Abstract
To investigate HIV-1 molecular epidemiology in Singapore, we sequenced portions of three regions of the HIV-1 genome (protease HXB2: 2163 to 2620, gp120 HXB2: 6904 to 7628, and gp41 HXB2: 7817 to 8264) from 212 plasma samples collected between February 2008 and August 2009. From these samples, 109 (51.4%) generated interpretable data in all regions. Sixty-one (56.0%) were identified as CRF01_AE, 26 (23.9%) as subtype B and 14 (12.8%) as possible novel recombinant forms. The main novel recombinant pattern, detected in 13 sequences, had subtype B in protease and gp41 and CRF01_AE in gp120. There was intermixing of subtypes within transmission risk groups. However, 85% of subjects infected with the novel recombinant forms self-identified as men who have sex with men or bisexuals compared with only 41% of individuals infected with CRF01_AE and 62% infected with subtype B (p = 0.001).
HIV-1 group M is subdivided into nine subtypes.1 Recombination between these subtypes has resulted in circulating recombinant forms (CRFs). Forty-eight intersubtype CRFs have been described to date.2 While this genetic diversity poses challenges for the prevention, diagnosis, and treatment of the disease, it also enables molecular epidemiology studies.1,3
In Singapore, HIV-1 subtyping of patient samples collected in 1996 revealed segregation of subtypes by the main transmission risk groups.4 Of men who have sex with men (MSM), 95% were infected with subtype B and 88% of heterosexual infections were with CRF01_AE. Among bisexuals, subtype B and CRF01_AE accounted for half of the infections each. A pattern of segregation by transmission risk group was also seen in the early epidemic in Thailand, with subtype B among injecting drug users (IDUs) and CRF01_AE among those heterosexually exposed.5 Subsequent studies from Thailand documented intermixing of these two main subtypes among the transmission risk groups and the increasing proportions of CRF01_AE infections.6 This was followed by the emergence of CRF01_AE/B CRFs in Thailand, mainly among IDUs and MSM.7 In Malaysia, CRF01_AE/B CRFs have also recently been identified among IDUs.8 In Singapore, a study of 60 recent seroconverters revealed subtype intermixing among transmission risk groups. However, no novel recombinant forms were detected.9 This study aimed to update the HIV-1 molecular epidemiology and to determine the presence of intersubtype recombinants in Singapore. The three genomic regions chosen for sequencing would allow detection of all subtypes and CRFs in Southeast Asia identified to date.
From February 2008 to August 2009, this cross-sectional study enrolled treatment-naive HIV-1 patients from the Singapore Communicable Disease Centre (CDC) outpatient clinic, the national HIV treatment reference center. Blood drawn for HIV-1 genotyping was centrifuged to obtain plasma samples, which were stored at −80°C. Demographic information (age, gender, and ethnicity) and transmission risk factor data were obtained from chart review. The National Healthcare Group ethics committee approved this study and written, informed consent was obtained from all patients.
Viral RNA was extracted from all plasma samples using the QIAmp Viral RNA Mini Kit (Qiagen, Valencia, CA) according to the manufacturer's protocol. The eluted RNA was amplified by reverse transcriptase polymerase chain reaction (RT-PCR) and additional nested PCR in two separate reactions to sequence portions of protease (HXB2 2163 to 2620), gp120 (6904 to 7628), and gp41 (7817 to 8264).10,11 Amplified products were purified with ExoSAP-IT (USB, Cleveland, OH) prior to Sanger sequencing using the Applied Biosystems 3730xl DNA Analyser (Life Technologies Corporation, Carlsbad, CA). Subtypes were assigned using the NCBI genotyping database and confirmed with phylogenetic analysis using reference sequences from the Los Alamos HIV Sequence Database.2,12 Phylogenetic analysis was performed using MrBayes3.2 and parameter convergence was determined by Tracer1.5.13,14 Breakpoint analysis was performed using bootscanning in RDP3.15 Subtype distribution was analyzed by age, gender, ethnicity, and transmission risk factor. Fisher's exact test was used to compare gender, ethnicity, and transmission risk grouping. The Wilcoxon rank-sum test was used to compare median age.
Two hundred and twelve patients participated in the study. The median age was 37 years (range: 18–82) with 192 (90.6%) males. One hundred and seventy patients (80.2%) were Chinese, 18 (8.5%) Malay, four (1.9%) Indian, and 20 (9.4%) were from other ethnic groups. Ninety-seven (45.8%) patients were heterosexual, 86 (40.6%) homosexual, and 19 (9.0%) bisexual. Six (2.8%) reported both IDU and heterosexual exposure and one patient reported only IDU (0.5%). One (0.5%) patient reported occupational risk and two (0.9%) denied any risk factors.
Samples from 109 (51.4%) patients generated interpretable sequence data in all three regions. Similar to previous studies, the main circulating subtypes continued to be CRF01_AE (n = 61, 56.0%) and subtype B (n = 26, 23.9%).4,9 The other strains detected were CRF02_AG (n = 1), CRF07_BC (n = 1), CRF15_01B (n = 2), CRF33_01B (n = 3), and CRF34_01B (n = 1). Fourteen (12.8%) of the 109 samples did not follow the pattern of any reported subtypes or CRFs, suggesting possible novel recombinant forms.
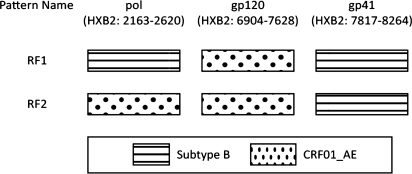
NCBI subtyping and phylogenetic analysis of the 14 novel recombinant forms revealed two possible patterns of intersubtype recombination (Fig. 1). Thirteen samples clustered with subtype B in the protease and gp41 regions and CRF01_AE in the gp120 region (Supplementary Fig. S1; Supplementary Data are available online at www.libertonline.com/aid). This pattern was labeled RF1. A second pattern was seen in one sample (RF2). RF2 clustered with CRF01_AE in the protease and gp120 regions and subtype B in the gp41 region. No breakpoints were detected in the regions analyzed.
FIG. 1.
Deduced pattern of recombination identified by NCBI genotyping and phylogenetic analysis of the gag–pol, gp120, and gp41 regions. No breakpoints were detected in the sequenced regions.
There was no significant difference in the age, gender, or ethnicity of patients between the main subtypes detected. There was intermixing of subtypes in the transmission risk groups (Table 1). This result was similar to the findings of the study involving 60 recent seroconverters.9 The transmission risk factor distribution comparing CRF01_AE, subtype B, and RF1 infection differed significantly (p = 0.001), with a higher proportion of RF1-infected patients compared to CRF01_AE or subtype B patients reporting MSM and bisexual risk (85% vs. 41% vs. 62%). The intermixing of subtypes within transmission risk groups and the higher prevalence of novel recombinants in high-risk groups mirror the developments observed in neighboring countries.7,8
Table 1.
Demographic Variables and Risk Factors by Infecting Subtype
| CRF01_AE (n = 61) | B (n = 26) | RF1 (n = 13) | |
|---|---|---|---|
| Age (median) (IQR) | 37 (29–47) | 30 (27–38) | 40 (30–45) |
| Gender | |||
| Males (%) | 51 (85) | 24 (92) | 12 (92) |
| Females (%) | 9 (15) | 2 (8) | 1 (8) |
| Ethnicity | |||
| Chinese (%) | 52 (85) | 20 (77) | 10 (77) |
| Others (%) | 9 (15) | 6 (23) | 3 (23) |
| Risk factor | |||
| Heterosexual (%) | 35 (57) | 8 (31) | 1 (8) |
| Homosexual (%) | 23 (38) | 13 (50) | 7 (54) |
| Bisexual (%) | 2 (3) | 3 (12) | 4 (31) |
| Others (%) | 1 (2) | 2 (8) | 1 (8) |
The chi-square test for comparing risk factor by different subtypes was significant with a p = 0.001. The only other significant test was for the comparison of median age between CRF01_AE and B where p = 0.017 (Wilcoxon rank-sum). One person in the CRF01_AE group was missing gender information.
Our study demonstrates the dynamic molecular epidemiology of HIV-1 in Singapore. These data could be used to inform public health measures and to monitor their effectiveness. Detection of circulating recombinant forms across borders may also facilitate the identification of trans-national networks of transmission.
Supplementary Material
Acknowledgments
A Singapore National Medical Research Training Fellowship grant provided salary support for O.T. Ng. We acknowledge the physicians, staff, and patients of the outpatient service at the Communicable Disease Centre, Singapore who made this study possible. Additional support was provided by the Division of Intramural Research, NIAID, NIH. The sequences analyzed in this study have been deposited in GenBank under accession numbers HQ538893 to HQ539411. Clinical chart review was conducted mainly by Dr. Ohnma Papa Seinn.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Taylor BS. Sobieszczyk ME. McCutchan FE. Hammer SM. The challenge of HIV-1 subtype diversity. N Engl J Med. 2008;358:1590–1602. doi: 10.1056/NEJMra0706737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.HIV Circulating Recombinant Forms (CRFs) Sep 29, 2010. [Dec 21;2010 ]. http://www.hiv.lanl.gov/content/sequence/HIV/CRFs/CRFs.html http://www.hiv.lanl.gov/content/sequence/HIV/CRFs/CRFs.html
- 3.Tovanabutra S. Watanaveeradej V. Viputtikul K, et al. A new circulating recombinant form, CRF15_01B, reinforces the linkage between IDU and heterosexual epidemics in Thailand. AIDS Res Hum Retroviruses. 2003;19:561–567. doi: 10.1089/088922203322230923. [DOI] [PubMed] [Google Scholar]
- 4.Kalish ML. Korber BT. Pillai S, et al. The sequential introduction of HIV-1 subtype B and CRF01AE in Singapore by sexual transmission: Accelerated V3 region evolution in a subpopulation of Asian CRF01 viruses. Virology. 2002;304:311–329. doi: 10.1006/viro.2002.1691. [DOI] [PubMed] [Google Scholar]
- 5.Ou CY. Takebe Y. Weniger BG, et al. Independent introduction of two major HIV-1 genotypes into distinct high-risk populations in Thailand. Lancet. 1993;341:1171–1174. doi: 10.1016/0140-6736(93)91001-3. [DOI] [PubMed] [Google Scholar]
- 6.Subbarao S. Vanichseni S. Hu DJ, et al. Genetic characterization of incident HIV type 1 subtype E and B strains from a prospective cohort of injecting drug users in Bangkok, Thailand. AIDS Res Hum Retroviruses. 2000;16:699–707. doi: 10.1089/088922200308693. [DOI] [PubMed] [Google Scholar]
- 7.Tovanabutra S. Kijak GH. Beyrer C, et al. Identification of CRF34_01B, a second circulating recombinant form unrelated to and more complex than CRF15_01B, among injecting drug users in northern Thailand. AIDS Res Hum Retroviruses. 2007;23:829–833. doi: 10.1089/aid.2006.0300. [DOI] [PubMed] [Google Scholar]
- 8.Li Y. Tee KK. Liao H, et al. Identification of a novel second-generation circulating recombinant form (CRF48_01B) in Malaysia: A descendant of the previously identified CRF33_01B. J Acquir Immune Defic Syndr. 2010;54:129–136. doi: 10.1097/QAI.0b013e3181d82ce5. [DOI] [PubMed] [Google Scholar]
- 9.Lee CC. Sun YJ. Barkham T. Leo YS. Primary drug resistance and transmission analysis of HIV-1 in acute and recent drug-naive seroconverters in Singapore. HIV Med. 2009;10:370–377. doi: 10.1111/j.1468-1293.2009.00698.x. [DOI] [PubMed] [Google Scholar]
- 10.Laeyendecker O. Zhang GW. Quinn TC, et al. Molecular epidemiology of HIV-1 subtypes in southern China. J Acquir Immune Defic Syndr. 2005;38:356–362. [PubMed] [Google Scholar]
- 11.Gray RR. Tatem AJ. Lamers S, et al. Spatial phylodynamics of HIV-1 epidemic emergence in east Africa. AIDS. 2009;23:F9–F17. doi: 10.1097/QAD.0b013e32832faf61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.NCBI Genotyping. [Dec 21;2010 ]. http://www.ncbi.nlm.nih.gov/projects/genotyping/formpage.cgi http://www.ncbi.nlm.nih.gov/projects/genotyping/formpage.cgi
- 13.Ronquist F. Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 14.Martin DP. Posada D. Crandall KA. Williamson C. A modified bootscan algorithm for automated identification of recombinant sequences and recombination breakpoints. AIDS Res Hum Retroviruses. 2005;21:98–102. doi: 10.1089/aid.2005.21.98. [DOI] [PubMed] [Google Scholar]
- 15.Rambaut A. Drummond AJ. Tracer v1.4. 2007. [Dec 21;2010 ]. http://beast.bio.ed.ac.uk/Tracer http://beast.bio.ed.ac.uk/Tracer
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.