Abstract
There is growing evidence to suggest that HIV may interact with several hepatic cell types; however, evaluation of HIV variability in liver tissue has not been addressed to date. Among 16 HIV-positive individuals examined, nine (56%) had detectable HIV RNA in the liver. The mean CD4 cell count for these nine individuals was 337 cells/mm3 (range: 0–601), while their mean plasma HIV RNA level was 106,974 copies/ml (range: 1200–320,740). Among individuals in this study with detectable HIV in both the plasma and the liver, the consensus gag nucleotide sequences for each tissue type were different for seven of seven (100%) individuals, while amino acid sequences were distinct for five of seven (71%). Consensus envelope (env) nucleotide and amino acid sequences were also distinct in the plasma and liver tissue for six of six (100%) individuals. Statistical evidence of compartmentalization between HIV in the plasma and in the liver was demonstrated, and multiple liver-specific amino acids were identified that may distinguish HIV variants replicating within the liver. These preliminary data demonstrate that HIV is frequently detectable in the liver of HIV-positive persons at various levels of immunosuppression. Possible compartmentalization may reflect tissue-specific selection pressures that drive viral adaptation to the liver microenvironment and may facilitate interactions with other hepatotropic viruses.
Introduction
Hepatic disease is increasingly recognized as a major cause of morbidity and mortality among HIV-positive individuals. Furthermore, HIV coinfection is associated with enhanced hepatitis C virus (HCV) replication, more advanced liver fibrosis and cirrhosis, higher rates of progressive liver disease and death, and decreased HCV treatment response.1 While liver biopsies represent the gold standard for detecting liver damage, their utilization in individuals with HIV remains infrequent despite high frequencies of both hepatomegaly and liver enzyme abnormalities.2–5 Even HIV-positive individuals with no evidence of viral hepatitis coinfection often exhibit mild-to-moderate increases in liver enzyme levels.6,7 A recent study also found that HIV RNA levels were positively associated with liver fibrosis in HIV monoinfected persons even after controlling for other confounders,8,9 thus supporting the involvement of HIV itself in hepatic disease.
Several lines of evidence suggest that HIV is present in the liver.1 For instance, HIV RNA and proviral DNA have been detected in liver biopsies from persons with HIV infection.10,11 Immunohistochemistry and in situ hybridization studies using liver specimens from HIV-infected individuals have also demonstrated HIV p24 protein and HIV RNA in Kupffer cells, inflammatory mononuclear cells, sinusoidal cells, and hepatocytes.10,12,13 Efficient activation of the HIV long terminal repeat has also been reported in hepatocytes.14,15 Importantly, we16 and others17–19 have demonstrated that HIV can infect hepatocyte-derived cell lines, as well as primary hepatocytes, although likely at lower levels than occurs during infection of lymphocytes. Collectively, these data would indicate that several distinct cell types within the liver might be permissive to HIV infection.
A hallmark of RNA viruses is their extreme variability. Within an individual, a population of viral variants termed the viral quasispecies exists. These variants may allow for the rapid, adaptive response of HIV to immunologic selection pressures and/or antiviral therapy.20 Several studies have demonstrated an association between quasispecies diversity and HIV disease progression.21–23 Importantly, HIV variability is not evenly distributed throughout the body, and distinct viral subpopulations may exist in different compartments within an infected individual.24 For example, the blood and male genital tract may represent distinct HIV compartments as viral diversity and/or the majority sequences are often discordant in the blood compared to the genital tract.25–29 Similarly, HIV compartmentalization may occur in the brain and cerebrospinal fluid,30–32 suggesting that viral adaptation is frequently necessary for efficient infection of and replication within a particular cell/tissue type. Currently, there are no published reports on HIV diversity within the liver despite the link between HIV and liver disease. Thus, it is not clear if all variants of HIV present within an individual are equally capable of infecting the liver or if selection of particular HIV variants with tropism for the liver is occurring. Therefore, we investigated the presence of HIV RNA in liver biopsies and addressed whether HIV variability in the liver differed from that in the plasma.
Materials and Methods
Study participants
For this pilot study, a convenience sampling of 12 HIV-infected individuals was randomly selected from those receiving routine clinical care at the University of Cincinnati College of Medicine or those being evaluated for the initiation of antiretroviral therapy (ART). All subjects signed informed consents permitting collection of tissue and blood. Liver tissue and plasma collected at the time of autopsy were available for an additional four individuals through the National Disease Research Interchange.
Reverse transcriptase polymerase chain reaction amplification of HIV
Viral RNA was extracted from 140 μl of patient plasma using the QIAamp Viral RNA kit or from homogenized liver biopsies (typically 1–2 mm in length) using the RNeasy Mini kit. HIV RNA was detected by nested reverse transcriptase–polymerase chain reaction (RT-PCR) for HIV gag (p24) and env as described previously.33 Briefly, to amplify a 485-nucleotide fragment of gag (nucleotides 1237–1721 of the HIV reference HXB2), first round primers were 5′–CCC TGR CAT GCT GTC ATC A–3′ and 5′–AGY CAA AAT TAY CCY ATA GT–3′ and second round primers were 5′–AGR ACY TTR AAY GCA TGG GT–3′ and 5′–TGT GWA GCT TGY TCR GCT C–3′. To amplify a 337-nucleotide fragment of env (nucleotides 7002–7338 of HXB2), first round primers were 5′–ATG GGA TCA AAG CCT AAA GCC ATG TG–3′ and 5′–ACT GCT TCC TGC TGC TCC CAA GAA CCC AAG–3′ and second round primers were 5′–CTG TTA AAT GGT AGT CTA GC–3′ and 5′–CAA TTT CTG GGT CCC CTC CTG AG–3′. All RT-PCR amplifications included one reaction containing no reverse transcriptase and a separate reaction containing no template as negative controls. The housekeeping gene, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), was also detected in liver tissues as previously described.34
PCR products were gel purified and ligated into a standard cloning vector (Promega; Madison, WI). Plasmids were propagated and purified prior to sequencing using dye terminator chemistry. Multiple plasmids per sample source were sequenced for gag (average 10.3 clones) and for env (average 10.8 clones) in the forward and reverse directions and edited using CodonCode Aligner 1.5.2 (CodonCode Corporation, Dedham, MA).
Phylogenetic and signature sequence analyses
All alignments were performed using the neighbor-joining (NJ) approach implemented in Clustal X.35 By aligning compartment-specific viral variants from each tissue/cell type, consensus sequences were generated. References available through the HIV sequence database (http://www.hiv.lanl.gov/content/sequence/NEWALIGN/align.html) were used to identify the HIV subtype (data not shown). Subsequent analyses were performed using the prototype HIV-1 subtype B sequences HXB2 and NL4-3 as references, and the statistical robustness and reliability of the branching order within each phylogenetic tree were confirmed by bootstrap analysis using 1000 replicates.36 Phylogenetic inference was also performed using a Bayesian Markov chain Monte Carlo (MCMC) approach as implemented in the BEAST v1.5.0 program37 under an uncorrelated log-normal relaxed molecular clock using the generalized time reversible (GTR) or Hasegawa, Kishino, and Yano (HKY) model with nucleotide site heterogeneity estimated using a gamma distribution. The MCMC analysis was run for a chain length of 50,000,000 with sampling every 5000th generation. Results were visualized in Tracer v1.4 to confirm chain convergence, and the effective sample size (ESS) was calculated for each parameter. All ESS values were >500 indicating sufficient sampling. The maximum clade credibility tree was selected from the posterior tree distribution after a 10% burn-in using TreeAnnotator v1.5.0. Consensus sequences have been submitted to GenBank under accession numbers HM365303–HM365331.
Intrapatient genetic distances were calculated by pairwise comparison of nucleotide sequences at each time point using the Kimura method of MegAlign (DNASTAR, Inc., Madison, WI). Shannon entropy [Sn=–∑(pi ln pi)/ln N], where pi is the frequency of each distinct nucleotide sequence and N is the total number of sequences analyzed, was also calculated.38 Entropy values vary from 0 (all sequences are identical) to 1 (all sequences are distinct). Nonsynonymous (dN) and synonymous (dS) mutations were calculated via the Nei–Gojobori method in MEGA.39 Amino acid translations and manual editing to preserve the open reading frame were performed in MacClade version 4.08.40 The Viral Epidemiology Signature Pattern Analysis (VESPA) program was used to determine the frequency of each amino acid in liver-derived versus serum-derived viral variants for each individual.41 Only amino acid signatures above a 70% threshold were considered significant. Envelope coreceptor utilization was determined for consensus and clonal sequences using the WetCat program at http://genomiac2.ucsd.edu:8080/wetcat/v3.html.
Compartmentalization analyses
Compartmentalization of viral variants was assessed using Mantel's test as previously described to explore HIV compartmentalization.42,43 Briefly, the Kimura two-parameter distance matrix that included all plasma and liver biopsy variants for an individual subject was compared to a similar matrix in which distances are replaced with 0 if the sequences are from the same compartment (e.g., plasma versus plasma or liver biopsy versus liver biopsy) and with 1 if the sequences are from distinct compartments (e.g., plasma versus liver biopsy or vice versa). The correlation coefficient was computed for the simple Mantel's test using the Permute! Software version 3.4α9 (www.bio.umontreal.ca/casgrain/en/labo/permute/index.html) with 9999 permutations. Compartmentalization was also assessed using the Slatkin–Maddison (S-M) test44 for population gene flow as implemented in the HyPhy program45 using at least 5000 permutations. p-values <0.05 were considered statistically significant evidence that viral sequences derived from liver biopsies were compartmentalized compared to those derived from the corresponding plasma.
Statistical analysis
CD4 cell counts and plasma HIV viral loads were compared in persons with and without detectable HIV in the liver using two-sample t tests, while measures of diversity were compared using the Wilcoxon rank sum test (Statistix 9.0; Analytical Software, Tallahassee, FL).
Results
Patient demographics and HIV detection
A convenience sample of 16 HIV-positive individuals was utilized for this pilot study (Table 1). The average CD4 cell count was 401 cells/mm3 (range: 0–764 cells/mm3). Plasma HIV viral loads were detectable in 12 of 13 individuals (mean: 68,131 copies/ml; range 463–320,740 copies/ml) with available data. ART utilization was not an exclusion criteria; however, only two individuals were receiving ART at the time of sample collection. Thirteen of 16 individuals were HCV seropositive, while two individuals were hepatitis B virus (HBV) surface antigen positive. The median ALT and AST values were 58 U/liter and 77 U/liter, respectively.
Table 1.
Patient Demographic and Clinical Characteristics
| Patient ID | HIV viral load (copies/ml) | CD4 cell count (cells/mm3) | HIV treatment at time of biopsy | Year of HIV diagnosis | ALTa(U/liter) | AST (U/liter) | HCV serostatus | HBV surface antigen | Risk factor |
|---|---|---|---|---|---|---|---|---|---|
| Cin 01 | 56,335 | 408 | No | 1995 | 91 | 91 | Positive | Negative | Sexual |
| Cin 02 | 218,106 | 423 | No | 1989 | 39 | 62 | Positive | Negative | Sexual |
| Cin 03 | 320,740 | 338 | No | 2005 | 53 | 77 | Positive | Negative | Sexual |
| Cin 04 | 98,919 | 265 | No | 2006 | 177 | 212 | Positive | Negative | Sexual |
| Cin 05 | 29,638 | 346 | No | 1987 | 33 | 185 | Positive | Negative | Sexual; drug use |
| 1347 | 17,813 | 569 | No | 1988 | NA | NA | Positive | Negative | Transfusion |
| 1370 | 5,481 | 764 | No | 1997 | NA | NA | Positive | Negative | Sexual |
| 1493 | 6,126 | 397 | No | 1990 | 59 | 42 | Positive | Negative | Hemophilia |
| 1580 | 1,200 | 410 | No | 2001 | NA | NA | Positive | Negative | Sexual; drug use |
| 1627 | 463 | 547 | NA | 1990 | 58 | 47 | Positive | Negative | Drug use |
| 1724 | <48 | 304 | Yes | 1999 | 56 | 54 | Positive | Negative | Drug use |
| 1756 | 66,370 | 601 | No | 1986 | 154 | 108 | Negative | Negative | Sexual; drug use |
| HV52b | 64,485 | 0 | No | 1992 | NA | NA | Negative | Positive | NA |
| HV67b | NA | NA | NA | 2005 | NA | NA | Negative | Positive | NA |
| HV104b | NA | NA | Yes | 1988 | NA | NA | Positive | Negative | Hemophilia |
| HV106b | NA | 238 | No | 2000 | NA | NA | Positive | NA | Drug use |
Normal values for healthy individuals are defined as <19 U/liter for women and <30 U/liter for men.
Denotes samples collected at autopsy.
PIDs in bold had detectable HIV gag and/or env sequences in liver biopsy. NA, not available; ALT, alanine aminotransferase; AST, aspartate aminotransferase; HCV, hepatitis C virus; HBV, hepatitis B virus.
Using RT-PCR, nine individuals (56.3%) had detectable HIV RNA in the liver. This included eight individuals with HIV gag detected in their livers, and six individuals with HIV env detected in their livers. To further confirm the quality of the extracted RNA, RT-PCR for GAPDH was attempted for 10 randomly selected liver biopsies included in the current study; all 10 were PCR positive (data not shown). The mean CD4 cell count for these nine individuals was 337 cells/mm3 (range: 0–601), while their mean plasma HIV RNA level was 106,974 copies/ml (range: 1200–320,740). Seven individuals were also coinfected with HCV, while one individual was HBV surface antigen positive. CD4 cell counts were lower in those individuals with HIV detected in the liver compared to those without detectable HIV in the liver, although this difference did not reach statistical significance (336.6 cells/mm3 versus 516.2 cells/mm3; p=0.080). Interestingly, plasma HIV viral loads were significantly higher in those individuals with HIV detected in the liver compared to those without detectable HIV in the liver (106,974 copies/ml versus 5982 copies/ml; p=0.033).
Intrapatient HIV variability
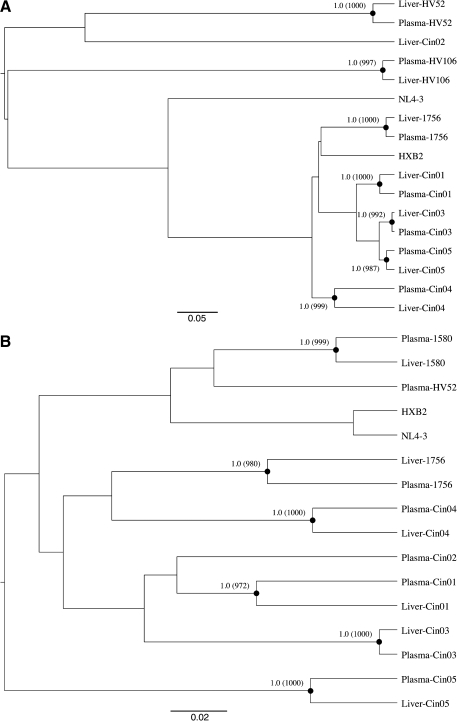
We performed a phylogenetic analysis of HIV gag and/or env sequences for the nine individuals with detectable HIV RNA in the liver. All were infected with HIV-1 subtype B, and sequences clustered by individual suggesting that there were no epidemiologically linked infections. The consensus gag nucleotide sequences were different for seven of seven (100%) individuals with matched liver biopsies and plasma using the NJ approach (data not shown). Similar analyses and results were obtained using a Bayesian inference approach based on a GTR or HKY substitution model (Fig. 1A). Consensus gag amino acid sequences were distinct for five of seven (71.4%) individuals, while two individuals had synonymous nucleotide changes not affecting the resultant amino acid sequence. Similarly, the consensus env nucleotide sequences were different for six of six (100%) individuals with matched liver biopsies and plasma using both NJ (data not shown) and Bayesian approaches (Fig. 1B). The consensus env amino acid sequences were distinct for all six individuals.
FIG. 1.
Consensus nucleotide sequences in gag (A) and env (B) from plasma and liver biopsy tissue using a Bayesian inference approach. Relevant posterior probabilities greater than 90% are shown. Relevant bootstrap values >700 out of 1000 from a neighbor-joining approach are shown in parentheses. The reference sequences HXB2 (accession number K03455) and NL4-3 (M19921) are also included.
To more accurately represent intrapatient diversity, individual viral variants derived from plasma and liver biopsies were compared. In individuals with available gag sequence data, the median intrapatient genetic distance was 1.36% in the plasma (range: 0.80–3.12%) and 1.09% in the liver biopsy tissue (range: 0.00–2.77%) as shown in Supplementary Fig. S1A (Supplementary Data are available online at www.liebertonline.com/aid) [p=not significant (NS)]. Median gag entropy, which represents both the number of distinct variants as well as their frequencies, was 1.00 in the plasma (range: 0.84–1.00) and 0.83 in the liver biopsy tissue (range: 0.00–1.00) as shown in Supplementary Fig. S1C (p=0.021). As an indicator of positive selection, dN-dS values were calculated for gag but were not greater than 0 for any plasma or liver biopsy tissue analyzed. Median dN-dS values were −0.045 (range: −0.082 to −0.026) and −0.30 (–0.053–0.00) in the plasma and liver biopsy tissue, respectively (Supplementary Fig. S1E; p=NS).
In individuals with available env sequence data, the median intrapatient genetic distance was 2.17% in the plasma (range: 0.00–6.47%) and 0.16% in the liver biopsy tissue (range: 0.00–1.85%) (Supplementary Fig. S1B; p=0.059). Median env entropy was 0.77 in the plasma (range: 0.18 – 1.00) and 0.36 in the liver biopsy tissue (range: 0.00–0.84) (Supplementary Fig. S1D; p=NS). Values for dN-dS were greater than 0 for one plasma sample (1580) and three liver biopsies (Cin05, 1580, and 1756). Median dN-dS values were −0.0032 (range: −0.0250–0.0234) and 0.0003 (–0.0205–0.0147) in plasma and liver biopsy samples, respectively (Supplementary Fig. S1F; p=NS).
Coreceptor utilization
Envelope coreceptor utilization was assessed with pretrained classifier algorithms using consensus and clonal envelope sequences. For viruses from individuals 1756, HV52, Cin01, Cin02, Cin03, Cin04, and Cin05, the consensus envelope sequences from plasma and liver were predicted to utilize the CCR5 coreceptor. However, for subject 1580, the envelope sequences from plasma and liver were predicted to utilize the CXCR4 coreceptor. When clonal sequences were analyzed, the majority (64–100%) of viruses from the plasma and liver were predicted to utilize CCR5 for subjects 1756, Cin01, Cin03, Cin04, and Cin05. In contrast, two algorithms (C4.5 and C4.5–only p8 and p12) predicted CCR5 in all clones for subject 1580 regardless of the cell/tissue type, while three algorithms (PART, SVM, and the Charge Rule) predicted that only 0–22% of plasma-derived clones and 11–44% of liver-derived clones utilized CCR5. These findings are further supported by in vitro studies suggesting that liver cell infection may utilize CCR5 or CXCR4.16,17,19
Analysis of compartmentalization
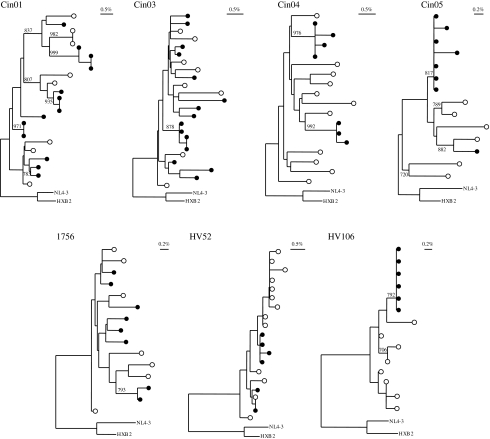
Phylogenetic trees were also reconstructed for each of the individuals with both plasma and liver biopsy variants (Fig. 2). In the analysis of intrapatient gag variability, three distinct patterns were observed. Pattern 1 included one individual (HV52) with plasma- and liver-derived variants that were intermingled with no statistically significant clustering of these variants by sample source. Pattern 2 included four individuals (Cin01, Cin03, Cin04, 1756) with plasma- and liver-derived variants that were intermingled but with at least one statistically significant clustering of variants by sample source. Pattern 3 included two individuals (Cin05 and HV106) with complete, or near complete, separation of plasma- and liver-derived variants into distinct groupings that were supported by high bootstrap values.
FIG. 2.
Patient-specific phylogenetic trees with gag viral variants from plasma (open circles) and liver biopsy tissue (closed circles). Shown in the upper right corner is a bar depicting the percent genetic distance for each tree. Only relevant bootstraps greater than 700 out of 1000 are shown. HXB2 (accession number K03455) and NL4-3 (M19921) are included as references.
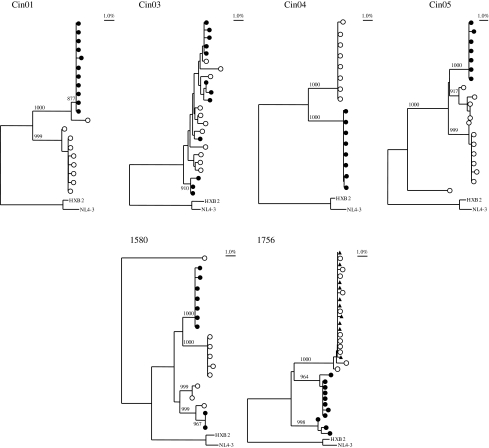
In the analysis of intrapatient env variability, only patterns 2 and 3 were observed (Fig. 3). For example, in individuals Cin03 and 1580, some intermingling of plasma- and liver-derived variants was observed, although there were also smaller groupings of variants that clustered by sample source. In contrast, for individuals Cin01, Cin04, Cin05, and 1756, viral variants from the liver clearly clustered separately from viral variants from the plasma and were supported by high bootstrap values. For 1756, env sequences from peripheral blood mononuclear cells (PBMCs) were also available for analysis and demonstrated clear clustering of PBMC- and plasma-derived variants that was distinct from the cluster of liver-derived variants. A replicate RT-PCR of env using a second RNA extraction from the same liver biopsy, plasma, and PBMCs samples of subject 1756 also demonstrated evidence of significant compartmentalization between the liver and plasma/PBMCs (data not shown).
FIG. 3.
Patient-specific phylogenetic trees with env viral variants from plasma (open circles) and liver biopsy tissue (closed circles). For subject 1756, viral variants from peripheral blood mononuclear cells (PBMCs) (closed triangles) were also included. Shown in the upper left corner is a bar depicting the percent genetic distance for each tree. Only relevant bootstrap values greater than 700 out of 1000 are shown. HXB2 (accession number K03455) and NL4-3 (M19921) are included as references.
We further explored the potential compartmentalization of HIV in the liver using Mantel's test as shown in Table 2. When comparing matched plasma- and liver biopsy-derived viral variants in the gag region of HIV, the results of Mantel's test were consistent with significant quasispecies compartmentalization for Cin05 (p=0.0009) and HV106 (p=0.0027), while Cin04 (p=0.066) and HV52 (p=0.091) showed a trend toward compartmentalization. No significant compartmentalization in gag was observed for 1756, Cin01, or Cin03. S-M test results demonstrated statistically significant evidence of gag compartmentalization for Cin04, Cin05, HV52, and HV106.
Table 2.
p-Values from Mantel's Test and Slatkin–Maddison Test for HIV Compartmentalization in the Liver Compared to the Plasma
| Patient ID | Gag Mantel's test | Gag Slatkin–Maddison test | Env Mantel's test | Env Slatkin–Maddison test |
|---|---|---|---|---|
| Cin01 | NSa | NS | 0.0001 | <0.0001 |
| Cin03 | NS | NS | 0.060 | 0.0049 |
| Cin04 | 0.066 | 0.0109 | 0.0005 | 0.0001 |
| Cin05 | 0.0009 | 0.0103 | 0.0002 | 0.0001 |
| 1580 | Not done | Not done | 0.0043 | 0.0104 |
| 1756 | NS | NS | 0.0001 | <0.0001 |
| HV52 | 0.091 | 0.0023 | Not done | Not done |
| HV106 | 0.0027 | 0.0003 | Not done | Not done |
NS, nonsignificant (p>0.10).
Evidence of significant compartmentalization in env was observed for 1580 (p=0.0043), 1756 (p=0.0001), Cin01 (p=0.0001), Cin04 (p=0.0005), and Cin05 (p=0.0002) using Mantel's test, while Cin03 (p=0.060) also showed a trend toward compartmentalization in env. S-M test results similarly demonstrated statistically significant evidence of env compartmentalization for 1580, 1756, Cin01, Cin02, Cin03, Cin04, and Cin05. Thus, all nine individuals with HIV detectable in the liver demonstrated evidence of gag and/or env compartmentalization between the plasma and liver biopsy with at least one statistical test of compartmentalization.
Signature sequence analysis of liver-specific amino acids
To identify specific amino acids associated with HIV in the liver, signature sequence analysis was performed using gag and/or env sequences for matched plasma-biopsy samples. A significant difference in amino acid frequency was identified in a single individual when analyzing gag variants in the liver biopsy compared to those in the corresponding plasma samples. For HV52, a 12 amino acid insertion was present in the liver biopsy that was present only at very low frequency among variants from the corresponding plasma. For the other five individuals with matched sequence data, no other liver-specific signature amino acids were identified in the region of gag analyzed.
In contrast, five of six individuals (Cin01, Cin04, Cin05, 1580, and 1756) had evidence of distinct amino acids frequencies when analyzing env data. For these five individuals, a total of 49 liver-specific signature amino acids (mean 9.8; range: 1–18) was identified (Fig. 4). Of these signature amino acids 14 (26.9%) were located within the V3 loop. Four amino acid signatures were shared by at least two individuals.
FIG. 4.
Signature sequence analysis showing amino acid positions at which the distributions of liver- and plasma-specific envelope variants are significantly different. Numbers in parentheses indicate the number of amino acid signatures present in the liver. Shown are the consensus amino acids at each position. In several instances, a given amino acid may appear to be identical between the two compartments; however, the frequency distribution of the viral variants that make up that consensus is different between the two compartments. Asterisks denote amino acid signatures that were shared by at least two individuals. All sequences are shown relative to their position within HXB2 with the single underlined amino acid residues denoting the V3 loop (amino acids 296–330) and the V3 tip (amino acids 312–315) double–underlined.
Discussion
To date, only one published study has examined HIV variability in the liver. Van't Wout et al. assessed HIV proviral DNA variability from a single individual who died of AIDS-related complications.46 They concluded that the presence of HIV in nonlymphoid tissues was likely the result of the late disease stage of the samples examined, although samples collected from earlier disease stages were not included. Similarly, Donaldson et al. reported that infection of nonlymphoid organs, such as the liver, occurred only in individuals with AIDS-defining illnesses and not among asymptomatic individuals11; however, HIV variability was not explored. In contrast, in the current study, HIV RNA was detected in nine individuals with a wide range of CD4 cell counts (0 to 601 cells/mm3) suggesting that HIV infection of the liver may occur at varying levels of immunosuppression.
Importantly, several lines of evidence suggest that the liver may represent a potential site of HIV compartmentalization. First, the presence of distinct consensus sequences in the liver compared to the corresponding plasma/PBMCs argues against simple contamination of biopsy tissues with peripheral lymphocytes and/or cell-free virions from the peripheral blood supply. Second, patient-specific clonal analysis frequently demonstrated distinct HIV variants in the liver compared to the corresponding plasma. These findings were further supported statistically by the results of both Mantel's and Slatkin–Maddison tests. Third, signature sequence analysis identified 49 amino acid residues in the region of env sequenced that were associated with HIV detection in the liver. These amino acids imply adaptation of HIV for infection of the liver and may impact HIV replication and/or cell tropism.
Despite frequent observation of liver enzyme abnormalities in those with HIV infection, HIV treatment providers rarely include liver biopsy in the evaluation process of these patients. Liver biopsy is more commonly employed in patients with HBV and/or HCV infection; therefore, few investigators have had access to samples that would permit assessment of the direct effects of HIV on the liver in vivo. While our methodology does not provide any information regarding the specific cell type(s) that may be infected by HIV or the overall level of HIV replication in the liver, there is considerable evidence suggesting that several liver cell types can support HIV replication as reviewed elsewhere.1 Thus, it is reasonable to assume that there is at least one cell type in the liver that is capable of supporting HIV replication. Although we cannot definitively rule out contamination by PBMC-derived HIV, this is unlikely for two reasons. First, several studies have shown little or no compartmentalization, or similar mutational patterns, in PBMCs compared to plasma/serum.47–49 Second, when PBMCs were included from subject 1756, PBMC- and plasma-derived viruses grouped together but were separate from liver-derived sequences.
Several limitations of the current study warrant further discussion. While the population size is modest, these data represent the largest study of HIV variability in the liver ever performed. Additionally, while several distinct methods to detect viral compartmentalization are available, there is no one gold standard or preferred approach. For example, Zarate et al. compared multiple methods for detecting HIV compartmentalization and found that discordant predictions by distinct methods may occur; therefore, utilizing several complementary methods, as performed here, provides the most reliable assessment of viral compartmentalization.50 Furthermore, the cross-sectional nature of this analysis does not permit a detailed examination of liver-specific HIV variants over time or provide important data on the possible trafficking of liver-specific HIVs into the peripheral circulation. Similarly, it is possible that our cloning strategy may not have amplified all minor variants present in a given tissue/cell type.
Our study is the first to explore HIV RNA variability in the liver and to demonstrate distinct HIV variants in liver biopsy tissues. However, it is important to note that HIV was not amplifiable in all samples examined, although we did not directly quantify intrahepatic levels of HIV in the current study. Thus, identification of the virologic, immunologic, and genetic factors that impact HIV detection in the liver will require additional study. Moreover, exploring the variability of additional HIV genomic regions amplified from the liver, as well as detecting low-frequency viral variants by single-genome amplification, is warranted and may enhance our understanding of HIV pathogenesis. Finally, additional studies are currently underway to determine the relative contributions of distinct liver cell types to HIV pathogenesis and viral diversity, as well as the impact of liver-derived HIV on liver damage and fibrosis progression.
Supplementary Material
Acknowledgments
This work has been supported in part by a Dean's Scholar Award from the University of Cincinnati College of Medicine to J.T.B. and by NIAID R01 (AI 065256) to K.E.S. This work was presented at the 15th Conference on Retroviruses and Opportunistic Infections held in Boston, Massachusetts from February 3 to 6, 2008 and the 5th International HIV and Hepatitis Co-Infection Workshop held in Lisbon, Portugal from June 4 to 6, 2009. We acknowledge use of liver tissue provided by the National Disease Research Interchange (NDRI), with support from NIH grant 5 U42 RR006042.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Blackard JT. Sherman K. HCV/HIV co-infection: Time to re-evaluate the role of HIV in the liver? J Viral Hepat. 2008;15(5):323–330. doi: 10.1111/j.1365-2893.2008.00970.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keaveny AP. Karasik M. Hepatobiliary and pancreatic infections in AIDS: Part one. AIDS Patient Care STDs. 1998;12(5):347–357. doi: 10.1089/apc.1998.12.347. [DOI] [PubMed] [Google Scholar]
- 3.Kahn J. Walker B. Acute human immunodeficiency virus type 1 infection. N Engl J Med. 1998;339(1):33–39. doi: 10.1056/NEJM199807023390107. [DOI] [PubMed] [Google Scholar]
- 4.Bonacini M. Hepatobiliary complications in patients with human immunodeficiency virus infection. Am J Med. 1992;92:404–411. doi: 10.1016/0002-9343(92)90271-c. [DOI] [PubMed] [Google Scholar]
- 5.Ingiliz P. Valantin MA. Duvivier C, et al. Liver damage underlying unexplained transaminase elevation in human immunodeficiency virus-1 mono-infected patients on antiretroviral therapy. Hepatology. 2009;49(2):436–442. doi: 10.1002/hep.22665. [DOI] [PubMed] [Google Scholar]
- 6.Sterling R. Wilson M. Sanyal A, et al. Impact of highly active antiretroviral therapy on the spectrum of liver disease in HCV-HIV coinfection. Clin Gastroenterol Hepatol. 2004;2(5):432–439. doi: 10.1016/s1542-3565(04)00129-6. [DOI] [PubMed] [Google Scholar]
- 7.Mata-Marin JA. Gaytan-Martinez JE. Grados-Chavarria BH. Fuentes-Allen JL. Arroyo-Anduiza CI. Alfaro-Mejia A. Correlation between HIV viral and aminotransferases as liver damage markers in HIV infected naive patients: A concordance cross-sectional study. Virol J. 2009;6:181. doi: 10.1186/1743-422X-6-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rhee MS. Sterling RK. McGovern B. Knox TA. Terrin N. Forrester J. The effect of HIV RNA on the severity of liver disease in HIV-infected hispanics; Paper presented at the American Association for the Study of Liver Diseases; Oct-Nov;2008 ; San Francisco, CA. [Google Scholar]
- 9.Blackard JT. Welge JA. Taylor LE, et al. HIV mono-infection is associated with FIB-4–a noninvasive index of liver fibrosis–in women. Clin Infect Dis. 2011;52(5):674–680. doi: 10.1093/cid/ciq199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao Y. Dieterich D. Thomas P. Huang Y. Mirabile M. Ho D. Identification and quantitation of HIV-1 in the liver of patients with AIDS. AIDS. 1992;6(1):65–70. doi: 10.1097/00002030-199201000-00008. [DOI] [PubMed] [Google Scholar]
- 11.Donaldson YK. Bell JE. Ironside JW, et al. Redistribution of HIV outside the lymphoid system with onset of AIDS. Lancet. 1994;343(8894):383–385. doi: 10.1016/s0140-6736(94)91222-x. [DOI] [PubMed] [Google Scholar]
- 12.Housset C. Lamas E. Brechot C. Detection of HIV1 RNA and p24 antigen in HIV-1-infected human liver. Res Virol. 1990;141:153–159. doi: 10.1016/0923-2516(90)90017-d. [DOI] [PubMed] [Google Scholar]
- 13.Housset C. Lamas E. Courgnaud V, et al. Presence of HIV-1 in human parenchymal and non-parenchymal liver cells in vivo. J Hepatol. 1993;19(2):252–256. doi: 10.1016/s0168-8278(05)80579-3. [DOI] [PubMed] [Google Scholar]
- 14.Zhu M. Duan L. Pomerantz R. TAR- and Tat-independent replication of human immunodeficiency virus type 1 in human hepatoma cells. AIDS Res Hum Retroviruses. 1996;12(12):1093–1101. doi: 10.1089/aid.1996.12.1093. [DOI] [PubMed] [Google Scholar]
- 15.Pizzella T. Banerjee R. Identification of a human immunodeficiency virus type 1 TAR binding protein in human hepatoblastoma HepG2 cells that trans-activates HIV-1 LTR-directed gene expression. DNA Cell Biol. 1994;13(1):67–74. doi: 10.1089/dna.1994.13.67. [DOI] [PubMed] [Google Scholar]
- 16.Ma G. Moreno M. Cardona Maya W, et al. HIV infection of an HCV-producing hepatocyte cell line—a model system for exploring HIV-HCV co-infection in vitro; Paper presented at the 13th International Symposium on Viral Hepatitis and Liver Disease; Mar 20–24;2009 ; Washington, DC. [Google Scholar]
- 17.Iser DM. Warner N. Revill PA, et al. Coinfection of hepatic cell lines with human immunodeficiency virus and hepatitis B virus leads to an increase in intracellular hepatitis B surface antigen. J Virol. 2010;84(12):5860–5867. doi: 10.1128/JVI.02594-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Banerjee R. Sperber K. Pizzella T. Mayer L. Inhibition of HIV-1 productive infection in hepatoblastoma HepG2 cells by recombinant tumor necrosis factor-a. AIDS. 1992;6(10):1127–1131. doi: 10.1097/00002030-199210000-00010. [DOI] [PubMed] [Google Scholar]
- 19.Xiao P. Usami O. Suzuki Y, et al. Characterization of a CD4-independent clinical HIV-1 that can efficiently infect human hepatocytes through chemokine (C-X-C motif) receptor 4. AIDS. 2008;22(14):1749–1757. doi: 10.1097/QAD.0b013e328308937c. [DOI] [PubMed] [Google Scholar]
- 20.Blackard JT. Cohen DE. Mayer K. Human immunodeficiency virus superinfection and recombination: Current state of knowledge and potential clinical consequences. Clin Infect Dis. 2002;34(8):1108–1114. doi: 10.1086/339547. [DOI] [PubMed] [Google Scholar]
- 21.Shioda T. Oka S. Xin X, et al. In vivo sequence variability of human immunodeficiency virus type 1 envelope gp120: Association of V2 extension with slow disease progression. J Virol. 1997;71(7):4871–4881. doi: 10.1128/jvi.71.7.4871-4881.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ganeshan S. Dickover RE. Korber BT. Bryson YJ. Wolinsky S. Human immunodeficiency virus type 1 genetic evolution in children with different rates of development of disease. J Virol. 1997;71(1):663–677. doi: 10.1128/jvi.71.1.663-677.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delwart EL. Pan H. Sheppard HW, et al. Slower evolution of human immunodeficiency virus type 1 quasispecies during progression to AIDS. J Virol. 1997;71(10):7498–7508. doi: 10.1128/jvi.71.10.7498-7508.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGrath KM. Hoffman NG. Resch W. Nelson JA. Swanstrom R. Using HIV-1 sequence variability to explore virus biology. Virus Res. 2001;76(2):137–160. doi: 10.1016/s0168-1702(01)00271-4. [DOI] [PubMed] [Google Scholar]
- 25.Liuzzi G. Chirianni A. Clementi M, et al. Analysis of HIV-1 load in blood, semen and saliva: Evidence for different viral compartments in a cross-sectional and longitudinal study. AIDS. 1996;10(14):F51–F56. doi: 10.1097/00002030-199612000-00001. [DOI] [PubMed] [Google Scholar]
- 26.Coombs RW. Speck CE. Hughes JP, et al. Association between culturable human immunodeficiency virus type 1 (HIV-1) in semen and HIV-1 RNA levels in semen and blood: evidence for compartmentalization of HIV-1 between semen and blood. J Infect Dis. 1998;177(2):320–330. doi: 10.1086/514213. [DOI] [PubMed] [Google Scholar]
- 27.Kiessling AA. Fitzgerald LM. Zhang D, et al. Human immunodeficiency virus in semen arises from a genetically distinct virus reservoir. AIDS Res Hum Retroviruses. 1998;14(Suppl 1):S33–S41. [PubMed] [Google Scholar]
- 28.Ping LH. Cohen MS. Hoffman I, et al. Effects of genital tract inflammation on human immunodeficiency virus type 1 V3 populations in blood and semen. J Virol. 2000;74(19):8946–8952. doi: 10.1128/jvi.74.19.8946-8952.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paranjpe S. Craigo J. Patterson B, et al. Subcompartmentalization of HIV-1 quasispecies between seminal cells and seminal plasma indicates their origin in distinct genital tissues. AIDS Res Hum Retroviruses. 2002;18(17):1271–1280. doi: 10.1089/088922202320886316. [DOI] [PubMed] [Google Scholar]
- 30.Ritola K. Robertson K. Fiscus SA. Hall C. Swanstrom R. Increased human immunodeficiency virus type 1 (HIV-1) env compartmentalization in the presence of HIV-1-associated dementia. J Virol. 2005;79(16):10830–10834. doi: 10.1128/JVI.79.16.10830-10834.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harrington PR. Haas DW. Ritola K. Swanstrom R. Compartmentalized human immunodeficiency virus type 1 present in cerebrospinal fluid is produced by short-lived cells. J Virol. 2005;79(13):7959–7966. doi: 10.1128/JVI.79.13.7959-7966.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang J. Jozwiak R. Wang B, et al. Unique HIV type 1 V3 region sequences derived from six different regions of brain: Region-specific evolution within host-determined quasispecies. AIDS Res Hum Retroviruses. 1998;14(1):25–30. doi: 10.1089/aid.1998.14.25. [DOI] [PubMed] [Google Scholar]
- 33.Jacobs GB. de Beer C. Fincham JE, et al. Serotyping and genotyping of HIV-1 infection in residents of Khayelitsha, Cape Town, South Africa. J Med Virol. 2006;78(12):1529–1536. doi: 10.1002/jmv.20735. [DOI] [PubMed] [Google Scholar]
- 34.Blackard JT. Hiasa Y. Smeaton L, et al. Compartmentalization of hepatitis C virus (HCV) during HCV/HIV coinfection. J Infect Dis. 2007;195(12):1765–1773. doi: 10.1086/518251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Larkin MA. Blackshields G. Brown NP, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;2007(23):21. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 36.Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 37.Drummond AJ. Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolution Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wolinksy S. Korber B. Neumann A, et al. Adaptive evolution of human immunodeficiency virus-type 1 during the natural course of infection. Science. 1996;272:537–542. doi: 10.1126/science.272.5261.537. [DOI] [PubMed] [Google Scholar]
- 39.Kumar S. Tamura K. Jakobsen I. Nei M. MEGA2: Molecular evolutionary genetics analysis software. Bioinformatics. 2001;17(12):1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- 40.MacClade 4: Analysis of phylogeny and character evolution [computer program]. Version. Sunderland, MA: Sinauer Associates; 2003. [Google Scholar]
- 41.Korber B. Myers G. Signature pattern analysis: A method for assessing viral sequence relatedness. AIDS Res Hum Retroviruses. 1992;8(9):1549–1560. doi: 10.1089/aid.1992.8.1549. [DOI] [PubMed] [Google Scholar]
- 42.Collins KR. Quiñones-Mateu ME. Wu M, et al. Human immunodeficiency virus type 1 (HIV-1) quasispecies at the sites of Mycobacterium tuberculosis infection contribute to systemic HIV-1 heterogeneity. J Virol. 2002;76(4):1697–1706. doi: 10.1128/JVI.76.4.1697-1706.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poss M. Rodrigo AG. Gosink JJ, et al. Evolution of envelope sequences from the genital tract and peripheral blood of women infected with clade A human immunodeficiency virus type 1. J Virol. 1998;72(10):8240–8451. doi: 10.1128/jvi.72.10.8240-8251.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Slatkin M. Maddison W. A cladistic measure of gene flow inferred from the phylogenies of alleles. Genetics. 1989;123:603–613. doi: 10.1093/genetics/123.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pond SL. Frost SD. Muse S. HyPhy: Hypothesis testing using phylogenetics. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 46.van't Wout A. Ran L. Kuiken C. Kootstra N. Pals S. Schuitemaker H. Analysis of the temporal relationship between human immunodeficiency virus type 1 quasispecies in sequential blood samples and various organs obtained at autopsy. J Virol. 1998;72(1):488–496. doi: 10.1128/jvi.72.1.488-496.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ince WL. Harrington PR. Schnell GL, et al. Major coexisting human immunodeficiency virus type 1 env gene subpopulations in the peripheral blood are produced by cells with similar turnover rates and show little evidence of genetic compartmentalization. J Virol. 2009;83(9):4068–4080. doi: 10.1128/JVI.02486-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sarmati L. Nicastri E. Uccella I, et al. Drug-associated resistance mutations in plasma and peripheral blood mononuclear cells of human immunodeficiency virus type 1-infected patients for whom highly active antiretroviral therapy is failing. J Clin Microbiol. 2003;41(4):1760–1762. doi: 10.1128/JCM.41.4.1760-1762.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Devereux HL. Loveday C. Youle M. Sabin CA. Burke A. Johnson M. Substantial correlation between HIV type 1 drug-associated resistance mutations in plasma and peripheral blood mononuclear cells in treatment-experienced patients. AIDS Res Hum Retroviruses. 2000;16(11):1025–1030. doi: 10.1089/08892220050075273. [DOI] [PubMed] [Google Scholar]
- 50.Zárate S. Pond SL. Shapshak P. Frost S. Comparative study of methods for detecting sequence compartmentalization in human immunodeficiency virus type 1. J Virol. 2007;81(12):6643–6651. doi: 10.1128/JVI.02268-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.