Abstract
Background
Reduced peroxisome proliferator-activated receptor-γ coactivator 1α (PGC1α) gene expression has been observed in striatal cell lines, transgenic mouse models of Huntington's disease (HD), and brain tissue from HD patients. As this protein is a key transcription regulator of the expression of many mitochondrial proteins, these observations strongly support the role of aberrant mitochondrial function in the pathogenesis of HD. The PGC1α protein undergoes posttranslational modifications that affect its transcriptional activity. The N-truncated splice variant of PGC1α (NT-PGC1α) is produced in tissues, but the role of truncated splice variants of PGC1α in HD and in the regulation of mitochondrial gene expression has not been elucidated.
Objective
To examine the expression and modulation of expression of NT-PGC1α levels in HD.
Methods and Results
We found that the NT-PGC1α protein, a splice variant of ∼38 kDa, but not full-length PGC1α is severely and consistently altered in human HD brain, human HD myoblasts, mouse HD models, and HD striatal cells. NT-PGC1α levels were significantly upregulated in HD cells and mouse brown fat by physiologically relevant stimuli that are known to upregulate PGC1α gene expression. This resulted in an increase in mitochondrial gene expression and cytochrome c content.
Conclusion
Our data suggest that NT-PGC1α is an important component of the PGC1α transcriptional network, which plays a significant role in the pathogenesis of HD.
Copyright © 2011 S. Karger AG, Basel
Key Words: Neurodegeneration, PGC1α, Mitochondrial gene expression, Alternative splicing, Huntington's disease, Striatal neurons
Introduction
Huntington's disease (HD) is a progressive neurological disorder caused by a mutation in the huntingtin gene resulting in an expanded polyglutamine repeat within exon 1 [1]. Aberrant transcriptional regulation, oxidative stress, and dysregulation of mitochondrial energy metabolism have been implicated in the pathogenesis of HD [2,3,4]. Recent findings [5,6] suggest that mutant huntingtin may disrupt normal mitochondrial function by inhibiting the expression of peroxisome proliferator-activated receptor-γ coactivator 1α (PGC1α), a transcriptional coactivator that has been shown to regulate the expression of nuclear-encoded mitochondrial proteins. cAMP signaling is a key activator of PGC1α transcription in many tissues, promoting the binding of cAMP response element (CRE)-binding protein (CREB) or the activating transcription factor-2 to a conserved DNA response element in the PGC1α promoter. Mutant huntingtin occupies the PGC1α promoter region and represses CRE-mediated transcription of PGC1α, by interfering with the CREB/TAF4 transcriptional pathway, in the brains and cells of transgenic HD mice [5]. Reduced CRE-dependent transcription has been previously observed as an early abnormality in the pathogenesis of HD [7,8,9]. Moreover, it has been shown that cAMP levels are reduced in a striatal cell model of HD, i.e. STHdhQ111/Q111 cells [7]; treatment of these cells with 8-bromo-cAMP significantly upregulated the activity of the PGC1α reporter [5].
Several PGC1α splice variants have been reported which might arise from differential internal splicing and/or alternative promoter usage [10,11,12,13]. The role of these variants in the regulation of transcription remains to be elucidated [14]. Zhang et al. [13] suggested that a truncated form of PGC1α (NT-PGC1α) may exhibit regulatory activity. This protein is produced by alternative 3′ splicing between exons 6 and 7 that introduces an in-frame stop codon into PGC1α mRNA. NT-PGC1α retains the N-terminal transcriptional activation and nuclear-receptor interacting domains but lacks all domains within 268–797 amino acids of the full-length protein. NT-PGC1α produces a cell context-specific subset of responses that complement, overlap, or prolong the actions of PGC1α [13].
Here, we report that the NT-PGC1α protein is the major variant of PGC1α that is severely altered in human HD brain and in mouse and cell models of HD. Stimulation of the expression of NT-PGC1α in HD cells results in upregulation of the expression of PGC1α-controlled mitochondrial genes. NT-PGC1α is responsive to cold stimuli (a well-established physiologic stimulus of PGC1α) in brown adipose tissue (BAT). Thus, NT-PGC1α appears to be an important component of the PGC1α transcriptional network that may play a role in the pathogenesis of HD.
Animals and Methods
Transgenic Animals
The N171-18Q mice, N171-82Q mice, and R6/2 mice were from Jackson Labs. The N171 mice have the N-terminal fragment (171 amino acids) of human huntingtin and either 82 or 18 glutamines on a B6C3F1 background [15]. The R6/2 mice were created with a 1.9-kb human genomic fragment containing promoter sequences and exon 1 carrying expansions of approximately 130 CAG repeats on a CBA × C57BL/6 background [16]. The animals were kept on a 12-hour light/dark cycle with food and water available ad libitum. All experiments were conducted in accordance with National Institutes of Health guidelines for animal research and were approved by the Weill Cornell Medical College Animal Care and Use Committee.
Striatal Cell Culture and Treatments
STHdhQ7/Q7 (Q7) and STHdhQ111/Q111 (Q111) cells, generated from striatal primordia of wild-type (WT) HdhQ7/Q7 and homozygous mutant HdhQ111/Q111 knock-in mouse embryos, respectively [17], were a gift from Dr. Marcy E. MacDonald. The cells were cultured in DMEM (33°C, 5% CO2, 10% FBS, 1% antibiotic-antimycotic, and 400 μg/ml G418) and treated with 10 μM forskolin + 50 μM 3-isobutyl-1-methylxanthine (IBMX) for 2 h.
Myoblast Cell Culture
Myoblasts were derived from muscle biopsies obtained from nondiseased subjects and HD patients (who gave their informed consent) and cultured as previously described [18]. Myoblasts were grown in HAM's F10 medium (GIBCO; Invitrogen, Carlsbad, Calif., USA) supplemented with 15% FBS (Invitrogen, San Diego, Calif., USA), 0.5 mg/ml bovine serum albumin, 4 ng/ml insulin, 10 ng/ml epidermal growth factor, 0.39 μg/ml dexamethasone, 0.1 mg/ml streptomycin, and 100 U/ml penicillin.
Human Brain Samples
Human postmortem brain specimens from the putamen region (HD grade 2, n = 5; grade 3, n = 4, and grade 4, n = 2; mean age ∼63 years, range 53–75; age-matched nondiseased samples (n = 7), mean age ∼65 years, range 57–76) were generously provided by the New York Brain Bank at Columbia University, Taub Institute.
Cold Challenge
WT and R6/2 mice were exposed to 4°C for 4 h [19] and then sacrificed by cervical dislocation; BAT was harvested and frozen immediately.
Gene Expression Analysis by RT-PCR
Striatal cells were scraped and total RNA was isolated using Trizol reagent (Invitrogen). Genomic DNA was removed using RNase-free DNase (Ambion) in RNA pellets resuspended in DEPC-treated water (Ambion). Total RNA purity and integrity was confirmed by a ND-1000 NanoDrop (NanoDrop Technologies) and a 2100 Bioanalyzer (Agilent), respectively, with average 260/280 ratios for all study samples ranging from 1.9 to 2.1 and average RNA integrity numbers ranging from 7.5 to 9.0. Equal amounts of RNA were diluted in nuclease-free water (Ambion) to a final concentration of 10 ng/μl and reverse transcribed using a RevertAid First Strand cDNA Synthesis Kit (Fermentas, USA). Real-time RT-PCR was performed using the ABI prism 7900 HT sequence detection system (Applied Biosystems, Foster City, Calif., USA) to detect changes in mRNA expression using various primer pairs (table 1) at a final volume of 20 μl. All qPCR plating was performed on ice. Expression of the gene cyclophilin B served as a control to normalize values. Thermal cycling conditions were 50°C for 2 min, 95°C for 10 min, and then 40 cycles of 95°C for 15 s and 60°C for 1 min. Relative expression was calculated using the ΔΔCt method.
Table 1.
Primer sequences used for real-time quantitative PCR
| Gene name | Forward primer | Reverse primer |
|---|---|---|
| NT-PGClα | tgccattgttaagaccga | ggtcactggaagatatgg |
| Tfam | agccaggtccagctcactaa | aaacccaagaaagcatgtgg |
| NRF-1 | tggtccagagagtgcttgtg | ttcctgggaagggagaagat |
| Cyclophilin B | ccatcgtgtcatcaaggacttcat | cttgccatccagccaggaggtctt |
Western Blot
Human brain tissues, striatum of WT and HD mice, myoblasts, and striatal cells were homogenized in cell extraction buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 2 mM EDTA, 1% SDS, 0.5% NP-40, and 0.5% deoxycholate supplemented with protease and phosphatase inhibitors (Sigma), and the protein concentration was determined using a BCA protein assay (Pierce, USA). Equal amounts of protein (40 μg) were loaded onto a 4–20% Tris-glycine gel (Invitrogen). The positive controls (cell lysates of COS1 cells transfected with the full-length PGC1α or NT-PGC1α) for full-length PGC1α and NT-PGC1α were obtained from Calbiochem. After transfer, membranes were blocked for 1h at room temperature in Tris-buffered saline/Tween-20 (TBST) [50 mM Tris-HCl, 150 mM NaCl (pH 7.4), and 1% Tween-20] containing 5% nonfat dried milk. The membranes were incubated overnight at 4°C for PGC1α (1:750; Calbiochem), uncoupling protein-1 (UCP-1) (1:1,000; Calbiochem), and α-tubulin (1:10,000; Sigma). The membranes were then washed 3 times with TBST and incubated for 1h with HRP-conjugated secondary antibody, and the immunoreactive proteins were detected using a chemiluminescent substrate (Pierce). Protein expression was quantified using Scion Image software (NIH, USA).
HPLC Assay for cAMP
The cultured cells in each well were washed twice with phosphate-buffered saline (PBS) and then lysed by adding 150 μl ice-embedded acetonitrile followed by 75 μl ice-cold sterile water. Lysates were centrifuged at 14,000 rpm and 4°C for 20 min. The supernatant was taken and diluted with an equal volume of water for HPLC assay. Fifty microliters of supernatant was applied to an HPLC system which comprises a Perkin Elmer M-250 binary LC pump, a Waters 717 plus autosampler, a Waters 2489 UV detector set at 260-nm wavelength, and an ESA 501 chromatography data process system. The gradient elution was built with 2 mobile phases at a rate of 1 ml/min through a 4.6 × 250 mm, 5-μm particle size TSK-GEL HPLC column. Mobile phase A contained 25 mM NaH2PO4 and 100 mg/l tetrabutylammonium (pH 5.5), and mobile phase B contained 10%(v/v) acetonitrile mixed in a buffer of 200 mM NaH2PO4 and 100 mg/l tetrabutylammonium (pH 4.0). Quantification was carried out using external standard calibration. The protein concentration was measured from the cell lysate.
Statistical Analysis
Data are presented as means ± SEM. Statistical analysis was performed using GraphPad InStat software (GraphPad Software, Inc., San Diego, Calif., USA). The mean significant difference was determined using a 1-way ANOVA followed by a Tukey-Kramer post hoc multiple comparisons test or a 2-tailed Student's t test where appropriate. p < 0.05 was considered statistically significant.
Results
The NT-PGC1α Protein Is Altered in HD Human and Mouse Brain
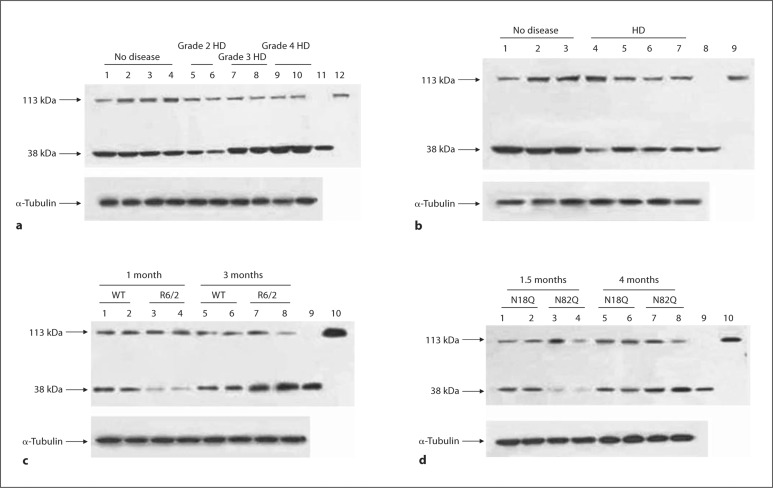
Using an antibody that recognizes both the full-length (‘PGC1α’, ∼113 kDa) form and the spliced form (‘NT-PGC1α’, ∼38 kDa) of PGC1α, we analyzed several samples of human brain, human myoblasts, and mouse brain (fig. 1a–d shows representative Western blots) and Q111 HD cells (fig. 2a). The depletion of the full-length PGC1α protein was not consistent in human HD myoblasts and human and mouse HD brain samples (fig. 1a–d) or in Q111 HD cells (fig. 2a). However, the NT-PGC1α protein was prominently and consistently depleted in human HD myoblasts and mouse Q111 striatal cells (fig. 1b, 2a). In human brain, NT-PGC1α was depleted in grade 2 HD patients, while it was upregulated in grade 3 and grade 4 HD patients (fig. 1a). NT-PGC1α was severely depleted in young HD transgenic mice, R6/2 mice, and N171-82Q mice, but it was upregulated in older phenotypic HD mice (fig. 1c, d). It appears that the amount of NT-PGC1α rather than the amount of the full-length PGC1α variant is severely altered in HD human and mouse brain and in cultured HD myoblasts and striatal cells.
Fig. 1.
Evidence of the altered protein levels of NT-PGC1 in the putamen of symptomatic HD patients (a), in human HD myoblasts (b), and in the striatum of 2 mouse models of HD, i.e. R6/2 (c), and N171-82Q (d). a-d Representative Western blots. Upper panels represent the full-length PGC1 (113 kDa) and NT-PGC1 (38 kDa) isoforms, and lower panels represent-tubulin as a loading control. a Lanes 1-4 contain no disease control samples, lanes 5 and 6 are samples from grade 2 HD patients, lanes 7 and 8 contain samples from grade 3 HD patients, lanes 9 and 10 are samples from grade 4 HD patients, and lanes 11 and 12 contain the positive controls for NT-PGC1 and full-length PGC1, respectively. Western blot analysis shows that the full-length isoform is downregulated in 1 out of 4 control samples and in all HD patients. The protein expression level of the short isoform is severely reduced in grade 2 HD patients and upregulated in grade 3 and grade 4 HD patients. b Lanes 1-3 contain no disease control (normal CAG repeat size) human myoblasts, and lanes 4-7 contain myoblasts from HD patients (CAG repeats: lane 4 = 40/46, lane 5 = 17/51, lane 6 = 18/48, and lane 7 = 10/42). In human myoblasts, there are no consistent changes in the full-length isoform, while the short isoform is downregulated in all of the HD myoblasts. Lanes 8 and 9 contain the positive controls for NT-PGC1α and full-length PGC1α, respectively. c Representative Western blots (total number of animals used per group = 6). Lanes 1-4 contain striatum samples from 1-month-old WT (lanes 1 and 2) and R6/2 (lanes 3 and 4) mice. Lanes 5-8 contain striatum samples from 3-monthold WT (lanes 5 and 6) and R6/2 (lanes 7 and 8) mice. Lanes 9 and 10 contain the positive controls for NT-PGC1α and full-length PGC1α, respectively. d Representative Western blots (total number of animals used per group = 6). Lanes 1-4 contain striatum samples from 1.5-month-old N171-18Q (lanes 1 and 2) and N171-82Q (lanes 3 and 4) mice. Lanes 5-8 contain striatum samples from 4-month-old N171-18Q (lanes 5 and 6) and N171-82Q (lanes 7 and 8) mice. Lanes 9 and 10 contain the positive controls for NTPGC1α and full-length PGC1α, respectively. In younger R6/2 and N171-82Q mice (c, d), NT-PGC1α is severely reduced compared to that in WT or N171-18Q mice, respectively, while it is upregulated in symptomatic R6/2 and N171-82Q mice.
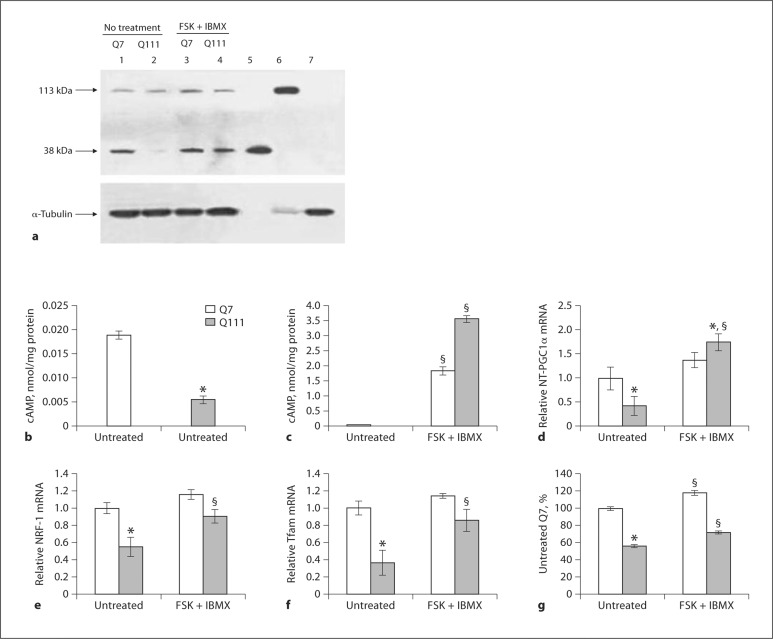
Fig. 2.
Inducibility of NT-PGC1 in striatal Q7 and Q111 cells upon chemical stimulation. a Representative Western blots (number of samples = 5 per group) showing protein expression levels for full-length PGC1α and NT-PGC1α (top panel) in untreated Q7 and Q111 cells (lanes 1 and 2, respectively) and in cells treated with a combination of forskolin + IBMX (FSK + IBMX) (lanes 3 and 4, respectively). Lanes 5, 6, and 7 contain, respectively, positive controls for NT-PGC1α, full-length PGC1α, and brain lysate from PGC1α knock-out mice. Lower panel indicates α-tubulin as a loading control. NT-PGC1α is severely suppressed in Q111 cells and responds to treatment with FSK + IBMX. b, c HPLC measurement of cAMP levels in Q7 and Q111 striatal cells. b cAMP levels in untreated Q7 and Q111 cells, expressed as nanomols per milligram of protein. cAMP levels are significantly lower in Q111 cells compared to Q7 cells at baseline. c FSK + IBMX produces a several-fold increase in the cAMP concentration in Q7 and Q111 cells. Data are expressed as means ± SEM from 3 experiments. * p < 0.01 compared to untreated Q7; § p < 0.001 compared to untreated Q7 or Q111, respectively. Quantitative real-time PCR analysis (number of samples = 3 per group) of relative NT-PGC1α (d), NRF-1 (e), and Tfam (f) mRNA expression normalized to cyclophilin B. The mRNA expression levels for NT-PGC1α, NRF-1, and Tfam are significantly downregulated in untreated Q111 cells compared to untreated Q7 cells, and treatment with FSK + IBMX (10 μM FSK and 50 μM, 2 h) significantly increases the expression levels for NT-PGC1α, NRF-1, and Tfam mRNA in Q111 cells. Data are presented as means ± SEM from 3 experiments. * p < 0.05 compared to untreated Q7; § p < 0.05 compared to untreated Q111 cells. g Changes in mitochondrial Cyt c in striatal neurons following treatment with FSK + IBMX detected by ELISA. The amount of Cyt c was calculated as nanograms per milligram of protein and expressed as a percent of that in untreated Q7 cells. Cyt c protein is significantly reduced in Q111 cells compared to Q7 cells. Treatment with FSK + IBMX significantly increases Cyt c levels in both Q7 and Q111 striatal cells. Data are expressed as means ± SEM from 3 experiments. * p < 0.01 compared to untreated Q7; § p < 0.001 compared to untreated Q7; or Q111 cells, respectively.
Elevated Intracellular cAMP Upregulates the Expression of Mitochondrial Genes and the NT-PGC1α Protein in HD Cells
PGC1α gene expression in HD cells can be significantly increased by cAMP [5]. We examined whether NT-PGC1α protein levels are affected by cAMP in Q111 HD cells. Consistent with earlier reports [7], cAMP was greatly reduced in the Q111 cells versus Q7 cells (fig. 2b). We treated Q7 and Q111 cells with forskolin + the phosphodiesterase inhibitor IBMX, which resulted in a 100-fold increase in cAMP levels in the Q7 and in a 710-fold increase in Q111 cells (fig. 2c). This treatment also resulted in a significant increase in NT-PGC1αprotein (fig. 2a) and mRNA (fig. 2d) levels in Q111 striatal cells. We also observed about a 3-fold increase in the protein expression of NT-PGC1α in Q111 cells treated with a cAMP analog (8-bromo-cAMP; data not presented), which corroborates the finding that the expression of NT-PGC1α is modulated by cAMP levels in these cells (fig. 2a, d).
PGC1α acts upstream of nuclear respiratory factor NRF-1, which regulates the expression of nuclear-encoded mitochondrial genes such as mitochondrial transcription factor A (Tfam) and cytochrome c (Cyt c). We examined whether cAMP-mediated upregulation of NT-PGC1α in Q111 HD cells modulates the expression of its downstream targets. The mRNA expression levels of NRF-1 and Tfam were significantly downregulated in Q111 cells versus Q7 cells (fig. 2e, f). Forskolin + IBMX produced similar changes in NRF-1 and Tfam mRNA expression (fig. 2e, f). Similarly, treatment with 8-Br-cAMP (data not presented) also resulted in an increase in NRF-1 and Tfam expression consistent with the expression of NT-PGC1α. Thus, it appears that NT-PGC1α is functionally important in HD cells.
To ensure that an increase in NT-PGC1α protein results in an actual increase in mitochondrial proteins, we quantified cAMP-induced changes in mitochondrial Cyt c by ELISA. The amount of Cyt c protein in Q111 cells before treatment was ∼50% of that in Q7 cells. Treatment of cells with forskolin + IBMX significantly increased Cyt c in HD (Q111) and Q7 cells (fig. 2g).
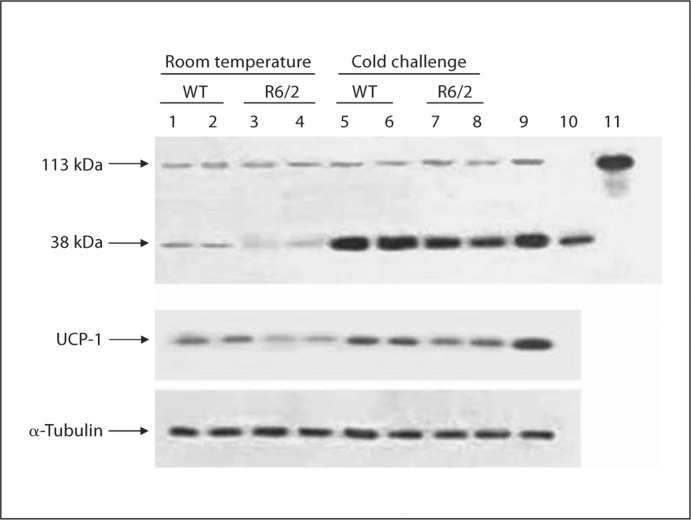
Cold Challenge Upregulated NT-PGC1α in Mouse BAT
PGC1α mRNA is strongly induced in BAT by cold exposure [19], resulting in enhanced mitochondrial heat generation during adaptive thermogenesis. We found that cold exposure of WT and R6/2 mice strongly increased NT-PGC1α protein levels rather than full-length PGC1α in BAT (fig. 3). Interestingly, the level of NT-PGC1α protein is also significantly upregulated in mice overexpressing PGC1α (no treatment or exposure to cold; fig. 3). It is known that PGC1α activates the thermogenic gene program of BAT through the control of UCP-1. We show here that UCP-1 protein levels are upregulated in concert with NT-PGC1α levels (fig. 3).
Fig. 3.
Inducibility of NT-PGC1α in BAT of R6/2 mice upon metabolic stimulation. Cold challenge produces an increase in the levels of NT-PGC1α and UCP-1 protein in the BAT of WT and R6/2 mice. Representative Western blots (n = 6 animals per group) showing protein expression levels for full-length PGC1α and NTPGC1α (top panel) and α-tubulin (lower panel). Lanes 1 and 2 represent untreated WT mice, lanes 3 and 4 represent untreated R6/2 mice, lanes 5 and 6 represent cold-exposed WT mice, lanes 7 and 8 represent cold-exposed R6/2 mice, lane 9 represents BAT from PGC-1α overexpressor mice, and lanes 10 and 11 contain positive controls for NT-PGC1α and full-length PGC1α.
Discussion
Reduced full-length PGC1α expression has been observed in transgenic mouse models of HD, in postmortem brain tissue from HD patients [5,6,20], in muscle biopsies from HD patients, and in human HD myoblast cultures [21]. Our data are in line with this finding (fig. 1a–d). All the studies on the role of PGC1α in HD to date have dealt with full-length PGC1α. Although splice variants of PGC1α have been reported earlier [10,11,12,13], their functional significance has not been elucidated. This is the first report to demonstrate that the amount of NT-PGC1α protein is severely altered in human HD brain, mouse HD models, and HD striatal neurons. We found that NT-PGC1α is significantly upregulated in HD cells by an increase in cAMP and by cold exposure in mouse BAT. An increase in NT-PGC1α was associated with upregulation of gene expression and increased mitochondrial protein content.
PGC1α has a complex structure with multiple domains. N-terminal leucine-rich domains (L2 and L3) mediate the interaction of PGC1α with the ligand-binding domains of nuclear receptors, whereas central and C-terminal domains mediate the interactions with PPARγ, NRF-1, and other transcription factors. PGC1α also has a strong activation domain at the N-terminus, which interacts with a histone acetyltransferase complex, including steroid receptor coactivator-1 (SRC-1) and CREB-binding protein (CBP)/p300. Adjacent to the N-terminal domain is an inhibitory region that spans ∼200 amino acids. The short variant of PGC1α, NT-PGC1α, lacks this inhibitory domain, which implies that NT-PGC1α is likely a bona fide activator of gene expression; this is consistent with our data.
Regulation of PGC1α expression includes a ‘positive feedback’ loop where an increase in expression causes a further increase in its expression [22]. The transcriptional activity of PGC1α is also regulated by interaction with deacetylases such as SIRT1 [23,24], which increases its transcriptional activity, and by changes in PGC1α stability [23,25].
Further studies are required to elucidate whether an increase in NT-PGC1α protein content induced by cAMP in striatal cells or cold challenge in BAT was due to an increase in the stability of NT-PGC1α or an increase in expression of the PGC1α gene. It is not clear why the NT-PGC1α protein amount is strongly decreased in young animals that do not yet show a HD phenotype and at early stages of HD in humans but becomes strongly and consistently elevated in older HD animal models and in human brains with advanced HD stages. This may represent an adaptive increase in NT-PGC1α as an attempt to compensate for the increasing severity of the pathology and loss of mitochondria which occurs with increasing pathologic grades [26]. It has been shown that the major ATP-buffering and redistributing system in cells, i.e. the creatine kinase/phosphocreatine system, is severely reduced in human HD [27]. In theory, this should put more demand on the ATP-producing systems, such as glycolysis and mitochondria. The grade-dependent increases in NT-PGC1α levels in human postmortem brain could also be explained by the occurrence of inflammation and gliosis accompanying the death of neurons in later disease stages in which the glial population preferentially survives. The full-length PGC1α protein is localized in the nucleus and has a fast turnover (t1/2 <30 min), while NT-PGC1α is predominantly cytoplasmic and relatively stable (t1/2 >7 h). It has recently been shown that PGC1α is primarily degraded in the nucleus via a ubiquitin proteasome pathway. It is evident that both isoforms are synthesized in HD tissues despite the presence of huntingtin on the PGC1α promoter. The accumulation of NT-PGC1α may therefore also be explained on the basis of its relatively higher stability and slower degradation, although further studies are required to investigate this phenomenon.
Conclusions
We showed here that the NT-PGC1α protein, a splice variant of ∼38 kDa, but not full-length PGC1α is severely and consistently altered in human HD brain, human HD myoblasts, mouse HD models, and HD striatal cells. The NT-PGC1α levels were significantly upregulated in HD cells and mouse brown fat by physiologically relevant stimuli that are known to upregulate PGC1α gene expression. This resulted in an increase in mitochondrial gene expression and Cyt c content. In summary, our data support the concept that NT-PGC1α is the major variant of PGC1α which is altered in HD brain and cells and that it is biologically active in controlling the expression of mitochondrial genes.
Acknowledgements
This study was supported by NIH grants NS39258 and P01AG14930 to M.F.B. and grant AG014930 to A.A.S. M.F.B. and A.J. are supported by the HDSA Coalition for the Cure. We would like to thank Dr. Bruce Spiegelman for providing knock-out samples.
References
- 1.Vonsattel JP, DiFiglia M. Huntington disease. J Neuropathol Exp Neurol. 1998;57:369–384. doi: 10.1097/00005072-199805000-00001. [DOI] [PubMed] [Google Scholar]
- 2.Beal MF. Mitochondria take center stage in aging and neurodegeneration. Ann Neurol. 2005;58:495–505. doi: 10.1002/ana.20624. [DOI] [PubMed] [Google Scholar]
- 3.Browne SE, Beal MF. The energetics of Huntington's disease. Neurochem Res. 2004;29:531–546. doi: 10.1023/b:nere.0000014824.04728.dd. [DOI] [PubMed] [Google Scholar]
- 4.Browne SE, Beal MF. Oxidative damage in Huntington's disease pathogenesis. Antioxid Redox Signal. 2006;8:2061–2073. doi: 10.1089/ars.2006.8.2061. [DOI] [PubMed] [Google Scholar]
- 5.Cui L, Jeong H, Borovecki F, Parkhurst CN, Tanese N, Krainc D. Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 2006;127:59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 6.Weydt P, Pineda VV, Torrence AE, Libby RT, Satterfield TF, Lazarowski ER, Gilbert ML, Morton GJ, Bammler TK, Strand AD, Cui L, Beyer RP, Easley CN, Smith AC, Krainc D, Luquet S, Sweet IR, Schwartz MW, La Spada AR. Thermoregulatory and metabolic defects in Huntington's disease transgenic mice implicate PGC-1alpha in Huntington's disease neurodegeneration. Cell Metab. 2006;4:349–362. doi: 10.1016/j.cmet.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 7.Gines S, Seong IS, Fossale E, Ivanova E, Trettel F, Gusella JF, Wheeler VC, Persichetti F, MacDonald ME. Specific progressive cAMP reduction implicates energy deficit in presymptomatic Huntington's disease knock-in mice. Hum Mol Genet. 2003;12:497–508. doi: 10.1093/hmg/ddg046. [DOI] [PubMed] [Google Scholar]
- 8.Sugars KL, Rubinsztein DC. Transcriptional abnormalities in Huntington disease. Trends Genet. 2003;19:233–238. doi: 10.1016/S0168-9525(03)00074-X. [DOI] [PubMed] [Google Scholar]
- 9.Sugars KL, Brown R, Cook LJ, Swartz J, Rubinsztein DC. Decreased cAMP response element-mediated transcription: an early event in exon 1 and full-length cell models of Huntington's disease that contributes to polyglutamine pathogenesis. J Biol Chem. 2004;279:4988–4999. doi: 10.1074/jbc.M310226200. [DOI] [PubMed] [Google Scholar]
- 10.Baar K, Wende AR, Jones TE, Marison M, Nolte LA, Chen M, Kelly DP, Holloszy JO. Adaptations of skeletal muscle to exercise: rapid increase in the transcriptional coactivator PGC-1. FASEB J. 2002;16:1879–1886. doi: 10.1096/fj.02-0367com. [DOI] [PubMed] [Google Scholar]
- 11.Miura S, Kai Y, Kamei Y, Ezaki O. Isoform-specific increases in murine skeletal muscle peroxisome proliferator-activated receptor-gamma coactivator-1alpha (PGC-1alpha) mRNA in response to beta2-adrenergic receptor activation and exercise. Endocrinology. 2008;149:4527–4533. doi: 10.1210/en.2008-0466. [DOI] [PubMed] [Google Scholar]
- 12.Yoshioka T, Inagaki K, Noguchi T, Sakai M, Ogawa W, Hosooka T, Iguchi H, Watanabe E, Matsuki Y, Hiramatsu R, Kasuga M. Identification and characterization of an alternative promoter of the human PGC-1alpha gene. Biochem Biophys Res Commun. 2009;381:537–543. doi: 10.1016/j.bbrc.2009.02.077. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, Huypens P, Adamson AW, Chang JS, Henagan TM, Boudreau A, Lenard NR, Burk D, Klein J, Perwitz N, Shin J, Fasshauer M, Kralli A, Gettys TW. Alternative mRNA splicing produces a novel biologically active short isoform of PGC-1alpha. J Biol Chem. 2009;284:32813–32826. doi: 10.1074/jbc.M109.037556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Handschin C, Spiegelman BM. Peroxisome proliferator-activated receptor gamma coactivator 1 coactivators, energy homeostasis, and metabolism. Endocr Rev. 2006;27:728–735. doi: 10.1210/er.2006-0037. [DOI] [PubMed] [Google Scholar]
- 15.Schilling G, Becher MW, Sharp AH, Jinnah HA, Duan K, Kotzuk JA, Slunt HH, Ratovitski T, Cooper JK, Jenkins NA, Copeland NG, Price DL, Ross CA, Borchelt DR. Intranuclear inclusions and neuritic aggregates in transgenic mice expressing a mutant N-terminal fragment of huntingtin. Hum Mol Genet. 1999;8:397–407. doi: 10.1093/hmg/8.3.397. [DOI] [PubMed] [Google Scholar]
- 16.Mangiarini L, Sathasivam K, Seller M, Cozens B, Harper A, Hetherington C, Lawton M, Trottier Y, Lehrach H, Davies SW, Bates GP. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 17.Trettel F, Rigamonti D, Hilditch-Maguire P, Wheeler VC, Sharp AH, Persichetti F, Cattaneo E, MacDonald ME. Dominant phenotypes produced by the HD mutation in STHdh(Q111) striatal cells. Hum Mol Genet. 2000;9:2799–2809. doi: 10.1093/hmg/9.19.2799. [DOI] [PubMed] [Google Scholar]
- 18.Ciammola A, Sassone J, Alberti L, Meola G, Mancinelli E, Russo MA, Squitieri F, Silani V. Increased apoptosis, Huntingtin inclusions and altered differentiation in muscle cell cultures from Huntington's disease subjects. Cell Death Differ. 2006;13:2068–2078. doi: 10.1038/sj.cdd.4401967. [DOI] [PubMed] [Google Scholar]
- 19.Lin J, Wu PH, Tarr PT, Lindenberg KS, St-Pierre J, Zhang CY, Mootha VK, Jager S, Vianna CR, Reznick RM, Cui L, Manieri M, Donovan MX, Wu Z, Cooper MP, Fan MC, Rohas LM, Zavacki AM, Cinti S, Shulman GI, Lowell BB, Krainc D, Spiegelman BM. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell. 2004;119:121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 20.McGill JK, Beal MF. PGC-1alpha, a new therapeutic target in Huntington's disease? Cell. 2006;127:465–468. doi: 10.1016/j.cell.2006.10.023. [DOI] [PubMed] [Google Scholar]
- 21.Chaturvedi RK, Adhihetty P, Shukla S, Hennessy T, Calingasan N, Yang L, Starkov A, Kiaei M, Cannella M, Sassone J, Ciammola A, Squitieri F, Beal MF. Impaired PGC-1alpha function in muscle in Huntington's disease. Hum Mol Genet. 2009;18:3048–3065. doi: 10.1093/hmg/ddp243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amat R, Planavila A, Chen SL, Iglesias R, Giralt M, Villarroya F. SIRT1 controls the transcription of the peroxisome proliferator-activated receptor-gamma Co-activator-1alpha (PGC-1alpha) gene in skeletal muscle through the PGC-1alpha autoregulatory loop and interaction with MyoD. J Biol Chem. 2009;284:21872–21880. doi: 10.1074/jbc.M109.022749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerhart-Hines Z, Rodgers JT, Bare O, Lerin C, Kim SH, Mostoslavsky R, Alt FW, Wu Z, Puigserver P. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. EMBO J. 2007;26:1913–1923. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 25.Rodgers JT, Lerin C, Gerhart-Hines Z, Puigserver P. Metabolic adaptations through the PGC-1 alpha and SIRT1 pathways. FEBS Lett. 2008;582:46–53. doi: 10.1016/j.febslet.2007.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim J, Moody JP, Edgerly CK, Bordiuk OL, Cormier K, Smith K, Beal MF, Ferrante RJ. Mitochondrial loss, dysfunction and altered dynamics in Huntington's disease. Hum Mol Genet. 2010;19:3919–3935. doi: 10.1093/hmg/ddq306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim J, Amante DJ, Moody JP, Edgerly CK, Bordiuk OL, Smith K, Matson SA, Matson WR, Scherzer CR, Rosas HD, Hersch SM, Ferrante RJ. Reduced creatine kinase as a central and peripheral biomarker in Huntington's disease. Biochim Biophys Acta. 2010;1802:673–681. doi: 10.1016/j.bbadis.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]