Abstract
In filamentous fungi, several lines of experimental evidence indicate that secondary metabolism is triggered by oxidative stress; however, the functional and molecular mechanisms that mediate this association are unclear. The basic leucine zipper (bZIP) transcription factor AtfB, a member of the bZIP/CREB family, helps regulate conidial tolerance to oxidative stress. In this work, we investigated the role of AtfB in the connection between oxidative stress response and secondary metabolism in the filamentous fungus Aspergillus parasiticus. This well characterized model organism synthesizes the secondary metabolite and carcinogen aflatoxin. Chromatin immunoprecipitation with specific anti-AtfB demonstrated AtfB binding at promoters of seven genes in the aflatoxin gene cluster that carry CREs. Promoters lacking CREs did not show AtfB binding. The binding of AtfB to the promoters occurred under aflatoxin-inducing but not under aflatoxin-noninducing conditions and correlated with activation of transcription of the aflatoxin genes. Deletion of veA, a global regulator of secondary metabolism and development, nearly eliminated this binding. Electrophoretic mobility shift analysis demonstrated that AtfB binds to the nor-1 (an early aflatoxin gene) promoter at a composite regulatory element that consists of highly similar, adjacent CRE1 and AP-1-like binding sites. The five nucleotides immediately upstream from CRE1, AGCC(G/C), are highly conserved in five aflatoxin promoters that demonstrate AtfB binding. We propose that AtfB is a key player in the regulatory circuit that integrates secondary metabolism and cellular response to oxidative stress.
Keywords: AP-1 Transcription Factor, Aspergillus, Chromatin Immunoprecipitation (ChiP), Reactive Oxygen Species (ROS), Transcription, Aflatoxin Biosynthesis, Aspergillus Parasiticus, AtfB, Oxidative Stress, Secondary Metabolism
Introduction
Cellular response to oxidative stress in vertebrates, plants, and fungi is of fundamental importance; it enables the cell to survive a variety of extra- and intracellular oxidative stressors. An uncontrolled increase in reactive oxygen species (ROS)4 in mammalian cells is associated with various pathological conditions such as inflammation and cardiovascular and neurodegenerative disorders, including hypertension, atherosclerosis, Parkinson disease, and Alzheimer disease; oxidative stress is also linked to premature aging and cancer (1–10). A detailed understanding of the regulatory network that coordinates the cellular response to oxidative stress will enable better control over its detrimental impacts on humans.
As a part of the response to oxidative stress, transcription factors activated directly or indirectly by ROS bind to the promoters of specific genes that trigger defense and signaling related activities. In mammalian cells, Drosophila, Caenorhabditis elegans, and yeast, response to oxidative stress is mediated by an evolutionarily conserved bZIP transcription factor Nrf2 that binds as a heterodimer with Maf or ATF4 to antioxidant-response elements in the promoters of more than 200 mammalian genes (11–16). Signaling pathways that involve PKC, PI3K, and MAPK participate in Nrf2 activation under ROS exposure. In Arabidopsis, 175 genes were demonstrated to be regulated by hydrogen peroxide, including genes with MYB and AP-1-response elements (17). 140 core stress-related genes were identified in Schizosaccharomyces pombe (18, 19).
Filamentous fungi in the genus Aspergillus must cope with ROS during their growth and development. Genetic and biochemical studies shed light on the role of ROS in fungal defense, pathogenicity, and development and suggest that fungi use similar stress response pathways as mammalian and plant cells (20–24). The transcription factors AP-1, AtfA, and AtfB (all members of the basic leucine zipper (bZIP) transcription factor family) have been identified as major players in providing conidia with resistance to oxidative stress in aspergilli. Saccharomyces cerevisiae yap-1 is an ortholog of mammalian AP-1 (25–27). A yap-1 ortholog in Aspergillus parasiticus (ApyapA) is reported to regulate the timing of ROS accumulation, conidiospore development, and stress tolerance in conidiospores (26, 27). AtfA, an ortholog of S. pombe Atf1, controls conidial response to heat and oxidative stress in Aspergillus nidulans and Aspergillus oryzae (28–31). AftB is required for oxidative stress tolerance in conidiospores of A. oryzae (31).
Recent studies support a functional interaction between oxidative stress and secondary metabolism. Secondary metabolites are natural products that serve specific biological functions for the producing organism and also have detrimental or beneficial properties for humans (41). Reducing the level of oxidative stress using antioxidants correlated with a reduction in aflatoxin biosynthesis suggesting that oxidative stress serves as a trigger of secondary metabolism (26, 27, 32–36). Other data suggest that secondary metabolism evolved as a response of the fungal cell to the changing environment and is tightly associated with conidiospore development (37, 38). However, the molecular mechanisms that connect secondary metabolism, oxidative stress, and development are not clear.
We study the molecular switch mechanism that orchestrates activation of secondary metabolism using aflatoxin biosynthesis in Aspergillus as a model system (39–44). Biosynthesis of aflatoxin (one of the most potent naturally occurring carcinogens known) involves 27 genes; these genes are tightly clustered in a 70-kb region in the Aspergillus genome and are coordinately regulated (39). Our goal is to better control the beneficial and detrimental impacts of secondary metabolites on humans.
In the course of our studies, we found cyclic AMP-response element (CRE)-like sites in the promoters of several aflatoxin genes in Aspergillus parasiticus (42). One novel element designated CRE1 (TGACATAA) found in the nor-1 (early aflatoxin gene) promoter helps activate this gene in vivo in response to addition of exogenous cAMP; this treatment down-regulates cAMP/PKA signaling (42). We also showed that a protein with an approximate molecular mass of 32 kDa binds CRE1 and physically interacts with a positive aflatoxin pathway regulator called AflR (31, 42).
In this study, we investigate the expression and activity of an A. parasiticus protein that exhibits 96% identity to the bZIP protein AtfB in A. oryzae and shares several physical/biochemical characteristics with p32 (see above). Using chromatin immunoprecipitation (ChIP), we demonstrate that under aflatoxin-inducing conditions in a rich growth medium, A. parasiticus AtfB binds promoters in seven genes in the aflatoxin gene cluster. Significantly, the binding occurs at promoters that carry CRE sites, but binding does not occur in promoters lacking a CRE site. In contrast, under aflatoxin noninducing conditions, binding of AtfB to the aflatoxin gene promoters was barely detected. Moreover, disruption of veA, a global regulator of secondary metabolism and development, also severely reduced AtfB binding to the aflatoxin gene promoters. Electrophoretic mobility shift analysis (EMSA) suggested that AtfB forms a heterodimer with AP-1 at the nor-1 promoter. These data provide new insights into the association between secondary metabolism and oxidative stress response at the molecular level.
EXPERIMENTAL PROCEDURES
Strains, Growth Media, and Growth Conditions
A. parasiticus isogenic strains used in this study were derived from SU-1 (ATCC 56775), a wild type aflatoxin producer. A. parasiticus ΔveA (TJW35.21) (ver-1 wh-1 pyrG ΔveA::pyrG) was a gift from Dr. A. Calvo (45). YES liquid medium (contains 2% yeast extract and 6% sucrose; pH 5.8) was used as an aflatoxin-inducing rich growth medium; YEP liquid medium (contains 2% yeast extract and 6% peptone; pH 5.8) was used as an aflatoxin-noninducing rich growth medium; each flask contained five 6-mm glass beads (Sigma) (43). A. parasiticus strains were grown in the dark at 30 °C and 150 rpm (standard conditions).
Construction of pMBP-AtfB and Expression of MBP::AtfB Fusion Protein
Plasmid pMBP-AtfB was constructed by cloning a cDNA fragment encoding AtfB into the BamHI-XbaI region of the expression vector pMAL-c2 (New England Biolabs, Beverly, MA). To facilitate cloning, the atfB cDNA was amplified by PCR with one primer containing a BamHI (GGATCC) restriction site and the other containing an XbaI (TCTAGA) restriction site (5′TCGGATCCATGTCGGTGGACCAAACCC3′ and 5′CGACTCTAGACTAAACATTAATCAGCTC3′) under the following conditions: denaturation at 95 °C for 5 min followed by 30 cycles of 95 °C for 1 min, 60 °C for 1 min (annealing), and 72 °C for 2 min (extension). The mixture was incubated at 72 °C for 10 min to complete the reaction. The resulting plasmid construct, pMBP-AtfB, was transformed into Escherichia coli DH5α. The proper construction of pMBP-AtfB in clones expressing MBP::AtfB was confirmed by sequence analysis of the purified plasmid DNA isolated by the Qiagen miniprep plasmid kit (Qiagen, Valencia, CA). The size of the fusion protein was determined by small scale expression studies. E. coli DH5α carrying pMBP-AtfB was incubated in 5 ml of Luria-Bertani broth containing ampicillin (100 μg per ml) for 16 h. One ml of bacterial culture was saved as uninduced control. The remaining 4 ml of culture was induced to express fusion protein by the addition of 0.3 mm isopropyl β-d-thiogalactopyranoside for 3 h. Large scale production and purification of MBP::AtfB fusion protein were conducted (amylose affinity column chromatography) using methods described previously (46).
Anti-peptide AtfB Antibodies
Design of Peptide Antigens
Peptide antigens YSRTPAAMADPTCAGPAAFT (amino acids 8–27; YSR) and PPFDDKKLQTPMGEMYPVAQ (amino acids 92–111; PPF) were designed using the Immune Epitope Database and Hydrophobicity Scale carried on the ProtScale, ExPASy Proteomics Server. Based on a BLAST search of the Aspergillus flavus genome, peptide YSR possessed an amino acid sequence that is unique to A. flavus AFLA_094010. Peptide PPF shares 25% identity to a hypothetical protein designated 77.m03816 (852 amino acids, molecular mass 92.1 kDa, pI 9.81).
Immunization of Rabbits
Synthetic peptides YSR and PPF carrying Cys on the C terminus (to help conjugate to the carrier protein keyhole limpet hemocyanin) were prepared in a highly pure form and analyzed using RP-HPLC and MS. The purity of both peptides was at least 95%, and the measured molecular masses for YSR (2172.2 Da) and PPF (2395.6 Da) agreed with the predicted masses. 5 mg of each purified peptide were coupled through the terminal cysteine thiol to keyhole limpet hemocyanin using the hetero-bifunctional cross-linking agent maleimidobenzoyl-N-hydroxysuccinimide, in a ratio of 1 part peptide to 1 part keyhole limpet hemocyanin (w/w). Equal masses of peptide YSR and PPF conjugates were mixed and used as a mixture for the immunization protocol. The antigen (1:1, conjugate mixture w/w) was suspended in PBS buffer (1 mg/ml), emulsified by mixing with an equal volume of Complete Freund's adjuvant, and injected into three to four subcutaneous dorsal sites in New Zealand White rabbits for the primary immunization. Subsequent boost immunizations (on days 14, 42, and 56) were performed using Incomplete Freund's adjuvant. Specificity and titer of sera were determined for each peptide using ELISA.
Immunoaffinity Antibody Purification
5 mg of each purified synthetic peptide were coupled to 5 ml of Sulfo-Link gel through the N-terminal cysteine residue. 5 ml of crude serum were applied to each peptide-linked gel. After washing with PBS buffer, pH 7.5, the absorbed antibodies (anti-AtfB) were eluted with 0.1 n glycine HCl buffer, pH 2.5, and collected in 1 m Tris buffer for neutralization.
Western Blot Analysis
A. parasiticus SU-1 and ΔveA were grown in YES liquid medium for 48 h and mycelia harvested by filtration through Miracloth. Mycelial samples for Western blot analysis were frozen in liquid N2. Frozen mycelium was ground in liquid N2 with a mortar with a pestle, and the powdered mycelium was resuspended 1:1 (w/v) in TSA buffer (0.01 m Tris, 0.15 m NaCl, 0.05% NaN3, pH 8.0) containing 1 tablet of Complete mini protease inhibitor mixture (Roche Diagnostics), 50 μl of proteinase inhibitor mixture (Sigma), and 0.125 mm phenylmethylsulfonyl fluoride per 10 ml. Total protein (80–90 μg/lane) was separated by electrophoresis on 12% SDS-polyacrylamide gels, transferred to PVDF membranes (PerkinElmer Life Sciences), and exposed to antibody specific to AtfB (PPF or YSR, 5 μg/ml). Filters were incubated with goat anti-rabbit secondary antibody conjugated to the fluorescent tag IRDye 800CW (Li-Cor Biosciences, Lincoln, NE). Visualization of protein bands was performed using an Odyssey infrared imaging system (Li-Cor) at 795 nm.
Immunoprecipitation Using Anti-AtfB PPF
To demonstrate the specificity of the anti-AtfB antibody to A. parasiticus AtfB, immunoprecipitation was performed with the use of a Dynabeads protein G immunoprecipitation kit (Invitrogen) following the manufacturer's instructions. A total protein extract from ∼4 g of mycelia (prepared as described previously (39)) was precipitated in three steps. First, the extract was precleared with Dynabeads only to remove proteins that bind nonspecifically to the beads. Second, the supernatant was precipitated with preimmune serum to remove proteins that nonspecifically bind to rabbit IgG. Third, the supernatant was precipitated with anti-AtfB PPF. All samples (20 μg/lane) were separated by electrophoresis on 12% SDS-polyacrylamide gels, transferred to PVDF membranes, and exposed to anti-AtfB PPF and visualized as described above.
Chromatin Immunoprecipitation (ChIP)
ChIP was performed essentially as described elsewhere (39) with minor modifications. Freshly harvested conidiospores of A. parasiticus SU-1 were inoculated at 104 spores per ml and incubated for 24, 30, or 40 h in 100 ml of aflatoxin-inducing YES or aflatoxin-noninducing YEP liquid media (with five 6-mm glass beads) under standard conditions (see above). An equal quantity of conidiospores of A. parasiticus ΔveA were inoculated, incubated, and harvested at the same times points in 100 ml of aflatoxin-inducing YES liquid medium. Proteins were cross-linked in the chromatin sample, which was then sheared using conditions described previously (39). Immunoprecipitation was performed overnight at 4 °C with anti-AtfB PPF or preimmune serum obtained from the same rabbit (as a negative control). Immunoprecipitated chromatin was washed and eluted, and cross-links were reversed as described previously (39). DNA was purified using a PCR purification kit (Qiagen, Valencia, CA). Semi-quantitative PCR analysis of immunoprecipitated DNA was performed using purified DNA as a template and primers to the designated loci as described previously (39). PCR products were separated on a 1.3% agarose gel containing ethidium bromide (0.5 μg/μl) and quantified by measuring absolute intensity of the DNA bands using Adobe Photoshop software as described previously (43). Fold enrichment was calculated as the ratio of absolute band intensity obtained from the DNA immunoprecipitated by anti-AtfB to that obtained from DNA immunoprecipitated by preimmune rabbit serum for each promoter region at each time point.
Enrichment of Nuclear Proteins and Electrophoretic Mobility Shift Analysis (EMSA)
Preparation of A. parasiticus SU-1 protein extracts for EMSA was performed essentially as described previously (42). Briefly, conidiospores (104/ml) were inoculated into 100 ml of liquid YES medium and incubated under standard conditions. The mycelium was harvested on Miracloth, ground in liquid nitrogen, and resuspended in lysis buffer. 5 ml of lysis buffer (25 mm HEPES-KOH, pH 7.5, 50 mm KCl, 5 mm MgCl2, 0.1 mm EDTA, 10% glycerol, 0.5 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride) and 50 μl of protease inhibitor mixture (Sigma) were added per g of ground mycelia. The proteins were fractionated by ammonium sulfate precipitation (10% and then 70%), pelleted by centrifugation (10,000 × g for 20 min at 4 °C), and resuspended in dialysis buffer (15% glycerol, 15 mm HEPES-KOH, pH 7.9, 100 mm KCl, 1 mm EDTA, 2 mm dithiothreitol, 0.5 mm phenylmethylsulfonyl fluoride, and protease inhibitor mixture (Sigma)) at 50 μl per 10 ml and dialyzed twice against the dialysis buffer. The dialyzed protein solution was frozen in aliquots and stored at −80 °C.
EMSA
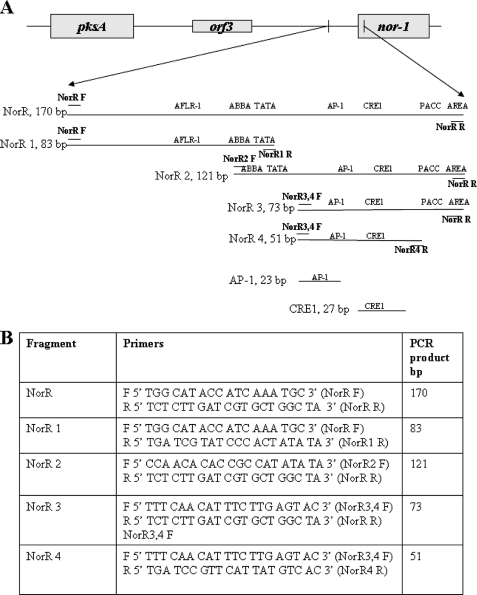
EMSA and competition EMSA were performed essentially as described previously (42). A 170-bp double-stranded NorR fragment (Fig. 4) derived from the nor-1 promoter region was generated by PCR and gel-purified using a Wizard SV gel and PCR clean-up system (Promega, Madison, WI). NorR was 5′ end-labeled with [γ-32P]ATP using Ready-to-Go T4 polynucleotide kinase (Amersham Biosciences), purified using a Micro Bio-Spin P-30 chromatography column (Bio-Rad), and used as a probe. Double-stranded (ds) DNA competitor fragments were generated by PCR (Fig. 1) and gel-purified prior to EMSA using a Wizard SV gel and PCR cleanup system (Promega, Madison, WI). Single-stranded synthetic oligonucleotides (complementary strands) were annealed to generate DNA fragments containing ds CRE1 or AP-1-like sites. Complementary strands for generation of a ds 27-bp CRE1-containing oligo were 5′TCTAAGCCGTGACATAATGAACGGATC3′ and 5′GATCCGTTCATTATGTCACGGCTTAGA3′. Complementary strands for generation of a ds 27-bp oligo containing CRE1 with five mutations were 5′TCTAAGCCGCTAGCTAGTGAACGGATC3′ and 5′GATCCGTTCACTAGCTAGCGGCTTAGA3′. Complementary strands for generation of a ds 23-bp oligo containing an AP-1 site were 5′CAACATTTCTTGAGTACTTTTCT3′ and 5′AGAAAAGTACTCAAGAAATGTTG3′. Complementary strands for generation of a ds 23-bp oligo containing an AP-1 site with five mutations were 5′CAACATTTCTCCGTCACTTTTCT3′ and 5′AGAAAAGTGACGGAGAAATGTTG3′.
FIGURE 4.
NorR and NorR subfragments used in EMSA. A, schematic of the pksA/nor-1 intergenic region showing the position of NorR in the nor-1 promoter. NorR was subdivided into smaller fragments NorR1, NorR2, NorR3, NorR4, AP-1, and CRE1; location and size (bp) of fragments are shown. The location of putative cis-acting sites are shown, including AflR1, TATA, CRE1, AP-1-like, ABBA, PACC, and AREA. B, primers used to generate NorR and NorR subfragments. Location of the primers is indicated by short horizontal lines.
FIGURE 1.
Alignment of AtfB amino acid sequences from aspergilli.
5% acrylamide (80:1 acrylamide/bisacrylamide) nondenaturing gels were used to separate DNA-protein complexes. 20 fmol of NorR probe were incubated for 15 min at 30 °C with 2 μg of poly(dI-dC), 7.5 μg of bovine serum albumin, and competitor (as appropriate) with 5 μg of cell protein extract (added last) in a final binding reaction volume of 25 μl. After separation, the gels were dried and exposed to x-ray film.
Polyclonal anti-AtfB antibodies (PPF or YSR) were used to block formation of DNA-protein complexes. 5 μg of fractionated protein extract was prepared from A. parasiticus SU-1 grown in YES liquid medium for 48 h (see above) and incubated in a DNA binding buffer (dialysis buffer) for 15 min at RT with 5 or 10 μl of anti-AtfB or preimmune serum obtained from the same rabbit as the antibody. Then the 32P-labeled NorR fragment was added and incubation continued for an additional 15 min at 30 °C. Finally, DNA-protein complexes were resolved by electrophoresis in a 5% native polyacrylamide gel.
Total RNA Isolation and Transcript Analysis
Transcript levels in cells were analyzed using real time PCR or semi-quantitative PCR (39). A. parasiticus SU-1 was grown in liquid YES medium, and the mycelia were harvested and frozen in liquid N2 at appropriate time points. Total RNA was isolated in duplicate samples by the TRIzol method (TRIzol Reagent; Invitrogen). Total RNA was treated with RNase-free DNase (Qiagen, Valencia, CA), and the RNA quality was examined by an Agilent 2100 Bioanalyzer (Agilent Technologies Santa Clara, CA). For real time PCR analyses, total RNA (1 μg) was treated with gDNA WipeOut and cDNA prepared with the QuantiTect reverse transcription kit (Qiagen). Primers were designed using Primer Express® software 3.0 (Applied Biosystems, Carlsbad, CA). Primer sequences used in this study are listed in Table 1.
TABLE 1.
Primer sequences used for quantitative real time PCR analysis
For real time PCR analyses, 1 μg of the reverse-transcribed RNA was amplified using ABI PRISM 7900HT sequence detection system with Power SYBR Green PCR Mastermix (Applied Biosystems). Reactions were carried out using the following parameters: an initial step for 2 min at 50 °C and 10 min at 95 °C followed by 40 cycles of 15 s at 95 °C and 1 min at 60 °C. Results were analyzed with a relative quantification method using SDS 2.1 software (Applied Biosystems). Samples were analyzed in triplicate. Relative gene expression levels were determined by the standard curve method. The standard curve was derived by plotting cycles to threshold values (Ct) versus the logarithm of known concentration of reverse-transcribed RNA between the ranges of 0.05 and 50 ng per reaction. For each gene, the relative level of mRNA at every time point is depicted by fold difference of mRNA levels of a target gene divided by mRNA levels of β-tubulin at the same time point.
For semi-quantitative PCR analyses, first-strand cDNA was synthesized using 1 μg of total RNA and Moloney murine leukemia virus reverse transcriptase (Invitrogen) according to the manufacturer's instructions. 2 μl of cDNA was used as a template in the subsequent PCR using a Robocycler Gradient 96 (Stratagene, La Jolla, CA) under the following conditions: initial denaturation at 94 °C for 5 min; followed by 30 cycles of 94 °C for 1 min, 60 °C for 1 min, and 72 °C for 2 min, with a final extension at 72 °C for 10 min. Primer pairs for PCR were as published previously (39). laeA primers were forward primer 5′ACTTACCGGACAGTGCAAGAAC3′ and reverse primer 5′TAGGACCAGGCAGAGAATCAAC3′; vbs primers were forward primer 5′GCTCGGTATTGCGACTGGTG3′ and reverse primer 5′TACGGTCTGCCTCGCTGTCC3′. The PCR products were separated by electrophoresis on a 1% agarose gel.
Statistical analysis was performed with the use of SigmaStat software, version 1.0 (Jandel Corp., San Rafael, CA). Quantitative real time PCR data are presented as mean ± S.E., n = 4, and were analyzed by Student's t test and one-way analysis of variance.
RESULTS
Cloning of AtfB from A. parasiticus
Previous EMSA and Southwestern blot analysis indicated that a 32-kDa A. parasiticus protein is a key component in a protein complex that binds a cAMP-response element (CRE1) in the nor-1 promoter in vivo and in vitro. To identify candidate genes that encode p32 in A. parasiticus, we searched the A. flavus genome using “cAMP-response element-binding protein” as a search landmark. The complete A. flavus genome sequence is available, although only specific locations (e.g. aflatoxin gene cluster) have been sequenced in A. parasiticus. The sequenced regions in A. parasiticus exhibit >90% identity to A. flavus in the coding regions and ∼40% identity in the intergenic regions. The search identified a single gene designated AFLA_094010 (92.m03394) encoding a transcription factor with bZIP and basic domains characteristic of the cyclic AMP-response element-binding protein (CREB) family; AFLA_094010 mapped to chromosome V. The deduced amino acid sequence of AFLA_094010 exhibited 96% identity to transcription factor AtfB in A. oryzae (a member of the ATF/CREB family) (31), 34% identity to atf21 in fission yeast, and 50% identity to a putative transcription factor Atf21 from A. fumigatus. Identity to genes from other organisms (A. nidulans, Neurospora crassa, mouse, and human) was less than 40% suggesting that that AFLA_094010 may be unique to A. oryzae, A. flavus, and A. parasiticus. AFLA_094010 encodes an intronless ORF of 957 bp. The deduced protein consists of 318 amino acids with a mass of 35.9 kDa, similar to the predicted molecular mass for the p32 we identified in previous work. Using PCR primers designed based on the AFLA_094010 sequence, the entire ORF from A. parasiticus was amplified, ligated into the pCR 2.1-TOPO plasmid vector, and transformed into competent E. coli cells (Invitrogen). The deduced protein sequence of the A. parasiticus ortholog shared 96% identity with the amino acid sequence of A. flavus AFLA_094010 and A. oryzae AtfB (Fig. 1). Based on the high degree of identity to A. flavus and A. oryzae proteins, we designated the cloned A. parasiticus protein AtfB. A. parasiticus AtfB possesses a bZIP basic domain KFLERNRLAASKCRQK (amino acids 165–180), two PKA phosphorylation sites (Ser-149 and Ser-294), and five PKC sites. The nucleotide sequence of AtfB was submitted to GenBankTM (accession number HQ396161).
Anti-peptide Polyclonal Antibodies to AtfB Specifically Recognize an MBP::AtfB Fusion
To analyze expression of AtfB in A. parasiticus and to localize this protein in the fungal cell, we generated rabbit polyclonal antibodies to two peptides derived from the AtfB amino acid sequence. We chose these sequences PPFDDKKLQTPMGEMYPVAQC (PPF) and YSRTPAAMADPTCAGPAAFTC (YSR) because they were exposed on the protein surface (based on a hydropathy profile), and they did not contain the bZIP or basic domains, and they did not carry either PKA phosphorylation site. To study the function of AtfB, we also generated a maltose-binding protein (MBP)::AtfB fusion in the vector pMalC2 (see “Experimental Procedures”). MBP::AtfB was expressed in E. coli, purified using a starch affinity column, and resolved by SDS-PAGE (Fig. 2A). Antibodies to peptide PPF (anti-AtfB PPF) consistently and specifically recognized the MBP::AtfB fusion in Western blot analysis (Fig. 2A). Because of this specificity, we utilized anti-AtfB (anti-peptide PPF) for experiments described below.
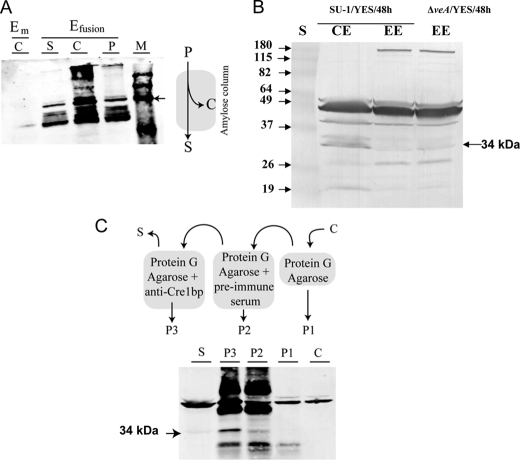
FIGURE 2.
Antibodies to AtfB (anti-AtfB) specifically recognize AtfB. A, Western blot analysis of MBP-AtfB fusion with anti-AtfB. Schematic illustrates the purification method for the fusion protein. Total protein extracts (P) obtained from E. coli expressing either MBP (Em) or recombinant MBP-AtfB (Efusion) were passed through an amylose affinity column according to manufacturer's instructions (see under “Experimental Procedures”). Western blot (left) was conducted using anti-AtfB PPF. The amount of protein loaded in each lane was 20 μg. Samples (from left) are as follows: C, protein eluted from the amylose column after passage of Em total protein extract; S and C, flow-through and protein eluted from the amylose column, respectively, after passage of Efusion total protein extract; P, Efusion total protein extract; M, BenchMark PreStained Protein Ladder, Invitrogen. Arrow, 78-kDa band in protein ladder. B, Western blot analysis of A. parasiticus protein extracts using anti-AtfB PPF. Conidiospores (106) were inoculated into 100 ml of YES liquid medium and incubated at 30 °C with shaking for 48 h. Proteins were extracted, and Western blot analysis was conducted as described under “Experimental Procedures.” 90 μg of total protein was loaded per lane. Enriched nuclear proteins were obtained as described under “Experimental Procedures.” CE, cell extract; EE, enriched nuclear proteins; S, BenchMark PreStained Protein Ladder. C, immunoprecipitation of AtfB from A. parasiticus SU-1 cell extracts (for details see “Experimental Procedures”). A cell extract (C) was obtained from 4 g of SU-1 grown in YES liquid medium. Extract C was precleared with protein G-agarose beads only to obtain precipitate P1 (obtained from beads), and supernatant was treated with protein G-agarose tagged with antibodies in preimmune serum (see “Experimental Procedures” for description). The precipitate obtained at this step, P2, was stored, and the supernatant was treated with protein G-agarose tagged with anti-AtfB. The immunoprecipitate, P3, and the supernatant, S, were stored separately. Western blot analysis was conducted on samples C, P1, P2, P3, and S using anti-AtfB. 20 μg of protein was loaded in each lane.
Anti-AtfB Specifically Detects a Native AtfB Protein in Cell Extracts
Anti-AtfB was sufficiently sensitive to detect a native 34-kDa protein directly in A. parasiticus cell extracts at 90 μg of total protein per lane (Fig. 2B). The antibody also enriched a 34-kDa native protein by immunoprecipitation, and this protein could then be detected by Western blot analysis using anti-AtfB and 20 μg of total protein per lane (Fig. 2C). These data strongly suggest that the 34-kDa protein in A. parasiticus cell extracts is AtfB, which we identified via analysis of the A. flavus genome data base.
Aflatoxin Gene Promoters Carrying CRE Bind AtfB under Aflatoxin-inducing Conditions
We conducted chromatin immunoprecipitation (ChIP) to determine whether AtfB binds to aflatoxin gene promoters carrying CRE sites. A. parasiticus SU-1 and A. parasiticus ΔveA were grown under aflatoxin-inducing (YES rich growth medium) or -noninducing conditions (YEP rich growth medium) and harvested at 24, 30, or 40 h after inoculation. Chromatin was prepared from harvested cells and subjected to ChIP using anti-AtfB. Aflatoxin gene promoters carrying CRE sites (pksA, nor-1, the fas2-fas1 intergenic region, omtA, ordA, ver-1, and aflR) were amplified using appropriate primers (see Table 1), and the degree of promoter enrichment was determined. The vbs promoter located in the aflatoxin gene cluster and the laeA promoter located outside of the cluster were amplified as controls; these promoters lacked CRE sites. Neither the vbs nor laeA promoter was enriched to significant levels at any time point under aflatoxin-inducing conditions (YES; Fig. 3A) or -noninducing conditions (YEP; Fig. 3B). In contrast, all seven promoters carrying at least one CRE site (see above) demonstrated a >2-fold enrichment at 30 and 40 h under aflatoxin-inducing conditions (YES; Fig. 3A), and these observed changes were statistically significant.
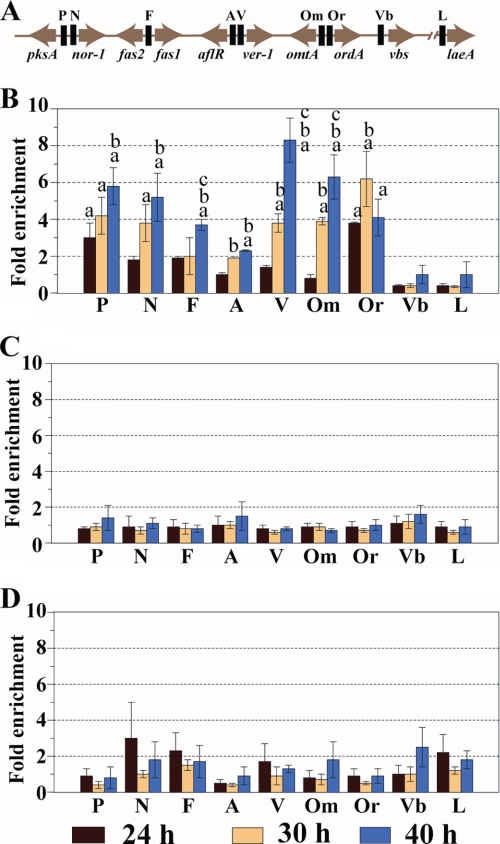
FIGURE 3.
ChIP analysis of AtfB binding to promoters in the A. parasiticus aflatoxin gene cluster. A. parasiticus conidiospores (104/ml) were inoculated into appropriate growth media and incubated at 30 °C with shaking. At 24, 30, and 40 h of growth, cultures were treated with 1% formaldehyde, and chromatin was prepared as described under “Experimental Procedures.” ChIP was performed with anti-AftB PPF. A, schematic representation of the relevant genes in the aflatoxin gene cluster and one gene, laeA, outside the cluster. Solid black bars indicate targets in the promoter region of the genes for PCR amplification in PCR analysis. Abbreviations: P, pksA; N, nor-1; FF, fas1/fas2 intergenic region; A, aflR; V, ver-1; Om, omtA; Or, ordA; Vb, vbs; L, laeA. B, relative fold enrichment (see “Experimental Procedures”) in AtfB binding at the promoters of the designated genes of A. parasiticus SU-1 grown in YES liquid medium. C, relative fold enrichment in AtfB binding at the promoters of the designated aflatoxin genes in A. parasiticus SU-1 grown in YEP liquid medium. D, relative fold enrichment in AtfB binding at the promoters of designated aflatoxin genes in A. parasiticus ΔveA grown in YES liquid medium. Two independent experiments were performed for each growth condition. Data are presented as mean ± S.E. for both experiments. Statistical analysis was performed by Student's t test. a, statistically significant 2-fold or more relative enrichment, p < 0.05; b, statistically significant difference in relative fold enrichment compared with 24 h, p < 0.05; c, statistically significant difference in fold enrichment compared with 30 h, p < 0.05.
None of the aflatoxin promoters or control promoters were enriched greater than 2-fold at 24 h in YES (prior to aflatoxin gene activation, see Fig. 4) or any time point in chromatin isolated from cells grown under aflatoxin-noninducing conditions (YEP; Fig. 3B). The data suggested that under aflatoxin-inducing conditions AtfB can bind CRE sites in aflatoxin gene promoters at increasing levels from 30 to 40 h. The one exception was ordA whose promoter binding was highest at 30 h with a trend downward by 40 h (still greater than 2-fold enrichment). ChIP was also conducted using chromatin prepared from A. parasiticus ΔveA that lacks a global regulator of secondary metabolism VeA (Fig. 3C). This strain does not synthesize aflatoxin, aflatoxin pathway intermediates, or aflatoxin enzymes. In contrast to A. parasiticus strain SU-1, anti-AtfB significantly enriched only two promoters (the vbs promoter at 40 h, 3-fold, and the nor-1 promoter at 24 h, 2-fold) in chromatin prepared from A. parasiticus ΔveA. These data strongly suggest that VeA directly or indirectly regulates AtfB binding to aflatoxin promoters. Chromatin prepared from SU-1 grown in YES and immunoprecipitated by anti-AtfB also was analyzed by real time PCR using primers specific for the nor-1 and ver-1 promoters. The enrichment values for these two promoters using semi-quantitative PCR (Fig. 3A) were similar to enrichment values generated by real time PCR (data not shown).
AtfB Binds at the Nor-1 Promoter
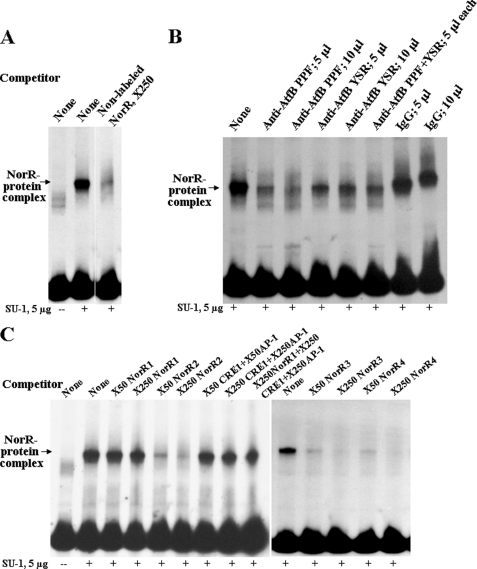
To obtain additional evidence that AtfB binds aflatoxin gene promoters and to elucidate the binding site for AtfB, EMSA was conducted on the nor-1 promoter. Previously, we detected protein(s) in cell extracts prepared from A. parasiticus grown for 48 h in minimal medium (GMS) that bind a novel cis-acting cyclic AMP-response element TGACATAA, CRE1, in the nor-1 promoter (42). In this study, a protein extract was prepared from A. parasiticus grown under aflatoxin-inducing conditions for 48 h in a rich growth medium (YES). This extract was enriched for nuclear proteins using fractionation by magnesium sulfate precipitation as described previously (see under “Experimental Procedures”) (42). The enriched extract was subjected to EMSA using a ds 170-bp nor-1 promoter fragment NorR (probe) that carries a CRE1 site (Fig. 4A). A NorR-protein complex was evident by EMSA using an A. parasiticus protein extract (Fig. 5A). Formation of this complex is specific because nonlabeled ds NorR competed efficiently with the labeled NorR probe for complex formation (Fig. 5A); however, single-stranded NorR did not compete (data not shown). Formation of NorR-protein complex was inhibited by pretreating the enriched protein extract with anti-AtfB prior to addition of the NorR fragment but not by preimmune serum obtained from the same rabbit that provided the antibody (Fig. 5B). Inhibition of NorR-protein complex formation by anti-AtfB PPF was more efficient as compared with anti-AtfB YSR; simultaneous addition of both antibodies to the protein extract did not result in improved inhibition compared with pretreatment with anti-AtfB PPF alone (Fig. 5B).
FIGURE 5.
EMSA analysis of AtfB binding at the nor-1 promoter. A. parasiticus SU-1 was grown for 48 h at 30 °C in the dark with shaking at 150 rpm. Enriched nuclear protein extracts were prepared as described under “Experimental Procedures.” 5 μg of protein in enriched nuclear protein extracts were added to the labeled NorR probe in each lane. A, competition EMSA. Nonlabeled NorR (50 and 250× molar excess) was added to compete for labeled NorR probe. B, shift inhibition EMSA. Anti-AtfB PPF or preimmune serum were added to determine whether these could block protein/DNA interaction. C, competition EMSA. NorR subfragments were added in 50 or 250× molar excess to compete for labeled NorR probes. Competitors were as follows: donors (83 bp), dsNorR2 (121 bp), ds NorR3 (73 bp), dsNorR4 (51 bp), dsCRE1 (27 bp), and dap-1 (23 bp) (see Fig. 4).
cis-Acting Binding Element in NorR Consists of CRE1 and AP-1-like Sites
To locate the site in NorR responsible for NorR-protein complex formation, competition EMSA was performed using NorR subfragments NorR1, NorR2, NorR3, and NorR4 (Fig. 4A). EMSA analysis demonstrated that only NorR fragments containing a CRE1 site competed at significant levels (Fig. 5C). However, neither double- nor single-stranded short DNA fragments (27 bp) carrying a CRE1 site only could effectively compete for NorR complex formation (data not shown). An imperfect AP-1 (47) site TGAGTAC was identified 12 nucleotides upstream from CRE1 (Fig. 4A) in the nor-1 promoter. NorR4, a 51-bp double-stranded DNA fragment containing both CRE1 and AP-1-like sites, competed very efficiently for complex formation (Fig. 5C), but ds oligonucleotides (23 bp) that carried either an AP-1-like site or a mutant AP-1-like (AP-1m5) (Fig. 4A) did not compete at significant levels (data not shown). Also, simultaneous addition of two oligonucleotides carrying independent CRE1 or AP-1-like sites or a combination of these oligonucleotides with NorR1 did not inhibit complex formation (Fig. 5C). The data strongly suggest that AtfB is a key element in the NorR-protein complex and that NorR-protein complex formation requires both CRE1 and AP-1-like sites.
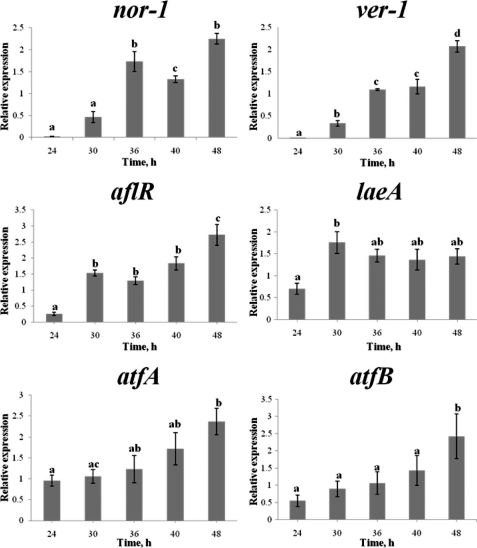
Expression of atfB Correlates with Aflatoxin Gene Expression
We analyzed accumulation of transcripts for aflatoxin cluster genes (nor-1, ver-1, and aflR), a global regulator of secondary metabolism laeA, and two transcription factors related to oxidative stress (atfB and atfA) in A. parasiticus SU-1 grown in a rich, liquid growth medium under aflatoxin inducing conditions (YES) at 24, 30, 36, 40, and 48 h using real time PCR. Transcript accumulation of aflatoxin genes steadily increased from 24 to 48 h of growth (Fig. 6), which paralleled an increase in aflatoxin synthesis and release of the toxin into the growth medium. atfB transcript accumulation showed a similar trend as aflatoxin gene transcripts. These data support the idea that p32 identified as part of a previous study (42) is AtfB and indicate that changes in aflatoxin promoter binding by AtfB observed by ChIP correlate with transcript levels of atfB and aflatoxin genes.
FIGURE 6.
Expression of atfB correlates with temporal pattern of expression of nor-1, ver-1, aflR, and atfA, but not of laeA. A. parasiticus SU-1 was grown in YES liquid medium for designated periods of time, and RNA extraction and real time PCR analyses were performed as described under “Experimental Procedures.” The relative level of mRNA is depicted as the mRNA level of the target gene divided by mRNA level of β-tubulin at the same time point. Bars represent mean ± S.E. (n = 4). Statistical analysis was performed by the Student's t test and one-way analysis of variance(see “Experimental Procedures”). Same lowercase letters indicate that there is no statistically significant difference between two measurements. Different lowercase letters specify a statistically significant difference between two measurements. p values are as follows: nor-1 (p < 0.001), ver-1 (p = 0.002), aflR (p < 0.001), laeA (p = 0.018), atfA (p = 0.016), atfB (p = 0.029). Two independent biological replicates were performed showing the same trend. Two duplicates samples were analyzed for each biological replicate.
Sequence Analysis of Aflatoxin Gene Promoters
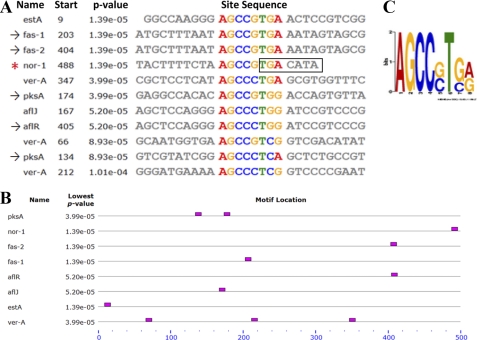
Several lines of evidence presented above indicated that aflatoxin gene promoters contain AtfB-binding sites. We applied MEME motif-based sequence analysis to search for random 8-mer motif pattern occurrence within the 500-bp promoter regions upstream from ATG in 17 aflatoxin genes (pksA, nor-1, fas-2, fas-1, aflR, aflJ, adhA, estA, ver-1, verA, avnA, verB, avfA, omtB, omtA, ordA, and vbs) and laeA. Exceptions included avnA and avfA, whose entire intergenic regions (367 and 173 bp correspondingly) were used for the analysis. A conserved motif AGCC(G/C)T(G/C)(A/G) (with the second highest frequency occurrence) was found 11 times in eight promoters (Fig. 7). Six of the eight promoters possessed only one motif. Promoters of divergently transcribed genes (fas-2/fas-1 and aflR/aflJ) shared the only motif that was located in the corresponding intergenic region. Two promoters contained multiple octamer motifs: pksA carried two motifs and verA possessed three motifs. Five of these eight promoters exhibited AtfB binding in ChIP analysis: nor-1, fas-1, fas-2, pksA, and aflR. The conserved octamer motif was not observed in the vbs promoter that also did not bind AtfB in ChIP analysis. In the nor-1 promoter, the motif was found one time and included a portion of the CRE1 site identified previously in the nor-1 promoter, TGACATAA (Fig. 7). Interestingly, the five nucleotides immediately upstream from TGACATAA were highly conserved in all eight promoters, suggesting that they may contribute to AtfB binding.
FIGURE 7.
Sequence analysis of aflatoxin gene promoters using MEME. 500-bp promoter regions upstream from ATG in 17 aflatoxin genes and laeA were analyzed for 8-mer motif pattern occurrence using MEME motif-based sequence analysis. The motif AGCC(G/C)T(G/C)(A/G) (second highest frequency occurrence) was found a total of 11 times within eight promoters. A, sites carrying the motif in eight promoters; *, the nor-1 promoter contained a functional CRE1 motif; →, promoters analyzed by ChIP. B, occurrence and location of the motif relative to start site ATG (denoted at the end of the scale). Divergently transcribed fas-2/fas-1 and aflR/aflJ share the only motif located in the corresponding intergenic region. C, sequence logo of the motif, which contains partial CRE1 site represented by position-specific probability matrices on the positive strand.
DISCUSSION
AtfB Binds Multiple Aflatoxin Gene Promoters under Aflatoxin-inducing Conditions
In this study, we demonstrated that AtfB, a protein that shares many characteristics with p32, binds aflatoxin promoters (nor-1 and six others) that carry at least one CRE site under aflatoxin-inducing conditions (YES) strongly suggesting that the CRE site drives interaction of AtfB with promoters within the aflatoxin gene cluster.
The actual role of the CRE site in directing complex formation in the promoters appears to be complicated. This work demonstrated that under aflatoxin-inducing conditions (YES), the CRE1 site alone is not sufficient for complex formation; CRE1 and the adjacent region, possibly an AP-1-like site, are involved in binding, and probably, a physical interaction between AtfB and AP-1 is required for promoter binding activity. In contrast, our previous work indicated that the CRE1 is the only site required for complex formation on the nor-1 promoter during growth in GMS, a chemically defined aflatoxin-inducing medium (42). Follow-up work will determine whether these two transcription factors interact in vivo and in vitro. Interestingly, the CRE1 site and the AP-1-like site show a high degree of sequence similarity; future work will also focus on determining the differential recognition by AtfB of highly similar binding sites. Another possible explanation for the lack of competition by the CRE1 site alone is that the protein complex binds to single-stranded DNA. However, competition EMSA experiments with the use of single-stranded DNA fragments carrying a CRE1 site demonstrated that this is not the case (data not shown).
AtfB Is an Important Component of the NorR-Protein Complex
We generated several independent lines of evidence that suggest that A. parasiticus AtfB is a key component of the NorR-protein complex (identified by Southwestern blot and EMSA analyses of the nor-1 promoter). (a) AtfB is member of the CREB/ATF family that binds CRE sites. (b) AtfB associates with aflatoxin promoters that carry at least one CRE site but not with promoters that lack CRE sites (ChIP). (b) Anti-AtfB specifically blocks DNA-protein complex formation in the nor-1 promoter using extracts from cells grown under aflatoxin-inducing conditions (EMSA). Together, these data confirm that AtfB is an important component of a protein complex that binds the nor-1 promoter under aflatoxin-inducing conditions.
AtfB Binding Is Associated with Activation of Aflatoxin Gene Expression
The patterns of AtfB binding to aflatoxin promoters and transcript analysis strongly support a direct role for the AtfB protein complex in the activation of several aflatoxin gene promoters. In previous work, we observed that A. parasiticus does not synthesize aflatoxin enzymes or aflatoxin until at least 24 h after conidiospore inoculation in either GMS or YES growth media (39, 48). Between 30 and 40 h post-inoculation, aflatoxin enzymes and aflatoxin accumulate at maximum rates. ChIP analysis demonstrated that histone H4 acetylation in aflatoxin promoters mirrored the pattern of activation of the aflatoxin genes at the level of transcript accumulation (39). In this study, we observed a similar temporal pattern of AtfB binding to aflatoxin promoters consistent with a role in activation. Moreover, the expression of atfB showed a similar trend as expression of nor-1, ver-1, and aflR.
How does AtfB promote gene activation? We hypothesize that AtfB promotes DNA unwinding and binding of the transcription factor AflR, a key positive regulator of aflatoxin gene expression, to aflatoxin promoters. Members of the CREB family of transcription factors associate with p300/CBP (CREB-binding protein) and PCAF (p300/CBP-associated factor) that possess histone acetyltransferase activity. If AtfB acts in an analogous manner, promoter binding may initiate the previously observed waves of histone H4 acetylation associated with gene activation (39).
AtfB May Play a Role in Integration of Aflatoxin Synthesis and Stress Response
The nucleotide sequence of AtfB exhibits 96% identity to atfB from A. oryzae (31). atfB (and atfA) encodes a bZIP type transcription factor in the ATF/CREB family and plays an important role in promoting resistance of conidiospores to heat shock, osmotic stress, and oxidative stress (31, 49). In response to oxidative stress, A. oryzae atfB directly activates the catalase gene, catA (31, 49). Consistent with this theme, atf1 and its ortholog aftA activate the catalase gene ctt1 in fission yeast S. pombe and catalase B in A. nidulans, respectively (28, 50). ATF/CREB family proteins also activate genes involved in osmotic stress response in S. cerevisiae and S. pombe. In our recent preliminary studies to identify proteins associated with AtfB, we performed mass spectrometric analysis of the immunoprecipitate obtained using anti-AtfB PPF and a total protein extract of A. parasiticus SU-1 grown for 40 h in YES liquid medium. The data revealed the presence of AP-1 and AtfA along with other proteins.5
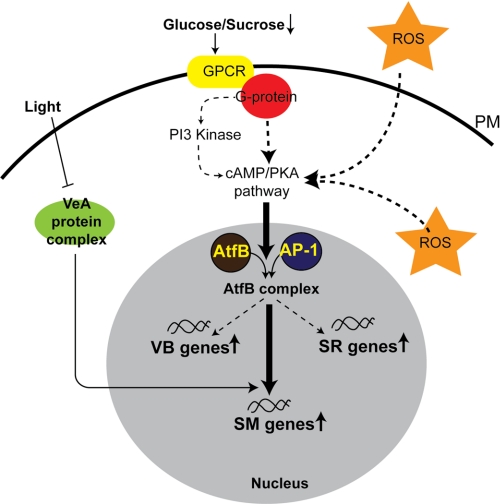
Data from our group and others indicate that aflatoxin biosynthesis is an important component in the cellular response to oxidative stress in A. parasiticus. Aflatoxin biosynthesis also requires simultaneous formation of specialized vesicles, aflatoxisomes, for completion of biosynthetic pathway and export of the toxin to the cell exterior. We therefore propose that an increase in intracellular ROS down-regulates the cAMP/PKA/PI3K signaling pathway, which facilitates formation of an AtfB·AP-1 complex (may also contain AtfA). This complex in turn activates stress-response genes and secondary metabolism genes and influences genes involved in formation of aflatoxisomes (see Fig. 8 for a schematic of this regulatory model). To address this hypothesis, in future work, we will analyze A. parasiticus promoters of genes involved in aflatoxin biosynthesis, heat shock (HSP70 and HSP90), oxidative stress (catalase and superoxide dismutases with three different metal affinities), and osmotic stress (trehalose synthase and ion transporters) to determine whether their promoters bind AtfB or AP-1.
FIGURE 8.
AtfB integrates secondary metabolism and oxidative stress response. Based on available experimental evidence, we propose that exposure of the fungal cell to intra- or extracellular ROS down-regulates a cAMP/PKA/PI3K signaling cascade. This promotes formation of an active AftB/AP-1 heterodimer on target gene promoters involved in stress response (SR), secondary metabolism (SM), and vacuole biogenesis (VB). Alternatively, ROS may directly facilitate heterodimer formation on the promoters of target genes by affecting the redox state of the transcription factor. Initiation of secondary metabolism triggered by ROS plays a protective and/or signaling role in the overall cellular response to oxidative stress. PKA, protein kinase A; PI3K, phosphatidylinositol 3-kinase; PM, plasma membrane.
Novel Predicted Motif in Aflatoxin Gene Promoters Overlaps CRE1
MEME motif-based sequence analysis of aflatoxin gene promoter regions revealed one novel motif AGCCG/CTG/CA/G that was highly conserved in eight aflatoxin promoters that belong to early (pksA, nor-1, fas-2, fas-1, and aflR) and middle (aflJ, estA, and verA) genes in the aflatoxin pathway. Importantly, in the nor-1 promoter this motif overlapped the CRE1 site. The discovery of five conserved nucleotides AGCCG immediately upstream from CRE1 suggests that the functional AtfB-binding site in aflatoxin promoters may consist of 13 or more nucleotides, which agrees with our EMSA data. Future studies are necessary to clarify the functional role of this novel motif.
Our observations support the idea that the in vivo interaction of AtfB with the aflatoxin promoters occurs at the extended recognition sites containing CRE motifs of variable sequence. Analogous phenomenon was noticed in S. cerevisiae during the study of in vivo binding of a transcription factor Sko1 (an ortholog of AtfA) to the target gene promoters in response to osmotic stress (51). In the course of this study it has been found that the 15 Sko1 target promoters contain 47 CRE motifs with variable near-consensus sequence, whereas only one was identical to the consensus. Moreover, the evolutionarily conserved CRE motifs also possess 2–5 flanking nucleotides upstream and/or downstream from the CRE site, and these nucleotides were conserved among the sensu stricto yeast species.
Conclusions
AtfB binds to multiple promoters in the aflatoxin gene cluster carrying at least one CRE-like site in vivo, and binding correlates with activation of transcription of the aflatoxin genes. AtfB is a crucial part of the protein complex formed in the nor-1 promoter and is necessary for activation of aflatoxin biosynthesis. In addition, the nor-1 promoter possesses a composite regulatory element that consists of two closely positioned and highly similar binding sites, which play different roles under aflatoxin-inducing and -noninducing conditions. Future experiments will focus on the significance of AtfB binding in other aflatoxin gene promoters and will determine the functional importance of the observed association between oxidative stress and aflatoxin synthesis.
Acknowledgment
We thank Dr. A. Calvo for the gift of A. parasiticus ΔveA.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1 52003-19 from NCI. This work was also supported by the Michigan Agricultural Experiment Station.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s) HQ396161.
J. E. Linz, unpublished data.
- ROS
- reactive oxygen species
- YES
- yeast extract sucrose medium
- GMS
- glucose minimal salts medium, an aflatoxin inducing chemically defined medium
- YEP
- yeast extract peptone medium
- MBP
- maltose-binding protein
- CRE
- cAMP-response element
- oligo
- oligonucleotide
- bZIP
- basic leucine zipper
- CREB
- cyclic AMP-response element-binding protein.
REFERENCES
- 1. Coisne C., Engelhardt B. (2011) Antioxid. Redox. Signal. 15, 1285–1303 [DOI] [PubMed] [Google Scholar]
- 2. DeWeese T. L., Shipman J. M., Larrier N. A., Buckley N. M., Kidd L. R., Groopman J. D., Cutler R. G., te Riele H., Nelson W. G. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 11915–11920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hu D., Cao P., Thiels E., Chu C. T., Wu G. Y., Oury T. D., Klann E. (2007) Neurobiol. Learn. Mem. 87, 372–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Leiser S. F., Miller R. A. (2010) Mol. Cell. Biol. 30, 871–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Meyer T. E., Liang H. Q., Buckley A. R., Buckley D. J., Gout P. W., Green E. H., Bode A. M. (1998) Int. J. Cancer 77, 55–63 [DOI] [PubMed] [Google Scholar]
- 6. Radunoviæ A., Porto W. G., Zeman S., Leigh P. N. (1997) Neurosci. Lett. 239, 105–108 [DOI] [PubMed] [Google Scholar]
- 7. Shen G. X. (2010) Can. J. Physiol. Pharmacol. 88, 241–248 [DOI] [PubMed] [Google Scholar]
- 8. Spooner R., Yilmaz O. (2011) Int. J. Mol. Sci. 12, 334–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Weseler A. R., Bast A. (2010) Curr. Hypertens. Rep. 12, 154–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wingler K., Hermans J., Schiffers P., Moens A., Paul M., Schmidt H. (2011) Br. J. Pharmacol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nguyen T., Huang H. C., Pickett C. B. (2000) J. Biol. Chem. 275, 15466–15473 [DOI] [PubMed] [Google Scholar]
- 12. Nguyen T., Nioi P., Pickett C. B. (2009) J. Biol. Chem. 284, 13291–13295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nguyen T., Yang C. S., Pickett C. B. (2004) Free Radic. Biol. Med. 37, 433–441 [DOI] [PubMed] [Google Scholar]
- 14. Lee J. S., Surh Y. J. (2005) Cancer Lett. 224, 171–184 [DOI] [PubMed] [Google Scholar]
- 15. Hur W., Gray N. S. (2011) Curr. Opin. Chem. Biol. 15, 162–173 [DOI] [PubMed] [Google Scholar]
- 16. Nguyen T., Sherratt P. J., Pickett C. B. (2003) Annu. Rev. Pharmacol. Toxicol. 43, 233–260 [DOI] [PubMed] [Google Scholar]
- 17. Desikan R., A-H-Mackerness S., Hancock J. T., Neill S. J. (2001) Plant Physiol. 127, 159–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen D., Toone W. M., Mata J., Lyne R., Burns G., Kivinen K., Brazma A., Jones N., Bähler J. (2003) Mol. Biol. Cell 14, 214–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen D., Wilkinson C. R., Watt S., Penkett C. J., Toone W. M., Jones N., Bähler J. (2008) Mol. Biol. Cell 19, 308–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Aguirre J., Ríos-Momberg M., Hewitt D., Hansberg W. (2005) Trends Microbiol. 13, 111–118 [DOI] [PubMed] [Google Scholar]
- 21. Gessler N. N., Aver'yanov A. A., Belozerskaya T. A. (2007) Biochemistry 72, 1091–1109 [DOI] [PubMed] [Google Scholar]
- 22. Miskei M., Karányi Z., Pócsi I. (2009) Fungal Genet. Biol. 46, S105–S120 [DOI] [PubMed] [Google Scholar]
- 23. Scandalios J. G. (2005) Braz. J. Med. Biol. Res. 38, 995–1014 [DOI] [PubMed] [Google Scholar]
- 24. Scott B., Eaton C. J. (2008) Curr. Opin. Microbiol. 11, 488–493 [DOI] [PubMed] [Google Scholar]
- 25. Karin M., Liu Z., Zandi E. (1997) Curr. Opin. Cell Biol. 9, 240–246 [DOI] [PubMed] [Google Scholar]
- 26. Reverberi M., Zjalic S., Punelli F., Ricelli A., Fabbri A. A., Fanelli C. (2007) Food Addit. Contam. 24, 1070–1075 [DOI] [PubMed] [Google Scholar]
- 27. Reverberi M., Zjalic S., Ricelli A., Punelli F., Camera E., Fabbri C., Picardo M., Fanelli C., Fabbri A. A. (2008) Eukaryot. Cell 7, 988–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Balázs A., Pócsi I., Hamari Z., Leiter E., Emri T., Miskei M., Oláh J., Tóth V., Hegedus N., Prade R. A., Molnár M., Pócsi I. (2010) Mol. Genet. Genomics 283, 289–303 [DOI] [PubMed] [Google Scholar]
- 29. Hagiwara D., Asano Y., Yamashino T., Mizuno T. (2008) Biosci. Biotechnol. Biochem. 72, 2756–2760 [DOI] [PubMed] [Google Scholar]
- 30. Lara-Rojas F., Sánchez O., Kawasaki L., Aguirre J. (2011) Mol. Microbiol. 80, 436–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sakamoto K., Arima T. H., Iwashita K., Yamada O., Gomi K., Akita O. (2008) Fungal Genet. Biol. 45, 922–932 [DOI] [PubMed] [Google Scholar]
- 32. Chang P. K., Scharfenstein L. L., Luo M., Mahoney N., Molyneux R. J., Yu J., Browm R. L., Campbell B. C. (2010) Toxins 3, 82–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Huang J. Q., Jiang H. F., Zhou Y. Q., Lei Y., Wang S. Y., Liao B. S. (2009) Int. J. Food Microbiol. 130, 17–21 [DOI] [PubMed] [Google Scholar]
- 34. Jayashree T., Subramanyam C. (2000) Free Radic. Biol. Med. 29, 981–985 [DOI] [PubMed] [Google Scholar]
- 35. Kim J. H., Yu J., Mahoney N., Chan K. L., Molyneux R. J., Varga J., Bhatnagar D., Cleveland T. E., Nierman W. C., Campbell B. C. (2008) Int. J. Food Microbiol. 122, 49–60 [DOI] [PubMed] [Google Scholar]
- 36. Narasaiah K. V., Sashidhar R. B., Subramanyam C. (2006) Mycopathologia 162, 179–189 [DOI] [PubMed] [Google Scholar]
- 37. Guzmán-de-Peña D., Ruiz-Herrera J. (1997) Fungal Genet. Biol. 21, 198–205 [DOI] [PubMed] [Google Scholar]
- 38. Hicks J. K., Yu J. H., Keller N. P., Adams T. H. (1997) EMBO J. 16, 4916–4923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Roze L. V., Arthur A. E., Hong S. Y., Chanda A., Linz J. E. (2007) Mol. Microbiol. 66, 713–726 [DOI] [PubMed] [Google Scholar]
- 40. Roze L. V., Chanda A., Laivenieks M., Beaudry R. M., Artymovich K. A., Koptina A. V., Awad D. W., Valeeva D., Jones A. D., Linz J. E. (2010) BMC Biochem. 11, 33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Roze L. V., Chanda A., Linz J. E. (2011) Fungal Genet. Biol. 48, 35–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Roze L. V., Miller M. J., Rarick M., Mahanti N., Linz J. E. (2004) J. Biol. Chem. 279, 27428–27439 [DOI] [PubMed] [Google Scholar]
- 43. Chanda A., Roze L. V., Kang S., Artymovich K. A., Hicks G. R., Raikhel N. V., Calvo A. M., Linz J. E. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 19533–19538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chanda A., Roze L. V., Linz J. E. (2010) Eukaryot. Cell 9, 1724–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Calvo A. M., Bok J., Brooks W., Keller N. P. (2004) Appl. Environ. Microbiol. 70, 4733–4739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lee L. W., Chiou C. H., Linz J. E. (2002) Appl. Environ. Microbiol. 68, 5718–5727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Abate C., Patel L., Rauscher F. J., 3rd, Curran T. (1990) Science 249, 1157–1161 [DOI] [PubMed] [Google Scholar]
- 48. Skory C. D., Chang P. K., Linz J. E. (1993) Appl. Environ. Microbiol. 59, 1642–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sakamoto K., Iwashita K., Yamada O., Kobayashi K., Mizuno A., Akita O., Mikami S., Shimoi H., Gomi K. (2009) Fungal Genet. Biol. 46, 887–897 [DOI] [PubMed] [Google Scholar]
- 50. Degols G., Russell P. (1997) Mol. Cell. Biol. 17, 3356–3363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Proft M., Gibbons F. D., Copeland M., Roth F. P., Struhl K. (2005) Eukaryot. Cell 4, 1343–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]