Abstract
The formyl peptide receptor gene family encodes G protein-coupled receptors for phagocyte chemoattractants, including bacteria- and mitochondria-derived N-formylpeptides. The human family has 3 functional genes, whereas the mouse family has 7 functional genes and 2 possible pseudogenes (ΨFpr-rs2 and ΨFpr-rs3). Here we characterize ΨFpr-rs2, a duplication of Fpr-rs2. Compared to Fpr-rs2, the ΨFpr-rs2 ORF is 186 nucleotides shorter but 98% identical. Due to a deletion and frame shift, the sequences lack homology from amino acid 219–289. Both transcripts were detected constitutively in multiple immune organs; however, ΨFpr-rs2 was consistently less abundant than Fpr-rs2. LPS induced expression of ΨFpr-rs2, but not Fpr-rs2, in spleen and bone marrow. Both transcripts were detected constitutively in thioglycollate-elicited peritoneal neutrophils, whereas only Fpr-rs2 was detected in thioglycollate-elicited peritoneal macrophages. Both transcripts were induced in LPS-stimulated macrophages. ΨFpr-rs2-GFP fusion protein appeared in cytoplasm but not plasma membrane of transfected HEK 293 cells, whereas Fpr-rs2-GFP labeled only plasma membrane. Survival of ΨFpr-rs2–/– mice was 33% shorter than that of wild-type and heterozygous littermates (p < 0.05), but no signature pathology was identified. Since ΨFpr-rs2 is expressed in phagocytes and regulated by bacterial products, and may affect longevity, we propose renaming it Fpr-rs8, an atypical member of the formyl peptide receptor gene family.
Copyright © 2011 S. Karger AG, Basel
Key Words: Chemotaxis, Inflammation, Formylpeptide, G protein-coupled receptor
Introduction
The formyl peptide receptor (FPR) gene family, a branch of the type A, rhodopsin-like, 7-transmembrane domain, G protein-coupled receptor (GPCR) superfamily, has three human members, FPR1, FPRL1 and FPRL2, encoding functional receptors named FPR1, FPR2/ALX and FPR3, respectively [1,2,3]. All three receptors bind N-formylpeptide agonists and mediate chemotactic responses in phagocytic leukocytes by activating Gi-type G proteins. N-formylpeptides are produced in nature by bacteria and mitochondria, both of which initiate protein synthesis with N-formylmethionine, and specific N-formylpeptides have been identified from these sources that have differential selectivity for the three family members [4,5,6]. Moreover, the N-formyl group has been shown to affect agonist activity [7]. Nevertheless, each of these receptors is also able to bind additional endogenous host ligands that lack not only an N-formyl group but also any obvious common structural feature, even among those ligands that bind to the same receptor. This is an unusual but poorly understood aspect of the family.
The FPR gene family has undergone a complex evolutionary history resulting in seven related mouse genes (Fpr1 and Fpr-rs1, Fpr-rs2, Fpr-rs3, Fpr-rs4, Fpr-rs6 and Fpr-rs7, where rs denotes ‘related sequence’) and two possible pseudogenes ΨFpr-rs2 and ΨFpr-rs3 (also known as Fpr-rs5), all of which are clustered on chromosome 17 A3.2 in a 2,700-kb region syntenic with the human gene cluster on chromosome 19 [2,8,9,10,11]. Fpr1 is the orthologue of human FPR1, whereas Fpr-rs1 and Fpr-rs2 are equally similar at the sequence level to human FPRL1 (encoding the FPR2/ALX receptor). Fpr-rs1 encodes a receptor named ALX, signifying its specificity for the anti-inflammatory lipid lipoxin A4 [10], a property it shares with human FPR2/ALX. Likewise, Fpr-rs2 encodes a receptor named Fpr2, signifying its specificity for N-formylpeptides, serum amyloid A, amyloid β and other ligands shared with human FPR2/ALX [3]. The remaining 4 genes are expressed in hematopoietic tissues and cells, but also have been reported to encode vomeronasal chemosensors and may be important in the sense of smell [12].
The ability to mediate chemotaxis in vitro is usually predictive of a host defense or pro-inflammatory role, and evidence for the former has been reported from a Listeria monocytogenes infection model in mice lacking Fpr1 [13]. However, an unexpected anti-inflammatory function in Fpr1-deficient mice, possibly mediated by the anti-inflammatory Fpr1 ligand annexin 1, has been reported in a peritonitis model [14]. A mechanistic explanation for how the same receptor can support both pro-inflammatory and anti-inflammatory functions in vivo has not yet been developed. In humans, functional FPR1 polymorphisms have been associated with localized juvenile periodontitis [15,16]. Biological roles for FPR2/ALX and FPR3 have not been established in man. In mice, genetic inactivation of Fpr-rs2 has been reported to reduce allergic airway inflammation [17] as well as to increase sensitivity to arthrogenic serum [18].
ΨFpr-rs2 was identified by the Mouse Genome Project 30 kb from Fpr-rs2, in the same orientation, on chromosome 17 A3.2, and has been referred to in a review article [2], but has not previously been structurally or functionally analyzed in the scientific literature. Here we provide the first detailed characterization of ΨFpr-rs2, and provide evidence that it is not a pseudogene, but rather a constitutively expressed and LPS-inducible gene that may affect longevity. As a result, we propose renaming ΨFpr-rs2 as Fpr-rs8.
Animals and Methods
Mice
All animal experiments were done in compliance with the Animal Care and Use Committee of the NIAID. The Genbank accession number for the ΨFpr-rs2 (Fpr-rs8) sequence is NG_019782, and the gene name listed in Genbank for ΨFpr-rs2 (Fpr-rs8) is Gm5966. This deposit contains an 81,466,437-bp contig for chromosome 17 from C57BL/6J mouse genomic DNA deposited by the Mouse Genome Project. The sequence of ΨFpr-rs2 (Fpr-rs8) is from 4,251,019 to 4,251,888 bp of this contig. In the targeting construct made by Deltagen (San Mateo, Calif., USA; construct No. 9995), nt 46-222 of the ΨFpr-rs2 (Fpr-rs8) ORF were replaced by a LacZ Neo cassette (fig. 1). Homologous recombination was then performed using ES cells from 129/Sv mice. Knockout mice were backcrossed 4 generations onto C57Bl/6 mice (Charles River, Wilmington, Mass., USA), and marketed as an Fpr-rs2 knockout. When we performed PCR analysis of genomic DNA from littermate progeny of the purchased mating pairs using primer sequences recommended by the manufacturer, we failed to identify a disrupted Fpr-rs2 gene. Upon further investigation, we discovered that the sequence of the targeting construct was not that of Fpr-rs2, but instead identical to ΨFpr-rs2 (Fpr-rs8), and that the primer sequences recommended for genotyping were for gene regions of Fpr-rs2 that are 100% identical to the sequence of ΨFpr-rs2, and therefore could not distinguish between the two genes. We designed new 5′ primers to pair with a common 3′ primer to selectively amplify the wild-type and targeted ΨFpr-rs2 (Fpr-rs8) alleles for genotyping littermates as follows (fig. 1): primer 1, 5′ primer specific for wild-type ΨFpr-rs2 (Fpr-rs8), 5′-CTCTCTGGAGTGTACAACTCATTCTTTG; primer 2, 5′ primer specific for disrupted ΨFpr-rs2 (Fpr-rs8) allele (Neo-specific sequence), 5′-GACGAGTTCTTCTGAGGGGATCG; primer 3, 3′ primer common to both Fpr-rs2 and Fpr-rs8, 5′-CCATTTCAACAAGAAGGAATGGTAG.
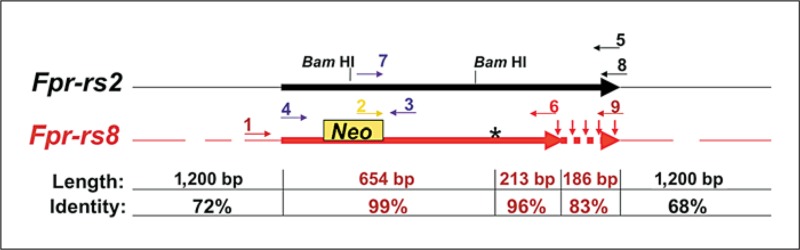
Fig. 1.
Fpr-rs8 is located on mouse chromosome 17 A3.2 and is most highly homologous to Fpr-rs2. The locations of stop codons are indicated in Fpr-rs8 as downward-pointing red arrows. Thickened horizontal lines indicate the coding region of Fpr-rs2 and colinear sequence of Fpr-rs8; thin horizontal lines represent the flanking regions. The 5′ flanking region of Fpr-rs8 has two major gaps relative to Fpr-rs2 at −424 to −483 bp and −527 to −626 bp relative to the start of the ORF. The 3′ -flanking region of Fpr-rs8 has one major gap relative to Fpr-rs2 at 515 to 586 bp downstream of the stop codon. The dashed horizontal red line indicates putative untranslated Fpr-rs8 sequence that is colinear with and highly homologous to the corresponding region of the Fpr-rs2 ORF. The length of Fprrs2 sequence in bp and percent identity of each segment with colinear Fpr-rs8 sequence are indicated at the bottom. The position of the Neo cassette used in constructing the targeting vector by Deltagen Inc. to create mouse line T631 is indicated by the yellow box. Short horizontal arrows indicate the position and sense of primers used for PCR analysis, and are numbered according to the names given in the Materials and Methods section. Primer colors indicate gene specificity: red = Fpr-rs8-specific; black = Fpr-rs2-specific; purple = common to both genes; yellow = specific to Neo. An asterisk demarcates a single base pair deletion in Fpr-rs8 relative to Fpr-rs2 that creates a frame shift and a premature stop codon. Note that the gene distances are not drawn to scale.
PCR amplification was performed using Platinum PCR Supermix and Platinum Taq Polymerase (Invitrogen, Carlsbad, Calif., USA): 95°C for 7 min, 30 cycles of 96°C for 10 s, 60°C for 30 s and 68°C for 90 s, 68°C for 7 min and hold samples at 4°C.
Plasmid Construction
Fpr-rs2 and Fpr-rs8 ORFs were amplified by PCR from genomic DNA of wild-type C57Bl/6 mice using the following primers that complemented the start and end of the coding regions (fig. 1): primer 4, 5′ primer common to both Fpr-rs2 and Fpr-rs8, 5′-ACGTACCTCGAGATGGAATCCAACTACTCCATCCAT, where the bold and underlined sequence is proposed to be the start codon and bold with dotted underline indicates a Xho I site; primer 5, 3′ primer specific for Fpr-rs2, 5′-GTACGTAAGCTTTGGGGCCTTTAACTCAATGTCTGC, where bold with dashed underline indicates a Hind III site; primer 6, 3′ primer specific for Fpr-rs8, 5′-GTACGTAAGCTTTGAACTTGTTGGATTAACAAACAT, where bold with dashed underline indicates a Hind III site. PCR products were subcloned into the topoisomerase site in the vector TOPO (Invitrogen). Random clones were selected and sequenced to ascertain the correct orientation and sequence. The Fpr-rs2 and Fpr-rs8 ORFs were excised from TOPO by Xho I and Hind III restriction enzyme digestion, and purified after electrophoretic separation on a 1% agarose in TBE gel using a GenElute Agarose Spin Column (Sigma, St. Louis, Mo., USA). Plasmids pEGFP-N1 or pDsRedExpress-1 (BD Biosciences, Palo Alto, Calif., USA) were cut with Xho I and Hind III, dephosphorylated, electrophoresed in a 1% agarose in TBE gel and purified using Sigma GenElute Agarose Spin Column. Gene fragments and plasmid vectors were ligated using NEB Quick T4 Ligase and subcloned using One Shot competent cells (Invitrogen). Colonies were selected on LB agarose containing 50 μg/ml kanamycin and analyzed for the correct insert by DNA sequencing.
Cell Transfections
The cell line HEK 293 was purchased from ATCC (Manassas, Va., USA) and grown in DMEM (Invitrogen) containing 10% heat-inactivated fetal bovine serum (Atlanta Biological, Atlanta, Ga., USA), 100 units/ml penicillin and 100 μg/ml streptomycin (Invitrogen). Cells were cultured at 37°C in 100% humidity and 5% CO2. Cells were transfected using the Nucleofector system (Amaxa, Gaithersburg, Md., USA) using Nucleofector Kit V following the manufacturer's optimized protocol. Cells were cultured in DMEM in 6-well tissue culture plates. The next day, transfected cells were selected in DMEM containing G418 at 2 mg/ml. Media was changed every 3 days until single colonies appeared in the wells, which were then expanded in DMEM containing G418. Cells were then plated on a Lab-Tek 2 chambered sterile cover glass slide (Nagle Nunc International, Naperville, Ill., USA) and cultured for 2 days. GFP expression was analyzed using a Leica TCS-NT/SP1 confocal fluorescence microscope.
Gene Expression Analysis
To define expression of Fpr-rs2 and Fpr-rs8 in tissues and cells we designed a gene-specific PCR-RFLP assay using the following primer pairs and Bam HI digestion of the PCR product (fig. 1): primer 7, 5′ primer common to both Fpr-rs2 and Fpr-rs8, 5′-TCTCAATGGTGGTTGTCTCCATCA; primer 8, 3′ primer specific for Fpr-rs2, 5′-TCACAGACTTCATGGGGCCTTTAA; primer 9, 3′ primer specific for Fpr-rs8, 5′-TCCCTATCTTCATGTAGTCTTCAT.
All primers were purchased from Invitrogen. The predicted PCR products are 980 bp for both genes. The PCR products for the two genes can be distinguished because the Fpr-rs2 PCR product, but not the Fpr-rs8 product, has a single internal Bam HI site in this bounded region, which when cut generates 522- and 458-bp fragments. mRNA expression was analyzed using these primers by PCR of multiple tissue cDNA panels from Clontech (mouse panel I, Cat. No. 636745; mouse panel III, Cat. No. 636757). Gene induction was tested in vivo by injecting mice in the peritoneal cavity with 150 μg of LPS. After 6 h, mice were sacrificed, organs and cells were harvested, and RNA was prepared using the MELT™ Total Nucleic Acid Isolation System following the manufacturer's directions (Cat. No. 1982; Ambion, Austin, Tex., USA). Specifically, spleens were harvested and disrupted to form single-cell suspensions, bone marrow cells were harvested from mouse femurs by rinsing with a syringe containing PBS, mouse neutrophils and macrophages were harvested from the peritoneal cavity following local thioglycolate installation for 3 h or 3 days, respectively, and RAW 264.7 cells, cultured with or without LPS, were harvested from tissue culture flasks. Each cell preparation was washed twice in PBS, and the cells were then placed in RNALater (Ambion) overnight at 4°C. RNA was prepared the following day using the RiboPure™ –Blood kit (Cat. No. 1928; Ambion) following the manufacturer's recommended protocol. Reverse transcription was performed using the RETROscript kit (Cat. No. 1710; Ambion). PCR amplification was performed as follows using Platinum PCR SuperMix and Platinum Taq DNA Polymerase (Invitrogen): 95°C for 7 min, 30 cycles of 96°C for 10 s, 60°C for 30 s, 68°C for 90 s, 68°C for 7 min, and hold samples at 4°C.
Calcium Flux Assay
Macrophages harvested 3 days after thioglycollate treatment of Fpr-rs8+/+, Fpr-rs8+/– and Fpr-rs8–/– mice were incubated in DMEM (Invitrogen) containing 10% heat-inactivated fetal bovine serum (Atlanta Biological), 100 units/ml penicillin/100 μg/ml streptomycin (Invitrogen) and 500 ng/ml LPS overnight at 37°C in a 5% CO2 chamber. Cells were then washed twice with PBS, and incubated in 20 ml PBS for 20 min. Cells were then released by vigorously shaking the flask and scraping the cells using a rubber policeman. After rinsing twice in PBS, cells were suspended at 2.5 × 106/ml in HBSS and added to a final density of 0.5 × 106/ml to a well of a 96-well clear bottom black plate (Greiner, St. Louis, Mo., USA). Cells were centrifuged onto the plate and 100 ml HBSS plus 100 ml reagent from a calcium 3 assay kit (Molecular Devices, Sunnyvale, Calif., USA) were added. Plates were incubated for 30 min at 37°C. Calcium flux was measured in a FlexStation instrument (Molecular Devices) using an excitation wavelength of 485 nm and an emission wavelength of 525 nm.
Phenotype Analysis
Body weight, organ weight, complete blood counts, serum electrolytes, liver function tests, kidney function tests and histological examination of organs (skin, lymph node, salivary gland, thymus, trachea, esophagus, lung, heart, ear, eye, pituitary gland, thyroid gland, spleen, liver, pancreas, all levels of gastrointestinal tract, reproductive tract, adrenal gland, nasal sinuses, urinary bladder, tongue, kidney, skeletal muscle, sciatic nerve, brain, spinal cord, femur, tibia, teeth, gall bladder, bone marrow, mammary gland and parathyroid gland) from Fpr-rs8+/+, Fpr-rs8+/– and Fpr-rs8–/– mice were carried out using mice between 7 and 19 months of age for each group. In addition, a cohort of Fpr-rs8+/+, Fpr-rs8+/– and Fpr-rs8–/– mice was allowed to age naturally until an illness developed. When an NIH veterinarian determined that a mouse was ill and showed signs of being moribund, it was sacrificed and sent to the NIH Division of Veterinary Resources for necropsy. Necropsy reports were collected over a 2-year period and analyzed.
Results
Identification of Fpr-rs8, a Novel Gene in the Mouse FPR Family
We and others have previously identified members of the mouse FPR gene family [2,8,9,10,11]. We have focused our interest on Fpr1 and Fpr-rs2, which we and others have previously shown both encode N-formylpeptide receptors [5,9,19]. With the aim of functionally characterizing the biological role of Fpr-rs2, we attempted repeatedly, but unsuccessfully, to create an Fpr-rs2 knockout mouse. When Deltagen listed an Fpr-rs2 knockout mouse in their catalogue (line t631), we purchased heterozygous mating pairs, but unexpectedly were only able to obtain homozygous wild-type and heterozygous offspring using the genotyping method recommended by the company. Completion of the mouse genome sequencing project allowed us to identify a sequence highly related to Fpr-rs2 that had inadvertently and unknowingly been disrupted by Deltagen in their knockout line t631 (Fpr-rs8 in fig. 1). This sequence is located 30 kb downstream from Fpr-rs2 on chromosome 17 A3.2 and is an apparent duplication of Fpr-rs2. For reasons to be detailed in the sections that follow, we refer to this sequence as a gene and we have named it Fpr-rs8. Fpr-rs8 refers to the same DNA sequence previously referred to as a pseudogene named ΨFpr-rs2 in a review article by Migeotte and colleagues [2], and was deposited in Genbank under accession number NG_019782 and under the gene name Gm5966. The region of the genome between Fpr-rs2 and Fpr-rs8 has no annotated genes.
Relative to the Fpr-rs2 sequence, there is a single base pair deletion in the Fpr-rs8 ORF 653 nucleotides downstream from the start site. This causes a frame shift in the coding sequence and a premature stop codon 213 nucleotides downstream from the deletion. Four other stop codons are also found in the last 186 nucleotides of Fpr-rs8 that are colinear with the ORF sequence of Fpr-rs2 (fig. 1, fig.2a). Overall, the aligned ORF sequences are 96% identical; however, the distribution of differences found is not homogeneous. In particular, the ORF of Fpr-rs8 is 867 nucleotides long and 98% identical to the corresponding sequence of Fpr-rs2, whereas the sequence of Fpr-rs8 after the premature stop codon that aligns with the ORF of Fpr-rs2 is 186 nucleotides long but only 83% identical to the corresponding sequence of Fpr-rs2. Also, the portion of the Fpr-rs8 ORF before the deletion is 654 nucleotides long and 99% identical to the corresponding region of Fpr-rs2, whereas the sequence after the deletion is 213 nucleotides long and 96% identical to the corresponding sequence of Fpr-rs2. Comparison of the corresponding upstream and downstream regions of the genes (approximately 1.2 kb 5′ and 3′ of the ORFs) revealed that these regions are 72 and 68% identical, respectively, between Fpr-rs2 and Fpr-rs8 (fig. 1). BLAST analysis of the human genome with the sequence of FPRL1, a human homolog of Fpr-rs2 [3,8], did not reveal a similar duplication in the human genome.
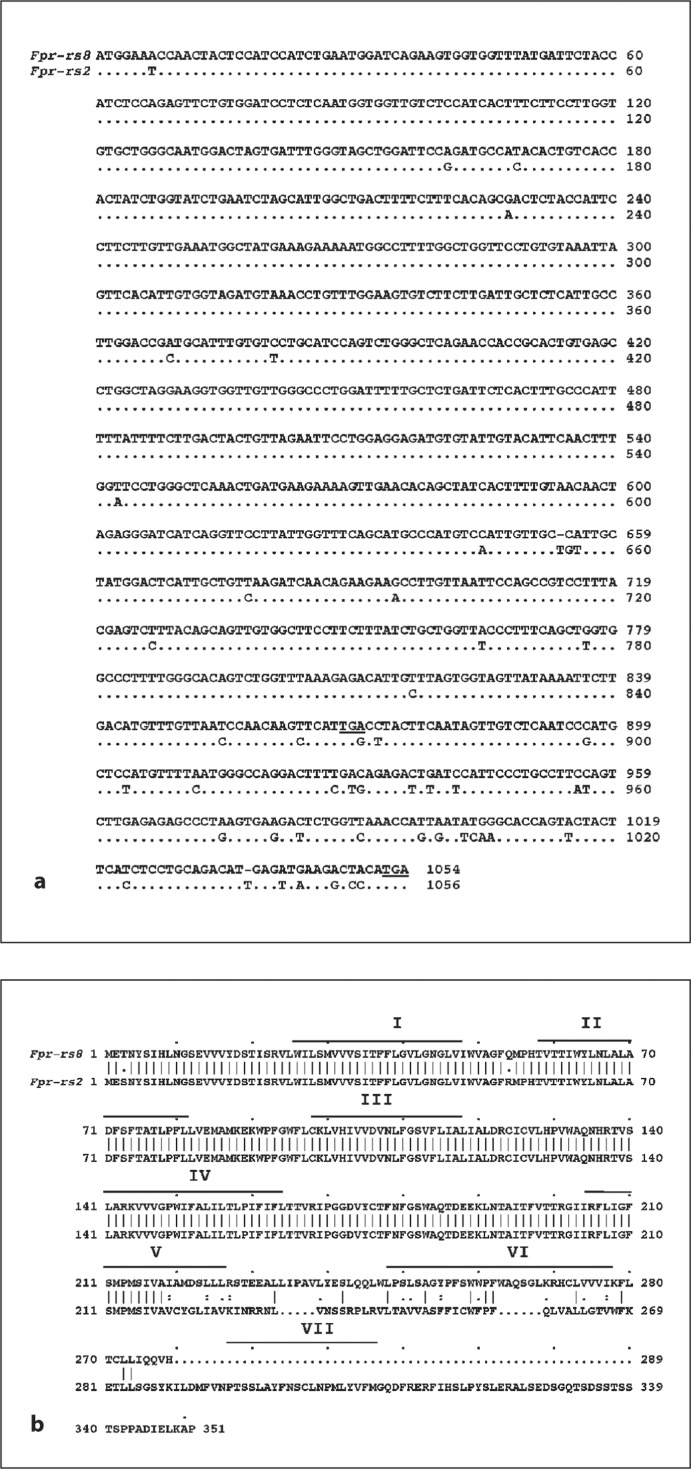
Fig. 2.
Alignment of DNA sequence (a) and predicted amino acid sequence (b) of Fpr-rs8 and Fpr-rs2. a Dots indicate positions of Fpr-rs2 where the sequence is identical to that of Fpr-rs8. The first and last stop codons in the Fpr-rs8 sequence are underlined. Gaps are inserted to optimize the alignment. b Roman numerals represent the 7 transmembrane regions predicted for the Fpr-rs2 gene product. Vertical lines indicate position identities for the two sequences. Individual letters represent the amino acid code. Gaps are inserted to optimize the alignment.
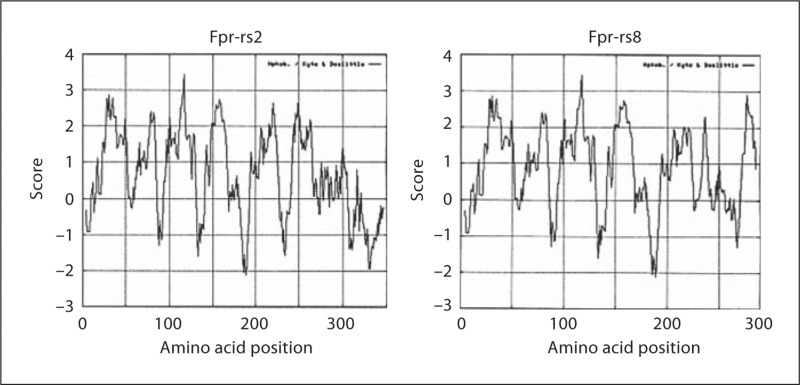
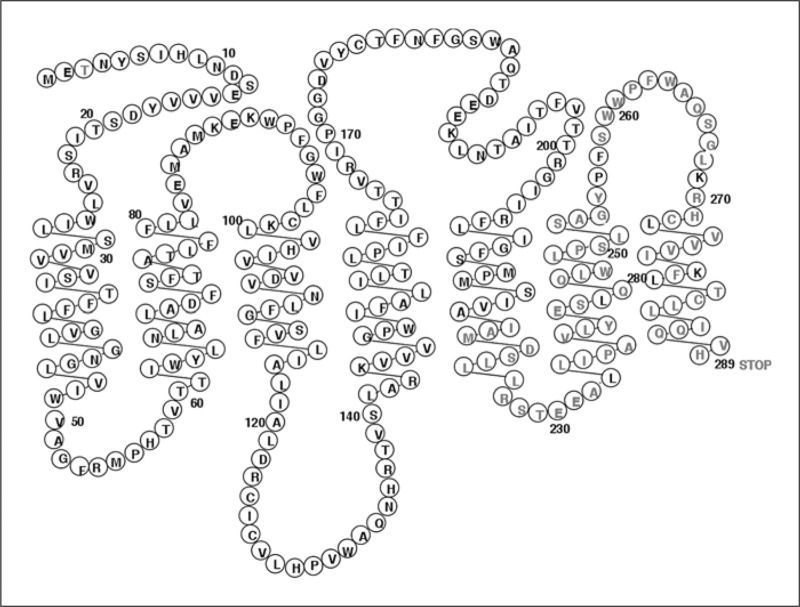
Translation of the Fpr-rs8 ORF sequence revealed a theoretical 289-amino acid protein (fig. 2). The first 218 amino acids are identical to the translated Fpr-rs2 gene product except for a change of serine to threonine at codon 3 and arginine to glutamine at codon 54. Following the frame shift, the amino acid sequences deduced from the two ORFs have no significant homology. Hydrophobicity analysis shown in figure 3 suggested that the predicted Fpr-rs8 product is unlikely to have 7 membrane-spanning domains, and that if it does, it would lack a cytoplasmic tail (fig. 4).
Fig. 3.
Kyte-Doolittle hydrophobicity plots for Fpr-rs2 and Fpr-rs8 predicted protein sequences.
Fig. 4.
The Fpr-rs8 gene product depicted as a potential 7-transmembrane receptor. Circles contain letter codes for amino acids. Black letters represent amino acids identical to the corresponding position in Fpr-rs2, and red represents the unique Fprrs8 sequence.
Fpr-rs2 and Fpr-rs8 Are Both Differentially Expressed and LPS-Inducible Genes in Mouse Tissues and Phagocytes
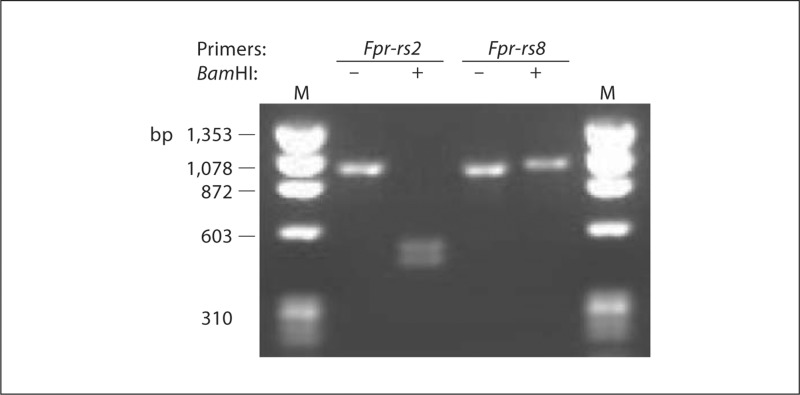
DNA sequence analysis revealed that 1 of 2 Bam HI sites found in the Fpr-rs2 ORF is absent in Fpr-rs8, and that the sequence near the 3′ end of the ORF of Fpr-rs2 is distinct from the corresponding sequence in Fpr-rs8. Hence, it was possible to develop a specific PCR-RFLP genotyping assay for each gene (fig. 5). PCR amplification using this assay produces a product of 980 bp for both genes, but only the PCR product from Fpr-rs2 can be digested by Bam HI, producing two fragments 522 and 458 bp long.
Fig. 5.
Development of a PCR-RFLP assay to distinguish Fpr-rs2 and Fpr-rs8 in genomic DNA. Genomic DNA from C57Bl/6 mice was amplified as described in Materials and Methods using primers specific for the genes indicated at the top of each lane, followed by Bam HI digestion (+) or mock digestion (-) for 1 h at 37°C. Samples were electrophoresed in a 1% agarose gel. M = ΦX174 Hae III fragment size markers.
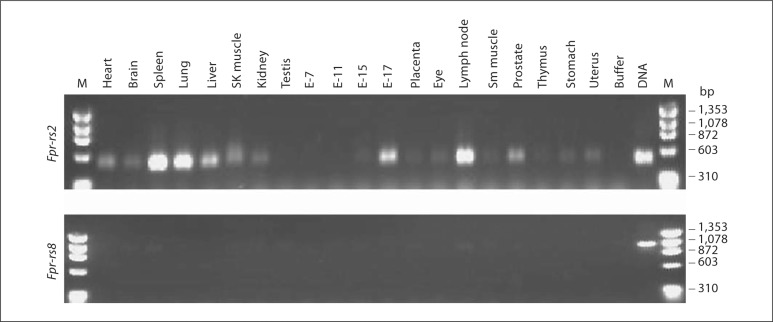
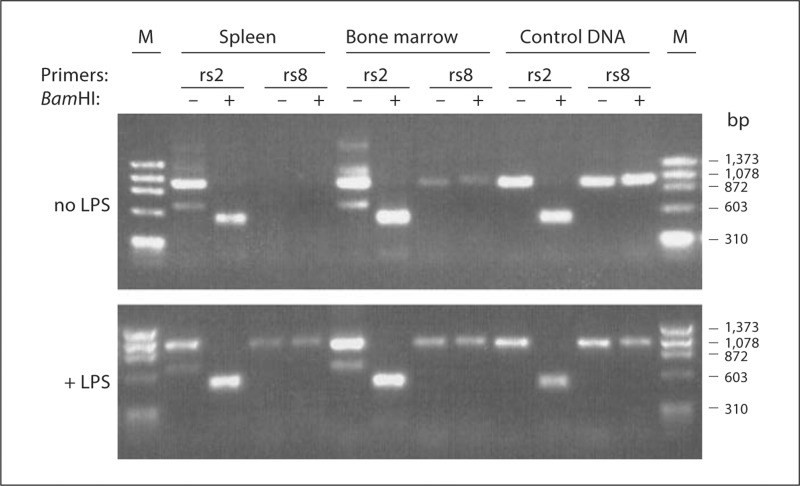
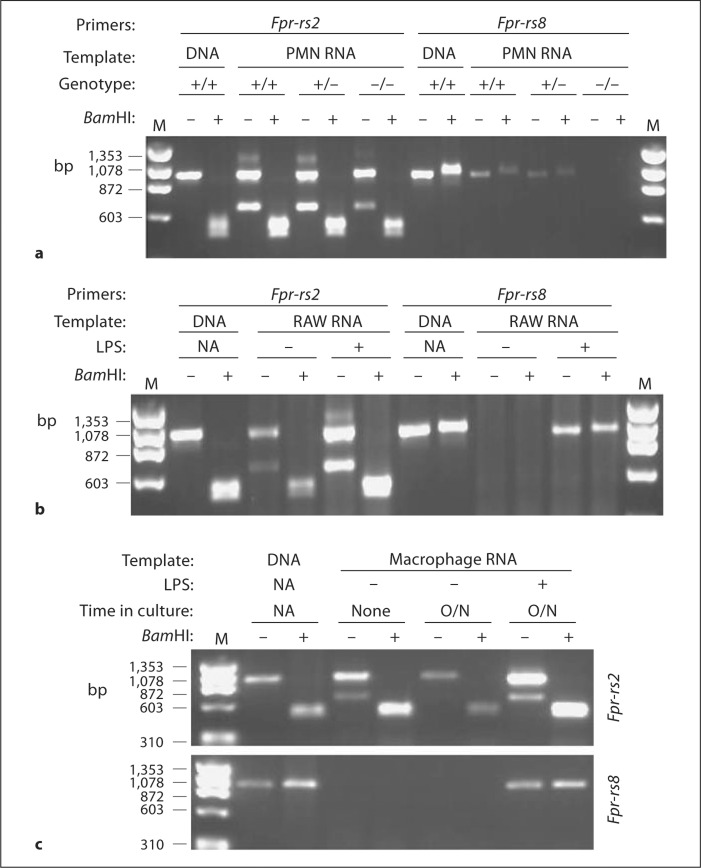
Using this assay and mouse tissue cDNA panels, constitutive Fpr-rs2 expression was detected at the mRNA level in most tissues as previously reported [8] (fig. 6). The highest expression was observed in spleen, lymph node, lung and liver. Expression was first observed in whole embryo RNA at embryonic day 15 and was strongly increased by day 17. Fpr-rs8 expression was also detected constitutively in most organs but only at very low levels. To test whether expression of Fpr-rs8 could be increased, for example during an infection, we analyzed wild-type C57Bl/6 mice injected subcutaneously with 150 μg of LPS. Six hours after injection, Fpr-rs8 expression was greatly increased in spleen and to a lesser extent in bone marrow (fig. 7). In contrast, LPS had a minor effect on expression of Fpr-rs2 in these organs. LPS-injected mice were found to have an increase in splenic neutrophils 6 h after injection, which correlated with increased expression of Fpr-rs8 in this organ (data not shown). We therefore next examined gene expression at the cell level in freshly isolated thioglycolate-elicited peritoneal neutrophils (fig. 8). Consistent with the in vivo observations, RNA for both Fpr-rs2 and Fpr-rs8 was detected constitutively in these cells, with Fpr-rs2 mRNA being much more abundant than Fpr-rs8 RNA (fig. 8a).
Fig. 6.
Differential tissue distribution of Fpr-rs2 and Fpr-rs8 mRNA. The same panel of cDNAs was analyzed by PCR using primers specific for the genes indicated to the left of each panel following the manufacturer's procedures. After PCR amplification the samples were digested using Bam H1 for 1 h at 37°C. E = Embryonic day; M = ΦX174 Hae III fragment markers; DNA = wild-type C57Bl/6 mouse control DNA.
Fig. 7.
LPS in vivo induces expression of Fpr-rs8 in spleen and bone marrow. Tissues were prepared from mock-injected (top panel) and LPS-injected (bottom panel) wild-type C57Bl/6 mice, and analyzed by PCR-RFLP, as described in Materials and Methods. rs2 = Fpr-rs2; rs8 = Fpr-rs8; M = ΦX174 Hae III fragment markers. The indicated primers and Bam HI treatments correspond to the lane directly beneath on both gels.
Fig. 8.
Constitutive and LPS-inducible expression of Fpr-rs2 and Fpr-rs8 in mouse phagocytic cells. a PMN harvested from the peritoneal cavity of mice with the indicated Fpr-rs8 genotypes following 3 h of thioglycolate treatment intraperitoneally. b Mouse monocytic cell line RAW 264.7 (RAW) cultured +/− LPS for 24 h. c Macrophages harvested from the peritoneal cavity of wild-type mice following 3 days thioglycolate treatment intraperitoneally were analyzed immediately or after overnight culture (O/N) +/− LPS. The indicated mRNA template in each panel was amplified by RT-PCR using primers from the indicated gene, then incubated +/− Bam H1. Wild-type mouse DNA (DNA) was used as positive control. M = Markers (ΦX174 DNA digested with Hae III): fragment sizes are indicated in base pairs on the left of each panel; NA = not applicable.
Since many FPR family members are expressed by macrophages and macrophages are present in most organs that have positive Fpr-rs2 and Fpr-rs8 mRNA signals, we next tested expression in this cell type focusing first on the mouse monocytic cell line RAW 264.7. We detected constitutive expression of Fpr-rs2 but not Fpr-rs8 mRNA in these cells (fig. 8b). However, LPS treatment (500 ng/ml overnight) resulted in expression of abundant Fpr-rs8 message, and in increased steady-state levels of Fpr-rs2 (fig. 8b). Consistent with this, freshly isolated macrophages elicited from the peritoneal cavities of mice irritated with thioglycolate intraperitoneally for 3 days also expressed Fpr-rs2, but not Fpr-rs8 (fig. 8c). However, as with the RAW 264.7 cell line, adding 500 ng/ml LPS to the culture medium overnight not only increased expression of Fpr-rs2 but also induced de novo expression of Fpr-rs8. In the absence of LPS, Fpr-rs8 mRNA was not detectable in macrophages cultured overnight, and Fpr-rs2 expression actually decreased (fig. 8c).
Fpr-rs8-GFP Fusion Protein Is Localized to the Cytoplasm in HEK 293 Cells
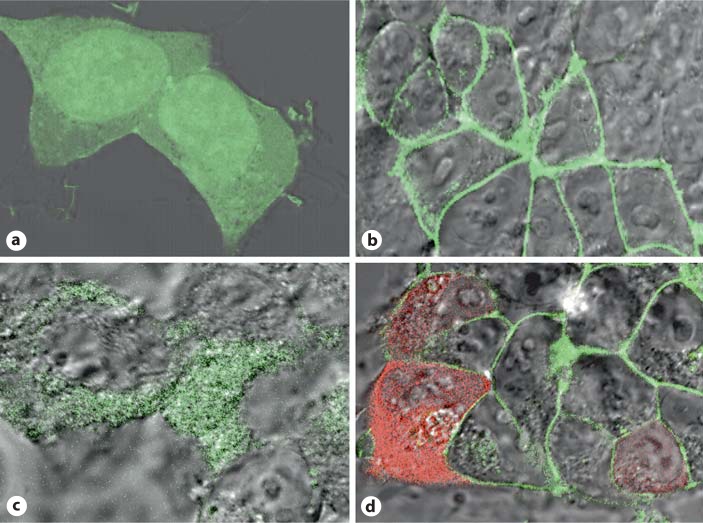
Lacking antibody reagents specific for the Fpr-rs2- and Fpr-rs8-encoded proteins, we have been unable to determine directly whether they are actually produced, and if so where. We have previously inferred expression of the Fpr-rs2 product Fpr2 using knockout mice and functional/pharmacologic criteria, specifically calcium flux and chemotaxis assays in wild-type versus Fpr1 knockout mice [19], since the only two known fMLF receptors Fpr1 and Fpr2 have reasonably well-defined pharmacologic signatures. To obtain some information about the potential for expression of the Fpr-rs2 and Fpr-rs8 protein products, we created C-terminal GFP and RFP fusion constructs and examined subcellular localization in transfected living HEK 293 cells. Cells transfected with control EGFP plasmid alone were found to have diffuse cytoplasmic distribution of EGFP (fig. 9a). Cells transfected with Fpr-rs2-EGFP fusion construct had green fluorescence localized specifically to the plasma membrane as expected (fig. 9b). In contrast, cells transfected with the Fpr-rs8-EGFP fusion construct displayed green fluorescence in the cytoplasm, and not the nucleus or plasma membrane (fig. 9c). Double transfection of HEK 293 cells with Fpr-rs2-EGFP and Fpr-rs8-RFP clearly displayed green fluorescence on the plasma membrane and red fluorescence in the cytoplasm (fig. 9d). These data suggest that the protein encoded by Fpr-rs8 may not be expressed on the plasma membrane, unlike other GPCRs, and that it does not physically interact constitutively in these cells with the product of Fpr-rs2 (Fpr2). We also conducted calcium flux experiments using wild-type and Fpr-rs8 knockout mouse neutrophils, but were unable to observe significant differences in fMLF-induced responses (data not shown).
Fig. 9.
An Fpr-rs8-GFP fusion protein localizes to the cytoplasm in transfected HEK 293 cells. Confocal microscopic images are shown for representative fields of living cells transfected with plasmids encoding EGFP (a), Fpr-rs2-EGFP fusion protein (b), Fpr-rs8-EGFP fusion protein (c) or both Fpr-rs2-EGFP and Fpr-rs8-RFP fusion proteins (d).
Fpr-rs8-Deficient Mice May Have Shortened Life Span
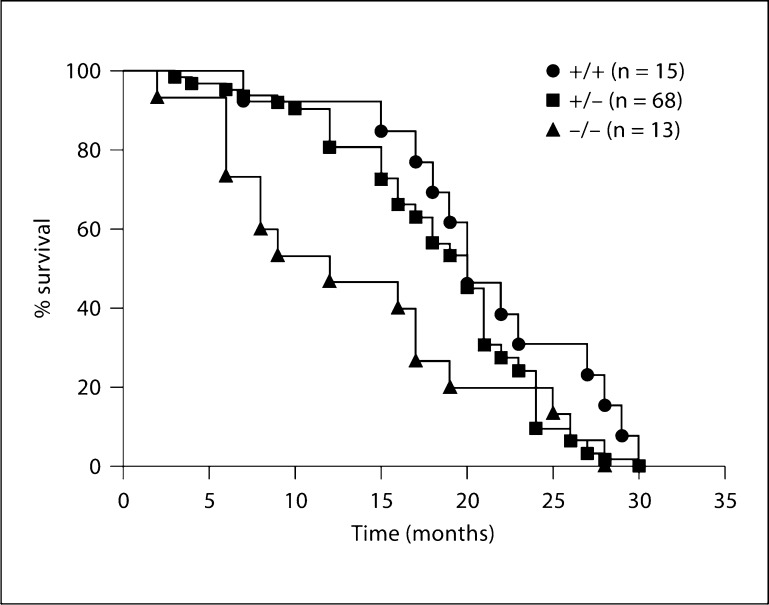
Fpr-rs8+/+, Fpr-rs8+/– and Fpr-rs8–/– mice were examined for developmental and immunologic phenotypes. Three mice of each sex were examined for each genotype at ages ranging from 7 to 19 months. The average age for each genotype group was 14 ± 3 months. No significant differences were found among the groups for body weight, organ weights (heart, liver, brain, thymus, spleen and kidney) or complete blood counts. Serum chemistries fell within the normal range for all groups. Histological examination of tissue samples (see Materials and Methods for full list) found no consistent differences between wild-type and knockout mice. Spleen, bone marrow and blood smears were similar among the three groups. We also established a cohort of Fpr-rs8+/+, Fpr-rs8+/– and Fpr-rs8–/– mice from littermates for an aging study. No differences among groups were observed with regard to types of illness or final diagnoses at death. However, knockout mice remained healthy for a shorter period of time (13.7 ± 8.2 months, mean ± SEM) compared with wild-type mice (21.2 ± 6.4 months; p = 0.014, Fpr-rs8–/– vs. Fpr-rs8+/+) or heterozygotes (18.6 ± 5.9 months; p = 0.168, Fpr-rs8+/− vs. Fpr-rs8+/+, and p = 0.009, Fpr-rs8+/− vs. Fpr-rs8–/–) (fig. 10).
Fig. 10.
Fpr-rs8 knockout mice have reduced longevity. Over a 2-year period, Fpr-rs8 knockout, heterozygous and wild-type littermates were weaned and left unstressed in their cages on normal protocol chow and water ad libitum. When mice showed signs of being moribund that met Animal Care and Use Committee approved criteria for euthanasia, they were sacrificed and sent to the NIH Division of Veterinary Resources for necropsy. Survival is shown by Kaplan-Meyer plot for each genotype (p = 0.014 and 0.009 for comparison of Fpr-rs8−/− vs. Fpr-rs8+/+ and Fpr-rs8+/−, respectively).
Since Fpr-rs8 is a member of a gene family known to be involved in the inflammatory response (including responses to LPS) [Gao et al., unpubl. observations], we challenged Fpr-rs8 knockout mice with LPS as described in the Materials and Methods section. However, no difference in clinical condition compared to wild-type controls was observed. All mice were sacrificed at 6 h after injection. Since formylpeptide receptors are generally thought to be involved in host defense, we have also tested Fpr-rs8 knockout mice by intravenous challenge with Trypanosoma cruzi, a protozoan which causes myocarditis due in part to phagocyte accumulation in the heart. Control C57Bl/6 mice are able to control parasitemia within 40 days and survive. No difference in survival was observed for the Fpr-rs8 knockouts (n = 5 in each group).
Discussion
In the present work, we have provided an initial characterization of Fpr-rs8, an orphan member of the mouse FPR gene family, and shown that it has unusual properties compared to other family members characterized to date. Fpr-rs8 is located in the FPR gene cluster on chromosome 17, approximately 30 kb downstream of Frp-rs2, from which it appears to have arisen by gene duplication. The atypical features relate specifically to the encoded theoretical protein product (see below).
Fpr-rs8 has typical features of a functional gene, including a large ORF, a favorable Kozak sequence and evidence of regulated mRNA expression. In particular, tissue and cell type-specific constitutive expression was detected at the RNA level by gene-specific PCR-RFLP analysis in spleen, lymph node, neutrophil and monocyte. Moreover, mRNA accumulated in macrophages stimulated with LPS in vitro, a property typical of many innate immunoregulatory genes, including other FPR family members [20], which suggests it may play a role in host defense and inflammation. Consistent with these in vitro findings, increased Fpr-rs8 expression was noted in spleen after LPS injection of wild-type C57Bl/6 mice, and this correlated with an increase in splenic neutrophils, which may express it constitutively.
The atypical properties of the putative Fpr-rs8 protein product include (1) small theoretical size (289 amino acids vs. approx. 350 amino acids for other chemotactic GPCRs), (2) a unique C-terminal sequence and (3) possible failure to traffic to the plasma membrane. Small size is the consequence of a single base pair deletion after codon 212 relative to the Fpr-rs2 sequence, which results in a premature stop codon after codon 289. Thus, the Fpr-rs8 ORF DNA sequence is not chimeric but rather homologous to that of Fpr-rs2 from start to stop. Interestingly, the sequence before the deletion is 98% identical to that of Fpr-rs2, but only 83% after the deletion, suggesting very different evolutionary pressures and histories of these regions. At a practical level, this feature made possible our development of a PCR-RFLP-based assay to discriminate the two genes at the genomic and mRNA levels, and allowed us to show that the deletion was not a somatic event, but instead occurred in the germline of the Deltagen C57Bl/6 mouse. Despite small size, secondary structure analysis suggested that a 7-transmembrane domain topography was still possible for the predicted Fpr-rs8 protein; however, this would consume the entire sequence leaving no amino acids to form a cytoplasmic tail.
After the frame shift, the remaining codons in the Fpr-rs8 ORF encode a unique 77-amino acid sequence with no significant relatedness to any FPR family member or any other protein in the Genbank database. Since prior to the frame shift Fpr-rs2 and Fpr-rs8 encode proteins that differ by only 2 amino acids, the unique 77-amino acid C-terminal sequence is most likely responsible for the failure of the Fpr-rs8-EGFP fusion protein we constructed to traffic to the plasma membrane. Since fluorescence was detected in the cytoplasm of HEK 293 cells transfected with the plasmid encoding this fusion protein, we infer that the protein was most likely produced.
The lack of surface expression precluded ligand screening for the theoretical receptor encoded by Fpr-rs8. However, since we detected EGFP in the cytoplasm, Fpr-rs8 protein could theoretically act as a cytoplasmic receptor, for example for formylpeptides of mitochondrial origin, to relay signals through G proteins. Another possibility is that it may regulate other FPR family members by heterodimerization. There are now many reports of GPCR heterodimers, including for chemokine receptors which are highly related to FPR family members [21]. In any case, in an analysis limited to calcium flux we were unable to detect any evidence of altered signaling of endogenous formylpeptide receptors in neutrophils lacking an endogenous Fpr-rs8 product (data not shown).
The presence of a single base pair deletion and frame shift relative to a known functional gene raises the important possibility that Fpr-rs8 might be transitioning from a gene to a pseudogene. Pseudogenes arise from gene duplication or retrotransposition. The gene then accumulates stop codons, frame shifts and base deletions or insertions [22,23]. After duplication, pseudogenes can conceivably be dead on arrival at the new site or else die by accumulating mutations over time. Some pseudogenes have been reported to be expressed, and are referred to as functional pseudogenes. Complete genome sequencing has shown that pseudogenes are common. It has been speculated that they are retained in order to maintain a reservoir or resource of genetic material to provide the organism adaptability to changing conditions and stress [22,23]. Whether Fpr-rs8 is in the process of inactivation, is a functional pseudogene or has a unique and important biological function is an important question for future research. However, to date, the only evidence we have been able to generate that suggests functionality is the intriguing observation that in a 2-year aging study Fpr-rs8 knockout mouse life span was markedly reduced (approx. 33%) compared with wild-type and heterozygous littermates. We did not detect a difference in the cause of death between wild-type and knockout animals.
In conclusion, Fpr-rs8 is a new member of the mouse FPR gene family which appears to have arisen by duplication of Fpr-rs2. Fpr-rs8 is a functional gene as defined by expression of a regulated mRNA. Like Fpr-rs2, Fpr-rs8 is differentially expressed in multiple immune system tissues and innate immune cells, and expression can be constitutive and/or upregulated by LPS, both in vitro and in vivo. Unlike Fpr-rs2, Fpr-rs8 has an atypical short ORF and the putative protein product is localized to the cytoplasm of transfected cells, not the plasma membrane. There is no cellular or biochemical evidence yet of functional activity for the putative Fpr-rs8 product; however, mice in which the gene has been disrupted have markedly reduced life span without a signature pathology. Whether this phenotype is caused by loss of an Fpr-rs8 function or by distal effects on expression of other genes remains to be determined.
Acknowledgements
The authors wish to thank Dr. Meggan Czapiga and Dr. Owen Schwartz of the Biological Imaging Facility, NIAID Research Technologies Branch for excellent assistance with the confocal microscopy. This work was supported by funding from the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health.
References
- 1.Le Y, Murphy PM, Wang JM. Formyl-peptide receptors revisited. Trends Immunol. 2002;23:541–548. doi: 10.1016/s1471-4906(02)02316-5. [DOI] [PubMed] [Google Scholar]
- 2.Migeotte I, Communi D, Parmentier M. Formyl peptide receptors: a promiscuous subfamily of G protein-coupled receptors controlling immune responses. Cytokine Growth Factor Rev. 2006;17:501–519. doi: 10.1016/j.cytogfr.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 3.Ye RD, Boulay F, Wang JM, et al. International Union of Basic and Clinical Pharmacology. LXXIII. Nomenclature for the formyl peptide receptor (FPR) family. Pharmacol Rev. 2009;61:119–161. doi: 10.1124/pr.109.001578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rabiet MJ, Huet E, Boulay F. Human mitochondria-derived N-formylated peptides are novel agonists equally active on FPR and FPRL1, while Listeria monocytogenes-derived peptides preferentially activate FPR. Eur J Immunol. 2005;35:2486–2495. doi: 10.1002/eji.200526338. [DOI] [PubMed] [Google Scholar]
- 5.Southgate EL, He RL, Gao JL, Murphy PM, Nanamori M, Ye RD. Identification of formyl peptides from Listeria monocytogenes and Staphylococcus aureus as potent chemoattractants for mouse neutrophils. J Immunol. 2008;181:1429–1437. doi: 10.4049/jimmunol.181.2.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ye RD, Cavanagh SL, Quehenberger O, Prossnitz ER, Cochrane CG. Isolation of a cDNA that encodes a novel granulocyte N-formyl peptide receptor. Biochem Biophys Res Commun. 1992;184:582–589. doi: 10.1016/0006-291x(92)90629-y. [DOI] [PubMed] [Google Scholar]
- 7.Showell HJ, Freer RJ, Zigmond SH, et al. The structure-activity relations of synthetic peptides as chemotactic factors and inducers of lysosomal secretion for neutrophils. J Exp Med. 1976;143:1154–1169. doi: 10.1084/jem.143.5.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao JL, Chen H, Filie JD, Kozak CA, Murphy PM. Differential expansion of the N-formylpeptide receptor gene cluster in human and mouse. Genomics. 1998;51:270–276. doi: 10.1006/geno.1998.5376. [DOI] [PubMed] [Google Scholar]
- 9.Gao JL, Murphy PM. Species and subtype variants of the N-formyl peptide chemotactic receptor reveal multiple important functional domains. J Biol Chem. 1993;268:25395–25401. [PubMed] [Google Scholar]
- 10.Takano T, Fiore S, Maddox JF, Brady HR, Petasis NA, Serhan CN. Aspirin-triggered 15-epi-lipoxin A4 (LXA4) and LXA4 stable analogues are potent inhibitors of acute inflammation: evidence for anti-inflammatory receptors. J Exp Med. 1997;185:1693–1704. doi: 10.1084/jem.185.9.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang ZG, Ye RD. Characterization of two new members of the formyl peptide receptor gene family from 129S6 mice. Gene. 2002;299:57–63. doi: 10.1016/s0378-1119(02)01012-0. [DOI] [PubMed] [Google Scholar]
- 12.Riviere S, Challet L, Fluegge D, Spehr M, Rodriguez I. Formyl peptide receptor-like proteins are a novel family of vomeronasal chemosensors. Nature. 2009;459:574–577. doi: 10.1038/nature08029. [DOI] [PubMed] [Google Scholar]
- 13.Gao JL, Lee EJ, Murphy PM. Impaired antibacterial host defense in mice lacking the N-formylpeptide receptor. J Exp Med. 1999;189:657–662. doi: 10.1084/jem.189.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perretti M, Getting SJ, Solito E, Murphy PM, Gao JL. Involvement of the receptor for formylated peptides in the in vivo anti-migratory actions of annexin 1 and its mimetics. Am J Pathol. 2001;158:1969–1973. doi: 10.1016/S0002-9440(10)64667-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gwinn MR, Sharma A, De Nardin E. Single nucleotide polymorphisms of the N-formyl peptide receptor in localized juvenile periodontitis. J Periodontol. 1999;70:1194–1201. doi: 10.1902/jop.1999.70.10.1194. [DOI] [PubMed] [Google Scholar]
- 16.Nanamori M, He R, Sang H, Ye RD. Normal cell surface expression and selective loss of functions resulting from Phe110 to Ser and Cys126 to Trp substitutions in the formyl peptide receptor. Immunol Invest. 2004;33:193–212. doi: 10.1081/imm-120034234. [DOI] [PubMed] [Google Scholar]
- 17.Chen K, Le Y, Liu Y, et al. A critical role for the G protein-coupled receptor mFPR2 in airway inflammation and immune responses. J Immunol. 2010;184:3331–3335. doi: 10.4049/jimmunol.0903022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dufton N, Hannon R, Brancaleone V, et al. Anti-inflammatory role of the murine formyl-peptide receptor 2: ligand-specific effects on leukocyte responses and experimental inflammation. J Immunol. 2010;184:2611–2619. doi: 10.4049/jimmunol.0903526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartt JK, Barish G, Murphy PM, Gao JL. N-formylpeptides induce two distinct concentration optima for mouse neutrophil chemotaxis by differential interaction with two N-formylpeptide receptor (FPR) subtypes. Molecular characterization of FPR2, a second mouse neutrophil FPR. J Exp Med. 1999;190:741–747. doi: 10.1084/jem.190.5.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tiffany HL, Lavigne MC, Cui YH, et al. Amyloid-beta induces chemotaxis and oxidant stress by acting at formylpeptide receptor 2, a G protein-coupled receptor expressed in phagocytes and brain. J Biol Chem. 2001;276:23645–23652. doi: 10.1074/jbc.M101031200. [DOI] [PubMed] [Google Scholar]
- 21.Springael JY, Urizar E, Parmentier M. Dimerization of chemokine receptors and its functional consequences. Cytokine Growth Factor Rev. 2005;16:611–623. doi: 10.1016/j.cytogfr.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 22.Balakirev ES, Ayala FJ. Pseudogenes: are they ‘junk’ or functional DNA? Annu Rev Genet. 2003;37:123–151. doi: 10.1146/annurev.genet.37.040103.103949. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Z, Gerstein M. Large-scale analysis of pseudogenes in the human genome. Curr Opin Genet Dev. 2004;14:328–335. doi: 10.1016/j.gde.2004.06.003. [DOI] [PubMed] [Google Scholar]