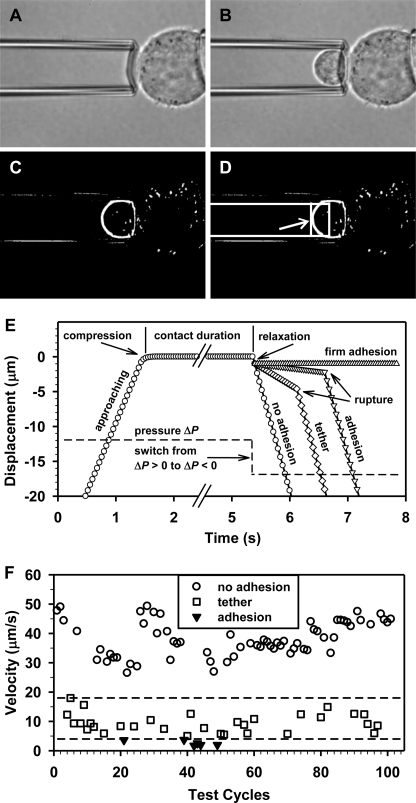
To determine the occurrence of an adhesion event, the time course of PMN movement inside the pipette was monitored in the entire approach-contact-retraction cycle (supplemental Movie S1). In contrast to the cross-correlation method previously applied to monitor the displacement of a bead (31–34), we developed here a simplified direct-search protocol to track the movement of a PMN cell from its left partial side during the retraction phase (Fig. 2, A–D). This process was repeated frame-by-frame, which resulted in the time course of PMN displacement (Fig. 2E). Such a displacement spectrum enabled us to identify the occurrence of an adhesion event for a nucleated cell because the membrane deflection on PMN tip was no longer visible when the PMN cell was withdrawn. Generally, the PMN displacement exhibited four distinct patterns of cell retraction: 1) it moved at a high velocity (circles), indicating that no interactions were detected when the two cells were separated (no adhesion); 2) it moved at a relatively low velocity over a distance before its free movement (diamonds), suggesting that the adhesion presumably occurred with a membrane tether (tether) (27); 3) it stayed still or yielded a very low velocity prior to its free movement (inverted triangles), implying that the adhesion appeared likely without a tether extraction (adhesion); and 4) it did not separate from another cell during entire retraction phase (triangles), illustrating that a firm adhesion was formed (firm adhesion). Note that the free PMN compressed a little when it was enforced by positive pressure to contact with another cell and the adhered PMN recoiled slightly ∼1.0 μm when a negative pressure was imposed with a quick response less than 0.04 s (one frame) (Fig. 2E). The relaxation length is mainly determined by the lengths of microvillus and receptor-ligand complex as well as the cell deformation generated by the impinging force.
FIGURE 2.
Determination of an adhesion event. A–D, a direct search protocol was employed to monitor PMN movement inside micropipette. An image without PMN was chosen as the background frame (A). Then an arbitrary image with moving PMN (B) was subtracted by the background frame to obtain the image of cell boundary (C). A rectangular monitoring zone was selected inside the micropipette (box in D) to minimize the impact of noises outside the micropipette, and the cell displacement was then calculated using the trajectory of its left partial side (indicated by arrow in D) within the rectangle at a threshold gray value of 25 (at least 5 pixel points). E, schematic of a test cycle with four independent trajectories superposed to define an event of no adhesion (circles), tether (diamonds), adhesion (inverted triangles), and firm adhesion (triangles). F, a typical velocity distribution of PMN movement. The data were collected from one PMN-WM9 pair with assays repeated 101 times, and three subgroups were presented at 0–4 μm/s for adhesion/firm adhesion (inverted triangles), 5–18 μm/s for tether (squares), and 26–50 μm/s for no adhesion (circles). Dashed lines denote the threshold velocities at 4 and 18 μm/s.
Thus, an adhesive event was able to be identified from the resulted velocity distribution of moving PMN. In a typical measurement of a PMN interacting with a quiescent WM9 cell (Fig. 2F), three velocity subgroups were presented where the first two were counted individually as an adhesive event for those with adhesion/firm adhesions (inverted triangles) and tether (squares), and the latter one was indicative for those without adhesion (circles). Noting that the threshold velocities (dashed lines) could vary slightly from one PMN to another, it is straightforward to discriminate those adhesive and nonadhesive events directly from the same pattern of velocity distribution for various PMNs, implying the feasibility and reliability of the identifying protocol applied in the current study.
Adhesion between PMN-WM9 and PMN-HPMEC Is Specific
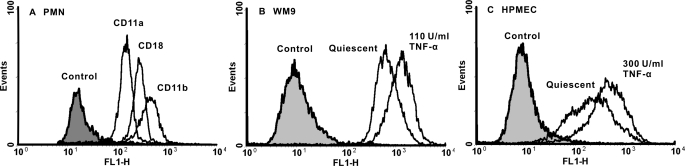
Previous studies have shown that β2-integrin expressed on PMN and ICAM-1 expressed on TCs or ECs form the major molecular pair that mediates PMN-TC and PMN-EC adhesions (9, 10). Here flow cytometry was used to measure the expressions of β2-integrin on PMNs and ICAM-1 on WM9 cells or HPMECs at least in triplet, and the site densities were then determined by calibration curves using the measured fluorescent intensity. It was found that the site densities of CD11a, CD11b, and CD18 were estimated to be mr = 52 ± 20, 231 ± 38, and 181 ± 34 μm−2, respectively (Fig. 3A). ICAM-1 expression was enhanced for WM9 cells from ml = 799 ± 146 to 1151 ± 13 μm−2 or HPMECs from 246 ± 196 to 478 ± 251 μm−2 when the cells were stimulated at the respective concentrations of 110 and 300 units/ml of TNF-α (Fig. 3, B and C).
FIGURE 3.
Expression of β2-integrin and ICAM-1 molecules on PMNs (A), WM9 cells (B), and HPMECs (C). The cells were incubated with the respective blocking mAbs followed by secondary FITC-conjugated mAbs (unfilled histograms), whereas the blank cells were used as controls (shaded histograms). Also plotted were for WM9 and HPMEC cells preincubated with 110 units/ml and 300 units/ml TNF-α, respectively, for 24 h.
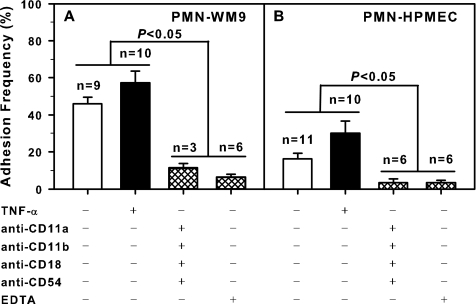
The specificity of cell adhesion was quantified by averaging the adhesion probabilities at sufficiently long contact duration t (> 10 s for specific adhesions or >6 s for nonspecific controls). For each cell pair, the approach-contact-retraction cycle was repeated ∼40–100 times (in some cases with long contact duration, e.g. >15 s, the cycle was repeated only 30–50 times) to obtain the running frequency at a preset cycle duration, and the frequency at the steady phase was used to estimate the adhesion probability (Pt) at that contact duration (t) (21). As shown in Fig. 4, the adhesion probability yielded Pt = ∼46 and 16% for respective PMN-WM9 and PMN-HPMEC pair when both β2-integrin and ICAM-1 molecules were present (open bars). By contrast, the adhesion was abolished to be Pt = ∼11 or 6% for the former and 3 or 3% for the latter, respectively, when anti-CD11a, anti-CD11b, anti-CD18, and anti-CD54 blocking mAbs or divalent cation chelator EDTA was present (hatched bars). It was also found that the adhesion frequency was enhanced when WM9 or HPMEC cells were stimulated by TNF-α to up-regulate ICAM-1 expression (solid bars). These results indicated that the adhesions measured between the nucleated cells were specifically mediated by β2-integrin-ICAM-1 binding.
FIGURE 4.
Binding specificity for PMN-WM9 (A) and PMN-HPMEC (B) pairs under quiescent (unfilled histograms) case or TNF-α stimulation (shaded histograms). The adhesion frequency was obtained by averaging all the points at sufficient long contact time (>10 s for specific or >6 s for nonspecific binding). The data are presented as the means ± S.E., and n denotes the sample size.
Binding of β2-integrin-ICAM-1 Follows a Simple Kinetics
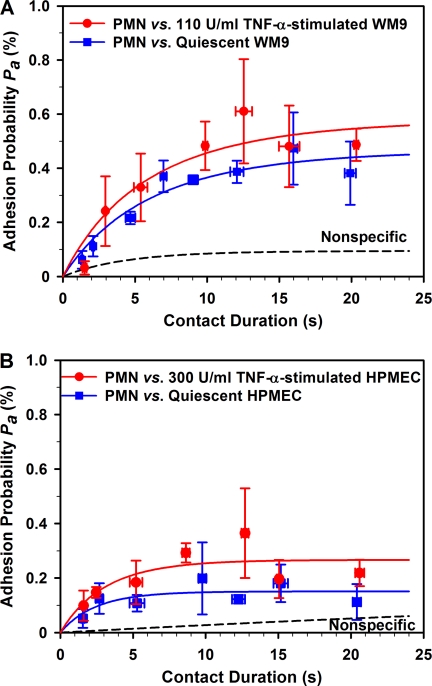
Cell adhesions mediated by β2-integrin-ICAM-1 bindings were systematically measured at the given contact duration t = ∼1–21 s. In a typical measurement of a PMN adhering to a quiescent WM9 cell (Fig. 5A), the specific adhesion frequencies (Pa) (blue squares) were calculated by removing the nonspecific adhesion frequency (Pn) (dash line, obtained by fitting directly the nonspecific adhesion frequencies to Equation 1) from total adhesion frequency (Pt) measured per cell pair and renormalized by Pa = (Pt − Pn)/(1 − Pn) (22, 23). The specific frequency Pa for all cell pairs was binned upon similar contact durations (the variance of contact duration was presented as error bars along x axis, whereas the variance of Pa was presented as error bars along the y axis), and a total of 24–30 cell pairs were used for each binding curve (Fig. 5A). It was evident that the adhesion exhibited a transition phase when Pa increased with t and a steady phase when Pa reached the equilibrium, following a simple kinetics of receptor-ligand binding. Time dependence of adhesion frequency or binding curve Pa versus t was then best fitted using Equation 1 to obtain two kinetic parameters: zero force reverse rate, kr0, and cellular binding affinity, AcmrmlKa0. The estimated kinetic parameters were then used to predict each Pa versus t curve (solid lines). Similar transition phases were also found for PMN adhering to TNF-α-stimulated WM9 cells (Fig. 5A, red circles) and PMN adhering to quiescent (blue squares) or TNF-α-stimulated HPMECs (Fig. 5B, red circles). These results demonstrated that the β2-integrin-ICAM-1 binding follows well with a simple kinetics.
FIGURE 5.
Binding curves of PMN-WM9 (A) and PMN-HPMEC (B) adhesions when WM9 or HPMEC cells were quiescent (blue squares) or stimulated by TNF-α (red circles). The data points were obtained by averaging cell pairs with similar contact durations and presented as the means ± S.E. for both adhesion probabilities and contact durations. Also plotted were the predictions fitted using Equation 1 (solid lines), whereas the dashed lines represented nonspecific binding, obtained by fitting Equation 1 to nonspecific data (not shown for the sake of clarity). The binding curves for quiescent and TNF-α-stimulated cells were compared using an F test to give the PF values for comparisons.
In contrast to the conventional MAT where the preset contact duration remains almost the same in the repeated test cycles for one cell pair (21–23, 26), it was noticed that the actual contact duration reported here varied slightly from cycle to cycle for a same cell pair. This is presumably due to the fact that the occurrence of one adhesive event in one cycle would shorten the moving distance of free cell in the following cycle and then result in the extended contact duration in the sequential cycles even at the same cycle duration preset. Experimentally the duration of each contact was recorded, and the zero pressure was timely checked and reset to minimize the variation of contact duration. To further validate the reliability of applying the novel approach to predict the binding kinetics of characteristic adhesions of nucleated cells, we tested the reasonability of our kinetics analysis upon data set per cell pair aforementioned. Here all the measured contact events from different cell pairs were pooled together to get the single value of Pa by binning the events with similar contact durations. Again, the model fits the data well for PMNs interacting with quiescent and TNF-α-stimulated WM9 cells or HPMEC cells (supplemental Fig. S1). No significant difference was found from the two analyses for both zero force reverse rate, kr0, and cellular binding affinity, AcmrmlKa0 (data not shown), supporting that the analysis of adhesion kinetics is reliable from the two methods. Taken together, the adhesion between two nucleated cells is reasonably determined by the novel GDMAT approach, imparting the reliability on predicted kinetic parameters.
PMN-WM9 Adhesion Is Stronger Than PMN-HPMEC Adhesion
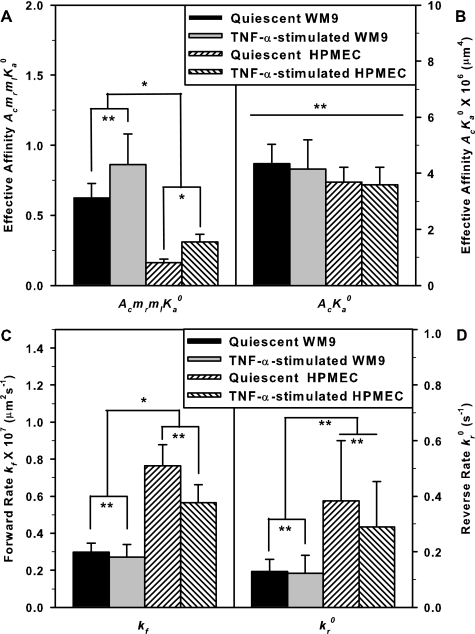
It has long been noticed that the PMN-EC adhesion is the first step to mediate the adhering of tumor cells to endothelium, suggesting that the adhesion of PMN to ECs might be stronger than that for PMN-TC interactions (10). To test this hypothesis, we compared the binding kinetics between PMN-WM9 and PMN-HPMEC pairs. Our results indicated that the PMN-WM9 adhesion (Fig. 5A, blue squares) was higher than that for PMN-HPMEC pair (Fig. 5B, blue squares), as seen that the adhesion frequency at sufficient long contact time (t → ∞), Pa(∞) = 1 − exp(−AcmrmlKa0), which serves as an indicative of cell adhesion, yielded 0.47 and 0.15, respectively. Comparing the two curves using an F test gave PF < 0.01. The binding curves were also fitted using Equation 1 to estimate the lumped affinity AcmrmlKa0 and zero force reverse rate kr0. The returned AcmrmlKa0 for PMN-WM9 adhesion (0.63 ± 0.10) was 3.9-fold higher than that for PMN-HPMEC adhesion (0.16 ± 0.02) (Pt < 0.05 using a t test; Fig. 6A). This observation was further supported when WM9 or HPMEC cells were preincubated with TNF-α. Similarly, the equilibrium adhesion frequency Pa(∞) was enhanced in PMN-WM9 (0.58) and PMN-HPMEC (0.27) binding (Fig. 5, A and B, red circles; PF < 0.01). A 2.8-fold difference in AcmrmlKa0 was found in between (0.86 ± 0.22 and 0.31 ± 0.05, Pt < 0.05) together with 1.4-fold (0.86 ± 0.22 and 0.63 ± 0.10, Pt = 0.35) and 1.9-fold (0.31 ± 0.05 and 0.16 ± 0.02, Pt < 0.05) up-regulations for PMN-WM9 and PMN-HPMEC pairs, respectively (Fig. 6A), when WM9 and HPMEC cells were activated by TNF-α.
FIGURE 6.
The comparison of cellular binding affinity AcmrmlKa0 (A), effective binding affinity AcKa0 (B), intrinsic forward rate kf (C), and zero force reverse rate kr0 (D). The data are presented for quiescent (solid bars) or TNF-α-stimulated (open bars) WM9 cells, and quiescent (leftwards-hatched bars) or TNF-α-stimulated (rightwards-hatched bars) HPMEC cells. The significant differences were accessed by unpaired t test, *, Pt < 0.05; **, Pt > 0.2.
We further tested the underlying molecular mechanisms for the distinct adhesions of two cell pairs. The effective binding affinity per molecular pair, AcKa0, was calculated using AcKa0 = (AcmrmlKa0)/(mr × ml), where the site density of CD18 subunit was set as mr. Interestingly, AcKa0 yielded the comparable values between PMN-WM9 ((4.34 ± 0.69) × 10−6 μm4) and PMN-HPMEC ((3.68 ± 0.5) × 10−6 μm4) adhesions (Fig. 6B, Pt = 0.81), suggesting that the adhesion differences between the two cell pairs be mainly attributed to 3.2-fold difference in the site density of ICAM-1 ligands on WM9 and HPMEC cells, whereas the effective binding affinity of β2-integrin-ICAM-1 interactions was not altered. To further test this, we also compared the AcKa0 values between quiescent and stimulated cells for each cell pair. Because an activated PMN induced by cytokines or chemoattractants prevented its steady movement inside the pipette (data not shown), only WM9 or HPMEC cells were preincubated with TNF-α. Again, although the expression of ICAM-1 ligand was up-regulated by TNF-α stimulation and yielded 2.4-fold difference between WM9 and HPMEC cells (Fig. 3, B and C), the effective binding affinity AcKa0 presented the similar values between PMN-WM9 ((4.15 ± 0.16) × 10−6 μm4) and PMN-HPMEC ((3.60 ± 0.61) × 10−6 μm4) adhesions (Fig. 6B; Pt = 0.66), supported that the stronger adhesion in PMN-WM9 pair is governed by the higher expression of ICAM-1 ligands. The effective binding affinity yielded very similar values for intact and cytokine-activated cells, indicating that TNF-α stimulation has no impact on the effective adhesion.
The Intrinsic Two-dimensional Kinetics per Molecular Pair Is Different between Two Cell Pairs
Although the effective binding affinities AcKa0 between PMN-WM9 and PMN-HPMEC were similar, the intrinsic kinetics per receptor-ligand bond may vary from one cell pair to another. Here we further compared the intrinsic forward rate kf and zero force reverse rate kr0 between two cell pairs, where kr0 was obtained by best fitting the binding curves using Equation 1 (Fig. 5), and kf was calculated by kf = (AcKa0 × kr0)/Ac. The apparent contact area, Ac, defined as the projected area between two cells for each contact, was calculated by Ac ≈ πD2/4, where D is the projected diameter of the contact zone, which can be measured from recorded images. Noting that the realistic contact between two cell surfaces is composed of discrete microvilli in three-dimensional configuration, this estimation still makes a sense to predict the molecular kinetics. Upon the calculated values of Ac = 18.9 ± 5.8 μm2 for PMN-WM9 pair (n = 5) and 18.5 ± 5.2 μm2 for PMN-HPMEC interaction (n = 5), the forward rate kf of β2-integrin-ICAM-1 bond yielded 2.6-fold higher for the binding of a PMN to a quiescent HPMEC than that to a quiescent WM9 cell ((0.77 ± 0.11 and 0.30 ± 0.05) × 10−7 μm2 s−1, respectively; Pt < 0.05), which is consistent with 2.1-fold difference in kf between two cell pairs when WM9 or HPMEC cells were treated with TNF-α ((0.57 ± 0.10 and 0.27 ± 0.07) × 10−7 μm2 s−1, respectively; Pt < 0.05) (Fig. 6C). Similarly, the zero force reverse rate kr0 was 2.9-fold higher for the binding of a PMN to a quiescent HPMEC than that to a quiescent WM9 (0.38 ± 0.22 and 0.13 ± 0.04 s−1, respectively), which is in accord with 2.4-fold difference in kr0 when WM9 or HPMEC cells were activated (0.29 ± 0.16 and 0.12 ± 0.06 s−1, respectively), although no significant difference was found between those values (Fig. 6D). These results demonstrated that, as compared with those expressed on WM9 cells, ICAM-1 expressed on HPMECs present a fast kinetics of binding to and detaching from β2-integrin. Our data also indicated that TNF-α stimulation relatively down-regulated the intrinsic kinetic rates (26% for kf and 24% for kr0) (Fig. 6, C and D), implying that TNF-α stimulation could, at least partially, alter the accessibility of and induce the conformational changes of ICAM-1 molecules.
Adhesion between Neutrophil and Breast Cancer Cell MDA-MB-231
To further demonstrate the biological relevance of the current work, we also measured the adhesion between human metastatic breast cell line MDA-MB-231 and PMN by GDMAT. Similar to melanoma cells, MDA-MB-231 cells are unable to adhere to endothelium directly in shear flow; instead, they express a high level of ICAM-1 and are able to bind to β2-integrin on PMNs to facilitate the tumor cell metastasis, indicating that PMN-mediated tumor cell migration was significantly reduced by blockage of β2-integrin on PMNs and of ICAM-1 on MDA-MB-231 cells (35). Here we further verified the observation using our GDMAT measurements when CD11a, CD11b, CD18, and ICAM-1 were blocked by their mAbs (Fig. 7A, Nonspecific line).
FIGURE 7.
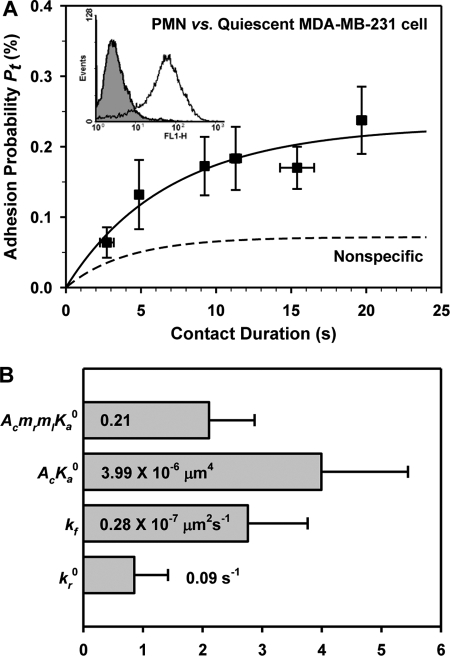
Binding curve of PMNs adhesion to quiescent MDA-MB-231 cells (A) and the two-dimensional binding kinetics (B). The data points were obtained by averaging cell pairs with similar contact durations and presented as the means ± S.E. for both adhesion probabilities and contact durations. Binding curves were calculated by PT = PN + Pa − PNPa based on the best fit parameters (54). Also plotted were the predictions fitted using Equation 1 (solid line), whereas the dashed line represented nonspecific binding, obtained by fitting Equation 1 to nonspecific data (not shown for the sake of clarity). ICAM-1 expression on MDA-MB-231 cells was measured by flow cytometer (A, insert).
The expression of ICAM-1 on MDA-MB-231 cells was quantified by flow cytometry (Fig. 7A, inset) as ml = 292 ± 67 μm−2 (n = 3), which is much lower than that on WM9 cells (799 ± 146 μm−2) but similar to that on HPMECs (246 ± 196 μm−2). The adhesion between MDA-MB-231 cells and PMNs (Fig. 7A) was lower than that for PMN-WM9 pair (PF < 0.01) but similar to that for PMN-HPMEC pair (PF > 0.1). This observation was further supported by the best fit cellular binding affinity (0.21 ± 0.07), which was lower than that for PMN-WM9 pair (0.63 ± 0.10, Pt < 0.05) but similar to that for PMN-HPMEC pair (0.16 ± 0.02, Pt = 0.57). However, the effective binding affinities per molecular pair, AcKa0, were consistent with each other ((4.34 ± 0.69, 3.68 ± 0.54, and 3.99 ± 1.45) × 10−6 μm4, respectively), indicating that the ICAM-1 expression level is the determinant for the cell adhesion. In contrast, the intrinsic forward rate kf and the zero force reverse rate kr0 at molecular level were similar to those of PMN-WM9 pair (Fig. 7B).
DISCUSSION
The goal of the current work is to determine the adhesions among tumor cells, neutrophils, and endothelial cells through direct quantification of the two-dimensional binding kinetics of interacting molecules on nucleated cells. A human melanoma cell line WM9, which has average metastatic, chemotactic and adhesion potentials (unpublished data in the lab of Dr. Cheng Dong) among melanoma cell lines, was chosen for the study. Upon the modified gas-driven micropipette aspiration technique, the interactions between β2-integrin and ICAM-1 expressed on the opposed nucleated cells were measured using an adhesion frequency approach, and the kinetic rates and binding affinities were estimated from a probabilistic model. We found that cell adhesion between PMN-WM9 pair was 3.9-fold higher than that for PMN-HPMEC pair, mainly because of the higher ICAM-1 expression level for quiescent and TNF-α-stimulated melanoma cells. We also demonstrated that the effective binding affinity was comparable between two cell pairs when WM9 or HPMEC cells were intact or cytokine-induced, whereas intrinsic forward and reverse rates of β2-integrin and ICAM-1 interactions varied significantly between the two cell pairs. We also chose another ICAM-1-expressed tumor cell line, the human metastatic breast adenocarcinoma cell MDA-MB-231, to quantify the binding kinetics of ICAM-1 to β2-integrin on PMNs. The results further supported our conclusion that the ICAM-1 expression level is the determinant for such β2-integrin-ICAM-1-mediated cell adhesion. Furthermore, it seems that the intrinsic kinetic rates per molecule pair (kf and kr0) of tumor cells are similar but lower than those of ECs. Therefore, our results provided a new insight into understanding the kinetics of interacting molecules to regulate the leukocyte-facilitated tumor cell adhesion to the endothelium.
Malignant melanoma is a potentially lethal melanocytic neoplasm with a propensity for distant metastasis. Our findings in the current work are physiologically relevant to test the hypothesis proposed in leukocyte-facilitated metastasis of melanoma cells (36–38). For example, two-dimensional kinetics measurements at the cellular level indicated that PMN-WM9 adhesion is much stronger than PMN-HPMEC adhesion, proposing a possible mechanism that WM9 cells may first interact with PMNs and then bind to HPMECs. This speculation is distinct from the “two-step adhesion” hypothesis where PMN was captured by ECs prior to its capture of the circulating TCs (10). Melanoma adhesion to the endothelium by the bridging of leukocytes is mainly mediated by β2-integrin-ICAM-1 bindings (Fig. 4). Although other cellular adhesion molecules, i.e. P-/E-selectin, sLex, and VCAM-1 on endothelial cells (39); PSGL-1, CD44, and VLA-4 on PMNs (39, 40); or sLex, CD44, and VLA-4 on tumor cells (1, 7, 10) are also able to support the adhesion between tumor cells, PMNs, and endothelial cells, our previous results showed that, with a 24-h TNF-α simulation, P-/E-selectin, sLex, and VCAM-1 expressions are really low on stimulated HPMECs, and those of sLex and VLA-4 are much lower than ICAM-1 on stimulated WM9 cells (7). Blockage of β2-integrin on PMNs and ICAM-1 on WM9 or HPMEC cells almost abolished PMN-WM9 and PMN-HPMEC adhesions (Fig. 4), indicating the dominant role of β2-integrin and ICAM-1 interaction in mediating cell adhesion. In a typical test, for example, the baseline expression of E-selectin was quite low for quiescent HPMECs, and no significant up-regulation was found with 24-h TNF-α-simulation (data not shown), which is consistent with previous results (30). Regardless of this, the impact of other adhesion molecules on cell adhesion should be taken into account in the future physiological studies. β2-Integrin and ICAM-1 interaction facilitated tumor cell adhesion, and extravasation is a common mechanism for tumor cells. This paradigm has been found in melanoma (9, 10, 17, 37, 38), breast cancer (35, 41, 42), and colon adenocarcinoma (1, 16). Thus, we also conducted a kinetics study between breast cancer cells MDA-MB-231 and PMNs. The nonspecific binding curve by blocking β2-integrin on PMNs (CD11a, CD11b, and CD18) and ICAM-1 on tumor cells showed the dominant role of β2-integrin and ICAM-1 in supporting the adhesion of breast cancer cells and PMNs. Furthermore, two-dimensional kinetic parameters so obtained are meaningful to understand the potential mechanisms of how a breast cancer tumor cell adheres to the endothelium.
The two-dimensional kinetics measurements at the molecular level reported in the current work are in good agreement with those reported previously (as summarized in Table 1). Here the binding of β2-integrin on PMNs to ICAM-1 ligand on WM9 or HPMEC cells yielded similar effective binding affinity AcKa0 (Fig. 6B), indicating that the difference at cellular level between PMN-WM9 and PMN-HPMEC adhesion is mainly attributed to the distinct expressions of ICAM-1 on WM9 cells and HPMECs (Fig. 3). In contrast, the high forward rate kf and reverse rate kr0 in PMN-HPMEC pair suggested that PMNs bind fast to and also detach quickly from ECs, as compared with WM9 cells (Fig. 6, C and D). A possible interpretation for this contrast is the different surface expressions of ICAM-1 molecules, because the combination of monomeric and dimeric ICAM-1 isoforms should not necessarily be identical between two types of cells, and the different presentation of those isoforms would yield distinct binding kinetics (43–45). The difference of body stiffness and surface microtopology between WM9 and HPMEC cells may also significantly affect the two-dimensional binding kinetics (22, 46).
TABLE 1.
Summary of two-dimensional kinetics studies for β2-integrin-ICAM-1 binding
AFM, atomic force microscopy.
| Receptor | AcKa0 × 106 | kr0 | Assay | Reference |
|---|---|---|---|---|
| μm4 | s−1 | |||
| β2-Integrin | 3.60–4.34 | 0.12–0.38 | GDMAT | This work |
| β2-Integrin | 0.33–1.97 | Cone-plate | Ref. 12 | |
| β2-Integrin | 0.40 | Cone-plate | Ref. 58 | |
| β2-Integrin | 0.31–0.50 | Flow chamber | Ref. 59 | |
| β2-Integrin | 0.70 | MAT | Ref. 56 | |
| LFA-1 | 0.25–0.44 | Flow chamber | Ref. 18 | |
| LFA-1 | 0.99–8570 | 0.19–2.51 | MAT | Ref. 60a |
| LFA-1 | 0.17–57 | AFM | Ref. 61b | |
| Mac-1 | 0.49–2.38 | AFM | Ref. 62 |
It has been known that LFA-1 and Mac-1, the two primary β2-integrin members that mediate the arrest and transmigration of leukocytes at inflammation site, play different roles to mediate the adhesion between PMNs and WM9 or HPMEC cells. On one hand, the expression of αL (CD11a), αM (CD11b), and β2 (CD18) subunits determined by flow cytometry are 10,418 ± 4,020, 46,501 ± 7,565, and 36,341 ± 6,871 per PMN, respectively. It is interesting that the site density of CD11b is higher than CD18 in the current work, as well as those reported previously (47, 48). Although the underlying mechanism of such the expression of β2-integrin is not clear, some studies hinted that, in addition to the dominant configuration of a heterodimer consisting of one-to-one matched α and β subunits, the transmembrane domains of both subunits tend to form homo-oligomers (αα or ββ) (49–51), so that the number of the β subunit is not necessarily equal to that of α subunit. On the other hand, the biological functions of LFA-1 and Mac-1 are quite distinct, where the former mediates the initial tethering on endothelium and the latter stabilizes the binding to form the firm adhesions (9, 10, 52, 53). Because the measurements of two-dimensional kinetics of LFA-1 or Mac-1 molecules alone are beyond the scope of the current work, we performed a simple analysis upon a concurrent binding model for multiple receptor-ligand species in cell adhesion (54, 55). To isolate the kinetic parameters of LFA-1 and Mac-1 binding to ICAM-1 respectively, the adhesion frequency for PMNs binding to WM9 or HPMEC cells mediated by LFA-1 and Mac-1 molecules is written as Pa = 1 − exp(−<n1> − <n2>), where <n1> and <n2> are the averaged numbers of bonds for respective LFA-1 and Mac-1 molecules and follow <ni> = AcmrimliKai0[1 − exp(−kri0t)] for species i. We tried to best fit the formulations using our data sets (Fig. 5), but unfortunately, no reliable kinetic parameters were collected. The line of reasoning lies in the following aspects (55): 1) the difference between kr10 and kr20 should be at least 1 order of magnitude different; 2) both species should have significant contribution to cell adhesion; and 3) the experimental data should have relative small deviations. Thus, further measurements for LFA-1 or Mac-1 are required to isolate their respective biological functions in mediating the cell adhesion.
Technically, the GDMAT assay modified in the current work is able to be widely applied in determining the binding kinetics of surface-bound molecules, as compared with the conventional MAT technique (21–23). First, GDMAT is more physiologically relevant to determine the binding kinetics of cell adhesion molecules constitutively expressed on the nucleated cells. In a conventional MAT assay, however, at least one species of interacting molecules needs to be purified from cell surface and then coupled on RBC surface, which reduces its biological significance by altering their intrinsic surface presentations. Second, the PMN cell in GAMAT assay is forced to move freely inside the pipette with relatively low impinging force, whereas the PMN serving as the force transducer in MAT assay needs to be mechanically aspirated by suction pressure and readily activated by the repeated mechanical contacts with high impinging force. Third, although a few works reported that the adhesion between an ICAM-1-coated bead and a β2-integrin-expressed PMN was determined by identifying the membrane deformation of the PMN when separating from the bead (56, 57), it is almost impossible in our tests to visualize directly the membrane deflection of PMN, TC, or EC, presumably because of the differences of mechanical features among PMN, RBC, or the bead. Thus, this new technique makes the in vitro adhesion frequency approach a more powerful tool in measuring surface-bound receptor-ligand interactions between physiological cells.
Finally, we applied a modified GDMAT assay to quantify the two-dimensional kinetics of β2-integrin-ICAM-1 binding that mediates PMN-TC and PMN-EC adhesion. Our data proposed that the ICAM-1 expression level is dominant for cell-cell adhesion. Furthermore, the intrinsic kinetic rates of β2-integrin and ICAM-1 per molecule pair were found to be different between PMN-TC and PMN-EC pairs. Combined with the other functional assays such as parallel flow chamber and even in vivo measurements, this approach is beneficial to understand the mechanisms in leukocyte-facilitated metastasis of melanoma cells.
Supplementary Material
Acknowledgments
We thank Dr. M. Herlyn (Wistar Institute, Philadelphia, PA) for the generous gifts of WM9 melanoma cells and Dr. C. J. Kirkpatrick (Institute of Pathology, Johannes-Gutenberg University, Mainz, Germany) for providing HPMEC ST1.6R. We are also grateful to Eric Weidert for critical reading, Dr. Jun Huang for helpful discussion in curve fitting, and Dr. Bing Li for suggestions in statistical analysis.
This work was supported, in whole or in part, by National Institutes of Health Grants CA-97306, CA-125707, M01-RR-010732, and C06-RR-016499. This work was also supported by National Natural Science Foundation of China Grants 30730032, 10702075, and 10902117; National Key Basic Research Foundation of China Grants 2006CB910303 and 2011CB710904; Strategic Priority Research Program of Chinese Academy of Science grants XDA01030102 and XDA04020219, Knowledge Innovation Program of Chinese Academy of Science Grant KJCX2-YW-L08 (to M. L.), and National Science Foundation Grants BES-0138474 and CBET-0729091 (to C. D.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and supplemental Movie S1.
- TC
- melanoma tumor cell
- EC
- endothelial cell
- HPMEC
- human pulmonary microvascular endothelial cell
- PMN
- polymorphonuclear neutrophil
- ICAM-1
- intercellular adhesion molecule 1
- MAT
- micropipette aspiration technique
- GDMAT
- gas-driven MAT
- RBC
- red blood cell.
REFERENCES
- 1. Konstantopoulos K., Thomas S. N. (2009) Annu. Rev. Biomed. Eng. 11, 177–202 [DOI] [PubMed] [Google Scholar]
- 2. Miles F. L., Pruitt F. L., van Golen K. L., Cooper C. R. (2008) Clin. Exp. Metastasis 25, 305–324 [DOI] [PubMed] [Google Scholar]
- 3. Miller A. J., Mihm M. C., Jr. (2006) N. Engl. J. Med. 355, 51–65 [DOI] [PubMed] [Google Scholar]
- 4. Ma Y. Q., Geng J. G. (2000) J. Immunol. 165, 558–565 [DOI] [PubMed] [Google Scholar]
- 5. Ludwig R. J., Boehme B., Podda M., Henschler R., Jager E., Tandi C., Boehncke W. H., Zollner T. M., Kaufmann R., Gille J. (2004) Cancer Res. 64, 2743–2750 [DOI] [PubMed] [Google Scholar]
- 6. Goetz D. J., el-Sabban M. E., Hammer D. A., Pauli B. U. (1996) Int. J. Cancer 65, 192–199 [DOI] [PubMed] [Google Scholar]
- 7. Liang S., Dong C. (2008) Am. J. Physiol. Cell. Physiol. 295, C701–C707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zetter B. R. (1993) Semin. Cancer Biol. 4, 219–229 [PubMed] [Google Scholar]
- 9. Slattery M. J., Liang S., Dong C. (2005) Am. J. Physiol. Cell. Physiol. 288, C831–C839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liang S., Slattery M. J., Dong C. (2005) Exp. Cell Res. 310, 282–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Slattery M. J., Dong C. (2003) Int. J. Cancer 106, 713–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liang S., Fu C., Wagner D., Guo H., Zhan D., Dong C., Long M. (2008) Am. J. Physiol. Cell. Physiol. 294, C743–C753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Springer T. A. (1994) Cell 76, 301–314 [DOI] [PubMed] [Google Scholar]
- 14. Muller W. A. (2002) Lab. Investig. 82, 521–533 [DOI] [PubMed] [Google Scholar]
- 15. Jadhav S., Bochner B. S., Konstantopoulos K. (2001) J. Immunol. 167, 5986–5993 [DOI] [PubMed] [Google Scholar]
- 16. Jadhav S., Konstantopoulos K. (2002) Am. J. Physiol. Cell. Physiol. 283, C1133–C1143 [DOI] [PubMed] [Google Scholar]
- 17. Liang S., Slattery M. J., Wagner D., Simon S. I., Dong C. (2008) Ann. Biomed. Eng. 36, 661–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vitte J., Pierres A., Benoliel A. M., Bongrand P. (2004) J. Leukocyte Biol. 76, 594–602 [DOI] [PubMed] [Google Scholar]
- 19. Rinker K. D., Prabhakar V., Truskey G. A. (2001) Biophys. J. 80, 1722–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Long M., Lü S., Sun G. (2006) Cell. Mol. Immunol. 3, 79–86 [PubMed] [Google Scholar]
- 21. Chesla S. E., Selvaraj P., Zhu C. (1998) Biophys. J. 75, 1553–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wu L., Xiao B., Jia X., Zhang Y., Lü S., Chen J., Long M. (2007) J. Biol. Chem. 282, 9846–9854 [DOI] [PubMed] [Google Scholar]
- 23. Huang J., Chen J., Chesla S. E., Yago T., Mehta P., McEver R. P., Zhu C., Long M. (2004) J. Biol. Chem. 279, 44915–44923 [DOI] [PubMed] [Google Scholar]
- 24. Chen W., Evans E. A., McEver R. P., Zhu C. (2008) Biophys. J. 94, 694–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Long M., Zhao H., Huang K. S., Zhu C. (2001) Ann. Biomed. Eng. 29, 935–946 [DOI] [PubMed] [Google Scholar]
- 26. Huang J., Zarnitsyna V. I., Liu B., Edwards L. J., Jiang N., Evavold B. D., Zhu C. (2010) Nature 464, 932–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shao J. Y., Hochmuth R. M. (1996) Biophys. J. 71, 2892–2901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shao J. Y., Ting-Beall H. P., Hochmuth R. M. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 6797–6802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Levin J. D., Ting-Beall H. P., Hochmuth R. M. (2001) Biophys. J. 80, 656–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Krump-Konvalinkova V., Bittinger F., Unger R. E., Peters K., Lehr H. A., Kirkpatrick C. J. (2001) Lab. Investig. 81, 1717–1727 [DOI] [PubMed] [Google Scholar]
- 31. Shao J. Y., Xu J. B. (2002) J. Biomech. Eng. 124, 388–396 [DOI] [PubMed] [Google Scholar]
- 32. Gelles J., Schnapp B. J., Sheetz M. P. (1988) Nature 331, 450–453 [DOI] [PubMed] [Google Scholar]
- 33. Sun G., Zhang Y., Huo B., Long M. (2009) Eur. Biophys. J. 38, 701–711 [DOI] [PubMed] [Google Scholar]
- 34. Sun G. Y., Zhang Y., Huo B., Long M. (2009) Cell. Mol. Bioeng. 2, 495–503 [Google Scholar]
- 35. Wu Q. D., Wang J. H., Condron C., Bouchier-Hayes D., Redmond H. P. (2001) Am. J. Physiol. Cell. Physiol. 280, C814–C822 [DOI] [PubMed] [Google Scholar]
- 36. Dong C., Robertson G. P. (2009) Biorheology 46, 265–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Huh S. J., Liang S., Sharma A., Dong C., Robertson G. P. (2010) Cancer Res. 70, 6071–6082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liang S., Sharma A., Peng H. H., Robertson G., Dong C. (2007) Cancer Res. 67, 5814–5820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ley K., Laudanna C., Cybulsky M. I., Nourshargh S. (2007) Nat. Rev. Immunol. 7, 678–689 [DOI] [PubMed] [Google Scholar]
- 40. McEver R. P., Zhu C. (2010) Annu. Rev. Cell Dev. Biol. 26, 363–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Welch D. R., Schissel D. J., Howrey R. P., Aeed P. A. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 5859–5863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Aeed P. A., Nakajima M., Welch D. R. (1988) Int. J. Cancer 42, 748–759 [DOI] [PubMed] [Google Scholar]
- 43. Miller J., Knorr R., Ferrone M., Houdei R., Carron C. P., Dustin M. L. (1995) J. Exp. Med. 182, 1231–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Reilly P. L., Woska J. R., Jr., Jeanfavre D. D., McNally E., Rothlein R., Bormann B. J. (1995) J. Immunol. 155, 529–532 [PubMed] [Google Scholar]
- 45. Jun C. D., Shimaoka M., Carman C. V., Takagi J., Springer T. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 6830–6835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Williams T. E., Nagarajan S., Selvaraj P., Zhu C. (2001) J. Biol. Chem. 276, 13283–13288 [DOI] [PubMed] [Google Scholar]
- 47. Bateman J., Parida S. K., Nash G. B. (1993) Cell Biochem. Funct. 11, 87–91 [DOI] [PubMed] [Google Scholar]
- 48. McCarthy D. A., Macey M. G., Cahill M. R., Newland A. C. (1994) Cytometry 17, 39–49 [DOI] [PubMed] [Google Scholar]
- 49. Fu G., Wang C., Wang G. Y., Chen Y. Z., He C., Xu Z. Z. (2006) Biochem. Biophys. Res. Commun. 351, 847–852 [DOI] [PubMed] [Google Scholar]
- 50. Parthasarathy K., Lin X., Tan S. M., Law S. K., Torres J. (2008) Protein Sci. 17, 930–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Solovjov D. A., Pluskota E., Plow E. F. (2005) J. Biol. Chem. 280, 1336–1345 [DOI] [PubMed] [Google Scholar]
- 52. Hentzen E. R., Neelamegham S., Kansas G. S., Benanti J. A., McIntire L. V., Smith C. W., Simon S. I. (2000) Blood 95, 911–920 [PubMed] [Google Scholar]
- 53. Neelamegham S., Taylor A. D., Burns A. R., Smith C. W., Simon S. I. (1998) Blood 92, 1626–1638 [PubMed] [Google Scholar]
- 54. Williams T. E., Selvaraj P., Zhu C. (2000) Biophys. J. 79, 1858–1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhu C., Williams T. E. (2000) Biophys. J. 79, 1850–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lomakina E. B., Waugh R. E. (2004) Biophys. J. 86, 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lomakina E. B., Waugh R. E. (2009) Biophys. J. 96, 276–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fu C. L., Tong C. F., Dong C., Long M. (2011) Cell. Mol. Bioeng. 10.1007/s12195-12011-10167-x [DOI] [Google Scholar]
- 59. Hoskins M. H., Dong C. (2006) Mol. Cell. Biomech. 3, 79–87 [PMC free article] [PubMed] [Google Scholar]
- 60. Zhang F., Marcus W. D., Goyal N. H., Selvaraj P., Springer T. A., Zhu C. (2005) J. Biol. Chem. 280, 42207–42218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhang X., Wojcikiewicz E., Moy V. T. (2002) Biophys. J. 83, 2270–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yang H., Yu J., Fu G., Shi X., Xiao L., Chen Y., Fang X., He C. (2007) Exp. Cell Res. 313, 3497–3504 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.