Abstract
Lipopolysaccharide (LPS)-induced production of tumor necrosis factor (TNF)-α requires the recruitment of two pairs of adaptors to the Toll-like receptor 4 cytoplasmic domain. The contribution of one pair – Toll-interleukin-1 receptor domain-containing adaptor inducing interferon-β (TRIF) and TRIF-related adaptor molecule (TRAM) – to TNF-α expression is not well understood. To clarify this issue, we studied TRAM knockout bone marrow-derived macrophages (BMDM). LPS-stimulated TRAM-deficient BMDM had decreased TNF-α protein expression even at times when TNF-α mRNA levels were normal, suggesting impaired translation. Consistent with this idea, knockdown of TRAM in RAW264.7 macrophages decreased translation of a reporter controlled by the TNF-α 3′ untranslated region, while transfection of TRAM in HEK293T cells increased translation of this reporter. Also consistent with a role for TRAM in TNF-α translation, LPS-induced activation of MK2, a kinase involved in this process, was impaired in TRAM-deficient BMDM. TRIF did not increase translation of the TNF-α 3′ untranslated region reporter when expressed in HEK293T cells. However, BMDM that lacked functional TRIF produced reduced levels of TNF-α protein in response to LPS despite normal amounts of the mRNA. Unlike BMDM, LPS-stimulated TRAM-deficient peritoneal macrophages displayed equivalent reductions in TNF-α protein and mRNA. Our results indicate that TRAM- and TRIF-dependent signals have a previously unappreciated, cell type-specific role in regulating TNF-α translation.
Copyright © 2011 S. Karger AG, Basel
Key Words: Macrophage, Lipopolysaccharide, Toll-like receptor, Inflammation, Signal transduction
Introduction
Toll-like receptor 4 (TLR4) is the major mammalian pattern recognition receptor for lipopolysaccharide (LPS) and is expressed on macrophages and other cells of the innate immune system. It plays an important role in the response to several Gram-negative bacterial pathogens, including Salmonella enterica, as we and others have shown previously [1,2,3,4]. TLR4 activates distinct signal transduction pathways by recruiting 2 sets of adaptor proteins – myeloid differentiation factor 88 (MyD88) paired with MyD88-like adaptor protein (Mal), and Toll-interleukin-1 receptor (TIR) domain-containing adaptor inducing interferon-β (TRIF) paired with TRIF-related adaptor molecule (TRAM) – to its cytoplasmic domain, with Mal and TRAM acting as bridges to MyD88 and TRIF, respectively [5,6]. All 4 of these proteins contain conserved TIR domains that are involved in interactions with the TIR domain of either TLR4, in the case of Mal and TRAM, or the corresponding bridging adaptor, in the case of MyD88 and TRIF [6].
The adaptor protein TRAM is utilized uniquely by TLR4 and is not recruited to the cytoplasmic domains of other TLRs [5,6]. It consists almost entirely of a TIR domain, which, in the mouse protein, extends from amino acid 75 to amino acid 232 at the carboxy terminus. Single amino acid substitutions within this domain (P116H, C117H) have been shown to disrupt the weak interaction with TRIF in a yeast two-hybrid assay and the more robust TLR4-TRAM-TRIF trimolecular interaction in coimmunoprecipitation assays with transfected HEK293T cells [7]. Although the amino-terminal portion of TRAM is less well conserved than the TIR domain, it contains several functionally important motifs, including a myristoylation signal required for membrane localization and signaling (residues 1–7), a poly-basic, phospholipid binding region (residues 8–20) that contributes to the normal pattern of distribution on the plasma membrane and endosomes, a protein kinase C∊ phosphorylation site required for signal transduction (serine 16), and potential proline, glutamic acid, serine-threonine-rich sequences that could be involved in degradation [7,8,9,10].
The Mal/MyD88 pathway leads to increased expression of inflammatory cytokines such as tumor necrosis factor (TNF)-α, whereas the major function of the TRAM/TRIF pathway is the transcriptional upregulation of innate interferons. However, TRAM- and TRIF-dependent signals are also known to contribute to LPS-induced TNF-α expression. Deficiency in either TRAM or TRIF results in a significant reduction in the amount of TNF-α secreted when macrophages are stimulated with LPS [11,12]. While it is generally accepted that the main role of the Mal/MyD88 pathway in the upregulation of inflammatory cytokine expression is to activate transcription factors such as nuclear factor (NF)-κB, the exact contribution of TRAM/TRIF-dependent signals to the expression of these cytokines is not well understood. The biosynthesis of cytokines such as TNF-α can be complex: expression can be regulated at the level of transcription, mRNA stability and translation, with the post-transcriptional effects being mediated by the interaction of regulatory proteins with cis-acting sequences, including AU-rich elements (AREs), in the 3′ untranslated region (UTR) of the mRNA [13,14]. One potential influence of TRAM/TRIF-dependent signals on TNF-α expression could be transcriptional, via the activation of NF-κB. However, this effect is likely to be minor given that NF-κB activation by the TRAM/TRIF pathway is much less prominent and more delayed compared to activation by the Mal/MyD88 pathway [11,12,15,16].
Earlier studies from our group raised the possibility that the TRAM/TRIF signaling pathway might be involved in promoting the translation of mRNAs encoding cytokines such as TNF-α [17,18]. We found that macrophages from the Hfe knockout mouse strain, a model of human type I hemochromatosis [19,20,21], had an impairment in the expression of TNF-α and IL-6 in response to LPS stimulation and Salmonella infection in vitro that correlated with a significant attenuation of Salmonella-induced enterocolitis in vivo [17]. We suggested that this impairment was at the level of translation based on analysis of multiple parameters, including cytokine protein and mRNA levels, and the differential association of cytokine transcripts with polyribosomes in macrophage cell lines that mimicked the wild-type and Hfe knockout phenotypes [17]. In subsequent studies, we showed that Hfe-deficient macrophages had an abnormality of TLR4 signaling that selectively affected TRAM/TRIF-dependent responses but not Mal/MyD88-dependent responses [18]. Taken together, our results are consistent with the novel idea that signals transduced by the TRAM/TRIF pathway specifically regulate cytokine mRNA translation. In the present work, we address this possibility directly and demonstrate that TRAM has a clear role in activating signals that enhance TNF-α translation, with TRIF also contributing to this function.
Materials and Methods
Macrophages
Bone marrow from wild-type C57BL/6J, TRAM knockout [12] and TRIF mutant (C57BL/6J-Ticam1Lps2/J, Jackson Laboratory, Bar Harbor, Me., USA) [22] strains of mice was cultured at 2 × 106 cells/ml per well in 24-well tissue culture plates in the presence of 10% L929 conditioned medium. After 6–7 days, the differentiated bone marrow-derived macrophages (BMDM) were stimulated with ultra-pure LPS (List Biological Laboratories, Campbell, Calif., USA) using the doses and times indicated in the individual experiments. Thioglycollate-elicited peritoneal macrophages were prepared as previously described [2] and stimulated with LPS in the same way as the BMDM. Supernatants were collected and analyzed by enzyme-linked immunosorbent assay (ELISA) to determine cytokine concentrations as previously described [17,18]. At the same time, total RNA was prepared from the cells using Trizol (Invitrogen, Carlsbad, Calif., USA) according to the manufacturer's recommendations and analyzed by quantitative RT-PCR to determine relative mRNA levels by the 2−ΔΔCt method, using the housekeeping transcripts glyceraldehyde phosphate dehydrogenase (GAPDH) or 36B4 for normalization. Previously published primer sequences and methods were used for these experiments [17,18]. All studies involving animals were approved by the Subcommittee on Research Animal Care of Massachusetts General Hospital.
Knockdown of TRAM in RAW264.7 Cells
The RAW264.7 murine macrophage cell line was transfected with 50 nM TRAM siRNA (Dharmacon, Chicago, Ill., USA; target sequence: 5′ CGAGAUGCCGUGCGGAAGA 3′) or a pool of irrelevant control siRNAs (provided by Dharmacon) using Lipofectamine 2000 (Invitrogen, Carlsbad, Calif., USA) according to the manufacturer's directions. TRAM expression was assessed by quantitative RT-PCR 48 h after transfection following stimulation of the cells for 6 h with 100 ng/ml of LPS. In experiments evaluating the effect of TRAM knockdown on translational efficiency, 20 ng of a plasmid encoding the TNF-α full-length 3′ UTR luciferase reporter (described below) and a constitutively expressed Renilla luciferase construct were mixed with siRNA at the time of transfection. Translational efficiency was determined as described below at 48 h after siRNA transfection following stimulation of the cells for 6 h with 100 ng/ml of LPS.
Analysis of Luciferase Expression and Translational Efficiency in HEK293T Cells
HEK293T cells were transfected using Lipofectamine 2000 with 20 ng of plasmids encoding the firefly luciferase open reading frame under the transcriptional control of a constitutively expressed cytomegalovirus promoter and connected to 1 of 3 different 3′ UTRs – the full-length, 795 nucleotide human TNF-α 3′ UTR, a control 3′ UTR of the same length, or the minimal 34 nucleotide ARE from the TNF-α 3′ UTR. Included in the transfection mix were 20 ng of a constitutively expressed Renilla luciferase reporter connected to a control 3′ UTR (to normalize for transfection efficiency) and 100 ng of a plasmid encoding a green fluorescent protein (GFP) fusion with the full-length TRAM protein [10]. The luciferase reporters were kindly provided by Drs. Shobha Vasudevan and Joan Steitz of Yale University and have been described in detail earlier [23]. Forty-eight hours after transfection, an aliquot of the transfected cells was used to measure firefly luciferase and Renilla luciferase activities on a Synergy 2 luminometer (BioTek, Winooski, Vt., USA) using the Dual-Luciferase Reporter Assay System (Promega, Madison, Wisc., USA) according to the manufacturer's instructions. Another aliquot of the transfected cells was used to prepare total RNA for measurement of firefly and Renilla luciferase mRNA levels by quantitative RT-PCR. As described by Vasudevan and Steitz [23], the ratio of firefly luciferase activity to firefly luciferase mRNA (normalized to the corresponding values of Renilla luciferase in each case) was used as an indicator of translational efficiency. The effect of coexpressing TRAM and other proteins on translational efficiency was calculated relative to cells that were transfected with the luciferase reporters plus empty vector plasmid.
Analysis of MK2 Activation
Wild-type and TRAM-deficient BMDM were stimulated with 100 ng/ml of LPS for 10, 20 or 60 min. The cells were washed with ice-cold PBS and lysed in buffer containing protease and phosphatase inhibitors as previously described [2]. The cleared lysate containing equivalent amounts of total protein were separated by polyacrylamide gel electrophoresis and transferred to nitrocellulose. Immunoblotting was carried out with primary antibodies specific to either the threonine 222 phosphorylated form of MK2 (Cell Signaling Technology, Danvers, Mass., USA) or β-actin (Sigma-Aldrich, St. Louis, Mo., USA), followed by the relevant fluorochrome-conjugated secondary antibodies. The blot was imaged using the Li-Cor Odyssey Infrared Imaging System (Li-Cor Biotechnology, Lincoln, Nebr., USA). The fluorescence intensities of the phospho-MK2 band were quantified, normalized to that of the corresponding actin bands and expressed relative to the unstimulated cells. To determine the role of MK2 in TNF-α expression, wild-type BMDM were stimulated with 100 ng/ml of LPS for 2 h in the presence of 1, 5 or 10 μM of the cell permeable MK2 inhibitor III (Calbiochem, EMD Chemicals, Gibbstown, N.J., USA) or an equivalent volume of the vehicle dimethylsulfoxide. TNF-α protein (secreted) and mRNA levels were determined by ELISA and quantitative RT-PCR, respectively.
Primer Sequences
Previously unpublished sequences of primers used in the various PCRs are indicated in table 1.
Table 1.
Sequences of primers used for quantitative RT-PCR
| Forward primer | Reverse primer | |
|---|---|---|
| TNF-α | CCCTCACACTCAGATCATCTTCT | GCTACGACGTGGGCTACAG |
| IL-6 | TAGTCCTTCCTACCCCAATTTCC | TTGGTCCTTAGCCACTCCTTC |
| TRAM | CGATCAAGACGGCCATGAGTC | CTCGTCGGTGTCATCTTCTGC |
| Firefly luciferase | TTCCATCTTCCAGGGATACGA | ATCATCCCCCTCGGGTGTA |
| Renilla luciferase | AAGGTGAAGTTCGTCGTCCAA | GTACAACGTCAGGTTTACCACCTTT |
| GAPDH | CCTGCACCACCAACTGCTT | ATGACCTTGCCCACAGCCT |
| 36B4 | AGATGCAGCAGATCC GCAT | GTTCTTGCCCATCAGCACC |
All sequences are indicated 5′ to 3′.
Statistical Analysis
Means ± standard deviations from multiple experiments are shown. Results were analyzed using the unpaired, 2-tailed Student t test. A p value <0.05 was considered significant.
Results
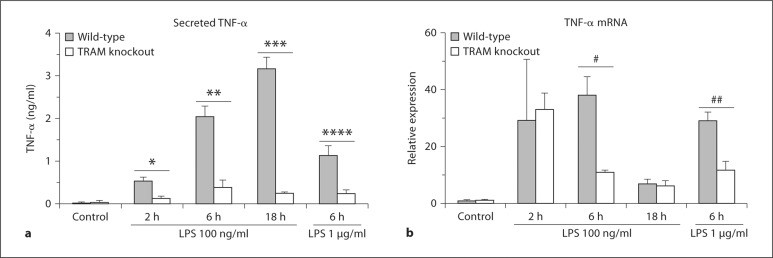
To elucidate the role of the TRAM/TRIF pathway in the regulation of TNF-α expression, we stimulated BMDM from wild-type and TRAM knockout mice with 100 ng/ml of LPS for 2, 6 and 18 h, and analyzed levels of secreted TNF-α protein and TNF-α mRNA. The TRAM-deficient cells produced significantly lower amounts of TNF-α protein than the wild-type controls at all time points (fig. 1a), in keeping with earlier reports [12]. This abnormality persisted even when the dose of LPS used was increased to 1 μg/ml. Interestingly, the levels of TNF-α mRNA were equivalent in the wild-type and mutant BMDM at the 2-hour time point, although the level fell significantly below that of the wild-type at the 6-hour time point (fig. 1b). The discordance between TNF-α protein and mRNA levels in the knockout macrophages at the 2-hour time point suggested that TRAM deficiency impaired translation of TNF-α transcripts.
Fig. 1.
Effect of TRAM deficiency on LPS-induced TNF-α protein and mRNA expression. BMDM from wild-type and TRAM knockout mice were stimulated with 100 ng/ml of LPS for 2, 6 or 18 h, or with 1 μg/ml for 6 h. Secreted TNF-α in the cell supernatants was measured by ELISA (a), while TNF-α mRNA in the corresponding cell homogenates was estimated by quantitative RTPCR (b). TNF-α mRNA levels are expressed relative to the control cells. * p < 0.0001, ** p = 0.0013, *** p = 0.0027, **** p = 0.0118, # p = 0.0169, ## p = 0.002 (n = 3-11 separate stimulations).
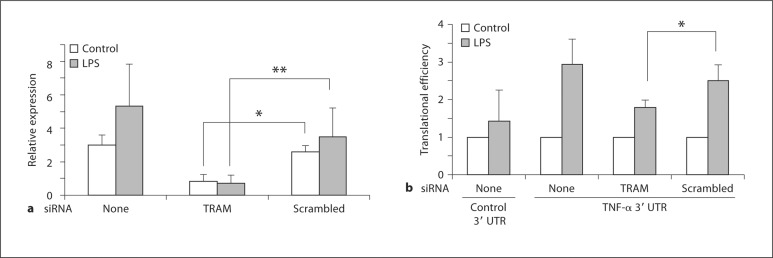
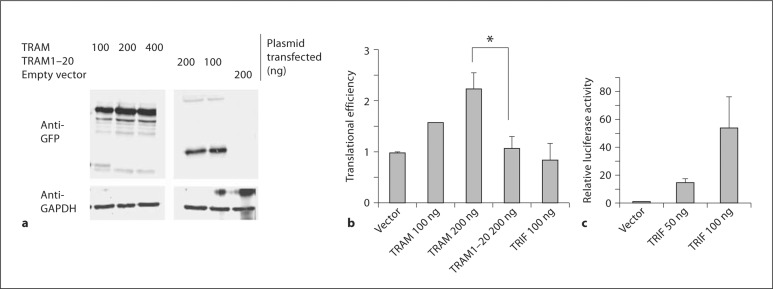
To substantiate this idea, we used siRNA-mediated knockdown and transfection approaches to evaluate the role of TRAM in TNF-α mRNA translation. In initial experiments, we transfected the RAW264.7 murine macrophage cell line with siRNA to knock down TRAM expression by 70–80% under basal and LPS-stimulated conditions (fig. 2a). As described under Materials and Methods, we then used an established assay of translational efficiency [23] to determine the effect of lowered TRAM levels on the translation of a luciferase reporter controlled by the 795 nucleotide TNF-α 3′ UTR, a region of the mRNA that contains an ARE and other important cis-acting elements that regulate translation [13]. The results of our experiment show that transfection with a TRAM-specific siRNA led to a significant attenuation of the LPS-induced increase in translational efficiency of the reporter when compared with cells transfected with an irrelevant, scrambled siRNA (fig. 2b). We also carried out the reciprocal experiment by transiently overexpressing a TRAM-GFP fusion protein in HEK293T cells (fig. 3a) to determine the effect on translation of the luciferase TNF-α 3′ UTR reporter. We found that expression of the full-length TRAM-GFP significantly enhanced the translational efficiency of the reporter (fig. 3b, c), whereas the equivalent expression of a truncated form of TRAM containing only the first 20 amino acids (fig. 3a) did not have any effect on the expression of the reporter (fig. 3b, c). Transient overexpression of TRIF did not alter the translation of the reporter either, although it did increase the expression of another luciferase reporter controlled by an interferon-sensitive response element (fig. 3b, c). The results of the knockdown and overexpression experiments, together with our observations on the TRAM-deficient macrophages, strongly support the notion that TRAM is involved in promoting the translation of TNF-α mRNA. Our findings also suggest that the TIR domain of TRAM is required for this function.
Fig. 2.
Effect of TRAM knockdown on the LPS-induced increase in TNF-α translational efficiency. a RAW264.7 cells were transfected with either TRAM siRNA or a scrambled control siRNA. Forty-eight hours later, total RNA was prepared, following 6 h of stimulation with 100 ng/ml LPS or without stimulation (control) as indicated in the figure, and TRAM expression was analyzed by quantitative RT-PCR. * p = 0.001, ** p = 0.02 (n = 5). b RAW264.7 cells were transfected with either TRAM siRNA or scrambled control siRNA, along with a constitutively expressed Renilla luciferase reporter and a firefly luciferase reporter linked to the TNF-α 3′ UTR or a control 3′ UTR. Forty-eight hours later, cell lysates were prepared, following 6 h of stimulation with 100 ng/ml LPS or without stimulation (control) as indicated in the figure, and luciferase activities and mRNA levels were determined. These values were used to calculate translational efficiency as described in the text and expressed relative to the value in unstimulated cells in each case. * p = 0.015 (n = 5).
Fig. 3.
Effect of TRAM overexpression on TNF-α 3′ UTR-dependent translational efficiency in HEK293T cells. a HEK293T cells were transfected with the indicated expression plasmids. Cell lysates were prepared 48 h later and analyzed by Western blotting with anti-GFP or anti-GAPDH antibodies. b HEK293T cells were transfected with the constitutively expressed Renilla luciferase reporter and the firefly luciferase reporter linked to the TNF-α 3′ UTR, along with the indicated TRAM expression plasmids or empty vector. Cell lysates were prepared 48 h later and used to measure luciferase activities and mRNA levels. These values were used to calculate translational efficiency as described in the text and expressed relative to cells transfected with empty vector. * p = 0.001 (n = 3-7). c To confirm TRIF function, HEK293T cells were transfected with a constitutively expressed Renilla luciferase reporter and a firefly luciferase reporter controlled by an interferon-sensitive response element, along with the indicated amounts of a TRIF expression plasmid or empty vector. Cell lysates were prepared 48 h later and luciferase activities determined. Firefly luciferase activity, normalized to Renilla luciferase, was expressed relative to cells transfected with empty vector.
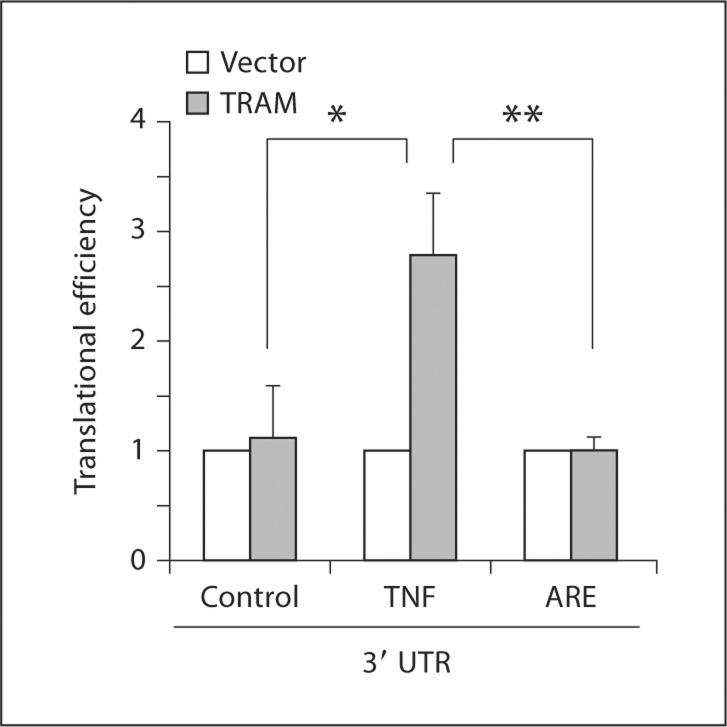
To further address the role of the TNF-α 3′ UTR in the TRAM-dependent increase in translational efficiency, we used firefly luciferase reporter constructs connected to 3 different 3′ UTRs – the full-length 795 nucleotide TNF-α 3′ UTR used in our initial experiments, a control 3′ UTR of similar length, or a minimal 34 nucleotide ARE found in the TNF-α 3′ UTR [23]. The effect of the full-length TRAM-GFP protein on translation of these constructs was determined following transfection into HEK293T cells. We found that TRAM increased translation of the reporter linked to the full-length TNF-α 3′ UTR but not of the reporters connected to the control 3′ UTR or to the minimal 34 nucleotide ARE, indicating that the latter element was not sufficient to mediate the effects of TRAM on translation (fig. 4).
Fig. 4.
Role of the TNF-α 3′ UTR in TRAM-dependent translational regulation. HEK293T cells were transfected with a constitutively expressed Renilla luciferase reporter, a firefly luciferase reporter linked to a control 3′ UTR, the full-length TNF-α 3′ UTR or the 34 nucleotide minimal ARE from the TNF-α 3′ UTR, along with either a TRAM expression plasmid or empty vector. Cell lysates were prepared after 48 h and luciferase activities and mRNA levels determined. Translational efficiency was calculated as described in the text and expressed relative to cells transfected with empty vector. * p = 0.018, ** p = 0.027 (n = 3 separate transfections each).
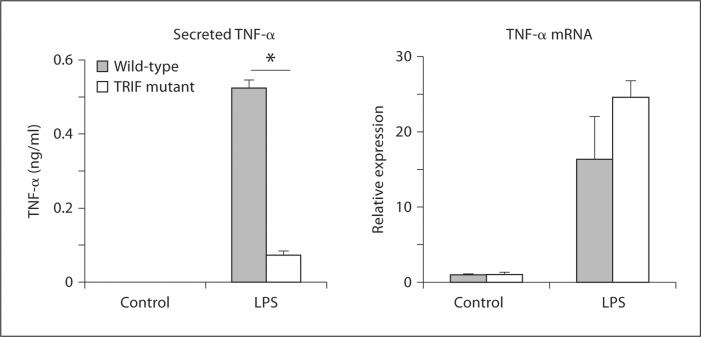
Other than for TLR4 and protein kinase C∊, TRIF is the only binding partner for TRAM that has been identified so far [24]. Accordingly, even though our data indicated that transient overexpression of TRIF was not sufficient to enhance TNF-α 3′ UTR-dependent translation (fig. 3b, c), it seemed likely that TRIF was involved in TRAM-dependent translational regulation. To address this possibility, we made use of the Lps2 strain of mice, which express a functionally inactive mutant form of TRIF [22]. BMDM from Lps2 mice were stimulated with 100 ng/ml of LPS for 2 h, and secreted TNF-α protein and TNF-α mRNA levels were determined by ELISA and quantitative RT-PCR, respectively. The results were very similar to those obtained with the TRAM knockout BMDM, i.e. the TRIF mutant macrophages produced significantly lower levels of TNF-α protein than the wild-type cells even though they expressed normal levels of the mRNA (fig. 5). The findings suggest that TRIF, like TRAM, is required for TNF-α translation in BMDM.
Fig. 5.
Role of TRIF in LPS-induced TNF-α protein and mRNA expression. BMDM from wild-type and Lps2 (TRIF-mutant) mice were stimulated for 2 h with 100 ng/ml of LPS. Secreted TNF-α in the cell supernatants was measured by ELISA, while TNF-α mRNA in the corresponding cell homogenates was estimated by quantitative RT-PCR. TNF-α mRNA levels are expressed relative to the control cells. * p < 0.0001 (n = 3 separate stimulations).
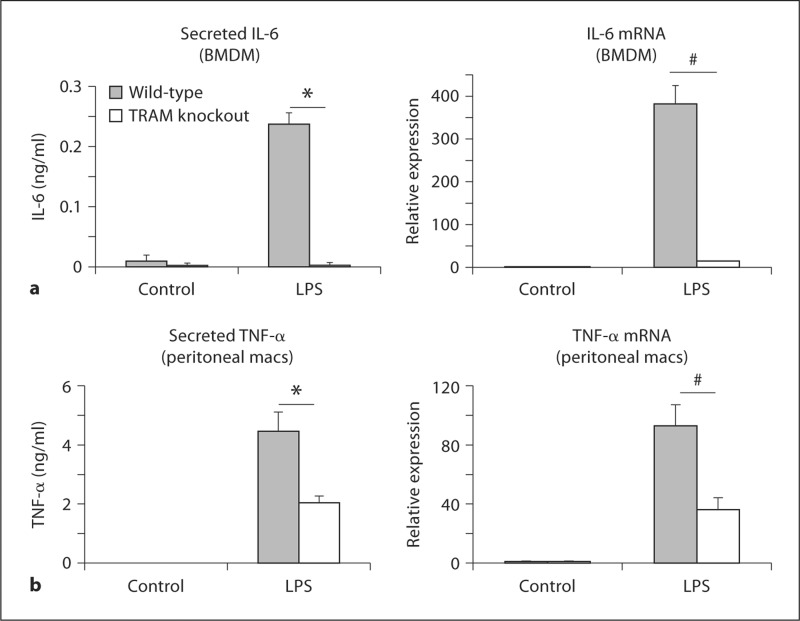
Interestingly, the effect of TRAM deficiency on LPS-induced IL-6 expression was different from the effect on TNF-α: IL-6 protein and mRNA levels were both significantly lower in the TRAM knockout BMDM than in the wild-type cells at all time points examined, an observation that was also seen with the TRIF mutant BMDM (fig. 6a; data not shown). We also examined TNF-α expression in thioglycollate-elicited peritoneal macrophages and found that TRAM deficiency led to significant decreases in both TNF-α protein and mRNA levels following 2 h of LPS stimulation, a result that was also observed with TRIF mutant peritoneal macrophages (fig. 6b; data not shown). Thus, the effect of TRAM on the translation of TNF-α appears to be specific to BMDM and does not appear to apply to IL-6.
Fig. 6.
Role of TRAM in LPS-induced IL-6 expression in BMDM and TNF-α expression in peritoneal macrophages. a BMDM from wild-type and TRAM knockout mice were stimulated for 6 h with 100 ng/ml of LPS. Secreted IL-6 in the cell supernatants was measured by ELISA, while IL-6 mRNA in the corresponding cell homogenates was estimated by quantitative RT-PCR. IL-6 mRNA levels are expressed relative to the control cells. * p = 0.0013, # p = 0.05 (n = 3 separate stimulations). b Thioglycollate-elicited peritoneal macrophages (macs) from wild-type and TRAM knockout mice were stimulated for 2 h with 100 ng/ml of LPS. Secreted TNF-α in the cell supernatants was measured by ELISA, while TNF-α mRNA in the corresponding cell homogenates was estimated by quantitative RT-PCR. TNF-α mRNA levels are expressed relative to the control cells. * p = 0.003, # p = 0.001 (n = 4 separate stimulations).
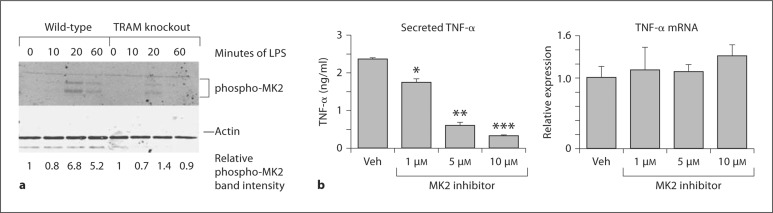
The MK2 kinase (also known as MAPKAP kinase 2) plays important roles in post-transcriptional regulation of TNF-α expression, including at the level of translation [25]. To determine if TRAM deficiency had any effect on activation of this kinase, we carried out Western blotting with an antibody specific for its phosphorylated (activated) form in LPS-stimulated wild-type and TRAM knockout BMDM. As shown in fig. 7a, LPS treatment of the wild-type cells led to an increase in MK2 activation that peaked at about 20 min but was still observable at 60 min. MK2 activation was appreciably reduced in the TRAM-deficient cells, particularly at the 60-min time point. The results thus indicate that TRAM is required for normal activation of MK2. To assess the functional relevance of this observation, we treated wild-type BMDM with an MK2 inhibitor. We found that the inhibitor produced a significant dose-dependent decrease in LPS-induced TNF-α protein expression with little or no effect on the mRNA (fig. 7b), a result that is consistent with involvement of MK2 in translation.
Fig. 7.
The role of the MK2 kinase in TRAM-dependent regulation of TNF-α translation. a Wild-type and TRAM knockout BMDM were stimulated with 100 ng/ml LPS for the times indicated. Cell lysates were immunoblotted with antibodies to phospho-MK2 or β-actin. b Wild-type BMDM were stimulated for 2 h with 100 ng/ml of LPS in the presence of the indicated concentrations of MK2 inhibitor or an equivalent volume of the vehicle (Veh). Secreted TNF-α in the cell supernatants was measured by ELISA, while TNF-α mRNA in the corresponding cell homogenates was estimated by quantitative RT-PCR. TNF-α mRNA levels are expressed relative to that in vehicle-treated cells. * p = 0.004, ** p = 0.0002, *** p < 0.00001, compared to the vehicle in each case (n = 3 separate stimulations).
Discussion
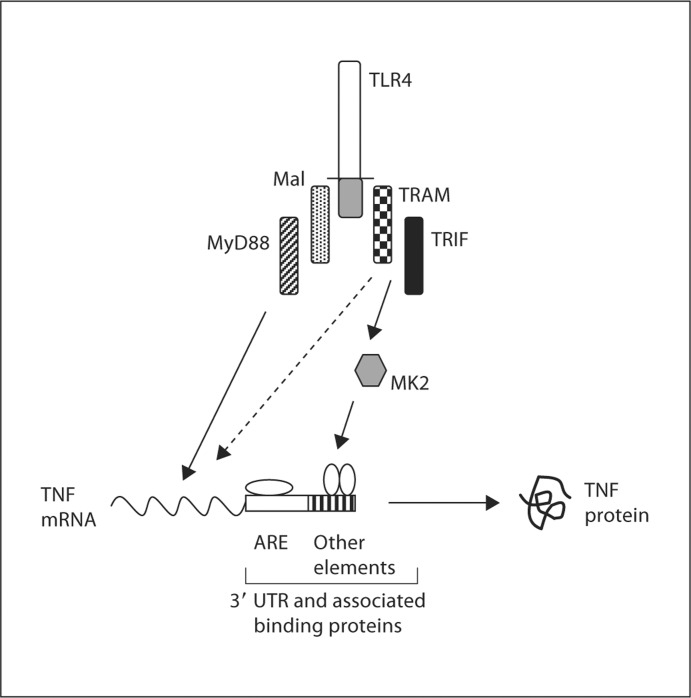
Our studies have revealed the ability of TRAM/TRIF-dependent signals to regulate TNF-α mRNA translation. Our findings help to explain why both Mal/MyD88- and TRAM/TRIF-dependent signals are required for normal TNF-α expression in response to stimulation with LPS [5,6,11,12]. Based on our data and existing information in the literature, the major consequence of Mal/MyD88-transduced signals appears to be the transcriptional upregulation of the TNF-α gene via effects on transcription factors such as NF-κB, while TRAM/TRIF-dependent signals act in a complementary fashion to activate the MK2 kinase and thus promote the translation of the TNF-α mRNA (fig. 8). The influence of TRAM/TRIF signaling on TNF-α translation appears to be mediated via effects on regulatory elements in the 3′ UTR. These elements are known to interact with RNA-binding proteins that modulate translation [13,26,27], and the signals transduced by the TRAM/TRIF pathway may alter the binding and/or function of these proteins. Our results also indicate that the 34 nucleotide minimal ARE in the TNF-α 3′ UTR is not sufficient to mediate the effects of TRAM on translation (fig. 4), suggesting the involvement of other regions of the 3′ UTR. Although the ARE plays a major role in translational regulation, there are other cis-acting elements in the TNF-α 3′ UTR that have been implicated in this process [13,26,27]. The effect of TRAM-dependent signals on TNF-α translation may be mediated by such elements. TRAM deficiency also probably affects aspects of TNF-α expression other than translation since we found that the levels of TNF-α mRNA in the TRAM knockout BMDM were significantly lower than those in wild-type BMDM 6 h after LPS stimulation (fig. 1). This would not be surprising given that several of the proteins that interact with the TNF-α 3′ UTR influence transcript stability in addition to translation, and that MK2 regulates the function of such proteins [13,25]. It should also be noted, as mentioned earlier, that the TRAM/TRIF pathway does contribute to NF-κB activation, albeit in a relatively minor and delayed fashion, and thus, could play a role in regulating TNF-α expression at the level of transcription [11,12,15,16]. Moreover, based on a combination of experimental and computational modeling data, Covert et al. [28] have suggested that TRAM/TRIF-dependent signals can function in an NF-κB-independent, IRF3-dependent fashion, to induce the production of small amounts of TNF-α, which then acts in an autocrine manner to cause the delayed activation of NF-κB and thereby promote further TNF-α transcription. Thus, the TRAM/TRIF pathway can influence TNF-α expression by multiple mechanisms.
Fig. 8.
A model of the role of TLR4 signaling pathways in regulating TNF-α expression. Mal/MyD88-dependent signals are required for the transcriptional upregulation of the TNF-α gene. In addition to a minor contribution to transcription of the TNF-α gene (dashed arrow), signals transduced via the TRAM/TRIF pathway are involved in promoting the translation of the TNF-α mRNA, probably by activating the MK2 kinase and thus influencing regulatory proteins that interact with cis-acting elements in the 3′ UTR outside the minimal 34 nucleotide ARE.
While our experiments were in progress, Gais et al. [29] reported that BMDM, as well as bone marrow-derived dendritic cells, from the TRIF mutant Lps2 mouse strain had impairments in LPS-induced activation of the MK2 kinase and TNF-α translation. Our findings complement those of Gais et al. [29] by showing that TRAM, which mediates recruitment of TRIF to the TLR4 cytoplasmic domain [6,7], is also required for LPS-induced MK2 activation and TNF-α translation. Furthermore, our data provide additional mechanistic insight by showing that TRAM influences translation in a TIR domain-dependent fashion (fig. 3b, c) and that cis-regulatory elements outside the minimal ARE in the TNF-α 3′ UTR are involved in the translational regulatory effects (fig. 4). The requirement for the TRAM TIR domain, which is needed for binding to TRIF [7], and the phenotype of the Lps2 BMDM (fig. 5) [29], strongly support the notion that regulation of TNF-α translation is carried out by TRAM and TRIF acting together (fig. 8).
Although our results (fig. 5), as well those of Gais et al. [29], indicate a requirement for TRIF in TNF-α translation, we were somewhat surprised to find that transfection of TRIF into HEK293T cells did not increase the translation of the TNF-α 3′ UTR-dependent luciferase reporter (fig. 3b, c). This observation indicates that transient expression of TRIF in HEK293T cells is not sufficient to activate TNF-α translation even though expression of TRAM under the same conditions is sufficient for this function. We do not have a definitive explanation for these findings. Given that TRAM and TRIF are both required for the effect on TNF-α translation and that HEK293T cells do not express transcripts for either of these proteins (E.T., B.J.C., unpublished data), one possible explanation is that HEK293T cells may express a protein that is able to substitute functionally for TRIF but no protein that can substitute for TRAM. Another possibility is that TRAM may be able to achieve the ‘activated’ conformation required to upregulate TNF-α translation when expressed in HEK293T cells, whereas TRIF may require additional factors (presumably present in the environment of LPS-activated macrophages but not in HEK293T cells) to attain this state. Further studies will be required to clarify these issues.
Although lack of TRAM or functional TRIF resulted in a reduction in TNF-α protein production without decrease in the mRNA (fig. 1, 5), the deficiencies of these proteins impaired IL-6 expression at the level of both protein and mRNA (fig. 6a). This result suggests that whereas a significant effect of TRAM/TRIF-transduced signals on TNF-α expression appears to be on translation, the effect on IL-6 expression may be largely at the level of transcription or post-transcriptional mRNA stability, with any influence on translation obscured by the changes in the amounts of mRNA. This idea would be consistent with the fact that although the IL-6 3′ UTR does contain an ARE, there are clear differences from the TNF-α 3′ UTR, indicative of the expression being influenced by distinct regulatory mechanisms [30]. Our observation that the ARE is not sufficient to mediate the translational effects of TRAM (fig. 4) would also be consistent with differential effects of TRAM/TRIF-dependent signals on TNF-α versus IL-6.
Like the observations of Gais et al. [29] on TRIF mutant macrophages, we found that TRAM deficiency had differential effects on LPS-induced TNF-α expression in peritoneal macrophages versus BMDM. While the absence of TRAM in BMDM significantly reduced TNF-α protein production without affecting mRNA levels (fig. 1), TRAM-deficient peritoneal macrophages showed equivalent reductions in both mRNA and protein (fig. 6b). The difference in behavior between the peritoneal macrophages and the BMDM could reflect a cell type-specific involvement of the TRAM/TRIF pathway in TNF-α translation. Alternatively, TRAM/TRIF-dependent signals could be required for TNF-α translation in both cell types, but an additional requirement for these signals in controlling levels of the mRNA in peritoneal macrophages could mask the effects on translation in these cells.
The requirement for TRAM and TRIF for normal production of TNF-α appears to be specific to TLR4: other TLRs, such as TLR2 and TLR9, are able to induce normal expression of TNF-α mRNA and protein even though they only activate MyD88-dependent signals and are not connected to the TRAM/TRIF pathway [15,22,31]. The explanation for this unique dependence of TLR4 on TRAM and TRIF for the induction of TNF-α is not clear. TLR4 is distinguished from other members of its family by the fact that it signals from 2 different subcellular locations: it activates the Mal/MyD88 pathway from the plasma membrane, while its interaction with TRAM and TRIF requires internalization into an endosomal compartment [10]. It has recently been suggested that these processes may involve separate populations of TLR4 molecules [32], and perhaps the existence of these 2 distinct receptor pools may make the signals activated by either one of them insufficient to induce normal expression of TNF-α. Further investigation will be required to clarify this issue and to elucidate the newly discovered role for the TRAM/TRIF pathway in cytokine translation.
Disclosure Statement
The authors do not have any competing financial interests.
Acknowledgements
This work was supported by grants from the Broad Medical Research Program (IBD-0253) and the National Institute of Allergy and Infectious Diseases (R56AI089700) to B.J.C., by an unrestricted educational grant from Wyeth Nutrition to L. Wang, and by National Institutes of Health award number R01AI067497 to K.A.F. and 5R00AI072955 to J.C.K. We are very grateful to Dr. Shobha Vasudevan and Dr. Joan Steitz of Yale University for kindly providing the luciferase reporter constructs used in this study.
References
- 1.Lembo A, Kalis C, Kirschning CJ, Mitolo V, Jirillo E, Wagner H, Galanos C, Freudenberg MA. Differential contribution of Toll-like receptors 4 and 2 to the cytokine response to Salmonella enterica serovar Typhimurium and Staphylococcus aureus in mice. Infect Immun. 2003;71:6058–6062. doi: 10.1128/IAI.71.10.6058-6062.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Q, Cherayil BJ. Role of Toll-like receptor 4 in macrophage activation and tolerance during Salmonella enterica serovar Typhimurium infection. Infect Immun. 2003;71:4873–4882. doi: 10.1128/IAI.71.9.4873-4882.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Royle M, Totemeyer S, Aldridge LC, Maskell DJ, Bryant CE. Stimulation of Toll-like receptor 4 by lipopolysaccharide during cellular invasion by live Salmonella typhimurium is a critical but not exclusive event leading to macrophage responses. J Immunol. 2003;170:5445–5454. doi: 10.4049/jimmunol.170.11.5445. [DOI] [PubMed] [Google Scholar]
- 4.Weiss DS, Raupach B, Takeda K, Akira S, Zychlinsky A. Toll-like receptors are temporally involved in host defense. J Immunol. 2004;172:4463–4469. doi: 10.4049/jimmunol.172.7.4463. [DOI] [PubMed] [Google Scholar]
- 5.Kawai T, Akira S. The role of pattern recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 6.Sheedy FJ, O'Neill LA. The troll in Toll: Mal and TRAM as bridges for TLR2 and TLR signaling. J Leukoc Biol. 2007;82:196–203. doi: 10.1189/jlb.1206750. [DOI] [PubMed] [Google Scholar]
- 7.Oshiumi H, Sasai M, Shida K, Fujita T, Matsumoto M, Seya T. TIR-containing adaptor molecule (TICAM)-2, a bridging adaptor recruiting to Toll-like receptor 4 TICAM-1 that induces interferon β. J Biol Chem. 2003;278:49751–49762. doi: 10.1074/jbc.M305820200. [DOI] [PubMed] [Google Scholar]
- 8.McGettrick AF, Brint EK, Palsson-McDermott EM, Rowe DC, Golenbock DT, Gay NJ, Fitzgerald KA, O'Neill LA. TRIF-related adaptor molecule is phosphorylated by PKC(∊) during Toll-like receptor 4 signaling. Proc Natl Acad Sci USA. 2006;103:9196–9201. doi: 10.1073/pnas.0600462103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rowe DC, McGettrick AF, Latz E, Monks BG, Gay NJ, Yamamoto M, Akira S, O'Neill LA, Fitzgerald KA, Golenbock DT. The myristoylation of TRIF-related adaptor molecule is essential for Toll-like receptor 4 signal transduction. Proc Natl Acad Sci USA. 2006;103:6299–6304. doi: 10.1073/pnas.0510041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kagan JC, Su T, Horng T, Chow A, Akira S, Medzhitov R. TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon β. Nat Immunol. 2008;9:361–368. doi: 10.1038/ni1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamamoto M, Sato S, Hemmi H, Hoshino K, Kaisho T, Sanjo H, Takeuchi O, Sugiyama M, Okabe M, Takeda K, Akira S. Role of adaptor TRIF in the MyD88-independent Toll-like receptor signaling pathway. Science. 2003;301:640–643. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- 12.Yamamoto M, Sato S, Hemmi H, Uematsu S, Hoshino K, Kaisho T, Takeuchi O, Takeda K, Akira S. TRAM is specifically involved in the Toll-like receptor 4-mediated MyD88-independent signaling pathway. Nat Immunol. 2003;4:1144–1150. doi: 10.1038/ni986. [DOI] [PubMed] [Google Scholar]
- 13.Anderson P. Post-transcriptional control of cytokine production. Nat Immunol. 2008;9:353–359. doi: 10.1038/ni1584. [DOI] [PubMed] [Google Scholar]
- 14.Falvo JV, Tsytsykova AV, Goldfeld AE. Transcriptional control of the TNF gene. Curr Dir Autoimmun. 2010;11:27–60. doi: 10.1159/000289196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 16.Yamamoto M, Sato S, Mori K, Hoshino K, Takeuchi O, Takeda K, Akira S. Cutting edge: a novel Toll/IL-1 receptor domain-containing adapter that preferentially activates the IFN-beta promoter in the Toll-like receptor signaling. J Immunol. 2002;169:6668–6672. doi: 10.4049/jimmunol.169.12.6668. [DOI] [PubMed] [Google Scholar]
- 17.Wang L, Johnson EE, Shi HN, Walker WA, Wessling-Resnick M, Cherayil BJ. Attenuated inflammatory responses in hemachromatosis reveal a role for iron in the regulation of macrophage cytokine translation. J Immunol. 2008;181:2723–2731. doi: 10.4049/jimmunol.181.4.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang L, Harrington L, Trebicka E, Shi HN, Kagan JC, Hong CC, Lin HY, Babitt JL, Cherayil BJ. Selective modulation of TLR4-activated inflammatory responses by altered iron homeostasis. J Clin Invest. 2009;119:3322–3328. doi: 10.1172/JCI39939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levy JE, Montross LK, Cohen DE, Fleming MD, Andrews NC. The C282Y mutation causing hereditary hemochromatosis does not produce a null allele. Blood. 1999;194:9–11. [PubMed] [Google Scholar]
- 20.Pietrangelo A. Hereditary hemochromatosis: pathogenesis, diagnosis and treatment. Gastroenterology. 2010;139:393–408. doi: 10.1053/j.gastro.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 21.Zhou XY, Tomatsu S, Flemin RE, Parkkila S, Waheed A, Jiang J, Fei Y, Brunt EM, Ruddy DA, Prass CE, Schatzman RC, O'Neill R, Britton RS, Bacon BR, Sly WS. HFE gene knock-out produces a mouse model of hereditary hemochromatosis. Proc Natl Acad Sci USA. 1998;95:2492–2497. doi: 10.1073/pnas.95.5.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoebe K, Du X, Georgel P, Janssen E, Tabeta K, Kim SO, Goode J, Mann N, Mudd S, Crozat K, Sovath S, Han J, Beutler B. Identification of Lps2 as a key transducer of MyD88-independent TLR signalling. Nature. 2003;424:743–748. doi: 10.1038/nature01889. [DOI] [PubMed] [Google Scholar]
- 23.Vasudevan S, Steitz JA. AU-rich element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell. 2007;128:1105–1118. doi: 10.1016/j.cell.2007.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carpenter S, O'Neill LA. Recent insights into the structure of Toll-like receptors and post-translational modifications of their associated signalling proteins. Biochem J. 2009;422:1–10. doi: 10.1042/BJ20090616. [DOI] [PubMed] [Google Scholar]
- 25.Ronkina N, Menon MB, Schwermann J, Tiedje C, Hitti E, Kotlyarov A, Gaestel M. MAPKAP kinases MK2 and MK3 in inflammation: complex regulation of TNF biosynthesis via expression and phosphorylation of tristetraprolin. Biochem Pharmacol. 2010;80:1915–1920. doi: 10.1016/j.bcp.2010.06.021. [DOI] [PubMed] [Google Scholar]
- 26.Han J, Brown T, Beutler B. Endotoxin-responsive sequences control cachectin/tumor necrosis factor biosynthesis at the translational level. J Exp Med. 1990;171:465–475. doi: 10.1084/jem.171.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hel Z, Di Marco S, Radzioch D. Characterization of the RNA binding proteins forming complexes with a novel putative regulatory region in the 3′ UTR of TNFα mRNA. Nucl Acids Res. 1998;26:2803–2812. doi: 10.1093/nar/26.11.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Covert MW, Leung TH, Gaston JE, Baltimore D. Achieving stability of lipopolysaccharide-induced NF-κB activation. Science. 2005;309:1854–1857. doi: 10.1126/science.1112304. [DOI] [PubMed] [Google Scholar]
- 29.Gais P, Tiedje C, Altmayr F, Gaestel M, Weighardt H, Holzmann B. TRIF signaling stimulates translation of TNFα mRNA via prolonged activation of MK2. J Immunol. 2010;184:5842–5848. doi: 10.4049/jimmunol.0902456. [DOI] [PubMed] [Google Scholar]
- 30.Neininger A, Kontoyiannis D, Kotlyarov A, Winzen R, Ecker R, Volk HD, Holtmann H, Kollias G, Gaestel M. MK2 targets AU-rich elements and regulates TNF and IL-6 independently at different post-transcriptional levels. J Biol Chem. 2002;277:3065–3068. doi: 10.1074/jbc.C100685200. [DOI] [PubMed] [Google Scholar]
- 31.Jiang Z, Georgel P, Du X, Shamel L, Sovath S, Mudd S, Huber M, Kalis C, Keck S, Galanos C, Freudenberg M, Beutler B. CD14 is required for MyD88-independent LPS signaling. Nat Immunol. 2005;6:565–570. doi: 10.1038/ni1207. [DOI] [PubMed] [Google Scholar]
- 32.Tseng PH, Matsuzawa A, Zhang W, Mino T, Vignali DAA, Karin M. Different modes of ubiquitination of the adaptor TRAF3 selectively activate the expression of type I interferons and proinflammatory cytokines. Nat Immunol. 2010;11:70–75. doi: 10.1038/ni.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]