Abstract
Background
Foodborne antimicrobial-resistant Escherichia coli may colonize and cause infections in humans, but definitive proof is elusive and supportive evidence is limited. Methods: Approximately contemporaneous antimicrobial-resistant (n = 181) and antimicrobial-susceptible (n = 159) E. coli isolates from retail meats and from human stool and clinical specimens from a single rural U.S. community were compared for polymerase chain reaction (PCR)-defined phylogenetic group (A, B1, B2, or D) and virulence genotype. Meat and human isolates from the same phylogenetic group with similar virulence profiles underwent sequential two-locus sequence analysis, random amplified polymorphic DNA (RAPD) analysis, and pulsed-field gel electrophoresis (PFGE) analysis. Results: According to phylogenetic distribution, resistant stool isolates were more similar to resistant meat isolates than to susceptible stool isolates. Overall, 19% of meat isolates satisfied molecular criteria for extraintestinal pathogenic E. coli (ExPEC). Nine sequence groups included meat and human isolates, and 17 of these 64 isolates demonstrated >80% RAPD profile similarity to an isolate from the alternate source group (meat vs. human). However, PFGE profiles of the 17 isolates were unique, excepting two stool isolates from the same household. Conclusion: Nearly 20% of meat-source resistant E. coli represented ExPEC. The observed molecular similarity of certain meat and human-source E. coli isolates, including antimicrobial-resistant and potentially pathogenic strains, supports possible foodborne transmission.
Introduction
Escherichia coli is a major cause of extra-intestinal infections in humans, contributing greatly to morbidity, mortality, and increased health care costs (Russo and Johnson, 2000). Resistance to commonly used antimicrobial agents, such as trimethoprim-sulfamethoxazole (TMP-SMZ), fluoroquinolones, and extended-spectrum cephalosporins (ESCs), is increasingly prevalent (Bacheller and Bernstein, 1997; Allen et al., 1999) The food supply, particularly meat, is suspected of contributing to this problem by providing a source of antimicrobial-resistant organisms to humans (Krumperman, 1983; Chulasiri and Suthienkul, 1989; Bazile-Pham-Khac et al., 1996; Johnson et al., 2003b). However, little is known regarding to what extent the drug-resistant E. coli strains that colonize healthy humans—especially those strains that go on to cause extraintestinal disease—resemble those found in retail meats. Resemblance would be expected if food-source transmission to human is common and poses a significant health risk (Johnson et al., 2006b).
Food-source E. coli could conceivably impact human health in two ways. First, the bacteria could colonize the host and subsequently cause extraintestinal disease themselves. This would be more likely if they possessed the specialized virulence factors (VF) characteristic of extraintestinal pathogenic E. coli (ExPEC), the E. coli strains that cause most extraintestinal infections in humans (Johnson et al., 2003a). If the infecting food-source strain exhibited antimicrobial resistance, the result would be a drug-resistant infection. Second, during passage through (or residence within) the human gut, low virulence drug-resistant food-source E. coli could transfer mobile resistance elements to more pathogenic, human-adapted strains that, in turn, could produce drug-resistant extraintestinal infection.
This study had two interrelated goals: 1) to assess the virulence potential of foodborne E. coli by comparing the virulence profiles of E. coli isolates from retail meats versus those from human clinical and stool specimens; and 2) to determine, within the same population, whether food-source and human-source antimicrobial-resistant E. coli are more similar genetically than either group is to susceptible E. coli from the same source, consistent with resistant isolates in humans possibly deriving from the food supply. We used multiple complementary molecular methods to address these questions, focusing on three antimicrobial classes: TMP-SMZ, quinolones (nalidixic acid [NA]), and ESCs, because these are therapeutically important in humans and are used in animal agriculture, which allows them to possibly serve as selectors for clinically significant foodborne antimicrobial resistance.
Materials and Methods
Samples and isolates
The study was conducted in a rural Idaho community (population approximately 35,000), and involved three ecological populations. For fecal isolates (n = 217), between March and May of 2002, a convenience sample of healthy community volunteers (and their household members) was recruited through local advertising and scheduled clinics at a drug store. Subjects used a CultureSwab™ (BD, Franklin Lakes, NJ) to collect fecal material. Samples were refrigerated and transported to the Idaho State Bureau of Laboratories (State Laboratory) in Boise, Idaho. For meat isolates (n = 231), between February and April of 2002, fresh and frozen ground beef and chicken samples were purchased from local markets once per month. For clinical isolates (n = 502), between March and September of 2002, consecutive, unique (i.e., one isolate per patient) resistant and susceptible E. coli isolates were obtained from the sole microbiology laboratory serving the local hospital and its ambulatory clinics. Details regarding sample collection, identification, and susceptibility testing have been reported previously (Hannah et al., 2005).
Phenotypic and molecular analysis strategy
For meat and fecal samples, at the State Laboratory, broth homogenates or rinsates from the primary samples were streaked across three MacConkey agar plates, each containing a screening antimicrobial agent (16 μg/mL of TMP, 16 μg/mL of NA, or 2 μg/mL of cefotaxime). All samples were also plated on a nonsupplemented blood agar plate. One representative of each phenotypically distinct colony type per plate was further analyzed. Putative E. coli colonies were confirmed using Microscan™ (Dade Behring, Deerfield, IL). Susceptibility was assessed via broth microdilution (Microscan) for cefpodoxime, ceftazadime, ceftriaxone, and TMP-SMZ and via Etest® (AB-BIODISK, Solna, Sweden) for NA. Manufacturer-specified procedures and reference strains were used, according to National Committee for Clinical Laboratory Standards/Clinical and Laboratory Standards Institute (NCCLS/CLSI) guidelines. NCCLS/CLSI interpretive criteria were used to classify isolates as resistant to TMP-SMZ (MIC > 4/76 μg/mL), NA (MIC ≥ 32 μg/ml), or ESCs, with the latter defined as resistance to ceftriaxone (MIC ≥ 64 μg/mL), ceftazadime (MIC ≥ 32 μg/mL), and/or cefpodoxime (MIC ≥ 8 μg/mL).
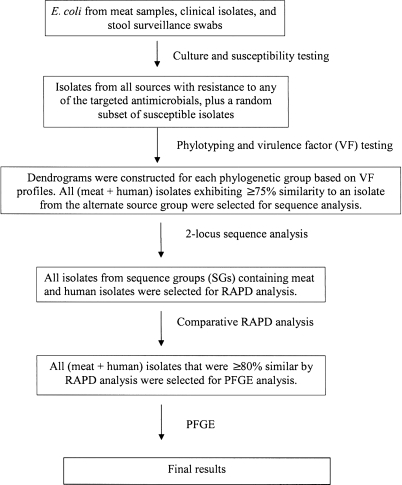
All meat, stool, and clinical isolates that were classified as resistant to one or more of the three targeted antimicrobials (TMP-SMZ, NA, and/or ESCs) underwent molecular analysis, as did a random sample of isolates susceptible to all three drug classes. The random sample was obtained by using a random number list (one per source group) to select from among all available susceptible isolates for the particular source group, stratified by whether the isolate was recovered from selective media or nonselective blood agar. The selected resistant and susceptible isolates then underwent a stepwise analysis using a series of molecular assays (described in greater detail below) according to a predetermined algorithm (Fig. 1) to assess commonality among meat- and human-source E. coli at increasing levels of similarity. First, isolates were categorized as to major E. coli phylogenetic group (A, B1, B2, and D) and underwent two-stage (i.e., screening and, selectively, extended) virulence genotyping. Second, within each phylogenetic group, any meat- and human-source isolates that exhibited ≥75% similarity (meat vs. human) according to screening or extended VF profiles were next compared using two-locus sequence analysis. Third, any of the meat and human isolates that exhibited identical sequence at both loci examined (i.e., belonged to the same sequence group [SG]) were compared by random amplified polymorphic DNA (RAPD) analysis, a whole-genome typing method. Finally, meat and human isolates with ≥80% similar RAPD profiles underwent pulsed-field gel electrophoresis (PFGE) analysis to identify individual clones.
FIG. 1.
Flow diagram of the laboratory processing, including the logic used to select isolates for molecular testing. RAPD, random amplified polymorphic DNA; PFGE, pulsed-field gel electrophoresis.
Phylogenetic analysis and virulence genotyping
Major E. coli phylogenetic groups (A, B1, B2, and D) (Herzer et al., 1990) were defined by polymerase chain reaction (PCR) (Clermont et al., 2000). All isolates were initially screened for hlyD (hemolysin) and five ExPEC-defining virulence markers: papA and papC (P fimbrial structural subunits; analyzed collectively), sfa/foc (S and F1C fimbriae), afa/dra (Dr-binding adhesins), iutA (aerobactin system), and kpsMT II (group 2 capsule). ExPEC status was defined by the presence of two or more of the five ExPEC-defining markers (Picard et al., 1999; Johnson et al., 2000; Johnson and Stell, 2000). All ExPEC isolates were tested for 35 ExPEC-associated VFs (including diverse adhesins, toxins, siderophores, polysaccharide coatings, invasins, protectins, and miscellaneous traits) and, if positive for any pap element, for 12 papA alleles, by using established PCR and dot-blot–based methods (Johnson et al., 2000, 2001, 2002b). Testing was done in duplicate using independently prepared boiled lysates of each isolate with appropriate positive and negative controls.
The screening ExPEC score was the number of the five ExPEC-defining markers detected in each lysate, plus hlyD (range, 0–6). Cluster analysis of the VF data (in which the VFs not tested in the non-ExPEC isolates were scored as being absent) was done using the unweighted pair group method with arithmetic averaging (UPGMA) (Johnson et al., 2006c).
Sequence analysis
Meat and human isolates that belonged to the same phylogenetic group and exhibited >75% similar VF profiles underwent two-gene sequence analysis. The genes used for sequence analysis (by phylogenetic group) were adk and metG (group A), icd and purA (group B1), recA and fumC (group B2), and adk and fumC (group D). These genes were selected on the basis of pilot data to provide maximal diversity within each phylogenetic group (Johnson et al., 2006c). Partial coding sequence for each gene was determined bidirectionally using primers from http://web.mpiib-berlin.mpg.de/mlst/dbs/Ecoli/documents/primersColi_html. For each phylogenetic group, concatenated sequences were separately aligned using CLUSTAL-X (Thompson et al., 1997), and trees were constructed according to Maximum Parsimony within PAUP* (Swofford, 2002). Sequence groups (SGs) were defined based on sequence identity across both loci examined.
RAPD and PFGE analysis
Members of SGs containing both meat- and human-source isolates were subjected to RAPD analysis, using (separately) arbitrary decamer primers 1247 and/or 1283 (Berg et al., 1994). Gel images were digitally captured and analyzed using the application Molecular Fingerprinting (Bio-Rad, Hercules, CA). Based on Pearson's correlation coefficient analysis of analog densitometric scans of the RAPD profiles, similarity trees were constructed according to UPGMA. Meat- and human-source isolates with ≥80% similar RAPD profiles, which approximates the reproducibility limit for replicate testing of the same isolate (J.R. Johnson, unpublished data), underwent PFGE analysis according to the PulseNet protocol (Ribot et al., 2006) with visual assessment of profile similarity.
Statistical methods
Proportions were compared using Fisher's exact test and McNemar's test for unpaired and paired comparisons, respectively (both, two-tailed). Comparisons of ExPEC scores and aggregate virulence scores were analyzed using the Wilcoxon-Mann Whitney test. The threshold for statistical significance was p < 0.05.
Results
Study population
Overall, 340 isolates were chosen for molecular analysis, including all available resistant isolates and a randomly selected subset of susceptible isolates distributed by source group. The 340 study isolates included 115 (50%) of 232 retail meat isolates, 122 (24%) of 517 human stool isolates, and 103 (21%) of 502 human clinical isolates. Of these, 181 (53%) (67 meat, 64 stool, and 50 clinical) were resistant to one or more of the three antimicrobial classes, whereas 159 (47%) (48 meat, 58 stool, and 53 clinical) were susceptible to all three antimicrobial classes. Resistance to TMP-SMZ was most common (n = 130), followed by NA resistance (n = 68), and ESC resistance (n = 4).
Phylogenetic analysis
Phylotyping of the 340 isolates showed that the three source populations (meat, stool, and clinical) exhibited substantially different phylogenetic distributions, along a continuum from meat (lowest prevalence for group B2, highest prevalence for groups A, B1, and D), through stool, to clinical (highest prevalence for group B2, lowest for groups A, B1, and D) (Table 1).Stool isolates, although intermediate overall between the meat and clinical isolates, were distributed more similarly to the meat isolates (Table 1).However, this relationship varied substantially by resistance status. For example, for stool isolates, resistant isolates were more likely than susceptible isolates to be from group D or B1 and less likely to be from group B2. Similarly, for meat isolates, resistant isolates were more likely than susceptible isolates to be from group D. Consequently, the phylogenetic distribution of resistant stool isolates more closely resembled that of resistant meat isolates than that of susceptible stool isolates (Table 1).
Table 1.
Phylogenetic Distribution and Extraintestinal Pathogenic Escherichia coli (ExPEC) Status of 340 E. coli Isolates from Retail Meats and from Human stool or Clinical Specimens, According to Resistance Status
| |
|
Phylogenetic group, no. (row%) |
|
|||
|---|---|---|---|---|---|---|
| Resistance status and sourcea | No. of isolates | A | B1 | B2 | D | ExPEC, no. (row%) |
| Resistant or susceptible | ||||||
| Meat | 115 | 46 (40) | 20 (17) | 10 (9) | 39 (34) | 16 (14) |
| Stool | 122 | 32 (26) | 17 (14) | 35 (29) | 38 (31) | 76 (61) |
| Clinical | 103 | 11 (11) | 4 (4) | 69 (67) | 19 (18) | 76 (74) |
| Resistant | ||||||
| Meat | 67 | 23 (34) | 9 (13) | 5 (8) | 30 (45)b | 13 (19) |
| Stool | 64 | 19 (29) | 14 (22)b | 3 (5)d | 28 (44)b | 41 (64) |
| Clinical | 50 | 8 (16) | 1 (2) | 26 (52) | 15 (30)c | 36 (72) |
| Susceptible | ||||||
| Meat | 48 | 23 (48) | 15 (31) | 1 (2) | 9 (19) | 3 (6) |
| Stool | 58 | 13 (23) | 3 (5) | 32 (55) | 10 (17) | 33 (57) |
| Clinical | 53 | 3 (6) | 3 (6) | 43 (81) | 4 (7) | 40 (76) |
Resistant defined as MIC > 4/76 mg/L to trimethoprim-sulfamethoxazole; MIC ≥ 32 mg/L to nalidixic acid; and/or MIC ≥ 64 μg/L to ceftriaxone, MIC ≥ 32 μg/ml to ceftazadime, or MIC ≥ 8 μg/ml to cefpodoxime. Susceptible defined as nonresistant to all three drug classes.
Significant difference between resistant and susceptible isolates, p < 0.05.
Significant difference between resistant and susceptible isolates, p < 0.01.
Significant difference between resistant and susceptible isolates, p < 0.001.
ExPEC screen
Overall, 166 (49%) of the 340 isolates satisfied molecular criteria for ExPEC. The prevalence of ExPEC varied by source group along a descending gradient from clinical isolates (67%), through stool isolates (61%), to meat isolates (14%) (Table 1).For meat isolates, the prevalence of ExPEC was significantly greater among resistant than susceptible isolates (19% vs. 6%, p = 0.04) (Table 1).No such relationship between ExPEC prevalence and resistance status was seen for stool isolates or clinical isolates.
Screening ExPEC scores likewise varied by source group, along a descending gradient as follows (group mean [range]): clinical (2.2 [0–5]) > stool (1.8 [0–4]) > meat (0.7 [0–2]). Resistant meat isolates had significantly higher screening ExPEC scores than did susceptible isolates (mean, 1.0 vs. 0.3; p < 0.001), whereas human resistant and susceptible isolates had similar screening ExPEC scores (mean, 2.4 vs. 2.3; non-significant). Thus, the screening ExPEC scores of resistant meat isolates were intermediate between those of susceptible meat isolates and those of (resistant or susceptible) human isolates.
Extended virulence genotypes
All 166 isolates that satisfied molecular criteria for ExPEC (16 meat, 74 stool, and 76 clinical) next underwent extended virulence genotyping for 35 VFs and, if pap positive, for 12 papA alleles. These extended VF profiles were combined with the screening VF profiles of the non-ExPEC isolates, and a similarity dendrogram was constructed for each phylogenetic group. Overall, 140 isolates exhibited >75% VF profile similarity to an isolate from the alternate host group (meat vs. human) within the same phylogenetic group. However, all but five of these isolates were non-ExPEC (i.e. had only one or no VF detected in the ExPEC screen). Consequently, the apparent similarity of VF profiles resulted primarily from the indeterminate nature of most of the studied markers (Table 2).
Table 2.
Extraintestinal Pathogenic Escherichia coli (ExPEC) Status and Virulence Genotypes Associated with the Nine Two-Locus Sequence Groups (SGs) That Contained Both Meat- and Human-Source E. coli Isolates
| |
|
No. of isolates per SGawithin each source and resistance status subgroupb |
|
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| |
|
|
|
Human |
|
|
|||
| |
|
Meat |
Stool |
Urine |
|
|
|||
| Phylogenetic group | SG | R | S | R | S | R | S | ExPECc | Extended virulence genotyped |
| A | A-1 | 4 | 7 | 6 | 0 | 0 | 0 | no | n/a |
| A | A-3 | 0 | 1 | 1 | 0 | 0 | 0 | no | n/a |
| A | A-5 | 9 | 5 | 2 | 2 | 0 | 0 | no | n/a |
| A | A-6 | 0 | 1 | 0 | 1 | 0 | 0 | no | n/a |
| A | A-7 | 0 | 1 | 0 | 0 | 0 | 1 | no | n/a |
| B1 | B1-2 | 0 | 1 | 0 | 2 | 0 | 0 | no | n/a |
| B1 | B1-4 | 4 | 4 | 1 | 0 | 0 | 0 | no | n/a |
| B2 | B2-1 | 2 | 0 | 0 | 0 | 0 | 1 | yes | papA, papC, sfa/foc, focG, fim, iutA |
| D | D-1 | 0 | 1 | 0 | 1 | 2 | 1 | yes | papA, papC, fim, kpsM II, iutA |
SGs were defined based on idenical sequence over both loci analyzed. A total of 21 SGs were identified; the nine shown are those that contained both meat and human isolates.
R, resistant to one or more of the three key antimicrobial classes, i.e., MIC > 4/76 mg/L to trimethoprim-sulfamethoxazole; MIC ≥ 32 mg/L to nalidixic acid; and/or MIC ≥ 64 μg/ml to ceftriaxone, MIC ≥ 32 μg/ml to ceftazadime, or MIC ≥ 8 μg/ml to cefpodoxime. S, susceptible to all three antimicrobial classes.
ExPEC, positive for two or more of the following five traits: papA and/or papC (P fimbriae structural subunit and assembly; analyzed jointly), sfa/foc (S and FIC fimbriae), afa/dra (Dr-binding adhesins), iutA (aerobactin system), and kpsM II (group 2 capsule).
Extended virulence genotypes determined only for isolates that tested as ExPEC in the initial screen. n/a, not applicable (non-ExPEC).
Sequence typing
The 140 isolates that exhibited >75% VF profile similarity to a representative of the alternate host group next underwent two-gene sequence analysis. Overall, 21 unique SGs were identified. Sixteen of these SGs contained multiple isolates, including nine SGs with both meat and human isolates (64 isolates total; 40 meat, 17 stool, 4 clinical, and 3 reference strains). Of the nine (meat + human) SGs, five were from phylogenetic group A, two were from group B1, and one each was from group B2 and D, respectively. All seven (meat + humans) SGs from phylogenetic groups A and B1 contained only non-ExPEC isolates, whereas the two (meat + human) SGs from groups B2 and D contained only ExPEC isolates, which exhibited variably robust VF profiles (Table 2).
RAPD analysis
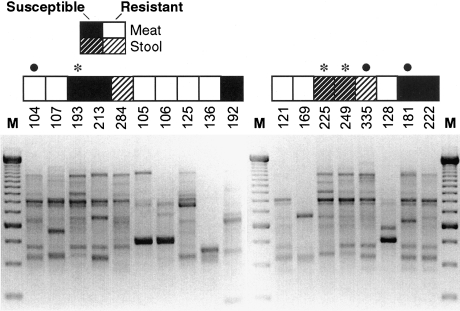
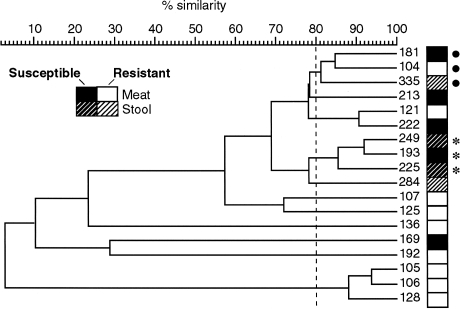
The 64 members of the nine (meat + human isolate) SGs next underwent composite RAPD analysis. The resulting nine RAPD dendrograms (one per SG) contained 12 clusters at the >80% profile similarity level, three of which included both meat and human study isolates. The three (meat + human isolate) RAPD clusters, each from a different SG, contained 17 isolates, including six meat, seven stool, and three clinical isolates, plus one human-source reference strain (ECOR 47). Figures 2 and 3 (showing RAPD profiles of isolates from SG A-5 and the corresponding similarity dendrogram) illustrate the considerable genetic diversity within even a single two-locus SG, but also the potential genomic similarity between certain meat- and stool-source E. coli isolates, as exemplified by isolates 104, 335, and 181 (bullets in Figs. 2 and 3).
FIG. 2.
Random amplified polymorphic DNA (RAPD) profiles of phylogenetic group A isolates of Escherichia coli from sequence group 5, phylogenetic group A. Profiles were generated using arbitrary decamer primer 1283, M, 250-bp marker. Strain identifiers are shown above each gel lane. Boxes above strain identifiers indicate each isolate's source. Considerable genomic diversity is evident, although some profiles look quite similar; e.g., isolates 181 (susceptible, meat), 104 (resistant, meat), and 335 (resistant, stool), as indicated by bullets, and isolates 249 (susceptible, meat), 193 (susceptible, stool), and 225 (susceptible, meat), as indicated by asterisks.
FIG. 3.
Random amplified polymorphic DNA (RAPD)-based dendrogram for isolates from sequence group 5, phylogenetic group A. Cluster analysis (by UPGMA) of RAPD profiles from Fig. 2., based on Pearson correlation coefficient analysis of analog densitometric scans of each gel lane. Two clusters of meat and human isolate (three isolates per cluster), as indicated by bullets and asterisks, respectively, exhibit >80% similarity of profiles (vertical dashed line).
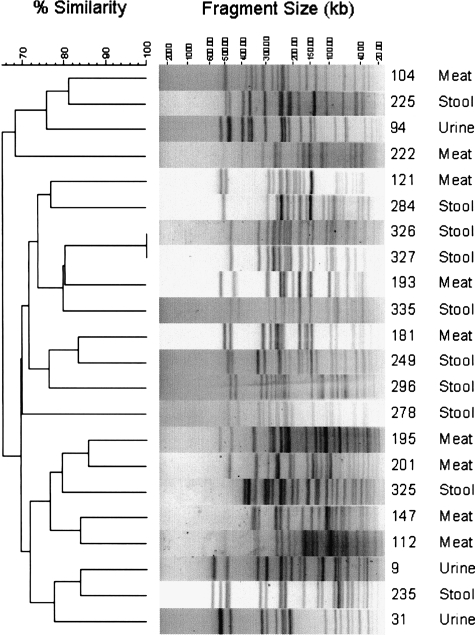
To assess genomic similarity more stringently, all isolates from the three RAPD clusters that included both meat and human isolates underwent PFGE analysis. All PFGE profiles were unique excepting those of two stool isolates from members of the same household, which were indistinguishable (Fig. 4). Notably, however, according to PFGE analysis several meat isolates were more similar (i.e., are closer in the PFGE-based dendrogram) to one or more human isolates than to another meat isolate.
FIG. 4.
Pulsed-field gel electrophoresis (PFGE) profiles of selected meat and human isolates of Escherichia coli from the same phylogenetic and sequence group, with similar virulence profiles and random amplified polymorphic DNA profiles. PFGE analysis was done on the selected meat and human isolates using XbaI. The dendrogram to the left of the gel strip images was inferred according to UPGMA, based on Dice similarity coefficients, and reflects the relatedness of the various isolates' PFGE profiles. Specimen identification number and source of the isolate are shown to the right of the gel strips.
Discussion
In this study we assessed the virulence potential of food-source E. coli and examined the extent of commonality between food-source and geographically and temporally matched human-source (commensal and clinical) E. coli with respect to genomic background and VF profiles, all in relation to antimicrobial resistance. Our findings support three main conclusions. First, an appreciable minority (19%) of antimicrobial-resistant retail meat isolates represented ExPEC, suggesting possible human pathogenic potential. However, the dissimilarity of these isolates' extended virulence profiles to those of human clinical and stool isolates leaves in question their direct human health relevance. Second, according to phylogenetic group distribution, the resistant stool isolates (as a group) resembled meat isolates, particularly resistant meat isolates, more closely than they did susceptible stool isolates. This is consistent with the resistant stool isolate population possibly having originated in meats, rather than by conversion of susceptible human-associated strains to resistance. Third, certain meat and human isolates exhibited considerable genomic similarity according to sequence analysis and RAPD analysis, further supporting the hypothesized food–human connection, whereas commonality by PFGE was scarcely detectable even among isolates from the same source group, reflecting the extreme genetic diversity of E. coli.
We found that only 14% of meat-source E. coli isolates (19% if resistant) qualified as ExPEC, compared with 61% of stool isolates and 74% of clinical isolates, whereas the few that did qualify exhibited VF profiles quite different from the VF profiles of human-source ExPEC isolates. This might be interpreted as evidence against a significant direct human health threat from meat-source E. coli, with their pathogenicity instead perhaps being limited to their respective associated host species (e.g., avian pathogenic E. coli in poultry) (T.J. Johnson et al., 2007). However, considerable diversity of VF profiles was evident even among the human-source ExPEC. This suggests that the absence of VF profile commonality between meat and human ExPEC isolates does not necessarily mean a lack of human pathogenicity for the meat isolates; experimental challenge studies would be needed to address this (Johnson et al., 2006a). Furthermore, even if meat-source ExPEC are not directly pathogenic for humans, they still could serve as VF donors (via horizontal gene transfer) to more human-adapted strains they encounter during passage through the human gut (Skyberg et al., 2007).
Among the meat-source E. coli, resistant isolates were significantly more likely to qualify as ExPEC, and exhibited significantly higher screening ExPEC scores, compared with susceptible isolates. This may reflect the genetic linkage on certain plasmids of resistance elements and the iuc/iut (aerobactin) operon, since we used iutA as one of the ExPEC-defining screening markers. This would underscore the potential threat posed by such strains and their resistance plasmids, some of which also carry other VFs such as iss, traT, and iroN (T.J. Johnson et al., 2007), since aerobactin-encoding resistance plasmids are often conjugally transferable and may provide a vehicle for coordinate horizontal transfer of VFs and resistance elements (Johnson et al., 2002a).
Two striking patterns of phylogenetic distribution were apparent. One was the predominance of phylogenetic group B2 among human isolates, compared with the predominance of non-B2 groups among meat isolates. This finding, which is consistent with previous work, seems to suggest that although human and meat source populations overlap to some extent, they are nonetheless distinct overall, with the human clinical isolates in particular representing an extreme of the “human” phenotype. However, the other pattern, which was evident within each source group, was the association of resistance with non-B2 phylogenetic groups, particularly group D. This suggests some degree of commonality among resistant isolates across source groups, for which one possible explanation would be strain transfer among source groups. For example, based on phylogenetic group distribution, resistant stool isolates could represent resistant meat isolates, imported into humans. Likewise, resistant clinical isolates could possibly represent resistant stool isolates, selectively enriched with group B2 and depleted of group B1. Such enrichment might occur because of the typical abundance (group B2) versus paucity (group B1) of VFs in these two groups and the selective advantage VFs confer within the pathogenic niche.
The basis for the association of resistance with group D, specifically among meat isolates, is unclear. Multiple studies have documented this phenomenon among human isolates, consistent with importation of resistant isolates from an external source, possibly food (Angulo et al., 2004; Kang et al., 2005; J.R. Johnson et al., 2007). However, in previous studies of food-source isolates, no differences have been noted in the phylogenetic distribution of resistant versus susceptible isolates within a given food type (e.g., poultry, beef, and pork). This is consistent with on-farm selection for resistance within host-specific, animal-associated commensal E. coli populations (Johnson et al., 2005, 2006b). In our study, for enhanced statistical power, different meat types were analyzed collectively. If different meats yielded phylogenetically distinct E. coli populations with different resistance prevalences, the observed phylogenetic differences between resistant and susceptible meat isolates may represent confounding by meat type. A larger sample size would be needed for a stratified analysis to test that hypothesis.
Regarding commonality among individual meat and human isolates, we used sequence analysis (based on only two loci, for economy) to screen for genomic similarity among isolates with similar VF profiles, followed by RAPD analysis to provide an independent whole-genome scan. Sequence analysis is reproducible and quantitative, but also resource intensive, and examines only the loci analyzed, which are subject to horizontal transfer, obliging inclusion of multiple loci for a valid phylogenetic signal and adequate discrimination. In contrast, RAPD analysis is quick and economical and surveys the entire genome, but it suffers from irreproducibility and subjective interpretation. By combining the two modalities we sought to capitalize on the strengths of each. This hybrid approach identified several groups of genomically similar meat and human isolates, supporting the hypothesis of commonality across populations.
The observed near-total diversity of PFGE profiles, in contrast to the considerable overlap according to phylogenetic group, SG, and RAPD profile, reflects the tremendous clonal heterogeneity of E. coli. This demonstrates the limited utility of PFGE, an extremely discriminating typing tool, as a sole method for identifying commonality among ecologically disparate E. coli populations. Commonality at the PFGE profile level between food- and human-source antimicrobial-resistant E. coli isolates has been reported only once before, with a single isolate pair (Johnson et al., 2006a). The present evidence, and previous examples, of E. coli isolates with the same PFGE profile from multiple members of a given household suggest that food–human commonality of E. coli strains at the PFGE level may best be sought by studying household members and the actual foods they consume or handle (Murray et al., 2004; Hannah et al., 2005; Johnson and Clabots, 2006).
This study has several limitations. Aggregation of isolates based on resistance to TMP-SMZ, NA, and/or ESCs and from different types of meat products could have blurred important differences between these resistance phenotypes and sources. We did not examine resistance elements and, consequently, do not know how similar these are in human vs. meat isolates. Applicability to other locales and times is unknown. Finally, we assessed the virulence potential of meat-source E. coli through molecular typing, not experimentation, although this has empirical support (Johnson et al., 2006a). Strengths include the prospective community-based study design, which allowed comparison of E. coli from meat items purchased in community grocery stores with contemporaneous clinical and fecal isolates from local residents, and the use of multiple state-of-the-art molecular approaches to estimate the similarity of meat and human isolates and to define the presumed virulence potential of meat isolates.
In summary, in this community-based comparison of human- and meat-source E. coli isolates, we found that some meat-source E. coli exhibited robust virulence profiles, suggesting a potential reservoir of virulent strains and/or transferable VFs in meat products. Furthermore, our phylogenetic analysis suggested some degree of commonality between the meat-source and human-source resistant E. coli populations, since each of the four E. coli phylogenetic groups contained clusters of genomically similar (albeit not indistinguishable) meat and human isolates. These data support a possible role for retail meats in promoting acquisition and dissemination of resistant and virulent E. coli in human populations, but they also indicate a need for further study of this important question.
Acknowledgments
Funding was provided by the Centers for Disease Control and Prevention grants RS1 CCR820631 (M.H.S.) and R01-CI000204 (J.R.J.), Office of Research and Development, Medical Research Service, Department of Veterans Affairs (J.R.J.), and National Research Initiative (NRI) Competitive Grants Program/United States Department of Agriculture grant 00-35212-9408 (J.R.J.). Special thanks to Vivian Lockary and Christopher Ball at the Idaho State Laboratory for their invaluable assistance on this project.
Disclosure Statement
No competing financial interests exist.
References
- Allen UD. MacDonald N. Fuite L, et al. Risk factors for resistance to “first-line” antimicrobials among urinary tract isolates of Escherichia coli in children. Can Med Assoc J. 1999;160:1436–1440. [PMC free article] [PubMed] [Google Scholar]
- Angulo FJ. Nargund VN. Chiller TC. Evidence of an association between use of anti-microbial agents in food animals and anti-microbial resistance among bacteria isolated from humans and the human health consequences of such resistance. J Vet Med B Infect Dis Vet Public Health. 2004;51:374–379. doi: 10.1111/j.1439-0450.2004.00789.x. [DOI] [PubMed] [Google Scholar]
- Bacheller CD. Bernstein JM. Urinary tract infections. Med Clin North Am. 1997;81:719–730. doi: 10.1016/s0025-7125(05)70542-3. [DOI] [PubMed] [Google Scholar]
- Bazile-Pham-Khac S. Truong QC. Lafont JP, et al. Resistance to fluoroquinolones in Escherichia coli isolated from poultry. Antimicrob Agents Chemother. 1996;40:1504–1507. doi: 10.1128/aac.40.6.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg DE. Akopyants NS. Kersulyte D. Fingerprinting microbial genomes using the RAPD or AP-PCR method. Methods Mol Cell Biol. 1994;5:13–24. [Google Scholar]
- Chulasiri M. Suthienkul O. Antimicrobial resistance of Escherichia coli isolated from chickens. Vet Microbiol. 1989;21:189–194. doi: 10.1016/0378-1135(89)90032-1. [DOI] [PubMed] [Google Scholar]
- Clermont O. Bonacorsi S. Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol. 2000;66:4555–4558. doi: 10.1128/aem.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannah EL. Angulo FJ. Johnson JR, et al. Drug-resistant Escherichia coli: epidemiology, rural Idaho. Emerg Infect Dis. 2005;11:1614–1617. doi: 10.3201/eid1110.050140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzer PJ. Inouye S. Inouye M, et al. Phylogenetic distribution of branched RNA-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J Bacteriol. 1990;172:6175–6181. doi: 10.1128/jb.172.11.6175-6181.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JR. Clabots CR. Sharing of virulent Escherichia coli clones among household members of a woman with acute cystitis. Clin Infect Dis. 2006;43:101–108. doi: 10.1086/508541. [DOI] [PubMed] [Google Scholar]
- Johnson JR. Clermont O. Menard M, et al. Experimental mouse lethality of Escherichia coli isolates in relation to accessory traits phylogenetic group, clinical source. J Infect Dis. 2006a;194:1141–1150. doi: 10.1086/507305. [DOI] [PubMed] [Google Scholar]
- Johnson JR. Delvari M. Kuskowski MA, et al. Phylogenetic distribution of extraintestinal virulence-associated traits in Escherichia coli. J Infect Dis. 2000;183:78–88. doi: 10.1086/317656. [DOI] [PubMed] [Google Scholar]
- Johnson JR. Delvari P. O'Bryan TT, et al. Contamination of retail foods, particularly turkey, from community markets (Minnesota, 1999–2000) with antimicrobial-resistant and extraintestinal pathogenic Escherichia coli. Foodborne Pathog Dis. 2005;2:38–49. doi: 10.1089/fpd.2005.2.38. [DOI] [PubMed] [Google Scholar]
- Johnson JR. Gajewski A. Lesse AJ, et al. Extraintestinal pathogenic Escherichia coli as a cause of invasive non-urinary infections. J Clin Microbiol. 2003a;41:5798–5802. doi: 10.1128/JCM.41.12.5798-5802.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JR. Kuskowski MA. Menard M, et al. Similarity of human, chicken-source Escherichia coli isolates in relation to ciprofloxacin resistance status. J Infect Dis. 2006b;194:71–78. doi: 10.1086/504921. [DOI] [PubMed] [Google Scholar]
- Johnson JR. Murray AC. Gajewski A, et al. Antimicrob Agents Chemother. 47; 2003b. Isolation, molecular characterization of nalidixic acid-resistant extraintestinal pathogenic Escherichia coli from retail chicken products; pp. 2161–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JR. O'Bryan TT. Kuskowski MA, et al. Ongoing horizontal and vertical transmission of virulence genes and papA alleles among Escherichia coli blood isolates from patients with diverse-source bacteremia. Infect Immun. 2001;69:5363–5374. doi: 10.1128/IAI.69.9.5363-5374.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JR. Owens KL. Clabots CR, et al. Phylogenetic relationships among clonal groups of extraintestinal pathogenic Escherichia coli as assessed by multi-locus sequence analysis. Microbes Infect. 2006c;8:1702–1713. doi: 10.1016/j.micinf.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Johnson JR. Sannes MR. Croy C, et al. Antimicrobial drug-resistant Escherichia coli from humans and poultry products, Minnesota and Wisconsin, 2002–2004. Emerg Infect Dis. 2007;13:838–846. doi: 10.3201/eid1306.061576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JR. Stell AL. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis. 2000;181:261–272. doi: 10.1086/315217. [DOI] [PubMed] [Google Scholar]
- Johnson JR. Stell AL. O'Bryan TT, et al. Global molecular epidemiology of the O15:K52:H1 extraintestinal pathogenic Escherichia coli clonal group: evidence of distribution beyond Europe. J Clin Microbiol. 2002a;40:1913–1923. doi: 10.1128/JCM.40.6.1913-1923.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JR. van der Schee C. Kuskowski MA, et al. Phylogenetic background, virulence profiles of fluoroquinolone-resistant clinical Escherichia coli isolates from the Netherlands. J Infect Dis. 2002b;186:1852–1856. doi: 10.1086/345767. [DOI] [PubMed] [Google Scholar]
- Johnson TJ. Karlyawasam S. Wannemuehler Y, et al. The genome sequence of avian pathogenic Escherichia coli strain O1:K1:H7 shares strong similarities with human extraintestinal pathogenic E. coli genomes. J Bacteriol. 2007;189:3228–3236. doi: 10.1128/JB.01726-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HY. Jeong YS. Oh JY, et al. Characterization of antimicrobial resistance and class 1 integrons found in Escherichia coli isolates from humans and animals in Korea. J Antimicrob Chemother. 2005;55:639–644. doi: 10.1093/jac/dki076. [DOI] [PubMed] [Google Scholar]
- Krumperman PH. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl Environ Microbiol. 1983;46:165–170. doi: 10.1128/aem.46.1.165-170.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray AC. Kuskowski MA. Johnson JR. Virulence factors predict Escherichia coli colonization patterns among human and animal household members. Ann Intern Med. 2004;140:848–849. doi: 10.7326/0003-4819-140-10-200405180-00032. [DOI] [PubMed] [Google Scholar]
- Picard B. Sevali Garcia J. Gouriou S, et al. The link between phylogeny and virulence in Escherichia coli extraintestinal infections. Infect Immun. 1999;67:546–553. doi: 10.1128/iai.67.2.546-553.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribot EM. Fair MA. Gautom R, et al. Standardization of pulsed-field gel electrophoresis protocols for the sub-typing of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis. 2006;3:59–67. doi: 10.1089/fpd.2006.3.59. [DOI] [PubMed] [Google Scholar]
- Russo TA. Johnson JR. Proposal for a new inclusive designation for extraintestinal pathogenic isolates of Escherichia coli: ExPEC. J Infect Dis. 2000;181:1753–1754. doi: 10.1086/315418. [DOI] [PubMed] [Google Scholar]
- Skyberg JA. Johnson TJ. Clabots CR, et al. Acquisition of APEC plasmids by a commensal Escherichia coli isolate enhances its abilities to kill chick embryos, grow in human urine, and colonize the murine kidney. Infect Immun. 2006;74:6287–6292. doi: 10.1128/IAI.00363-06. Epub 2006 Sep 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. PAUP* Phylogenetic Analysis Using Parsimony (* and other methods) Sunderland, MA: Sinauer Associates; 2002. [Google Scholar]
- Thompson JD. Gibson TJ. Plewniak F, et al. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]