Abstract
Global burdens from existing or emerging infectious diseases emphasize the need for point-of-care (POC) diagnostics to enhance timely recognition and intervention. Molecular approaches based on PCR methods have made significant inroads by improving detection time and accuracy but are still largely hampered by resource-intensive processing in centralized laboratories, thereby precluding their routine bedside- or field-use. Microfluidic technologies have enabled miniaturization of PCR processes onto a chip device with potential benefits including speed, cost, portability, throughput, and automation. In this review, we provide an overview of recent advances in microfluidic PCR technologies and discuss practical issues and perspectives related to implementing them into infectious disease diagnostics.
Keywords: PCR, microfluidic, diagnostic methods, infectious disease, point-of-care
1. Introduction
POC testing is defined as analytical testing performed outside the central laboratory using devices that can be easily transported to the vicinity of the patient (College-of-American-Pathologists, 2001). The value of near-patient testing for routine infectious disease diagnosis is well recognized given that real-time test results can direct timely therapeutic interventions and improve patients’ clinical outcomes. With the increasing threat of accelerated epidemic-to-pandemic transitions of new or reemerging infectious disease outbreaks owing to globalization, decentralizing diagnostic testing closer to frontline clinical settings could facilitate earlier implementations of public health responses to contain and mitigate such events (Nichols, 2007, Rajan and Glorikian, 2009). In the developing countries where high infectious disease burden is compounded by diagnostic challenges due to poor clinical laboratory infrastructure and cost constraints, the potential utility for POC testing is even greater (Urdea et al. , 2006, Yager et al. , 2008, Yager et al. , 2006). In fact, among the ‘Grand Challenges for Global Health’ identified by the Bill and Melinda Gates Foundation and the NIH, one of the major priorities involves developing POC technologies for diagnosing infectious diseases (Mabey et al. , 2004).
Culture remains the mainstay of microbiological diagnosis, and enables antimicrobial susceptibilities to be determined; however, it is time-consuming, expensive, and poorly sensitive in cases of fastidious organisms or prior antibiotics exposure. Moreover, culture requires well-maintained laboratory-based equipment, a constant supply of reagents and electricity, and adequately trained and supervised technologists. Meanwhile, molecular approaches to amplify microbial nucleic acids have clear theoretical advantages over culture methods in terms of detection accuracy and turnaround time (Cuchacovich, 2006, Yang and Rothman, 2004). Among the various nucleic acid amplification methods, polymerase chain reaction (PCR) has been the most mature and popular due to its simplicity (Fredricks and Relman, 1999, Yang and Rothman, 2004). Variations of the method have been modified to expand its utility and versatility. Multiplex PCR enables simultaneous detection of several target sequences by incorporation of multiple sets of primers. Nested PCR, a double amplification step with appropriately designed primers, can increase sensitivity and specificity (Fredricks and Relman, 1999). Reverse transcript PCR (RT-PCR) enables the detection of RNA, which is converted into a complementary DNA copy and amplified (Erlich et al. , 1991). Real-time PCR allows simultaneous amplification and product detection without additional post-PCR processing. Digital PCR enables direct quantification of nucleic acids through parallel single-molecule amplification (Vogelstein and Kinzler, 1999). With the increasing number of genomes of infectious pathogens being sequenced, catalogues of genes can be exploited to serve as amplification targets fundamental to the design of clinically useful diagnostic tests. As a result, the number of PCR assays developed to identify either specific pathogen or classes of pathogens by amplifying unique or conserved sequences, respectively, continues to expand. Widespread clinical applications, particularly in detecting difficult-to-culture, limb- or life-threatening, biothreat or emerging infectious pathogens, have demonstrated significant performance and cost advantages over traditional methods (Rothman et al. , 2010, Yang et al. , 2008a, Yang and Rothman, 2004, Yang et al. , 2008b). While commercial development of PCR-based diagnostics has progressed significantly in recent years, all FDA-cleared PCR test kits to-date are still categorized as high or moderate complexity under Clinical Laboratory Improvement Amendments (CLIA) (Holland and Kiechle, 2005). Inefficient nucleic acid preparation from complex sample types (e.g. whole blood, stool, etc.) requires highly skilled personnel to manually perform multiple processing steps in a dedicated laboratory space, batched testing with next-day reporting of results, and costly reagents and instrumentation. These features are some of the major hurdles which still preclude PCR-based assays from being classified as a “simple test” based on FDA recommendations for POC use (Table 1) (Holland and Kiechle, 2005).
Table 1.
FDA guidance for simple test
| Is a fully automated instrument or a unitized or self-contained test. |
| Uses direct unprocessed specimens, such as capillary blood (fingerstick), venous whole blood, nasal swabs, throat swabs, or urine. |
| Needs only basic, non-technique-dependent specimen manipulation, including any for decontamination. |
| Needs only basic, non-technique-dependent reagent manipulation, such as “mix reagent A and reagent B.” |
| Needs no operator intervention during the analysis steps. |
| Needs no technical or specialized training with respect to troubleshooting or interpretation of multiple or complex error codes. |
| Needs no electronic or mechanical maintenance beyond simple tasks, e.g., changing a battery or power cord. |
| Produces results that require no operator calibration, interpretation, or calculation. |
| Produces results that are easy to determine, such as ‘positive’ or ‘negative,’ a direct readout of numerical values, the clear presence or absence of a line, or obvious color gradations. |
| Provides instructions in the package insert for obtaining and shipping specimens for confirmation testing in cases where such testing is clinically advisable. |
| Has test performance comparable to a traceable reference method as demonstrated by studies in which intended operators perform the test. If a reference method is not available for a test you are proposing for waiver, please contact OIVD to discuss your proposed plan prior to submitting your application. |
| Contains a quick reference instruction sheet that is written at no higher than a 7th grade reading level. |
| Sample manipulation should NOT be required to perform the assay. (For example, tests that use plasma or serum are not considered simple.) Sample manipulation includes processes such as centrifugation, complex mixing steps, or evaluation of the sample by the operator for conditions such as hemolysis or lipemia. |
| Measurement of an analyte should NOT be affected by conditions such as sample turbidity or cell lysis. |
Recommendations: Clinical Laboratory Improvement Amendments of 1988 (CLIA) Waiver Applications for Manufacturers of In Vitro Diagnostic Devices (http://www.fda.gov/downloads/MedicalDevices/DeviceRegulationandGuidance/GuidanceDocuments/ucm070890.pdf)
The emerging field of microfluidics, in combination with micro-electro-mechanical systems (MEMS) technology, to create so called lab-on-a-chip (LOC) or micro total analysis system (μTAS), promises exciting solutions to realize PCR-based, POC diagnostics to the POC. Microfluidics involves the behavior, precise control and manipulation of fluids in the micro-scale environment. Bioanalytical systems based on microfluidics allow miniaturization and integration of processes that were previously done at larger scale in separate operations with enhanced speed and efficiency (Whitesides, 2006). By exploiting the strengths of microfluidics, miniaturization of PCR devices can be achieved with significant advantages, including shorter assay time, lower reagent consumption, rapid heating/cooling, portability, as well as the possibility of integrating multiple pre- and post-PCR processing modules in a self-contained system with full automation. This unique set of capabilities has the potential to bring about decentralization of PCR-based diagnostic testing for use in resource-limited settings.
In this review, we concisely discuss the latest advancement in microfluidic PCR technologies for infectious disease diagnostics. We first review the current status of on-chip PCR technologies. Key aspects of the recent advancements toward a fully integrated POC platform based on microfluidic technologies are discussed, including pre-PCR sample preparation, post-PCR amplicon analysis, system integration/automation, adaptation for use in developing countries, as well as current challenges and future perspectives in this field. It is beyond our scope to provide an exhaustive review of all the related technologies in this domain. Instead, our goal is to present a snapshot of the promising technical advances with a focus on practical concerns appropriate for infectious disease applications.
2. On-chip PCR
PCR is an enzyme-driven process for amplifying short regions of DNA in vitro. It can create millions of DNA copies by cycling between different temperatures to allow repeating steps (denaturation, annealing and elongation) of DNA replication to take place. Despite the simplicity and amplification power of PCR chemistry, limitations in its supporting hardware still hinder PCR from reaching its full potential. In particular, improvements in thermal cycling speed, instrument size, and reaction volume are still much needed. The bulky instrumentation and large reaction volume required in conventional bench-top thermal cyclers lead to large thermal mass which reduces the temperature transition speed and reaction efficiency. These shortcomings can all be directly addressed through miniaturization of the PCR device. Rapid thermal cycling can be achieved through rapid heat transfer in microfluidic-based PCR due to the small reaction mass and the high surface to volume ratio of the small reactor. The small length scale can also lead to a more uniform temperature distribution and enhance the yield and integrity of PCR.
2.1 Device materials and fabrication
Miniaturizing the PCR device onto a single-use, self-contained, disposable microchip, which can reduce cross-contamination and biohazard risks, would be most practical for infectious disease testing. To that end, cost and performance are important considerations when selecting substrate materials to create the chip device. Silicon and glass substrates are popular with well-established microfabrication methods commonly used in the microelectronics fields. However, expensive material and fabrication costs associated with these substrates preclude them from disposable use. Recently, polymeric materials such as polydimethylsiloxane (PDMS) (Thorsen et al. , 2002, Unger et al. , 2000), polymethylmethacrylate (PMMA) (Hataoka et al. , 2004, Lee et al. , 2004), and polycarbonate (PC) (Hashimoto et al. , 2004, Liu et al. , 2002) have become more popular owing primarily to their lower material and fabrication costs. In particular, PDMS has emerged as the most promising substrate given its inert and non-toxic properties, improved optical transparency, high thermal stability, and PCR compatibility (Melin and Quake, 2007, Thorsen, Maerkl, 2002, Whitesides, 2006). Soft lithography, which refers to a replica molding process on micro-fabricated master templates, provides a simple, low cost, and rapid prototyping of micron scale fluidic circuits on elastomeric polymers such as PDMS, and thus has become one of the most popular approaches to fabricate microfluidic PCR system (Xia and Whitesides, 1998). Other thermoplastics including PMMA and PC can be fabricated either in a batch process by using hot embossing and injection molding techniques, or in a sequential process by using laser ablation. Many substrates bonding technologies such as adhesive, thermal, and plasma treatment bonding are available for making enclosed fluidic environments (Dittrich et al. , 2006, Sun and Kwok, 2006).
2.2 Chip designs
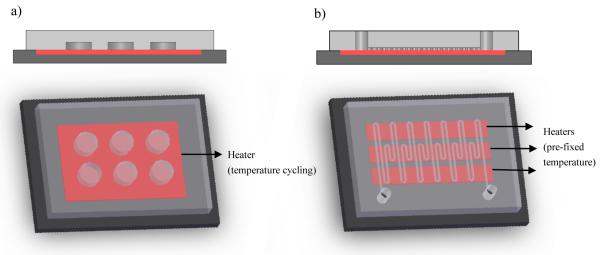
Two distinct design approaches, namely the stationary reaction chamber system and the continuous flow system, have been adopted for on-chip PCR devices to reduce the reaction volume. The stationary reaction chamber system works in the same manner as conventional PCR devices in which the PCR mixture is kept stationary inside the reaction chambers while the thermal cycling is done by alternating the temperatures of the heating units (Figure 1a). Since the first PCR chip based on this format was introduced by Northrup et al (1993) (Northrup, 1993), substantial improvements have been made, including implementing a multi-chamber design to increase throughput. Stationary systems generally allow a small reaction volume and simple system configuration. Detection of micro-organisms in pL scale chamber volumes with chip configurations of up to 1176 parallel reaction chambers have been reported (Marcus et al. , 2006b, Ottesen et al. , 2006, Pal et al. , 2005). Precise sample handling and processing, in addition to ensuring temperature uniformity between chambers, in the setting of increasing numbers of them still pose challenges. Further miniaturization of the reaction volume may also lead to nonspecific absorption of PCR samples on the walls of the chamber due to increased surface-to-volume ratio.
Figure 1.
Schematic illustrations of types of microfluidic PCR chip designs. a) stationary chamber system b) continuous flow system
A continuous flow system transports the PCR mixture through different pre-fixed temperature zones for PCR thermal reactions (Figure 1b). Compared to a stationary chamber based format, the continuous system approach provides faster thermal cycling in general because thermal inertia depends only on the thermal mass of the sample, rather than the chip. In this system, the required heating and cooling sequence and the residence time are controlled by channel routing and flow speed (Kopp et al. , 1998). Multiple closed-loop microfluidic channels through different temperature zones can be effectively used for continuous cycling of small volumes of reaction mixtures while reducing cross-contamination risks (Jian et al. , 2002). Higher fabrication cost and fixed cycle number dictated by the channel layout are some of the drawbacks as compared to the stationary chamber system. Nonetheless, the dynamic nature of fluidic transport in this format may facilitate integration with other functional components towards μTAS.
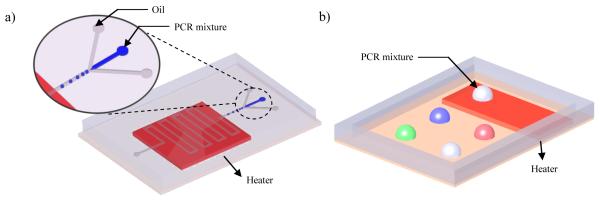
Recently, microfluidic droplet technology dramatically decreased the thermal inertia of the microfluidic PCR system. Droplet emulsion PCR system, which utilizes discrete aqueous droplets dispersed in a continuous oil phase as illustrated in Figure 2a, enables high throughput testing at the single copy level with automated generation and control of multiple droplets, providing a digital PCR platform. Many microfluidic technologies are available for integrating the required microfluidic functions on a single chip, such as rapid generation of water-in-oil droplets by using electric field, flow focusing, and T-junction methods (Huebner et al. , 2008, Rane et al. , 2010, Teh et al. , 2008), and fusion or sorting of multiple droplets by applying localized electric field, droplet surface modification, and variation of channel geometries (Agresti et al. , 2010, Chabert and Viovy, 2008, Mazutis et al. , 2009b). An integrated platform that can detect single copy RNA and virion through RT-PCR in a pL droplet system has been reported (Beer et al. , 2007, Beer et al. , 2008). High throughput amplification and quantification in millions of droplets have been demonstrated for large scale genomic sequencing (Kiss et al. , 2008, Mazutis et al. , 2009a, Tewhey et al. , 2009) and digital detection of pathogenic E. coli O157 cells in a high background of normal K12 cells (Zeng et al. , 2010). Despite high throughput testing capability of emulsion PCR system with reduced risk of cross-contamination by compartmentalizing samples and reagents into pL or nL size droplets, the development of a compact and portable system for POC testing is still limited because external instruments and tubings are required for continuous actuation of fluid and droplet manipulation.
Figure 2.
Schematic illustrations of droplet based microfluidic PCR chips. a) Droplet emulsion system b) Open surface droplet based system
Another major form of droplet microfluidic device manipulates droplets in open space (Figure 2b). Free aqueous droplets containing PCR mixtures on an open surface platform provide simple stationary reaction vessels (Juergen et al. , 2008, Pipper et al. , 2007, Zhang et al. , 2009b, Zhang et al. , 2011, Zhang et al. , 2010). The reaction droplet encapsulated by immiscible oil is insensitive to vapor generation during thermal cycling, thus avoiding the problem of micro bubble formation and expansion often found in conventional microfluidic PCR chambers of fixed volume. Moreover, given that each discrete droplet can function both as a reaction chamber and a fluid transportation unit (Guttenberg, Muller, 2005, Hsieh, Zhang, 2006, Juergen, Yi, 2008, Pipper, Inoue, 2007), no additional microfluidic components are required and the fluid handling is relatively simple. As a result, the open surface droplet platform is particularly advantageous to POC applications because the valve- and pump-less droplet manipulation enable easy implementation of upstream sample preparation into a fully functional μTAS without added complexity such as bulky external fluid couplings.
2.3 Thermal management
As the temperature ramping rate of the PCR system limits the speed of the reaction, a compact and efficient heater design with reduced thermal mass is essential to achieve fast target amplification. Various on-chip heaters have demonstrated improved speed of temperature cycling as compared to that of conventional bench top systems. In general, on-chip heaters can be categorized into contact and non-contact types. Many contact type heaters such as thin films, metal heating blocks, and Peltier units are commercially available at low cost and are thus widely adopted in on-chip PCR systems, while the temperature cycling speed can be limited by the thermal loss due to external coupling (Zhang and Xing, 2007). Despite the complexity and cost of fabrication, micro-fabricated thin film heaters are also widely used as a contact type method to achieve high speed reactions as fast as 6 minutes for 40 thermal cycles at the heating speed of 175°C/sec and cooling speed of −125°C/sec (Neuzil et al. , 2006). On the other hand, non-contact methods are more favorable for simple chip designs as they provide more flexibility for system integration. Several researchers have successfully integrated IR-mediated (Huhmer and Landers, 2000, Legendre et al. , 2006) or laser assisted heater units (Slyadnev et al. , 2001) with amplicon detection components. Recently, investigators used microwaves to achieve local heating with a 15 msec response time in conjunction with a droplet based microfluidic system (Issadore et al. , 2009). Although its reported heating speed is lower than that of common contact methods, the benefit from the simplified device configuration and the capability of selective heating of aqueous phase of droplets while maintaining surrounding oil streams at room temperature more than offset this deficiency.
3. Post PCR amplicon analysis
As part of the effort to miniaturize PCR-based bioassays for POC testing, developing chip-scale amplicon detection units capable of automated operations with the reduced risk of cross-contamination have become one of the major challenges towards realizing a fully integrated system. In conventional bench top PCR assays, amplicons are either collected or analyzed off-line in a separate analysis device (e.g., gel-electrophoresis, capillary electrophoresis), or monitored directly in the same reaction chambers using an on-line method (e.g. fluorescent probe or intercalating dye for real-time detection). Although on-chip PCR products can also be detected off-line using the same methods which require transferring of samples, on-line detection would be more ideal in improving throughput and reducing risk of contamination. To this end, much effort has been made to miniaturize and integrate conventional techniques as well as explore alternative approaches capable of highly sensitive detection within the appropriate volume range of on-chip PCR (down to subnanoliter level) and without the need for any off-chip apparatus such as external illumination sources or imaging devices. In this section, we discuss several relatively mature on-chip detection techniques for microfluidic PCR amplicon analysis.
3.1 Electrophoresis
With the aid of microfabrication technology, existing detection assays have been miniaturized into chip-scale devices. Among them, capillary electrophoresis (CE) and capillary gel electrophoresis (CGE) in microfluidic channels are the most common on-chip detection methods (Burns et al. , 1998, Govind et al. , 2006, Lagally et al. , 2001, Lagally et al. , 2004). In these electrophoretic separation methods, PCR amplicons are first labeled with intercalating dyes before they are loaded into a microfluidic channel. Upon the application of an electric field, amplicons migrate along the microfluidic channel filled with gel matrix. DNA molecules of different sizes migrate at different speeds and are subsequently separated. Advantages of electrophoretic-based detection include simple, fast analysis and high amplicon separation efficiency. Easley et al. reported an integrated on-chip PCR coupled CE which successfully detected Bacillus anthracis, Bordetella pertussis, and Salmonella typhimurium in as early as 12 minutes (Easley et al. , 2006b). A similar approach has been applied for detection of BK virus in unprocessed urine samples from renal transplant patients with limit of detection of 1-2 viral copies (Govind, Ryan, 2006). Given its size-based approach for amplicon analysis, one of the major limitations of electrophoretic separation is the inability to differentiate amplicons of similar size but different sequence.
3.2 DNA hybridization microarray
The DNA hybridization microarray is a powerful amplicon sequence analysis method. Advances in microfabrication technology has made it possible to immobilize large numbers of oligonucleotide probes, each having complementary sequences against a specific target pathogen’s DNA/RNA, all on a predefined, array-patterned microchip. Fluorescent patterns generated through hybridization with labeled PCR amplicons can be analyzed to extract sequence information. By taking advantage of microfluidic technology, PCR has been successfully coupled with microarrays on the same platform. For example, various combined PCR-microarray devices have been developed for genotyping HIV (Anderson et al. , 2000), Chinese medicinal plants (Trau et al. , 2002), and point mutations in human genomic DNA (Hashimoto et al. , 2006, Hashimoto et al. , 2005). While providing high throughput analysis of multiple amplicon sequences, microarray technology is limited by the prolonged assay time for hybridization reaction and the need for sensitive fluorescent pattern scanning and analysis methods.
3.3 Fluorescence based real-time detection
Real-time PCR combines amplification with detection in synchrony without post-PCR processing. Using fluorescently labeled probes or intercalating dyes, increasing fluorescent signals generated from PCR amplification can be measured during the assay run and greatly reduce overall assay time. The number of cycles required to reach a detectable threshold (Ct) of PCR products can be used to further assess PCR efficiency and determine the initial template concentration or pathogen load. To ensure accurate acquisition and analysis of the emitted fluorescent signal after each PCR cycle, the choice of excitation light source and optical detection apparatus requires much attention. Unfortunately, conventional bench-top reading instruments are limited to complex laser optics and imaging components, such as charge-coupled devices (CCD), with large footprints. To develop a POC device requires use of miniaturized and low cost external light sources and photo-detecting units. Light emitting diode (LED) and photo-diode (Cady et al. , 2005) are inexpensive alternatives although the illumination power, bandwidth, and the detection sensitivity are limited compared to the conventional laser optic system. Various microfluidic-based real-time PCRs using either SYBR green, ethidium bromide dye, or TaqMan probes for specific pathogen detection have been reported (Belgrader et al. , 1998, Lee et al. , 2006, Northrup et al. , 1998, Pipper, Inoue, 2007).
3.4 Electrochemical method
Despite the use of relatively miniaturized external fluorescence detectors such as LEDs and photodiodes for on-line amplicon detection, a fully integrated detection system microfabricated as part of the chip device would be most ideal. Electrochemical based detection offers a promising alternative with inherent features including high sensitivity, adjustable selectivity, low power, low cost, independence from external optical components, and compatibility with microfabrication technology. Electrochemical methods require sensor electrodes, which are often functionalized with probes or other chemicals, to generate electrical signals which can be correlated with the concentration of target amplicons. In most cases, the detected electrical signals such as voltage, current, and impedance need to be amplified or processed. Few researchers have reported electrochemical detection of PCR amplicons. An integrated chip device that can detect E. coli from whole blood samples based on DNA hybridization and electrochemical sensing has been developed with a reported limit of detection of 103 cells/mL (Liu et al. , 2004). Alternatively, Yeung et al. demonstrated an integrated system combining sample preparation, DNA amplification and subsequent multiplexed electrochemical detection of E. coli and B. subtilis in a single silicon-glass microchamber (Yeung et al. , 2006a). They further advanced the technology to develop the first electrochemical real-time PCR (ERT-PCR) (Yeung et al. , 2006b, 2007). Conductivity-based electrical DNA detection using functionalized gold nanoparticles has also reported significant improvement in detection limit down to as low as 500 femtomolar of target DNA, which is comparable to the sensitivity of an optical fluorescence detection system (Park et al. , 2002).
4. Pre-PCR sample preparation
Direct pre-processing of crude biological samples with efficient purification and extraction of target analytes is a key prerequisite of a POC device with sample-in answer-out capability. In general, sample preparation is a multi-stage process including sample collection, cell separation and concentration, cell lysis and nucleic acids extraction. Depending on the sample type, the complexity of the sample preparation process may differ significantly. In samples with ample quantities of target organisms in relatively inert sample matrices, one can lyse the target cells and directly analyze the nucleic acids from the crude samples (Govind, Ryan, 2006, Legendre, Bienvenue, 2006). In the majority of cases, however, tests are utilized to establish early diagnosis by detecting pathogens which may be present in extremely low quantities. Large sample volumes are often required to ensure sufficient capture of pathogens for detection. As a result, most samples need a concentration step to increase the pathogen load. Samples with a complex matrix, such as whole blood, which contain complex constituents, including large amount of blood cells and PCR inhibitory anticoagulant additives, may require more than centrifugation to concentrate target pathogens (Lim et al. , 2005, Toner and Irimia, 2005). In addition, the large human genomic content from the blood cells contributes to the PCR background and can significantly compromise the amplification efficiency of the target sequence (Al-Soud and Radstrom, 2001). Depending on the target organism, the specific blood fractions in which the microbe preferentially resides may also differ. For example, malaria diagnosis relies on the separation of parasite infected RBCs from uninfected cells. Thus, prior separation of the target pathogens from background cells is essential for the accurate and sensitive PCR detection downstream. Moreover, effective lysis of the target organisms, which may have variable protective outer layers, to release their genetic contents for analysis also plays an important role in sample preparation. In the following sections, we shall discuss in detail how microfluidic technology contributes to these key aspects of sample preparation.
4.1 Cell separation and concentration
As mentioned earlier, a typical microfluidic device handles a liquid volume in the range of nanoliter to microliter. In contrast, the sample volume required for infectious disease detection ranges from microliter to milliliter depending on the disease and type of sample under investigation. The volume mismatch may raise the issue of stochastic sampling resulting in false negative results (Mariella, 2008, Rådström et al. , 2004). Therefore sample pre-concentration as well as additional cell separation steps to exclude unwanted cell types and matrix constituents are often required to enhance the accuracy of detection. Disparities in the intrinsic properties of the different cell populations can be exploited to achieve cell separation. Physical and mechanical properties (e.g. size, density, shape, deformity) are popular parameters for differentiation. Conventional target cell concentration is done by pelleting the cells using centrifugation. Although some microfluidic platforms utilize centrifugal force, it is mainly used as means for fluidic transport instead (Gorkin et al. , 2010, Mark et al. , 2010). Filtration is one of the common concentration techniques implemented in microfluidic devices. Obstacle structures created inside microfluidic channels serve as selective filters based on cellular size and rigidity have been shown to separate white blood cells from other blood cells (Lee and Tai, 1999, Panaro et al. , 2005, Yuen et al. , 2001). In combination with hydrodynamic flow and variations in channel geometry, bacterial cells and human blood cells can be effectively separated (Wu et al. , 2009). Flow field fractionation is another common technique for continuous-flow sample concentration. In this method, cells are first confined within a thin flow stream with the application of a lateral force field, and the narrow cell stream is then directed to a split outlet, resulting in a reduction of fluid volume and sample concentration (Laurell et al. , 2007, Peter and Jody, 2002). Acoustic standing waves have been demonstrated as an efficient force field for cell separation and concentration based on size (Evander et al. , 2007, Petersson et al. , 2007). Additional force fields applied to enhance separation based on polarizability and magnetic characteristics of different cells are gaining popularity owing to their high sensitivity and efficiency. Dielectrophoresis (DEP) has been used to discriminate bacterial species and cell viability based on their distinctive dielectric properties (Bhattacharya et al. , 2008, Cheng et al. , 1998, Cho et al. , 2010, Lapizco-Encinas et al. , 2004, Park and Beskok, 2008, Park et al. , 2009a). External magnetic fields have also been applied to separate a heterogeneous population of cells through a process called magnetophoresis (Blakemore et al. , 1979, Pamme and Wilhelm, 2006). Alternatively, using antibodies conjugated on a solid substrate, such as magnetic particles, cell sorting and concentration can also be achieved through immunoaffinity mechanisms (Cho et al. , 2007, Juergen, Yi, 2008, Kang-Yi et al. , 2008, Kell et al. , 2008, Shih et al. , 2008).
4.2 Cell lysis
Effective cell lysis to liberate microbial genetic content for downstream PCR detection can improve the overall sensitivity of the assay. Many microfluidic devices have adopted various traditional methods of cell lysis, which may be mechanical, chemical/enzymatic, thermal or electrical (Belgrader et al. , 1999, Marentis et al. , 2005). Minisonication has been shown to induce chaotropic disruption of bacterial spores despite their rigid coats. Knife-like nanostructures fabricated inside the microfluidic channels can create shearing effects on cell membranes (Di Carlo et al. , 2003). A microfluidic counterpart of ball milling has been used to lyse cells by packing the cells and beads together into a microfluidic CD (Kim et al. , 2004). The addition of chemicals (e.g. chaotropic salt, detergent, alcohols) to induce osmotic pressure, or enzymes (e.g. lysozyme, lysostaphin, protease) for cell wall/membrane digestion, can be incorporated in the cell medium as further augmenting measures (Irimia et al. , 2004, Schilling et al. , 2002, Sethu et al. , 2004). Recently, a laser-irradiated magnetic bead heating system has been proposed as a convenient and efficient thermal cell lysis platform (Cho, Kim, 2010, Cho, Lee, 2007, Lee, Cheong, 2006). Electroporation of cell membranes can be induced via a high-intensity pulsed electric field or high frequency AC current (Lee and Tai, 1999). To achieve the optimal result, various combinations of the aforementioned cell lysis methods may need to be adjusted based on the microbial pathogen of interest and the strength of its outer protective layer (Mahalanabis et al. , 2009, Stachowiak et al. , 2007).
4.3 Nucleic acids extraction
The last step before performing PCR is to extract and purify the nucleic acids from cell lysates. This process usually involves multiple manual operations with specific instruments, especially when the number of target copies is low, or the sample is in the form of complex mixtures. The cell lysates may contain PCR-inhibitory materials that limit the efficiency of the amplification process or even cause complete failure of the amplification. Some of the common PCR inhibitors in blood include heme, leukocyte DNA, immunoglobulin G or anticoagulant additives such as EDTA and heparin (Al-Soud and Radstrom, 2001). Although much of the PCR inhibitors are removed during the earlier cell separation and concentration stages, residual can remain without a proper nucleic acids purification step. Various on-chip nucleic acid purification methods have been developed, such as electrophoretic (Vulto et al. , 2009) and hybridization based purification in which poly(dT) conjugated beads are used to capture target mRNA (Hong et al. , 2004, Marcus et al. , 2006a). Nonetheless, silica based solid phase extraction (SPE) is the mainstream approach. The mechanism of the silica based SPE is believed to be the combined effects of dehydrating nucleic acids molecules, hydrogen bonding and electrostatic interactions (Melzak et al. , 1996). The purification process starts with binding of nucleic acids to the silica surface in high-ionic-strength chaotropic salt, followed by several washing steps to remove debris before eluting in low ionic strength buffers. When implementing SPE in microfluidic devices, the solid substrate is usually in the form of micropillars/microposts (Cady et al. , 2003, West et al. , 2007) or immobilized silica beads/particles (Breadmore et al. , 2003, Gijs, 2004, Juergen, Yi, 2008, Pipper, Inoue, 2007). Several microfluidic SPE devices have been reported to have high extraction efficiencies of up to 80% with small elution volumes in the microliter range, however, issues with sample volume capacity and flexibility, as well as process automation with minimal contamination, still require substantial improvement (Gijs et al. , 2010, Wen et al. , 2008).
5. System integration and automation
A unique advantage of microfluidic PCR is the feasibility of integrating pre-PCR sample preparation with post-PCR analysis into a streamlined and automated system for POC applications. System integration and automation are critical for avoiding manual operation errors, minimizing cross-contamination risks, and reducing sample loss caused by multiple sample transfers between instruments (Mariella, 2008). However, to develop a fully integrated μTAS capable of completing an assay from sample-in to answer-out in clinical settings remains challenging mainly because operation of these platforms still requires complex and bulky peripherals. Large gas tanks and syringe pumps are often used as the pressure source for fluid actuation. Fluid control requires incorporation of micro-pumps, micro-valves, and micro-mixers with extensive external tubings. Bulky electrical instruments and microscopy are often required for signal detection and imaging. All these accessories are not portable. Therefore, current platforms are often considered as chip-in-lab rather than lab-on-chip. A truly POC platform should have compact actuating and controlling modules with universal interfaces for easy operation and high portability. In the past decade, great effort has been put into creating a μTAS by integrating a PCR module with the aforementioned pre- and post-PCR modules. PCR based whole blood sample preparation and amplification platforms that utilize micro-filter separation (Munchow et al. , 2005, Panaro, Lou, 2005, Wilding et al. , 1998, Yuen, Kricka, 2001), embedded SPE (Legendre, Bienvenue, 2006), and immuno-magnetic beads (Easley et al. , 2006a, Juergen, Yi, 2008, Kang-Yi, Chien-Ju, 2008, Liu, Yang, 2004, Pipper, Inoue, 2007) have been reported. Other types of crude samples including oral fluid (Legendre, Bienvenue, 2006, Pipper, Inoue, 2007), nasal aspirate (Easley, Karlinsey, 2006a, Legendre, Bienvenue, 2006), urine (Govind, Ryan, 2006), semen (Legendre, Bienvenue, 2006), and serum (Anderson, Su, 2000, Lee et al. , 2009) have also been reported. Table 2 summarizes examples of such on-chip biological sample preparation devices integrated with downstream analysis modules. Many of these devices utilize magnetic beads based nucleic acid purification and transportation mainly because the nucleic acids can be precisely handled via manipulating the magnetic bead with syringe pumps and magnets.
Table 2.
Microfluidic PCR devices with on-chip crude sample processing capability
| Ref. | Sample Type | Sample Volume |
Detection Target | Sample Pre-processing |
On-chip Detection |
|---|---|---|---|---|---|
| (Panaro, Lou, 2005, Wilding, Kricka, 1998, Yuen, Kricka, 2001) |
Whole blood | < 3.5 μl | WBC | Filtration (micropillar) |
N/A |
| (Liu, Yang, 2004) | Whole blood | 1 ml | E. coli | SPE (bead) |
Electrochemical |
| (Juergen, Yi, 2008, Pipper, Inoue, 2007) |
- Throat swab - Whole blood |
25 μl | - influenza virus H5N1 - THP-1 cells |
SPE (bead) |
Real-time (Taqman) |
| (Easley, Karlinsey, 2006a) | - Whole blood - Nasal aspirate |
1 μl | - Bacillus anthracis spores - Bordetella pertussis |
SPE (bead) |
Electrophoresis |
| (Munchow, Dadic, 2005) | Whole blood | 8 μl | WBC | Filtration (fiber matrix) |
N/A |
| (Legendre, Bienvenue, 2006) | - Whole blood -Nasal swab - Semen |
4~200 μl | - Human gene - Anthrax spores |
SPE (packed bed) |
N/A |
| (Kang-Yi, Chien-Ju, 2008, Lee, Lien, 2009) | - Whole blood - Serum |
100~ 200 μl |
- leucocytes - Dengue virus |
SPE (bead) |
N/A |
| (Govind, Ryan, 2006) | Urine | 0.24 μl | BK virus | N/A | Electrophoresis |
| (Anderson, Su, 2000) | Serum | 100 μl | HIV RNA | SPE (Cellulose) |
Hybridization microarray |
Recently, open surface droplet based microfluidic devices have attracted increasing attention due to their benefits for system integration with pre- and post-PCR modules (Fan et al. , 2009, Guttenberg et al. , 2005, Hsieh et al. , 2006, Juergen, Yi, 2008, Lehmann et al. , 2006, Ohashi et al. , 2007, Pipper, Inoue, 2007, Shastry et al. , 2005, Teh, Lin, 2008, Velev et al. , 2003, Zhang et al. , 2009a, Zhang, Bailey, 2009b, Zhang, Park, 2011, Zhang, Park, 2010). The on-chip actuation methods of free droplets, either active actuation such as surface acoustic wave (Guttenberg, Muller, 2005), electrowetting (Fan, Hsieh, 2009), dielectrophoresis (Fan, Hsieh, 2009, Velev, Prevo, 2003), magnetic force (Hsieh, Zhang, 2006, Juergen, Yi, 2008, Lehmann, Vandevyver, 2006, Ohashi, Kuyama, 2007, Pipper, Inoue, 2007, Zhang, Park, 2011) or passive actuation (Shastry, Case, 2005, Zhang, Cheng, 2009a) are being extensively investigated. Recent optoelectronic approaches demonstrated functional flexibility for electrowetting or dielectrophoresis based droplet manipulation by employing a light-activated reconfigurable electrode (Chiou et al. , 2003, Park et al. , 2009b, Valley et al. , 2011). The magnet-actuated droplet is especially preferred as the entire operation can be performed through a single magnet, enabling valveless and pumpless POC sample preparation and analysis with reduced cost and complexity. In addition, the magnetic particles used for droplet actuation are often functionalized to serve as carriers for biomolecules, such as silica superparamagnetic particles (SSP) for nucleic acid binding and transfer. This approach has been applied for DNA preparation module for whole blood samples (Sista et al. , 2008) and a fully integrated droplet based system for detecting H5N1 avian flu virus from crude throat swab sample (Pipper, Inoue, 2007). Advances in an open surface droplet based system have enabled automated droplet control with relatively simple device configurations, but large-scale integration for high throughput applications with such devices is still limited (Mohamed and Aaron, 2009).
6. Design constraints in developing countries
Although use of POC devices is feasible in a number of settings (e.g. physician’s office, emergency first responder, home, etc.) with different design constraints, for maximum range of use, the ideal POC device should be designed for remote testing in developing countries. The outskirts of the medical care system typically lack laboratory infrastructure. Electrical power, clean water, and cold storage are intermittently present or absent. Given such physical limitations, devices should ideally be battery- or solar-powered, and not rely on external water if a high quality supply is needed. Also, reagents must be able to withstand large fluctuations in temperatures. The lack of trained personnel requires devices to be simple to use with easy-to-interpret results. Self-calibration with controls along with test samples should be incorporated into the design. Turnaround time ranging from a few minutes up to 1 h would be ideal to implement treatment during the same health-care encounter given lack of resources to ensure patient follow up. To ensure affordability of the end product, single-use, disposable microfluidic chips composed of inexpensive polymer materials through advanced fabrication methods would reduce the cost of production. The small quantities of sample and reagents consumed should be retained within the disposable to avoid contamination with the external instrument and minimize spread of biohazards. Based upon the mathematical modeling analysis of disease impact as reported by Global Health Diagnostic Forum, the sensitivity and specificity requirements for diagnostic tests should range from 85%-95% and 76%-97% respectively depending on various clinical diagnostic considerations (Urdea, Penny, 2006). The WHO has developed a list of general characteristics that make a diagnostic test appropriate for resource-limited sites, which are abbreviated using the acronym ASSURED (Table 3) (Mabey, Peeling, 2004). Striving for the most accurate test, however, should not prevent the development of the most useful test. A test that is less sensitive but rapid may result in more infected people receiving treatment, as not all patients return for the results of tests. So, the best test is not always the most useful test; the context of the end-user’s setting needs to be considered.
Table 3.
Characteristics of the ideal diagnostic test – ASSURED (Mabey, Peeling, 2004)
| Affordable by those at risk of infection |
| Sensitive (few false-negatives) |
| Specific (few false-positives) |
| User-friendly (simple to perform and requiring minimal training) |
| Rapid (to enable treatment at first visit) and Robust (does not require refrigerated strorage) |
| Equipment-free |
| Delivered to those who need it |
7. Current challenges and future perspectives
7.1 Multiplex detection
From the clinical perspective, the same clinical symptom can be caused by infections from many etiologic agents; therefore, a POC test which can simultaneously screen/detect multiple pathogens from a single specimen would be highly desirable. Test design strategies include comprehensive identification of all clinically-relevant pathogens, or a flexible platform that detects a panel of suspected pathogens which can be readily changed based on symptomatology and local epidemiology. Assay approaches that split the initial specimen for parallel simplex PCRs, each with a single primer set specific for a pathogen, are often not possible due to limited quantity of target DNA. Alternatively, multiplex PCR using multiple primer sets in the same reaction can decrease specimen and reagent consumption but is limited in detection sensitivity due to uneven amplification efficiencies of the different primer sets (Elnifro et al. , 2000). PCR using a single primer set targeting phylogenetically conserved sequences (e.g. 16S rRNA) can allow broad-range pathogen detection without compromising sensitivity (Yang et al. , 2002, Yang and Rothman, 2004). Moreover, broad-range PCR can also serve as a “molecular petri dish” to identify unsuspected, mutated, or emerging infectious causes of diseases. It is also ideal for use to determine the presence of any infection in an otherwise sterile specimen. RT-PCR amplification of 16S rRNA has also been found to positively correlate with microbial viability, which may provide indications of active infection and prove useful in assessing efficacy of antimicrobial treatment (Keer and Birch, 2003). Sequence analysis of the amplified product from broad-range PCR using differentially labeled fluorogenic probes has been incorporated into real-time PCR assays to identify a small panel of pathogens. Oligonucleotide arrays can greatly expand the number of pathogens identifiable and the integration of PCR with microarray on a microfluidic platform has been developed (Liu, Yang, 2004). Nonetheless, the principle shortcoming of all probe-based approaches to amplicon analysis is still limited to a pre-defined set of pathogens identifiable. Direct sequencing of amplicons would allow identification of emerging or unsuspected pathogens, and a microfluidic array device integrated with an on-chip sequencing assay has recently been developed to identify mutant strains of influenza virus (Liu et al. , 2006). Alternatively, high-resolution melt analysis (HRMA), which detects sequence variations within the amplicons based on their melting curve profiles, offers a simple, low-cost, PCR-compatible solution to broad-scale pathogen identification (Yang et al. , 2009).
7.2 Multiple analytes analysis
Assisting clinicians in the differential diagnosis of diseases that produce common symptoms, identifying multiple classes of microbes (e.g. virus, bacteria, fungi, etc.) which may be involved, determining disease severity and predicting clinical outcomes based on host response, a having the capacity to analyze multiple samples and biomarker types are all attributes of an ideal assay. For example, acute lower respiratory infection with complex etiologies may require testing multiple sample types, including sputum, nasopharyngeal aspirate, and blood, to ensure adequate detection of all relevant respiratory pathogens while determining if infection has progressed into the bloodstream. Detection of inflammatory or pathogen biomarkers derived from various types of analytes (e.g. DNA, RNA, proteins, toxin, etc.) would require integration of different assay formats, including PCR, RT-PCR, and immunoassays, onto the same platform. While conserved or specific DNA targets can be detected in conventional PCR, RT-PCR allows detection of RNA viruses as well as microbial rRNA/mRNA targets to assess microbial viability; however, given the highly labile nature of RNA molecules with short half-life, additional measures must be taken in sample preparation and reverse transcription of RNA to avoid degradation (Keer and Birch, 2003). Many microfludic-based devices have been developed for testing various analyte types individually (Chen et al. , 2007, Roper et al. , 2005), but combined testing on a single platform has yet to be achieved. On the other hand, a droplet-based microfluidic device has been reported to simultaneously analyze multiple sample types using the electrowetting effect for fluid actuation (Srinivasan et al. , 2004). Future research in this direction can significantly increase the utility of new tests.
7.3 Antibiotic susceptibility
A POC diagnostic for infectious pathogens will not supplant traditional culture-based methods if the former cannot rapidly characterize the antimicrobial susceptibility profile of the detected pathogen to help direct early therapy. Traditional laboratory approaches for susceptibility testing, which rely mostly on phenotypic assays utilizing in vitro growth inhibition of a microorganism in the presence of antibiotics, are slow and resource demanding. Although genotypic-based (e.g. PCR) detection of specific antimicrobial resistance genes may offer a more rapid alternative, the genetic mechanisms for resistance are diverse and the presence of genetic marker may not always confer resistance. Therefore, antimicrobial susceptibility will continue to be more accurately determined by phenotypic methods.
Microfluidics is an attractive approach for susceptibility testing by providing a miniaturized fluidic environment suitable for monitoring cell growth in the presence of antibiotics (Balaban et al. , 2004, Orit and Nathalie, 2009). High surface-to-volume ratio in microfluidic channels has been demonstrated to enhance bacterial oxygenation and reproduction, reducing susceptibility testing time to 2 hours as compared to days using traditional methods (Chen et al. , 2010). Droplet-based microfluidic platforms can potentially allow single cell testing with multiple antibiotics (Boedicker et al. , 2008). Monitoring physiologic stress responses, instead of cell count, through measurable changes in specific cellular properties (e.g. morphology, membrane potential, dielectric properties, metabolites, etc.) may offer more rapid alternative approaches for assessing drug susceptibility (Kohanski et al. , 2010, Mann and Mikkelsen, 2008).
8. Conclusions
With the pressing need for developing new technologies toward near-patient testing for infectious diseases, microfluidic PCR is well-positioned to contribute to this challenge by leveraging its inherent advantages and recent advances in many aspects of the field, including device material, microfabrication, thermal management, amplicon detection and sample preparation. A paradigm switch from component and device based research to a system and product oriented approach is currently underway in order to face the next challenge: translating technologies into reliable and cost-effective clinical diagnostics with flexible capabilities adaptable to the context of end users. Advanced design considerations based on clinical context may incorporateetiologic pathogens suspected, patient symptomatologies, resource limitations, and treatment decisions. The new challenge will necessitate new ways of thinking in order to realize the full potential of microfluidic PCR technologies.
Acknowledgments
The authors would like to thank The Hartwell Foundation and NIAID-MARCE (grant U54-AI057168-07) for their generous support of this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agresti JJ, Antipov E, Abate AR, Ahn K, Rowat AC, Baret JC, et al. Ultrahigh-throughput screening in drop-based microfluidics for directed evolution. Proc Natl Acad Sci U S A. 2010;107:4004–9. doi: 10.1073/pnas.0910781107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Soud WA, Radstrom P. Purification and Characterization of PCR-Inhibitory Components in Blood Cells. J Clin Microbiol. 2001;39:485–93. doi: 10.1128/JCM.39.2.485-493.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson RC, Su X, Bogdan GJ, Fenton J. A miniature integrated device for automated multistep genetic assays. Nucleic Acids Res. 2000;28:e60. doi: 10.1093/nar/28.12.e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial Persistence as a Phenotypic Switch. Science. 2004;305:1622–5. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- Beer NR, Hindson BJ, Wheeler EK, Sara B, Rose KA, Kennedy IM, et al. On-chip, real-time, single-copy polymerase chain reaction in picoliter droplets. Anal Chem. 2007;79:8471–5. doi: 10.1021/ac701809w. [DOI] [PubMed] [Google Scholar]
- Beer NR, Wheeler EK, Lee-Houghton L, Watkins N, Nasarabadi S, Hebert N, et al. On-chip single-copy real-time reverse-transcription PCR in isolated picoliter droplets. Anal Chem. 2008;80:1854–8. doi: 10.1021/ac800048k. [DOI] [PubMed] [Google Scholar]
- Belgrader P, Benett W, Hadley D, Long G, Mariella R, Jr., Milanovich F, et al. Rapid pathogen detection using a microchip PCR array instrument. Clin Chem. 1998;44:2191–4. [PubMed] [Google Scholar]
- Belgrader P, Hansford D, Kovacs GTA, Venkateswaran K, Mariella R, Milanovich F, et al. A minisonicator to rapidly disrupt bacterial spores for DNA analysis. Anal Chem. 1999;71:4232–6. doi: 10.1021/ac990347o. [DOI] [PubMed] [Google Scholar]
- Bhattacharya S, Salamat S, Morisette D, Banada P, Akin D, Liu Y-S, et al. PCR-based detection in a microfabricated platform. Lab Chip. 2008;8:1130–6. doi: 10.1039/b802227e. [DOI] [PubMed] [Google Scholar]
- Blakemore RP, Maratea D, Wolfe RS. ISOLATION AND PURE CULTURE OF A FRESHWATER MAGNETIC SPIRILLUM IN CHEMICALLY DEFINED MEDIUM. J Bacteriol. 1979;140:720–9. doi: 10.1128/jb.140.2.720-729.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boedicker JQ, Li L, Kline TR, Ismagilov RF. Detecting bacteria and determining their susceptibility to antibiotics by stochastic confinement in nanoliter droplets using plug-based microfluidics. Lab Chip. 2008;8:1265–72. doi: 10.1039/b804911d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breadmore MC, Wolfe KA, Arcibal IG, Leung WK, Dickson D, Giordano BC, et al. Microchip-based purification of DNA from biological samples. Anal Chem. 2003;75:1880–6. doi: 10.1021/ac0204855. [DOI] [PubMed] [Google Scholar]
- Burns MA, Johnson BN, Brahmasandra SN, Handique K, Webster JR, Krishnan M, et al. An Integrated Nanoliter DNA Analysis Device. Science. 1998;282:484–7. doi: 10.1126/science.282.5388.484. [DOI] [PubMed] [Google Scholar]
- Cady NC, Stelick S, Batt CA. Nucleic acid purification using microfabricated silicon structures. Biosens Bioelectron. 2003;19:59–66. doi: 10.1016/s0956-5663(03)00123-4. [DOI] [PubMed] [Google Scholar]
- Cady NC, Stelick S, Kunnavakkam MV, Batt CA. Real-time PCR detection of Listeria monocytogenes using an integrated microfluidics platform. Sensors Actuators B. 2005;107:332–41. [Google Scholar]
- Chabert M, Viovy JL. Microfluidic high-throughput encapsulation and hydrodynamic self-sorting of single cells. Proc Natl Acad Sci U S A. 2008;105:3191–6. doi: 10.1073/pnas.0708321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CH, Lu Y, Sin MLY, Mach KE, Zhang DD, Gau V, et al. Antimicrobial Susceptibility Testing Using High Surface-to-Volume Ratio Microchannels. Anal Chem. 2010;82:1012–9. doi: 10.1021/ac9022764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Manz A, Day PJR. Total nucleic acid analysis integrated on microfluidic devices. Lab Chip. 2007;7:1413–23. doi: 10.1039/b708362a. [DOI] [PubMed] [Google Scholar]
- Cheng J, Sheldon EL, Wu L, Uribe A, Gerrue LO, Carrino J, et al. Preparation and hybridization analysis of DNA/RNA from E. coli on microfabricated bioelectronic chips. Nat Biotechnol. 1998;16:541–6. doi: 10.1038/nbt0698-541. [DOI] [PubMed] [Google Scholar]
- Chiou PY, Moon H, Toshiyoshi H, Kim CJ, Wu MC. Light actuation of liquid by optoelectrowetting. Sensors Actuators A. 2003;104:222–8. [Google Scholar]
- Cho YK, Kim TH, Lee JG. On-chip concentration of bacteria using a 3D dielectrophoretic chip and subsequent laser-based DNA extraction in the same chip. J Micromech Microeng. 2010;20 065010. [Google Scholar]
- Cho YK, Lee JG, Park JM, Lee BS, Lee Y, Ko C. One-step pathogen specific DNA extraction from whole blood on a centrifugal microfluidic device. Lab Chip. 2007;7:565–73. doi: 10.1039/b616115d. [DOI] [PubMed] [Google Scholar]
- College-of-American-Pathologists . Point-of-care testing. College of American Pathologists; Northfield, IL: 2001. section 30. [Google Scholar]
- Cuchacovich R. Clinical Applications of the Polymerase Chain Reaction: An Update. Infect Dis Clin North Am. 2006;20:735–58. doi: 10.1016/j.idc.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Di Carlo D, Jeong KH, Lee LP. Reagentless mechanical cell lysis by nanoscale barbs in microchannels for sample preparation. Lab Chip. 2003;3:287–91. doi: 10.1039/b305162e. [DOI] [PubMed] [Google Scholar]
- Dittrich PS, Tachikawa K, Manz A. Micro total analysis systems. Latest advancements and trends. Anal Chem. 2006;78:3887–908. doi: 10.1021/ac0605602. [DOI] [PubMed] [Google Scholar]
- Easley CJ, Karlinsey JM, Bienvenue JM, Legendre LA, Roper MG, Feldman SH, et al. A fully integrated microfluidic genetic analysis system with sample-in-answer-out capability. Proc Natl Acad Sci U S A. 2006a;103:19272–7. doi: 10.1073/pnas.0604663103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easley CJ, Karlinsey JM, Landers JP. On-chip pressure injection for integration of infrared-mediated DNA amplification with electrophoretic separation. Lab Chip. 2006b;6:601–10. doi: 10.1039/b600039h. [DOI] [PubMed] [Google Scholar]
- Elnifro EM, Ashshi AM, Cooper RJ, Klapper PE. Multiplex PCR: Optimization and Application in Diagnostic Virology. Clin Microbiol Rev. 2000;13:559–70. doi: 10.1128/cmr.13.4.559-570.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlich HA, Gelfand D, Sninsky JJ. Recent advances in the polymerase chain reaction. Science. 1991;252:1643–51. doi: 10.1126/science.2047872. [DOI] [PubMed] [Google Scholar]
- Evander M, Johansson L, Lilliehorn T, Piskur J, Lindvall M, Johansson S, et al. Noninvasive acoustic cell trapping in a microfluidic perfusion system for online bioassays. Anal Chem. 2007;79:2984–91. doi: 10.1021/ac061576v. [DOI] [PubMed] [Google Scholar]
- Fan SK, Hsieh TH, Lin DY. General digital microfluidic platform manipulating dielectric and conductive droplets by dielectrophoresis and electrowetting. Lab Chip. 2009;9:1236–42. doi: 10.1039/b816535a. [DOI] [PubMed] [Google Scholar]
- Fredricks DN, Relman DA. Application of Polymerase Chain Reaction to the Diagnosis of Infectious Diseases. Clin Infect Dis. 1999;29:475–86. doi: 10.1086/598618. [DOI] [PubMed] [Google Scholar]
- Gijs MAM. Magnetic bead handling on-chip: new opportunities for analytical applications. Microfluid Nanofluid. 2004;1:22–40. [Google Scholar]
- Gijs MAM, Lacharme Fdr, Lehmann U. Microfluidic Applications of Magnetic Particles for Biological Analysis and Catalysis. Chem Rev. 2010;110:1518–63. doi: 10.1021/cr9001929. [DOI] [PubMed] [Google Scholar]
- Gorkin R, Park J, Siegrist J, Amasia M, Lee BS, Park J-M, et al. Centrifugal microfluidics for biomedical applications. Lab Chip. 2010;10:1758–73. doi: 10.1039/b924109d. [DOI] [PubMed] [Google Scholar]
- Govind VK, Ryan JH, Jutta P, Xiao-Li P, Linda MP, Christopher JB. Automated screening using microfluidic chip-based PCR and product detection to assess risk of BK virus-associated nephropathy in renal transplant recipients. Electrophoresis. 2006;27:3753–63. doi: 10.1002/elps.200600061. [DOI] [PubMed] [Google Scholar]
- Guttenberg Z, Muller H, Habermuller H, Geisbauer A, Pipper J, Felbel J, et al. Planar chip device for PCR and hybridization with surface acoustic wave pump. Lab Chip. 2005;5:308–17. doi: 10.1039/b412712a. [DOI] [PubMed] [Google Scholar]
- Hashimoto M, Barany F, Soper SA. Polymerase chain reaction/ligase detection reaction/hybridization assays using flow-through microfluidic devices for the detection of low-abundant DNA point mutations. Biosensors Bioelectron. 2006;21:1915–23. doi: 10.1016/j.bios.2006.01.014. [DOI] [PubMed] [Google Scholar]
- Hashimoto M, Chen P-C, Mitchell MW, Nikitopoulos DE, Soper SA, Murphy MC. Rapid PCR in a continuous flow device. Lab Chip. 2004;4:638–45. doi: 10.1039/b406860b. [DOI] [PubMed] [Google Scholar]
- Hashimoto M, Hupert ML, Murphy MC, Soper SA, Cheng Y-W, Barany F. Ligase Detection Reaction/Hybridization Assays Using Three-Dimensional Microfluidic Networks for the Detection of Low-Abundant DNA Point Mutations. Anal Chem. 2005;77:3243–55. doi: 10.1021/ac048184d. [DOI] [PubMed] [Google Scholar]
- Hataoka Y, Zhang L, Mori Y, Tomita N, Notomi T, Baba Y. Analysis of Specific Gene by Integration of Isothermal Amplification and Electrophoresis on Poly(methyl methacrylate) Microchips. Anal Chem. 2004;76:3689–93. doi: 10.1021/ac035032u. [DOI] [PubMed] [Google Scholar]
- Holland CA, Kiechle FL. Point-of-care molecular diagnostic systems -- past, present and future. Curr Opin Microbiol. 2005;8:504–9. doi: 10.1016/j.mib.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Hong JW, Studer V, Hang G, Anderson WF, Quake SR. A nanoliter-scale nucleic acid processor with parallel architecture. Nat Biotechnol. 2004;22:435–9. doi: 10.1038/nbt951. [DOI] [PubMed] [Google Scholar]
- Hsieh T-M, Zhang Y, Pipper J, Neuzil P. PCR by moving a free droplet over different temperature zones. microTAS 2006 conference proceeding.2006. [Google Scholar]
- Huebner A, Sharma S, Srisa-Art M, Hollfelder F, Edel JB, deMello AJ. Microdroplets: A sea of applications? Lab Chip. 2008;8:1244–54. doi: 10.1039/b806405a. [DOI] [PubMed] [Google Scholar]
- Huhmer AFR, Landers JP. Noncontact Infrared-Mediated Thermocycling for Effective Polymerase Chain Reaction Amplification of DNA in Nanoliter Volumes. Anal Chem. 2000;72:5507–12. doi: 10.1021/ac000423j. [DOI] [PubMed] [Google Scholar]
- Irimia D, Tompkins RG, Toner M. Single-cell chemical lysis in picoliter-scale closed volumes using a microfabricated device. Anal Chem. 2004;76:6137–43. doi: 10.1021/ac0497508. [DOI] [PubMed] [Google Scholar]
- Issadore D, Humphry KJ, Brown KA, Sandberg L, Weitz DA, Westervelt RM. Microwave dielectric heating of drops in microfluidic devices. Lab Chip. 2009;9:1701–6. doi: 10.1039/b822357b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian L, Markus E, Stephen Q. A nanoliter rotary device for polymerase chain reaction. Electrophoresis. 2002;23:1531–6. doi: 10.1002/1522-2683(200205)23:10<1531::AID-ELPS1531>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Juergen P, Yi Z, Pavel N, Tseng-Ming H. Clockwork PCR Including Sample Preparation. Angew Chem. 2008;47:3900–4. doi: 10.1002/anie.200705016. [DOI] [PubMed] [Google Scholar]
- Kang-Yi L, Chien-Ju L, Gwo-Bin L. Magnetic-bead-based microfluidic systems for detection of genetic diseases. Micro Electro Mechanical Systems, 2008 MEMS 2008 IEEE 21st International Conference on2008.pp. 66–9. [Google Scholar]
- Keer JT, Birch L. Molecular methods for the assessment of bacterial viability. J Microbiol Methods. 2003;53:175–83. doi: 10.1016/s0167-7012(03)00025-3. [DOI] [PubMed] [Google Scholar]
- Kell AJ, Stewart G, Ryan S, Peytavi R, Boissinot M, Huletsky A, et al. Vancomycin-modified nanoparticles for efficient targeting and preconcentration of Gram-positive and Gram-negative bacteria. Acs Nano. 2008;2:1777–88. doi: 10.1021/nn700183g. [DOI] [PubMed] [Google Scholar]
- Kim J, Jang SH, Jia GY, Zoval JV, Da Silva NA, Madou MJ. Cell lysis on a microfluidic CD (compact disc) Lab Chip. 2004;4:516–22. doi: 10.1039/b401106f. [DOI] [PubMed] [Google Scholar]
- Kiss MM, Ortoleva-Donnelly L, Beer NR, Warner J, Bailey CG, Colston BW, et al. High-Throughput Quantitative Polymerase Chain Reaction in Picoliter Droplets. Anal Chem. 2008;80:8975–81. doi: 10.1021/ac801276c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohanski MA, Dwyer DJ, Collins JJ. How antibiotics kill bacteria: from targets to networks. Nat Rev Microbiol. 2010;8:423–35. doi: 10.1038/nrmicro2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp MU, Mello AJ, de, Manz A. Chemical Amplification: Continuous-Flow PCR on a Chip. Science. 1998;280:1046–8. doi: 10.1126/science.280.5366.1046. nbsp. [DOI] [PubMed] [Google Scholar]
- Lagally ET, Emrich CA, Mathies RA. Fully integrated PCR-capillary electrophoresis microsystem for DNA analysis. Lab Chip. 2001;1:102–7. doi: 10.1039/b109031n. [DOI] [PubMed] [Google Scholar]
- Lagally ET, Scherer JR, Blazej RG, Toriello NM, Diep BA, Ramchandani M, et al. Integrated portable genetic analysis microsystem for pathogen/infectious disease detection. Anal Chem. 2004;76:3162–70. doi: 10.1021/ac035310p. [DOI] [PubMed] [Google Scholar]
- Lapizco-Encinas BH, Simmons BA, Cummings EB, Fintschenko Y. Dielectrophoretic concentration and separation of live and dead bacteria in an array of insulators. Anal Chem. 2004;76:1571–9. doi: 10.1021/ac034804j. [DOI] [PubMed] [Google Scholar]
- Laurell T, Petersson F, Nilsson A. Chip integrated strategies for acoustic separation and manipulation of cells and particles. Chem Soc Rev. 2007;36:492–506. doi: 10.1039/b601326k. [DOI] [PubMed] [Google Scholar]
- Lee D-S, Park SH, Yang H, Chung K-H, Yoon TH, Kim S-J, et al. Bulk-micromachined submicroliter-volume PCR chip with very rapid thermal response and low power consumption. Lab Chip. 2004;4:401–7. doi: 10.1039/b313547k. [DOI] [PubMed] [Google Scholar]
- Lee J-G, Cheong KH, Huh N, Kim S, Choi J-W, Ko C. Microchip-based one step DNA extraction and real-time PCR in one chamber for rapid pathogen identification. Lab Chip. 2006;6:886–95. doi: 10.1039/b515876a. [DOI] [PubMed] [Google Scholar]
- Lee SW, Tai YC. A micro cell lysis device. Sensors Actuators A. 1999;73:74–9. [Google Scholar]
- Lee Y-F, Lien K-Y, Lei H-Y, Lee G-B. An integrated microfluidic system for rapid diagnosis of dengue virus infection. Biosensors Bioelectron. 2009;25:745–52. doi: 10.1016/j.bios.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre LA, Bienvenue JM, Roper MG, Ferrance JP, Landers JP. A Simple, Valveless Microfluidic Sample Preparation Device for Extraction and Amplification of DNA from Nanoliter-Volume Samples. Anal Chem. 2006;78:1444–51. doi: 10.1021/ac0516988. [DOI] [PubMed] [Google Scholar]
- Lehmann U, Vandevyver C, Parashar VK, Gijs MAM. Droplet-based DNA purification in a magnetic lab-on-a-chip. Angew Chem. 2006;45:3062–7. doi: 10.1002/anie.200503624. [DOI] [PubMed] [Google Scholar]
- Lim DV, Simpson JM, Kearns EA, Kramer MF. Current and Developing Technologies for Monitoring Agents of Bioterrorism and Biowarfare. Clin Microbiol Rev. 2005;18:583–607. doi: 10.1128/CMR.18.4.583-607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu RH, Lodes MJ, Nguyen T, Siuda T, Slota M, Fuji HS, et al. Validation of A Fully Integrated Microfluidic Array Device for Influenza A Subtype Identification and Sequencing. Anal Chem. 2006;78:4184–93. doi: 10.1021/ac060450v. [DOI] [PubMed] [Google Scholar]
- Liu RH, Yang J, Lenigk R, Bonanno J, Grodzinski P. Self-Contained, Fully Integrated Biochip for Sample Preparation, Polymerase Chain Reaction Amplification, and DNA Microarray Detection. Anal Chem. 2004;76:1824–31. doi: 10.1021/ac0353029. [DOI] [PubMed] [Google Scholar]
- Liu Y, Rauch CB, Stevens RL, Lenigk R, Yang J, Rhine DB, et al. DNA Amplification and Hybridization Assays in Integrated Plastic Monolithic Devices. Anal Chem. 2002;74:3063–70. doi: 10.1021/ac020094q. [DOI] [PubMed] [Google Scholar]
- Mabey D, Peeling RW, Ustianowski A, Perkins MD. Tropical infectious diseases: Diagnostics for the developing world. Nat Rev Microbiol. 2004;2:231–40. doi: 10.1038/nrmicro841. [DOI] [PubMed] [Google Scholar]
- Mahalanabis M, Al-Muayad H, Kulinski MD, Altman D, Klapperich CM. Cell lysis and DNA extraction of gram-positive and gram-negative bacteria from whole blood in a disposable microfluidic chip. Lab Chip. 2009;9:2811–7. doi: 10.1039/b905065p. [DOI] [PubMed] [Google Scholar]
- Mann TS, Mikkelsen SR. Antibiotic Susceptibility Testing at a Screen-Printed Carbon Electrode Array. Anal Chem. 2008;80:843–8. doi: 10.1021/ac701829c. [DOI] [PubMed] [Google Scholar]
- Marcus JS, Anderson WF, Quake SR. Microfluidic Single-Cell mRNA Isolation and Analysis. Anal Chem. 2006a;78:3084–9. doi: 10.1021/ac0519460. [DOI] [PubMed] [Google Scholar]
- Marcus JS, Anderson WF, Quake SR. Parallel Picoliter RT-PCR Assays Using Microfluidics. Anal Chem. 2006b;78:956–8. doi: 10.1021/ac0513865. [DOI] [PubMed] [Google Scholar]
- Marentis TC, Kusler B, Yaralioglu GG, Liu SJ, Haeggstrom EO, Khuri-Yakub BT. Microfluidic sonicator for real-time disruption of eukaryotic cells and bacterial spores for DNA analysis. Ultrasound Med Biol. 2005;31:1265–77. doi: 10.1016/j.ultrasmedbio.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Mariella R. Sample preparation: the weak link in microfluidics-based biodetection. Biomed Microdevices. 2008;10:777–84. doi: 10.1007/s10544-008-9190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark D, Haeberle S, Roth G, Stetten Fv, Zengerle R. Microfluidic lab-on-a-chip platforms: requirements, characteristics and applications. Chem Soc Rev. 2010;39:1153–82. doi: 10.1039/b820557b. [DOI] [PubMed] [Google Scholar]
- Mazutis L, Araghi AF, Miller OJ, Baret JC, Frenz L, Janoshazi A, et al. Droplet-based microfluidic systems for high-throughput single DNA molecule isothermal amplification and analysis. Anal Chem. 2009a;81:4813–21. doi: 10.1021/ac900403z. [DOI] [PubMed] [Google Scholar]
- Mazutis L, Baret JC, Griffiths AD. A fast and efficient microfluidic system for highly selective one-to-one droplet fusion. Lab Chip. 2009b;9:2665–72. doi: 10.1039/b903608c. [DOI] [PubMed] [Google Scholar]
- Melin J, Quake SR. Microfluidic Large-Scale Integration: The Evolution of Design Rules for Biological Automation. Annu Rev Biophys Biomol Struct. 2007;36:213–31. doi: 10.1146/annurev.biophys.36.040306.132646. [DOI] [PubMed] [Google Scholar]
- Melzak KA, Sherwood CS, Turner RFB, Haynes CA. Driving forces for DNA adsorption to silica in perchlorate solutions. J Colloid Interface Sci. 1996;181:635–44. [Google Scholar]
- Mohamed A, Aaron RW. The Digital Revolution: A New Paradigm for Microfluidics. Adv Mater. 2009;21:920–5. [Google Scholar]
- Munchow G, Dadic D, Doffing F, Hardt S, Drese K-S. Automated chip-based device for simple and fast nucleic acid amplification. Expert Rev Mol Diagn. 2005;5:613–20. doi: 10.1586/14737159.5.4.613. [DOI] [PubMed] [Google Scholar]
- Neuzil P, Zhang C, Pipper J, Oh S, Zhuo L. Ultra fast miniaturized real-time PCR: 40 cycles in less than six minutes. Nucleic Acids Res. 2006;34:e77. doi: 10.1093/nar/gkl416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols JH. Point of Care Testing. Clin Lab Med. 2007;27:893–908. doi: 10.1016/j.cll.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Northrup MA, Benett B, Hadley D, Landre P, Lehew S, Richards J, et al. A Miniature Analytical Instrument for Nucleic Acids Based on Micromachined Silicon Reaction Chambers. Anal Chem. 1998;70:918–22. doi: 10.1021/ac970486a. [DOI] [PubMed] [Google Scholar]
- Northrup MA, Ching MT, White RM, Watson RT. DNA amplification in a microfabricated reaction chamber. Proceedings of the 7th International Conference on Solid-State Sensors and Actuators (Transducers ‘93); Yokohama, Japan. 1993. pp. 924–6. [Google Scholar]
- Ohashi T, Kuyama H, Hanafusa N, Togawa Y. A simple device using magnetic transportation for droplet-based PCR. Biomed Microdevices. 2007;9:695–702. doi: 10.1007/s10544-007-9078-y. [DOI] [PubMed] [Google Scholar]
- Orit G, Nathalie QB. The importance of being persistent: heterogeneity of bacterial populations under antibiotic stress. FEMS Microbiol Rev. 2009;33:704–17. doi: 10.1111/j.1574-6976.2008.00156.x. [DOI] [PubMed] [Google Scholar]
- Ottesen EA, Hong JW, Quake SR, Leadbetter JR. Microfluidic Digital PCR Enables Multigene Analysis of Individual Environmental Bacteria. Science. 2006;314:1464–7. doi: 10.1126/science.1131370. [DOI] [PubMed] [Google Scholar]
- Pal R, Yang M, Lin R, Johnson BN, Srivastava N, Razzacki SZ, et al. An integrated microfluidic device for influenza and other genetic analyses. Lab Chip. 2005;5:1024–32. doi: 10.1039/b505994a. [DOI] [PubMed] [Google Scholar]
- Pamme N, Wilhelm C. Continuous sorting of magnetic cells via on-chip free-flow magnetophoresis. Lab Chip. 2006;6:974–80. doi: 10.1039/b604542a. [DOI] [PubMed] [Google Scholar]
- Panaro NJ, Lou XJ, Fortina P, Kricka LJ, Wilding P. Micropillar array chip for integrated white blood cell isolation and PCR. Biomol Eng. 2005;21:157–62. doi: 10.1016/j.bioeng.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Park S-J, Taton TA, Mirkin CA. Array-Based Electrical Detection of DNA with Nanoparticle Probes. Science. 2002;295:1503–6. doi: 10.1126/science.1067003. [DOI] [PubMed] [Google Scholar]
- Park S, Beskok A. Alternating Current Electrokinetic Motion of Colloidal Particles on Interdigitated Microelectrodes. Anal Chem. 2008;80:2832–41. doi: 10.1021/ac7024859. [DOI] [PubMed] [Google Scholar]
- Park S, Koklu M, Beskok A. Particle Trapping in High-Conductivity Media with Electrothermally Enhanced Negative Dielectrophoresis. Anal Chem. 2009a;81:2303–10. doi: 10.1021/ac802471g. [DOI] [PubMed] [Google Scholar]
- Park SY, Kalim S, Callahan C, Teitell MA, Chiou EPY. A light-induced dielectrophoretic droplet manipulation platform. Lab Chip. 2009b;9:3228–35. doi: 10.1039/b909158k. [DOI] [PubMed] [Google Scholar]
- Peter RCG, Jody V. Particle separation by dielectrophoresis. Electrophoresis. 2002;23:1973–83. doi: 10.1002/1522-2683(200207)23:13<1973::AID-ELPS1973>3.0.CO;2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersson F, Aberg L, Sward-Nilsson AM, Laurell T. Free flow acoustophoresis: Microfluidic-based mode of particle and cell separation. Anal Chem. 2007;79:5117–23. doi: 10.1021/ac070444e. [DOI] [PubMed] [Google Scholar]
- Pipper J, Inoue M, Ng LFP, Neuzil P, Zhang Y, Novak L. Catching bird flu in a droplet. Nat Med. 2007;13:1259–63. doi: 10.1038/nm1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rådström P, Knutsson R, Wolffs P, Lövenklev M, Löfström C. Pre-PCR processing. Mol Biotechnol. 2004;26:133–46. doi: 10.1385/MB:26:2:133. [DOI] [PubMed] [Google Scholar]
- Rajan A, Glorikian H. Point-of-care diagnostics: market trends and growth drivers. Expert Opin Med Diagn. 2009;3:1–4. doi: 10.1517/17530050802651579. [DOI] [PubMed] [Google Scholar]
- Rane TD, Puleo CM, Liu KJ, Zhang Y, Lee AP, Wang TH. Counting single molecules in sub-nanolitre droplets. Lab Chip. 2010;10 doi: 10.1039/b917503b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper MG, Easley CJ, Landers JP. Advances in Polymerase Chain Reaction on Microfluidic Chips. Anal Chem. 2005;77:3887–94. doi: 10.1021/ac050756m. [DOI] [PubMed] [Google Scholar]
- Rothman R, Ramachandran P, Yang S, Hardick A, Won H, Kecojevic A, et al. Use of Quantitative Broad-based Polymerase Chain Reaction for Detection and Identification of Common Bacterial Pathogens in Cerebrospinal Fluid. Acad Emerg Med. 2010;17:741–7. doi: 10.1111/j.1553-2712.2010.00790.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling EA, Kamholz AE, Yager P. Cell lysis and protein extraction in a microfluidic device with detection by a fluorogenic enzyme assay. Anal Chem. 2002;74:1798–804. doi: 10.1021/ac015640e. [DOI] [PubMed] [Google Scholar]
- Sethu P, Anahtar M, Moldawer LL, Tompkins RG, Toner M. Continuous Flow Microfluidic Device for Rapid Erythrocyte Lysis. Anal Chem. 2004;76:6253. doi: 10.1021/ac049429p. [DOI] [PubMed] [Google Scholar]
- Shastry A, Case MJ, Bohringer KF. Engineering surface roughness to manipulate droplets in microfluidic systems. MEMS 2005 Miami: Technical Digest. 2005:694–7. [Google Scholar]
- Shih PH, Shiu JY, Lin PC, Lin CC, Veres T, Chen P. On chip sorting of bacterial cells using sugar-encapsulated magnetic nanoparticles. J Appl Phys. 2008;103 [Google Scholar]
- Sista R, Hua Z, Thwar P, Sudarsan A, Srinivasan V, Eckhardt A, et al. Development of a digital microfluidic platform for point of care testing. Lab Chip. 2008;8:2091–104. doi: 10.1039/b814922d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slyadnev MN, Tanaka Y, Tokeshi M, Kitamori T. Photothermal Temperature Control of a Chemical Reaction on a Microchip Using an Infrared Diode Laser. Anal Chem. 2001;73:4037–44. doi: 10.1021/ac010318p. [DOI] [PubMed] [Google Scholar]
- Srinivasan V, Pamula VK, Fair RB. An integrated digital microfluidic lab-on-a-chip for clinical diagnostics on human physiological fluids. Lab Chip. 2004;4:310–5. doi: 10.1039/b403341h. [DOI] [PubMed] [Google Scholar]
- Stachowiak JC, Shugard EE, Mosier BP, Renzi RF, Caton PF, Ferko SM, et al. Autonomous microfluidic sample preparation system for protein profile-based detection of aerosolized bacterial cells and spores. Anal Chem. 2007;79:5763–70. doi: 10.1021/ac070567z. [DOI] [PubMed] [Google Scholar]
- Sun Y, Kwok YC. Polymeric microfluidic system for DNA analysis. Anal Chim Acta. 2006;556:80–96. doi: 10.1016/j.aca.2005.09.035. [DOI] [PubMed] [Google Scholar]
- Teh S-Y, Lin R, Hung L-H, Lee AP. Droplet microfluidics. Lab Chip. 2008;8:198–220. doi: 10.1039/b715524g. [DOI] [PubMed] [Google Scholar]
- Tewhey R, Warner JB, Nakano M, Libby B, Medkova M, David PH, et al. Microdroplet-based PCR enrichment for large-scale targeted sequencing. Nat Biotechnol. 2009;27:1025–31. doi: 10.1038/nbt.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsen T, Maerkl SJ, Quake SR. Microfluidic Large-Scale Integration. Science. 2002;298:580–4. doi: 10.1126/science.1076996. [DOI] [PubMed] [Google Scholar]
- Toner M, Irimia D. BLOOD-ON-A-CHIP. Annu Rev Biomed Eng. 2005;7:77–103. doi: 10.1146/annurev.bioeng.7.011205.135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trau D, Lee TMH, Lao AIK, Lenigk R, Hsing IM, Ip NY, et al. Genotyping on a Complementary Metal Oxide Semiconductor Silicon Polymerase Chain Reaction Chip with Integrated DNA Microarray. Anal Chem. 2002;74:3168–73. doi: 10.1021/ac020053u. [DOI] [PubMed] [Google Scholar]
- Unger MA, Chou H-P, Thorsen T, Scherer A, Quake SR. Monolithic Microfabricated Valves and Pumps by Multilayer Soft Lithography. Science. 2000;288:113–6. doi: 10.1126/science.288.5463.113. [DOI] [PubMed] [Google Scholar]
- Urdea M, Penny LA, Olmsted SS, Giovanni MY, Kaspar P, Shepherd A, et al. Requirements for high impact diagnostics in the developing world. Nature. 2006 doi: 10.1038/nature05448. [DOI] [PubMed] [Google Scholar]
- Valley JK, NingPei S, Jamshidi A, Hsu HY, Wu MC. A unified platform for optoelectrowetting and optoelectronic tweezers. Lab Chip. 2011;11:1292–7. doi: 10.1039/c0lc00568a. [DOI] [PubMed] [Google Scholar]
- Velev OD, Prevo BG, Bhatt KH. On-chip manipulation of free droplets. Nature. 2003;426:515–6. doi: 10.1038/426515a. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Kinzler KW. Digital PCR. Proc Natl Acad Sci U S A. 1999;96:9236–41. doi: 10.1073/pnas.96.16.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vulto P, Hermann C, Zahn P, Maier U, Dame G, Urban GA. Transducer 2009. Denver: 2009. A microchip for automated extraction of rna from gram-positive bacteria. [Google Scholar]
- Wen J, Legendre LA, Bienvenue JM, Landers JP. Purification of Nucleic Acids in Microfluidic Devices. Anal Chem. 2008;80:6472–9. doi: 10.1021/ac8014998. [DOI] [PubMed] [Google Scholar]
- West J, Boerlin M, Jadhav AD, Clancy E. Silicon microstructure arrays for DNA extraction by solid phase sample contacting at high flow rates. Sensors Actuators B. 2007;126:664–71. [Google Scholar]
- Whitesides GM. The origins and the future of microfluidics. Nature. 2006;442:368–73. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- Wilding P, Kricka LJ, Cheng J, Hvichia G, Shoffner MA, Fortina P. Integrated Cell Isolation and Polymerase Chain Reaction Analysis Using Silicon Microfilter Chambers. Anal Biochem. 1998;257:95–100. doi: 10.1006/abio.1997.2530. [DOI] [PubMed] [Google Scholar]
- Wu Z, Willing B, Bjerketorp J, Jansson JK, Hjort K. Soft inertial microfluidics for high throughput separation of bacteria from human blood cells. Lab Chip. 2009;9:1193–9. doi: 10.1039/b817611f. [DOI] [PubMed] [Google Scholar]
- Xia Y, Whitesides GM. Soft lithography. Annu Rev Mater Sci. 1998;28:153–84. [Google Scholar]
- Yager P, Domingo GJ, Gerdes J. Point-of-Care Diagnostics for Global Health. Annu Rev Biomed Eng. 2008;10:107–44. doi: 10.1146/annurev.bioeng.10.061807.160524. [DOI] [PubMed] [Google Scholar]
- Yager P, Edwards T, Fu E, Helton K, Nelson K, Tam MR, et al. Microfluidic diagnostic technologies for global public health. Nature. 2006;442:412–8. doi: 10.1038/nature05064. [DOI] [PubMed] [Google Scholar]
- Yang S, Lin S, Kelen GD, Quinn TC, Dick JD, Gaydos CA, et al. Quantitative Multiprobe PCR Assay for Simultaneous Detection and Identification to Species Level of Bacterial Pathogens. J Clin Microbiol. 2002;40:3449–54. doi: 10.1128/JCM.40.9.3449-3454.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Ramachandran P, Hardick A, Hsieh Y-H, Quianzon C, Kuroki M, et al. Rapid PCR-Based Diagnosis of Septic Arthritis by Early Gram-Type Classification and Pathogen Identification. J Clin Microbiol. 2008a;46:1386–90. doi: 10.1128/JCM.02305-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Ramachandran P, Rothman R, Hsieh Y-H, Hardick A, Won H, et al. Rapid Identification of Biothreat and Other Clinically Relevant Bacterial Species Using Universal PCR Coupled with High Resolution Melting Analysis. J Clin Microbiol. 2009 doi: 10.1128/JCM.00033-09. JCM.00033-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Rothman RE. PCR-based diagnostics for infectious diseases: uses, limitations, and future applications in acute-care settings. Lancet Infect Dis. 2004;4:337–48. doi: 10.1016/S1473-3099(04)01044-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Rothman RE, Hardick J, Kuroki M, Hardick A, Doshi V, et al. Rapid Polymerase Chain Reaction-based Screening Assay for Bacterial Biothreat Agents. Acad Emerg Med. 2008b;15:388–92. doi: 10.1111/j.1553-2712.2008.00061.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung S-W, Lee TM-H, Cai H, Hsing IM. A DNA biochip for on-the-spot multiplexed pathogen identification. Nucleic Acids Res. 2006a;34:e118. doi: 10.1093/nar/gkl702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung SSW, Lee TMH, Hsing IM. Electrochemical Real-Time Polymerase Chain Reaction. J Am Chem Soc. 2006b;128:13374–5. doi: 10.1021/ja065733j. [DOI] [PubMed] [Google Scholar]
- Yeung SSW, Lee TMH, Hsing IM. Electrochemistry-Based Real-Time PCR on a Microchip. Anal Chem. 2007;80:363–8. doi: 10.1021/ac071198+. [DOI] [PubMed] [Google Scholar]
- Yuen PK, Kricka LJ, Fortina P, Panaro NJ, Sakazume T, Wilding P. Microchip module for blood sample preparation and nucleic acid amplification reactions. Genome Res. 2001;11:405–12. doi: 10.1101/gr.155301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Novak R, Shuga J, Smith MT, Mathies RA. High-performance single cell genetic analysis using microfluidic emulsion generator arrays. Anal Chem. 2010;82:3183–90. doi: 10.1021/ac902683t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Xing D. Miniaturized PCR chips for nucleic acid amplification and analysis: latest advances and future trends. Nucleic Acids Res. 2007;35:4223–37. doi: 10.1093/nar/gkm389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JH, Cheng ZJ, Zheng YM, Jiang L. Ratchet-induced anisotropic behavior of superparamagnetic microdroplet. Appl Phys Lett. 2009a;94 [Google Scholar]
- Zhang Y, Bailey V, Puleo CM, Easwaran H, Griffiths E, Herman JG, et al. DNA methylation analysis on a droplet-in-oil PCR array. Lab Chip. 2009b;9:1059–64. doi: 10.1039/b821780g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Park S, Liu K, Tsuan J, Yang S, Wang T-H. A surface topography assisted droplet manipulation platform for biomarker detection and pathogen identification. Lab Chip. 2011;11:398–406. doi: 10.1039/c0lc00296h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Park S, Yang S, Wang T-H. An all-in-one microfluidic device for parallel DNA extraction and gene analysis. Biomed Microdevices. 2010;12:1043–9. doi: 10.1007/s10544-010-9458-6. [DOI] [PubMed] [Google Scholar]