Abstract
The frequency of dengue virus (DENV) infection has increased dramatically in the last few decades, and the lack of a vaccine has led to significant morbidity and mortality worldwide. To date, a convenient murine system to study human T cell responses to DENV has not been available. Mice transgenic for human leukocyte antigens (HLA) are widely used to model human immune responses and it has been shown that mouse-passaged DENV is able to replicate to significant levels in IFN-α/βR−/− mice. To cover a wide range of HLA phenotypes, we backcrossed IFN-α/βR−/− mice with HLA A*0201, A*0101, A*1101, B*0702 and DRB1*0101 transgenic mice. A DENV proteome-wide screen identified a total of 42 epitopes across all HLA-transgenic IFN-α/βR−/− strains tested. In contrast only 8 of these elicited responses in the corresponding IFN-α/βR+/+ mice. We were able to identify T cell epitopes from 9 out of the 10 DENV proteins. However, the majority of responses were derived from the highly conserved nonstructural proteins NS3 and NS5. The relevance of this model is further demonstrated by the fact that most of the epitopes identified in our murine system are also recognized by PBMC from DENV exposed human donors, and a dominance of HLA B*0702 restricted responses has been detected in both systems. Our results provide new insights into HLA-restricted T cell responses against DENV, and we herein describe a novel murine model, which allows the investigation of T cell-mediated immune mechanisms relevant to vaccine design.
Introduction
Dengue virus (DENV) is the etiologic agent of dengue fever (DF), the most prevalent arthropod-borne viral illness in humans. Infections with DENV can be asymptomatic, or result in a febrile illness (DF) characterized by headache, retro-orbital pain, myalgia and rash. More severe forms of the infection, like dengue hemorrhagic fever and dengue shock syndrome (DHF/DSS), are additionally accompanied by increased plasma leakage, thrombocytopenia and hemorrhagic manifestations which may be fatal (1). Demographic changes, urbanization and international travel have contributed to the rapid spreading of all four serotypes (DENV1–4). DENV is now endemic in over 100 tropical and subtropical countries worldwide, placing 2.5 billion people at risk for infection and it has been estimated that 50 million new cases of DF, and 250,000–500,000 cases of DHF/DSS occur each year (2). At present, no effective antiviral therapy or licensed vaccine exists, and treatment is largely supportive in nature. Therefore, the development of a vaccine against DENV is of global public health interest.
DENV is a single-stranded RNA virus and its genome is translated as a single polyprotein which is cleaved into three structural proteins, capsid (C), pre-membrane (prM/M), envelope (E), and seven non-structural (NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS5) proteins by both viral and host proteases (3). Numerous studies have demonstrated a major role for T cells during DENV infection (4–7). Beyond the protective role of CD4+ and CD8+ T cells in mice (8, 9) several lines of evidence suggest that T cells are also involved in resolving DENV infection in humans. First, activated serotype-specific CD4+ and CD8+ T cell responses are observed in humans with primary DENV infection (10, 11). Second, DENV-specific human CD4+ T cells specific for NS3 proliferate, produce IFN-γ, and can lyse infected target cells (11, 12). Finally, DENV-specific CD8+ T cells that recognize viral proteins NS1, NS3, and E can kill infected target cells (13). Furthermore, it has been demonstrated that IFNγ levels correlate with protection after viral challenge in human donors, suggesting that protection against DENV involves a TH1 response (14). These studies indicate that serotype-specific T cells are activated and functional in humans with primary DENV infection, thereby suggesting a role for T cells in protection against the virus. Infection with one DENV serotype presumably affords life-long serotype-specific immunity, but does not protect against other serotypes. In fact, the severe forms of DENV disease are most often observed in individuals experiencing a secondary infection with a different serotype (15, 16). One hypothesis to explain this phenomenon is that serotype cross-reactive antibodies enhance infection of FcγR bearing cells during a secondary infection, resulting in higher viral loads and more severe disease via a phenomenon known as antibody-dependent enhancement (ADE) (17, 18). Another hypothesis postulates that T cells raised against the original infecting serotype dominate during a secondary heterologous infection in a phenomenon termed “original antigenic sin.” (19, 20) In sequential DENV infections, due to the 67–75% sequence homology between different serotypes (21), memory T cells from a primary DENV infection with one serotype respond to a secondary infection by a different serotype. However, these memory cells have a lower threshold for activation than naïve cells (22), and possibly a lower affinity to antigens from the current infection that might lead to an altered response, including delayed viral clearance and immunopathology (19).
This suspected dual role of T cells in protection and pathogenesis is difficult to study in humans, since in most donor cohorts the time point of initial infection and, in case of secondary infections, the sequence of infection, is unknown, and thus does not allow direct correlations with T cell responses. A mouse model, which allows the investigation of adaptive immune responses restricted by human major histocompatibility complex (MHC) molecules to DENV infection, would greatly contribute to shed light on the role of T cells in protection and/or pathogenesis. Mice transgenic for human leukocyte antigens (HLA) are widely used to study T cell responses restricted by human MHC molecules and studies in other viral systems have shown the valuable impact of HLA transgenic mice in epitope identification (23–25). The host specificity of DENV is extremely narrow and thus DENV does not replicate in wild-type mice. However, it has been shown that mice lacking the IFN - α/β receptor support a productive infection and allow the study of T cell responses after DENV infection (8, 9, 26). To cover a wide range of HLA phenotypes, we backcrossed IFN-α/βR−/− mice with HLA, A*0201, A*0101, A*1101, B*0701 and DRB1*0101 transgenic mice and determined the T cell response against infection with DENV. We applied bioinformatics predictions on HLA affinity to identify T cell targets in the DENV proteome.
By combining the bioinformatics approach and the HLA transgenic mouse model, we were able to identify 42 T cell epitopes derived from 9 of the 10 DENV proteins. The majority of responses however were directed against epitopes from the nonstructural proteins NS3 and NS5. 80% of the epitopes identified in HLA transgenic mice were also able to elicit a T cell response in human donors, previously exposed to DENV. Thus, the mouse model described herein reflects the response patterns observed in humans making it a valuable model to study epitopes of human relevance during infection with dengue virus.
Materials and Methods
Viral stocks
S221 is a plaque-purified DENV2 strain, which was derived from the clinical isolate PL046 (27), by passaging through IFN-α/βR−/−; IFN-γR−/− and mosquito cells as previously described (9, 28). Viral stocks were amplified in C6/36 mosquito cells and purified over a sucrose gradient, as described previously (29). Infectious doses were determined based on genomic equivalents (GE), which were quantified by RT-PCR (29). There are ~ 5×104 GE/PFU for S221, based on a plaque assay on baby hamster kidney cells as described previously (30).
Mice and infections
HLA-A*0201/Kb, A*1101/Kb, A*0101, B*0702 and DRB1*0101 transgenic mice were bred at the La Jolla Institute for Allergy and Immunology facility (La Jolla, CA) as previously described (23, 25, 31, 32). IFN-α/βR−/− mice on the C57BL/6 background were originally obtained from Dr. W. Yokoyama (Washington University, St. Lois, MO) and subsequently bred and maintained in-house. All transgenic mouse strains were subsequently backcrossed with the IFN-α/βR−/− mice at the animal facility at the La Jolla Institute for Allergy and Immunology. Mice were used between 6 and 10 weeks of age. For all experiments mice were infected retro-orbitally with 1010 GE of S221 (~ 2×105 PFU) in 100 μl PBS. On day 7 post-infection, the mice were sacrificed and splenic CD8+ or CD4+ T cells were used in mouse IFNγ ELISPOT assays. All mouse experiments were performed following Institutional Animal Care and Use Committee-approved animal protocols.
Bioinformatic analyses
All 9 and 10mer peptides encoded in the DENV2 proteome were predicted for their binding affinity to MHC class I alleles A*0201, A*0101, A*1101 and B*0702. Binding predictions were performed using the command-line version of the MHC class I consensus prediction tool available on the Immune Epitope Database (IEDB; www.iedb.org) web site (33). Peptides were selected if they were in the top 1% of binders in a given strain. For MHC class II binding predictions all 15mer peptides were predicted for their binding affinity to the DRB1*0101 allele using the consensus approach as previously described (34). The top 2% of predicted binders were then selected for synthesis. All peptides tested in this study were derived from the DENV2 virus strain S221, which was also used as the infectious agent in this study, as described above.
For the conservancy analysis full-length DENV polyprotein sequences were retrieved for each serotype from the NCBI Protein database using the following query: txid11053 AND 3000:5000[slen]. The number of isolates from any one country was limited to 10 to eliminate geographical bias. Sequences were considered unique if they varied by at least 1 amino acid from all other sequences. In summary we retrieved 162 DENV1, 171 DENV2, 169 DENV3 and 53 DENV4 sequences from the NCBI Protein database and investigated the conservancy of the identified epitopes within the sequences of the respective serotypes.
Peptide synthesis
All peptides used in this study were synthesized by Mimotopes (Victoria, Australia). A total of 431 9-mer and 10-mer peptides were identified by MHC class I predictions, as described above and made as crude material. Peptides were combined into pools of 10 individual peptides, according to their predicted HLA restriction. MHC class II predictions resulted in the synthesis of 12 15-mer peptides, which were tested individually.
MHC peptide-binding and restriction assays
Purification of HLA A*0201, A*0101, A*1101, B*0702 and DRB1*0101 MHC molecules and quantitative assays to measure the binding affinity of peptides to purified MHC were performed as described elsewhere (35, 36). Binding assays are based on competitive inhibition of binding of a radiolabeled standard peptide. Briefly, after a 2-day incubation, inhibition of binding of the radiolabeled probe peptide to the corresponding MHC molecule, in the presence of various doses of a pest peptide, was determined by capturing MHC/peptide complexes on Greiner Lumitrac 600 microplates (Greiner Bio-One, Monroe, NC) coated with either the W6/32 (HLA class I specific) or L243 (HLA DR specific) monoclonal antibodies. Bound cpm were then measured using the Topcount microscintillation counter (Packard Instrument, Meriden, CT). The concentration of peptide yielding 50% inhibition of the binding of the radiolabeled probe peptide (IC50) was then calculated.
The tumor cell line 721.221 (37), which lacks expression of HLA -A, -B and C class I genes, was transfected with the HLA-A*0201/Kb or HL-A*1101 chimeric genes, and used as antigen-presenting cells (APC) in the restriction assays. The non-transfected cell line was used as a negative control.
Human blood samples
Peripheral blood samples were obtained from healthy adult blood donors from the National Blood Center, Ministry of Health, Colombo, Sri Lanka in an anonymous fashion. Donors are of both sexes and between 18 and 60 years of age. PBMC were purified by density gradient centrifugation (Ficoll-Paque Premium, GE Healthcare Biosciences, Kowloon, Hong Kong). Cells were suspended in fetal bovine serum (Gemini Bio-products, Sacramento, CA) containing 10% dimethyl sulfoxide, and cryo-preserved in liquid nitrogen. DENV seropositivity was determined by dengue IgG ELISA as previously described (38). Flow cytometry-based neutralization assays were performed for further characterization of seropositve donors, as previously described (39).
Genomic DNA isolated from PBMC of the study subjects by standard techniques (QIAmp. Qiagen, Valencia, CA) was use for HLA typing. High resolution Luminex-based typing for HLA Class I and Class II was utilized according the manufacturer’s protocol (Sequence-Specific Oligonucleotides (SSO) typing; One Lambda, Canoga Park, CA). Where needed, PCR based methods were used to provide high resolution sub-typing. (Sequence-Specific Primer (SSP) typing; One Lambda, Canoga Park, CA). Based on the HLA phenotype 15 DENV-seropositive and 5 DENV seronegative donors have been selected for the validation experiments described in this study.
IFNγ ELISPOT assay
For all murine experiments splenic CD4+ or CD8+ T cells were isolated by magnetic bead positive selection (Miltenyi Biotec, Bergisch Gladbach, Germany) 7 days after infection with the S221 strain of DENV2.. 2 × 105 T cells were stimulated with 1 × 105 uninfected splenocytes as APCs and 10 μg/ml of individual DENV peptides in 96-well flat-bottom plates (Immobilon-P; Millipore, Bedford, MA) coated with anti-IFNγ mAb (clone AN18; Mabtech, Stockholm, Sweden). Each peptide was tested in triplicates. Following a 20-h incubation at 37°C, the wells were washed with PBS/0.05% Tween 20 and then incubated with biotinylated IFNγ mAb (clone R4-6A2; Mabtech) for 2 h. The spots were developed using Vectastain ABC peroxidase (Vector Laboratories, Burlingame, CA) and 3-amino-9-ethylcarbazole (Sigma-Aldrich, St. Lois, MO) and counted by computer-assisted image analysis (KS-ELISPOT reader, Zeiss, Munich, Germany). Responses against peptides were considered positive if the net spot-forming cells (SFC) per 106 were ≥20, had a stimulation index of ≥2, and a p<0.05 in a t test comparing replicates with those from the negative control.
For ELISPOT with human cells, 2×106 PBMC/ml were stimulated in the presence of 1 μg/ml individual peptide for 7 days. Cells were cultured at 37°C, 5% CO2, and recombinant IL2 (10U/mL, eBiosciences, San Diego, CA) was added 3 days after antigenic stimulation. After 7 days, PBMC were harvested and 1×105 cells were added to each well of a 96-well flat-bottom plate (Millipore). The IFNγ ELISPOT assaywas performed as described above, except the mAb 1-D1K (Mabtech) and mAb 7-B6-1 (Mabtech) were used as coating and biotinylated secondary Ab, respectively. All epitopes identified in the murine study have been tested in all of the donors and subsequently assigned to the HLA-matched or non-HLA matched group according to the actual HLA phenotype of the donors. To be considered positive, IFNg responses needed to exceed the threshold set as the mean responses of HLA non-matched and DENV seronegative donors plus 3 times the standard deviation.
Results
A novel system to identify DENV specific HLA*0201 epitopes
Mice transgenic for HLA are widely used to study T cell responses restricted by MHC molecules, and it has been shown that mouse-passaged DENV is able to replicate to significant levels in IFN-α/βR−/− mice. Herein, we backcrossed HLA*0201 transgenic and IFN-α/βR−/− mice strains as a model system to study DENV-specific HLA restricted T cell responses. These mice were then infected with the mouse adapted DENV2 strain S221 and purified splenic T cells were used to study the anti-DENV CD8+ T cell responses.
To this end, a panel of 116 predicted A*0201 binding peptides were generated using bioinformatics predictions previously used to define HLA restricted CD8+ responses in the vaccinia virus system (40). Predicted HLA A*0201 binding peptides were combined into pools of 10 individual peptides and tested in an IFNγ ELISPOT assay using CD8+ T cells from HLA transgenic IFN-α/βR−/− and IFN-α/βR+/+, DENV2 infected mice, respectively (data not shown). Positive pools were deconvoluted and the individual peptides were tested in two independent experiments. Using this approach we identified a single peptide in the HLA*A0201 IFN-α/βR+/+ mice (NS53058–3066, Fig. 1A, white bars) whereas screening in HLA*A0201 IFN-α/βR−/− mice lead to identification of this epitope and ten additional epitopes (Fig. 1a, black bars). The CD8+ T cell responses were higher in the knockout mice than the wild-type mice. Even increasing the infectious dose 10-fold did not lead to an increase in magnitude or repertoire in the HLA*A0201 IFN-α/βR+/+ mice (data not shown). These results demonstrate that the use of HLA A*0201 transgenic IFN-α/βR−/− mice allows for the discovery of a stronger and broader T cell response.
Fig. 1. Identification of DENV-derived epitopes recognized by CD8+ T cells.
DENV specific epitope identification was performed in four different HLA transgenic mouse strains (A) A*0201; (B) A *1101; (C) A*0101; (D) B*0702. For all strains tested, IFNγ ELISPOT was performed using spleenic T cells isolated from HLA transgenic IFN-α/βR−/− mice (black bars) and HLA transgenic IFN-α/βR+/+ mice (white bars). Mice were infected retro-orbitally with 1×1010 GE of DENV2. Seven days post-infection, CD8+ T cells were purified and tested against a panel of DENV2 predicted peptides. The data are expressed as mean number of SFC/106 CD8+ T cells of two independent experiments. Error bars represent SEM. Responses against peptides were considered positive if the stimulation index (SI) exceeded double the mean negative control wells (effector cells plus APCs without peptide) and net spots were above the threshold of 20 SFCs/106 CD8+ T cells in two independent experiments. Asterisks indicate peptides, which were able to elicit a significant IFNγ response in each individual experiment, according to the criteria described above.
Broad population coverage by additional HLA transgenic IFN-α/βR−/− mouse strains
We next addressed whether similar observations could be made by measuring T cell responses in other HLA-transgenic IFN-α/βR−/− and IFN-α/βR+/+ mice. For this purpose, we backcrossed IFN-α/βR−/− mice with HLA A*0101, A*1101, and B*0702 transgenic mice. These alleles were chosen as representatives of three additional HLA class I supertypes (A1, A3 and B7, respectively).
Screening in HLA A*0101 and A*1101 transgenic IFN-α/βR−/− mice revealed 9 HLA A*0101 restricted (Fig. 1B, black bars), and 16 A*1101 restricted epitopes (Fig. 1C, black bars), respectively. In the case of the HLA A*0101 transgenic wildtype mice, no epitope could be detected whereas the HLA A*1101 transgenic mice showed an overlap of 5 epitopes with the corresponding IFN-α/βR−/− strain (M111–120, NS31608–1617, NS4B2288–2296, NS4B2315–2323 and NS53112–3121). Two of these epitopes were able to elicit a stronger response in the HLA A*1101 IFN-α/βR+/+ mice compared to the IFN-α/βR−/− strain (M111–120 and NS4B2287–2296). All other responses observed were more robust in the IFN-α/βR−/− mice. It should be noted that for several epitopes in NS5 inconsistent responses were observed in the HLA A*1101 IFN-α/βR+/+ mice. In these cases, positive responses were noted only in one out of two independent experiments, as also reflected by the large standard deviation. Our stringent criteria of positivity requires consistent responses in two out of two independent experiment, as described in the Figure legend. For the same reason, we have excluded some of the inconsistent responses observed in the IFN-α/βR−/− mice (Figure 1A NS4A2150–2159 ff. and Figure 1B NS32079–2087). In our experience the application of these stringent criteria is important for defining the most robust, and thereby most relevant, epitopes.
To extend our observations to mice transgenic for an HLA B allele we infected HLA B*0702 transgenic IFN-α/βR−/− and IFN-α/βR+/+ mice and compared epitope recognition between the two strains. We identified 15 B*0702 restricted epitopes in the IFN-α/βR−/− strain (Fig. 1D, black bars), with 1 of these also being detected in the corresponding IFN-α/βR+/+ mice (NS4B2280–2289; Fig 1D, white bars). Similar to the other HLA transgenic mouse strains, the responses observed in the HLA B*0702 transgenic IFN-α/βR−/− mice were not only broader but also more than ten-fold higher in magnitude. The single epitope recognized in the IFN-α/βR+/+ strain elicited an IFNγ response of 50 SFC/106 CD8+ T cells compared to an average of 857 SFC/106 CD8+ T cells in the IFN-α/βR−/− mice.
Dengue specific T cell responses in a MHC class II transgenic mouse model
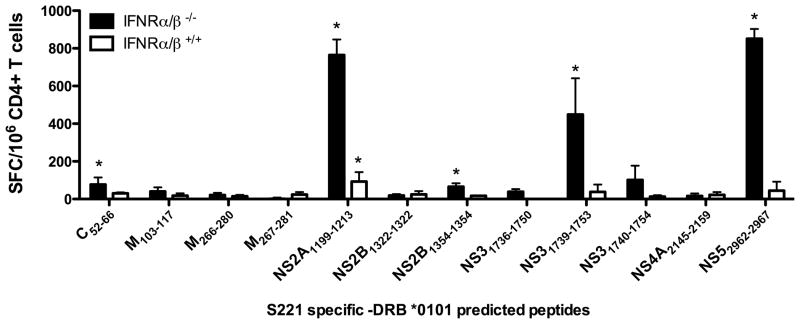
To determine if the observations made in the case of MHC class I transgenic mice were also applicable to MHC class II restricted responses we determined the antigenicity of HLA DRB1*0101 predicted binding peptides in HLA DRB1*0101, IFN-α/βR−/− and IFN-α/βR+/+ mice, respectively. Using the same experimental conditions described above for the MHC class I transgenic mice, we infected HLA DRB1*0101, IFN-α/βR−/− and IFN-α/βR+/+ mice with DENV2 and isolated CD4+ T cells 7 days post infection. A panel of 12 predicted S221 specific peptides predicted to bind DRB1*0101 was then tested for their ability to elicit IFNγ production by ELISPOT. Five epitopes which could be identified in the DRB1*0101, IFN-α/βR−/− mice in two independent experiments (Fig. 2; black bars) were identified. Similarly to the observation in MHC class I transgenic mice described above, only a single peptide could be identified in the corresponding DRB1*0101, IFN-α/βR+/+ mice (NS2A1199–1213; Figure 2, white bars). This epitope in the IFN-α/βR+/+ did not represent a novel epitope as it was also observed in the corresponding IFN-α/βR−/− mice. Similarly to the MHC class I transgenic mice, all observed responses were stronger in the IFN-α/βR−/− mice than their wild-type counterparts.
Fig. 2. Identification of DENV-derived epitopes recognized by CD4+ T cells.
IFNγ ELISPOT was performed using CD4+ T cells isolated from DRB1*0101 transgenic IFN-α/βR−/− (black bars) and IFN-α/βR+/+ (white bars) mice. Mice were infected retro-orbitally with 1×1010 GE of DENV2. Seven days post-infection, CD4+ T cells were purified and tested against a panel of DENV2 predicted peptides. The data are expressed as mean number of SFC/106 CD4+ T cells of two independent experiments. Error bars represent SEM. Responses against peptides were considered positive if the stimulation index (SI) exceeded double the mean negative control wells (effector cells plus APCs without peptide) and net spots were above the threshold of 20 SFCs/106 CD4+ T cells in two individual experiments. Asterisks indicate peptides, which were able to elicit a significant IFNγ response, according to the criteria described above.
In summary we identified a total of 55 epitopes in the HLA transgenic IFN-α/βR−/− mice whereas the same screen in HLA transgenic IFN-α/βR+/+ mice only revealed 8 epitopes. Importantly, these 8 epitopes were also detected in the HLA transgenic IFN-α/βR−/− mice. The broader repertoire seen in IFN-α/βR−/− mice as well as the stronger and more robust IFNγ responses, suggest that HLA transgenic mice, backcrossed with IFN-α/βR−/− mice represent a more suitable model to study T cell responses to DENV infection than HLA transgenic wildtype mice, presumably because the latter do not establish a productive infection.
Mapping optimal epitope in respect to peptide length
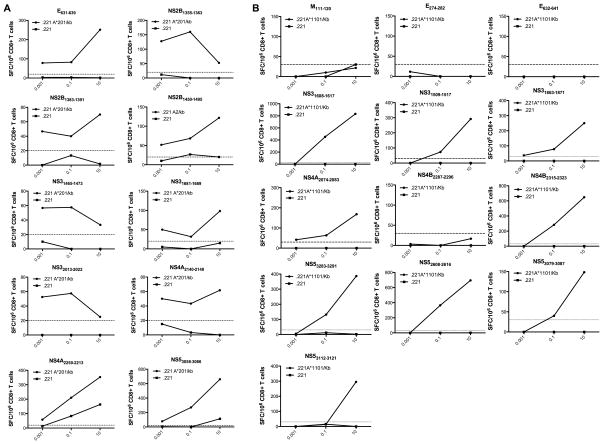
Within the 50 MHC class I restricted epitopes identified, 9 pairs of nested epitopes were identified, where the 10-mer as well as a nested 9-mer peptide was able to elicit an immune response (Figure 1A–D). To determine which peptide in each nested epitope pair was the optimal epitope, we employed peptide titration assays as shown in Figure 3A. For one epitope (NS4A2205–2213) both the 9- and the 10-mer displayed similar kinetics upon peptide titration (Fig. 3A). Since the 9-mer was able to elicit slightly higher responses in all conditions tested, we considered the 9-mer version of this epitope for further experiments. In all other cases an optimal epitope length peptide could be unequivocally identified.
Fig. 3. Determination of the optimal epitope.
To determine the optimal epitope, HLA-transgenic IFN-α/βR−/− mice were infected with 1 × 1010 GE of DENV2 and spleens were harvested seven days post-infection. CD8+ T cells were purified and incubated for 24 hours with ascending concentrations of nested peptides. Panel A shows pairs of nested epitopes where the 10-mer as well as a nested 9-mer peptide was able to elicit an immune response. Panel B shows two B*0702 restricted epitopes where 8- and 9-mer peptides carrying alternative dominant B7 motifs were synthesized and tested for T cell recognition. The peptides, which were able to elicit stronger IFNγ responses at various concentrations, were then considered the optimal epitope.
Similarly, for two of the B*0702 restricted epitopes (NS4B2296–2305 and NS52646–2655) 8- and 9-mer peptides carrying alternative dominant B7 motifs were synthesized and tested for T cell recognition. In one case the corresponding 8-mer (NS4B2296–2304) showed dominant IFNγ responses compared to the 9-mer. In the other case the 10mer originally identified (NS52646–2655) was able to elicit higher responses than the newly synthesized 8- and 9mer. In both cases the optimal epitope length could be identified and was considered further in the study, as shown in Figure 3B. Thus, taken together, our studies have identified 10 A*0201, 6 A*0101, 13 A*1101 and 12 B*0702 unique epitopes.
Further Characterization of the identified epitopes
Of the five HLA transgenic mouse strains tested, only the A*0201 and the A*1101 transgenic strains, co-expressed murine MHC molecules together with the respective HLA molecule. Thus it was necessary to assure that the observed responses were restricted by the human HLA class I molecule and not by murine Class I. We therefore tested purified T cells for their capacity to recognize the specific epitopes when pulsed on antigen presenting cells expressing only human class I. For this purpose we utilized the tumor cell line 721.221, which is negative for expression of any human or murine Class I molecule, and transfected it with either HLA A*0201 or HLA*1101. As shown in Figure 4A, all ten HLA*A0201 restricted epitopes were recognized when presented by APC exclusively expressing HLA*A0201 molecules. Nine out of thirteen of the HLA*A1101 restricted epitopes identified, stimulated a CD8+ T cell response when presented exclusively on HLA*1101 molecules (Fig. 4B). When the four remaining epitopes were tested in non-HLA transgenic IFN-α/βR−/− mice as described above, all elicited a significant T cell response (data not shown). Furthermore, one of the epitopes has already been described to be recognized by T cells from DENV2 infected Balb/c mice (E633–642 (41)). We therefore consider these 4 epitopes (M111–120, E274–282, E633–642, NS4B2287–2296) as solely mouse MHC restricted and excluded them from our further studies. Among those epitopes were also the two epitopes which were able to elicit a stronger response in the HLA A*1101 IFN-α/βR+/+ mice compared to the IFN-α/βR−/− strain (M111–120 and NS4B2287–2296).
Fig. 4. MHC-restriction of identified epitopes.
HLA A*0201 (A) and HLA A*1101 (B) transfected 721.221 cells as well as the non-transfected cell line as a control were used as antigen presenting cells in titration experiments to determine MHC restriction. Mice were infected retro-orbitally with 1×1010 GE of DENV2. Seven days post-infection purified CD8+ T cells from DENV2 infected HLA A*A0201 and HLA A*1101 transgenic IFN-α/βR−/− mice were incubated with ascending concentrations of peptides and tested for IFNγ production in an ELISPOT assay. Representative graphs of CD8+ T cell responses are shown, when incubated with HLA transfected cell lines (A and B; black lines) and non-tranfected cell lines (A and B, grey lines) are shown. The dotted line indicates the 25 net SFCs/106 cells threshold used to define positivity.
To further confirm the MHC restriction of the identified epitopes inferred on the basis of the bioinformatic predictions, we measured their MHC-binding capacity to their predicted allelic molecule using purified HLA molecules in an in-vitro binding assay. The results of these assays are also shown in Table 1. 32 of the 42 tested peptides (67%) bound the corresponding predicted allele with high affinity as indicated by an IC50< 50nM. 16 out of these even showed an IC50< 10 nM and can therefore be considered as very strong binders. Of the remaining peptides, 7 (17%) were able to bind the predicted allele with intermediate affinities, as indicated by IC50< 150 nM. Only three of the identified epitopes (7%) bound with low affinity, showing an IC50 value between 500nM and 1500nM. A summary of all 42 epitopes identified, after conclusion of these experiments and elimination of redundancies is shown in Table 1.
Table 1.
Identified DENV2 derived epitopes in HLA-transgenic IFN-α/βR−/− mice
| Epitope | Sequence | Restriction | T cell responses [SFC]
|
frequency in humans | HLA binding [IC50] | Conservancy within serotypes [%]
|
References | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| mouse | human | DENV2 | DENV1 | DENV3 | DENV4 | ||||||
| E451–459 | ITEAELTGY | A*0101 | 327 | 67 | 20% (1 out of 5) | 25 | 85 | 0 | 0 | 0 | |
| NS11090–1099 | RSCTLPPLRY | 228 | 104 | 20% (1 out of 5) | 5.9 | 100 | 0 | 100 | 0 | ||
| NS2A1192–1200 | MTDDIGMGV | 430 | 163 | 20% (1 out of 5) | 19 | 84 | 0 | 0 | 0 | ||
| NS2A1251–1259 | LTDALALGM | 465 | 143 | 40% (2 out of 5) | 129 | 91 | 0 | 0 | 0 | ||
| NS4B2399–2407 | VIDLDPIPY | 153 | 92 | 20% (1 out of 5) | 17 | 53 | 0 | 0 | 0 | ||
| NS53375–3383 | YTDYMPSMK | 495 | 143 | 20% (1 out of 5) | 37 | 98 | 0 | 0 | 0 | ||
|
|
|||||||||||
| mean | 350 | 119 | |||||||||
|
| |||||||||||
| E631–639 | RLITVNPIV | A*0201 | 265 | 393 | 43% (3 out of 7) | 2.8 | 98 | 0 | 0 | 0 | |
| NS2B1355–1363 | IMAVGMVSI | 503 | 417 | 43% (3 out of 7) | 1.9 | 92 | 0 | 0 | 0 | ||
| NS2B1383–1391 | GLLTVCYVL | 519 | 434 | 57% (4 out of 7) | 6.0 | 100 | 0 | 0 | 0 | ||
| NS2B1450–1459 | LLVISGLFPV | 361 | 588 | 43% (3 out of 7) | 26 | 50 | 0 | 0 | 0 | ||
| NS31465–1473 | AAAWYLWEV | 207 | 495 | 57% (4 out of 7) | 0.39 | 92 | 0 | 0 | 0 | ||
| NS31681–1689 | YLPAIVREA | 299 | 401 | 71% (5 out of 7) | 18 | 99 | 0 | 0 | 0 | [81] | |
| NS32013–2022 | DLMRRGDLPV | 417 | 312 | 71% (5 out of 7) | 6.3 | 92 | 0 | 0 | 0 | ||
| NS4A2140–2148 | ALSELPETL | 384 | 297 | 14% (1 out of 7) | 61 | 99 | 0 | 0 | 0 | [82] | |
| NS4A2205–2213 | IILEFFLIV | 336 | 301 | 28% (2 out of 7) | 18 | 99 | 0 | 0 | 0 | ||
| NS53058–3066 | KLAEAIFKL | 353 | 597 | 43% (3 out of 7) | 2.2 | 95 | 0 | 0 | 0 | [82] | |
|
|
|||||||||||
| mean | 365 | 423 | |||||||||
|
| |||||||||||
| NS31509–1517 | SQIGAGVYK | A*1101 | 436 | 0 | 0% (0 out of 5) | 33 | 98 | 0 | 0 | 0 | |
| NS31608–1617 | GTSGSPIIDK | 1003 | 880 | 20% (1 out of 5) | 12 | 30 | 0 | 0 | 0 | [83] | |
| NS31863–1871 | KTFDSEYVK | 208 | 0 | 0% (0 out of 5) | 140 | 75 | 0 | 0 | 0 | [81] | |
| NS4A2074–2083 | RIYSDPLALK | 148 | 3087 | 20% (1 out of 5) | 51 | 89 | 0 | 0 | 0 | [81] | |
| NS4B2315–2323 | ATVLMGLGK | 712 | 0 | 0% (0 out of 5) | 16 | 98 | 0 | 0 | 0 | ||
| NS52608–2616 | STYGWNLVR | 1030 | 0 | 0% (0 out of 5) | 22 | 100 | 0 | 0 | 0 | ||
| NS53079–3087 | TVMDIISRR | 105 | 0 | 0% (0 out of 5) | 71 | 91 | 0 | 0 | 0 | ||
| NS53112–3121 | RQMEGEGVFK | 284 | 0 | 0% (0 out of 5) | 118 | 43 | 0 | 0 | 0 | ||
| NS53283–3291 | RTTWSIHAK | 358 | 800 | 20% (1 out of 5) | 83 | 65 | 0 | 0 | 0 | ||
|
|
|||||||||||
| mean | 476 | 530 | |||||||||
|
| |||||||||||
| NS2A1212–1221 | RPTFAAGLLL | B*0702 | 400 | 335 | 20% (1 out of 5) | 4.8 | 92 | 0 | 0 | 0 | |
| NS31682–1690 | LPAIVREAI | 1293 | 207 | 20% (1 out of 5) | 6.5 | 100 | 98 | 96 | 0 | [81] | |
| NS31700–1709 | APTRVVAAEM | 1064 | 1426 | 40% (2 out of 5) | 4.6 | 99 | 0 | 100 | 100 | [81] | |
| NS31753–1761 | VPNYNLIIM | 509 | 410 | 20% (1 out of 5) | 43 | 100 | 0 | 89 | 0 | [81] | |
| NS31808–1817 | APIMDEEREI | 364 | 232 | 20% (1 out of 5) | 572 | 77 | 0 | 0 | 0 | ||
| NS31978–1987 | TPEGIIPSMF | 194 | 1825 | 20% (1 out of 5) | 589 | 99 | 0 | 0 | 0 | [81] | |
| NS32070–2078 | KPRWLDARI | 1853 | 1633 | 40% (2 out of 5) | 6.8 | 91 | 0 | 0 | 0 | [81] | |
| NS4B2280–2289 | RPASAWTLYA | 1539 | 0 | 0% (0 out of 5) | 7.4 | 100 | 37 | 0 | 100 | ||
| NS4B2296–2304 | TPMLRHSI | 1013 | 460 | 20% (1 out of 5) | 1.1 | 100 | 0 | 0 | 0 | ||
| NS52646–2655 | SPNPTVEAGR | 994 | 0 | 0% (0 out of 5) | 1332 | 54 | 0 | 0 | 0 | ||
| NS52885–2894 | TPRMCTREEF | 811 | 1341 | 60% (3 out of 5) | 13 | 89 | 0 | 0 | 0 | ||
| NS53077–3085 | RPTPRGTVM | 487 | 390 | 40% (2 out of 5) | 1.5 | 97 | 0 | 0 | 0 | ||
|
|
|||||||||||
| mean | 877 | 688 | |||||||||
|
| |||||||||||
| C53–67 | AFLRFLTIPPTAGIL | DRB1*0101 | 77 | 314 | 75% (3 out of 4) | 9.7 | 99 | 0 | 0 | 0 | [84] |
| NS2A1199–1213 | GVTYLALLAAFKVRP | 764 | 249 | 75% (3 out of 4) | 10 | 91 | 0 | 0 | 0 | ||
| NS2B1356–1370 | MAVGMVSILASSLLK | 65 | 279 | 75% (3 out of 4) | 34 | 100 | 0 | 0 | 0 | ||
| NS31742–1756 | TFTMRLLSPVRVPNY | 448 | 336 | 75% (3 out of 4) | 1.5 | 70 | 100 | 99 | 0 | [81] | |
| NS52966–2980 | SRAIWYMWLGARFLE | 851 | 729 | 75% (3 out of 4) | 17 | 100 | 99 | 0 | 100 | ||
|
|
|||||||||||
| mean | 441 | 381 | |||||||||
Validation of the identified epitopes in human DENV seropositive donors
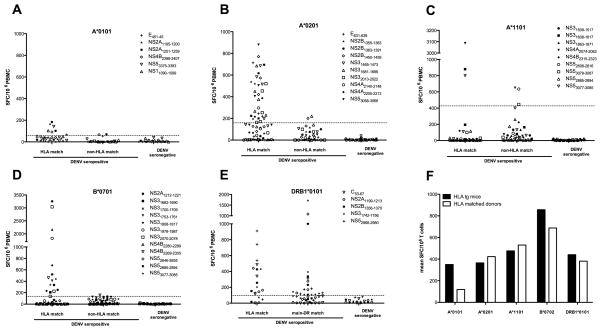
To validate the epitopes identified in the HLA-transgenic IFN-α/βR−/− mice we tested the capacity of these epitopes to stimulate PBMC from human donors previously exposed to DENV as determined ELISA (IgG OD405 mean = 1.5; range 0.8–1.9). The limited sample size did not allow us to detect a significant correlation between the level of seropositivity and the robustness of T cell response. Since the IFNγ response to these peptides was not detectable directly ex vivo (data not shown), we re-stimulated HLA-matched PBMC for 7 days in the presence of the respective peptides and IL2. As a control we re-stimulated PBMC from donors that expressed neither the exact HLA-molecule nor one from the same supertype, as well as PBMC from DENV seronegative donors. The average IFNγ response from these control donors, plus 3 times the standard deviation (SD) was set as a threshold of positivity. Figures 5A–D (HLA A*0101, A*0201, A*1101, and B*0702) show the capacity of the identified epitopes to stimulate PBMC from the various donor categories. Each of the A*0101 and A*0201 epitopes was antigenic in at least one HLA matched donor, although the magnitude as well as the frequency of responses was higher for the A*0201 restricted epitopes (Figs. 5A–B and Table 1). Out of the 9 A*1101 restricted epitopes 3 have been detected once in HLA matched donors. These three epitopes though have been able to stimulate a robust IFNγ response, as indicated by net SFCs > 800 (Fig. 5C). In the case of the B*0702 restricted epitopes, 10 out of the 12 were antigenic in one or more HLA matched donors as shown in Fig. 5D and Table 1. Interestingly the two epitopes, not detected in human donors were also demonstrated to require high concentrations in the HLA*B0702 IFN-α/βR−/− mice (NS4B2280–2289, Fig. 3A and NS52646–2655, Fig. 3B), which suggests that these epitopes might be more weakly stimulatory and maybe less relevant. No significant response could be detected in non-HLA matched donors tested. In contrast, all five restricted DRB1*0101 epitopes have been detected in 3 out of 4 HLA matched donors tested and were also able to elicit significant IFNγ responses in non-HLA matched donors. This is in accordance with recent reports demonstrating a high degree of repertoire sharing across MHC class II molecules (42). Overall, we could detect responses to 34 of the 42 epitopes in at least one donor, which corresponds to an overlap of 81% between the murine and human system. In addition to the experimental approach we also performed an IEDB query (43) with the epitopes identified in the mouse model. By this analysis we found that 13 of our 42 epitopes were previously described to elicit an IFNγ response in DENV seropositive individuals, as indicated in Table 1. 30% overlap with known epitopes contributes to the validation of our mouse model and shows on the other hand that 70% of the epitopes identified are novel, contributing to an extended knowledge of T cell mediated responses to DENV.
Fig. 5. Antigenicity of identified epitopes in human donors.
To validate the epitopes identified in the HLA-transgenic IFN-α/βR−/− mice, we tested their capacity to stimulate PBMC from human donors. Thus PBMC [2×106/ml] were stimulated in the presence of 1 μg/ml individual peptide for 7 days and then tested in an IFNγ ELISPOT assay. Figures A-E show IFNγ responses/106 PBMC after stimulation with A*0101, A*0201, A*1101, B*0702 and DRB1*0101 restricted peptides, respectively. Donors, seropositive for DENV, were grouped in HLA matched and non-HLA matched cohorts, as shown in panels 1 and 2 of each figure. All epitopes identified were also tested in DENV seronegative individuals. The average IFNγ responses elicited by PBMC from DENV seropositve non-HLA matched and DENV seronegative donors plus 3 times the standard deviation (SD) was set as a threshold for positivity, as indicated by the dashed line. Figure F shows the mean IFNγ response/106 T cells from HLA transgenic mice (black bars) and HLA matched donors (white bars) grouped by HLA restriction of the epitopes tested.
Dominance of B7 responses
A especially notable observation here was that out of all HLA transgenic mouse strains tested the strongest CD8+ T cell responses were detected in the B*0702 transgenic IFN-α/βR−/− mice. Four B*0702 restricted epitopes were able to elicit an IFNγ response above a 1000 SFC/106 CD8+ T cells (Table 1, Column 4). On average B*0702 epitopes were able to elicit an IFNγ response of 877 SFC/106 CD8+ T cells, compared to an average of 350, 365, and 476 SFC/106 CD8+ T cells for the HLA A*0101, A*0201 and A*1101 restricted epitopes, respectively (Fig. 5F. black bars). Most interestingly, the exact same response pattern could be observed testing PBMC from HLA matched donors previously exposed to DENV (Fig. 5F, white bars). As seen in mice, out of all epitopes tested, B*0702 restricted epitopes were able to elicit the strongest IFNγ responses when tested in HLA matched human donors, reaching an average of 688 SFC/106 PBMC, followed by an average of 530, 423 and 119 SFC/106 PBMC for HLA*1101, A*0202 and A*0101 restricted epitopes, respectively (Table 1, Column 5). Considering that the human PBMCs were stimulated for 7 days in the presence of IL-2, compared to 1 day for murine cells, the magnitude of responses observed with the two approaches cannot be compared. However, it is interesting that the hierarchy of responses observed in both systems is the same. The highest responses are seen in B*0702, followed by A*1101, DRB1*0101 and A*0201, and the weakest responses in both systems are observed for A*0101. The fact that the mouse model described herein reflects response patterns observed in humans makes it an even more valuable model to study epitopes of human relevance during infection with dengue virus.
Protein location of identified epitopes
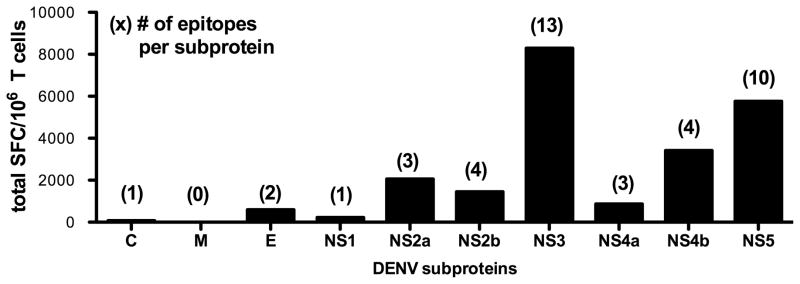
As shown in Figure 6, the identified epitopes are derived from 9 of the 10 DENV proteins, with the membrane protein being the only protein for which no epitope could be detected. The majority of epitopes however were derived from the seven nonstructural proteins. 39 out of 42 of the identified epitopes (93%) originate from the 7 nonstructural proteins, accounting for 97% of the total IFNγ response observed. Within the nonstructural proteins NS3 and NS5 alone account for 67% of the total response, representing a total of 23 epitopes detected from these two proteins. Furthermore NS5 was the only protein where at least one epitope was identified in all five HLA transgenic mouse strains. These results support the previously reported high immunogenicity of NS3, and also suggest NS5 as a major target of T cell responses.
Fig. 6. Protein location of identified epitopes.
All identified epitopes were grouped according to the DENV protein they are derived from. Black bars show the total IFNγ response of all epitopes from a given protein. Numbers in parenthesis indicate the number of epitopes that have been detected for this protein.
Conservancy of identified epitopes within the DENV2 serotype
Cross-reactivity of T cells is a well-described phenomenon in the context of DENV infection (5). To circumvent this issue, we have exclusively tested T cell reactivity to S221 derived peptides, which was also used as infectious agent in this study. However, to asses the relevance for infections with other DENV2 strains we investigated the conservancy of the S221 derived epitopes within other strains of the DENV2 serotype. We retrieved 171 full-length DENV2 polyprotein sequences from the NCBI Protein database and used them for conservancy analysis. Of the 42 epitopes identified, 30 were conserved in >90% of all DENV2 strains with 8 being conserved in all 171 strains analyzed. Of the remaining 12 epitopes, 6 were conserved in >75% of all strains analyzed and 6 were found in the 30–65% range. This accounts for an average conservancy of 92% for the epitopes identified, which is significantly higher than the average conservancy of non-epitopes (73%; p< 0.001). We then asked if the epitopes identified were also conserved in serotypes other than DENV2. Accordingly, we retrieved 162 DENV1, 169 DENV3 and 53 DENV4 sequences from the NCBI protein database and investigated the conservancy of the identified epitopes within the respective serotypes. In contrast to the high degree of conservancy within the DENV2 serotpye, 35 out of the 42 epitopes did not occur in any of the 384 DENV-1, 3 and 4 sequences tested, and only 7 epitopes had sequence homologues in one or more of the other serotypes. Interestingly, most of the epitopes which showed conservancy across serotypes have been identified in the B*0702 transgenic mice. 4 of the identified B*0702 restricted epitopes (NS31682–1690, NS31700–1709, NS31753–1761, NS4B2280–2892) were additionally conserved in 89–100% of sequences derived from serotypes other than DENV2. The same was observed for two DRB1*0101 restricted epitopes which were conserved across serotypes (NS31742–1756, NS52966–2980). Here, the epitopes were conserved in >99% of polyprotein sequences of two serotypes other than DENV2. Finally, one of the A*0101 restricted epitopes (NS11090–1099) was also conserved in 100% of DENV3 sequences. The results from this analysis are shown in Table 1.
Discussion
There have been several attempts to develop mouse models that allow studying of T cell responses in DENV disease in humans. Wild-type mice are resistant to DENV-induced disease. As such, development of mouse models for DENV infection to date has been challenging and has had to rely on infection of immunocompromised mice, non-physiologic routes of infection, and mouse-human chimeras (reviewed in (44)). Due to the importance of the IFN system in the host antiviral response, mice lacking IFN-α/βR support a productive infection. A mouse-passaged DENV2 strain, S221, is highly immunogenic and also replicates to high levels in IFN-α/βR−/− mice. A possible concern is that the lack of type I interferon may alter the T cell responses observed in IFN-α/βR−/− mice. It has been previously shown that the specificity of T cell responses after infection with DENV is similar in wild-type and IFN-α/βR−/− mice, thus allowing the study of CD4+ and CD8+ T cell responses in DENV infection. It was further demonstrated in this murine model that vaccination with T cell epitopes prior to DENV2 infection provided significant protection (8, 9). However, a small animal model in which epitope specificity can be linked with elements of the human immune system to study T-cell responses to DENV antigens, would add greatly to the relevance of the model. Therefore we backcrossed IFN-α/βR−/− mice with mice transgenic for HLA A*0101, A*0201, A*1101, B*0702 and DRB1*0101. While significant differences exist between human and murine TCR repertoires and processing pathways, HLA transgenic mice are fairly accurate models of human immune responses, especially when peptide immunizations are utilized. Numerous studies to date show that these mice develop T cell responses that mirror the HLA restricted responses observed in humans in the context of various pathogens (45–53).
The findings presented herein demonstrate that HLA transgenic IFN-α/βR−/− mice represent a valuable model to systematically map the epitopes recognized in humans. In fact, we could not only identify a number of HLA-restricted T cell responses, but our genome wide screen also provided further insight into the proteins targeted by T cells during DENV infection. Whereas we identified T cell epitopes from 9 out of 10 DENV proteins, the majority of responses (97%) were derived from the nonstructural proteins. More than half of the epitopes identified originated from the NS3 and NS5 protein. Thus we were able to confirm the suggested highly immunogenic role of the conserved NS3 and NS5 proteins as a major target of T cell responses (54, 55). Interestingly, the proteins previously described as antibody targets (prM, E and NS1) (56) accounted for less than 5% of all T cell responses, with only 3 epitopes identified from these proteins. The observation that T and B cell epitopes after primary DENV infection are not derived from the same proteins may factor in future vaccine design strategies, since immunizing with antigens containing NS3 and NS5 T cell epitopes would induce a robust T cell response without the risk of antibody-dependent-enhancement (ADE).
Another unique challenge in vaccine development is the high degree of sequence variation in a pathogen, characteristically associated with RNA viruses. This is of particular relevance in the case of DENV infections, where it is well documented that prior exposure to a different serotype may lead to more severe disease and immunopathology (16). The fact that there is also significant genetic variation within each serotype adds to the complexity of this topic (57, 58). It is hypothesized that in certain cases, peptide variants derived from the original antigen in the primary infection, with substitutions at particular residues, can induce a response that is qualitatively different from the response induced by the original antigen (for example inducing a different pattern of lymphokine production; partial agonism), or even actively suppress the response (TCR antagonism). Variants associated with this phenotype are often collectively referred to as altered peptide ligands (APLs(59)). During secondary infections, the T cell response directed at the APL may lead to altered or aberrant patterns of lymphokine production, and TCR antagonist mediated inhibition of T cell responses (60). Therefore, a balanced immunity to all four serotypes is required for a suitable vaccine candidate and it is necessary to explore DENV vaccine concepts that might overcome these potential problems. It is generally recognized that conserved protein sequences represent important functional domains (61), and that mutations at these important protein sites could be detrimental to the survival of the virus. T cells that target highly conserved regions of a protein are therefore likely to target the majority of genetic variants of a pathogen (62). Most interestingly in this context we could show that epitopes that are highly conserved within the DENV2 serotype are the major target for T cells. These data suggest, that immunization with peptides from a given serotype would protect from the majority of genotypes within this serotype. In contrast to the conservancy of epitopes within a serotype our analyses showed that most DENV2 derived epitopes identified were not conserved in other serotypes. These findings point to a tetravalent immunization strategy with a collection of multiple non-crossreactive epitopes derived from each of the four DENV serotypes. The induction of separate non-crossreactive responses would avoid issues arising from incomplete crossreactivity and APL/TCR antagonism effects.
In addition to sequence variation, HLA polymorphism adds to the complexity of studying T cell responses to DENV. Human MHC molecules are extremely polymorphic, with several hundred different variants known at most loci (63, 64). Therefore, selecting multiple peptides matching different MHC binding specificities will increase coverage of the patient population for basic investigations and diagnostic or vaccine applications alike. However, different MHC types are expressed at dramatically different frequencies in different ethnicities. To address this issue we backcrossed IFN-α/βR−/− mice with mice transgenic for HLA A*0101, A*0201, A*1101, B*0702 and DRB1*0101. These four MHC class I alleles were chosen as representatives of four supertypes (A1, A2, A3 and B7, respectively) allowing a combined coverage of approximately 90% of the worldwide population (65), and with more than 50% expressing at least one of the specific alleles selected. HLA supertypes are not limited to class I molecules. Several studies have demonstrated the existence of HLA class II supertypes (66–68) and functional classification has revealed a surprising degree of repertoire sharing across class II supertypes (42). This is in accordance with our data since we could identify the DRB1*0101 restricted epitopes in almost every donor, regardless if the donor expressed the DRB1*0101 allele (Fig. 5E). The fact that some of the DRB1*0101 restricted epitopes were able to elicit an even higher response in HLA non-matched donors suggests that theses epitopes might additionally be restricted by MHC class II molecule(s) other than DRB1*0101. In the case of the MHC class I restricted epitopes, all of the A*0101 and A*0201 restricted, and 10 out of 12 B*0702 restricted, epitopes have been able to elicit an IFNγ response when tested in human PBMC (Fig. 5, A, B and D). In the case of the A*1101 restricted epitopes, only 3 out of 9 were detected in both experimental systems (Fig. 5C). Because of the fact that responses to the two B*0702 peptides not detected in humans required higher amounts of peptide in the HLA B*0702 IFN-α/βR−/− mice, we hypothesized that a similar phenomenon might explain the low overlap in A*1101 epitopes. To test this hypothesis we have performed titrations for all the A*1101 restricted epitopes not recognized in the corresponding HLA matched donors (data not shown). However, we did not detect any correlation between the epitope dose response in HLA transgenic IFN-α/βR−/− mice and recognition in humans. Another possible explanation for this low overlap could be the substrate specificity of the murine TAP transporter. Murine TAP molecules prefer peptides with hydrophobic C-termini (69, 70), whereas the HLA A*1101 preference is for positively charged residues (71). Accordingly, it is possible that the optimal A*1101 peptides recognized in humans have not been presented and identified in the HLA A*1101 transgenic mice. Overall we could demonstrate that the mouse model significantly reflects the response pattern observed in humans and that HLA B restricted responses seem to be dominant in transgenic mice as well as in human donors (Fig 5F). The dominance of HLA B responses has been also shown in the context of several other viruses, such as HIV, EBV, CMV, and Influenza (72–75). These studies suggest that this observation is not limited to RNA viruses. In fact it has even been described for an intracellular bacterial pathogen, Mycobacterium tuberculosis (76, 77). Furthermore, HLA B restricted T cell responses have been described to be of higher magnitude (73) and have been described to influence infectious disease course and outcome. In the case of DENV it has been shown that one particular B*07 epitope was able to elicit higher responses in patients with DHF compared to patients suffering from DF only and could therefore be associated with disease outcome (78). Other studies suggest a role for HLA B44, B62, B76 and B77 alleles in protection against developing clinical disease after secondary DENV infection whereas other alleles were associated with contribution to pathology (79, 80). This suggests that HLA alleles are associated with clinical outcome of exposure to DENV, in previously exposed and immunologically primed individuals. All of this is in agreement with the observations we made in this study. Similarly, the fact that the stronger B*07 response occurs in our human samples as well as in our mouse model of DENV infection, underscores the value of this mouse model, since it seems to mimic patterns of immuno-dominance observed in humans.
In conclusion, the exact role of DENV sequence variation in protection and immunpathology has not been fully elucidated, and addressing this issue is key to the development of vaccines against DENV infection and DHF/DSS. Here we provide a novel model to study the mechanisms of T cell mediated mechanisms of protection and pathogenesis, relevant for vaccine design against DENV. In addition, our studies have led to the identification of 24 novel human epitopes, which contributes to an extended knowledge of T cell mediated responses against DENV infection. Future experiments will continue the evaluation of these novel epitopes to provide a more detailed examination of the epitope-specific responses, such as analysis of multi-cytokine production, CTL activity, and the persistence of these epitope-specific responses into memory. Further studies will also test the efficacy of vaccination strategies using the epitopes identified herein.
Acknowledgments
We thank the Blood Center, Ministry of Health, Colombo, Sri Lanka for providing buffy coat samples used in this study.
Footnotes
This work was supported by National Institutes of Health contract Nr. HHSN272200900042C (to A.S.) and National Institutes of Health grant Nr. U54AI057517 from the Southeastern Regional Center of Excellence for Emerging Infections and Biodefense (to S.S)
References
- 1.Kyle JL, Harris E. Global spread and persistence of dengue. Annu Rev Microbiol. 2008;62:71–92. doi: 10.1146/annurev.micro.62.081307.163005. [DOI] [PubMed] [Google Scholar]
- 2.WHO. Dengue: Guidelines for Diagnosis, Treatment, Prevention and Control. Geneva, Switzerland: 2009. [PubMed] [Google Scholar]
- 3.Burke D, Monath T. Flaviviruses. In: Knipe aPHD., editor. Field’s Virology. William and Wilkins; Philadelphia: 2001. pp. 1043–1126. [Google Scholar]
- 4.Rothman AL. Cellular immunology of sequential dengue virus infection and its role in disease pathogenesis. Curr Top Microbiol Immunol. 2010;338:83–98. doi: 10.1007/978-3-642-02215-9_7. [DOI] [PubMed] [Google Scholar]
- 5.Mathew A, Rothman AL. Understanding the contribution of cellular immunity to dengue disease pathogenesis. Immunol Rev. 2008;225:300–313. doi: 10.1111/j.1600-065X.2008.00678.x. [DOI] [PubMed] [Google Scholar]
- 6.Nielsen DG. The relationship of interacting immunological components in dengue pathogenesis. Virol J. 2009;6:211. doi: 10.1186/1743-422X-6-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fink J, Gu F, Vasudevan SG. Role of T cells, cytokines and antibody in dengue fever and dengue haemorrhagic fever. Rev Med Virol. 2006;16:263–275. doi: 10.1002/rmv.507. [DOI] [PubMed] [Google Scholar]
- 8.Yauch LE, Prestwood TR, May MM, Morar MM, Zellweger RM, Peters B, Sette A, Shresta S. CD4+ T cells are not required for the induction of dengue virus-specific CD8+ T cell or antibody responses but contribute to protection after vaccination. J Immunol. 2010;185:5405–5416. doi: 10.4049/jimmunol.1001709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yauch LE, Zellweger RM, Kotturi MF, Qutubuddin A, Sidney J, Peters B, Prestwood TR, Sette A, Shresta S. A protective role for dengue virus-specific CD8+ T cells. J Immunol. 2009;182:4865–4873. doi: 10.4049/jimmunol.0801974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Livingston PG, Kurane I, Dai LC, Okamoto Y, Lai CJ, Men R, Karaki S, Takiguchi M, Ennis FA. Dengue virus-specific, HLA-B35-restricted, human CD8+ cytotoxic T lymphocyte (CTL) clones. Recognition of NS3 amino acids 500 to 508 by CTL clones of two different serotype specificities. J Immunol. 1995;154:1287–1295. [PubMed] [Google Scholar]
- 11.Kurane I, Meager A, Ennis FA. Dengue virus-specific human T cell clones. Serotype crossreactive proliferation, interferon gamma production, and cytotoxic activity. J Exp Med. 1989;170:763–775. doi: 10.1084/jem.170.3.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gagnon SJ, Ennis FA, Rothman AL. Bystander target cell lysis and cytokine production by dengue virus-specific human CD4(+) cytotoxic T-lymphocyte clones. J Virol. 1999;73:3623–3629. doi: 10.1128/jvi.73.5.3623-3629.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mathew A, Kurane I, Rothman AL, Zeng LL, Brinton MA, Ennis FA. Dominant recognition by human CD8+ cytotoxic T lymphocytes of dengue virus nonstructural proteins NS3 and NS1.2a. J Clin Invest. 1996;98:1684–1691. doi: 10.1172/JCI118964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gunther VJ, Putnak R, Eckels KH, Mammen MP, Scherer JM, Lyons A, Sztein MB, Sun W. A human challenge model for dengue infection reveals a possible protective role for sustained interferon gamma levels during the acute phase of illness. Vaccine. 2011 doi: 10.1016/j.vaccine.2011.03.038. [DOI] [PubMed] [Google Scholar]
- 15.Burke DS, Nisalak A, Johnson DE, Scott RM. A prospective study of dengue infections in Bangkok. Am J Trop Med Hyg. 1988;38:172–180. doi: 10.4269/ajtmh.1988.38.172. [DOI] [PubMed] [Google Scholar]
- 16.Sangkawibha N, Rojanasuphot S, Ahandrik S, Viriyapongse S, Jatanasen S, Salitul V, Phanthumachinda B, Halstead SB. Risk factors in dengue shock syndrome: a prospective epidemiologic study in Rayong, Thailand. I. The 1980 outbreak. Am J Epidemiol. 1984;120:653–669. doi: 10.1093/oxfordjournals.aje.a113932. [DOI] [PubMed] [Google Scholar]
- 17.Morens DM. Antibody-dependent enhancement of infection and the pathogenesis of viral disease. Clin Infect Dis. 1994;19:500–512. doi: 10.1093/clinids/19.3.500. [DOI] [PubMed] [Google Scholar]
- 18.Halstead SB, Mahalingam S, Marovich MA, Ubol S, Mosser DM. Intrinsic antibody-dependent enhancement of microbial infection in macrophages: disease regulation by immune complexes. Lancet Infect Dis. 2010;10:712–722. doi: 10.1016/S1473-3099(10)70166-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mongkolsapaya J, Dejnirattisai W, Xu XN, Vasanawathana S, Tangthawornchaikul N, Chairunsri A, Sawasdivorn S, Duangchinda T, Dong T, Rowland-Jones S, Yenchitsomanus PT, McMichael A, Malasit P, Screaton G. Original antigenic sin and apoptosis in the pathogenesis of dengue hemorrhagic fever. Nat Med. 2003;9:921–927. doi: 10.1038/nm887. [DOI] [PubMed] [Google Scholar]
- 20.Halstead SB, Rojanasuphot S, Sangkawibha N. Original antigenic sin in dengue. Am J Trop Med Hyg. 1983;32:154–156. doi: 10.4269/ajtmh.1983.32.154. [DOI] [PubMed] [Google Scholar]
- 21.Fu J, Tan BH, Yap EH, Chan YC, Tan YH. Full-length cDNA sequence of dengue type 1 virus (Singapore strain S275/90) Virology. 1992;188:953–958. doi: 10.1016/0042-6822(92)90560-c. [DOI] [PubMed] [Google Scholar]
- 22.Veiga-Fernandes H, Walter U, Bourgeois C, McLean A, Rocha B. Response of naive and memory CD8+ T cells to antigen stimulation in vivo. Nat Immunol. 2000;1:47–53. doi: 10.1038/76907. [DOI] [PubMed] [Google Scholar]
- 23.Kotturi MF, Botten J, Maybeno M, Sidney J, Glenn J, Bui HH, Oseroff C, Crotty S, Peters B, Grey H, Altmann DM, Buchmeier MJ, Sette A. Polyfunctional CD4+ T cell responses to a set of pathogenic arenaviruses provide broad population coverage. Immunome Res. 2010;6:4. doi: 10.1186/1745-7580-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kotturi MF, Assarsson E, Peters B, Grey H, Oseroff C, Pasquetto V, Sette A. Of mice and humans: how good are HLA transgenic mice as a model of human immune responses? Immunome Res. 2009;5:3. doi: 10.1186/1745-7580-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pasquetto V, Bui HH, Giannino R, Banh C, Mirza F, Sidney J, Oseroff C, Tscharke DC, Irvine K, Bennink JR, Peters B, Southwood S, Cerundolo V, Grey H, Yewdell JW, Sette A. HLA-A*0201, HLA-A*1101, and HLA-B*0702 transgenic mice recognize numerous poxvirus determinants from a wide variety of viral gene products. J Immunol. 2005;175:5504–5515. doi: 10.4049/jimmunol.175.8.5504. [DOI] [PubMed] [Google Scholar]
- 26.Shresta S, Kyle JL, Snider HM, Basavapatna M, Beatty PR, Harris E. Interferon-dependent immunity is essential for resistance to primary dengue virus infection in mice, whereas T- and B-cell-dependent immunity are less critical. J Virol. 2004;78:2701–2710. doi: 10.1128/JVI.78.6.2701-2710.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin YL, Liao CL, Chen LK, Yeh CT, Liu CI, Ma SH, Huang YY, Huang YL, Kao CL, King CC. Study of Dengue virus infection in SCID mice engrafted with human K562 cells. J Virol. 1998;72:9729–9737. doi: 10.1128/jvi.72.12.9729-9737.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perry ST, Prestwood TR, Lada SM, Benedict CA, Shresta S. Cardif-mediated signaling controls the initial innate response to dengue virus in vivo. J Virol. 2009;83:8276–8281. doi: 10.1128/JVI.00365-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prestwood TR, Prigozhin DM, Sharar KL, Zellweger RM, Shresta S. A mouse-passaged dengue virus strain with reduced affinity for heparan sulfate causes severe disease in mice by establishing increased systemic viral loads. J Virol. 2008;82:8411–8421. doi: 10.1128/JVI.00611-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diamond MS, Edgil D, Roberts TG, Lu B, Harris E. Infection of human cells by dengue virus is modulated by different cell types and viral strains. J Virol. 2000;74:7814–7823. doi: 10.1128/jvi.74.17.7814-7823.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alexander J, Oseroff C, Sidney J, Wentworth P, Keogh E, Hermanson G, Chisari FV, Kubo RT, Grey HM, Sette A. Derivation of HLA-A11/Kb transgenic mice: functional CTL repertoire and recognition of human A11-restricted CTL epitopes. J Immunol. 1997;159:4753–4761. [PubMed] [Google Scholar]
- 32.Alexander J, Oseroff C, Sidney J, Sette A. Derivation of HLA-B*0702 transgenic mice: functional CTL repertoire and recognition of human B*0702-restricted CTL epitopes. Hum Immunol. 2003;64:211–223. doi: 10.1016/s0198-8859(02)00786-3. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Q, Wang P, Kim Y, Haste-Andersen P, Beaver J, Bourne PE, Bui HH, Buus S, Frankild S, Greenbaum J, Lund O, Lundegaard C, Nielsen M, Ponomarenko J, Sette A, Zhu Z, Peters B. Immune epitope database analysis resource (IEDB-AR) Nucleic Acids Res. 2008;36:W513–518. doi: 10.1093/nar/gkn254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang P, Sidney J, Dow C, Mothe B, Sette A, Peters B. A systematic assessment of MHC class II peptide binding predictions and evaluation of a consensus approach. PLoS Comput Biol. 2008;4:e1000048. doi: 10.1371/journal.pcbi.1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sidney J, Assarsson E, Moore C, Ngo S, Pinilla C, Sette A, Peters B. Quantitative peptide binding motifs for 19 human and mouse MHC class I molecules derived using positional scanning combinatorial peptide libraries. Immunome Res. 2008;4:2. doi: 10.1186/1745-7580-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sidney J, Southwood S, Oseroff C, del Guercio MF, Sette A, Grey HM. Measurement of MHC/peptide interactions by gel filtration. Curr Protoc Immunol. 2001;Chapter 18(Unit 18):13. doi: 10.1002/0471142735.im1803s31. [DOI] [PubMed] [Google Scholar]
- 37.Shimizu Y, DeMars R. Production of human cells expressing individual transferred HLA-A,-B,-C genes using an HLA-A,-B,-C null human cell line. J Immunol. 1989;142:3320–3328. [PubMed] [Google Scholar]
- 38.Kanakaratne N, Wahala WM, Messer WB, Tissera HA, Shahani A, Abeysinghe N, de-Silva AM, Gunasekera M. Severe dengue epidemics in Sri Lanka, 2003–2006. Emerging infectious diseases. 2009;15:192–199. doi: 10.3201/eid1502.080926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kraus AA, Messer W, Haymore LB, de Silva AM. Comparison of plaque- and flow cytometry-based methods for measuring dengue virus neutralization. J Clin Microbiol. 2007;45:3777–3780. doi: 10.1128/JCM.00827-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moutaftsi M, Peters B, Pasquetto V, Tscharke DC, Sidney J, Bui HH, Grey H, Sette A. A consensus epitope prediction approach identifies the breadth of murine T(CD8+)-cell responses to vaccinia virus. Nat Biotechnol. 2006;24:817–819. doi: 10.1038/nbt1215. [DOI] [PubMed] [Google Scholar]
- 41.Roehrig JT, Johnson AJ, Hunt AR, Beaty BJ, Mathews JH. Enhancement of the antibody response to flavivirus B-cell epitopes by using homologous or heterologous T-cell epitopes. J Virol. 1992;66:3385–3390. doi: 10.1128/jvi.66.6.3385-3390.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Greenbaum J, Sidney J, Chung J, Brander C, Peters B, Sette A. Functional classification of class II human leukocyte antigen (HLA) molecules reveals seven different supertypes and a surprising degree of repertoire sharing across supertypes. Immunogenetics. 2011;63:325–335. doi: 10.1007/s00251-011-0513-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vita R, Zarebski L, Greenbaum JA, Emami H, Hoof I, Salimi N, Damle R, Sette A, Peters B. The immune epitope database 2.0. Nucleic acids research. 2010;38:D854–862. doi: 10.1093/nar/gkp1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yauch LE, Shresta S. Mouse models of dengue virus infection and disease. Antiviral Res. 2008;80:87–93. doi: 10.1016/j.antiviral.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gianfrani C, Oseroff C, Sidney J, Chesnut RW, Sette A. Human memory CTL response specific for influenza A virus is broad and multispecific. Hum Immunol. 2000;61:438–452. doi: 10.1016/s0198-8859(00)00105-1. [DOI] [PubMed] [Google Scholar]
- 46.Wentworth PA, Vitiello A, Sidney J, Keogh E, Chesnut RW, Grey H, Sette A. Differences and similarities in the A2.1-restricted cytotoxic T cell repertoire in humans and human leukocyte antigen-transgenic mice. Eur J Immunol. 1996;26:97–101. doi: 10.1002/eji.1830260115. [DOI] [PubMed] [Google Scholar]
- 47.Shirai M, Arichi T, Nishioka M, Nomura T, Ikeda K, Kawanishi K, Engelhard VH, Feinstone SM, Berzofsky JA. CTL responses of HLA-A2.1-transgenic mice specific for hepatitis C viral peptides predict epitopes for CTL of humans carrying HLA-A2.1. J Immunol. 1995;154:2733–2742. [PubMed] [Google Scholar]
- 48.Ressing ME, Sette A, Brandt RM, Ruppert J, Wentworth PA, Hartman M, Oseroff C, Grey HM, Melief CJ, Kast WM. Human CTL epitopes encoded by human papillomavirus type 16 E6 and E7 identified through in vivo and in vitro immunogenicity studies of HLA-A*0201-binding peptides. J Immunol. 1995;154:5934–5943. [PubMed] [Google Scholar]
- 49.Vitiello A, Marchesini D, Furze J, Sherman LA, Chesnut RW. Analysis of the HLA-restricted influenza-specific cytotoxic T lymphocyte response in transgenic mice carrying a chimeric human-mouse class I major histocompatibility complex. J Exp Med. 1991;173:1007–1015. doi: 10.1084/jem.173.4.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Diamond DJ, York J, Sun JY, Wright CL, Forman SJ. Development of a candidate HLA A*0201 restricted peptide-based vaccine against human cytomegalovirus infection. Blood. 1997;90:1751–1767. [PubMed] [Google Scholar]
- 51.Firat H, Garcia-Pons F, Tourdot S, Pascolo S, Scardino A, Garcia Z, Michel ML, Jack RW, Jung G, Kosmatopoulos K, Mateo L, Suhrbier A, Lemonnier FA, Langlade-Demoyen P. H-2 class I knockout, HLA-A2.1-transgenic mice: a versatile animal model for preclinical evaluation of antitumor immunotherapeutic strategies. Eur J Immunol. 1999;29:3112–3121. doi: 10.1002/(SICI)1521-4141(199910)29:10<3112::AID-IMMU3112>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 52.Le AX, Bernhard EJ, Holterman MJ, Strub S, Parham P, Lacy E, Engelhard VH. Cytotoxic T cell responses in HLA-A2.1 transgenic mice. Recognition of HLA alloantigens and utilization of HLA-A2.1 as a restriction element. J Immunol. 1989;142:1366–1371. [PubMed] [Google Scholar]
- 53.Man S, Newberg MH, Crotzer VL, Luckey CJ, Williams NS, Chen Y, Huczko EL, Ridge JP, Engelhard VH. Definition of a human T cell epitope from influenza A non-structural protein 1 using HLA-A2.1 transgenic mice. Int Immunol. 1995;7:597–605. doi: 10.1093/intimm/7.4.597. [DOI] [PubMed] [Google Scholar]
- 54.Rothman AL. Immunology and immunopathogenesis of dengue disease. Adv Virus Res. 2003;60:397–419. doi: 10.1016/s0065-3527(03)60010-2. [DOI] [PubMed] [Google Scholar]
- 55.Duangchinda T, Dejnirattisai W, Vasanawathana S, Limpitikul W, Tangthawornchaikul N, Malasit P, Mongkolsapaya J, Screaton G. Immunodominant T-cell responses to dengue virus NS3 are associated with DHF. Proc Natl Acad Sci U S A. 2010;107:16922–16927. doi: 10.1073/pnas.1010867107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rothman AL. Dengue: defining protective versus pathologic immunity. J Clin Invest. 2004;113:946–951. doi: 10.1172/JCI21512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Twiddy SS, Farrar JJ, Vinh Chau N, Wills B, Gould EA, Gritsun T, Lloyd G, Holmes EC. Phylogenetic relationships and differential selection pressures among genotypes of dengue-2 virus. Virology. 2002;298:63–72. doi: 10.1006/viro.2002.1447. [DOI] [PubMed] [Google Scholar]
- 58.Holmes EC, Burch SS. The causes and consequences of genetic variation in dengue virus. Trends Microbiol. 2000;8:74–77. doi: 10.1016/s0966-842x(99)01669-8. [DOI] [PubMed] [Google Scholar]
- 59.Yachi PP, Ampudia J, Zal T, Gascoigne NR. Altered peptide ligands induce delayed CD8-T cell receptor interaction--a role for CD8 in distinguishing antigen quality. Immunity. 2006;25:203–211. doi: 10.1016/j.immuni.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 60.Kast WM, Brandt RM, Sidney J, Drijfhout JW, Kubo RT, Grey HM, Melief CJ, Sette A. Role of HLA-A motifs in identification of potential CTL epitopes in human papillomavirus type 16 E6 and E7 proteins. J Immunol. 1994;152:3904–3912. [PubMed] [Google Scholar]
- 61.Valdar WS. Scoring residue conservation. Proteins. 2002;48:227–241. doi: 10.1002/prot.10146. [DOI] [PubMed] [Google Scholar]
- 62.Khan AM, Miotto O, Heiny AT, Salmon J, Srinivasan KN, Nascimento EJ, Marques ET, Jr, Brusic V, Tan TW, August JT. A systematic bioinformatics approach for selection of epitope-based vaccine targets. Cell Immunol. 2006;244:141–147. doi: 10.1016/j.cellimm.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Klein J. Natural History of the Major Histocompatibility Complex. Wiley; New York: 1986. [Google Scholar]
- 64.Hughes AL, Nei M. Maintenance of MHC polymorphism. Nature. 1992;355:402–403. doi: 10.1038/355402b0. [DOI] [PubMed] [Google Scholar]
- 65.Sette A, Sidney J. Nine major HLA class I supertypes account for the vast preponderance of HLA-A and -B polymorphism. Immunogenetics. 1999;50:201–212. doi: 10.1007/s002510050594. [DOI] [PubMed] [Google Scholar]
- 66.Doolan DL, Southwood S, Chesnut R, Appella E, Gomez E, Richards A, Higashimoto YI, Maewal A, Sidney J, Gramzinski RA, Mason C, Koech D, Hoffman SL, Sette A. HLA-DR-promiscuous T cell epitopes from Plasmodium falciparum pre-erythrocytic-stage antigens restricted by multiple HLA class II alleles. J Immunol. 2000;165:1123–1137. doi: 10.4049/jimmunol.165.2.1123. [DOI] [PubMed] [Google Scholar]
- 67.Wilson CC, Palmer B, Southwood S, Sidney J, Higashimoto Y, Appella E, Chesnut R, Sette A, Livingston BD. Identification and antigenicity of broadly cross-reactive and conserved human immunodeficiency virus type 1-derived helper T-lymphocyte epitopes. J Virol. 2001;75:4195–4207. doi: 10.1128/JVI.75.9.4195-4207.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Southwood S, Sidney J, Kondo A, del Guercio MF, Appella E, Hoffman S, Kubo RT, Chesnut RW, Grey HM, Sette A. Several common HLA-DR types share largely overlapping peptide binding repertoires. J Immunol. 1998;160:3363–3373. [PubMed] [Google Scholar]
- 69.Momburg F, Roelse J, Howard JC, Butcher GW, Hammerling GJ, Neefjes JJ. Selectivity of MHC-encoded peptide transporters from human, mouse and rat. Nature. 1994;367:648–651. doi: 10.1038/367648a0. [DOI] [PubMed] [Google Scholar]
- 70.Heemels MT, Ploegh HL. Substrate specificity of allelic variants of the TAP peptide transporter. Immunity. 1994;1:775–784. doi: 10.1016/s1074-7613(94)80019-7. [DOI] [PubMed] [Google Scholar]
- 71.Kubo RT, Sette A, Grey HM, Appella E, Sakaguchi K, Zhu NZ, Arnott D, Sherman N, Shabanowitz J, Michel H, et al. Definition of specific peptide motifs for four major HLA-A alleles. Journal of immunology. 1994;152:3913–3924. [PubMed] [Google Scholar]
- 72.Kiepiela P, Leslie AJ, Honeyborne I, Ramduth D, Thobakgale C, Chetty S, Rathnavalu P, Moore C, Pfafferott KJ, Hilton L, Zimbwa P, Moore S, Allen T, Brander C, Addo MM, Altfeld M, James I, Mallal S, Bunce M, Barber LD, Szinger J, Day C, Klenerman P, Mullins J, Korber B, Coovadia HM, Walker BD, Goulder PJ. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature. 2004;432:769–775. doi: 10.1038/nature03113. [DOI] [PubMed] [Google Scholar]
- 73.Bihl F, Frahm N, Di Giammarino L, Sidney J, John M, Yusim K, Woodberry T, Sango K, Hewitt HS, Henry L, Linde CH, Chisholm JV, 3rd, Zaman TM, Pae E, Mallal S, Walker BD, Sette A, Korber BT, Heckerman D, Brander C. Impact of HLA-B alleles, epitope binding affinity, functional avidity, and viral coinfection on the immunodominance of virus-specific CTL responses. J Immunol. 2006;176:4094–4101. doi: 10.4049/jimmunol.176.7.4094. [DOI] [PubMed] [Google Scholar]
- 74.Boon AC, De Mutsert G, Fouchier RA, Sintnicolaas K, Osterhaus AD, Rimmelzwaan GF. Preferential HLA usage in the influenza virus-specific CTL response. J Immunol. 2004;172:4435–4443. doi: 10.4049/jimmunol.172.7.4435. [DOI] [PubMed] [Google Scholar]
- 75.Lacey SF, Villacres MC, La Rosa C, Wang Z, Longmate J, Martinez J, Brewer JC, Mekhoubad S, Maas R, Leedom JM, Forman SJ, Zaia JA, Diamond DJ. Relative dominance of HLA-B*07 restricted CD8+ T-lymphocyte immune responses to human cytomegalovirus pp65 in persons sharing HLA-A*02 and HLA-B*07 alleles. Hum Immunol. 2003;64:440–452. doi: 10.1016/s0198-8859(03)00028-4. [DOI] [PubMed] [Google Scholar]
- 76.Lewinsohn DA, Winata E, Swarbrick GM, Tanner KE, Cook MS, Null MD, Cansler ME, Sette A, Sidney J, Lewinsohn DM. Immunodominant tuberculosis CD8 antigens preferentially restricted by HLA-B. PLoS Pathog. 2007;3:1240–1249. doi: 10.1371/journal.ppat.0030127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Axelsson-Robertson R, Weichold F, Sizemore D, Wulf M, Skeiky YA, Sadoff J, Maeurer MJ. Extensive major histocompatibility complex class I binding promiscuity for Mycobacterium tuberculosis TB10.4 peptides and immune dominance of human leucocyte antigen (HLA)-B*0702 and HLA-B*0801 alleles in TB10.4 CD8 T-cell responses. Immunology. 2010;129:496–505. doi: 10.1111/j.1365-2567.2009.03201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zivna I, Green S, Vaughn DW, Kalayanarooj S, Stephens HA, Chandanayingyong D, Nisalak A, Ennis FA, Rothman AL. T cell responses to an HLA-B*07-restricted epitope on the dengue NS3 protein correlate with disease severity. J Immunol. 2002;168:5959–5965. doi: 10.4049/jimmunol.168.11.5959. [DOI] [PubMed] [Google Scholar]
- 79.Stephens HA, Klaythong R, Sirikong M, Vaughn DW, Green S, Kalayanarooj S, Endy TP, Libraty DH, Nisalak A, Innis BL, Rothman AL, Ennis FA, Chandanayingyong D. HLA-A and -B allele associations with secondary dengue virus infections correlate with disease severity and the infecting viral serotype in ethnic Thais. Tissue Antigens. 2002;60:309–318. doi: 10.1034/j.1399-0039.2002.600405.x. [DOI] [PubMed] [Google Scholar]
- 80.Appanna R, Ponnampalavanar S, Lum Chai See L, Sekaran SD. Susceptible and protective HLA class 1 alleles against dengue fever and dengue hemorrhagic fever patients in a Malaysian population. PLoS One. 2010;5 doi: 10.1371/journal.pone.0013029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Simmons CP, Dong T, Chau NV, Dung NT, Chau TN, Thao le TT, Hien TT, Rowland-Jones S, Farrar J. Early T-cell responses to dengue virus epitopes in Vietnamese adults with secondary dengue virus infections. J Virol. 2005;79:5665–5675. doi: 10.1128/JVI.79.9.5665-5675.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Appanna R, Huat TL, See LL, Tan PL, Vadivelu J, Devi S. Cross-reactive T-cell responses to the nonstructural regions of dengue viruses among dengue fever and dengue hemorrhagic fever patients in Malaysia. Clin Vaccine Immunol. 2007;14:969–977. doi: 10.1128/CVI.00069-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mongkolsapaya J, Duangchinda T, Dejnirattisai W, Vasanawathana S, Avirutnan P, Jairungsri A, Khemnu N, Tangthawornchaikul N, Chotiyarnwong P, Sae-Jang K, Koch M, Jones Y, McMichael A, Xu X, Malasit P, Screaton G. T cell responses in dengue hemorrhagic fever: are cross-reactive T cells suboptimal? J Immunol. 2006;176:3821–3829. doi: 10.4049/jimmunol.176.6.3821. [DOI] [PubMed] [Google Scholar]
- 84.Wen JS, Jiang LF, Zhou JM, Yan HJ, Fang DY. Computational prediction and identification of dengue virus-specific CD4(+) T-cell epitopes. Virus Res. 2008;132:42–48. doi: 10.1016/j.virusres.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]