Abstract
Mutations in IRAK4 have been associated with recurrent Gram-positive infections in children. Given the central role of IRAK4 in innate immunity signaling, we hypothesized that common genetic variants of IRAK4 may be associated with prevalence of Gram-positive infection in critically ill adults. Haplotype clade tag single nucleotide polymorphisms (SNPs) of the IRAK4 gene were selected and genotyped in a cohort of 1,029 critically ill patients with systemic inflammatory response syndrome (SIRS). We found that a haplotype clade tagged by the A allele of the htSNP G29429A (Ala428Thr) was associated with increased relative risk of Gram-positive infection at admission to ICU (RR = 1.2, p < 0.05). Furthermore, the 29429A allele was associated with decreased lymphoblastoid cell response to CpG (as measured by IL-6 production) (raw values ± 95% CI 40.3 ± 32.3 vs. 85.8 ± 29.4 pg/ml; log-transformed values ± 95% CI 1.13 ± 0.37 vs. 1.55 ± 0.18, p < 0.04). We also found that IRAK4-deficient fibroblasts transfected with an IRAK4 expression plasmid containing the 29429A allele produced less IL-6 in response to lipopolysaccharide (p = 0.07). Our data suggest that the IRAK4 haplotype clade marked by 29429A (428Thr) alters susceptibility to Gram-positive bacteria, by decreasing cellular response to TLR ligands.
Copyright © 2011 S. Karger AG, Basel
Key Words: Bacterial infections, Protein kinase, Inflammation
Introduction
The innate immune system is a first line of defense against invading micro-organisms. Pattern recognition receptors of the innate immune system, most importantly Toll-like receptors (TLRs), recognize conserved domains of pathogens, or pathogen-associated molecular patterns (PAMPs) [1,2]. Upon interaction with a PAMP, TLRs interact with a network of intracellular signaling molecules through their Toll-like/interleukin-1 receptor (TIR) domain leading to the activation of the inflammatory response. The inflammatory response is critical for isolation and clearance of pathogens; however, excessive inflammation may result in the clinical phenotype of systemic inflammatory response syndrome (SIRS) and multi-system organ failure. There is considerable interindividual variability in the degree of activation of the innate immunity and inflammatory responses to infection [3,4]. Genotype has been shown to contribute significantly to outcome in diseases involving innate immunity [5,6]. For example, the genetic contribution to the outcome from infectious diseases exceeded the genetic contribution to cancer risk by 5-fold [7]. Genetic variation within innate immunity genes may play a role in an individual's susceptibility to and outcome from severe infection.
IL-1 receptor-associated kinase 4 (IRAK4) is a critical intracellular signaling molecule in the MyD88-dependent pathway of Toll-like/IL-1-receptor (TIR) signaling and is essential for all TLR signaling with the exception of TLR3 [8]. IRAK4 is a serine-threonine kinase that interacts with MyD88 and then phosphorylates downstream molecules, ultimately leading to the activation and nuclear translocation of nuclear factor-κB (NF-κB) and the subsequent transcription of inflammatory mediators such as IL-6 [8]. Pediatric patients with rare mutations of IRAK4 experience recurrent Gram-positive infections and leukocytes from these children are hyporesponsive to TLR ligands in vitro [9,10,11,12,13,14]. To date there have been no studies of the role of common genetic variants of IRAK4 in susceptibility to infection. We hypothesized that common genetic variants of IRAK4 may be associated with the prevalence of Gram-positive infection in critically ill adults.
In order to test our hypotheses, we recruited a prospective cohort of critically ill patients with systemic inflammatory response syndrome (SIRS). We inferred haplotypes of IRAK4 and grouped them into clades, or evolutionarily related sets of haplotypes [15]. We then selected haplotype tag SNPs (htSNPs) that uniquely defined all haplotype clades of IRAK4 to be genotyped and associated to phenotype in the cohort of critically ill patients [16]. We found that a haplotype clade tagged by the A allele of the htSNP G29429A (Ala428Thr) was associated with increased prevalence of Gram-positive infection at admission to ICU. In order to elucidate the mechanism behind this association we tested for a difference of response (IL-6 production) to CpG (a TLR9 ligand) or CpG with a cytokine mixture (including IL-1) in lymphoblastoid cell lines of known genotype and also tested for a difference of response to lipopolysaccharide (LPS) in IRAK4-deficient fibroblasts transfected with an IRAK4 expression plasmid containing the 29429A or G allele.
Methods and Materials
This study was approved by the Providence Health Care/University of British Columbia Research Ethics Board.
Patient Cohort
1,029 consecutive patients admitted to the tertiary mixed medical-surgical Intensive Care Unit (ICU) of St. Paul's Hospital were screened for inclusion into this study. We only report the results for the Caucasian patients (n = 820) in order to decrease the potential confounding influence of population admixture secondary to ethnic diversity on associations between genotype and phenotype [17]. Patients were screened daily and were included in the study cohort (n = 775) if they met at least two of four SIRS criteria and were successfully genotyped at all three htSNPs of IRAK4. The SIRS criteria were (1) fever (>38°C) or hypothermia (<36°C), (2) tachycardia (>100 beats/min in the absence of beta blockers), (3) tachypnea (>20 breaths/min) or need for mechanical ventilation, and (4) leukocytosis (total leukocyte count >12,000/μl) or leukopenia (total leukocyte count <4,000/μl) [18]. Patients were included in this cohort on the calendar day that 2/4 SIRS criteria were met.
Clinical Phenotypes
We assessed the risk of the presence of a positive bacterial culture at admission to ICU (primary outcome variable) by IRAK4 haplotype clade. Bacterial cultures were taken as part of routine medical care for all patients at admission to the ICU. Cultures that were judged to be positive due to contamination or colonization by the attending physician were excluded. Positive bacterial cultures were categorized as Gram-positive, Gram-negative, fungal, or other. The source of a positive culture was categorized as respiratory (sputum), gastrointestinal (peritoneal fluid, abscess drainage, biliary tract), skin (soft tissues or wounds), genitourinary (urine), endovascular (blood) or other. Secondary outcome variables were sepsis upon admission to the ICU, septic shock upon admission to the ICU, and 28-day mortality. Sepsis was defined as the presence of two or more SIRS criteria plus the presence of a known or suspected infection during the 24-hour period. Septic shock was defined by sepsis plus significant hypotension (systolic blood pressure <90 mm Hg or the need for vasopressors). Patients were followed for 28 days for survival. Baseline demographics recorded were age, gender, medical or surgical diagnosis on admission to the ICU (based on the APACHE III diagnostic codes) [19], and the admission APACHE II score [20].
Blood Collection and Processing for Genomic DNA
Discarded blood from routine clinical laboratory tests was collected. Samples were centrifuged at 400 g for 15 min at room temperature. The buffy coat was collected and transferred into 2.0 ml cryotubes and stored at −80°C. DNA was extracted from the buffy coat using the Qiagen DNA Blood Mini Kit (Qiagen Inc., Mississauga, Ont., Canada).
Haplotypes and Selection of htSNPs
We used unphased genotypic data from 23 Caucasians from the Coriell Cell Repository (from pga.mbt.washington.edu) to infer haplotypes of the IRAK4 gene using PHASE software (fig. 1) [15,21]. We then used MEGA 2 software to infer a phylogenetic tree to identify major haplotype clades [22]. Haplotypes were sorted into clades according to this phylogenetic tree and this haplotype structure was inspected to choose a minimum set of ‘haplotype tag’ single nucleotide polymorphisms (htSNPs) (fig. 1) [23,24]. We chose 3 htSNPs that identified 4 major haplotype clades of IRAK4 in Caucasians. The first SNP was an intronic C-to-G substitution at nucleotide 23,338 relative to the start of sequencing (NCBI ID: rs4251513). The second SNP was an intronic T-to-C substitution at nucleotide 24,472 relative to the start (rs4251520). The third SNP was a G-to-A substitution at nucleotide 29,429 (rs4251545) (NCBI IRAK4 accession number AF155118) that results in an alanine at amino acid position 428 in exon 11 being replaced with a threonine. These three SNPs were then genotyped in our 775 patient cohort and PHASE was used to identify haplotypes of each patient.
Fig. 1.
Haplotype structure of the IL-1 receptor-associated kinase 4 (IRAK4) gene. Haplotypes of the IRAK4 gene were inferred from unphased genotype data from 23 Caucasians using PHASE software. Columns are polymorphic sites of IRAK4. Rows are haplotypes of IRAK4 ordered by phylogenetic relationship. Light grey/yellow boxes are minor alleles and dark grey/blue boxes are major alleles. Cladistic relationships of IRAK4 haplotypes were determined using MEGA2 phylogenetic software. There are 4 major haplotype clades of IRAK4, marked clades 1 through 4. C23338G, T24472C, and G29429A were chosen as a haplotype clade tag single nucleotide polymorphisms (htSNP) to distinguish between the 4 major haplotype clades.
Genotyping
The genotypic analysis was performed in a blinded fashion, without clinical information. Patients’ genotypes were determined by real-time polymerase chain reaction (PCR) assay using specific fluorescence-labeled hybridization probes in the ABI Prism 7900HT Sequence Detection System (Applied Biosystems, Inc., Foster City, Calif., USA) [25] (table 1). 5 ng of patients’ genomic DNA was used per genotyping reaction in a 384-well plate. We genotyped DNA with known genotype from 23 lymphoblastoid cells lines from the Coriell Cell Repository using ABI Prism 7900HT Sequence Detection System and found complete concordance between our genotyping and the SeattleSNPs genotyping at all three positions of the IRAK4 gene [21].
Table 1.
Primer and probe sequences used for genotyping IRAK4 htSNPs in the ABI prism 7900HT sequence detection system
| htSNP | |
|---|---|
| C23338G | |
| Primer L | TGTGCCTATAGGAAGGATCCAGATT |
| Primer R | CCCTACTGAACACATCATCTCATTTCT |
| Probe VIC | CTCTTCCATAGTATCCTC-VIC |
| Probe FAM | CTCTTCCATACTATCCTC-FAM |
| T24472C | |
| Primer L | CATTTGCCTGGAGTGCCTTTC |
| Primer R | CAGAGGGTGAAAAGTGTGCTTAGTA |
| Probe VIC | TTTCCCTCATATAACTATTAAC |
| Probe FAM | CCTCATATAACCATTAAC |
| G29429A | |
| Primer L | GAAGATTATATTGATAAAAAGATGAATGATGCTGATTCC |
| Primer R | GCAGACATTGACTAGCAACAGAGT |
| Probe VIC | ACTTCAGTTGAAGCTATGT |
| Probe FAM | ACTTCAGTTGAAACTATGT |
VIC and FAM are fluorophores used to label the allele-specific hybridization probes.
Functional Studies
We measured and compared activation of the immune response in cells of different IRAK4 genotype after stimulation with bacterial products to elucidate the mechanism behind the association of the non-synonymous SNP IRAK4 G29429A (Ala428Thr) with Gram-positive infection.
Forty-three lymphoblastoid cell lines (B lymphocytes immortalized by Epstein-Barr virus) from the individuals sequenced by the SeattleSNPs Programs for Genomic Applications were purchased from the Coriell Cell Repository (appendix 1), since these cell lines have been extensively genotyped and in particular were genotyped for genes in the pathway of interest. Because B-lymphocytes most highly express TLR9 in humans, we decided to stimulate them with CpG, which is recognized by TLR9 [26,27]. CpG oligonucleotide type B is a specific sequence with high GC content found uniquely in bacterial DNA. CpG signals through intracellular TLR9 and activates the MyD88-IRAK1-IRAK4-NF B signaling pathway. TLR9 is an important modulator of response to sepsis [26,28,29,30]. To ensure that any lymphoblastoid cells immune response was specific to CpG, and not simply a result of internalizing foreign DNA, NCpG oligonucleotide was used as a negative control. NCpG oligonucleotide control is the same size as CpG oligonucleotide type B and contains the same amount of each nucleotide, but in a scrambled sequence (table 2). Earlier work that suggested that the phosphorothioate modification of CpG and NCpG oligonucleotides (to increase their stability) may cause a nonspecific immune response. To avoid a nonspecific immune response, we obtained phosphodiester-bonded CpG and NCpG sequences to which the lymphoblastoid cells had no nonspecific immune response.
Table 2.
Synthetic bacterial DNA sequences (CpG) used for stimulation of lymphoblastoid cells
| Oligonucleotide | Sequence |
|---|---|
| CpG Oligonucleotide type B | 5′ TCGTCGTTTTGTCGTTTTGTCGTT 3′ (24mer) |
| NCpG (oligonucleotide control) | 5′ TGCTGCTTTTGTGCTTTTGTGCTT 3′ (24mer) |
The lymphoblastoid cells were cultured in suspension in RPMI with 10% fetal bovine serum (FBS) and 50 μg/ml gentamicin. For CpG stimulation, cells were washed and gentamicin-free media was added. 1-ml aliquots of the lymphoblastoid cells containing half a million cells were placed in a 12-well plate. The lymphoblastoid cells were then stimulated in triplicate with 10 or 50 μM synthetic CpG oligonucleotide type B or NCpG oligonucleotide control for 24 h. Following CpG or NCpG stimulation, IL-6 secretion in the lymphoblastoid cells supernatant was determined as a measure of the immune response using ELISA (R&D Systems, Minneapolis, Minn., USA). To validate the genotypic difference in lymphoblastoid cell response to CpG (n = 43), we next tested for a difference in response to the well-studied stimulus ‘cytomix’ (cytokine mixture, IL-1β, TNF-α, IFN-γ, 2.5 ng/ml) [31,32] with CpG (12.5 μM) [33] in lymphoblastoid cells (n = 88, appendix 1B). Finally, we tested for a difference in IRAK4 gene expression by genotype measured in 88 lymphoblastoid cells under control conditions using the Human WG-6 v.3 expression chip (Illumina, San Diego, Calif., USA) (Génome Québec Innovation Centre, Montréal, Que., Canada).
The lymphoblastoid cells lines from the Coriell Cell Repositories are from different individuals and so differ at many loci throughout the genome. In order to determine that the difference in immune response observed was solely attributable to the IRAK4 G29429A (Ala428Thr) SNP, we obtained a wild-type IRAK4 expression plasmid from Amgen Inc. This was used to transfect IRAK4-deficient fibroblasts (IDFs) (a gift from our collaborators, Dr. David Speert and Dr. Stuart Turvey at British Columbia Children's and Women's Hospital). These IRAK4-deficient fibroblasts come from a patient with a homozygous substitution of thymidine for cytidine at position 877 in exon 8 of IRAK4 resulting in replacement of glutamine by a stop codon (Q293X) [14]. This results in defective expression of full-length IRAK4 mRNA and do not activate NF-κB or produce cytokines in response to TLR ligands. To test whether the Ala428Thr polymorphism was associated with altered immune response the A allele was exchanged for the G allele at nucleotide 29429 in the IRAK4 expression plasmid using the Quick-Change site-directed mutagenesis kit (Stratagene, La Jolla, Calif., USA) so that all other SNP sites were the same between the two plasmids. Thus, the IDFs transfected with either the wild-type or mutated IRAK4 expression plasmids differed at only nucleotide 249249 of the IRAK4 gene and were otherwise genetically identical. Sequence-specific primers were designed containing the mutation based on the IRAK4 wild-type cDNA sequence according to guidelines provided by Stratagene (table 3). 5 ng of the IRAK4 expression plasmid was used as template DNA for the reaction. Following transformation of the mutated plasmid into XL1-blue supercompetent cells for nick repair the cells were plated onto agar plates containing 100 μg/ml ampicillin. Individual colonies were then picked from the plates and grown at 37°C overnight in 4 ml of LB broth with 100 μg/ml ampicillin. The mutated IRAK4 plasmids were purified using the QIAprep Miniprep kit (Qiagen Inc.) with a microcentrifuge according to the manufacturer's instructions. The DNA was eluted in water for sequencing at the University of British Columbia Nucleic Acid Protein Service Unit using a custom-designed primer (table 3). The sequencing results were compared to the IRAK4 wild-type cDNA sequence using the Basic Local Alignment Search Tool (BLAST) bl2seq to align two sequences.
Table 3.
Primers used for site-directed mutagenesis (SDM) and sequencing of the IRAK4 expression plasmid
| Primer | Sequence |
|---|---|
| SDM-sense | 5′ CCACTTCAGTTGAAACTATGTACTCTGTTGCTAGTC 3′ |
| SDM-antisense | 5′ GACTAGCAACAGAGTACATAGTTTCAACTGAAGTGG 3′ |
| Sequencing primer | 5′ CGGGCTTCTGAGAAGTTTGC 3′ |
Once the IRAK4 expression plasmid was successfully mutated to carry an A allele at nucleotide 294249, the IDFs were transfected with the IRAK4 29429G plasmid, the IRAK4 29429A plasmid or the pGL3 empty plasmid as a control. The IDFs were grown in T75 flasks in DMEM with 10% FBS and 50 μg/ml gentamicin. The day before transfection, the IDFs were seeded to 12-well plates in media containing no antibiotics. On the day of transfection, the cells were washed and 1 ml of serum-free media was added to each well. There were approximately half a million cells per well. 2 μg of the appropriate plasmid and 0.5 μg of lipofectamine 2000 (Invitrogen, Burlington, Ont., Canada) in 200 μl of Opti-MEM (Invitrogen) were added to each well. Four hours post-transfection, the media was removed and fresh media with serum and 50 μg/ml of gentamicin was added. The cells were incubated overnight at 37°C. Following transfection, the cells were washed with sterile PBS and 1 ml of fresh serum-free media was added. 0.1 μg/ml of LPS or serum-free media in triplicate was then added to the appropriate wells and the plates were incubated for 6 h at 37°C. After 6 h, the supernatant was collected. IL-6 concentration in the supernatant was measured using ELISA (R&D Systems, Minneapolis, Minn., USA).
Statistical Analysis
We used a cohort study design. Long and Langley [34] performed an extensive simulation study to determine the power of association-based studies when a dense, but not exhaustive, set of SNPs is available over a candidate gene region. A hidden polymorphism was assumed to contribute to the variance of a quantitative trait and the number of nearby SNPs was varied in an estimation of the number of subjects necessary to detect the causative locus. The power of SNP and haplotype association studies was also compared to the quantitative trait TDT (TDT-Q5). Under the assumptions of these models, a sample size of 500 or more is required to detect association with 80% power when 10% or less of the variance in the trait is explained by the locus [34]. Our study of 775 Caucasians had >80% power to detect an absolute difference of 10% in prevalence of positive bacterial cultures at admission to ICU, assuming a 40% baseline prevalence of positive bacterial cultures at admission with significance set at p < 0.05.
Rates of dichotomous outcomes (positive cultures at admission to ICU, sepsis and shock at onset of SIRS, 28-day survival) were compared between clades using Fisher's exact test. Differences in continuous outcome variables (age, APACHE II score) between haplotype clades were tested using ANOVA. 28-day survival was further compared between the two groups of patients while adjusting for other confounders (age, sex, and medical vs. surgical diagnosis) using a Cox regression analysis as well as a Kaplan-Meier analysis of censored survival data. Haplotype clade relative risk was calculated. Genotype distributions were tested for Hardy-Weinberg equilibrium.
Supernatant IL-6 concentrations following CpG stimulation of lymphoblastoid cells were compared among IRAK4 haplotype clades by ANOVA. Supernatant IL-6 concentrations after stimulation with LPS were compared among IDFs transfected with the wild-type IRAK4 plasmid or the PGL3 empty plasmid and normal adult fibroblasts using ANOVA. Supernatant IL-6 concentrations after stimulation with LPS were compared among IDFs transfected with the IRAK4 29429G plasmid, the IRAK4 29429A plasmid and the pGL7 empty plasmid by ANOVA.
We report the mean and 95% confidence intervals. Statistical significance was set at p < 0.05. The data were analyzed using SPSS 14.0 for Windows (SPSS Inc., Chicago, Ill., USA, 2003).
Results
Genotype-Clinical Phenotype Associations
We were able to infer haplotypes from complete sequencing of IRAK4 for 23 Caucasians in the Coriell Cell Repository [21] using PHASE software, and identified four major haplotype clades using MEGA2 software (fig. 1) [15,22]. These 4 clades could be resolved by genotyping three htSNPs: C23338G, T24472C, and G29429A [24]. 775 Caucasian patients with SIRS were successfully genotyped for all three htSNPs. All three SNPs were in Hardy-Weinberg equilibrium. The haplotype clade defined by 23338C/24472T/29429G (C/T/G) occurred with a frequency of 30.7%, the C/T/A haplotype clade occurred with a frequency of 8.6%, the C/C/G haplotype clade occurred with a frequency of 10.1%, and the G/T/G haplotype clade occurred with a frequency of 50.6% (table 4). These frequencies were similar to frequencies deduced from HapMap Caucasian data (table 4).
Table 4.
IRAK 4 haplotype frequencies and patient baseline characteristics by haplotype
| Clade | Frequency (homo-/heterozygote) | Frequency HapMapa | Baseline characteristics (mean ± SD) |
|||
|---|---|---|---|---|---|---|
| age, years | female | medical Dx | APACHE II | |||
| C/T/G | 30.7% (9.2/21.5%) | 32.1% | 57±16 | 37% | 80% | 23±9 |
| C/T/A | 8.6% (0.6/8.0%) | 8.6% | 59±16 | 36% | 74% | 23±9 |
| C/C/G | 10.1% (1.0/9.1%) | 9.4% | 58±18 | 32% | 76% | 22±9 |
| G/T/G | 50.6% (24.4/26.2%) | 50.8% | 58±17 | 37% | 73% | 23±9 |
| P | 0.4 | 0.7% | 0.05% | 0.7 | ||
Haplotype frequency of HapMap CEU data.
For the 775 successfully genotyped individuals of the cohort of Caucasian patients who had at least 2 of 4 SIRS criteria, there were no differences in age, gender (percent female), or severity of illness at the time of admission (as estimated by the APACHE II score) by IRAK4 haplotype clade (table 4). Of 775 patients, 357 patients had positive cultures [main pathogens, Gram-positive (46.0%), Gram-negative (34.2%); main site, respiratory (152), endovascular (128)] (table 5). The C/T/G haplotype clade was associated with a slightly increased prevalence of medical diagnoses for admission to the ICU.
Table 5.
Pathogens and sources of positive cultures
| Case of culture positive | 357 (46.0%) |
| Gram-positive | 177 (49.6%) |
| Gram-negative | 122 (34.2%) |
| Mixed | 40 (11.2%) |
| Fungal | 10 (2.8) |
| Other | 8 (2.2) |
| Source of a positive culture | |
| Respiratory | 152 |
| Gastrointestinal | 23 |
| Skin | 27 |
| Genitourinary | 18 |
| Endovascular | 128 |
| Other | 4 |
On initial analysis, the IRAK4 C/T/A haplotype clade was associated with increased prevalence of positive bacterial cultures at admission to the ICU compared to the other three haplotype clades (fig. 2a). For subsequent analysis, we compared the C/T/A haplotype clade to all other clades grouped. There was no difference in the prevalence of medical diagnoses for admission to ICU between the C/T/A clade versus all the other clades (p > 0.8).
Fig. 2.
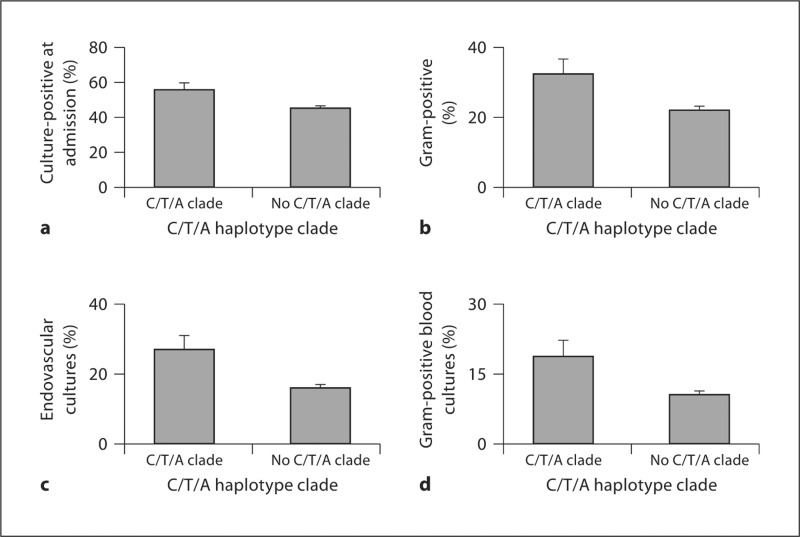
Prevalence of positive bacterial cultures at ICU admission by IRAK4 haplotype. Bacterial cultures were taken as part of routine medical care for any patients who were suspected of having an infection. Patients were genotyped at the IRAK4 htSNPs C23338G, T24472C, and G29429A. Association of IRAK4 haplotype clades with prevalence of positive bacterial cultures was tested by ×2 analysis and the C/T/A clade appeared to be associated with increased prevalence of positive cultures at admission (a). Furthermore, the C/T/A haplotype clade was specifically associated with increased prevalence of Gram-positive infections (b) and endovascular cultures (c). Specifically, the IRAK4 C/T/A clade was associated with increased prevalence of Gram-positive endovascular cultures (d).
The C/T/A haplotype clade was associated with significantly greater prevalence of positive bacterial cultures at admission to ICU (p < 0.03) (fig. 2a), and specifically with increased prevalence of Gram-positive cultures (p < 0.01) (fig. 2b). Patients with at least one copy of the C/T/A haplotype clade had a relative risk of 1.2 for a Gram-positive culture at admission to ICU. The prevalence of positive bacterial cultures from an endovascular source (i.e. positive blood cultures) was also significantly increased in patients who carried at least one copy of the C/T/A haplotype clade (p = 0.001) (fig. 2c). It is clinically significant to note that patients who carried at least one copy of the C/T/A clade had relative risk of 1.8 for a positive Gram-positive bacterial blood culture (p = 0.006) (fig. 2d).
IRAK4 haplotypes were not associated with a significantly different prevalence of sepsis or septic shock upon admission to the ICU, or with 28-day survival (table 6). IRAK4 haplotype clades were not associated with 28-day survival after adjusting for age, gender, APACHE II score, and surgical diagnosis for admission to ICU in a Cox proportional hazards regression analysis. Our study had >80% power (α = 0.05) to detect an absolute difference of survival of 10% assuming that the cohort had an average survival rate of 60%.
Table 6.
Prevalence of sepsis and septic shock at admission to ICU, and 28-day survival by IRAK4 haplotype clade
| Clade | Sepsis at admission | Septic shock at admission | 28-day survival | Days alive (mean ± SD) |
|---|---|---|---|---|
| C/T/G | 81% | 55% | 66% | 21 |
| C/T/A | 81% | 53% | 63% | 20 |
| C/C/G | 74% | 52% | 63% | 21 |
| G/T/G | 81% | 54% | 65% | 21 |
| P | 0.2% | 0.9% | 0.8% | 1.0 |
Mechanism
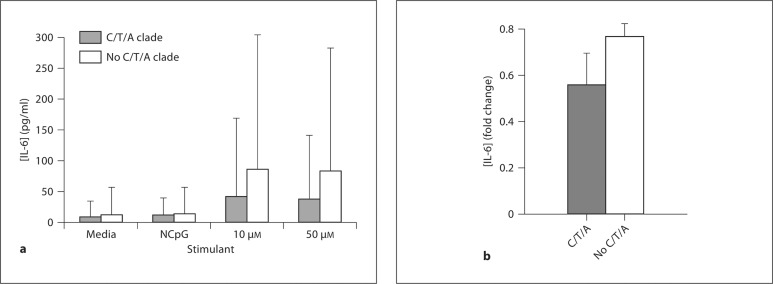
With the purpose of explaining the association of the IRAK4 C/T/A haplotype clade with increased prevalence of Gram-positive infections we compared lymphoblastoid cells immune response to CpG by IRAK4 haplotype clade. The measured IL-6 concentrations were logarithmically (log10) transformed prior to statistical analysis as the data was not normally distributed. The IRAK4 C/T/A haplotype clade was associated with significantly decreased IL-6 secretion compared to all other clades following stimulation of lymphoblastoid cells with 10 μM CpG (raw values ± 95% CI 40.3 ± 32.3 vs. 85.8 ± 29.4 pg/ml; log-transformed values ± 95% CI 1.13 ± 0.37 vs. 1.55 ± 0.18, p < 0.04) or 50 μM CpG (raw values ± 95% CI 38.3 ± 103.3 vs. 81.6 ± 201.3 pg/ml; log-transformed values ± 95% CI 1.15 ± 0.41 vs. 1.55 ± 0.16, p < 0.04) (fig. 3a). In a validation experiment of lymphoblastoid cells (n = 88), the cells with the IRAK4 C/T/A clade had decreased inflammatory response to cytokine mixture (IL-1β, TNF-α, IFN-γ) with CpG compared to all other clades (IL-6 log-transformed fold change to baseline ± 95% CI 0.56 ± 0.14 vs. 0.77 ± 0.06, p < 0.01) (fig. 3b). IRAK4 gene expression under control conditions was not different between cells with the C/T/A clade and cells with all other clades of 88 lymphoblastoid cells (p = 0.37).
Fig. 3.
Lymphoblastoid cells immune response to CpG or cytokine mixture with CpG (as measured by IL-6 supernatant concentration) by IRAK4 haplotype clade. B-lymphocytes of known IRAK4 genotype were stimulated with media, scrambled CpG (NCpG), 10 or 50 μM CpG, a TLR9 ligand, for 6 h (a) or cytokine mixture (IL-1β, TNF-α, IFN-γ, 2.5 ng/ml) with CpG (12.5 μM) for 24 h (b). Each stimulation was performed in triplicate. IL-6 supernatant concentration was measured using ELISA. Mean IL-6 concentrations were logarithmically (log10) transformed and compared between the C/T/A clade and all other IRAK4 clades using Student's t test. The C/T/A clade was associated with significantly decreased IL-6 concentration (a, p < 0.04; b, p < 0.01). Graph displays mean IL-6 concentration ± 95% CI.
In order to isolate the difference in immune response caused by the IRAK4 G29429A (Ala428Thr) SNP, we transfected IDFs with IRAK4 expression plasmids containing either the 29429G or A allele and measured their immune response (as measured by supernatant IL-6 concentration) to TLR ligands. We used site-directed mutagenesis to replace the IRAK4 29429G allele in the plasmid with an A at position 29429. We confirmed that we had exchanged the G for an A using the bl2seq tool of NCBI BLAST. We successfully transfected IDFs with expression plasmids containing either the 29,429G (IDF-G, n = 3) or A allele (IDF-A, n = 3) or with the empty expression vector (IDF-null, n = 3). IDFs transfected with IRAK expression plasmids containing either the 29429G or A allele produced significantly higher levels of IL-6 in response to stimulation with LPS than IDFs transfected with the empty vector (IL-6 production ± 95% CI of IDF-G vs. IDF-A vs. IDF-null in response to LPS: 1,580.6 ± 381.9 vs. 1,292 ± 116.1 vs. 61.6 ± 25.6 pg/ml, p < 0.05) (fig. 4). There was a nonsignificant trend to the IDF-A cell lines producing less IL-6 than the IDF-G cell line in response to stimulation with LPS (p = 0.07) (fig. 4).
Fig. 4.
TLR-ligand stimulation of IRAK4-deficient fibroblasts (IDFs) transfected with IRAK4 expression plasmids containing either the 29429G or A alleles or with the empty vector. IDFs transfected with IRAK4 expression plasmids containing either the IRAK4 29429G (IDF-G) (n = 3) or A (IDF-A) (n = 3) alleles or the empty vector PGL3 (IDF-null) (n = 3) were stimulated for 6 h with 0.1 μg/ml lipopolysaccharide (LPS). Poststimulation concentrations of IL-6 in the supernatant were measured using ELISA. Mean IL-6 concentration was compared among IDF-G, IDF-A and IDF-null cell lines by ANOVA. IDF-G and IDF-A cell lines had significantly greater supernatant IL-6 concentrations compared to IDF-null cell lines (p < 0.05). There was a trend to decreased IL-6 concentration in the IDF-A cell lines compared to the IDF-G cell lines in response to LPS stimulation (p = 0.07). Graph displays mean IL-6 concentration ± 95% CI.
Discussion
We found that the IRAK4 haplotype clade marked by 23338C/24472T/29429A (C/T/A) was associated with increased prevalence of positive bacterial cultures on admission to the ICU in a cohort of 775 critically ill Caucasians with SIRS. Specifically, the C/T/A clade was associated with increased prevalence of Gram-positive cultures and increased prevalence of positive blood cultures. No IRAK4 haplotype clade was associated with prevalence of sepsis or septic shock at admission, or with 28-day survival. Furthermore, we have demonstrated that the IRAK4 C/T/A clade is associated with decreased lymphoblastoid cell immune response to CpG (as measured by IL-6), and that fibroblasts transfected with the 29429A allele that marks the C/T/A clade show a trend to decreased IL-6 secretion in response to stimulation with LPS compared to IRAK4 29429G fibroblasts. To our knowledge, this is the first report of an association of genetic polymorphisms of IRAK4 with prevalence and type of bacterial cultures in a cohort of critically ill patients and supported by mechanistic evidence of decreased cellular immune response to TLR ligands.
Haplotypes within the C/T/A clade are uniquely distinguished from haplotypes in the other four clades of IRAK4 by 29429A. The substitution of an A for a G at position 29429 results in an alanine, a nonpolar aliphatic amino acid, being replaced by a threonine, a polar uncharged amino acid, at amino acid position 428 of the IRAK4 protein [21]. The rs4251545 (Ala428Thr) IRAK 4 polymorphism could alter the kinase activity since it is located in the kinase domain of the protein (154–460) [35]. This amino acid substitution may disrupt IRAK4 signaling and result in a less-effective immune response to invading pathogens so that critically ill patients carrying the C/T/A clade have an increased risk of positive bacterial cultures. Our finding that the C/T/A haplotype clade of IRAK4 is associated with increased prevalence of positive bacterial cultures at admission to the ICU is consistent with our previous studies [36], and with recent animal studies [37,38,39] which suggest that polymorphisms of innate immunity genes could be associated with impaired clearance of bacteria. A number of studies have shown that rare germline mutations in IRAK4 cause recurring bacterial infections in children and deficiencies in cytokine production in response to a range of microbial-derived TLR agonists and to recombinant IL-1β or IL-18 [10,11,12,13,40,41]. Our finding that the IRAK4 C/T/A clade is associated with decreased lymphoblastoid cell immune response to CpG and with a trend to decreased immune response to LPS in IDFs transfected with the IRAK4 expression plasmid carrying the 29429A allele suggests that the replacement of Ala428 with a threonine decreases the cellular response to TLR ligands, perhaps impairing the host's ability to clear an infection.
There are several strengths of our gene association study that minimized limitations of genetic association studies. Firstly, our large cohort of critically ill patients (n = 775) reduced the risk of type I error (finding a spurious association) compared to other studies of smaller sample size. Our large sample size also ensured that we had adequate power to determine that there was truly no association of IRAK4 haplotype clades with prevalence of sepsis and septic shock, or 28-day survival in our cohort of critically ill patients. Second, we adjusted for confounders in our survival analysis (age, gender, APACHE II score, surgical diagnosis) and still found no association of IRAK4 haplotypes with 28-day survival. Third, to avoid spurious associations we included only Caucasians in our cohort of critically ill adults, thus limiting the risk of positive associations due to ethnic heterogeneity [17]. Finally, by testing the immune response of cell lines of different IRAK4 haplotypes to TLR ligands, we have provided plausible mechanistic data to support our clinical association.
An important strength of the design of our association study is our use of haplotypes and haplotype clades as the unit of genetic variation. Haplotype analysis is more effective in determining association of genotype with phenotype than is individual SNP analysis [42,43]. Haplotypes serve as markers of unidentified polymorphisms that may be the cause of phenotypic variation [42]. The phylogenetic history of haplotypes within a population may be determined and used to group haplotypes into clades using cladistic analysis [16,34,44,45,46]. Cladistic analysis has two unique strengths. First, grouping haplotypes into clades decreases the degrees of freedom, thereby increasing the statistical power to associate genotype with phenotype [43]. Second, grouping haplotypes into clades facilitates identification of causal SNPs. Another practical strength of our approach is that a small number of ‘haplotype tag’ SNPs (htSNPs) can be used to distinguish haplotype clades, eliminating the need to genotype all SNPs within a gene [24,47]. Associations of altered phenotypes with polymorphisms of key genes such as innate immune receptors, second messengers in signaling pathways and cytokines have been extensively studied in critically ill patients [36,48,49,50]. Our study shows that variants of a second messenger, IRAK4, create a partial innate immune response defect leading to increased risk of Gram-positive infection while polymorphisms of receptors and signaling proteins show increased risk of sepsis (TLR1 [48], TLR2, CD14 and MBL [36]), and polymorphisms of receptors, second-messenger molecules and cytokines are associated with an increased risk of death (TLR1 [48], TNF, IL-6 [49], IL-10 [50]). We did not find an association of IRAK4 294249G/A and altered risk of sepsis, perhaps because this IRAK4 variant causes only a partial defect in NF-κB pathway signaling as described in case reports of IRAK4 deficiency [14]. Mutations that cause IRAK4 deficiency create dysfunctional signaling with partially intact NF-κB but defective MAPK signaling and dysregulated complex function that affects transcriptional and post-transcriptional control of TLR/IL-1R response in cell- and cytokine-specific ways. Thus, the IRAK4 294249G/A may alter the risk of acquiring Gram-positive infection, but not the risk of sepsis, because of a redundancy in the innate immune response.
There are several limitations of our gene association study. We have not examined how the 29429G/A (A428T) polymorphism affects the function or expression of the IRAK4 protein, so we do not know the functional consequences of the C/T/A haplotype clade in the systemic inflammatory response syndrome. We also do not know how the amino acid change at position 428 may affect the interaction of IRAK4 with other proteins in the MyD88-IRAK1-IRAK4 signaling pathway. Additionally, we were only able to show a trend to decreased IL-6 secretion in response to LPS stimulation of IRAK4-deficient fibroblasts transfected with the IRAK4 29429A allele (compared to the IRAK4 29429G allele). As this trend is in line with our finding of an association of the C/T/A clade with decreased IL-6 secretion in lymphoblastoid cells, we believe that it is an important result, although limited by the experimental design. The Ala428Thr amino acid change may only mildly disrupt IRAK4 function, and thus cause only minor changes in IL-6 secretion in response to stimulus with TLR ligands. Although we treated all cells identically at the same time, because we introduced IRAK4 into the IDFs using transient transfection, it is possible that varying numbers of cells in each well of the transfection and stimulation experiments were viable and expressing IRAK4. This experimental variability may affect our ability to accurately measure the genetic variability in the immune response due to the IRAK4 G29429A polymorphism. In the future, it may be appropriate to use stable transfection to introduce the IRAK4 gene into IDFs. Furthermore, although involved in wound repair, fibroblasts are not the ideal cells in which to be measuring variability of the innate immune response. The IRAK4-deficient fibroblasts were a generous gift to us from our collaborators and served to limit genetic heterogeneity at other loci. However, we may be able to observe greater genetic variability of the immune response to TLR ligands in monocytes or macrophages, cells that are much more sensitive to TLR ligands.
In summary, we have demonstrated a novel association between a haplotype clade of IRAK4 marked by G29429A (Ala428Thr) and increased risk of Gram-positive infection in a cohort of critically ill adults. We have shown that a possible explanation for this association is decreased cellular response to TLR ligands through the IRAK4 signaling pathway, potentially inhibiting the host's ability to clear an infection. Future studies in a separate clinical cohort and in other cellular models will be needed to confirm these findings.
Appendix 1
Coriell Cell Repository IDs, SeattleSNP IDs and IRAK4 G29429A genotypes of lymphoblastoid cell lines used in mechanistic studies.
Table A1a.
43 lymphoblastoid cells
| Coriell cell line ID | Seattle SNPs ID | G29429A genotype |
|---|---|---|
| GM17109 | D009 | AA |
| GM17134 | D034 | AA |
| GM07349 | E009 | AG |
| GM10844 | E014 | AG |
| GM10854 | E015 | AG |
| GM10858 | E011 | AG |
| GM17103 | D003 | AG |
| GM17104 | D004 | AG |
| GM17106 | D006 | AG |
| GM17106 | D006 | AG |
| GM17111 | D011 | AG |
| GM17113 | D013 | AG |
| GM17114 | D014 | AG |
| GM17133 | D033 | AG |
| GM17140 | D040 | AG |
| GM06990 | E022 | GG |
| GM07019 | E023 | GG |
| GM07348 | E020 | GG |
| GM10831 | E017 | GG |
| GM10842 | E007 | GG |
| GM10843 | E018 | GG |
| GM10843 | E018 | GG |
| GM10845 | E003 | GG |
| GM10848 | E012 | GG |
| GM10850 | E019 | GG |
| GM10851 | E008 | GG |
| GM10852 | E021 | GG |
| GM10853 | E004 | GG |
| GM10857 | E010 | GG |
| GM10860 | E005 | GG |
| GM12547 | E002 | GG |
| GM12548 | E013 | GG |
| GM12560 | E001 | GG |
| GM17107 | D007 | GG |
| GM17108 | D008 | GG |
| GM17110 | D010 | GG |
| GM17112 | D012 | GG |
| GM17115 | D015 | GG |
| GM17116 | D016 | GG |
| GM17135 | D035 | GG |
| GM17136 | D036 | GG |
| GM17138 | D038 | GG |
| GM17139 | D039 | GG |
Table A1b.
88 lymphoblastoid cells
| GM10830 | GM10831 | GM10835 | GM10838 | GM10839 | GM10846 | GM10847 | GM10851 |
| GM10854 | GM10855 | GM10857 | GM10859 | GM10860 | GM10861 | GM10863 | GM11829 |
| GM11830 | GM11831 | GM11832 | GM11839 | GM11840 | GM11881 | GM11882 | GM11992 |
| GM11993 | GM11994 | GM11995 | GM12003 | GM12004 | GM12005 | GM12006 | GM12043 |
| GM12044 | GM12056 | GM12057 | GM12144 | GM12145 | GM12146 | GM12154 | GM12155 |
| GM12156 | GM12234 | GM12236 | GM12239 | GM12248 | GM12249 | GM12264 | GM12707 |
| GM12717 | GM12740 | GM12750 | GM12751 | GM12752 | GM12753 | GM12760 | GM12761 |
| GM12762 | GM12763 | GM12801 | GM12802 | GM12812 | GM12813 | GM12814 | GM12815 |
| GM12864 | GM12865 | GM12872 | GM12873 | GM12874 | GM12875 | GM12878 | GM12891 |
| GM12892 | GM6985 | GM6991 | GM6993 | GM6994 | GM7000 | GM7019 | GM7022 |
| GM7029 | GM7034 | GM7048 | GM7055 | GM7056 | GM7345 | GM7348 | GM7357 |
Acknowledgements
We would like to thank Dr. David Speert and Dr. Stuart Turvey for providing us with IRAK4-deficient fibroblasts. We would like to thank Amgen for providing us with the IRAK4 expression plasmid. This work was supported by NIH grant.
References
- 1.Akira S, Takeda K, Kaisho T. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat Immunol. 2001;2:675–680. doi: 10.1038/90609. [DOI] [PubMed] [Google Scholar]
- 2.Beutler B, Hoebe K, Du X, Ulevitch RJ. How we detect microbes and respond to them: the Toll-like receptors and their transducers. J Leukoc Biol. 2003;74:479–485. doi: 10.1189/jlb.0203082. [DOI] [PubMed] [Google Scholar]
- 3.Burgner D, Levin M. Genetic susceptibility to infectious diseases. Pediatr Infect Dis J. 2003;22:1–6. doi: 10.1097/00006454-200301000-00002. [DOI] [PubMed] [Google Scholar]
- 4.Choi EH, Zimmerman PA, Foster CB, Zhu S, Kumaraswami V, Nutman TB, Chanock SJ. Genetic polymorphisms in molecules of innate immunity and susceptibility to infection with Wuchereria bancrofti in south India. Genes Immun. 2001;2:248–253. doi: 10.1038/sj.gene.6363767. [DOI] [PubMed] [Google Scholar]
- 5.Majetschak M, Obertacke U, Schade FU, Bardenheuer M, Voggenreiter G, Bloemeke B, Heesen M. Tumor necrosis factor gene polymorphisms, leukocyte function, and sepsis susceptibility in blunt trauma patients. Clin Diagn Lab Immunol. 2002;9:1205–1211. doi: 10.1128/CDLI.9.6.1205-1211.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mira JP, Cariou A, Grall F, Delclaux C, Losser MR, Heshmati F, Cheval C, Monchi M, Teboul JL, Riche F, Leleu G, Arbibe L, Mignon A, Delpech M, Dhainaut JF. Association of TNF2, a TNF-alpha promoter polymorphism, with septic shock susceptibility and mortality: a multicenter study. JAMA. 1999;282:561–568. doi: 10.1001/jama.282.6.561. [DOI] [PubMed] [Google Scholar]
- 7.Sorensen TI, Nielsen GG, Andersen PK, Teasdale TW. Genetic and environmental influences on premature death in adult adoptees. N Engl J Med. 1988;318:727–732. doi: 10.1056/NEJM198803243181202. [DOI] [PubMed] [Google Scholar]
- 8.Suzuki N, Suzuki S, Yeh WC. IRAK-4 as the central TIR signaling mediator in innate immunity. Trends Immunol. 2002;23:503–506. doi: 10.1016/s1471-4906(02)02298-6. [DOI] [PubMed] [Google Scholar]
- 9.Yang K, Puel A, Zhang S, Eidenschenk C, Ku CL, Casrouge A, Picard C, von Bernuth H, Senechal B, Plancoulaine S, Al-Hajjar S, Al-Ghonaium A, Marodi L, Davidson D, Speert D, Roifman C, Garty BZ, Ozinsky A, Barrat FJ, Coffman RL, Miller RL, Li X, Lebon P, Rodriguez-Gallego C, Chapel H, Geissmann F, Jouanguy E, Casanova JL. Human TLR-7-, −8-, and −9-mediated induction of IFN-alpha/beta and -lambda is IRAK-4 dependent and redundant for protective immunity to viruses. Immunity. 2005;23:465–478. doi: 10.1016/j.immuni.2005.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Currie AJ, Davidson DJ, Reid GS, Bharya S, MacDonald KL, Devon RS, Speert DP. Primary immunodeficiency to pneumococcal infection due to a defect in Toll-like receptor signaling. J Pediatr. 2004;144:512–518. doi: 10.1016/j.jpeds.2003.10.034. [DOI] [PubMed] [Google Scholar]
- 11.Chapel H, Puel A, von Bernuth H, Picard C, Casanova JL. Shigella sonnei meningitis due to interleukin-1 receptor-associated kinase-4 deficiency: first association with a primary immune deficiency. Clin Infect Dis. 2005;40:1227–1231. doi: 10.1086/428733. [DOI] [PubMed] [Google Scholar]
- 12.Medvedev AE, Lentschat A, Kuhns DB, Blanco JC, Salkowski C, Zhang S, Arditi M, Gallin JI, Vogel SN. Distinct mutations in IRAK-4 confer hyporesponsiveness to lipopolysaccharide and interleukin-1 in a patient with recurrent bacterial infections. J Exp Med. 2003;198:521–531. doi: 10.1084/jem.20030701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Picard C, Puel A, Bonnet M, Ku CL, Bustamante J, Yang K, Soudais C, Dupuis S, Feinberg J, Fieschi C, Elbim C, Hitchcock R, Lammas D, Davies G, Al-Ghonaium A, Al-Rayes H, Al-Jumaah S, Al-Hajjar S, Al-Mohsen IZ, Frayha HH, Rucker R, Hawn TR, Aderem A, Tufenkeji H, Haraguchi S, Day NK, Good RA, Gougerot-Pocidalo MA, Ozinsky A, Casanova JL. Pyogenic bacterial infections in humans with irak-4 deficiency. Science. 2003;299:2076–2079. doi: 10.1126/science.1081902. [DOI] [PubMed] [Google Scholar]
- 14.Davidson DJ, Currie AJ, Bowdish DM, Brown KL, Rosenberger CM, Ma RC, Bylund J, Campsall PA, Puel A, Picard C, Casanova JL, Turvey SE, Hancock RE, Devon RS, Speert DP. IRAK-4 mutation (q293x): rapid detection and characterization of defective post-transcriptional TLR/IL-1r responses in human myeloid and non-myeloid cells. J Immunol. 2006;177:8202–8211. doi: 10.4049/jimmunol.177.11.8202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Templeton AR, Weiss KM, Nickerson DA, Boerwinkle E, Sing CF. Cladistic structure within the human lipoprotein lipase gene and its implications for phenotypic association studies. Genetics. 2000;156:1259–1275. doi: 10.1093/genetics/156.3.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cardon LR, Bell JI. Association study designs for complex diseases. Nat Rev Genet. 2001;2:91–99. doi: 10.1038/35052543. [DOI] [PubMed] [Google Scholar]
- 18.Bone RC. The sepsis syndrome: definition and general approach to management. Clin Chest Med. 1996;17:175–181. doi: 10.1016/s0272-5231(05)70307-5. [DOI] [PubMed] [Google Scholar]
- 19.Knaus WA, Wagner DP, Draper EA, et al. The apache iii prognostic system: risk prediction of hospital mortality for critically ill hospitalized adults. Chest. 1991;100:1619–1636. doi: 10.1378/chest.100.6.1619. [DOI] [PubMed] [Google Scholar]
- 20.Knaus WA, Draper EA, Wagner DP, Zimmerman JE. Apache II: a severity of disease classification system. Crit Care Med. 1985;13:818–829. [PubMed] [Google Scholar]
- 21.Rieder M. IRAK-4 Sequence Variation, SeattleSNPs. NHLBI Program for Genomic Applications HL66682, 2002.
- 22.Kumar S, Tamura K, Jakobsen IB, Nei M. Mega2: molecular evolutionary genetics analysis software. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- 23.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 24.Johnson GC, Esposito L, Barratt BJ, Smith AN, Heward J, Di Genova G, Ueda H, Cordell HJ, Eaves IA, Dudbridge F, Twells RC, Payne F, Hughes W, Nutland S, Stevens H, Carr P, Tuomilehto-Wolf E, Tuomilehto J, Gough SC, Clayton DG, Todd JA. Haplotype tagging for the identification of common disease genes. Nat Genet. 2001;29:233–237. doi: 10.1038/ng1001-233. [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ. Allelic discrimination using fluorogenic probes and the 5′ nuclease assay. Genet Anal. 1999;14:143–149. doi: 10.1016/s1050-3862(98)00019-9. [DOI] [PubMed] [Google Scholar]
- 26.Baiyee EE, Flohe S, Lendemans S, Bauer S, Mueller N, Kreuzfelder E, Grosse-Wilde H. Expression and function of Toll-like receptor 9 in severely injured patients prone to sepsis. Clin Exp Immunol. 2006;145:456–462. doi: 10.1111/j.1365-2249.2006.03160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dasari P, Nicholson IC, Hodge G, Dandie GW, Zola H. Expression of Toll-like receptors on B lymphocytes. Cell Immunol. 2005;236:140–145. doi: 10.1016/j.cellimm.2005.08.020. [DOI] [PubMed] [Google Scholar]
- 28.Plitas G, Burt BM, Nguyen HM, Bamboat ZM, DeMatteo RP. Toll-like receptor 9 inhibition reduces mortality in polymicrobial sepsis. J Exp Med. 2008;205:1277–1283. doi: 10.1084/jem.20080162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sjolinder H, Mogensen TH, Kilian M, Jonsson AB, Paludan SR. Important role for Toll-like receptor 9 in host defense against meningococcal sepsis. Infect Immun. 2008;76:5421–5428. doi: 10.1128/IAI.00615-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yasuda H, Leelahavanichkul A, Tsunoda S, Dear JW, Takahashi Y, Ito S, Hu X, Zhou H, Doi K, Childs R, Klinman DM, Yuen PS, Star RA. Chloroquine and inhibition of Toll-like receptor 9 protect from sepsis-induced acute kidney injury. Am J Physiol Renal Physiol. 2008;294:F1050–F1058. doi: 10.1152/ajprenal.00461.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu S, Stolz DB, Sappington PL, Macias CA, Killeen ME, Tenhunen JJ, Delude RL, Fink MP. Hmgb1 is secreted by immunostimulated enterocytes and contributes to cytomix-induced hyperpermeability of Caco-2 monolayers. Am J Physiol Cell Physiol. 2006;290:C990–C999. doi: 10.1152/ajpcell.00308.2005. [DOI] [PubMed] [Google Scholar]
- 32.Farley KS, Wang L, Mehta S. Septic pulmonary microvascular endothelial cell injury: role of alveolar macrophage NADPH oxidase. Am J Physiol Lung Cell Mol Physiol. 2009;296:L480–L488. doi: 10.1152/ajplung.90201.2008. [DOI] [PubMed] [Google Scholar]
- 33.Nakada TA, Russell JA, Boyd JH, Aguirre-Hernandez R, Thain KR, Thair SA, Nakada E, McConechy M, Walley KR. Beta2-adrenergic receptor gene polymorphism is associated with mortality in septic shock. Am J Respir Crit Care Med. 2010;181:143–149. doi: 10.1164/rccm.200903-0332OC. [DOI] [PubMed] [Google Scholar]
- 34.Long AD, Langley CH. The power of association studies to detect the contribution of candidate genetic loci to variation in complex traits. Genome Res. 1999;9:720–731. [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Z, Liu J, Sudom A, Ayres M, Li S, Wesche H, Powers JP, Walker NP. Crystal structures of irak-4 kinase in complex with inhibitors: a serine/threonine kinase with tyrosine as a gatekeeper. Structure. 2006;14:1835–1844. doi: 10.1016/j.str.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 36.Sutherland AM, Walley KR, Russell JA. Polymorphisms in CD14, mannose-binding lectin, and Toll-like receptor-2 are associated with increased prevalence of infection in critically ill adults. Crit Care Med. 2005;33:638–644. doi: 10.1097/01.ccm.0000156242.44356.c5. [DOI] [PubMed] [Google Scholar]
- 37.Tasaka S, Ishizaka A, Yamada W, Shimizu M, Koh H, Hasegawa N, Adachi Y, Yamaguchi K. Effect of CD14 blockade on endotoxin-induced acute lung injury in mice. Am J Respir Cell Mol Biol. 2003;29:252–258. doi: 10.1165/rcmb.2002-0132OC. [DOI] [PubMed] [Google Scholar]
- 38.Opal SM, Palardy JE, Parejo N, Jasman RL. Effect of anti-CD14 monoclonal antibody on clearance of Escherichia coli bacteremia and endotoxemia. Crit Care Med. 2003;31:929–932. doi: 10.1097/01.CCM.0000054870.25767.EE. [DOI] [PubMed] [Google Scholar]
- 39.Echchannaoui H, Frei K, Schnell C, Leib SL, Zimmerli W, Landmann R. Toll-like receptor 2-deficient mice are highly susceptible to Streptococcus pneumoniae meningitis because of reduced bacterial clearing and enhanced inflammation. J Infect Dis. 2002;186:798–806. doi: 10.1086/342845. [DOI] [PubMed] [Google Scholar]
- 40.Day N, Tangsinmankong N, Ochs H, Rucker R, Picard C, Casanova JL, Haraguchi S, Good R. Interleukin receptor-associated kinase (IRAK-4) deficiency associated with bacterial infections and failure to sustain antibody responses. J Pediatr. 2004;144:524–526. doi: 10.1016/j.jpeds.2003.11.025. [DOI] [PubMed] [Google Scholar]
- 41.Enders A, Pannicke U, Berner R, Henneke P, Radlinger K, Schwarz K, Ehl S. Two siblings with lethal pneumococcal meningitis in a family with a mutation in interleukin-1 receptor-associated kinase 4. J Pediatr. 2004;145:698–700. doi: 10.1016/j.jpeds.2004.06.065. [DOI] [PubMed] [Google Scholar]
- 42.Akey J, Jin L, Xiong M. Haplotypes vs. single marker linkage disequilibrium tests: what do we gain? Eur J Hum Genet. 2001;9:291–300. doi: 10.1038/sj.ejhg.5200619. [DOI] [PubMed] [Google Scholar]
- 43.Zhang K, Calabrese P, Nordborg M, Sun F. Haplotype block structure and its applications to association studies: power and study designs. Am J Hum Genet. 2002;71:1386–1394. doi: 10.1086/344780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Templeton AR, Crandall KA, Sing CF. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics. 1992;132:619–633. doi: 10.1093/genetics/132.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Templeton AR. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping or DNA sequencing. V. Analysis of case/control sampling designs: Alzheimer's disease and the apoprotein E locus. Genetics. 1995;140:403–409. doi: 10.1093/genetics/140.1.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Braaten O, Rodningen OK, Nordal I, Leren TP. The genetic algorithm applied to haplotype data at the LDL receptor locus. Comput Methods Programs Biomed. 2000;61:1–9. doi: 10.1016/s0169-2607(99)00025-5. [DOI] [PubMed] [Google Scholar]
- 47.Weiss KM, Clark AG. Linkage disequilibrium and the mapping of complex human traits. Trends Genet. 2002;18:19–24. doi: 10.1016/s0168-9525(01)02550-1. [DOI] [PubMed] [Google Scholar]
- 48.Wurfel MM, Gordon AC, Holden TD, Radella F, Strout J, Kajikawa O, Ruzinski JT, Rona G, Black RA, Stratton S, Jarvik GP, Hajjar AM, Nickerson DA, Rieder M, Sevransky J, Maloney JP, Moss M, Martin G, Shanholtz C, Garcia JG, Gao L, Brower R, Barnes KC, Walley KR, Russell JA, Martin TR. Toll-like receptor 1 polymorphisms affect innate immune responses and outcomes in sepsis. Am J Respir Crit Care Med. 2008;178:710–720. doi: 10.1164/rccm.200803-462OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sutherland AM, Walley KR, Manocha S, Russell JA. The association of interleukin 6 haplotype clades with mortality in critically ill adults. Arch Intern Med. 2005;165:75–82. doi: 10.1001/archinte.165.1.75. [DOI] [PubMed] [Google Scholar]
- 50.Wattanathum A, Manocha S, Groshaus H, Russell JA, Walley KR. Interleukin-10 haplotype associated with increased mortality in critically ill patients with sepsis from pneumonia but not in patients with extrapulmonary sepsis. Chest. 2005;128:1690–1698. doi: 10.1378/chest.128.3.1690. [DOI] [PubMed] [Google Scholar]