Abstract
Perilipin (PLIN1) is a constitutive adipocyte lipid droplet coat protein. N-terminal amphipathic helices and central hydrophobic stretches are thought to anchor it on the lipid droplet, where it appears to function as a scaffold protein regulating lipase activity. We recently identified two different C-terminal PLIN1 frame shift mutations (Leu-404fs and Val-398fs) in patients with a novel subtype of partial lipodystrophy, hypertriglyceridemia, severe insulin resistance, and type 2 diabetes (Gandotra, S., Le Dour, C., Bottomley, W., Cervera, P., Giral, P., Reznik, Y., Charpentier, G., Auclair, M., Delépine, M., Barroso, I., Semple, R. K., Lathrop, M., Lascols, O., Capeau, J., O'Rahilly, S., Magré, J., Savage, D. B., and Vigouroux, C. (2011) N. Engl. J. Med. 364, 740–748.) When overexpressed in preadipocytes, both mutants fail to inhibit basal lipolysis. Here we used bimolecular fluorescence complementation assays to show that the mutants fail to bind ABHD5, permitting its constitutive coactivation of ATGL, resulting in increased basal lipolysis. siRNA-mediated knockdown of either ABHD5 or ATGL expression in the stably transfected cells expressing mutant PLIN1 reduced basal lipolysis. These insights from naturally occurring human variants suggest that the C terminus sequesters ABHD5 and thus inhibits basal ATGL activity. The data also suggest that pharmacological inhibition of ATGL could have therapeutic potential in patients with this rare but metabolically serious disorder.
Keywords: Cell Biology, Lipase, Lipid Droplets, Lipid Metabolism, Lipodystrophy
Introduction
Lipid droplets (LD)4 are increasingly recognized as dynamic cell organelles rather than inert fat droplets. Although all eukaryotic cells can form lipid droplets to accommodate surplus energy, white adipocytes are uniquely adapted to store energy for the whole organism in a large unilocular lipid droplet that occupies approximately 90% of the volume o the cell. As well as buffering diet-derived free fatty acids, they also need to supply other tissues with energy in the form of non-esterified fatty acids during exercise or when fasting. Unregulated lipolysis leads to excess delivery of lipid to other tissues where it accumulates and is strongly implicated in the pathogenesis of insulin resistance and highly prevalent metabolic diseases such as type 2 diabetes (1). A recent genomic screen in drosophilaS2 cells suggested that as many as 200 proteins could be involved in regulating the formation and degradation of LDs (2). However, a more select subgroup of LD proteins appear to be specifically expressed in white adipocytes (3). Of these, perilipin (PLIN1) is the most abundant constitutive LD coat protein. Originally cloned by Greenberg et al. (4) in 1990, PLIN1 was the first mammalian LD protein to be recognized. Since then, several loss of function and domain-selective deletion studies have suggested that both the N and C terminals of PLIN1 are required to restrain basal lipase activity and to facilitate stimulated lipolysis (5–10). Amphipathic helices in the N terminus of PLIN1 are thought to facilitate its binding to the surface phospholipid monolayer surrounding the LD (11). Another study suggests that central hydrophobic stretches are crucial for lipid droplet anchoring (12). PLIN1 plays a key role in the regulation of hormone-stimulated lipolysis. As well as phosphorylating and activating hormone-sensitive lipase (HSL), protein kinase A (PKA) also phosphorylates PLIN1, the N-terminal of which then binds to HSL at the surface of the lipid droplet. This interaction is essential for HSL to access its substrate (6, 8).
Following the realization that lipase activity was still present in HSL-null mice, another lipase was identified in 2004 (13–15) and named “adipose triglyceride lipase” (ATGL). In contrast to the mode of activation of HSL, ATGL activity appears to be primarily regulated by an inhibitor, G0S2, and a coactivator, ABHD5 (16–18). Elegant work by Granneman et al. (19) has suggested that PLIN1binds and sequesters ABHD5 in the basal state. Lipolytic stimuli trigger PLIN1 phosphorylation and the subsequent release of ABHD5, enabling it to activate ATGL-mediated triglyceride hydrolysis (19, 20). The resultant diacylglycerol is in turn hydrolyzed by HSL (21). These findings have led to a model in which PLIN1functions as a regulatory scaffold protein, facilitating activation of lipases and their access to core triglycerides. In the case of HSL, the lipase is active while bound to the N-terminal of PLIN1, whereas ATGL is activated by ABHD5 after the latter has been released by PLIN1.
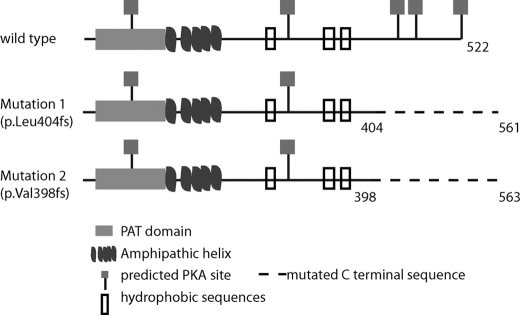
We recently identified the first loss-of-function mutations in PLIN1 in patients with a novel subtype of familial partial lipodystrophy, hypertriglyceridemia, severe insulin resistance, and type 2 diabetes. Both mutations encode proteins that retain the N-terminal amphipathic helices and are correctly targeted at the LD surface (1) but alter the C-terminal tail of PLIN1, resulting in an elongated C-terminal with a significantly altered amino acid sequence (Fig. 1). We hypothesized that these sequence changes would impair binding to ABHD5, enabling it to coactivate ATGL and increase basal lipolysis. We used bimolecular fluorescence to explore the impact of the PLIN mutants on protein trafficking and interactions at the LD surface and siRNA-mediated knockdown to validate the role of ABHD5 and ATGL in PLIN1-dependent inhibition of basal lipolysis.
FIGURE 1.
Schematic illustration of human PLIN1. Key domains and phosphorylation sites are highlighted, as are the consequences of the C-terminal PLIN1 frame shift mutations (p.Leu-404fs and p.Val-398fs). PAT, Perilipin-Adipophilin-TIP47.
EXPERIMENTAL PROCEDURES
Materials
Forskolin, isobutylmethylxanthine (IBMX), fatty acid-free BSA, oleic acid/albumin, neonatal calf serum (NCS), DMEM (4500 mg/dl glucose), G418, and puromycin were purchased from Sigma. TriacsinC was purchased from Alomone Labs, Ltd. FBS was purchased from Hyclone. Alexa-tagged secondary antibodies, ProLong Gold anti-fade mounting medium, and LipidTOX Deep Red were purchased from Molecular Probes (Invitrogen).
Cell Culture
3T3L1 cells were cultured in DMEM with 10% NCS, 400 μg/ml G418, and 4 μg/ml puromycin. COS7 cells were cultured in DMEM with 10% FBS.
Establishment of 3T3L1 Cells Stably Expressing PLIN1
3T3L1 cells were stably transfected with either pLXSN or pLXSN-FLAGPLIN1WT. Both cell lines were stably transfected with pBABE, pBABE-MycPLIN1WT, pBABE-MycPLIN1-404fs, or pBABE-MycPLIN1-398fs. Retroviruses were titrated using the RetroX virus titration kit (Clontech) to achieve an equal multiplicity of infection for all constructs. The generation of these mutants has been described previously (1).
Generation of Bimolecular Fluorescence Complementation Constructs
Split YFP fragments, Yn and Yc, were generated using primer pairs Yn1F_HindIII (5′-CGTCAGATCCAAGCTTGCTACCGGTCGCCACCATG-3′) + Yn1R (5′-CGCCGCTGGTACCGCACCACCACCGCTCCCACCACCACCGGCCATGATATAGACG-3′) and YcF2 (5′-CTCGAGTGGCCGACAAGCAGAAGAACG-3′) +YcR2 (5′-CGGGCCCTCACTTGTACAGCTCGTCCATGCC-3′). Yn was cloned into pcDNA3.1 using HindIII-KpnI, and Yc was cloned into pcDNA3.1 using XhoI-XbaI/SpeI, creating pcDNA3.1-Yn and pcDNA3.1-Yc, respectively. Human ABHD5 cDNA was subcloned from pcDNA4HisMaxABHD5 (kind gift of Dr. Rudolf Zechner) using KpnI-XbaI into pcDNA3.1-Yn, resulting in pcDNA3.1-Yn-ABHD5 (Yn fused in-frame with the N terminus of ABHD5). Human perilipin was PCR-amplified from pcDNA3.1-MycPLIN1 (WT)/404fs/398fs using primers MycPlpnFwd (5′-GATATCGTCGACATGGAACAGAAACTCATCTCAGAAGAGGATCTGGCAGTCAACAAAGGCCTCACCTTGCTGGATG-3′) and BiFcPlpnWTrev (5′-GCGGCCGCGCTCTTCTTGCGCAGCTGGC-3′) or BiFcPlpnMutrev (5′-GCGGCCGCGGATGAGGCTGAGCTCC-3′) and cloned in-frame with the C-terminal of Yc in pcDNA3.1-Yc using EcoRV-NotI, resulting in pcDNA3.1-MycPLIN-WT-Yc, pcDNA3.1- MycPLIN-404fs, and Yc pcDNA3.1-MycPLIN-398fs-Yc. Human ATGL was PCR-amplified from human adipose tissue cDNA using primers hATGLBiFcfwd (5′-CGGGATATCATGTTTCCCCGCGAGAAGACGTG-3′) and hATGLBifCrev (5′-GCGGCCGCCAGCCCCAGGGCCCC-3′) and cloned into the pCRIITOPO cloning vector. Active site Ser-47 was mutagenized to Ala using site-directed mutagenesis primers ATGLS47Atop (5′-CCACGCACATCTACGGCGCCGCGGCCGGGGCGCTCACGGCCAC-3′) and ATGLS47Abottom (5′-GTGGCCGTGAGCGCCCCGGCCGCGGCGCCGTAGATGTGCGTGG-3′).The resultant human ATGL(S47A) cDNA was subcloned into pcDNA3.1-Yc using EcoRV-NotI, resulting in pcDNA3.1-ATGL(S47A)-Yc (Yc fused in-frame with the C terminus of ATGL).
Bimolecular Fluorescence Complementation
To study the interaction between Yn-ABHD5 and PLIN1-Yc, a plasmid ratio of 200:200 ng was used for WT PLIN1, and 500:500 ng was used for the C-terminal frame shift mutants. To study the interaction between Yn-ABHD5 and ATGL(S47A)-Yc, a plasmid ratio of 100:100 ng:100 ng with MycPLIN1WT and 200:200 ng with MycPLIN1 p.Leu-404fs and MycPLIN1 p.Val-398fs was used. Plasmids were transfected using Lipofectamine 2000, cells were incubated at 37 °C, and the medium was changed to 400 μm oleic acid, at which point cells were incubated at 32 °C for 20 h. For BiFc between Yn-ABHD5 and ATGL(S47A)-Yc, cells were incubated for another 2 h at 30 °C. Cells were fixed or harvested at a total of 24 h post-transfection.
Microscopy and Image Analysis
Intracellular localization of PLIN1 (Myc-tagged) and ABHD5 was studied in confluent 3T3L1 preadipocytes or transiently transfected COS7 cells incubated in 400 μm oleic acid/albumin (Sigma) supplemented 10% NCS containing DMEM (25 mm glucose) for 60–70 h. Cells were fixed in 4% formaldehyde solution, permeabilized, and stained using anti-Myc mouse monoclonal 4A6 (Millipore) and anti-ABHD5 rabbit polyclonal (kind gift of Dawn Brasaemle), followed by detection antibodies (goat anti-rabbit highly cross adsorbed (HCA)-Alexa 594 (Molecular Probes, A11037) and goat anti-mouse (HCA)-Alexa 488 (Molecular Probes, A11029), respectively). Lipid droplets were stained with LipidTOX Deep Red at a 1:1000 dilution, and DAPI was present in the mounting medium. 0.7-μm confocal sections were imaged under ×63 magnification with additional ×2 or ×3 zoom or ×40 (without additional zoom) using a Zeiss LSM 510 Meta confocal microscope. Images were acquired using the sequential mode with Argon 458/477/488/514 nm, HeNe 594, HeNe 633, and Diode 405 lasers. Images were acquired using the Zen software package (Zeiss).
BiFc Calculations
BiFc between Yn-ABHD5 and PLIN1-Yc
Cells were imaged using a ×40 objective. The sum of YFP fluorescence intensity above the background (set to the same value across all images) was computed using Volocity 5 (PerkinElmer Life Sciences). An average of 10 randomly selected images per group was used for analysis, and the experiment was repeated six times.
BiFc between Yn-ABHD5 and ATGL(S47A)-Yc
Cells were imaged using a ×40 objective. Ten randomly chosen fields were imaged, and the sum of YFP fluorescence intensity above the background (set to the same value across all images) was computed using Volocity 5 (PerkinElmer Life Sciences). The sum of YFP fluorescence from the 10 randomly chosen fields of PLIN1-coexpressing cells was normalized to that of cells only expressing Yn-ABHD5 and ATGL(S47A)-Yc. The sum of YFP fluorescence from cells cotransfected with 100 ng of wild-type PLIN1 was normalized to that from cells cotransfected with 200 ng of Yn-ABHD5 and ATGL(S47A)-Yc, whereas the sum of YFP fluorescence from cells cotransfected with 200 ng of mutant PLIN1was normalized to that from cells transfected with 100 ng Yn-ABHD5 and ATGL(S47A)-Yc to compare data from cells with similar levels of ABHD5. The difference between the groups was also statistically significant when both test groups were normalized to cells transfected with 100 ng of Yn-ABHD5 and ATGL(S47A)-Yc. This experiment was repeated three times to obtain means and S.E.
Measurement of Lipolysis
Confluent monolayers of 3T3L1 preadipocytes, grown in 12- or 24-well plates, were treated with 0.0148 MBq of 14C-oleic acid/ml in combination with 400 μm oleic acid/albumin (Sigma) supplemented with 10% NCS containing DMEM (25 mm glucose) for 16 h. Cells were washed four times with prewarmed PBS and incubated for 4 h in 2% fatty acid-free BSA containing low-glucose DMEM (5 mm glucose) and 6 μm TriacsinC. For stimulated lipolysis, 10 μm forskolin and 250 μm IBMX were added to the chase medium. To assess lipolysis, 14C release in the chase medium after 4 h was normalized to total 14C accumulated in cells over 16 h at the end of the pulse period.
ABHD5 and ATGL Knockdown
Confluent monolayers of 3T3L1 cells were transfected with 50 nm of mouse ABHD5 or ATGL siRNA (Applied Biosystems, Inc., s85121 and s83965, respectively) using N-TER peptide nanoparticle-based transfection reagent (Sigma). 4 h post-transfection, cells were treated with 400 μm oleic acid in 10% NCS containing DMEM for 44 h and then harvested for RNA extraction. For lipolysis assays, 28 h post-transfection, cells were treated with 14C-oleic acid for 16 h, and lipolysis was measured as described above.
Western Detection of Proteins
Confluent monolayers of 3T3L1 cells were washed twice in ice-cold PBS and lysed in ice-cold lysis buffer (40 mm HEPES (pH7.4), 150 mm NaCl, 2 mm EDTA, 10 mm pyrosphosphate, 10 mm glycerophosphate, 0.3% CHAPS, complete EDTA-free protease inhibitor mixture (Roche)). Cell extracts so obtained were then centrifuged at 16,000 × g for 20 min at 4 °C. Supernatants so obtained were used for protein estimation. 10 μg of protein from COS7 cells and 50 μg of protein from 3T3L1 cells were used for SDS-PAGE. Proteins were electroblotted onto nitrocellulose membranes using the iBlot apparatus (Invitrogen). Membranes were blocked with either 5% skimmed milk or 3% BSA. Primary antibodies were used as follows: anti-Myc (4A6, Millipore), 1:1000 in 3% BSA-TBS 0.1% Tween 20; anti-FLAG (M2, Sigma), 1:1000 in 5% skimmed milk-TBST; anti-ABHD5 (kind gift of Dr. Dawn Brasaemle), 1:2500 for 3T3L1 cell lysates and 1:50,000 for COS7 cell lysates; anti-ATGL (Cell Signaling Technologies, Inc.), 1:1000 in 5% BSA-TBST.
mRNA Quantification
Confluent monolayers of 3T3L1 cells were treated with oleic acid/albumin for 48 h. Total RNA was used to generate cDNA using SuperscriptII (Invitrogen), and then mRNA expression was measured by real-time quantitative RT-PCR using an ABI Taqman master mix according to the manufacturer's protocols and analyzed on ABI PRISM 7900HT (Applied Biosystems, Inc.). The following primer sets were used: mABHD5, 5′-CATAGATGGCAACTCTGGAACCA-3′ (forward) and 5′-CCGAGGATGGCAATTGTCTT-3′ (reverse); probe, 5′FAM-CCAGTCACTGCGACCGAAGTCCTACG-TAMRA-3′); peptidyl prolyl isomerase A, 5′-TTCCTCCTTTCACAGAATTATTCCA-3′ (forward) and 5′-CCGCCAGTGCCATTATGG-3′ (reverse); probe, 5′FAM-6ATTCATGTGCCAGGGTGGTGACTTTACAC-TAMRA-3′); and ATGL (Applied Biosystems, Inc., Mm00503040_m1).
Statistics
Quantitative data are presented as mean ± S.E. One-way or two-way analysis of variance (effect of stimulation and effect of ABHD5/ATGL siRNA experiments) with post hoc Bonferroni analyses were performed on data at a minimum p < 0.05 threshold.
RESULTS
Perilipin C-terminal Frameshift Mutants Do Not Inhibit Basal Lipolysis
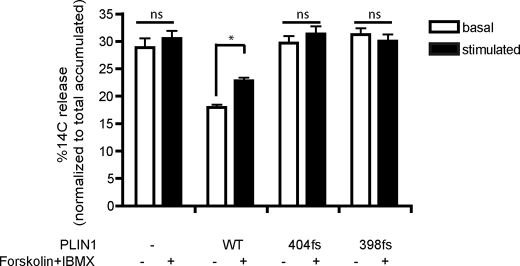
We have recently described C-terminal frame shift mutations in PLIN1associated with lipodystrophy and insulin resistance (1) (Fig. 1). We found previously that the inability of the mutants to promote triglyceride storage is due, at least in part, to their inability to suppress basal lipolysis. Next we wanted to test whether lipolytic stimulation would further enhance lipolysis in 3T3L1 cells stably expressing WT or mutant PLIN1 (p.Leu-404fs or p.Val-398fs). In this system, basal lipolysis was suppressed approximately 40% by wild-type PLIN1 but not by the mutants (p < 0.001, Fig. 2). Lipolysis increased in response to stimulation with forskolin and IBMX in cells expressing WT PLIN1 but was unchanged in cells expressing the PLIN1mutant proteins (Fig. 2). These data suggest that in 3T3L1 cells, the C terminus of human PLIN1(from amino acid 398 onwards) is not only essential for hormonally activated lipolysis but also for suppression of basal lipolysis.
FIGURE 2.
Impact of PLIN1 C terminus frame shift mutants on lipolysis. 3T3L1 cells stably expressing the control (empty) vector, wild-type PLIN1, PLIN1- 404fs, or PLIN1- 398fs were treated with 14C-oleic acid to label the triglyceride pool, and release of incorporated radioactivity was measured over 4 h in the presence of TriascinC without (basal lipolysis, white bars) or with (stimulated lipolysis, black bars) forskolin and IBMX. Data are mean ± S.E. from three independent experiments with three replicate wells per experiment (n = 9). *, p < 0.05. ns, not significantly different.
The C Terminus of Perilipin Is Required for ABHD5 Protein Stabilization
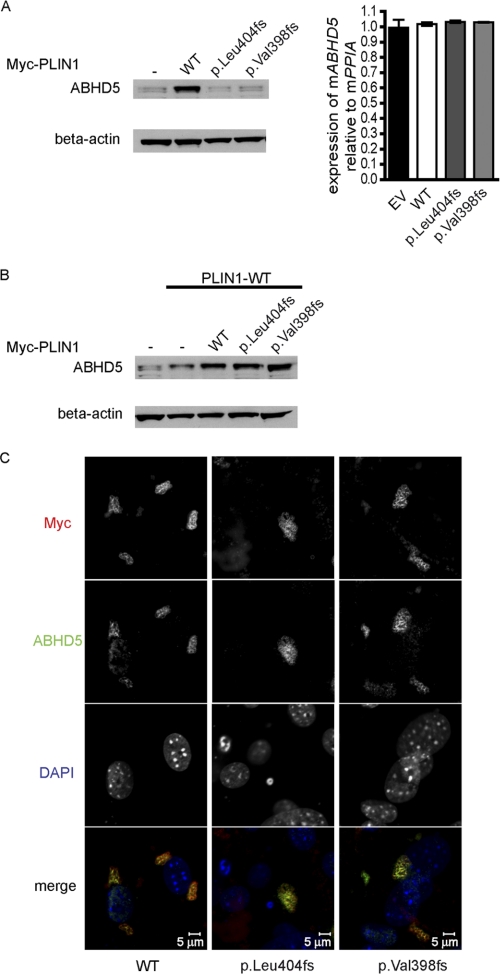
ATGL is the first enzyme required for triglyceride hydrolysis (13), whereas HSL exhibits a higher activity toward diglyceride substrates (21). Thus, we hypothesized that the C terminus of PLIN1 beyond Val-398 may harbor a stretch of amino acids required for preventing ATGL-dependent lipolysis in the basal state. ABHD5 is a known activator of ATGL and is proposed to bind to the C terminus of mouse PLIN1 (18, 20). We explored the expression and localization of endogenously expressed ABHD5 in 3T3L1 preadipocytes stably expressing WT and C-terminal frame shift mutants of human PLIN1. We observed that endogenous ABHD5 protein levels were consistently higher (approximately 5-fold) in cells expressing WT PLIN1compared with either the control cells or the 404fs and 398fs PLIN1 mutant expressing cells, whereas ABHD5 mRNA levels were similar in all cases (Fig. 3A). However, ABHD5 protein levels were comparable in cells coexpressing the 404fs/398fs mutant (Myc-tagged) and WT PLIN1(FLAG-tagged) when compared with cells transfected with double the amount of WT PLIN1 (Fig. 3B). In this cell system, we observed colocalization of ABHD5 and Myc-tagged PLIN1 irrespective of the C terminus sequence (Fig. 3C). This suggests that in patients with these heterozygous mutations in the C terminus of PLIN1, ABHD5 is likely to be present at the lipid droplet surface. To establish whether or not other LD proteins, such as PLIN2, which is thought to at least partly compensate for the loss of PLIN1, could rescue ABHD5 protein levels in the complete absence of PLIN1 in vivo, we checked ABHD5 expression in white adipose tissue samples from PLIN1-null mice (kindly provided by Constantine Londos (22)). ABHD5 protein was essentially undetectable in these samples, in keeping with our observations from cultured cells (supplemental Fig. 1).
FIGURE 3.
The C terminus of PLIN1 is required to stabilize ABHD5 on lipid droplets. Expression of endogenous ABHD5 protein detected in total cell lysates from 3T3L1 cells stably expressing WT or mutant PLIN1 (Myc-tagged) (A) and from cells cotransfected with both wild-type (FLAG-tagged) and mutant (Myc-tagged) PLIN1 (B). A, right panel, transcript (mRNA) levels of endogenous ABHD5 normalized to PPIA (n = 3). EV, empty vector. C, ABHD5 localizes to lipid droplets in 3T3L1 cells coexpressing mutant and wild-type PLIN1. Gray scale intensity profiles of Myc/Alexa 488 (pseudo-colored red) and ABHD5/Alexa 594 (pseudo-colored green) were merged to generate the colored panels at the bottom of each cell line. PPIA, peptidyl prolyl isomerase A.
PLIN1-independent localization of ABHD5 on the lipid droplet has been demonstrated by Gruber et al. (23). We found that transient coexpression of COS7 cells with WT PLIN1 and ABHD5 increased expression of ABHD5 protein when compared with cells transfected with ABHD5 alone (supplemental Figs. 2A and Fig. 4, A and B). In contrast, the C-terminal frame shift mutants of PLIN1 failed to increase the amount of ABHD5 protein. We tested whether the localization of ABHD5 was affected in this model. Although WT PLIN1and ABHD5 colocalized uniformly, the colocalization of ABHD5 and mutant PLIN1 (p.Leu-404fs and p.Val-398fs) was partial (supplemental Fig. 2B). Our data suggest that although ABHD5 localization on the lipid droplet is not fully dependent upon PLIN1 in this cellular system, interactions with the C terminus region may stabilize ABHD5 on the surface of the lipid droplet.
The C Terminus of Perilipin Binds ABHD5
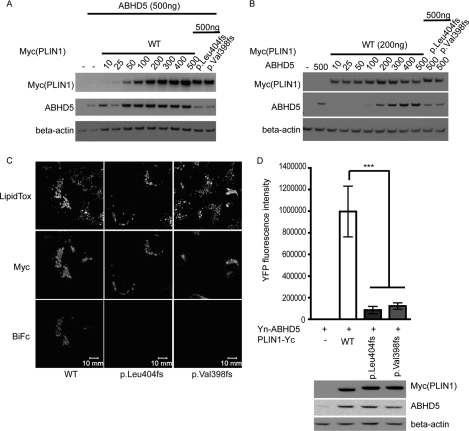
Although both WT and mutant PLIN1 and ABHD5 colocalize on the lipid droplet, this does not necessarily imply that they interact with each other. To assess binding between ABHD5 and the C terminus PLIN1 mutants, we employed bimolecular fluorescence complementation assays in COS7 cells (24). WT PLIN1(-Yc) and (Yn-)ABHD5 expression plasmids were titrated against a fixed amount of plasmids of mutant PLIN1(-Yc) and the cotransfected (Yn-)ABHD5 plasmid to obtain comparable levels of ABHD5 and PLIN1 protein levels in WT or mutant PLIN1-expressing cells (Fig. 4, A and B). YFP fluorescence could be detected in cells coexpressing ABHD5 and WT PLIN1, suggesting a direct interaction between these two proteins (Fig. 4C). PLIN1mutants (404fs and 398fs) only generated approximately 10% of the YFP fluorescence observed in cells expressing WT PLIN1, even though the cells were transfected so as to achieve equal levels of PLIN1 and ABHD5 protein in all cases (Fig. 4D).
FIGURE 4.
The C terminus of PLIN1 interacts with ABHD5 on lipid droplets. A, titration of PLIN1(WT)-Yc against a fixed amount of Yn-ABHD5 (200 ng of plasmid DNA) indicates that transfection of 200 ng of plasmid encoding wild-type PLIN1 results in PLIN1 levels equivalent to those seen in cells transfected with 500 ng of plasmids encoding the PLIN1 mutants. B, titration of Yn-ABHD5 against 200 ng of plasmid encoding PLIN1 (WT)-Yc indicates that 200 ng of plasmid encoding Yn-ABHD5 results in ABHD5 levels equivalent to those seen in cells transfected with 500 ng of plasmid encoding Yn-ABHD5 and 500 ng of plasmids encoding the PLIN1 mutants. C, direct interaction between PLIN1and ABHD5 was assessed using BiFc in COS7 cells after normalizing for equal expression of each protein. Plasmids encoding Yn-ABHD5 and Myc-PLIN1-Yc were cotransfected in COS7 cells and stained with anti-Myc and anti-rabbit-Alexa 594 antibodies and LipidTOX Deep Red to label lipid droplets. The presence of BiFc indicates direct interaction between PLIN1 and ABHD5. D, the BiFc signal was quantified as described under “Experimental Procedures.” Data are mean ± S.E. from ten 76.5 μ × 76.5 μ frames in one experiment and are representative of six independent experiments. The lower panel confirms equal expression levels of PLIN1 and ABHD5 in data sets being compared in the bar graph. ***, p < 0.001.
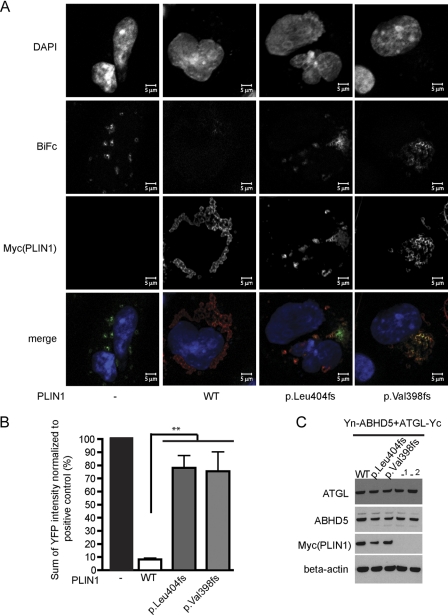
Impaired ABHD5 Binding by Perilipin C-terminal Mutants Facilitates Enhanced ATGL Activation in the Basal State
We hypothesized that the presence of ABHD5 on the lipid droplet and its inability to bind to the PLIN1 404fs and 398fs mutants would facilitate increased interaction between ABHD5 and ATGL and thereby enhance basal lipolysis (19). To test this hypothesis, we compared the ability of Yn-ABHD5 and ATGL-Yc to interact and generate bimolecular fluorescence in the presence of the WT or C-terminal frame shift mutant PLIN1. We observed that in the absence of PLIN1, ATGL-Yc and Yn-ABHD5 colocalize and interact on the lipid droplet (data not shown and Fig. 5A). Localization of WT PLIN1 to the lipid droplet suppresses this interaction between ATGL and ABHD5 (low BiFc, Fig. 5A, second panel from left). Although p.Leu-404fs and p.Val-398fs PLIN1 also localize to the lipid droplet, they fail to reduce the interaction between ATGL and ABHD5 at the lipid droplet surface. Although WT PLIN1 inhibits YFP complementation by approximately 90% compared with the PLIN1-negative control, p.Leu-404fs and p.Val-398fs PLIN1 only inhibit YFP complementation by approximately 20% (p < 0.01, Fig. 5B). Collectively, these results suggest that the C-terminal region of perilipin is required for suppressing basal ATGL-dependent lipolysis.
FIGURE 5.
PLIN p.Leu-404fs and p.Val-398fs mutants fail to fully suppress the interaction between ABHD5 and ATGL. The interaction between ABHD5 (Yn-ABHD5) and ATGL (ATGLS47A-Yc) was measured by BiFc in the absence or presence of PLIN1. A, Yn-ABHD5 and ATGL-Yc interact on lipid droplets in the absence of PLIN1, whereas an interaction between them is suppressed by wild-type PLIN1. The PLIN1mutants (p.Leu-404fs and p.Val-398fs) do not suppress interaction between ABHD5 and ATGL as efficiently as wild-type PLIN1. Each panel is a representative image of cells stained with antibodies against Myc (Alexa 594). The top four panels are gray scale intensity profiles, merged in the bottom panel, where blue indicates nuclei (DAPI), red indicates PLIN1, and green indicates the BiFc signal. B, the interaction between ABHD5 and ATGL was quantified as described under “Experimental Procedures.” Data are mean ± S.E. from three independent experiments. Data are normalized in each case to a positive control, i.e. cells expressing YnABHD5 and ATGL S47A-Yc in the absence of exogenous PLIN1. **, p < 0.01. Data shown are YFP fluorescence intensities in the test samples normalized to YFP fluorescence intensities in the positive control, i.e. Yn-ABHD5 and ATGL-Yc cotransfected in the absence of PLIN1. Normalization was done so that samples of equal ABHD5 expression levels were being compared. In the case of wild-type perilipin, the positive control used was 200:200 ng of plasmids encoding Yn-ABHD5:ATGL-Yc, and in the case of the mutants it was 100:100 ng of plasmids encoding Yn-ABHD5:ATGL-Yc. However, normalization was not required to observe a phenotypic difference, i.e. 5-fold greater suppression by wild-type perilipin compared with mutant perilipin. C, representative protein expression of PLIN1, ABHD5, and ATGL in sample sets being compared in the bar graph. _1, a ratio of 100:100 ng; _2, a ratio of 200:200 ng of plasmids encoding YnABHD5:ATGL S47A-Yc.
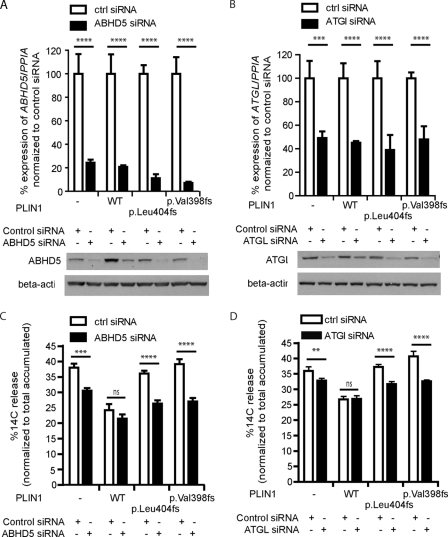
ABHD5 or ATGL Knockdown Rescues Impaired Lipid Accumulation by Perilipin C-terminal Mutants
To confirm that the defect in suppressing basal lipolysis was due to enhanced ATGL activation by ABHD5 in the 3T3L1 cell system, we tested whether the defect could be rescued by reducing ABHD5 or ATGL expression. Transient transfection of mouse-specific ABHD5 siRNA or ATGL siRNA led to a significant knockdown of endogenous ABHD5 or ATGL, respectively (Fig. 6, A and B), and significantly reduced basal lipolysis in cells expressing either of the perilipin mutants (C and D). In contrast, basal lipolysis was not altered by knocking down ABHD5 or ATGL in cells expressing WT PLIN1 perilipin, suggesting that maximum suppression of endogenous ATGL activity was achieved by the expression of WT PLIN1 in this preadipocyte model. Together, these data suggest that the C terminus of PLIN1 is required for effective suppression of basal ATGL activity and that human mutations that affect this region of the protein impair triglyceride storage because of their inability to sequester ABHD5 and thereby to suppress ATGL activity in the basal state.
FIGURE 6.
The impact of ABHD5 or of ATGL knockdown on basal lipolysis. The indicated 3T3L1 cell lines were transfected with either control siRNA, ABHD5 siRNA, or ATGL siRNA. A and C, ABHD5 (A) and ATGL (C) mRNA transcript levels were substantially depleted (to approximately 20–40% of the levels present in control (white bars) cells) (top panel) using ABHD5 (A) and ATGL (C) siRNA and resulting in significant depletion of ABHD5 and ATGL protein levels (lower panel, A and C, respectively). B and D, basal lipolysis was determined in cells treated with either control siRNA (white bars) or ABHD5- (B) or ATGL-specific (D) siRNA (black bars). Data are mean ± S.E. from three independent experiments, each with triplicate wells (n = 9). ns, p > 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
DISCUSSION
Accumulating experimental evidence suggests that rather than simply “coating” and “shielding” the lipid droplet in white adipocytes, PLIN1 orchestrates the access of lipases to core lipids, particularly triglyceride and diglyceride hydrolases, each of which predominantly hydrolyzes a specific class of glycerolipids. Early work by Brasaemle and co-workers (12, 25, 26) suggested that both the N and C termini of PLIN1 were required for optimal triglyceride storage, whereas central hydrophobic domains appeared to anchor PLIN1 to the LD surface. Hickenbottom et al. (11) solved the three-dimensional structure of the C terminus of PLIN3 and, on the basis of sequence homology, suggested that the central hydrophobic domains appear to be embedded in a four-helix bundle that is not likely to be lipid-associated. They instead proposed that N-terminal amphipathic helices in all PLINs are likely to be responsible for lipid droplet targeting. HSL appears to interact directly with PLIN1 and to depend upon PLIN1 for localization to the lipid droplet (6–9, 27). These interactions require an intact N-terminal of PLIN1, although they might also involve some interaction with the C terminus (8). Further investigations by the Greenberg and Zechner groups (5, 10, 13) suggested that ATGL, rather than HSL, is the major determinant of basal lipolysis in adipocytes. These data also suggested that the effects of PLIN1 on basal lipolysis are predominantly mediated via inhibition of ATGL. Granneman et al. (19) provided a molecular explanation for these observations by demonstrating the ability of WT PLIN1 to effectively sequester ABHD5 from ATGL, thereby inhibiting basal lipolysis.
We sought to shed further light on these findings by characterizing the molecular consequences of naturally occurring human PLIN1 mutations. We previously identified two different frame shift mutations affecting the C terminus of PLIN1 in patients with a novel familial partial lipodystrophy phenotype. Although the fact that the patients were already diabetic and at least relatively insulin-deficient precluded us from definitively characterizing lipolysis in vivo in these patients. Their phenotype, particularly the overall reduction in fat mass, elevated liver fat content, and elevated plasma triglyceride levels, is consistent with increased basal lipolysis (1). Increased basal lipolysis has been demonstrated in adipocytes from PLIN1-null mice (22, 28). We deliberately characterized the cellular effects of the PLIN1 mutants in cells in which endogenous PLIN1 is not expressed to allow us to compare the behavior of WT versus mutant PLIN1. The fact that the PLIN1 mutants failed to suppress basal lipolysis prompted further molecular studies aimed at addressing the consequences of the mutants on ATGL activation. Subramanian et al. (20) used several deletion mutants of mouse PLIN1 to identify a stretch of amino acids (382–429) indispensable for binding of ABHD5. Our data are consistent with these findings insofar as both the Leu-404fs and Val-398fs mutants alter the homologous region of human PLIN1 and fail to bind ABHD5 in transfected cells. Interestingly our data suggest that this does not prevent ABHD5 localization to the lipid droplet, which is consistent with the work of Gruber et al. (23), who proposed that an N-terminal Trp-rich region of ABHD5 is the major requirement for LD localization. These data are also consistent with subsequent binding to ATGL, which appears to be contingent upon ABHD5 localization to the lipid droplet (19). In addition, our data provide evidence that stabilization of ABHD5 requires the C-terminally intact perilipin.
The C terminus of mouse PLIN3 (TIP47) is the only PAT (Perilipin Adipophilin TIP47) domain-containing protein for which the crystal structure has been solved (11). PLIN1 and PLIN3 belong to a gene/protein family of five members sharing remote homology. Conserved residues at the C terminus map to a hydrophobic cleft formed between a four-helix bundle and a terminal α/β fold. In PLIN3, the α/β fold is contributed in part by two α helices and two β sheets at the N terminus and two β sheets at the C terminus of the four-helix bundle (11). The cleft was proposed to bind either a hydrophobic peptide, protein, or small monomeric lipid (11). PLIN1a, unlike the other PLINs, contains in addition an extended C-terminal tail with two putative PKA phosphorylation sites (11). Although the sequence similarity of PLINs in the C-terminal domain is low, their sequences can be aligned with confidence thanks to the modern profile-to-profile methods (29) and availability of hundreds of homologous PLIN sequences from different species in the databases (supplemental Fig. 3A). Despite clear homologies, many differences exist between the C termini, e.g. 1) several insertions and deletions between the elements of the regular secondary structure, of which the most prominent is the stretch of acidic amino acids between positions 302 and 316; 2) the alteration of the predicted irregular helix 4 with prolines at positions 326 and 328; and 3) altered residues in the putative groove.
Based on the currently available structural data of PLIN3, work by Subramanian et al. (20), and the available human PLIN1 mutants associated with lipolytic dysfunction, we propose an important role for this unique C-terminal region of PLIN1 in the control of ATGL-mediated lipolysis. The frame shift replaces the C-terminal tail and the two predicted β-strands with a long missense chain (supplemental Fig. 3B) with no predicted secondary structure (31).
ABHD5 is known to bind both PLIN5 and PLIN2 (24). Recent work by Chong et al. (31) demonstrates that the C terminus of PLIN2 (residues 172–425) forms an independently folding α-helical structure. In contrast to PLIN1, truncation of this C-terminal domain resulted in larger lipid droplets in mouse mammary epithelial cells. Together, these data and our findings suggest that a secondary structure, most likely unique to PLIN1, is required for efficiently sequestering ABHD5 and thereby promoting efficient fat packaging.
One of the more striking and unexpected findings of our work was the dramatic reduction in ABHD5 protein levels in cells expressing mutant forms of PLIN1. Our observations from cultured cells are consistent with similarly reduced protein levels of ABHD5 in white adipose tissue from PLIN1-null mice (supplemental Fig. 1). These data suggest that as well as sequestering ABHD5 from ATGL and thereby inhibiting basal lipolysis, PLIN1 stabilizes ABHD5 protein levels, potentially priming the cell to activate ATGL in response to lipolytic stimuli. Alternatively, the decrease in ABHD5 protein level may be due to increased association with ATGL. The relatively lower levels of ABHD5 seen in cells expressing the PLIN1 mutants and in tissue from Plin-null mice might also be contributing to the observed failure to further increase lipolysis in response to forskolin/IBMX in cultured cells or to in vivo lipolytic stimuli in the knockout mice. In the case of the human mutants, coexpressing WT and mutant PLIN isoforms in cells significantly increases ABHD5 levels. An alternative potential explanation for the reduced protein levels of ABHD5 is the reduction in lipid droplet size and cellular triglyceride content observed in these cells. Protein levels of PLIN1 itself and other lipid droplet proteins such as PLIN2 are known to be stabilized by interaction with the lipid droplet (30, 32).
As well as providing a molecular explanation for the observed increase in basal lipolysis associated with two novel human PLIN1 mutants, these data largely corroborate the emerging impression of PLIN1 as an essential lipid droplet coat protein whose C terminus regulates ATGL activity and basal lipolysis indirectly by binding and stabilizing its coactivator, ABHD5 (see supplemental Fig. 4 for a schematic illustration of this concept). This scaffolding function is also critical for HSL activation, enabling PLIN1 to precisely regulate both tri- and diglyceride hydrolysis. Finally, molecular understanding of the consequences of the PLIN1 mutants together with the observed response to ATGL knockdown suggests that ATGL inhibitors, which are as yet unavailable, may be therapeutically useful.
Supplementary Material
Acknowledgments
We are very grateful to Dawn Brasaemle for anti-CGI58 polyclonal rabbit serum, the late Constantine Londos for adipose tissue samples from PLIN1-null mice, and Rudolf Zechner for the hABHD5 cDNA clone.
This work was supported by grants from the Wellcome Trust (to S. G., S. O., and D. B. S.), the United Kingdom National Institute for Health Research Cambridge Biomedical Research Centre, and the Medical Research Council Centre for Obesity and Related Metabolic Disease.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
- LD
- lipid droplet
- ABHD5
- alpha beta hydrolase domain containing 5
- ATGL
- adipose triglyceride lipase
- HSL
- hormone sensitive lipase
- PKA
- protein kinase A
- PPIA
- peptidyl prolyl isomerase A
- PLIN1
- perilipin.
REFERENCES
- 1. Gandotra S., Le Dour C., Bottomley W., Cervera P., Giral P., Reznik Y., Charpentier G., Auclair M., Delépine M., Barroso I., Semple R. K., Lathrop M., Lascols O., Capeau J., O'Rahilly S., Magré J., Savage D. B., Vigouroux C. (2011) N. Engl. J. Med. 364, 740–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Guo Y., Walther T. C., Rao M., Stuurman N., Goshima G., Terayama K., Wong J. S., Vale R. D., Walter P., Farese R. V. (2008) Nature 453, 657–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wolins N. E., Brasaemle D. L., Bickel P. E. (2006) FEBS Lett. 580, 5484–5491 [DOI] [PubMed] [Google Scholar]
- 4. Greenberg A. S., Egan J. J., Wek S. A., Garty N. B., Blanchette-Mackie E. J., Londos C. (1991) J. Biol. Chem. 266, 11341–11346 [PubMed] [Google Scholar]
- 5. Miyoshi H., Perfield J. W., 2nd, Souza S. C., Shen W. J., Zhang H. H., Stancheva Z. S., Kraemer F. B., Obin M. S., Greenberg A. S. (2007) J. Biol. Chem. 282, 996–1002 [DOI] [PubMed] [Google Scholar]
- 6. Miyoshi H., Souza S. C., Zhang H. H., Strissel K. J., Christoffolete M. A., Kovsan J., Rudich A., Kraemer F. B., Bianco A. C., Obin M. S., Greenberg A. S. (2006) J. Biol. Chem. 281, 15837–15844 [DOI] [PubMed] [Google Scholar]
- 7. Moore H. P., Silver R. B., Mottillo E. P., Bernlohr D. A., Granneman J. G. (2005) J. Biol. Chem. 280, 43109–43120 [DOI] [PubMed] [Google Scholar]
- 8. Shen W. J., Patel S., Miyoshi H., Greenberg A. S., Kraemer F. B. (2009) J. Lipid Res. 50, 2306–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sztalryd C., Xu G., Dorward H., Tansey J. T., Contreras J. A., Kimmel A. R., Londos C. (2003) J. Cell Biol. 161, 1093–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang H. H., Souza S. C., Muliro K. V., Kraemer F. B., Obin M. S., Greenberg A. S. (2003) J. Biol. Chem. 278, 51535–51542 [DOI] [PubMed] [Google Scholar]
- 11. Hickenbottom S. J., Kimmel A. R., Londos C., Hurley J. H. (2004) Structure 12, 1199–1207 [DOI] [PubMed] [Google Scholar]
- 12. Garcia A., Sekowski A., Subramanian V., Brasaemle D. L. (2003) J. Biol. Chem. 278, 625–635 [DOI] [PubMed] [Google Scholar]
- 13. Zimmermann R., Strauss J. G., Haemmerle G., Schoiswohl G., Birner-Gruenberger R., Riederer M., Lass A., Neuberger G., Eisenhaber F., Hermetter A., Zechner R. (2004) Science 306, 1383–1386 [DOI] [PubMed] [Google Scholar]
- 14. Villena J. A., Roy S., Sarkadi-Nagy E., Kim K. H., Sul H. S. (2004) J. Biol. Chem. 279, 47066–47075 [DOI] [PubMed] [Google Scholar]
- 15. Jenkins C. M., Mancuso D. J., Yan W., Sims H. F., Gibson B., Gross R. W. (2004) J. Biol. Chem. 279, 48968–48975 [DOI] [PubMed] [Google Scholar]
- 16. Lu X., Yang X., Liu J. (2010) Cell Cycle 9, 2719–2725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yang X., Lu X., Lombès M., Rha G. B., Chi Y. I., Guerin T. M., Smart E. J., Liu J. (2010) Cell Metab. 11, 194–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lass A., Zimmermann R., Haemmerle G., Riederer M., Schoiswohl G., Schweiger M., Kienesberger P., Strauss J. G., Gorkiewicz G., Zechner R. (2006) Cell Metab. 3, 309–319 [DOI] [PubMed] [Google Scholar]
- 19. Granneman J. G., Moore H. P., Krishnamoorthy R., Rathod M. (2009) J. Biol. Chem. 284, 34538–34544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Subramanian V., Rothenberg A., Gomez C., Cohen A. W., Garcia A., Bhattacharyya S., Shapiro L., Dolios G., Wang R., Lisanti M. P., Brasaemle D. L. (2004) J. Biol. Chem. 279, 42062–42071 [DOI] [PubMed] [Google Scholar]
- 21. Schweiger M., Schreiber R., Haemmerle G., Lass A., Fledelius C., Jacobsen P., Tornqvist H., Zechner R., Zimmermann R. (2006) J. Biol. Chem. 281, 40236–40241 [DOI] [PubMed] [Google Scholar]
- 22. Tansey J. T., Sztalryd C., Gruia-Gray J., Roush D. L., Zee J. V., Gavrilova O., Reitman M. L., Deng C. X., Li C., Kimmel A. R., Londos C. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 6494–6499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gruber A., Cornaciu I., Lass A., Schweiger M., Poeschl M., Eder C., Kumari M., Schoiswohl G., Wolinski H., Kohlwein S. D., Zechner R., Zimmermann R., Oberer M. (2010) J. Biol. Chem. 285, 12289–12298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Granneman J. G., Moore H. P., Mottillo E. P., Zhu Z. (2009) J. Biol. Chem. 284, 3049–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Garcia A., Subramanian V., Sekowski A., Bhattacharyya S., Love M. W., Brasaemle D. L. (2004) J. Biol. Chem. 279, 8409–8416 [DOI] [PubMed] [Google Scholar]
- 26. Subramanian V., Garcia A., Sekowski A., Brasaemle D. L. (2004) J. Lipid Res. 45, 1983–1991 [DOI] [PubMed] [Google Scholar]
- 27. Clifford G. M., McCormick D. K., Vernon R. G., Yeaman S. J. (1997) Biochem. Soc. Trans. 25, S672 [DOI] [PubMed] [Google Scholar]
- 28. Zhai W., Xu C., Ling Y., Liu S., Deng J., Qi Y., Londos C., Xu G. (2010) Horm. Metab. Res. 42, 247–253 [DOI] [PubMed] [Google Scholar]
- 29. Kemena C., Notredame C. (2009) Bioinformatics 25, 2455–2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Brasaemle D. L., Barber T., Kimmel A. R., Londos C. (1997) J. Biol. Chem. 272, 9378–9387 [DOI] [PubMed] [Google Scholar]
- 31. Chong B. M., Russell T. D., Schaack J., Orlicky D. J., Reigan P., Ladinsky M., McManaman J. L. (2011) J. Biol. Chem. 286, 23254–23265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hall A. M., Brunt E. M., Chen Z., Viswakarma N., Reddy J. K., Wolins N. E., Finck B. N. (2010) J. Lipid Res. 51, 554–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.