Abstract
The malignancy of a tumor depends on the capability of cancer cells to metastasize. The process of metastasis involves cell invasion through connective tissue and transmigration through endothelial monolayers. The expression of the glycosylphosphatidylinositol-anchored receptor CD24 is increased in several tumor types and is consistently associated with increased metastasis formation in patients. Furthermore, the localization of β1-integrins in lipid rafts depends on CD24. Cell invasion is a fundamental biomechanical process and usually requires cell adhesion to the extracellular matrix (ECM) mainly through β1 heterodimeric integrin receptors. Here, we studied the invasion of A125 human lung cancer cells with different CD24 expression levels in three-dimensional ECMs. We hypothesized that CD24 expression increases cancer cell invasion through increased contractile forces. To analyze this, A125 cells (CD24 negative) were stably transfected with CD24 and sorted for high and low CD24 expression. The invasiveness of the CD24high and CD24low transfectants was determined in three-dimensional ECMs. The percentage of invasive cells and their invasion depth was increased in CD24high cells compared with CD24low cells. Knockdown of CD24 and of the β1-integrin subunit in CD24high cells decreased their invasiveness, indicating that the increased invasiveness is CD24- and β1-integrin subunit-dependent. Fourier transform traction microscopy revealed that the CD24high cells generated 5-fold higher contractile forces compared with CD24low cells. To analyze whether contractile forces are essential for CD24-facilitated cell invasion, we performed invasion assays in the presence of myosin light chain kinase inhibitor ML-7 as well as Rho kinase inhibitor Y27632. Cell invasiveness was reduced after addition of ML-7 and Y27632 in CD24high cells but not in CD24neg cells. Moreover, after addition of lysophosphatidic acid or calyculin A, an increase in pre-stress in CD24neg cells was observed, which enhanced cellular invasiveness. In addition, inhibition of the Src kinase or STAT3 strongly reduced the invasiveness of CD24high cells, slightly reduced that of CD24low cells, and did not alter the invasiveness of CD24neg cells. Taken together, these results suggest that CD24 enhances cell invasion through increased generation or transmission of contractile forces.
Keywords: Biophysics, Cell Migration, Integrin, siRNA, Tumor, CD24, Contractile Forces, Cytoskeletal Remodeling Dynamics, Invasion, Matrix Metatalloproteinases
Introduction
Metastasis is the primary cause of death in cancer patients. The process of metastasis formation depends on cell adhesion, transendothelial migration of lymphoid or blood vessels, and invasion of tumor cells that spread from the primary tumor (1–4).
All these steps involve integrins, a family of transmembrane, heterodimeric adhesion receptors, composed of noncovalently linked α and β subunits (5–7). Integrins facilitate transmembrane connections between the actomyosin cytoskeleton of the cell and the connective tissue (8–10). These connections are organized in discrete clusters as focal adhesions (11). Focal adhesions anchor cells in their environment and transmit contractile forces generated by the actomyosin cytoskeleton (12–15).
The actomyosin cytoskeleton carries the contractile pre-stress that is necessary for controlling cell morphology and for providing mechanical properties (16, 17). Besides the mechanical link between contractile pre-stress and focal adhesions, they are also connected through signaling processes (11, 18–20).
The activation of the integrins leads to conformational changes of the receptor and is critical for its function during cell adhesion, transendothelial migration, and cell invasion (7, 21, 22). The clustering of integrins on the cell surface (e.g. the alterations of the lateral localization) is also associated with their function (23).
Several adhesion receptors have been identified to act either as negative (E-cadherin) or positive factors (αvβ3- and α5β1-integrin) of tumor invasion and metastasis formation (24–26). The localization of the β1-integrins on the cell surface is regulated by the small, heavily glycosylated GPI2-anchored cell-surface protein CD24 that consists of 31 amino acids in humans (27).
Several studies reported that CD24 expression is a marker for poor prognosis in numerous tumor types (28–32). CD24 expression is increased in several tumor types and is consistently associated with increased metastasis formation in patients and rodent metastasis models (28, 29, 33–36). CD24 has been reported to increase tumor growth and motility in a Boyden chamber assay as well as in three-dimensional collagen fiber matrices (27, 37–39). The mechanism that underlies CD24-facilitated cancer cell motility and invasion still remains unrevealed. It is solely known that CD24 is located in the lipid rafts and plays a role in regulating the entrance of CXCR4 and β1-integrins into this specific membrane domain (27, 39).
The aim of this study was to analyze the role of the CD24 for cancer cell invasion under controlled in vitro conditions and to characterize the biomechanical invasion strategy that is activated by CD24. We used 2.4 mg/ml synthetic three-dimensional ECMs with pores under the size of a cell for the invasion assays (26, 40, 41). The invasiveness and the speed of migration in such a system depend mainly on biomechanical processes including the following: (i) cell adhesion and de-adhesion (42); (ii) cytoskeletal remodeling (41); (iii) protrusive force generation (42, 43), and (iv) matrix properties such as stiffness, pore size, ECM protein composition, and enzymatic degradation (15). In addition to the invasiveness, the cell invasion strategies, mesenchymal or amoeboid migration, depend on the balance of these biomechanical parameters (44, 45).
In this study, we investigated whether the expression of CD24 increased the invasiveness of cancer cells in a three-dimensional collagen fiber matrix by increasing the generation or transmission of contractile forces. Transfectants of lung carcinoma cell lines selected for high CD24 expression displayed an increased invasiveness in three-dimensional collagen matrices, whereas knockdown of the CD24 decreased cancer cell invasion. We systematically tested the CD24 receptor specificity of the invasion-enhancing effect and measured cell adhesion strength, cytoskeletal remodeling, and traction force generation. In addition, we blocked enzymatic matrix degradation. We found that the CD24 receptor contributes substantially to the invasiveness of lung cancer cells by promoting the transmission and generation of contractile forces.
EXPERIMENTAL PROCEDURES
Cells and Cell Culture
The lung adenocarcinoma cell line A125 was stably transfected with a CD24 expression plasmid using JetPEI (Biomol, Hamburg, Germany) or calcium-phosphate, respectively. Transfectants were enriched by G418 selection and cell sorting by FACS and magnetic cell sorting using mAb SWA-11 against human CD24 (46). A125 cells were maintained in Dulbecco's modified Eagle's medium (1 g/liter glucose) containing 10% fetal calf serum (FCS; low endotoxin, <0.1 enzyme units/ml), 2 mm l-glutamine, and 100 units/ml penicillin/streptomycin (Biochrom, Berlin, Germany). 80% confluent cells were used in passages 6–40. Accutase was used for cell harvesting (<1% dead cells). Mycoplasma contamination was excluded using a mycoplasma detection kit (Roche Diagnostics). All other chemicals used were purchased from Sigma.
Isolation of Subcell Lines
The human breast cancer cell line MDA-MB-231, which expresses endogenous CD24, was used for the generation of subcell lines with high (231endCD24high) and low endogenous expression of CD24 (231endCD24low). To isolate highly and lowly expressing subcell lines, MDA-MB-231 cells were stained with a mouse antibody against human CD24 (clone SWA11) for 30 min at 4 °C and then with a secondary (R)-PE-labeled goat anti-mouse-IgG (F(ab)2 fragment; Dianova) antibody for 30 min at 4 °C. Single cells of both subgroups were sorted in 10 96-well plates using a cell sorter and grown to colonies and finally subcell lines at 95% humidity, 37 °C, and 5% CO2 in an incubator. Subcell lines high and low endogenous CD24 expression were analyzed at least three times for each clone and remained stable over the whole investigation time.
Three-dimensional ECM Invasion Assay
A three-dimensional collagen invasion assay was used to study invasion of cancer cells. For a 6-well plate, 3.5 ml of collagen R (Serva, Heidelberg, Germany) and 3.5 ml of collagen G (Biochrom, Berlin, Germany) were mixed, and 0.8 ml of 278 mm sodium bicarbonate (end concentration 26.5 mm) and 0.8 ml of 10× DMEM (Biochrom) was added. After each addition to the collagen solution, air bubbles were avoided during the mixture process. Polymerization started after the solution was neutralized with 1 n sodium hydroxide and incubated at 37 °C, 95% humidity, and 5% CO2. 1.2 ml of collagen solution was added to each well of a 6-well plate and then polymerized. Polymerized collagen gels were normally 500 μm thick. The three-dimensional collagen matrices were incubated overnight with 2 ml of DMEM (41). 100,000 cancer cells were seeded on top of the three-dimensional ECMs and cultured for 72 h at 37 °C, 5% CO2, and 95% humidity in DMEM containing 10% FCS. At this time period, differences in the invasiveness of cells were clearly visible. For serum-free cell invasion, cancer cells were cultured 24 h before and during the invasion assay in EX-cell 293 medium (SAFC BioSciences, Lenexa, KS) with 100 units/ml penicillin/streptomycin. After fixation with 2.5% glutaraldehyde solution in PBS buffer, the percentage of invasive cells and their invasion depths were determined in 12 randomly selected fields of view. The calculation of the percentage of invasive cells was performed 3 days after invasion assay start. The percentage of invasive cells (cells inside the three-dimensional ECMs) was determined from total cells, which means adhesive cells on top of the three-dimensional ECMs and invasive cells inside of the three-dimensional ECMs) (26).
Modulation of Cell Invasion
To inhibit or modulate cell invasion, we added 15 μm myosin light chain kinase inhibitor ML-7 (Calbiochem), 100 μm Rho kinase inhibitor Y27632 (Sigma), 2 μm latrunculin B (sequesters G-actin and promotes depolymerization, Sigma), or 1 nm calyculin A (inhibits serine/threonine phosphatases and induces contraction, Calbiochem) to the three-dimensional ECM invasion assay prior to cell seeding. Furthermore, to analyze the CD24-mediating signaling, we inhibited or modulated cell invasion by addition of 30 μm Src tyrosine kinase inhibitor (catalog no. 567805, Calbiochem), 30 μm STAT3 inhibitor peptide (catalog no. 573096, Calbiochem), which blocks the STAT3 activation and suppresses constitutive STAT3-dependent Src transformation, or 100 μm tyrosine kinase inhibitor herbimycin A (catalog no. 375670, Calbiochem).
Inhibition of Enzymatic Degradation
To inhibit enzymatic degradation of the three-dimensional collagen matrices, we added a protease inhibitor (PI) mixture, which also blocks the matrix metalloproteinases, including MT1-MMP, before the start of the invasion assay. The PI mixture contains 50 μm GM6001, 250 μm E-64, 2 μm leupeptin, 100 μm pepstatin A, and 2.2 μm aprotinin (all from Calbiochem) (26, 44, 47, 48).
Flow Cytometry
80% confluent cancer cells were harvested with Accutase (PAA) and resuspended in HEPES buffer (20 mm HEPES, 125 mm NaCl, 45 mm glucose, 5 mm KCl, 0.1% albumin, pH 7.4). Cells were incubated with mouse antibodies against human CD24 (clone SWA11) (27), α1 (FB12), α2 (P1E6), β1 (Biozol), and MT1-MMP (128527, R&D Systems), all purchased from Chemicon unless otherwise stated. Isotype-matched antibodies were used as controls (Caltag, Burlingame, CA). After 30 min at 4 °C, cells were washed and stained with (R)-PE-labeled goat anti-mouse IgG (F(ab)2 fragment; Dianova) and for siRNA-treated cells with Cy2-labeled anti-mouse IgG antibodies (Dianova). Flow cytometry was performed using a FACSCalibur System (BD Biosciences). CellQuest software (BD Biosciences) and WinMDI2.9 software were used for data analysis.
siRNA Transfection
200,000 80% confluent CD24high cells or 231endCD24high cells were seeded in 3.5-cm inner diameter dishes and cultured in 2 ml of DMEM complete medium. 5 μl of a 20 μm CD24 (target sequences for CD24, siCD24-1 AAACAACAACTGGAACTTCAA and siCD24-2 AACATGTGAGAGGTTTGACTA), β1-integrin (target sequence for β1, siβ1 AGGGCCATTAGACCTGATAAA), α1-integrin (target sequence for α1, siα1 TTGGACTTTAATCTTACCGAT), α2-integrin (target sequence for α2, siα2 TCGCTAGTATTCCAACAGAAA), or Allstar-control RNAi solution (control siRNA), 12 μl of HiPerFect reagent (Qiagen), and 100 μl of DMEM were mixed (41). RNAi-mediated β1-, α1-, and α2-integrin knockdowns were confirmed by flow cytometry using anti-β1-integrin, anti-α1-integrin, or anti-α2-integrin and Cy2-labeled anti-mouse IgG antibodies (Dianova). Transfection efficiency was determined by flow cytometry to be >99% using 20 μm Alexa-Fluor546-labeled siRNA.
Quantitative Real Time PCR
The quantitative RT-PCR was performed as described (49). 10 ng of total cDNA were analyzed in triplicate. The primers for the quantitative RT-PCR were designed using the IDT primer quest program and produced by MWG (Ebersberg, Germany). The PCR was performed using a SYBR Green MasterMix (Applied Biosystems, Darmstadt, Germany) in an ABI 7300 analyzer. β-Actin was used as an internal standard. The sequences of primers were as follows: CD24 forward, TGCCTCGACACACATAAACC, and CD24 reverse, GTGACCATGCGAACAAAAGA; MT1-MMP forward, TGATGGATGGATACCCAATGCCC, and MT1-MMP reverse, CGCCTCATCAAACACCCAATGCT.
Immunofluorescence
20,000 cells were seeded on glass coverslips (Menzel, Braunschweig, Germany) coated with 50 mg/ml collagen type I or 100 μg/ml fibronectin (Roche Diagnostics) for 2 h at 37 °C. After 16 h of culture time, cells were fixed using 3% paraformaldehyde (15 min, 20 °C), permeabilized for 5 min with 0.1% Triton X-100, stained for 30 min at 20 °C with 66 nm Alexa-Fluor546-phalloidin (Molecular Probes, Eugene, OR).
Magnetic Tweezer
Using magnetic tweezers, a staircase-like sequence of step forces ranging from 0.5 to 10 nN was applied to superparamagnetic epoxylated 4.5-μm beads coated with 100 μg/ml fibronectin from human plasma (Roche Diagnostics, catalog no. 11080938001) (41, 45). Fibronectin was present in the three-dimensional ECMs, because it was embedded from the 10% fetal calf serum in the cell culture medium and from cultured cancer cells, which also secrete fibronectin. 2·105 beads were sonicated, added to 105 cells, and incubated for 30 min at 37 °C and 5% CO2. The measurements were performed at 37 °C with an inverted microscope (DMI-Leica) after at least 30 min of bead binding, which is sufficient to connect the beads to the cytoskeleton. Once the beads are firmly connected to the cytoskeleton, the molecular details of the connection matter little and had no impact on the motion of beads (50). The creep response J(t) of cells during force application followed a power law in time, J(t) = a(t/t0)b, where the pre-factor a and the power law exponent b were force-dependent, and the reference time t0 was set to 1 s. Bead displacement in response to a staircase-like force followed a superposition of power laws (51) from which the force dependence of a and b was determined by a least squares fit (45). The parameter a (units of μm/nN) characterizes the elastic cell properties and corresponds to a compliance (inverse of stiffness) (45). The force/distance relationship in units of nN/μm is related to cell stiffness in units of Pa by a geometric factor that depends on the contact area between the bead and the cell (or the degree of bead internalization) and the cell height. If those parameters are known, the geometric factor can be estimated from a finite element analysis (52). Without knowledge of cell height and bead internalization, one can still estimate the typical strain ϵ as the bead displacement d divided by the bead radius r, and the typical stress σ as the applied force F divided by the bead cross-sectional area πr2 such that the cell stiffness G = σ/ϵ = r/d·F/(πr2) (53). For 4.5-μm beads as used in our study, the geometric factor is 0.14 μm−1, and a cell with an apparent stiffness of 1 nN/μm would have a “proper” stiffness of 140 Pa.
The power law exponent b reflects the stability of force-bearing cellular structures connected to beads. A value for b = 1 and b = 0 indicates Newtonian-viscous and elastic behavior, respectively (54, 55). A non-zero power law exponent implies that part of the deformation energy during magnetic force application is not elastically stored in the cytoskeleton but is dissipated in the form of heat because the cytoskeletal structures to which the bead is connected remodel (56). Hence, dissipation is directly linked to the rate at which the elastic bonds in the cytoskeleton break up and turn over. The turnover of actomyosin bonds contributes to the dissipative properties (57), and although this is not considered a remodeling event, it enables contractility driven shape changes in the cytoskeleton.
In cells, the power law exponent usually falls in the range between 0.1 and 0.5, whereby higher values have been linked to a higher turnover rate of cytoskeletal structures (54, 58). The a and b values are averaged over all applied forces and all beads measured and are expressed as means ± S.E.
Traction Microscopy
4.7% acrylamide, 0.24% bisacrylamide matrices were cast on nonelectrostatic silane-coated glass slides (59). The Young's modulus of matrices was 5.4 kPa. Yellow-green fluorescent 0.5-μm carboxylated beads (Invitrogen) were embedded into the matrices and centrifuged toward the surface of the matrices during polymerization at 4 °C (41, 45). These embedded beads served as markers for matrix deformations. The matrix surface was activated with sulfo-SANPAH (Pierce) and coated with 100 μg/ml fibronectin. Fibronectin was used because it is a component of our three-dimensional ECMs; the coating is well suited for traction microscopy, and the fibronectin-binding α5β1-integrins are expressed with equal amounts on CD24neg, CD24low, and CD24high cells. After cells adhered to the matrices, cell tractions were computed from the surface displacements of the matrix measured before and after force relaxation and detachment of cells with 8 μm cytochalasin D and 15 μm ML-7 in trypsin/EDTA (60). Matrix deformations were measured using a Fourier-based difference with interpolation image analysis (61). All experiments were performed at 37 °C, 5% CO2 and 95% humidity in DMEM containing 10% FCS in a microscope stage incubation chamber.
Statistical Analysis
All data were expressed as mean values ± S.D. Statistical analyses were performed using the two-tailed Student's t test. A p < 0.05 was considered to be statistically significant.
RESULTS
CD24 Expression Increases the Invasiveness of Cancer Cells
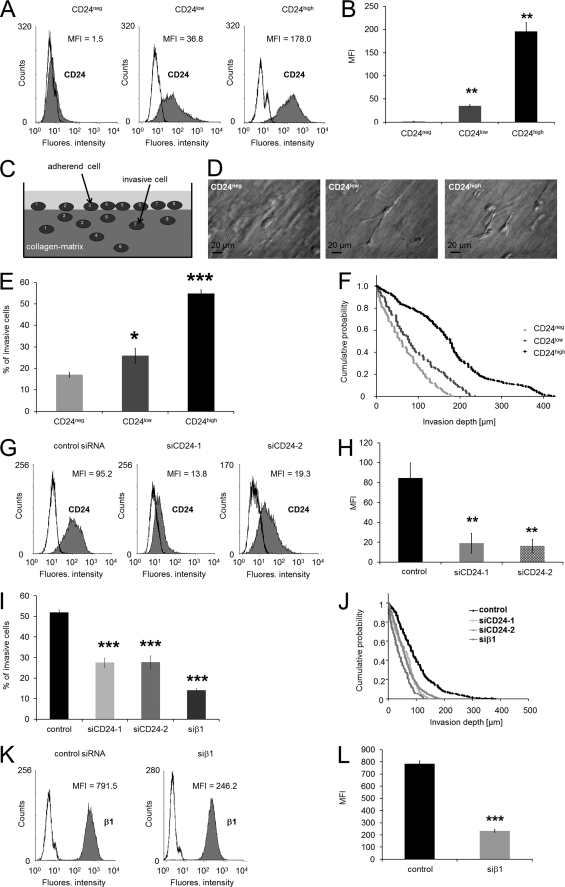
To investigate the effect of CD24 on tumor cell invasion, the CD24-negative lung carcinoma cell line A125 was stably transfected with CD24. Transfectants with high CD24 expression (CD24high) and low CD24 expression (CD24low) were isolated using fluorescence-activated cell sorting, and CD24 expressions were analyzed using flow cytometry (Fig. 1A). CD24high cells expressed 5-fold more CD24 on their cell surface compared with CD24low cells (Fig. 1B). As a control, an A125 transfectant with an empty expression vector (CD24neg) was established. In a previous study, we reported that the expression of CD24 stimulates the migration of cancer cells (27).
FIGURE 1.
High CD24 expression increased human lung carcinoma cell invasion. A, flow cytometric analysis ((R)-PE-labeled-labeled antibodies) of CD24 cell-surface expression on CD24neg, CD24low, and CD24high cells. B, data (MFI) of CD24 expression on CD24neg, CD24low, and CD24high cells are presented as means ± S.D. (n = 3). C, schematic image of the invasion assay. D, modulation contrast images of representative invasive lung carcinoma cells wild type (left), stably transfected with CD24 and sorted for low expression (middle), and high expression (right) that invaded into three-dimensional ECMs. Scale bars, 20 μm. E, percentage of invasive cells and invasion profiles (F) of CD24neg, CD24low, and CD24high cells are shown that invaded into three-dimensional ECMs after 3 days of culture. G, flow cytometric analysis (Cy2-labeled antibodies) of CD24 cell-surface expression on CD24high cells transfected with control siRNA (left), CD24-specific (siCD24-1) siRNA (middle), or CD24-specific (siCD24-2) siRNA (right). One representative experiment of three is shown. H, MFI (mean ± S.D., n = 3) of CD24 expression on CD24high cells treated with control siRNA, siCD24-1, or siCD24-2. I, percentage of invasive cells and invasion profiles (J) of CD24high cells that were treated with control siRNA, CD24-specific siRNA (siCD24-1 and siCD24-2), or β1-integrin subunit-specific siRNA that invaded into three-dimensional ECMs after 3 days of culture. K, flow cytometric analysis of the β1-integrin subunit cell-surface expression on CD24high cells transfected with control siRNA (left) or β1-integrin-specific (siβ1) siRNA (right). One representative experiment of three is shown. L, MFI (mean ± S.D., n = 3) of β1-integrin expression on CD24high cells treated with control siRNA or siCD24-1 or siβ1. (*, p < 0.05; **, p < 0.01; ***, p < 0.001.)
The invasiveness of cancer cells can be investigated in in vitro three-dimensional ECMs (Fig. 1C). The invasiveness is characterized by the percentage of invasive cells, and the invasion profile is expressed as cumulative probability of invasive cells. To evaluate whether the invasiveness of stably CD24-transfected cells was enhanced, invasion assays into three-dimensional ECMs with an average pore size of 1.3 μm were performed (Fig. 1, E and F) (41, 61). Typically representative invasive cells of CD24neg, CD24low, and CD24high cells are shown in Fig. 1D. The percentage of cells that were able to invade into a three-dimensional collagen matrix was 2.5-fold higher in CD24high cells compared with CD24low or CD24neg cells (Fig. 1E). In addition, the invasion profile (cumulative probability) of the invasive cells shows that CD24high cells invaded deeper into the ECM (Fig. 1F). Specific knockdown of CD24 in CD24high cells was 22.2 ± 5.7% (n = 3) for siCD24-1 and 18.6 ± 2.9% (n = 3) for siCD24-2 after 2 days (Fig. 1, G and H; note: the CD24 level differs from Fig. 1, A and B, because (R)-PE-labeled (Fig. 1, A and B) and Cy2-labeled secondary antibodies (Fig. 1, G and H) were used). Knockdown of CD24 using two specific siRNAs (siCD24-1 and siCD24-2) in CD24high cells decreased the number of invasive cells in three-dimensional ECMs compared with control siRNA-treated cells (Fig. 1I). The invasion profiles of CD24 knockdown cells show that the cells only invade less than 200 μm (Fig. 1J), and their average invasion depth was reduced from 98.0 ± 2.7 (n = 665) to 58.6 ± 2.5 (n = 225) for siCD24-1 and 60.0 ± 1.9 (n = 482) for siCD24-2 treated CD24high cells (Fig. 1J). These results indicate that CD24 expression leads to enhanced lung cancer cell invasion in three-dimensional collagen matrices.
CD24-facilitated Invasion Depends on β1-Integrin Subunit Expression
Previously, we reported that the β1-integrin subunit expression on CD24-negative and CD24-expressing cells was not altered (27). As CD24 is located in the lipid rafts and regulates the entrance of β1-integrins into this specific membrane domain (27), we tested whether β1-integrin subunits are involved in CD24-facilitated cell invasion. Thus, we knocked down the β1-integrin (siβ1) subunit in CD24high cells and analyzed their invasiveness in three-dimensional collagen matrices. A specific knockdown of the β1-integrin subunit of 29.8 ± 1.0% (n = 3) was reproducibly archived after 2 days (Fig. 1, K and L). We found that the number of invasive cells is reduced by one-fourth (Fig. 1G). In addition, the invasion profile shows that β1 knockdown cells invade only 125 μm deep into the three-dimensional ECMs, and their average invasion depth was reduced from 98.0 ± 2.7 (n = 665) to 41.5 ± 2.7 (n = 135) for siβ1 (Fig. 1H). These results indicate that the β1-integrin subunit plays a role in CD24-mediated cancer cell invasion.
Knockdown of Collagen-binding Integrin Subunits Reduced CD24-mediated Invasiveness
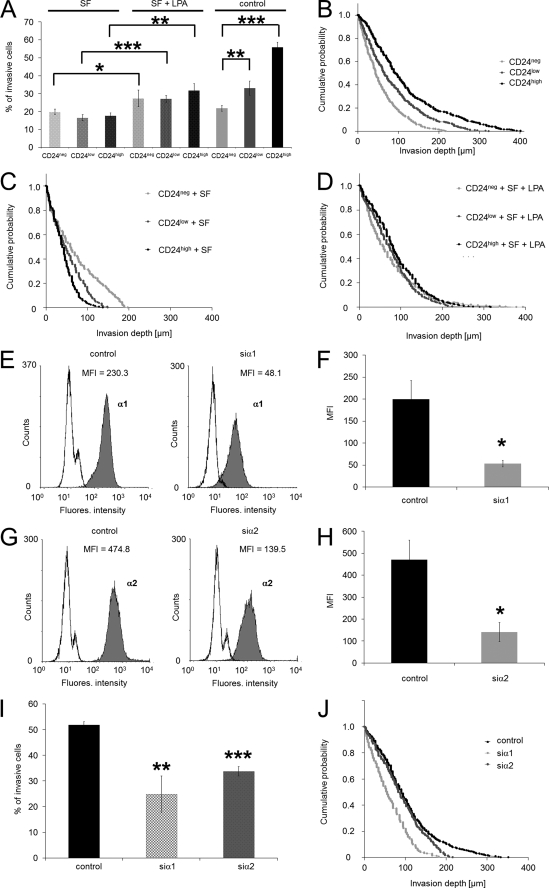
To investigate whether the enhanced invasiveness seen in CD24high cells results from increased expression of collagen-binding integrins or other ECM-binding integrins, we measured their cell-surface expression on CD24neg, CD24low, and CD24high cells. All three cell lines expressed α1 and α2 on their cell surface (27). The invasiveness into three-dimensional ECMs, expressed as percentage of invasive cells and the invasion profile, is reduced to nearly the same level as CD24neg cells (control) in the absence of serum components by using serum-free medium compared with serum-containing 10% FCS (Fig. 2, A–C). These results indicate that a component within the serum may increase cancer cell invasion and that this substance facilitates the differences in motility between the three cell lines. The reduced invasiveness in the absence of serum could be increased by addition of lysophosphatidic acid (LPA) in CD24neg cells to the original levels (Fig. 2, A and D). In contrast, the invasiveness of CD24low and CD24high was augmented but did not reach original levels (Fig. 2, A and D). These results show that LPA is in the absence of serum able to slightly enhance the invasiveness of the CD24-evoked increase in invasiveness indicating that an increase of contractile forces in cancer cells mediates increased invasiveness. The addition of LPA did not alter the expression levels of CD24 on the cell surface of CD24neg, CD24low, and CD24high (data not shown).
FIGURE 2.
Knockdown of the α1- and α2-integrin subunits in CD24-facilitated invasiveness of cancer cells and effect of serum on cell invasion. A, percentage of invasive cells and invasion profiles of CD24neg, CD24low, and CD24high cells cultured in serum-free medium (SF) (C), serum-free and LPA-stimulated (SF + LPA) (D), and in medium with serum (B) that invaded into three-dimensional ECMs after 3 days of culture. E, flow cytometric analysis of α1 cell-surface CD24high cells transfected with control siRNA (left) or α1-specific siRNA (siα1, right panel). One representative experiment of three is shown. F, MFI (mean ± S.D., n = 3) of α1-integrin expression on CD24high cells treated with control siRNA or siα1. G, flow cytometric analysis of α2 cell-surface CD24high cells transfected with control siRNA (left panel) or α2-specific siRNA (siα2, right panel). One representative experiment of three is shown. H, MFI (mean ± S.D., n = 3) of α2-integrin expression on CD24high cells treated with control siRNA or siα2. I, percentage of invasive cells and invasion profiles of CD24high cells (J) treated with control siRNA, α1-integrin subunit-specific siRNA (siα1) and α2-integrin subunit-specific siRNA (siα2) that invaded into three-dimensional ECMs after 3 days of culture. (*, p < 0.05, **, p < 0.05; ***, p < 0.001.)
Furthermore, the residual invasiveness may depend on collagen-binding integrins. Knockdown of the α1- (28.1% ± 6.3, n = 3) and the α2 (30.3% ± 4.8, n = 3)-integrin subunit reduced significantly the number of invasive cells, and the invasion profile showed that the invasion depth is also reduced (Fig. 2, E–H). These data imply that the expression of integrin subunits may not have contributed to the increased invasiveness of CD24high cells supporting our previous results (27).
Role of Matrix Degradation for CD24-mediated Cell Invasiveness
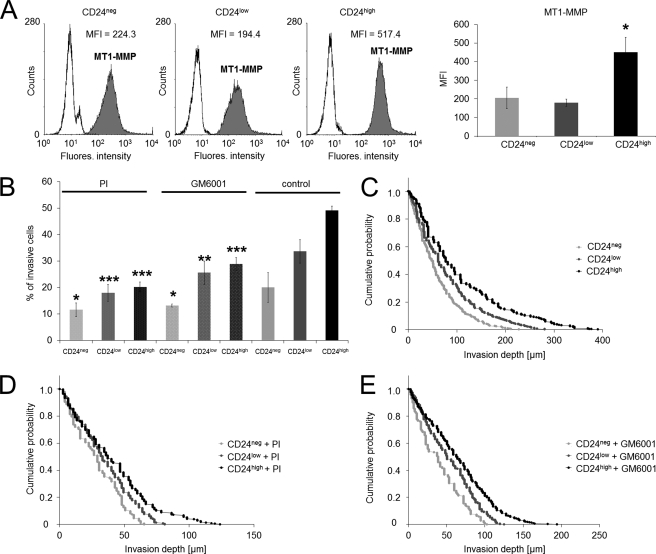
Cancer cell invasion has been shown to be associated with enhanced expression of the membrane type 1 matrix metalloproteinase (MT1-MMP), a major collagen-degrading proteinase (44). Therefore, we analyzed whether alterations in the cell-surface expression of MT1-MMP are responsible for differences in cell invasiveness between CD24neg, CD24low, and CD24high cells. Indeed, the expression of MT1-MMP was highest in CD24high cells, followed by CD24low and CD24neg cells (Fig. 3, A and B). These results indicate that matrix degradation can be involved in CD24-facilitated increased cell invasiveness. The expression of MT1-MMP mRNA was not enhanced in CD24high cells compared with CD24neg and CD24low cells (supplemental Fig. S1).
FIGURE 3.
MT1-MMP expression of CD24neg, CD24low, and CD24high cells and effect of proteinase inhibition on three-dimensional ECM invasion. A, left panel, flow cytometric analysis of the MT1-MMP receptor expression on the cell surface of CD24neg, CD24low, and CD24high cells. One representative experiment of three is shown. Right panel, MFI (mean ± S.D., n = 3) of MT1-MMP receptor expression on CD24neg, CD24low, and CD24high cells. B, percentage of invasive cells in the presence of an protease inhibitor mixture (PI), GM6001 inhibitor, or DMSO buffer control and invasion profiles of CD24neg, CD24low, and CD24high cells cultured in the presence of PI (D), GM6001 inhibitor (E), or DMSO buffer control (C) that invaded into three-dimensional ECMs after 3 days of culture. (*, p < 0.05; **, p < 0.05; ***, p < 0.001.)
The overall activity of proteolytic enzymes was investigated by adding a PI mixture or solely the matrix-metalloproteinase inhibitor GM6001 to CD24neg, CD24low, and CD24high cells prior to the start of the invasion assay. We found that the percentage of invasive cells after PI treatment was decreased in all three cell lines (Fig. 3B), and these cells did not migrate beyond 110 μm into the three-dimensional ECMs (Fig. 3D). The average migration depth of CD24high, CD24low, and CD24neg cells into three-dimensional ECMs in the presence of the PI mixture is now only 41.2 ± 2.0 μm (n = 213), 34.7 ± 1.2 μm (n = 322), and 28.7 ± 1.4 μm (n = 173), respectively (Fig. 3E). The GM6001 inhibitor results showed that the number of invasive CD24high, CD24low, and CD24neg cells into three-dimensional ECMs is reduced (Fig. 3B) as the average migration depth is only 65.4 ± 2.3 μm (n = 325), 53.5 ± 1.5 μm (n = 494), and 40.1 ± 2.0 μm (n = 206), respectively (Fig. 3E). Nonetheless, the differences in cell invasiveness between CD24neg, CD24low, and CD24high cells after protease inhibition still remained. Taken together, these data show that the enzymatic digestion of the three-dimensional ECMs plays a role in cancer cell invasion but is not a prerequisite for invasion of these cells. Thus, differences in the protease activity can only to a slight degree account for CD24-mediated cell invasion.
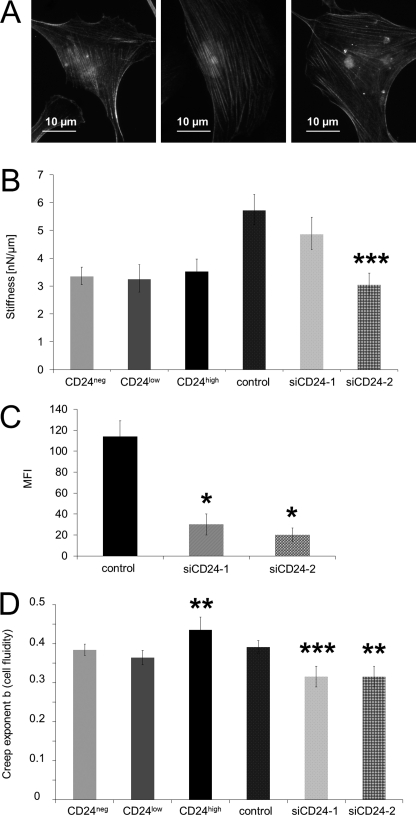
Effect of CD24 on Cancer Cell Stiffness
The similarities in stress fiber formation between CD24neg, CD24low, and CD24high cells (Fig. 4A) suggest that these three cell lines may not differ in cell stiffness. Cell stiffness was measured using magnetic tweezer microrheology (Fig. 4B). Forces of up to 10 nN were applied to super-paramagnetic beads coated with fibronectin (also a component of the three-dimensional ECMs). The bead displacement during a stepwise increase in force (creep measurement) followed a power law as described elsewhere (45). The stiffness measurements (averaged over all forces from 0.5 to 10 nN) of all three cell lines showed nearly the same values expressed as mean values (Fig. 4B). These results indicate that CD24 did not alter cell stiffness or stress-stiffening of the cancer cells. To analyze whether there are differences independent of alterations between the three cell lines, we used a CD24-specific siRNA approach. The CD24-specific siRNA reduced the MFI on CD24high cells after 2 days in the case of siCD24-1 to 29.3 ± 1.7% and of siCD24-2 to 19.5 ± 0.6% (Fig. 4C). These stiffness results showed a slight reduction in CD24high cells that had been treated with the CD24-specific siRNA siCD24-1 for 2 days and a significant reduction with siCD24-2 compared with control siRNA-treated cells (Fig. 4C). The stiffness results for the knockdown experiments suggest that contractile forces may play a role in CD24-facilitated cell invasion.
FIGURE 4.
Effect of CD24 expression on cell stiffness and cytoskeletal remodeling dynamics. A, representative fluorescence images of CD24neg (left panel), CD24low (middle panel), and CD24high cells (right panel) stained for actin with Alexa-Fluor546-conjugated phalloidin showed equal stress fiber formation on fibronectin-coated glass after 16 h. B, stiffness of CD24neg, CD24low, and CD24high cells as well as of CD24high cells transfected with control siRNA (control) and two CD24-specific (siCD24-1 and siCD24-2) siRNAs was measured after force application to fibronectin-coated beads using magnetic tweezers. C, flow cytometric analysis (MFI as mean ± S.D., n = 3) of CD24 expression on the cell surface of CD24high cells transfected with control siRNA (left panel) or CD24-specific siRNA (siCD24-1 and siCD24-2, respectively). D, creep exponent b (cell fluidity and cytoskeletal remodeling dynamics) of CD24neg, CD24low, and CD24high cells as well as of CD24high cells transfected with control siRNA (control) and two CD24-specific (siCD24-1 and siCD24-2) siRNAs was also determined using magnetic tweezers. The values are expressed as mean ± S.D. 27–60 cells were measured for each condition. (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
Effect of CD24 on Cytoskeletal Remodeling Dynamics
During cell invasion into a dense three-dimensional collagen fiber matrix with pore sizes smaller than the cell's diameter, the invasive cell might change its shape and restructure its cytoskeleton to move forward in a dense three-dimensional collagen fiber matrix. The dynamics of cytoskeletal remodeling processes can also be measured with magnetic tweezer microrheology. The power law exponent b characterizes the viscoelastic response of cells and can assume values between 0 for an elastic solid and 1 for a viscous fluid. b was significantly increased in CD24high cells at all external forces compared with CD24low and CD24neg cells, indicating that these cells were more fluid-like (Fig. 4D) and that the cytoskeletal remodeling dynamics was increased in CD24high cells. These results could not be confirmed by CD24high cells that had been treated with two CD24-specific siRNAs (siCD24-1 and siCD24-2) for 2 days, as these cells displayed significantly decreased cellular fluidity and hence decreased cytoskeletal remodeling dynamics compared with control siRNA-treated cells (Fig. 4D).
Contractile Force Generation Is Increased in the Presence of CD24
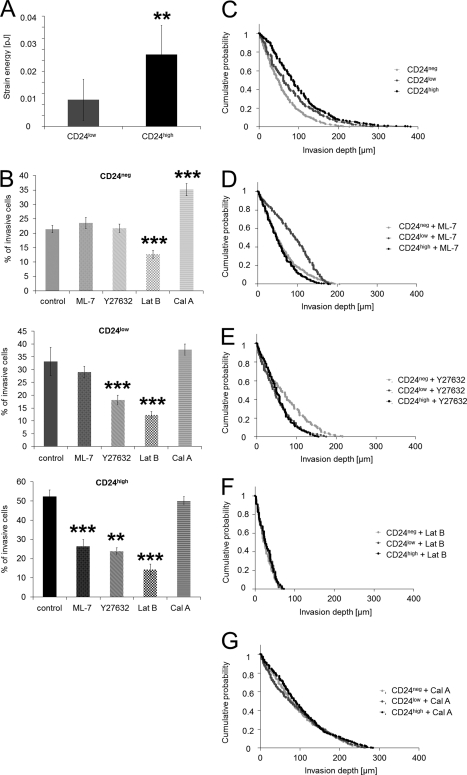
To analyze whether differences in the invasiveness of CD24 transfectants and control cells could be explained by their altered ability to generate contractile forces, their tractions on fibronectin-coated polyacrylamide gels (E = 5.4 kPa) were determined. To characterize the contractile forces of each cell, the elastic strain energy stored in the polyacrylamide gel due to cell tractions was calculated as the product of local tractions and deformations, integrated over the spreading area of the cells (59). The strain energy of CD24high cells was 3-fold higher compared with CD24low cells (Fig. 5A). After normalization for differences in the spreading area, the strain energy of CD24high cells was even 5-fold higher compared with CD24low cells.
FIGURE 5.
Increased contractile force generation of CD24high cells and inhibition of contractile force-mediated cell invasion. A, strain energy per cell (mean ± S.D.) of CD24high cells (n = 11) was 3-fold increased compared with CD24low cells (n = 5). B, percentage of invasive cells (mean ± S.D.) of CD24neg (top panel), CD24low (middle panel), and CD24high cells (bottom panel) determined after 3 days in the presence of myosin contraction inhibitors (15 μm ML-7 or 100 μm Y27632), the actin polymerization inhibitor (2 μm latrunculin B), the myosin phosphatase inhibitor (1 nm calyculin A), or DMSO as control. Invasion profiles of CD24neg, CD24low, and CD24high cells treated with DMSO control (C), ML-7 (D), Y26732 (E), latrunculin B (Lat B) (F), or calyculin A (Cal A) (G). (**, p < 0.05; ***, p < 0.001.)
The addition of the myosin light chain kinase inhibitor ML-7 or the Rho kinase inhibitor Y27632 to CD24high cells reduced significantly the percentage of invasive cells into three-dimensional collagen fiber matrices (Fig. 5B). In CD24low cells, only Y27632 but not ML-7 caused a decrease in the percentage of invasive cells (Fig. 5B). The Rho kinase and myosin light chain kinase inhibitors did not alter the invasion profile of CD24low cells (Fig. 5, C–E), whereas both inhibitors reduced invasion of CD24high cells into deeper regions of the three-dimensional collagen fiber matrices (Fig. 5, C–E). The addition of the actin polymerization inhibitor latrunculin B (2 μm) reduced the percentage of invasive CD24high, CD24low, and CD24neg cells and reduced their invasion depths (Fig. 5F). Taken together, these results indicate that CD24high cells are sensitive to changes in myosin light chain kinase-mediated myosin light chain phosphorylation and dephosphorylation as well as actomyosin contraction.
Subcell line-specific dependence of cell invasion on myosin light chain phosphorylation and hence on increased contractility was further investigated by the addition of the serine/threonine phosphatase inhibitor calyculin A (1 nm), which reduces the dephosphorylation of myosin light chain and thereby increases contractility (62). Calyculin A treatment increased the percentage of invasive CD24neg and CD24low cells (Fig. 5B) and also increased their invasion depths but had little effect on CD24high cells (Fig. 5G). This indicates that an increase in contractility of CD24neg and CD24low cells enhances their invasiveness, whereas the contractility of CD24high cells was already at a level optimal for cell invasion that could not be further increased by calyculin A. Finally, these data demonstrate that the CD24-mediated increased invasiveness is due to increased contractile force generation.
Increased CD24 Facilitated Invasiveness Depends on Src and STAT-3 Signaling
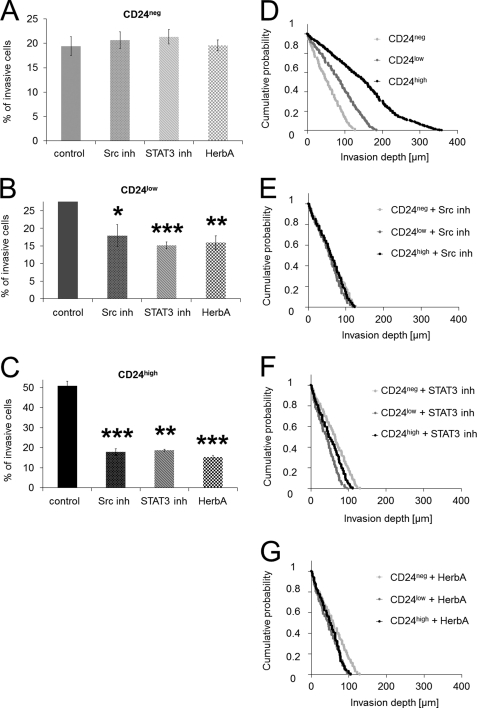
The addition of an Src kinase inhibitor (Src inh) to CD24high cells and CD24low cells reduced significantly the percentage of invasive cells into three-dimensional collagen fiber matrices, although it has no effect on the invasiveness of CD24neg cells (Fig. 6, A–C). Furthermore, the addition of a STAT3 inhibitory peptide (STAT3 inh) to CD24high cells and CD24low cells reduced significantly the percentage of invasive cells into three-dimensional collagen fiber matrices, although it has no effect on the invasiveness of CD24neg cells (Fig. 6, A–C). In addition, the tyrosine kinase inhibitor herbimycin A reduced significantly the percentage of invasive cells into three-dimensional collagen fiber matrices of CD24high cells and CD24low cells, although it has no effect on the invasiveness of CD24neg cells (Fig. 6, A–C). The invasion profiles of CD24neg, CD24low, and CD24high cells show that the invasion depths are also reduced after addition of the Src inhibitor, STAT3 inhibitor, and the tyrosine kinase inhibitor herbimycin A in CD24low and CD24high cells but not in CD24neg cells (Fig. 6, D–G). These results suggest that the increased invasiveness may be facilitated by an Src kinase and STAT3 pathway.
FIGURE 6.
Inhibition of Src kinase and STAT3 of CD24-mediated cell invasion. A–C, percentage of invasive cells (mean ± S.D.) of CD24neg (A), CD24low (B), and CD24high cells (C) determined after 3 days in the presence of an Src inhibitory peptide (Src inh 30 μm), STAT3 inhibitor (STAT3 inh 30 μm), the tyrosine kinase inhibitor herbimycin A (HerbA, 100 μm), or DMSO as control. Invasion profiles of CD24neg, CD24low, and CD24high cells treated with DMSO control (D), Src inhibitory (E), (*, p < 0.05; (F), or herbimycin A (G). (*, p < 0.05; **, p < 0.05; ***, p < 0.001).
Endogenous Expression of CD24 Increased Invasiveness of Human Breast Cancer Cells
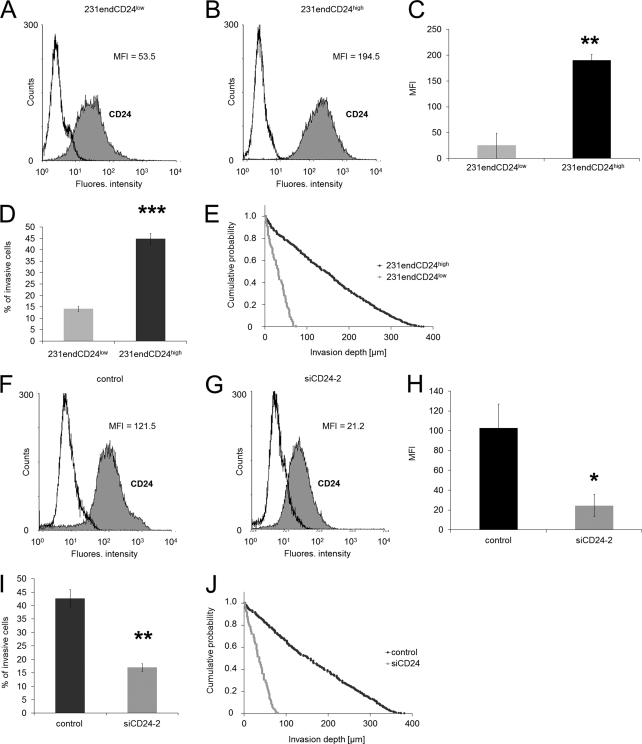
To investigate whether CD24 increased endogenously the invasiveness of cancer cells, we analyzed whether low expressing (231endCD24low cells, Fig. 7A) or high expressing CD24 (231endCD24high cells, Fig. 7B) breast cancer cells (MDA-MB-231 cells) showed altered invasiveness into three-dimensional ECMs indicated by the percentage of invasive cells (Fig. 7D) or their invasion depths (Fig. 7E). In addition, we knocked down the endogenous expression of CD24 in 231endCD24high cells to 25.1 ± 1.1% (Fig. 7, F–H) and showed that the invasiveness is reduced after CD24 knockdown in terms of percentage of invasive cells (Fig. 7I) as well as invasion depths (Fig. 7J). These results indicate that the results obtained with CD24-transfected human lung cancer cells are transferable to endogenous CD24-expressing human breast cancer cells.
FIGURE 7.
High endogenous CD24 expression increased human breast carcinoma cell invasion. A and B, flow cytometric analysis ((R)-PE-labeled-labeled antibodies) of CD24 cell-surface expression on 231endCD24low (A) and 231endCD24high subcell lines (B). C, data (MFI) of CD24 expression on 231endCD24low and 231endCD24high cells are presented as means ± S.D. (n = 3). D, percentage of invasive cells and invasion profiles (E) of 231endCD24low and 231endCD24high cells are shown that invaded into three-dimensional ECMs after 3 days of culture. F and G, flow cytometric analysis (Cy2-labeled antibodies) of CD24 cell-surface expression on 231endCD24high cells transfected with control siRNA (F) and CD24-specific (siCD24-2) siRNA (G). H, MFI (mean ± S.D., n = 3) of CD24 expression on 231endCD24high cells treated with control siRNA or siCD24-2. I, percentage of invasive cells and invasion profiles (J) of 231endCD24high cells that were treated with control siRNA and CD24-specific siRNA (siCD24-2) that invaded into three-dimensional ECMs after 3 days of culture. (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
DISCUSSION
Recently it has been reported that CD24 is involved in tumor growth and metastasis formation (28–31). Consistent with these studies, we showed that high CD24 expression (MFI = 196.1 ± 10.9, n = 3) increased lung carcinoma cell invasiveness into three-dimensional ECMs. Moreover, we showed that highly endogenous CD24-expressing MDA-MB-231 breast cancer cell subclones increased the invasiveness into three-dimensional ECMs compared with lowly endogenous CD24-expressing subclones. These results indicate that the CD24-facilitated invasiveness is not restricted to one cancer cell type, as it is transferable to other cancer types.
The invasion of cancer cells into connective tissue is a complex event and depends on the mechanical and biochemical properties of the microenvironment. Here, we demonstrate that the increased expression of CD24 enhances invasiveness of cancer cells into three-dimensional ECMs through the transmission and generation of higher contractile forces and increases cytoskeletal remodeling dynamics. We focused in this study on the mechanism that leads to higher invasiveness of cancer cells with high expression of the GPI-anchored protein CD24.
The mechanisms promoting the invasion of cancer cells are only fragmentarily investigated, but there is a general agreement that biomechanical factors determine the speed of cell migration in dense three-dimensional ECMs (15, 42, 45). These factors include adhesion forces, degradation of the ECM through secretion of matrix-degrading enzymes, cytoskeletal dynamics, cellular stiffness and fluidity, and contractile force generation (15, 44, 45, 63). To investigate which of these biomechanical factors contribute to the higher invasiveness of cells with high CD24 expression, they were isolated from transfected cell lines with high and low CD24 expression from A125 lung carcinoma cells. In each case we found that cells with high CD24 expression were highly invasive. The knockdown of CD24 using CD24-specific siRNA dramatically decreased their invasiveness into three-dimensional collagen fiber matrices. We further showed that the effect was not mediated through proteolytic enzyme activity between CD24neg, CD24low, and CD24high cells.
Integrins are known to be involved in cell adhesion, transmigration, and invasion processes. The activation of the integrin receptors could be through conformational changes after ligand binding (22). Integrin activation could be regulated through either β1-integrin translocation into lipid rafts or an increased affinity to ligands by enhancing the number of activated integrins and total integrins on the cell surface. The latter we recently reported not to be crucial because alterations in the expression levels of the β1-integrin were not observed in CD24 mutants using FACS analysis (27). Consistently, we did not observe alterations of the β1-integrin expression on the cell surface of CD24neg, CD24low, and CD24high cells (data not shown). Besides the unaltered expression of the β1-integrin subunit, its knockdown in CD24high cells reduced their invasiveness into three-dimensional ECMs indicating that its altered localization into lipid rafts may increase cell invasion. In addition, we found that knockdown of the α1- and α2-integrin subunits, which form both together with the β1 subunit a collagen-binding integrin receptor, decreased the number of invasive cells as well as their invasion depths into three-dimensional ECMs, indicating that these integrins play a role in CD24-mediated cell invasion, but only to a certain degree. Taken together, these results indicate that the CD24 is a key player in facilitating cell invasion into three-dimensional collagen fiber matrices by regulating the localization of the β1-integrin subunit.
The signal transduction pathways that connect integrin adhesion events with traction force generation and cell invasion are still elusive, although important components have been studied in detail, such as the activation of α5β1-integrins by ECM ligands (20, 64), the formation of focal adhesions following integrin activation (65), and the connection between focal adhesion assembly and contractile forces (14, 18, 20) and between contractile forces and three-dimensional cell invasion (41, 66). Here, we show that increased expression of CD24 leads to increased contractile forces that help the cell to overcome the steric hindrance of the ECM. These findings are consistent with our previously published study that showed increased contractile forces of highly invasive CXCR2high cells compared with the weakly invasive CXCR2low cancer cells (41).
Blocking of the myosin contraction through the myosin light chain kinase inhibitor ML-7 or the Rho kinase inhibitor Y27632 diminished the invasiveness of CD24high cells indicating that the CD24-facilitated increased invasiveness is contractile force-dependent. In CD24low cells only the inhibitor Y27632 reduced the invasiveness, whereas in CD24neg cells the invasiveness was not further diminished suggesting CD24neg cells can employ invasion strategies that do not rely on the generation of contractile forces. Nonetheless, by increasing the contractility of CD24neg cells using calyculin A, they show increased invasion, even higher than the original levels. These results indicate that contractile forces are needed for increased cell invasion. In line with this, addition of the actin polymerization inhibitor latrunculin B inhibited the invasiveness of all three cell lines indicating that the invasiveness is sensitive to alteration in the actomyosin cytoskeleton that is necessary to transmit and generate contractile forces. Furthermore, we showed that the inhibition of the Src kinase and its downstream target STAT3 reduced significantly the invasiveness of CD24high cells, but hardly the invasiveness of CD24low cells, and had no effect on the invasiveness of CD24neg cells. These results suggest that Src kinase signaling pathways may be involved in the CD24-mediated invasiveness.
Finally, we conclude that the generation of contractile forces is the driving factor for increased CD24-facilitated cell invasion into three-dimensional ECMs, and we propose that the measurement of biomechanical properties may be a novel factor in determining and explaining the malignancy of tumors.
Supplementary Material
Acknowledgments
We thank Philip Kollmannsberger and Navid Bonakdar for help with the magnetic tweezer experiments, Ben Fabry and Wolfgang H. Goldmann for helpful discussions, and Barbara Reischl and Christine Albert for technical assistance.
This work was supported by Deutsche Krebshilfe Grant 109432.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- GPI
- glycosylphosphatidylinositol
- ECM
- extracellular matrix
- MFI
- mean fluorescence intensity
- LPA
- lysophosphatidic acid
- (R)-PE
- (R)-phycoerythrin
- nN
- nanonewton
- Pa
- pascal
- PI
- protease inhibitor.
REFERENCES
- 1. Liotta L. A., Steeg P. S., Stetler-Stevenson W. G. (1991) Cell 64, 327–336 [DOI] [PubMed] [Google Scholar]
- 2. Al-Mehdi A. B., Tozawa K., Fisher A. B., Shientag L., Lee A., Muschel R. J. (2000) Nat. Med. 6, 100–102 [DOI] [PubMed] [Google Scholar]
- 3. Steeg P. S. (2006) Nat. Med. 12, 895–904 [DOI] [PubMed] [Google Scholar]
- 4. Langley R. R., Fidler I. J. (2007) Endocr. Rev. 28, 297–321 [DOI] [PubMed] [Google Scholar]
- 5. Horwitz A. F. (1997) Sci. Am. 276, 68–75 [DOI] [PubMed] [Google Scholar]
- 6. Giancotti F. G. (2000) Nat. Cell Biol. 2, E13–E14 [DOI] [PubMed] [Google Scholar]
- 7. Hynes R. O. (2002) Cell 110, 673–687 [DOI] [PubMed] [Google Scholar]
- 8. Neff N. T., Lowrey C., Decker C., Tovar A., Damsky C., Buck C., Horwitz A. F. (1982) J. Cell Biol. 95, 654–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Damsky C. H., Knudsen K. A., Bradley D., Buck C. A., Horwitz A. F. (1985) J. Cell Biol. 100, 1528–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hemler M. E., Huang C., Schwarz L. (1987) J. Biol. Chem. 262, 3300–3309 [PubMed] [Google Scholar]
- 11. Geiger B., Bershadsky A., Pankov R., Yamada K. M. (2001) Nat. Rev. Mol. Cell Biol. 2, 793–805 [DOI] [PubMed] [Google Scholar]
- 12. Loftus J. C., Liddington R. C. (1997) J. Clin. Invest. 99, 2302–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Palecek S. P., Loftus J. C., Ginsberg M. H., Lauffenburger D. A., Horwitz A. F. (1997) Nature 385, 537–540 [DOI] [PubMed] [Google Scholar]
- 14. Balaban N. Q., Schwarz U. S., Riveline D., Goichberg P., Tzur G., Sabanay I., Mahalu D., Safran S., Bershadsky A., Addadi L., Geiger B. (2001) Nat. Cell Biol. 3, 466–472 [DOI] [PubMed] [Google Scholar]
- 15. Zaman M. H., Trapani L. M., Sieminski A. L., Siemeski A., Mackellar D., Gong H., Kamm R. D., Wells A., Lauffenburger D. A., Matsudaira P. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 10889–10894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Elson E. L. (1988) Annu. Rev. Biophys. Biophys. Chem. 17, 397–430 [DOI] [PubMed] [Google Scholar]
- 17. Giannone G., Sheetz M. P. (2006) Trends Cell Biol. 16, 213–223 [DOI] [PubMed] [Google Scholar]
- 18. Gallant N. D., Michael K. E., García A. J. (2005) Mol. Biol. Cell 16, 4329–4340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Paszek M. J., Zahir N., Johnson K. R., Lakins J. N., Rozenberg G. I., Gefen A., Reinhart-King C. A., Margulies S. S., Dembo M., Boettiger D., Hammer D. A., Weaver V. M. (2005) Cancer Cell 8, 241–254 [DOI] [PubMed] [Google Scholar]
- 20. Friedland J. C., Lee M. H., Boettiger D. (2009) Science 323, 642–644 [DOI] [PubMed] [Google Scholar]
- 21. Calderwood D. A., Yan B., de Pereda J. M., Alvarez B. G., Fujioka Y., Liddington R. C., Ginsberg M. H. (2002) J. Biol. Chem. 277, 21749–21758 [DOI] [PubMed] [Google Scholar]
- 22. Wei Y., Czekay R. P., Robillard L., Kugler M. C., Zhang F., Kim K. K., Xiong J. P., Humphries M. J., Chapman H. A. (2005) J. Cell Biol. 168, 501–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guan J. L. (2004) Science 303, 773–774 [DOI] [PubMed] [Google Scholar]
- 24. Frixen U. H., Behrens J., Sachs M., Eberle G., Voss B., Warda A., Löchner D., Birchmeier W. (1991) J. Cell Biol. 113, 173–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bauer K., Mierke C., Behrens J. (2007) Int. J. Cancer 121, 1910–1918 [DOI] [PubMed] [Google Scholar]
- 26. Mierke C. T., Frey B., Fellner M., Herrmann M., Fabry B. (2011) J. Cell Sci. 124, 369–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Runz S., Mierke C. T., Joumaa S., Behrens J., Fabry B., Altevogt P. (2008) Biochem. Biophys. Res. Commun. 365, 35–41 [DOI] [PubMed] [Google Scholar]
- 28. Kristiansen G., Denkert C., Schlüns K., Dahl E., Pilarsky C., Hauptmann S. (2002) Am. J. Pathol. 161, 1215–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kristiansen G., Winzer K. J., Mayordomo E., Bellach J., Schlüns K., Denkert C., Dahl E., Pilarsky C., Altevogt P., Guski H., Dietel M. (2003) Clin. Cancer Res. 9, 4906–4913 [PubMed] [Google Scholar]
- 30. Jacob J., Bellach J., Grützmann R., Alldinger I., Pilarsky C., Dietel M., Kristiansen G. (2004) Pancreatology 4, 454–460 [DOI] [PubMed] [Google Scholar]
- 31. Chou Y. Y., Jeng Y. M., Lee T. T., Hu F. C., Kao H. L., Lin W. C., Lai P. L., Hu R. H., Yuan R. H. (2007) Ann. Surg. Oncol. 14, 2748–2758 [DOI] [PubMed] [Google Scholar]
- 32. Yang X. R., Xu Y., Yu B., Zhou J., Li J. C., Qiu S. J., Shi Y. H., Wang X. Y., Dai Z., Shi G. M., Wu B., Wu L. M., Yang G. H., Zhang B. H., Qin W. X., Fan J. (2009) Clin. Cancer Res. 15, 5518–5527 [DOI] [PubMed] [Google Scholar]
- 33. Sagiv E., Memeo L., Karin A., Kazanov D., Jacob-Hirsch J., Mansukhani M., Rechavi G., Hibshoosh H., Arber N. (2006) Gastroenterology 131, 630–639 [DOI] [PubMed] [Google Scholar]
- 34. Agrawal S., Kuvshinoff B. W., Khoury T., Yu J., Javle M. M., LeVea C., Groth J., Coignet L. J., Gibbs J. F. (2007) J. Gastrointest. Surg. 11, 445–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fukushima T., Tezuka T., Shimomura T., Nakano S., Kataoka H. (2007) J. Biol. Chem. 282, 18634–18644 [DOI] [PubMed] [Google Scholar]
- 36. Li C., Heidt D. G., Dalerba P., Burant C. F., Zhang L., Adsay V., Wicha M., Clarke M. F., Simeone D. M. (2007) Cancer Res. 67, 1030–1037 [DOI] [PubMed] [Google Scholar]
- 37. Aigner S., Ramos C. L., Hafezi-Moghadam A., Lawrence M. B., Friederichs J., Altevogt P., Ley K. (1998) FASEB J. 12, 1241–1251 [DOI] [PubMed] [Google Scholar]
- 38. Baumann P., Cremers N., Kroese F., Orend G., Chiquet-Ehrismann R., Uede T., Yagita H., Sleeman J. P. (2005) Cancer Res. 65, 10783–10793 [DOI] [PubMed] [Google Scholar]
- 39. Schabath H., Runz S., Joumaa S., Altevogt P. (2006) J. Cell Sci. 119, 314–325 [DOI] [PubMed] [Google Scholar]
- 40. Mierke C. T. (2008) J. Biophys. 2008, 183516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mierke C. T., Zitterbart D. P., Kollmannsberger P., Raupach C., Schlotzer-Schrehardt U., Goecke T. W., Behrens J., Fabry B. (2008). Biophys. J. 94, 2832–2846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Friedl P., Bröcker E. B. (2000) Cell. Mol. Life Sci. 57, 41–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Webb D. J., Donais K., Whitmore L. A., Thomas S. M., Turner C. E., Parsons J. T., Horwitz A. F. (2004) Nat. Cell Biol. 6, 154–161 [DOI] [PubMed] [Google Scholar]
- 44. Wolf K., Mazo I., Leung H., Engelke K., von Andrian U. H., Deryugina E. I., Strongin A. Y., Bröcker E. B., Friedl P. (2003) J. Cell Biol. 160, 267–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mierke C. T., Rösel D., Fabry B., Brábek J. (2008) Eur. J. Cell Biol. 87, 669–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jackson D., Waibel R., Weber E., Bell J., Stahel R. A. (1992) Cancer Res. 52, 5264–5270 [PubMed] [Google Scholar]
- 47. Bloom R. J., George J. P., Celedon A., Sun S. X., Wirtz D. (2008) Biophys. J. 95, 4077–4088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. McNulty A. L., Weinberg J. B., Guilak F. (2009) Clin. Orthop. Relat. Res. 467, 1557–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Riedle S., Kiefel H., Gast D., Bondong S., Wolterink S., Gutwein P., Altevogt P. (2009) Biochem. J. 420, 391–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Metzner C., Raupach C., Mierke C. T., Fabry B. (2010) J. Phys. Condens. Matter 22, 194105 [DOI] [PubMed] [Google Scholar]
- 51. Hildebrandt J. (1969) Bull. Math. Biophys. 31, 651–667 [DOI] [PubMed] [Google Scholar]
- 52. Mijailovich S. M., Kojic M., Zivkovic M., Fabry B., Fredberg J. J. (2002) J. Appl. Physiol. 93, 1429–1436 [DOI] [PubMed] [Google Scholar]
- 53. Kasza K. E., Nakamura F., Hu S., Kollmannsberger P., Bonakdar N., Fabry B., Stossel T. P., Wang N., Weitz D. A. (2009) Biophys. J. 96, 4326–4335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Fabry B., Maksym G. N., Butler J. P., Glogauer M., Navajas D., Fredberg J. J. (2001) Phys. Rev. Lett. 87, 148102 [DOI] [PubMed] [Google Scholar]
- 55. Kollmannsberger P., Mierke C. T., Fabry B. (2011) Soft Matter 7, 3127–3132 [Google Scholar]
- 56. Kollmannsberger P., Fabry B. (2009) Soft Matter RSC 5, 1771–1774 [Google Scholar]
- 57. Fredberg J. J., Jones K. A., Nathan M., Raboudi S., Prakash Y. S., Shore S. A., Butler J. P., Sieck G. C. (1996) J. Appl. Physiol. 81, 2703–2712 [DOI] [PubMed] [Google Scholar]
- 58. Fabry B., Fredberg J. J. (2003) Respir. Physiol. Neurobiol. 137, 109–124 [DOI] [PubMed] [Google Scholar]
- 59. Pelham R. J., Jr., Wang Y. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 13661–13665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Butler J. P., Toliæ-Nørrelykke I. M., Fabry B., Fredberg J. J. (2002) Am. J. Physiol. Cell Physiol. 282, C595–C605 [DOI] [PubMed] [Google Scholar]
- 61. Mickel W., Münster S., Jawerth L. M., Vader D. A., Weitz D. A., Sheppard A. P., Mecke K., Fabry B., Schröder-Turk G. E. (2008) Biophys. J. 95, 6072–6080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Inutsuka A., Goda M., Fujiyoshi Y. (2009) Biochem. Biophys. Res. Commun. 390, 1160–1166 [DOI] [PubMed] [Google Scholar]
- 63. Mierke C. T., Kollmannsberger P., Zitterbart D. P., Smith J., Fabry B., Goldmann W. H. (2008) Biophys. J. 94, 661–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Huveneers S., Truong H., Fässler R., Sonnenberg A., Danen E. H. (2008) J. Cell Sci. 121, 2452–2462 [DOI] [PubMed] [Google Scholar]
- 65. Burridge K., Wennerberg K. (2004) Cell 116, 167–179 [DOI] [PubMed] [Google Scholar]
- 66. Rösel D., Brábek J., Tolde O., Mierke C. T., Zitterbart D. P., Raupach C., Bicanová K., Kollmannsberger P., Panková D., Vesely P., Folk P., Fabry B. (2008) Mol. Cancer Res. 6, 1410–1420 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.