Abstract
Bacteriophages belonging to the Caudovirales order possess a tail acting as a molecular machine used during infection to recognize the host and ensure high-efficiency genome delivery to the cell cytoplasm. They bear a large and sophisticated multiprotein organelle at their distal tail end, either a baseplate or a tail-tip, which is the control center for infectivity. We report here insights into the baseplate assembly pathways of two lactoccocal phages (p2 and TP901–1) using electrospray ionization-mass spectrometry. Based on our “block cloning” strategy we have expressed large complexes of their baseplates as well as several significant structural subcomplexes. Previous biophysical characterization using size-exclusion chromatography coupled with on-line light scattering and refractometry demonstrated that the overproduced recombinant proteins interact with each other to form large (up to 1.9 MDa) and stable assemblies. The structures of several of these complexes have been determined by x-ray diffraction or by electron microscopy. In this contribution, we demonstrate that electrospray ionization-mass spectrometry yields accurate mass measurements for the different baseplate complexes studied from which their stoichiometries can be discerned, and that the subspecies observed in the spectra provide valuable information on the assembly mechanisms of these large organelles.
Two siphophages, p2 and TP901–1, have been shown to infect different strains of the low GC content gram+ lactic acid bacterium Lactococcus lactis. They possess a large multiprotein organelle (1–2 MDa) at their distal tail end, termed the baseplate, which is in charge of specific host recognition, attachment, and initiation of infection. In these long-tailed phages, gene expression is tightly and timely controlled during the replication cycle. Their open reading frames (ORFs)1 are organized in at least three sequentially transcribed clusters among which the late gene cluster encodes mostly proteins involved in final virion assembly including the tail and the baseplate morphogenesis module as well as that of the host lysis (1, 2).
We previously reported crystal structures of several receptor-binding proteins (RBPs) from lactococcal phages p2, TP901–1, and bIl170 (3–6). We also successfully implemented a “block cloning” strategy using whole phage genomic segments encoding two or more structural proteins for overexpression in Escherichia coli. This strategy yielded outstanding expression levels of structural protein complexes from three L. lactis phages (p2, TP901–1 and Tuc2009) and allowed us to characterize them as well as their individual components (when available) using size exclusion chromatography coupled with on-line static light scattering and refractometry (7–9). Finally, we were able to determine x-ray and/or electron microscopy (EM) structures of the complete baseplates of bacteriophages p2 (8) and TP901–1 (10) using these overexpressed samples along with baseplate EM reconstructions obtained from intact virions (8, 10). However, although we know the baseplate assembly process outcome for these two phages, the pathways yielding these huge complexes remain poorly characterized. Indeed, to date few reports have addressed baseplate morphogenesis for two lactococcal phages TP901–1 (11) and Tuc2009 (12) using genetic studies and EM, as well as for T4 using analytical ultracentrifugation and EM (13).
Although x-ray crystallography and EM have the potential to provide highly detailed information on large structures, these techniques require stable and homogeneous samples (14). Therefore, complementary techniques are needed to analyze individual components within heterogeneous solutions of macromolecular complexes, subcomplexes, and higher order assemblies of a wide range in both size and population. Such large noncovalent protein assemblies offer an interesting challenge to investigation by electrospray ionization-mass spectrometry (ESI-MS). Their size and sometimes fragile nature can make their transfer to the gas phase difficult and their subsequent mass measurement is complicated because of the greater likelihood of heterogeneity and the presence of small molecules and ions trapped within their quaternary structures, which can cause a significant but variable increase in molecular mass. However, recent instrumental developments and sample ionization optimization have made the analysis of noncovalently bound MDa complexes a reality and ESI-MS has become a powerful tool for studying protein interactions within macromolecular assemblies (15–20). This rapid and highly sensitive analytical method can provide mass measurements from which stoichiometries can be confidently ascertained for individual biomolecular complexes within a heterogeneous mix. Furthermore, the integration of ion mobility spectrometry (IMS) devices with mass spectrometry instrumentation has allowed the physical shape of large protein complexes to be assessed, thus providing information on their topologies and interaction networks (17, 20–22). Therefore, the marriage of IMS with MS can now be considered the ideal companion to complement three-dimensional structural studies.
In this report, we have applied ESI-MS and ESI-IMS-MS to study the mass and stoichiometry of the intact, noncovalently bound p2 and TP901–1 baseplate complexes as well as of the corresponding lower-energy subspecies to follow the pathway from large organelles to the smallest entities. From these data, and integrating previous information (7, 8, 10–12, 23, 24), we have been able to propose plausible baseplate assembly intermediates and pathways for these two lactoccocal phages. We then compared the resulting pathways to the one deduced from the in vivo studies carried out on the formation of TP901–1/Tuc2009 baseplates and pinpointed the similarities and differences observed.
EXPERIMENTAL PROCEDURES
Cloning and Protein Preparation
All the details related to cloning, protein overexpression and purification of the different complexes described here have been extensively described elsewhere and relied on our “block-cloning” strategy (7). Briefly, plasmids encoding either “natural” or “artificial” operons of the proteins of interest were transformed in Rosetta(DE3)pLysS (Novagen) or T7 Express Iq pLysS (New England Biolabs, Ipswich, MA). Cells were grown at 37 °C in Terrific Broth until the OD reached 0.6, and then expression was induced with 0.5 mm isopropyl β-d-thiogalactoside overnight at 25 °C or 17 °C. Protein purification was performed in two steps on a ÄKTA fast-performance liquid chromatography system: a Ni2+-NTA column (HisTrap Ni 5 ml, GE Healthcare, Amersham Biosciences, Bucks., UK) with a step gradient of 250 mm imidazole, followed by a preparative Superdex 200 HR 26/60 gel filtration. For the constructs bearing a thioredoxin fusion, a cleavage by recombinant TEV protease was performed after the Ni affinity chromatography.
Mass Spectrometry
Electrospray Ionization-Mass Spectrometry
Baseplate complexes and subcomplexes were dialyzed against 200 mm ammonium acetate solution (pH 6.8) before noncovalent ESI-MS analysis. Denaturing spectra were acquired in a solvent system of 50% acetonitrile, 30–40% water, 20–10% formic acid (v/v/v). Mass spectra were acquired using an LCT Premier time-of-flight instrument with collisional cooling capabilities (Micromass UK Ltd., Waters Corp, Manchester, UK) interfaced to a NanoMate automated sample infusion device (Advion Biosciences Inc., Ithaca, NY). ESI capillary voltages of 1.7–1.9 kV and sample cone voltages of 40–100 V were used. Tandem mass spectra (MS/MS) were acquired with a Synapt HDMS quadrupole-traveling wave oaTOF mass spectrometer (Micromass UK Ltd., Waters Corp, Manchester, UK). Samples were analyzed by nano-ESI ionization from platinum/gold-plated borosilicate capillaries fabricated in-house using a P-97 micropipette puller (Sutter Instrument Company, Novato, CA) and a sputter coater (Polaron SC7620; Quorum Technologies Ltd, Kent, UK). ESI capillary voltages of 1.7–1.9 kV and sample cone voltages of 40–100 V were used. The Synapt HDMS was operated with a source pressure of 5.5 mbar. The quadrupole analyzer was used to select the most abundant precursor charge state ions. In cases where the precursor signal had a low signal-to-noise, the entire charge state distribution of a particular species was mass-selected by the quadrupole analyzer to increase transmission. Collision voltages of 80–180 V were applied in the argon-filled Trap region of the Triwave device to dissociate macromolecular complexes and to reduce salt adducting. Transfer voltages of 10–30 V were used to optimize transmission of large species exiting the traveling wave device to the Tof analyzer. Charge state assignment of large complexes was carried out using established methodology followed by mass measurement taken as an average over all charge states and adjusted for adduct formation (supplementary Fig. S1) (25, 26). All spectra were calibrated using cluster ions generated from a separate introduction of cesium iodide solution.
Ion Mobility Spectrometry-Mass Spectrometry
IMS-MS was conducted on the Synapt HDMS instrument. For complexes in excess of 1 MDa, a ramped traveling wave height of 8–25 V was used; for complexes below 1 MDa, a fixed wave height of 10 V was found to be optimum. The wave velocity was 250 ms−1 in both cases. IMS drift times were calibrated using denatured equine cytochrome C and horse heart myoglobin (50% MeCN, 40% H2O, 10% acetic acid, v/v/v) and GroEL (200 mm aqueous ammonium acetate). Subsequent cross-sectional area measurements were carried out as described previously (20, 27).
Collision Cross-sectional Area Calculations
Theoretical collision cross-sectional areas for the p2 baseplate and its subcomplexes were estimated from PDB structures using an in-house projection approximation algorithm (28, 29) with the crystal structure 2WZP. PyMOL Molecular Graphics (Schrodinger, LLC, Camberley, UK) was used to build the subcomplex models from the crystal structure of the baseplate. RH-derived cross-sections were calculated from reported RH values (10) by the simple operation πRH2.
RESULTS
Phage p2 Baseplate
The phage p2 baseplate has been shown previously to be assembled from three different gene products: ORF15, ORF16, and ORF18 (7, 8). In the present study, we used ESI-MS to analyze intact p2 baseplate samples, in addition to a range of purified subcomplexes. The native baseplate was analyzed either directly after preparative gel-filtration without further modification (BP1) or after addition of an excess of the VHH5 neutralizing llama antibody fragment (BP2) (5, 30). This latter complex is a very stable assembly from which well-diffracting crystals have been grown leading to structural characterization (8). The other two samples were purified baseplate subcomplexes formed from either ORF15 and ORF16 (named SBP1) or ORF15 and ORF18 (named SBP2). Denaturing ESI-MS was initially conducted on each of the above-mentioned samples to confirm the masses of the individual p2 baseplate constituents with the highest possible accuracy. Thereafter, we carried out noncovalent ESI-MS analyses to detect the presence and decipher the nature and stoichiometry of the intact complexes and their subspecies. A complete list of measured and theoretical masses is presented in Table I.
Table I. Masses of the phage p2 baseplate and subcomponents. All the masses reported in this table are expressed in Da. N.D., not determined. Numbers underlined indicate masses determined using the method reported in McKay et al., J. Am. Chem. Soc. 2006, 128, 11433–11442.
| P2 | |||||||
|---|---|---|---|---|---|---|---|
| BP1 | |||||||
| Protein | ORF18 | ORF15 | ORF16 | ORF18 Trimer | ORF15 + ORF18 Trimer | Baseplate | |
| Observed Mass/Da | 29,337 | 34,478 | N.D. | 88,015 | 122,499 (121,006,120,874,121,281) | 862,727 | |
| Calculated Mass/Da | 29,337 | 34,475a (34,344) | 88,011 | 122,486a (122,355) | 862,638 | ||
| BP2 | |||||||
| Protein | ORF18 | ORF15 | ORF16 | ORF18 Trimer | vhh camelid antibody | Baseplate + 18 vhh | |
| Observed Mass/Da | 29,337 | N.D. | N.D. | 88,012 | 13,913 | 1,114,961 | |
| Calculated Mass/Da | 29,337 | 88,010 | (VHH residues 3–130) 13,914 | 1,113,069 | |||
| SBP1 | |||||||
| Protein | ORF15 | ORF16 | ORF15 degradation product | ORF16 Dimer | ORF16 Trimer | SBP1 Complex | SBP1 Dimer |
| Observed Mass/Da | 37,724 | 42,838 | 35,965 | 85,672 | 128,662 | 354,862 | 709,670 |
| Calculated Mass/Da | 37,723 | 42,837 | 35,970 | 85,674 | 128,511 | 354,854 | 709,708 |
| SBP2 | |||||||
| Protein | ORF15 | ORF18 | ORF15 + ORF18 Trimer | ORF18 trimer variant | |||
| Observed Mass/Da | N.D. | 29,337 | 122,508 (121,323,121,023) | 86,701 | |||
| Calculated Mass/Da | 34,475 | 29,337 | 122,486 |
a With (without) ORF15 N-terminal Met.
Denaturing ESI-MS Analysis of the p2 Baseplate Complexes
The BP1 denaturing ESI-MS spectrum (supplementary Fig. S2, Table I) was dominated by a charge state distribution of ions corresponding to a mass of 29,337 Da, which is consistent with the expected ORF18 monomer mass (RBP) lacking the N-terminal methionine residue (calculated mass 29,337 Da). The spectrum also showed evidence for the presence of ORF18 dimers and trimers with masses of 58,674 Da and 88,015 Da, respectively. ORF18 was the only protein for which a mass could be determined accurately from this sample, probably because of its high abundance (∼60% of the mass of the p2 baseplate) and its high propensity to ionize. The denaturing spectrum of the baseplate-VHH5 complex (BP2) was dominated by signals arising from the antibody together with signals corresponding to the ORF18 monomer and dimer (supplementary Fig. S2, Table I). Mass determination for the other baseplate constituents proved to be more tractable with the SBP1 subcomplex because of the absence of ORF18. In this case, we detected peaks corresponding to species of mass 37,724 and 42,838 Da, which were consistent with ORF15 (theoretical mass: 37,723 Da without the N-terminal methionine residue) and ORF16 (theoretical mass: 42,837 Da), respectively (supplementary Fig. S2, Table I).
Noncovalent ESI-MS of the p2 Baseplate Complexes
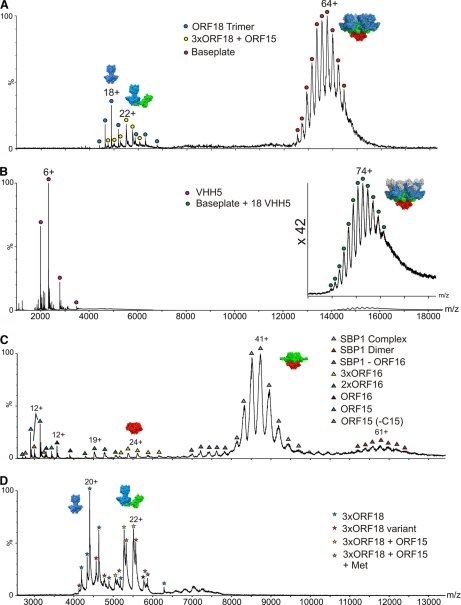
The BP1 noncovalent ESI-MS analysis, acquired under conditions that favor the preservation of quaternary biomolecular complexes (see Supplementary Information for a description of the mass measurements of the noncovalently bound complexes (25, 26)), confirmed the presence of the intact p2 baseplate with a measured mass of 862,727 Da (Fig. 1A). This result lies within 0.06% of the baseplate theoretical mass expected from the stoichiometry observed in the crystal structure [6×ORF15 + 3×ORF16 + 18×ORF18] (8), and illustrates that the intact BP1 complex can indeed be preserved in the gas phase. We also detected a species of mass ∼122,499 Da, which is consistent with a complex formed by an ORF18 trimer associated with one ORF15 monomer (theoretical mass 122,486 Da), as well as trimeric ORF18 (Fig. 1A).
Fig. 1.
ESI-MS spectra of p2 baseplates and sub-complexes. A, p2 baseplate (BP1). The measured mass of the intact complex centered at m/z 14,000 (red circles) is 862,727 Da, consistent with a stoichiometry of [6×ORF15 + 3×ORF16 + 18×ORF18]. Also present are ORF18 trimers (blue circles; 88,015 Da) and an [3× ORF18 + ORF15] complex (yellow circles; 122,499 Da). B, p2 baseplate in complex with the VHH5 llama antibody (BP2). Signals for the p2 baseplate complex with 18 VHH domains [6×ORF15 + 3×ORF16 + 18×ORF18 + 18×VHH5] are centered at m/z 15,000 with a measured mass of 1,114,961 Da (green circles; the inset shows a x42 magnification of this m/z region). VHH5 monomer can also be identified (magenta circles; m/z 3000; 13,913 Da). C, p2 baseplate subcomplex SBP1 [6×ORF15 + 3×ORF16]; 354,862 Da) centered at m/z 8500 (gray triangles). Also identified are a species corresponding to the dimer of SBP1 (m/z 12,000; red triangles), ORF16 monomers (42,838 Da), dimers and trimers (navy blue, green, and yellow triangles at m/z 3500, 4500, and 5500, respectively), and ORF15 monomer (37,724 Da) (pale blue triangles; m/z 3000). An ORF15 truncation product is observed (35,965 Da; m/z 3000; orange triangles), which is consistent with the loss of the first 15 C-terminal residues. D, p2 baseplate sub-complex SBP2 showing ORF18 trimers (88,015 Da) (blue stars; m/z 4500) and the [3×ORF18 + ORF15] complex with a mass of 122,508 Da (orange stars; m/z 5500). Both species are accompanied by the corresponding complexes containing a truncated ORF18 sub-unit.
The BP2 noncovalent ESI-MS analysis provided a mass of 1,114,961 Da (Fig. 1B), in close agreement with that expected for the p2 baseplate complexed with eighteen VHH5 (as seen in the x-ray structure) i.e. 1,113,069 Da. The <0.2% accuracy of this measurement achieved on a large assembly can be correlated to the homogeneity and stability of this four-protein, multisubunit complex.
The major species in the SBP1 subcomplex ESI-MS spectrum yielded a mass measurement of 354,862 Da, within 0.002% of the theoretical mass (354,854 Da) corresponding to a stoichiometry of [6×ORF15 + 3×ORF16] (Fig. 1C). Evidence for a dimer of this complex was also apparent, leading to a nonbiological species associated in the same way as the Dit hexamer of phage SPP1 (31). Monomers, dimers and trimers of ORF16 were also observed (Table I), the trimer being the biologically relevant oligomeric state, as seen in the p2 baseplate x-ray structure. Finally, we also detected ORF15 monomers (37,724 Da) together with a corresponding ORF15 C-terminal truncation product (35,965 Da).
The SBP2 spectrum showed the presence of the 122,508 Da species (Fig. 1D) observed in the BP1 sample, supporting the existence of a stable intermediary complex formed between one ORF18 trimer and one ORF15 monomer. We also detected the presence of isolated ORF18 trimers in this sample.
Collision induced dissociation using MS/MS was also applied to analyze these complexes and confirmed the presence of their respective subunits. The complexes all dissociated by loss of a monomeric subunit, leaving a charge-stripped complex less the monomer, as expected (32, 33) (supplementary Fig. S3).
The TP901–1 Baseplate
The TP901–1 baseplate was previously demonstrated to be assembled from several copies of ORF46 (Dit), ORF47 (Tal) as well as the two peripheral proteins ORF48 (BppU) and ORF49 (BppL, the RBP) (11). We extended this characterization using a combination of x-ray crystallography, EM, and light scattering to propose that this baseplate is likely formed by [12×ORF46 + 18×ORF48 + 54×ORF49 + 3×ORF47] (7, 10). Here, samples of the virtually complete TP901–1 baseplate and several selected subcomplexes were analyzed: the complex formed by ORF46, ORF48, and ORF49 (Dit-BppU-BppL) designated as TP1; the complex composed of ORF48 and ORF49 (termed Tripod), the complex containing ORF46 and ORF48 (Dit-BppU), and the ORF47 N-terminal region (residues 1–386, Tal-N).
Denaturing ESI-MS of the TP901–1 Baseplate Complexes
The denaturing spectra from both the TP1 and the Tripod samples were dominated by ORF49, the BppL monomer, which is understandable as this species is the most abundant one in the two complexes (supplementary Fig. S4 and Table II). Low intensity signals consistent with ORF48, the BppU monomer, were also observed in both samples. The Dit-BppU complex spectrum exhibited several peaks including the Dit monomer (ORF46) with a mass of 29,197 Da along with corresponding dimers, trimers, and hexamers. It is interesting to note that these oligomers remained associated under the denaturing conditions employed (low pH and organic solvent), thus suggesting robust noncovalent binding among individual Dit subunits.
Table II. Masses of the phage TP901–1 baseplate and subcomponents. All the masses reported in this table are expressed in Da. N.D., not determined. Numbers underlined indicate masses determined using the method reported in McKay et al., J. Am. Chem. Soc. 2006, 128, 11433–11442.
| TP901–1 | ||||||||
|---|---|---|---|---|---|---|---|---|
| Tripod | ||||||||
| Protein | BppU | BppL | BppL Trimer | BppU Trimer | BppU Trimer + BppL Trimer | BppU Trimer + 2xBppL Trimer | Tripod | |
| Observed Mass/Da | 33,972 | 17,934 (17,840) | 53,802 (53,522) | 101,918 | 155,470 | 209,764 | 264,088 | |
| Calculated Mass/Da | 33,969 | 17,970a (17,839) | 53,910a (53,517) | 101,907 | 155,817a (155,424) | 209,727a (208,941) | 263,637b (262,458) | |
| TP1 | ||||||||
| Protein | Dit | BppU | BppL | BppL Trimer | 2× BppL Trimer | 3× BppL Trimer | Tripod | Baseplate |
| Observed Mass/Da | 29,053b | 33,843 | 18,216 | 54,646 | 109,290 | 164,001 | 265,995 | 1,769,204 |
| Calculated Mass/Da | 29,051 | 33,841 | 18,214 | 54,643 | 109,286 | 163,929 | 265,449 | 1,767,016 |
| Dit-BppU | ||||||||
| Protein | Dit | BppU | BppU Trimer | Dit x 6 | Dit x 12 | 6xDit + 6xBppU-Trimers | 12xDit + 6xBppU-Trimers | |
| Observed Mass/Da | 29,197 | 33,855 | 101,584 | 175,173 | 350,348 | 784,979 | 959,852 | |
| Calculated Mass/Da | 29,051 | 33,841 | 101,524 | 175,184 | 350,369 | 784,571 | 959,755 | |
| Tal-N (ORF47 residues 1 to 386) | Tal-N monomer | Tal-N dimer | Tal-N trimer | |||||
| Observed Mass/Da | 47,425 | 94,852 | 142,623 | |||||
| Calculated Mass/Da | 47,523 | 95,046 | 142,569 |
a With (without) ORF48 N-terminal Met.
b Observed in native experiment.
Noncovalent ESI-MS of the TP901–1 Baseplate Complexes
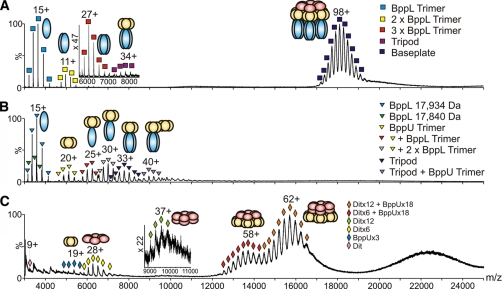
When analyzed under noncovalent conditions, the TP1 ESI-MS spectrum was dominated by significant peaks at m/z ∼18,000 corresponding to a mass of 1,769,204 Da (Fig. 2A). This measurement suggests a stoichiometry of [6×ORF46 + 18×ORF48 + 54×ORF49] (theoretical mass 1,767,016 Da, Table II). Therefore, a discrepancy exists between our ESI-MS results and the model we previously proposed (7, 10) regarding the presence or not of a second Dit hexamer in the TP901–1 baseplate. We analyzed again the TP901–1 baseplate by size exclusion chromatography coupled with on-line static light scattering and refractometry, and we found that the species with two Dit hexamers is transient and displaced within a few days toward the species with a single Dit molecule. Despite varying the ESI-MS conditions (instrumental parameters and ionic strength of the buffer) systematically the TP901–1 baseplate with two Dit hexamers was not detected. This is likely because of the fact that the second hexamer was lost between purification and spectrum recording. We do not know, however, which baseplate form exists in the phage particle. We have shown that Dit exists as a single hexamer in phage SPP1 (31), whereas phage p2 ORF15 (the Dit equivalent) is found as two hexamers (8). In addition to the baseplate species, the spectrum disclosed peaks corresponding to the Tripod (measured mass: 265,995 Da), assembled from a BppU trimer in complex with [9xBppL], as well as BppL trimers, the native state of ORF49 (measured mass: 54,646 Da).
Fig. 2.
ESI-MS spectra of TP901–1 baseplate and sub-complexes. A, TP901–1 baseplate (TP1). The measured mass of the intact complex centered at m/z 18,000 (navy blue squares) is 1,769,204 Da, consistent with a stoichiometry of [6×ORF46 + 18×ORF48 + 54×ORF49]. Also identified are Tripods with a mass of 265,995 Da (magenta squares; m/z 8000) as well as BppL trimers (54,646 Da), dimer of trimers and trimer of trimers (blue, yellow, and red squares centered at m/z 3500, 5000, and 6000, respectively). Inset: m/z 6000–8000 highlighting the Tripod complex and the BppL trimer of trimers. B, Tripod subcomplex. The Tripod measured mass was 264,088 Da (navy blue triangles; m/z 7500), consistent with a stoichiometry of [3×BppU + 9×BppL]. The presence of BppL trimers and BppU trimers was also detected (blue and yellow triangles centered at m/z 3500 and 5250, respectively) in addition to Tripods lacking one or two BppL trimers and complete Tripods associated with a BppU trimer (gray, red, and violet triangles centered at m/z 7000, 6250, and 9000, respectively). C, Dit-BppU complex. The signals centered at m/z 16,000 (orange diamonds) correspond to a mass of 959,852 Da consistent with a stoichiometry of [12× ORF46 + 18×ORF48]. Another observed species was centered at m/z 14,000 (red diamonds) and had a mass of 784,979 Da consistent with a complex of stoichiometry [6×ORF46 + 18×ORF48]. Also identified are Dit (ORF46) hexamers (measured mass: 175,173 Da, yellow diamonds; m/z 6500), dodecamers (350,348 Da, green diamonds; m/z 9500) and BppU (ORF48) trimers (101,584 Da, pale blue diamonds; m/z 5500). Inset: m/z 9000–11,000 highlighting the Dit dodecamers.
The noncovalent ESI-MS spectrum of the Tripod featured signals compatible with the presence of the complete Tripod complex [3×ORF48 + 9×ORF49] with a mass of 264,088 Da, that corresponds within 0.2% to the theoretical mass of 263,637 Da, thus confirming its already established stoichiometry (7, 10) (Fig. 2B). The ESI-MS spectrum distinguishes among copopulated species representing a range of Tripod-related complexes. Other species assigned in this spectrum included trimeric BppL (ORF49), trimeric BppU (ORF48), and complexes consistent with the Tripod lacking one or two BppL trimers (Table II).
The Dit-BppU (ORF46 + ORF48) complex exhibited several peaks in the high m/z region centered at 22,000 (unresolved), 16,000 and 14,000 (Fig. 2C). A mass of 959,852 Da was measured for the species centered at m/z 16,000, which is consistent (0.2%) with the expected mass for a complex of stoichiometry [12xORF46 + 18xORF48] (theoretical mass 957,755 Da). The signal at m/z 14,000 corresponded to a mass of 784,979 Da, which is within 0.05% of the theoretical mass of 784,571 Da for a complex of stoichiometry [6×ORF46 + 18×ORF48]. Interestingly, in contrast with the observation made above for the TP1 sample, a second Dit hexameric ring could be detected in a fraction of the Dit-BppU complex. This species seems, therefore, more stable in this complex as compared with the whole baseplate. Noteworthy, we also observed peaks in the low m/z region for the hexameric and dodecameric Dit with masses of 175,173 Da (6×ORF46) and 350,348 Da (12×ORF46), in good agreement (error <0.01%) with the respective expected masses of 175,184 Da and 350,369 Da, along with ORF46 monomers and ORF48 trimers (Table II). Finally, the ESI-MS spectrum of Tal-N acquired under nondenaturing conditions showed identical species as those observed in the denaturing spectrum, i.e. monomers, dimers, and trimers (Table II), in agreement with the expected trimeric state for the full-length TP901–1 ORF47.
Again, MS/MS analyses of the principal complexes were carried out to confirm the presence of the individual subunits: TP1, Tripod, [12×Dit + 18×BppU], and [6×Dit + 18×BppU] (supplementary Fig. S5).
Further Characterization of the Baseplate Complexes by IMS-MS
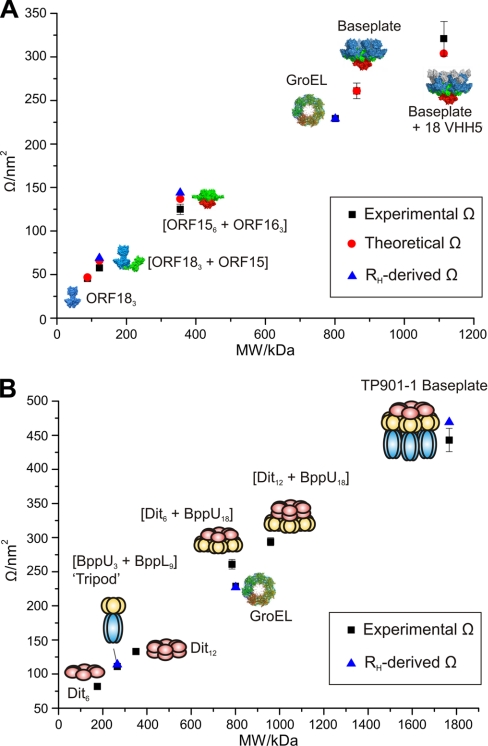
IMS-MS analysis with drift time calibration allowed the measurement of the collision cross-sectional areas (Ω) of the p2 and TP901–1 baseplate and subcomplex ions in the gas phase (Fig. 3 and supplementary Table S1). A Ω of 261 ± 10 nm2 was measured for the p2 baseplate, which is in agreement with the value of 261 nm2 calculated using an in-house projection approximation algorithm (28, 29). The Ω of BP2 was measured as 321 ± 20 nm2, also in close agreement with the calculated value (304 nm2) whereas the measured Ω of the ORF18 trimer agrees within 2% of the calculated value. The [3×ORF18 + ORF15] complex has a measured Ω of 57.7 ± 0.5 nm2, some 12% lower than the theoretical value (66 nm2) calculated from the arrangement of the ORF15 and ORF18 subunits as they appear in the baseplate structure. This discrepancy could mean that the 'L-shaped' structure (Fig. 3A) has collapsed somewhat, to a more 'U-shaped' conformation, on ionization (supplementary Table S1). Indeed, when such a U-shaped structure is modeled, the theoretical Ω is calculated as 58.5 nm2, in much better agreement with the observed value. If the above values are compared with the RH-derived cross-sectional areas determined from dynamic light scattering experiments, the proposed structural collapse is corroborated. The RH-derived Ω is 69.4 ± 0.2 nm2, which is in better agreement with the 'L-shaped' structure than the collapsed one, suggesting the L-shape is indeed present in solution. This shape change would rely on the flexibility of the ORF15 protein, which has been observed previously and was shown to facilitate cell adhesion (8). The measured Ω of the SBP1 complex (6×ORF15 + 3×ORF16) is 127 ± 6 nm2. The agreement with theory (137 nm2) is ∼7%. The RH-derived cross-sectional area for this complex is 144 ± 1 nm2 which is 12% larger than the value measured in the gas phase. There are no crystal structure coordinates available with which to compare the measured Ω values for the TP901–1 baseplate complexes and subcomplexes. However, the accuracy of the measurements for p2, together with the fact that the previously reported Ω value for the chaperonin complex GroEL (225 nm2) (20) agrees well with its published RH value (35) provides confidence in the IMS-MS methodology. Although it is difficult to infer structural features from the measurements alone, the observed Ω values can be compared with RH-derived cross-sectional area values (Fig. 3B). The TP901–1 baseplate has a RH-derived cross section of 471 ± 4 nm2, in reasonable agreement (6%) with the gas phase value of 443 ± 17 nm2. The Ω measured for the Tripod complex (111 ± 8 nm2) also agrees well with its RH-derived cross-section (113 ± 1 nm2).
Fig. 3.
A comparison of molecular mass with cross-sectional area measurements for phage complexes. Molecular mass (kDa) versus cross-sectional areas measured by IMS-MS (black squares), calculated using an in-house projection approximation algorithm (red circles) and derived from RH (light scattering) measurements (blue triangles) for (A) p2 baseplate and a range of subcomplexes: ORF18 (blue), ORF16 (red), ORF15 (green), and VHH5 (gray); and (B) TP901–1 baseplate and a range of subcomplexes: Dit (pink), BppU (yellow), and BppL (blue). The chaperonin, GroEL, is shown on both graphs for reference. Error bars on the ESI-IMS-MS measurements represent the standard deviation of three measurements whenever available (supplementary Table S1).
It is interesting to compare the [(Dit)6+BppU)18] complex (molecular mass 785 kDa) with the intact, homotetradecameric GroEL chaperonin (mass 801 kDa). Although very similar in mass, the cross-sectional area of the [(Dit)6+BppU)18] complex is 261 nm2, some 16% larger than that measured for GroEL (225 nm2), indicating a less compact structure, possibly as this complex has to be able to undertake further binding to multiple copies of the BppL trimer.
As a further note of interest, the ratio of the cross-sectional area of the Dit hexamer (82 nm2) compared with the Dit dodecamer (133 nm2) is 61%. This value can be compared with the ratio of 64% calculated from the cross-sectional areas of the single-ring GroEL heptamer (145 nm2) (16) and the double-ring GroEL tetradecamer (225 nm2) (20). The stacked ring system of the GroEL 14-mer is a well established feature of its quaternary structure and the similarity in ratios of the two Dit multimers with the two GroEL multimers may well be characteristic of ring-like multimers stacked in this fashion.
The comparison of gas-phase collision cross-section measurements with solution-phase data adds another dimension to structural analyses by IMS-MS. Crystal structures tend to be static snap-shots of a protein in crystal form and do not necessarily reflect its functional conformation. IMS-MS has the ability to monitor individual components within heterogeneous ensembles of protein conformers and macromolecular complexes and sub-complexes, each of which can be populated only transiently. The comparison of gas-phase IMS-MS data with solution-phase dynamic light scattering data reported here strongly supports the notion that the structure of a protein or protein complex is maintained significantly in the gas-phase. The superior mass accuracy and resolution of IMS-MS provides advantages over current solution-phase measurements.
DISCUSSION
Tail and Distal Tail Structure Morphogenesis in Siphoviridae
The wealth of data accumulated on several siphophages (e.g. λ, TP901–1, Tuc2009, etc) has been used previously to propose a pathway for tail and baseplate/tip morphogenesis (11, 12, 36, 37). Interestingly, common rules appear to apply to many Siphoviridae infecting Lactococcus lactis despite their morphological differences and the paucity of sequence similarity within their structural proteins. Three components, being the Tape Measure Protein (TMP), Dit, and Tal, are essential for tail formation, as TP901–1 mutant phages lacking any of these proteins are tail-less (11, 37). Therefore, it has been proposed that tail morphogenesis begins by the formation of an initiation complex made of these three components and forming a proteinaceous core priming tail and baseplate/tip assembly (11, 12). Considering the narrow time window during which Dit can be detected in Tuc2009 infected cells (a 10 min period late in the infection), its low expression level and the structural results we previously reported for p2, SPP1, and TP901–1 (8, 10, 38), it becomes clear that Dit-like proteins act as wide-spread maestros orchestrating tail and baseplate/tip morphogenesis. In phages bearing a sophisticated RBP-accommodating baseplate (e.g. TP901–1, Tuc2009, or p2) or a simplified tail-tip (e.g. SPP1 or λ), peripheral proteins forming these organelles can assemble onto the initiation complex before, during, or after tail formation (39). Indeed, assembly of TP901–1 or Tuc2009 phage mutants lacking selected components demonstrated that baseplate formation is not required for tail polymerization (11, 12).
Phage p2 Baseplate Assembly Pathway
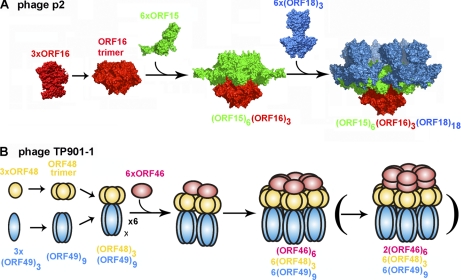
Noncovalent ESI-MS analysis of the BP2 complex (the p2 baseplate with bound VHH5) indicates that this species is highly stable, as proposed previously based on the outstanding diffraction power of the crystals we obtained from this sample (8). Besides, we demonstrated that BP1 has lost its second ORF15 ring after purification suggesting that the VHH5 subunits have complexed a pre-existing species lacking the upper ORF15 hexameric ring instead of expelling it, in contrast to the hypothesis we formulated previously (8). Of note, the presence of lower molecular weight species was detected in the BP1 and SBP2 noncovalent ESI-MS spectra, such as a complex formed between a trimeric ORF18 and an ORF15 monomer as well as ORF18 trimers. In the SBP1 spectrum we observed a complex corresponding to the central baseplate core, consisting of one hexameric ORF15 associated with an ORF16 trimer, that also constitutes a significant portion of the initiation complex. Also, the SBP1 spectrum contained peaks corresponding to ORF16 trimer, dimers, and monomers as well as monomeric ORF15. Collating all these data, we are able to pinpoint pre-assembled species of each polypeptide type including the RBPs (ORF18 trimer), ORF16 trimer, and ORF15 hexamer as well as other intermediary complexes such as (6×ORF15 + 3×ORF16) and (1×ORF15 + 3×ORF18). These building blocks assemble further to lead ultimately to the phage p2 baseplate (Fig. 4A). First, we propose that an ORF16 trimer is assembled and serves as a nucleus promoting oligomerization of ORF15 to yield a hexameric ring of ORF15 subunits bound to the ORF16 trimer. The data presented here did not allow us to distinguish which of the two possible pathways involved in the following steps of baseplate assembly occurs first. A second ORF15 hexamer might oligomerize on the top of the already formed moiety (resulting in a complex able to interact with the TMP) and then with the eighteen ORF18s or it can initially bind with the ORF18s before stacking the second ORF15 hexamer. It should be noted that the lower ORF15 hexamer has a key role in baseplate assembly, acting as a central hub onto which all other components are plugged. The fact that we reproducibly observed a complex formed by an ORF18 trimer with an ORF15 monomer, in several baseplate samples investigated, strongly suggests that association of the RBPs is independent of the baseplate assembly state. Indeed, we believe that tail morphogenesis can be dichotomized in its vertical and horizontal growing directions and that both processes are independent. This hypothesis is supported by the fact that ORF15 is able to form a stable complex with an ORF16 trimer in the absence of the RBPs but is also able to interact in its naïve monomeric state with the RBP, before the establishment of any contact with ORF16 or other tail components.
Fig. 4.
Proposed pathway assembly of phages p2 and TP901–1 baseplates. A, Putative p2 baseplate assembly pathway. The initial interactions of ORF15 (green) subunits with a preassembled ORF16 (red) trimer result in the formation of a complex consisting of an ORF16 trimer noncovalently bound to an ORF15 hexamer. This is followed by capture of six ORF18 (blue) trimers. There is no evidence for a complex composed of an ORF15 hexamer bound to ORF 18. B, Schematic representation of one plausible TP901–1 baseplate assembly pathway (see the Discussion section). ORF48 (yellow) and ORF49 (blue) initially form homotrimers and then associate to form Tripods, each composed of [3×ORF48 + 9×ORF49] subunits. ORF46 (pink) oligomerization is then mediated by interactions with ORF48 resulting in the formation of one or two ORF46 hexamers. Overall the TP901–1 baseplate is formed by 6×ORF46 (or 12×ORF46, depicted between parentheses), 18×ORF48 and 54×ORF49.
An interesting point would be to understand why the upper ORF15 hexamer is so weakly bound to the remaining structure because virions are supposed to be robust objects able to withstand extreme conditions. This issue might be resolved by considering the fact that all the studied complexes are characterized by the absence of a crucial component of the phage baseplate, the TMP, which controls the overall tail length (36, 37). This unusual protein is predicted to be structured as a long central helix flanked by one small globular domain at each extremity and is thought to be hexameric when present into the tail channel, based on the EM results reported for the bacteriophage SPP1 (40). In mature virions, the TMP C-terminal domains would be embedded within the central channel of the baseplate, stabilizing the whole assembly and therefore locking the second ORF15 hexameric ring. If we consider the possibility that the TMP is expressed first during morphogenesis (and stabilized by its chaperone (41)), it is possible to envision that the TMP binds to the intermediate of (12×ORF15 + 3×ORF16), before ORF18 fixation.
TP901–1 Baseplate Assembly Pathway
Our noncovalent ESI-MS analysis of the TP1 sample clearly demonstrates the presence of the TP901–1 baseplate constituted by one hexameric Dit ring associated with six peripheral Tripods, each of them containing a trimeric ORF48 and nine BppL organized as three RBPs. This result points to the presence of a single ORF46 hexamer in this sample whereas we previously proposed that two such rings can assemble in the baseplate, based on EM and light scattering results (10). Furthermore, the MS analysis of the Dit-BppU complex showed that either a dimer of back-to-back hexamers or a single hexamer of ORF46 can be found in isolation and in complex with the eighteen ORF48s. As stated above, the lability of the second Dit hexamer in the complete baseplate can explain this discrepancy. This lability may be because of a weakly bound second Dit hexamer to the baseplate in the absence of the stabilizing effect of TMP, a feature also found in the p2 baseplate (8). Moreover, the TP1 spectrum revealed the presence of several subspecies such as intact Tripods and trimeric BppL. We observed the same subspecies in the Tripod MS spectrum with additional Tripods lacking one or two RBP and isolated BppU trimers. Based on the fact that we detected several of the above-mentioned subcomplexes by light scattering in solution and EM, we believe that they most likely correspond to building-blocks and assembly intermediates relevant to the tail morphogenesis pathway taking place in TP901–1.
Similarly to the p2 situation, the TP901–1 baseplate is assembled onto a central platform made of one (or two) Dit hexamer(s) that ensures interaction with most of the tail/baseplate components. Our results here are suggestive that Dit oligomerization might be promoted by interactions with ORF48 at its periphery as we detected only the presence of ORF46 hexamers from a mixture of ORF46 and ORF48 and we have reported that isolated Dit purified in excess from the baseplate overexpression is monomeric in solution (7). However, it has been demonstrated previously that an effective initiation complex can be formed in the absence of both ORF48 and ORF49 allowing proper tail growth in its vertical direction (11). This latter fact seems to preclude the absolute requirement of ORF48 for Dit hexamerization. We thus propose that in an in vivo context, Dit oligomerization might be triggered either by ORF48 or by ORF47 (Tal). The putative involvement of this latter protein relies on high structural conservation between Dit and Tal-N proteins among lactococcal phages (34) and thus is proposed by analogy with the bacteriophage p2 assembly pathway. Interestingly, the requirement of an external assistance from proteinaceous partners for Dit folding is reminiscent of several other phage proteins that adopt their quaternary structures only on binding to their partners with a regulatory scheme relying on the folding of unstructured regions inducing a disorder-to-order transition to allow a strict control of tail morphogenesis (42, 43). Another point to consider is that we were able to overproduce and purify the assembled TP901–1 baseplate in the absence of two out of three components of the initiation complex (TMP and Tal) further confirming that tail and baseplate formation can proceed independently. Combining all these data, we speculate that during in vivo phage assembly, tail morphogenesis starts with the formation of the initiation complex and that association of the baseplate peripheral proteins can take place at any moment by addition of performed Tripods at the Dit periphery or by fixation of BppU onto Dit followed by anchoring of the RBPs (Fig. 4B).
CONCLUSIONS
The first example of a noncovalent ESI-MS investigation of large bacteriophage baseplate complexes has been described here, thus complementing and building on a large array of structural data available for lactococcal phages. The outcomes of this current study have allowed us to confirm the previously proposed stoichiometries for bacteriophage p2 and TP901–1 baseplates as well as to further our knowledge of these assemblies. In addition, the thorough characterization of baseplate subcomplexes associated with available data provided insights into the pathways followed during assembly of these organelles. These results along with those from future studies will help to obtain a comprehensive view of these fascinating complexes and to understand at the molecular level their implications in bacteriophage infections.
Furthermore, this work has provided a framework to develop and optimize specific methods for the study of heterogeneous, megaDalton, macromolecular baseplate complexes by noncovalent ESI-MS yielding mass measurements with an error of within 0.2%. This paves the way for the study of any multiprotein biological system with virtually no size limitation using picomolar amounts of sample within heterogeneous mixtures under near-physiological conditions. We thus believe that noncovalent ESI-MS bears the promise to be an essential tool in modern structural biology. In addition, the integration of IMS with MS has generated a technique that can produce insights into the tertiary and quaternary structure of copopulated proteins and protein complexes in terms of stoichiometry, shape, and dynamics.
Footnotes
* This work was supported, in part, by grants from the Marseille-Nice Génopole (to J.L.), the CNRS, the ANR (ANR-07-BLAN-0095), and by a PhD grant from the “Ministère Français de l'Enseignement Supérieur et de la Recherche” to D.V. (ref. no. 22976-2006). D.A.S. is funded by an EPSRC White Rose PhD Studentship and cosupervised by Dr Nicola J. Stonehouse at the University of Leeds. We also thank the Biotechnology and Biological Sciences Research Council (BBSRC; BB/E012558/1) and the Wellcome Trust (WT 075099/Z/04/Z) for funding the purchase of the Synapt HDMS and the LCT Premier mass spectrometers, respectively.
 This article contains supplemental Figs. S1 to S6 and Table S1.
This article contains supplemental Figs. S1 to S6 and Table S1.
1 The abbreviations used are:
- ORF
- open reading frame
- BppL
- baseplate lower protein
- BppU
- baseplate upper protein
- Dit
- distal tail protein
- EM
- electron microscopy
- ESI-MS
- electrospray ionization mass spectrometry
- RBP
- receptor binding protein
- TMP
- Tape Measure Protein.
REFERENCES
- 1. Madsen P. L., Hammer K. (1998) Temporal transcription of the lactococcal temperate phage TP901–1 and DNA sequence of the early promoter region. Microbiology 144, 2203–2215 [DOI] [PubMed] [Google Scholar]
- 2. Duplessis M., Russell W. M., Romero D. A., Moineau S. (2005) Global gene expression analysis of two Streptococcus thermophilus bacteriophages using DNA microarray. Virology 340, 192–208 [DOI] [PubMed] [Google Scholar]
- 3. Ricagno S., Campanacci V., Blangy S., Spinelli S., Tremblay D., Moineau S., Tegoni M., Cambillau C. (2006) Crystal structure of the receptor-binding protein head domain from Lactococcus lactis phage bIL170. J. Virol. 80, 9331–9335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Spinelli S., Campanacci V., Blangy S., Moineau S., Tegoni M., Cambillau C. (2006) Modular structure of the receptor binding proteins of Lactococcus lactis phages. The RBP structure of the temperate phage TP901–1. J. Biol. Chem. 281, 14256–14262 [DOI] [PubMed] [Google Scholar]
- 5. Spinelli S., Desmyter A., Verrips C. T., de Haard H. J., Moineau S., Cambillau C. (2006) Lactococcal bacteriophage p2 receptor-binding protein structure suggests a common ancestor gene with bacterial and mammalian viruses. Nat. Struct. Mol. Biol. 13, 85–89 [DOI] [PubMed] [Google Scholar]
- 6. Tremblay D. M., Tegoni M., Spinelli S., Campanacci V., Blangy S., Huyghe C., Desmyter A., Labrie S., Moineau S., Cambillau C. (2006) Receptor-binding protein of Lactococcus lactis phages: identification and characterization of the saccharide receptor-binding site. J. Bacteriol. 188, 2400–2410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Campanacci V., Veesler D., Lichière J., Blangy S., Sciara G., Moineau S., van Sinderen D., Bron P., Cambillau C. (2010) Solution and electron microscopy characterization of lactococcal phage baseplates expressed in Escherichia coli. J. Struct. Biol. 172, 75–84 [DOI] [PubMed] [Google Scholar]
- 8. Sciara G., Bebeacua C., Bron P., Tremblay D., Ortiz-Lombardia M., Lichière J., van Heel M., Campanacci V., Moineau S., Cambillau C. (2010) Structure of lactococcal phage p2 baseplate and its mechanism of activation. Proc. Natl. Acad. Sci. U.S.A. 107, 6852–6857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sciara G., Blangy S., Siponen M., Mc Grath S., van Sinderen D., Tegoni M., Cambillau C., Campanacci V. (2008) A topological model of the baseplate of lactococcal phage Tuc2009. J. Biol. Chem. 283, 2716–2723 [DOI] [PubMed] [Google Scholar]
- 10. Bebeacua C., Bron P., Lai L., Vegge C. S., Brøndsted L., Spinelli S., Campanacci V., Veesler D., van Heel M., Cambillau C. (2010) Structure and molecular assignment of lactococcal phage TP901–1 baseplate. J. Biol. Chem. 285, 39079–39086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vegge C. S., Brøndsted L., Neve H., Mc Grath S., van Sinderen D., Vogensen F. K. (2005) Structural characterization and assembly of the distal tail structure of the temperate lactococcal bacteriophage TP901–1. J. Bacteriol. 187, 4187–4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mc Grath S., Neve H., Seegers J. F., Eijlander R., Vegge C. S., Brøndsted L., Heller K. J., Fitzgerald G. F., Vogensen F. K., van Sinderen D. (2006) Anatomy of a lactococcal phage tail. J. Bacteriol. 188, 3972–3982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yap M. L., Mio K., Leiman P. G., Kanamaru S., Arisaka F. (2010) The baseplate wedges of bacteriophage T4 spontaneously assemble into hubless baseplate-like structure in vitro. J. Mol. Biol. 395, 349–360 [DOI] [PubMed] [Google Scholar]
- 14. Poliakov A., van Duijn E., Lander G., Fu C. Y., Johnson J. E., Prevelige P. E., Jr, Heck A. J. (2007) Macromolecular mass spectrometry and electron microscopy as complementary tools for investigation of the heterogeneity of bacteriophage portal assemblies. J. Struct. Biol. 157, 371–383 [DOI] [PubMed] [Google Scholar]
- 15. Uetrecht C., Watts N. R., Stahl S. J., Wingfield P. T., Steven A. C., Heck A. J. (2010) Subunit exchange rates in Hepatitis B virus capsids are geometry- and temperature-dependent. Phys. Chem. Chem. Phys. 12, 13368–13371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Uetrecht C., Versluis C., Watts N. R., Wingfield P. T., Steven A. C., Heck A. J. (2008) Stability and shape of hepatitis B virus capsids in vacuo. Angew Chem. Int. Ed. Engl. 47, 6247–6251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Uetrecht C., Versluis C., Watts N. R., Roos W. H., Wuite G. J., Wingfield P. T., Steven A. C., Heck A. J. (2008) High-resolution mass spectrometry of viral assemblies: molecular composition and stability of dimorphic hepatitis B virus capsids. Proc. Natl. Acad. Sci. U.S.A. 105, 9216–9220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhou M., Sandercock A. M., Fraser C. S., Ridlova G., Stephens E., Schenauer M. R., Yokoi-Fong T., Barsky D., Leary J. A., Hershey J. W., Doudna J. A., Robinson C. V. (2008) Mass spectrometry reveals modularity and a complete subunit interaction map of the eukaryotic translation factor eIF3. Proc. Natl. Acad. Sci. U.S.A. 105, 18139–18144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stockley P. G., Rolfsson O., Thompson G. S., Basnak G., Francese S., Stonehouse N. J., Homans S. W., Ashcroft A. E. (2007) A simple, RNA-mediated allosteric switch controls the pathway to formation of a T=3 viral capsid. J. Mol. Biol. 369, 541–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Knapman T. W., Morton V. L., Stonehouse N. J., Stockley P. G., Ashcroft A. E. (2010) Determining the topology of virus assembly intermediates using ion mobility spectrometry-mass spectrometry. Rapid Commun. Mass Spectrom. 24, 3033–3042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bush M. F., Hall Z., Giles K., Hoyes J., Robinson C. V., Ruotolo B. T. (2010) Collision cross sections of proteins and their complexes: a calibration framework and database for gas-phase structural biology. Anal. Chem. 82, 9557–9565 [DOI] [PubMed] [Google Scholar]
- 22. Wang S. C., Politis A., Di Bartolo N., Bavro V. N., Tucker S. J., Booth P. J., Barrera N. P., Robinson C. V. (2010) Ion mobility mass spectrometry of two tetrameric membrane protein complexes reveals compact structures and differences in stability and packing. J. Am. Chem. Soc. 132, 15468–15470 [DOI] [PubMed] [Google Scholar]
- 23. Veesler D., Blangy S., Siponen M., Vincentelli R., Cambillau C., Sciara G. (2009) Production and biophysical characterization of the CorA transporter from Methanosarcina mazei. Anal. Biochem. 388, 115–121 [DOI] [PubMed] [Google Scholar]
- 24. Veesler D., Dreier B., Blangy S., Lichière J., Tremblay D., Moineau S., Spinelli S., Tegoni M., Plückthun A., Campanacci V. et al. (2009) Crystal Structure and Function of a DARPin Neutralizing Inhibitor of Lactococcal Phage TP901–1: comparison of DARPin and CAMELID VHH binding mode. J. Biol. Chem. 284, 30718–30726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McKay A. R., Ruotolo B. T., Ilag L. L., Robinson C. V. (2006) Mass measurements of increased accuracy resolve heterogeneous populations of intact ribosomes. J. Am. Chem. Soc. 128, 11433–11442 [DOI] [PubMed] [Google Scholar]
- 26. Liepold L., Oltrogge L. M., Suci P. A., Young M. J., Douglas T. (2009) Correct charge state assignment of native electrospray spectra of protein complexes. J. Am. Soc. Mass Spectrom. 20, 435–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Smith D. P., Giles K., Bateman R. H., Radford S. E., Ashcroft A. E. (2007) Monitoring copopulated conformational states during protein folding events using electrospray ionization-ion mobility spectrometry-mass spectrometry. J. Am. Soc. Mass Spectrom. 18, 2180–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Smith D. P., Knapman T. W., Campuzano I., Malham R. W., Berryman J. T., Radford S. E., Ashcroft A. E. (2009) Deciphering drift time measurements from travelling wave ion mobility spectrometry-mass spectrometry studies. Eur. J. Mass Spectrom. 15, 113–130 [DOI] [PubMed] [Google Scholar]
- 29. Knapman T., Berryman J. T., Campuzano I., Harris S. A., Ashcroft A. E. (2010) Considerations in experimental and theoretical collision cross-section measurements of small molecules using travelling wave ion mobility spectrometry-mass spectrometry. Int. J. Mass Spectrom. 298, 17–23 [Google Scholar]
- 30. De Haard H. J., Bezemer S., Ledeboer A. M., Müller W. H., Boender P. J., Moineau S., Coppelmans M. C., Verkleij A. J., Frenken L. G., Verrips C. T. (2005) Llama antibodies against a lactococcal protein located at the tip of the phage tail prevent phage infection. J. Bacteriol. 187, 4531–4541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Veesler D., Robin G., Lichière J., Auzat I., Tavares P., Bron P., Campanacci V., Cambillau C. (2010) Crystal Structure of Bacteriophage SPP1 Distal Tail Protein (gp19.1): a baseplate hub paradigm in gram-positive infecting phages. J. Biol. Chem. 285, 36666–36673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Benesch J. L., Aquilina J. A., Ruotolo B. T., Sobott F., Robinson C. V. (2006) Tandem mass spectrometry reveals the quaternary organization of macromolecular assemblies. Chem. Biol. 13, 597–605 [DOI] [PubMed] [Google Scholar]
- 33. Jurchen J. C., Garcia D. E., Williams E. R. (2004) Further studies on the origins of asymmetric charge partitioning in protein homodimers. J. Am. Soc. Mass Spectrom. 15, 1408–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Veesler D., Blangy S., Lichière J., Ortiz-Lombarda M., Tavares P., Campanacci V., Cambillau C. (2010) Crystal structure of Bacillus subtilis SPP1 phage gp23.1, a putative chaperone. Protein Sci. 19, 1812–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Walters C., Errington N., Rowe A. J., Harding S. E. (2002) Hydrolysable ATP is a requirement for the correct interaction of molecular chaperonins cpn60 and cpn10. Biochem. J. 364, 849–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Katsura I., Hendrix R. W. (1984) Length determination in bacteriophage lambda tails. Cell 39, 691–698, doi: 0092-8674(84)90476-8 [DOI] [PubMed] [Google Scholar]
- 37. Pedersen M., Ostergaard S., Bresciani J., Vogensen F. K. (2000) Mutational analysis of two structural genes of the temperate lactococcal bacteriophage TP901–1 involved in tail length determination and baseplate assembly. Virology 276, 315–328 [DOI] [PubMed] [Google Scholar]
- 38. Veesler D., Blangy S., Spinelli S., Tavares P., Campanacci V., Cambillau C. (2010) Crystal structure of Bacillus subtilis SPP1 phage gp22 shares fold similarity with a domain of lactococcal phage p2 RBP. Protein Sci. 19, 1439–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cardarelli L., Pell L. G., Neudecker P., Pirani N., Liu A., Baker L. A., Rubinstein J. L., Maxwell K. L., Davidson A. R. (2010) Phages have adapted the same protein fold to fulfill multiple functions in virion assembly. Proc. Natl. Acad. Sci. U.S.A. 107, 14384–14389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Plisson C., White H. E., Auzat I., Zafarani A., São-José C., Lhuillier S., Tavares P., Orlova E. V. (2007) Structure of bacteriophage SPP1 tail reveals trigger for DNA ejection. EMBO J. 26, 3720–3728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Siponen M., Sciara G., Villion M., Spinelli S., Lichière J., Cambillau C., Moineau S., Campanacci V. (2009) Crystal structure of ORF12 from Lactococcus lactis phage p2 identifies a tape measure protein chaperone. J. Bacteriol. 191, 728–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pell L. G., Liu A., Edmonds L., Donaldson L. W., Howell P. L., Davidson A. R. (2009) The X-ray crystal structure of the phage lambda tail terminator protein reveals the biologically relevant hexameric ring structure and demonstrates a conserved mechanism of tail termination among diverse long-tailed phages. J. Mol. Biol. 389, 938–951 [DOI] [PubMed] [Google Scholar]
- 43. Pell L. G., Kanelis V., Donaldson L. W., Howell P. L., Davidson A. R. (2009) The phage lambda major tail protein structure reveals a common evolution for long-tailed phages and the type VI bacterial secretion system. Proc. Natl. Acad. Sci. U.S.A. 106, 4160–4165 [DOI] [PMC free article] [PubMed] [Google Scholar]