Abstract
Human glutathione synthetase (hGS) catalyzes the second ATP-dependent step in the biosynthesis of glutathione (GSH) and is negatively cooperative to the γ-glutamyl substrate. The hGS active site is composed of three highly conserved catalytic loops, notably the alanine rich A-loop. Experimental and computational investigations of the impact of mutation of Asp458 are reported, and thus the role of this A-loop residue on hGS structure, activity, negativity cooperativity and stability is defined. Several Asp458 hGS mutants (D458A, D458N, D458R) were constructed using site-directed mutagenesis and their activities determined (10, 15 and 7% of wild-type hGS, respectively). The Michaelis-Menten constant (Km) was determined for all three substrates (glycine, GAB, ATP): glycine Km increased by 30 - 115 fold, GAB Km decreased by 8 - 17 fold, and the ATP Km was unchanged. All Asp458 mutants display a change in cooperativity from negative cooperativity to non-cooperative. All mutants show similar stability as compared to wild-type hGS, as determined by differential scanning calorimetry. The findings indicate that Asp458 is essential for hGS catalysis and that it impacts the allostery of hGS.
Keywords: glutathione synthetase, glutathione, negative cooperativity
Introduction
Enzyme activity is regulated in many ways: substrate and feedback inhibition, post-translational modification and allosteric modulation. Cooperativity through allosteric activation, is a common form of enzyme/protein regulation, e.g., in hemoglobin [1], the binding of O2 to one subunit increases affinity of an adjacent subunit for dioxygen. Conversely, an enzyme may exhibit negative cooperativity (allosteric inhibition) when binding of a substrate to one active site reduces affinity for ligand binding in adjacent subunits [2]. One example of negative cooperativity is the insulin-like growth factor I of the tyrosine kinase family, which shows negative cooperativity when binding insulin [3]. Cooperativity for allosteric enzymes is typically quantified by the Hill coefficient (H): Hill coefficients of > 1 indicate positive cooperativity; H < 1 indicates negative cooperativity, and H = 1 defines a non-cooperative enzyme. Though there are many forms of regulation, negative cooperativity is rare and poorly understood [4,5].
Mammalian glutathione synthetase (rat and human GS) displays negative cooperativity for γ–glutamyl substrate: γ–glutamyl-α-aminobutyrate [6-8] or γ-glutamylcysteine [6]. Also, the Arabidopsis thaliana GS enzyme displays negative cooperativity to γ-glutamylcysteine [9]. The human GS (hGS) enzyme catalyzes formation of glutathione via ligation of glycine to γ–glutamylcysteine using ATP [10,11]. The compound γ-glutamyl-α-aminobutyrate is also a natural GS substrate, and in fact the product of its GS reaction, ophthalmic acid is found in the lens and may be a hepatic oxidative stress biomarker [12]. Homodimeric hGS is a member of the ATP-grasp superfamily of carboxylate-amine ATP-dependent ligases, which catalyze formation of peptide linkages via an acyl-phosphate intermediate [13,14]. Each hGS monomer has three conserved loops – G-loop, S-loop and A-loop – in the vicinity of the active sites of each subunit; notably, the A-loop is located near the glycine substrate and ATP (Fig. 1a).
Figure 1.
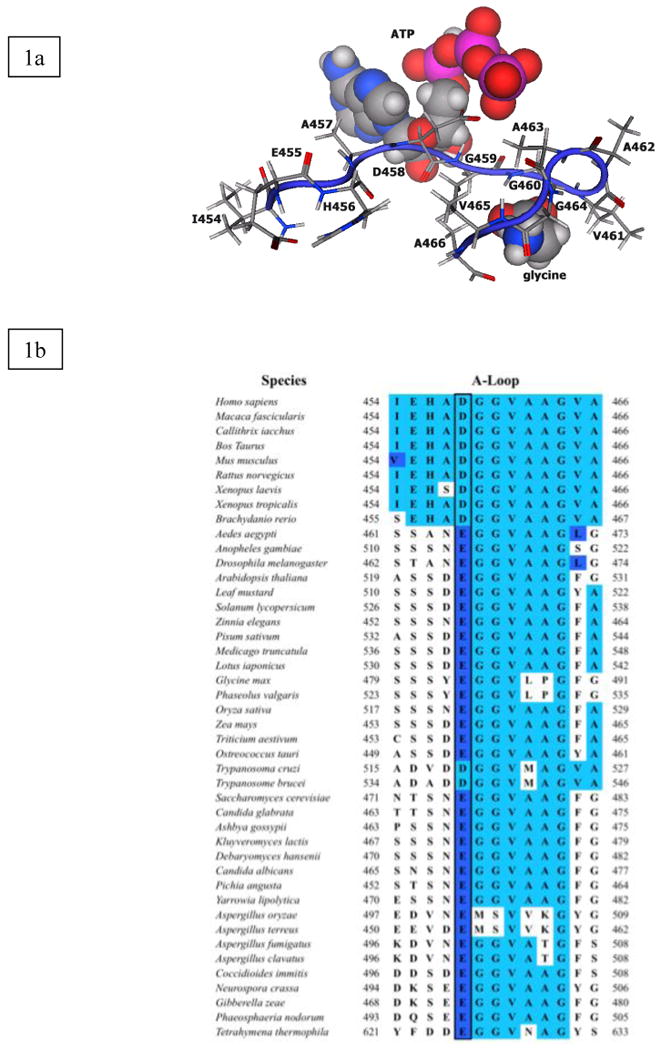
A-loop of human glutathione synthetase. (1a) Model of A-loop in reactant form with glycine and ATP. (1b) ClustalW sequence alignment of A-loop of hGS. The dark box indicates charge conserved Asp458.
Our previous hGS research suggests that the G-, A- and S-loops are essential for hGS activity [15]. A strong ionic bond between the Gly371 (G-loop) backbone and the Asp458 (A-loop) carboxyl is present in both reactant binding and product release, and facilitates active site closure [15]. In the present research, A-loop residue Asp458 was examined for its hydrogen bonding potential in inter-, intra- and ligand-loop interactions and to delineate its role in hGS catalysis.
Methods and Materials
Materials
Oligonucleotide primers for mutagenesis and sequencing were synthesized by Integrated DNA Technologies. The QuikChange™ site-directed mutagenesis kit was from Stratagene. γ-Glutamyl-α-aminobutyrate (GAB) was synthesized[16]. Isopropyl-1-thio-β-D-galactopyranoside (IPTG) was from American Bioanalytical. Other reagents were from Sigma (St. Louis, MO).
Construction of hGS Mutant Enzymes
The cDNA that encodes the wild-type human glutathione synthetase was subcloned into pET-15b expression vector (Novagen), which provides a His tag at the N-terminus [17]. Site-directed mutagenesis was performed on double-stranded plasmid (dsDNA) by PCR using the QuickChange® Site-Directed Mutagenesis Kit (Stratagene). The internal primers used for hGS mutant enzymes are shown in Table 1. All mutations were confirmed by sequencing the hGS DNA (Genewiz, Inc.).
Table 1.
Primers for site-directed mutagenesis of human glutathione synthetase
| Enzyme | DNA sequencea |
|---|---|
| D458A | 5′-CGAGCATGCAGCTGGTGGTGTGGC-3′ |
| 5′-GCCACACCACCAGCTGCATGCTCG-3′ | |
| D458R | 5′-CGAGCATGCACGTGGTGGTGTGGC-3′ |
| 5′-GCCACACCACCACGTGCATGCTCG-3′ | |
| D458N | 5′-CCATCGAGCATGCAAATGGTGGTGTGGC-3′ |
| 5′-GCCACACCACCATTTGCATGCTCGATGG-3′ |
Underlined, bold bases indicate changed nucleotide positions.
Glutathione Synthetase Growth and Purification
The overexpression and protein purification protocol used for the recombinant wild-type and mutant hGS enzymes, as previously described [17], except cell lysis occurred with pressure, (15 kPSI, One Shot Model, Constant Systems, Inc.). All purification steps were carried out at 4 °C [17]. The purity of all hGS enzymes was confirmed by SDS-PAGE.
Enzyme Assays and Kinetic Analysis
Analyses were carried out in duplicate using purified recombinant hGS as described [17,18]. In brief, the enzyme activity was measured using the pyruvate kinase (PK)/lactate dehydrogenase (LDH) coupled assay. γ-Glutamyl-α-aminobutyrate (GAB; a γ-glutamylcysteine analog) was used in place of γ-glutamylcysteine to avoid oxidation of the thiol group [7]. The reaction was initiated adding GS to a pre-incubated (37 °C, 5 min) standard reaction mixture (0.2 mL final volume) containing: 100 mM Tris-HCl pH 8.2, 50 mM KCl, 20 mM MgCl2, 5 mM phospho(enol)pyruvate, 10 mM ATP, 10 mM glycine, 10 U/mL LDH (type II rabbit muscle), 10 U/mL PK (type II rabbit muscle), 0.2 mM NADH, GAB (20 mM for specific activity assays). The disappearance of NADH was monitored continuously (340 nm for 3 min). The ΔOD/min for all hGS enzymes was calculated (3 min time course), except D458N, which did not exhibit linear disappearance of NADH throughout (first min used for calculations). One unit of activity is the amount that catalyzes 1 μmol of product per min.
Differential Scanning Calorimetry
All hGS enzymes were concentrated (∼1 mg/mL) with micro concentrators (Amicon). Samples (1 mL) were degassed (room temperature; 15 min), then loaded into the differential scanning calorimeter (Calorimetry Sciences Nano Series III) (scan rate, 1 °C/min, 1 atm).
Circular Dichroism
Each mutant was dialyzed overnight against phosphate buffer (10 mM, pH 7, 4 °C) followed by a second dialysis for approximately 6 h. Sample (500 μL) was loaded into a round quartz cuvette (0.2 cm). Measurements were carried out on an OLIS RSM 1000 with DSM CD attachment from 320 to 195 nm. The Savitsky-Golay algorithm was used to smooth data (RC filter - 13; digital filter - 15) during acquisition and analysis. OLIS GlobalWorks software was used for all data acquisition and analysis. For mutant comparison, all samples were converted to molar ellipticity, and then overlaid.
Computational Methods
Enzyme models were subjected to the molecular dynamics (MD) protocol described previously [15,17] using MOE [19]. The AMBER94 force field [20] and PEOE charges [21] were used. The X-ray structure [22] was used to initiate MD simulations. Pictures were rendered in PyMOL [23].
MD simulations with explicit water molecules were performed for wild-type and D458N mutant enzymes using GROMACS 4.0.7 [24,25]. Hydrogen atoms were added automatically with GROMACS, and each enzyme centered in a rhombic dodecahedron periodic cell surrounded by a 10 Å shell of simple point charge (SPC [26]) waters, neutralized with random Na+ and Cl- ions. The geometry was optimized using a conjugate gradient algorithm until forces acting on any atom were < 10 kJ mol-1 nm-1. For NVT MD runs, the temperature was raised from 0 to 300 K in 1 ps; a 0.5 fs time step was used for a 1 ns simulation, with coordinates saved every 0.5 ps. Long-range electrostatic interactions were evaluated using the Particle Mesh Ewald [24] approach.
Results
Sequence alignment
Glutathione synthetase is found in most eukaryotes [10,11,14]. The A-loop of hGS has thirteen, mostly hydrophobic, amino acid residues (Ile454 - Ala466). Multiple sequence alignment with ClustalW [27] (Fig. 1b) shows the A-loop is highly conserved among eukaryotes, and other portions invariant (Fig. 1b). Glu455 and Asp458 are the only charged residues in the A-loop, and Glu455 is only conserved in a few species. However, Asp458 is strictly conserved in mammals and amphibians, and is charge-conserved (Asp or Glu) in all other eukaryotes. The location of Asp458 among hydrophobic residues, and conservation of the carboxylate side chain, led to the hypothesis that Asp458 is important for loop closure and is a critical residue within the hGS A-loop.
To investigate these hypothesis, Asp458 was evaluated in detail. First, we modeled the different states (reactant, product) of wild-type hGS using MD simulations; the resulting structures were analyzed. Each residue was studied for intra-loop, inter-loop and loop-ligand interactions involving the A-loop. Next, three separate mutations of Asp458 were studied: D458A, D458N and D458R. Computational studies were performed for each mutant to predict how mutation would alter the active site relative to wild-type hGS. Finally, experiments (site-directed mutagenesis, enzyme kinetics, CD spectra, DSC analysis of melting points) were performed to substantiate and extend the modeling predictions.
Modeling
A- and G-loop (inter-loop) interactions
Analysis was performed on the lowest energy structure of hGS obtained from MD simulation with attention paid to Asp458 given its high degree of conservation (Fig. 1b). In wild-type hGS, the A- and G-loops are in close proximity and interact via hydrogen bonds (H-bonds) during the catalytic cycle (reactant and product) [15]. In reactant hGS (Fig. 2a; Supplementary Data, Fig. S1, Table S1), Ala462 has an H-bond with Gly369 (G-loop), and Asp458 has two H-bonds with G-loop residues (Gly371, Asn373). In product hGS, H-bonds between Ala462 and Gly369 are lost and one new H-bond is gained between Asp458 and Gly371, one more than in reactant form. Of the two A-loop residues that H-bond with the G-loop, only H-bonds involved with Asp458 are conserved in reactant and product hGS, and therefore these interactions serve to “latch” the conformation of the G- and A-loop during catalysis.
Figure 2.
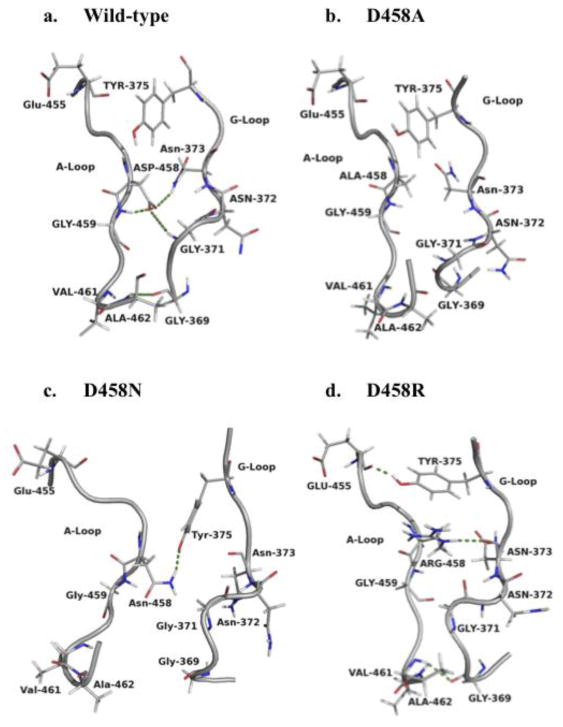
A- and G-loop interactions in reactant hGS. H-bonds are shown with dashes. Product form in Fig. S1, Supplementary Data.
When Asp458 is mutated to alanine, asparagine, or arginine, most of the above H-bonds are lost in both reactant and product form. No new H-bonds are gained in D458A to compensate for the loss of H-bonds present in wild-type hGS. However, in reactant and product, D458N gains one H-bond to the Tyr375 side chain. Interestingly, D458R is the only mutant that retains Asp458-Asn373 interaction, and reactant D458R gains two different new H-bonds (Tyr375-Glu455; Gly369-Val461). The network of inter-loop interactions is altered, and mostly missing for Asp458 mutant hGS enzymes. As a result of losing wild-type H-bonding between the A- and G-loops, both the shape of the G- and A-loops and the active site is altered upon mutation of Asp458.
A-loop (intra-loop) interactions
The A-loop has intra-loop interactions as well, i.e., interactions with itself (Fig. 3a; Table S2.1 and S2.2, Supplementary Data). Glu455 has one H-bond with itself, three with His456, and one with Ala457 that are conserved in reactant and product wild-type hGS. Gly460 H-bonds with Ala463 in the reactant and changes to a Gly460-Ala462 in product hGS. Notably, Asp458 side chain oxygens of the carboxylate form H-bonds with the backbone nitrogen of Gly459, in both reactant (1.91 and 2.64 Å) and product (1.76 Å) hGS. In summary, the A-loop of wild-type hGS, notably Asp458, has the ability to form H-bonds within the A-loop. In total, there are eight H-bonds in reactant and nine in product form, which creates a large network of intra-loop interactions that shape the active site.
Figure 3.
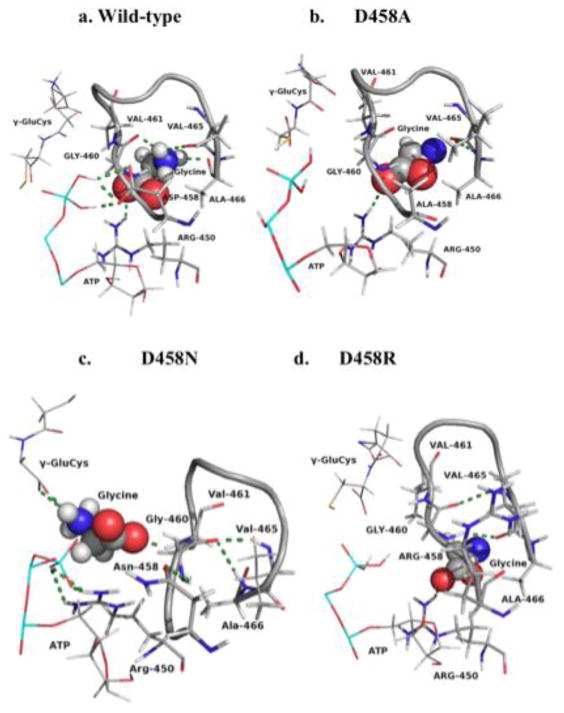
Wild-type and Asp458 mutant enzymes intra-loop and loop-ligand interactions in reactant form. H-bonds are shown in dashes. Partial ATP and γ-glu-cys used in some views. Full intra-loop figures in Fig. S2, Supplementary Data. Full figures with ligands in Fig. S3, Supplementary Data.
When Asp458 is mutated to alanine or arginine, some intra-loop H-bonds remain, or new ones are made; however, all H-bonds with Asp458 are lost. All but two intra-loop H-bonds of wild-type are lost in D458R (reactant), but it gains two new A-loop H-bonds between Val465 and Gly460. In the D458R product form, three bonds from wild-type are retained, and one new bond with Arg458-His456 is gained. Interestingly, reactant D458N retains one H-bond between the 458 side-chain and the Gly459 backbone, and gains five new H-bonds in reactant and three new H-bonds in product. Mutations of the Asp458 carboxylate alter or abolish the H-bonds within the A-loop. Also, the loss of intra-loop interactions, in conjunction with changes to the G- and A-loop inter-loop interactions (discussed above), alter the backbone shape of the A- and G-loops and, thus, of the active site.
A-loop and ligand (loop-ligand) interactions
The γ-glutamylcysteine (γ-glu-cys) substrate is near Val461 in the A-loop (2.4 Å); however, γ-glu-cys does not directly interact with the A-loop in wild-type hGS (Fig. 3). The A-loop interacts with substrates ATP via three H-bonds [15] and with glycine through three A-loop residues (Gly460, Val465, Ala466) (Table S3, Supplementary Data). Additionally, glycine substrate interacts with a beta sheet residue (Arg450). The four residues that interact with glycine secure the substrate on all sides. Arg450 interacts with the carboxylate of the glycine substrate, while the three A-loop residues, in the glycine pocket, orient glycine's amino group in the optimum position for nucleophilic attack on γ-glu-cys.
The Asp458 mutants lose all H-bonds with ATP and glycine, except for the H-bond between glycine and Arg450, which is seen in D458A and D458R, but not D458N. Though all direct bonds between the A-loop and ATP are lost, ATP has H-bonds with other hGS residues [17]. The orientation of the glycine substrate within the A-loop of wild-type is similar in D458A, but removal of the carboxylate side chain prohibits side chain H-bonds with glycine. Glycine is completely excluded from the A-loop in D458R and D458N by creation of new intra-loop interactions (Fig. 3). In D458N, glycine is repositioned and gains new H-bonds, one with Val461 and two with γ-glu-cys. This new orientation of glycine substrate allows for direct contact with γ-glu-cys, which is not seen in wild-type or any other Asp458 mutant and provides proximity and stabilization of glycine compared to D458A and D458R. Loop-ligand interactions, especially those in which glycine substrate is secured, are a direct result of the H-bond network engendered by the Asp458 carboxylate. Thus, mutation of Asp458 disrupts, or rearranges, many loop-ligand interactions necessary for glycine binding and hGS catalysis.
In Vitro Experiments
Enzyme activity, stability and secondary structure
Experimentally, mutation of Asp458 to a small, uncharged alanine (D458A) leads to a drastic (90%) decrease in activity. Mutation to asparagine (D458N), which is similar in terms of side chain size but different in electrostatic properties, loses 85% activity. Introduction of bulkier and oppositely charged arginine (D458R) also decreases activity (93%). All Asp458 mutant enzymes lose activity as a result of mutating the carboxyl side chain of aspartate, supporting the hypothesis derived from ClustalW analysis and MD simulations that the acidic carboxyl group plays an essential role in hGS catalysis.
Enzyme stability (unfolding temperature, Tm) was determined using differential scanning calorimetry (DSC). All mutant and wild-type hGS have essentially the same Tm (Table 2). Circular dichroism (CD) spectra (Fig. S4, Supplementary Data) for all hGS enzymes are similar, showing that their secondary structure (Fig. S5, Supplementary Data) is similar to wild-type hGS.
Table 2.
Activity, turnover number (kcat) and thermal stability (Tm) for wild-type and mutant hGSa
| Enzyme | kcat (s-1)a | % activity | Tm (°C) |
|---|---|---|---|
| wild-type | 15.68 ± 0.07 | 100 | 60.7 ± 0.3 |
| D458A | 1.60 ± 0.13 | 10 | 60.2 ± 0.7 |
| D458N | 2.31 ± 0.43 | 15 | 60.7 ± 0.4 |
| D458R | 1.17 ± 0.11 | 7 | 60.6 ± 0.0 |
Results are averages of at least two enzyme preparations, each assayed in duplicate. Control rates (minus GAB) were subtracted to obtain kcat values. Lower limit of detection is 0.008 s-1.
Kinetics
The Michaelis constants (Km) of Asp458 mutant hGS (D458A, D458N and D458R) were experimentally determined for the glycine, ATP and GAB substrates (Table 3). ATP affinity was similar between all Asp458 mutant and wild-type hGS (∼25 μM). Glycine affinity was reduced dramatically (67-, 30- and 115-fold for D458A, D458N and D458R, respectively). Initial Km determinations for GAB show a decrease for D458A and D458R (8-fold) compared to wild-type hGS, while D458N has a larger decrease in Km (17-fold decrease). All Asp458 enzymes have a Hill coefficient (H) of ∼1, corresponding to a loss of negative cooperativity compared to wild-type hGS (H ∼ 0.67). All Asp458 mutants thus have decreased affinity for glycine, increased affinity for GAB and loss of cooperativity.
Table 3.
Experimental kinetic parameters for wild-type and mutant hGS.
| Enzymea | Glycine Km (mM) | GAB Km (mM) | Hill Coeff., (H) |
|---|---|---|---|
| wild-type | 0.58 ± 0.07 | 1.41 ± 0.05 | 0.68 ± 0.01 |
| D458A | 35 ± 0 | 0.25 ± 0.02 | 1.00 ± 0.12 |
| D458N | 16 ± 3 | 0.12 ± 0.04 | 1.09 ± 0.05 |
| D458R | 51 ± 12 | 0.29 ± 0.04 | 0.99 ± 0.10 |
GAB and glycine Km were determined using saturating conditions of other substrates. Results are from duplicate preparations.
Temporal analysis
When D458N mutant hGS was re-assayed after two weeks, activity increased (∼20%) compared to initial activity. The unusual increase in activity was verified by performing the kinetics in triplicate on three separate enzyme preparations. A time course of activity with other Asp458 mutants - D458A and D458R - shows no changes in activity as a function of time. D458N activity doubled in activity by 6 months (Fig. S6, Supplementary Data), but by 12 months, activity returned to initial levels.
Michaelis-Menten constants were again determined when D458N was at peak activity. The Km values for D458A and D458R did not change for any substrates from initial determination. However, the glycine Km for D458N is 0.77 mM (Fig. S6, Supplementary Data), a 20-fold decrease from the initial determination (Table 3) and now is similar to wild-type hGS. There was no change measured for either GAB Km or Hill coefficient for D458N. Moreover, secondary structure was unchanged as determined by CD spectra, suggesting that localize active site perturbations improved glycine affinity, thus increasing D458N activity.
Discussion
The high level of charge-conservation of Asp458 in human glutathione synthetase (hGS) and our previous MD simulations [15] led to the hypothesis that this residue is important in hGS catalysis. Several significant conclusions relevant to the impact of this A-loop residue vis-à-vis intra-loop, inter-loop, and loop-ligand relationships and their impact on enzyme catalysis are discussed here.
The G- and A-loop of wild-type hGS have several inter-loop interactions in both reactant and product forms (Fig. 2) of hGS, most notably from the Asp458 side chain. While D458N and D458R gain different inter-loop bonding interactions involving Asp458, the shape of these loops, especially the G-loop, is distorted (Figs. 2 and 4). Mutation to alanine (D458A) only causes slight conformational changes to the backbone of the G- and A-loop; however, the Ala458 side chain is unable to H-bond to the G-loop. The hGS mutants with these changes in the inter-loop interactions have little activity, suggesting that in wild-type hGS, A- and G-inter-loop interactions likely bring substrates together and shield the catalytic intermediates from hydrolysis. Thus, inter-loop interactions near Asp458 are critical for active site conformation, providing a latching mechanism for loop closure during catalysis.
Figure 4.
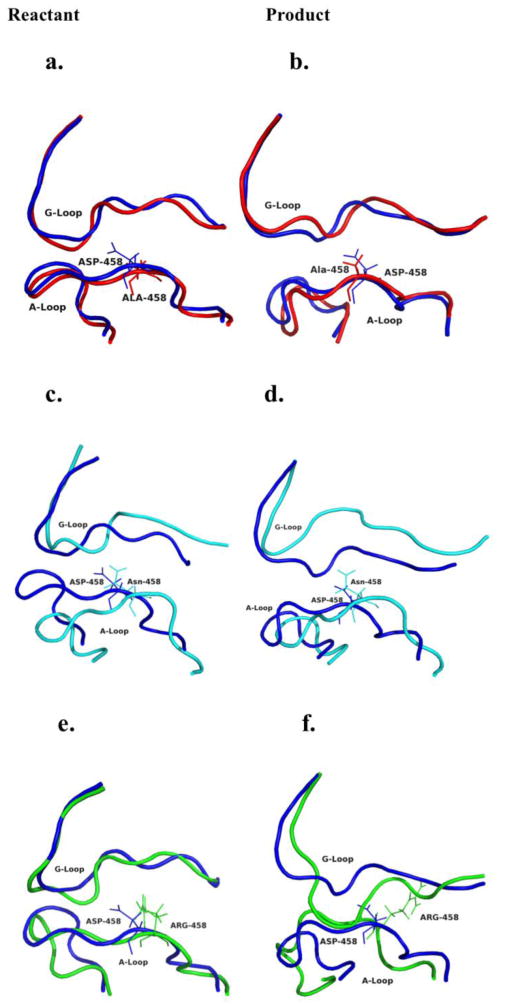
Modeled backbone comparison of A- and G-loop wild-type with each Asp458 mutant hGS enzymes. All wild-type are shown in blue. D458A (red) (a) reactant and (b) product; D458N (cyan) (c) reactant and (d) product; D458R (green) (e) reactant and (f) product.
The A-loop has a number of intra-A-loop H-bonds (e.g., Glu455-Asp458) (Fig. 3, Table S2, Fig. 2S). Mutant hGS lose many of their H-bonds (reactant and product), while gaining new H-bonds. For mutants studied, substrate (especially glycine) locations are altered. For both D458N and D458R, glycine substrate is partly excluded from the A-loop. Interestingly, for the D458N, glycine is now H-bonded to γ-glu-cys and Arg450 is bonded to ATP, not glycine. “Improved” substrate location may explain why D458N activity is somewhat greater than D458R. All mutants have greatly increased Km values for glycine, Table 3, which suggests that A-loop conformation, stabilized by Asp458, is important for binding and orienting glycine for its attack on the putative acyl-phosphate intermediate [13,14].
In wild-type hGS, the A-loop has direct interactions with glycine and ATP. Upon mutation, all Asp458 mutants lose their interactions with ATP. However, ATP has other interactions, therefore, no change in ATP Km is measured. The glycine Km shows that lack of loop-ligand interactions are responsible for decreased glycine affinity for all Asp458 mutants as most loop-ligand interactions are lost. Though the A-loop in wild-type hGS has no direct interactions with γ-glu-cys, the A-loop is repositioned in D458N and glycine is stabilized by interaction with γ-glu-cys (Fig. 3). Moreover, Gly371 [18], which has a strong H-bond to Asp458 in wild-type hGS, shows increased GAB affinity when mutated to valine, just as Asp458 does when the carboxylate is mutated. Thus, loop-ligand interactions in wild-type hGS are necessary for stabilizing the glycine conformation and promoting substrate binding.
Asp458 mutant hGS enzymes lost negative cooperativity for binding of GAB (H ∼ 1). While this is not fully understood, MD simulations suggest that single point mutations at residue 458 induce local changes in A- and G-loop structure that impact substrate binding. Further examples of small loop changes having a great impact on enzyme activity are seen with D458N, whereby hGS activity increased over time due to an increase in glycine affinity. Such a dramatic change in substrate affinity and activity is due to small changes near the active site, not global changes to the secondary structure of the enzyme (CD, MD analyses and Fig. S5). Thus, we conclude that small G- and A-loop changes impact hGS allostery, providing important new insight into the allosteric pathway of this negatively cooperative enzyme. Taken together, the present research indicates that Asp458 is necessary for maintaining active site shape, substrate binding, and cooperativity of hGS.
Future studies are needed to ascertain the role of Arg450 in wild-type hGS along with its proposed role in glycine binding. The change in cooperativity for Asp458 mutant enzymes suggests that active sites separated by a great distance can communicate, but further studies on hGS are needed.
Supplementary Material
Research highlights.
Mutations of Asp458 to alanine, asparagine and arginine have a large impact on human glutathione synthetase (hGS) activity.
The affinities of GAB and glycine substrates are different in Asp458 mutant hGS.
Mutations at Asp458 impact the allosteric pathway of hGS causing a loss of cooperativity.
Asp458Asn (D458N) exhibits an increase in activity and glycine affinity after prolonged storage.
Asp458 mutant enzymes show no loss in thermal stability and secondary structure is unaffected.
Acknowledgments
The authors acknowledge the Chemical Computing Group, Inc. for providing the MOE package; Mark Britt and Richard Sheardy (TWU) for assistance; Margarita DeJesus, Amy Graves, Sara Hernandez and Bisesh Shrestha (TWU) for technical assistance.
Funding: M.E.A. acknowledges the NIH (R15GM086833), NSF CHE-0820845, a Welch Foundation Grant to Texas Woman's University (TWU), and by a TWU Faculty REP Award. T.R.C acknowledges a UNT faculty research grant. A.S.C. acknowledges the North Texas Transition Program in Biomedical Science (5R25GM058397-08; PI: McIntire, S. A. (TWU)).
Abbreviations
- CD
circular dichroism
- DSC
differential scanning calorimetry
- GAB
γ-glutamyl-α-aminobutyrate
- hGS
human glutathione synthetase
- IPTG
isopropyl-1-thio-β-D-galactopyranoside
- MCAC
metal chelate affinity chromatography
- MOE
molecular operating environment
- PEOE
Partial Equalization of Orbital Electronegativity
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Adair GS. The hemoglobin system. VI. The oxygen dissociation curve of hemoglobin. J Biol Chem. 1925;63:529–545. [Google Scholar]
- 2.Ferrell JE., Jr Q&A: Cooperativity James E Ferrell, Jr. Journal of Biology. 2009;8:1–8. doi: 10.1186/jbiol157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Surinya KH, Forbes BE, Occhiodoro F, Booker GW, Francis GL, Siddle K, Wallace JC, Cosgrove LJ. An investigation of the ligand binding properties and negative cooperativity of soluble insulin-like growth factor receptors. J Biol Chem. 2008;283:5355–5363. doi: 10.1074/jbc.M707054200. [DOI] [PubMed] [Google Scholar]
- 4.Levitzki A, Koshland D. Negative cooperativity in regulatory enzymes. Proc Natl Acad Sci U S A. 1969;62:1121–1128. doi: 10.1073/pnas.62.4.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koshland DE, Hamadani K. Proteomics and models for enzyme cooperativity. J Biol Chem. 2002;277:46841–46844. doi: 10.1074/jbc.R200014200. [DOI] [PubMed] [Google Scholar]
- 6.Luo JL, Huang CS, Babaoglu K, Anderson ME. Novel Kinetics of Mammalian Glutathione Synthetase: Characterization of [gamma]-Glutamyl Substrate Cooperative Binding. Biochem Biophys Res Commun. 2000;275:577–581. doi: 10.1006/bbrc.2000.3337. [DOI] [PubMed] [Google Scholar]
- 7.Oppenheimer L, Wellner VP, Griffith OW, Meister A. Glutathione synthetase Purification from rat kidney and mapping of the substrate binding sites. J Biol Chem. 1979;254:5184–5189. [PubMed] [Google Scholar]
- 8.Njalsson R, Norgren S, Larsson A, Huang CS, Anderson ME, Luo JL. Cooperative Binding of [gamma]-Glutamyl Substrate to Human Glutathione Synthetase. Biochem Biophys Res Commun. 2001;289:80–84. doi: 10.1006/bbrc.2001.5961. [DOI] [PubMed] [Google Scholar]
- 9.Jez JM, Cahoon RE. Kinetic mechanism of glutathione synthetase from Arabidopsis thaliana. J Biol Chem. 2004;279:42726–42731. doi: 10.1074/jbc.M407961200. [DOI] [PubMed] [Google Scholar]
- 10.Meister A, Anderson ME. Glutathione. Annu Rev Biochem. 1983;52:711–760. doi: 10.1146/annurev.bi.52.070183.003431. [DOI] [PubMed] [Google Scholar]
- 11.Anderson ME. Glutathione: an overview of biosynthesis and modulation. Chem Biol Interact. 1998;111-112:1–14. doi: 10.1016/s0009-2797(97)00146-4. [DOI] [PubMed] [Google Scholar]
- 12.Soga T, Baran R, Suematsu M, Ueno Y, Ikeda S, Sakurakawa T, Kakazu Y, Ishikawa T, Robert M, Nishioka T. Differential metabolomics reveals ophthalmic acid as an oxidative stress biomarker indicating hepatic glutathione consumption. J Biol Chem. 2006;281:16768–16776. doi: 10.1074/jbc.M601876200. [DOI] [PubMed] [Google Scholar]
- 13.Galperin MY, Koonin EV. A diverse superfamily of enzymes with ATP-dependent carboxylate—amine/thiol ligase activity. Protein Science. 1997;6:2639–2643. doi: 10.1002/pro.5560061218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meister A. Glutathione synthesis. In: Boyer PD, editor. The Enzymes. Third. Academic Press; New York: 1974. pp. 671–697. [Google Scholar]
- 15.Dinescu A, Anderson ME, Cundari TR. Catalytic loop motion in human glutathione synthetase: A molecular modeling approach. Biochem Biophys Res Commun. 2007;353:450–456. doi: 10.1016/j.bbrc.2006.12.050. [DOI] [PubMed] [Google Scholar]
- 16.Anderson ME, Meister A. Preparation of gamma-glutamyl amino acids by chemical and enzymatic methods. Methods Enzymol. 1985;113:555–564. doi: 10.1016/s0076-6879(85)13074-0. [DOI] [PubMed] [Google Scholar]
- 17.Dinescu A, Cundari TR, Bhansali VS, Luo JL, Anderson ME. Function of Conserved Residues of Human Glutathione Synthetase. J Biol Chem. 2004;279:22412–22421. doi: 10.1074/jbc.M401334200. [DOI] [PubMed] [Google Scholar]
- 18.Dinescu A, Brown TR, Barelier S, Cundari TR, Anderson ME. The role of the glycine triad in human glutathione synthetase. Biochem Biophys Res Commun. 2010;400:511–516. doi: 10.1016/j.bbrc.2010.08.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chemical Computing Group, Inc. Montreal, Quebec, Canada, Molecular Operating Environment (MOE), 2003.02. 2003. [Google Scholar]
- 20.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J Am Chem Soc. 1995;117:5179–5197. [Google Scholar]
- 21.Gasteiger J, Marsili M. Iterative partial equalization of orbital electronegativity--a rapid access to atomic charges. Tetrahedron. 1980;36:3219–3228. [Google Scholar]
- 22.Polekhina G, Board PG, Gali RR, Rossjohn J, Parker MW. Molecular basis of glutathione synthetase deficiency and a rare gene permutation event. EMBO J. 1999;18:3204–3213. doi: 10.1093/emboj/18.12.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schrodinger L. The PyMOL Molecular Graphics System, 1.3. [Google Scholar]
- 24.Hess B, Kutzner C, Van Der Spoel D, Lindahl E. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. Journal of chemical theory and computation. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 25.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC. GROMACS: fast, flexible, and free. Journal of computational chemistry. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 26.Berendsen H, Postma J, Van Gunsteren W, Hermans J. Interaction models for water in relation to protein hydration. Intermolecular forces. 1981;331:331–338. [Google Scholar]
- 27.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


