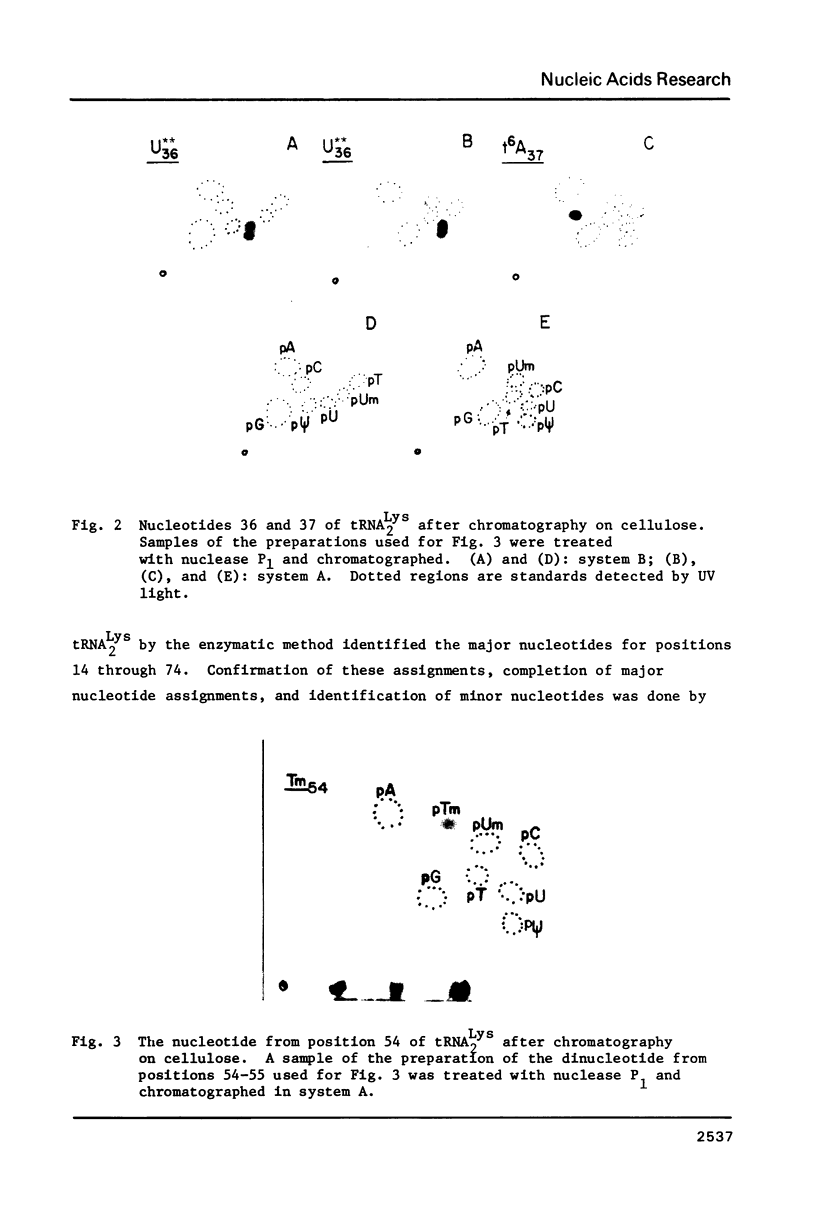
Abstract
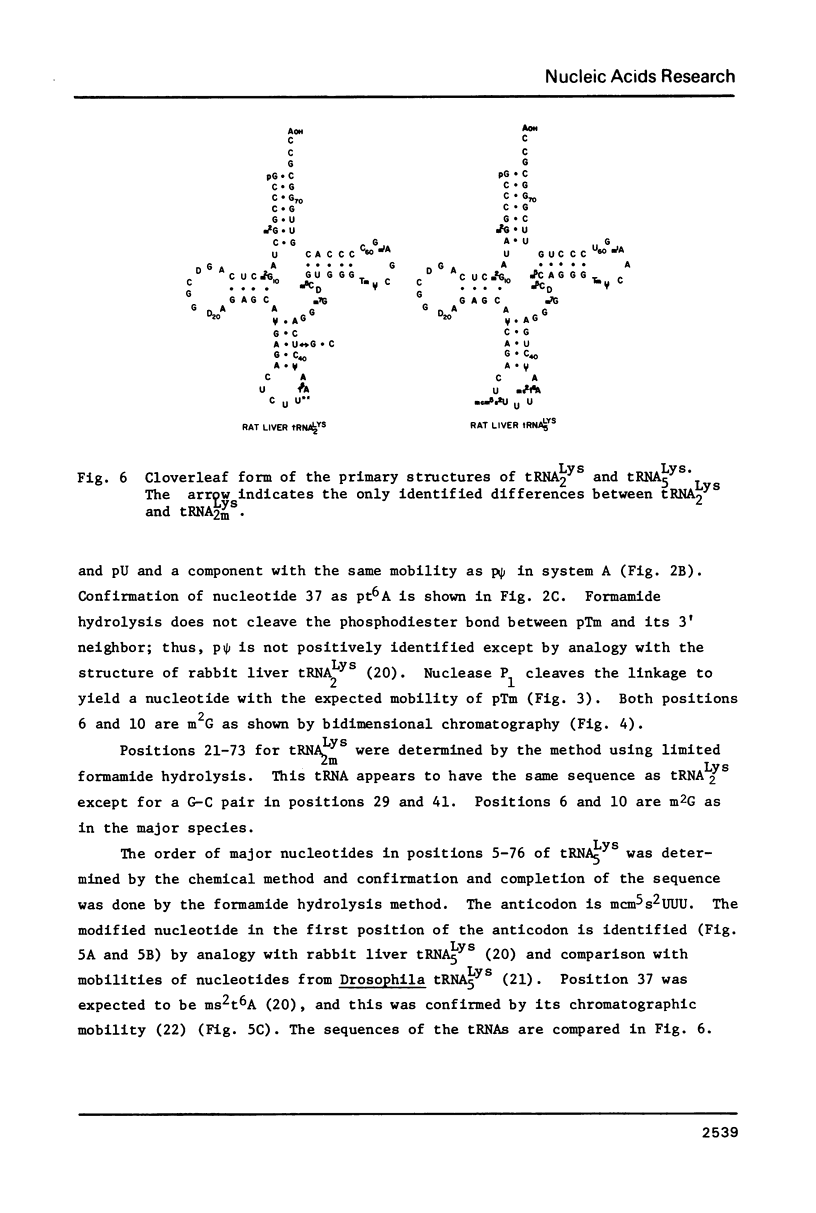
The two major lysine tRNAs from rat liver, tRNA2Lys and tRNA5Lys, were sequenced by rapid gel or chromatogram readout methods. The major tRNA2Lys differs from a minor form only by a base pair in positions 29 and 41; both tRNAs have an unidentified nucleotide, U**, in the third position of the anticodon. Although highly related, the major tRNA2Lys and tRNA5Lys differ in four base pairs and four unpaired nucleotides, including the first position of the anticodons, but have the same base pair in positions 29 and 41. The three tRNAs maintain a m2G-U pair in the acceptor stem. Detection of this m2G is in contrast to other reports of lysine tRNAs. Sequences of lysine tRNAs are strongly conserved in higher eukaryotes.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Conlon-Hollingshead C., Ortwerth B. J. Lys-tRNA levels and cell division in mouse 3T3 cells. Exp Cell Res. 1980 Jul;128(1):171–180. doi: 10.1016/0014-4827(80)90400-0. [DOI] [PubMed] [Google Scholar]
- Cribbs D. L., Gillam I. C., Tener G. M. The structure of tRNA 5 Lys from Drosophila melanogaster. Nucleic Acids Res. 1982 Oct 25;10(20):6393–6399. doi: 10.1093/nar/10.20.6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R. C., Randerath K. Rapid print-readout technique for sequencing of RNA's containing modified nucleotides. Nucleic Acids Res. 1979 Aug 10;6(11):3443–3458. doi: 10.1093/nar/6.11.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J. H., Harding J. D. Using iodinated single-stranded M13 probes to facilitate rapid DNA sequence analysis--nucleotide sequence of a mouse lysine tRNA gene. Nucleic Acids Res. 1983 Apr 11;11(7):2053–2064. doi: 10.1093/nar/11.7.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedgcoth C., Hayenga K., Scheets K., Thomas K. R., Harrison M., Lin V. K., Ortwerth B. J. Perturbation of the mitochondrial lysine tRNA population by virus-induced transformation or stress of mammalian cells: functional properties and nucleotide sequence of a mitochondrially associated lysine tRNA. Recent Results Cancer Res. 1983;84:171–183. doi: 10.1007/978-3-642-81947-6_12. [DOI] [PubMed] [Google Scholar]
- Juarez H., Juarez D., Hedgcoth C., Ortwerth B. J. Amounts of isoaccepting lysine tRNAs change with the proliferative state of cells. Nature. 1975 Mar 27;254(5498):359–360. doi: 10.1038/254359a0. [DOI] [PubMed] [Google Scholar]
- Katze J. R. Relation of cell type and cell density to the degree of post-transcriptional modification of tRNALys and tRNAPhe. Biochim Biophys Acta. 1975 Nov 4;407(4):392–398. doi: 10.1016/0005-2787(75)90291-9. [DOI] [PubMed] [Google Scholar]
- Levy C. C., Karpetsky T. P. The purification and properties of chicken liver RNase: An enzyme which is useful in distinguishing between cytidylic and uridylic acid residues. J Biol Chem. 1980 Mar 10;255(5):2153–2159. [PubMed] [Google Scholar]
- Lin V. K., Ortwerth B. J. Competence and progression growth factors stimulate different tRNAlys modification reactions in BALB/C 3T3 cells. Biochem Biophys Res Commun. 1983 Sep 15;115(2):598–605. doi: 10.1016/s0006-291x(83)80186-7. [DOI] [PubMed] [Google Scholar]
- Lockard R. E., Alzner-Deweerd B., Heckman J. E., MacGee J., Tabor M. W., RajBhandary U. L. Sequence analysis of 5'[32P] labeled mRNA and tRNA using polyacrylamide gel electrophoresis. Nucleic Acids Res. 1978 Jan;5(1):37–56. doi: 10.1093/nar/5.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura S. Minor components in transfer RNA: their characterization, location, and function. Prog Nucleic Acid Res Mol Biol. 1972;12:49–85. [PubMed] [Google Scholar]
- Ortwerth B. J. Isoaccepting transfer ribonucleic acids in specialized mammalian tissues. Biochemistry. 1971 Nov;10(23):4190–4197. doi: 10.1021/bi00799a006. [DOI] [PubMed] [Google Scholar]
- Ortwerth B. J., Lin V. K. The effects of growth factors on tRNALys modification. Recent Results Cancer Res. 1983;84:160–170. doi: 10.1007/978-3-642-81947-6_11. [DOI] [PubMed] [Google Scholar]
- Ortwerth B. J., Liu L. P. Correlation between a specific isoaccepting lysyl transfer ribonucleic acid and cell division in mammalian tissues. Biochemistry. 1973 Sep 25;12(20):3978–3984. doi: 10.1021/bi00744a030. [DOI] [PubMed] [Google Scholar]
- Ortwerth B. J., Wolters J., Nahlik J., Conlon-Hollingshead C. Cell growth and tRNA-lys4 synthesis in mouse 3T3 cells. Exp Cell Res. 1982 Apr;138(2):241–250. doi: 10.1016/0014-4827(82)90173-2. [DOI] [PubMed] [Google Scholar]
- Peattie D. A. Direct chemical method for sequencing RNA. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1760–1764. doi: 10.1073/pnas.76.4.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peattie D. A., Gilbert W. Chemical probes for higher-order structure in RNA. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4679–4682. doi: 10.1073/pnas.77.8.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raba M., Limburg K., Burghagen M., Katze J. R., Simsek M., Heckman J. E., Rajbhandary U. L., Gross H. J. Nucleotide sequence of three isoaccepting lysine tRNAs from rabbit liver and SV40-transformed mouse fibroblasts. Eur J Biochem. 1979 Jun;97(1):305–318. doi: 10.1111/j.1432-1033.1979.tb13115.x. [DOI] [PubMed] [Google Scholar]
- Roy K. L., Cooke H., Buckland R. Nucleotide sequence of a segment of human DNA containing the three tRNA genes. Nucleic Acids Res. 1982 Nov 25;10(22):7313–7322. doi: 10.1093/nar/10.22.7313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya T., Nishizawa R., Matsuda K., Taya Y., Nishimura S. A rat tRNA gene cluster containing the genes for tRNAPro and tRNALys. Analysis of nucleotide sequences of the genes and the surrounding regions. Nucleic Acids Res. 1982 Oct 25;10(20):6411–6419. doi: 10.1093/nar/10.20.6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberklang M., Gillum A. M., RajBhandary U. L. Use of in vitro 32P labeling in the sequence analysis of nonradioactive tRNAs. Methods Enzymol. 1979;59:58–109. doi: 10.1016/0076-6879(79)59072-7. [DOI] [PubMed] [Google Scholar]
- Silverman S., Gillam I. C., Tener G. M., Söll D. The nucleotide sequence of lysine tRNA2 from Drosophila. Nucleic Acids Res. 1979 Feb;6(2):435–442. doi: 10.1093/nar/6.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley J., Vassilenko S. A different approach to RNA sequencing. Nature. 1978 Jul 6;274(5666):87–89. doi: 10.1038/274087a0. [DOI] [PubMed] [Google Scholar]
- Vold B. S., Keith D. E., Jr, Buck M., McCloskey J. A., Pang H. Lysine tRNAs from Bacillus subtilis 168: structural analysis. Nucleic Acids Res. 1982 May 25;10(10):3125–3132. doi: 10.1093/nar/10.10.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittig B., Reuter S., Gottschling H. Purification of the four lysine-specific transfer ribonucleic acids from chick embryos. Biochim Biophys Acta. 1973 Dec 7;331(2):221–230. doi: 10.1016/0005-2787(73)90435-8. [DOI] [PubMed] [Google Scholar]