Abstract
Error-free chromosome segregation depends on the precise regulation of phosphorylation to stabilize kinetochore-microtubule attachments (K-fibers) on sister chromatids that have attached to opposite spindle poles (bi-oriented)1. In many instances, phosphorylation correlates with K-fiber destabilization2–7. Consistent with this, multiple kinases, including Aurora B and Plk1, are enriched at kinetochores of mal-oriented chromosomes compared to bi-oriented chromosomes, which have stable attachments2, 8. Paradoxically, however, these kinases also target to prometaphase chromosomes that have not yet established spindle attachments and it is therefore unclear how kinetochore-microtubule interactions can be stabilized when kinase levels are high. Here we show that generation of stable K-fibers depends on the B56-PP2A phosphatase, which is enriched at centromeres/kinetochores of unattached chromosomes. When B56-PP2A is depleted, K-fibers are destabilized and chromosomes fail to align at the spindle equator. Strikingly, B56-PP2A depletion increases the phosphorylation of Aurora B and Plk1 kinetochore substrates as well as Plk1 recruitment to kinetochores. Consistent with increased substrate phosphorylation, we find that chemical inhibition of Aurora or Plk1 restores K-fibers in B56-PP2A depleted cells. Our findings reveal that PP2A, an essential tumor suppressor9, tunes the balance of phosphorylation to promote chromosome-spindle interactions during cell division.
Serine/threonine phosphorylation marks generated by ~ 400 kinases are reversed by a handful of phosphatases that are targeted to substrates via dozens of regulatory subunits10. For most signaling networks, including those required for the establishment of chromosome-microtubule attachments during prometaphase, it remains unclear which, or even if, a specific phosphatase regulatory subunit is important. In mammals, PP1 and PP2A, the most abundant eukaryotic serine/threonine phosphatases, target to kinetochores (PP1)11 or centromeres (PP2A)12–14 during mitosis. PP1, however, localizes to kinetochores only after chromosomes have bi-oriented and preventing PP1 targeting to the kinetochore does not impair chromosome alignment15, 16, suggesting it is not essential for establishment of kinetochore-microtubule interactions in prometaphase. Therefore, we decided to examine if PP2A plays a role in this process.
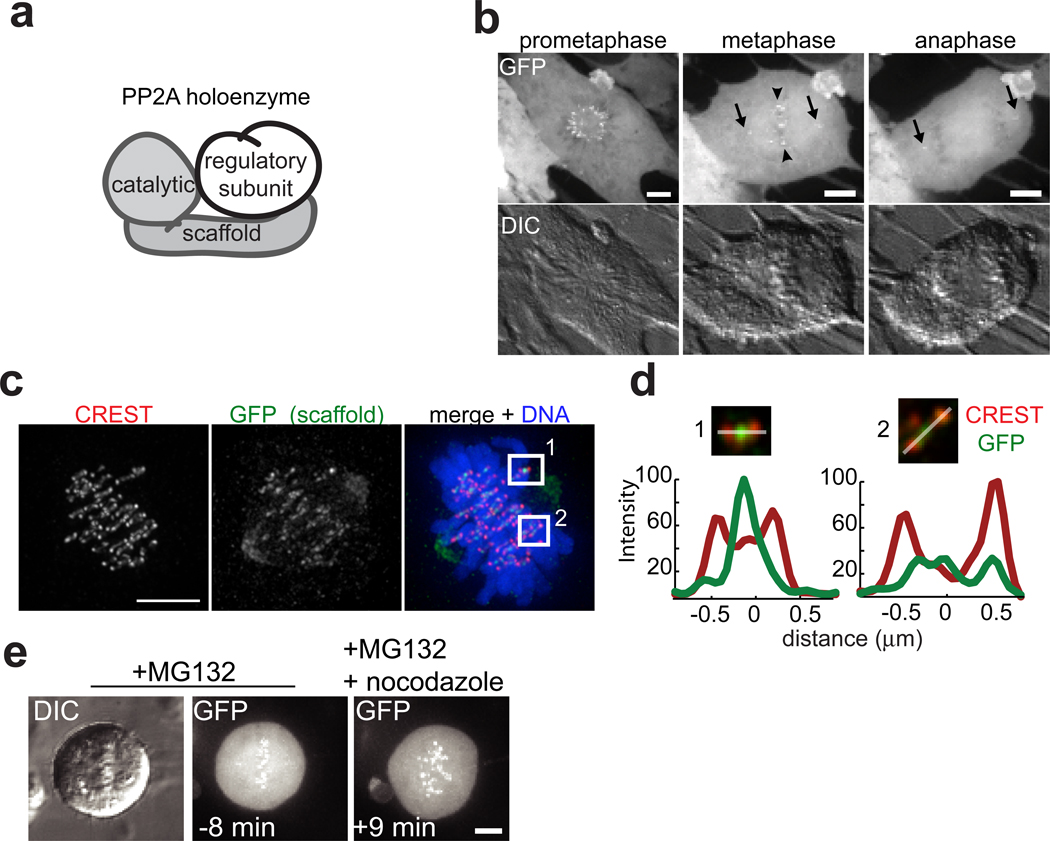
We reasoned that a high-resolution live-cell localization analysis might inform on PP2A functions at centromeres. PP2A holoenzymes are composed of a common catalytic and scaffold subunit, and a variable regulatory subunit17 (Fig. 1a). To analyze the dynamics of PP2A localization during mitosis we fused the core subunits to green fluorescent protein (GFP). As expected, this approach was unsuccessful for the catalytic subunit, an abundant protein that cannot be over-expressed18. Therefore, we examined the localization dynamics of GFP-scaffold stably expressed human RPE1 cells. Near-simultaneous differential interference contrast (DIC) and real-time confocal microscopy of mitotic cells revealed that the scaffold was enriched at centrosomes (Fig. 1b, arrows) and to discrete regions on chromosomes (Fig. 1b, arrowheads), which corresponded to centromere/kinetochore targeting (Fig. 1c, d). Unexpectedly, we found that scaffold centromere/kinetochore targeting decreased between prometaphase and metaphase in RPE1 (Fig. 1b) and HeLa cells (Fig. S1a). GFP-scaffold signal was enriched on centromeres of chromosomes that had not congressed to the metaphase plate (Fig. 1c–d, compare centromere 1 and 2), suggesting that its targeting may be sensitive to chromosome-microtubule attachment status. To test this, we arrested cells at metaphase and tracked distribution of GFP-scaffold before and after depolymerization of microtubules by nocodazole. Strikingly, within minutes of nocodazole addition, GFP-scaffold became enriched on centromeres (Fig. 1e), confirming attachment-sensitive localization.
Figure 1.
Microtubule-sensitive targeting PP2A to centromeres/kinetochores during cell division. (a) Schematic showing PP2A’s scaffold, catalytic, and regulatory subunits. (b) Maximum intensity confocal projections show distributions of GFP-scaffold expressed in an RPE1 cell at mitosis (top). Centrosome (arrow) and centromere (arrowhead) localizations are indicated. DIC images (below) show chromosomes in same cell. (c) Immunofluorescence images of a maximum intensity projection of an RPE1 cell expressing GFP-scaffold fixed and stained for kinetochores (CREST, red), GFP (green) and DNA (blue, only shown in overlay). (d) Maximum intensity projection of the optical sections spanning the boxed regions in (c) enlarged 2x with DNA omitted. Plotted is the intensity profile of the CREST (red) and GFP (green) signal measured along a line (white) drawn across the centromere. (e) Maximum intensity confocal projections of GFP-scaffold distribution and chromosomes (DIC) in a cell arrested in metaphase (10 µM MG132), and imaged live at the indicated times relative to addition of nocodazole (3.2 µM, time zero). Scale bars, 5 µm.
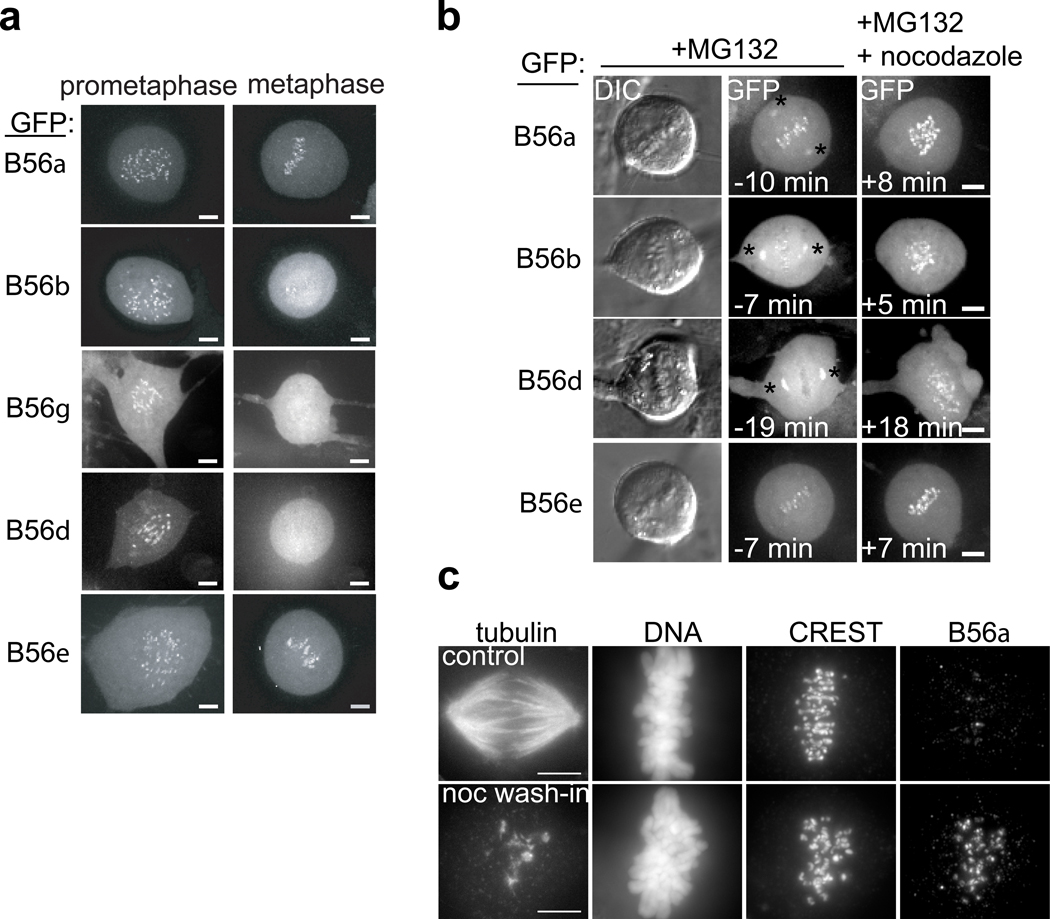
To identify regulatory subunits that also reveal a microtubule attachment-dependent localization, we performed a comprehensive localization analysis for all regulatory subunit genes, using stable cell lines expressing GFP fusions of individual subunits. In humans, there are at least fifteen regulatory subunits, distributed over four evolutionarily conserved families17 but the localization dynamics of only two subunits have been reported thus far19. By live-imaging, only the five members of the B56 (B’) family of regulatory subunits were observed at centromeres (Fig. 2a, and Fig. S1b–e). Similar to the scaffold, B56 targeting was highest in prometaphase, and reduced (B56α, ε) or undetectable (B56β, γ, δ) by metaphase (Fig. 2a). Furthermore, when we repeated the nocodazole wash-in assay, GFP-B56α, β, δ, and ε targeting to centromeres increased within minutes (Fig. 2b), confirming microtubule-attachment sensitive targeting. We did not consistently detect targeting of B56γ to centromeres in this assay (data not shown). Microtubule-sensitive localization of endogenous B56α to centromeres/kinetochores was confirmed by immunofluorescence in unperturbed cells (Fig. S2), and in a nocodazole wash-in assay (Fig. 2c). Together, these data indicate that B56-PP2A targeting to centromeres/kinetochores is sensitive to kinetochore-microtubule attachment status.
Figure 2.
Microtubule-sensitive targeting of B56 regulatory subunits to centromeres/kinetochores. (a) Maximum intensity confocal projections show distributions of GFP in different cell lines stably expressing GFP-B56α-ε proteins. (b) RPE1 cells stably expressing a GFP-fusion of the indicated B56 regulatory subunit were arrested in metaphase (10 µM MG132) and imaged live before and after addition of nocodazole (3.2 µM, time zero). Maximum intensity confocal projections show GFP distribution, and DIC images show chromosomes before nocodazole addition. Spindle pole targeting was observed in MG132-arrested cells (asterisk). (c) Cells in MG132 (10 µM) were treated with nocodazole (3.2 µM, bottom) or control solvent (DMSO, top) for 5 min and processed for immunofluorescence. Equivalently scaled maximum intensity projections of tubulin, DNA, kinetochores (CREST), and B56α staining are shown. Scale bars, 5 µm.
Microtubule-attachment sensitive kinetochore targeting is a hallmark of proteins that regulate microtubule binding (e.g. dynein, Cenp-E20) and/or mitotic checkpoint signaling (e.g. Mad221). However, we considered a requirement for B56-PP2A in the spindle checkpoint unlikely because depletion of the scaffold results in a mitotic arrest in human cells12, 14.
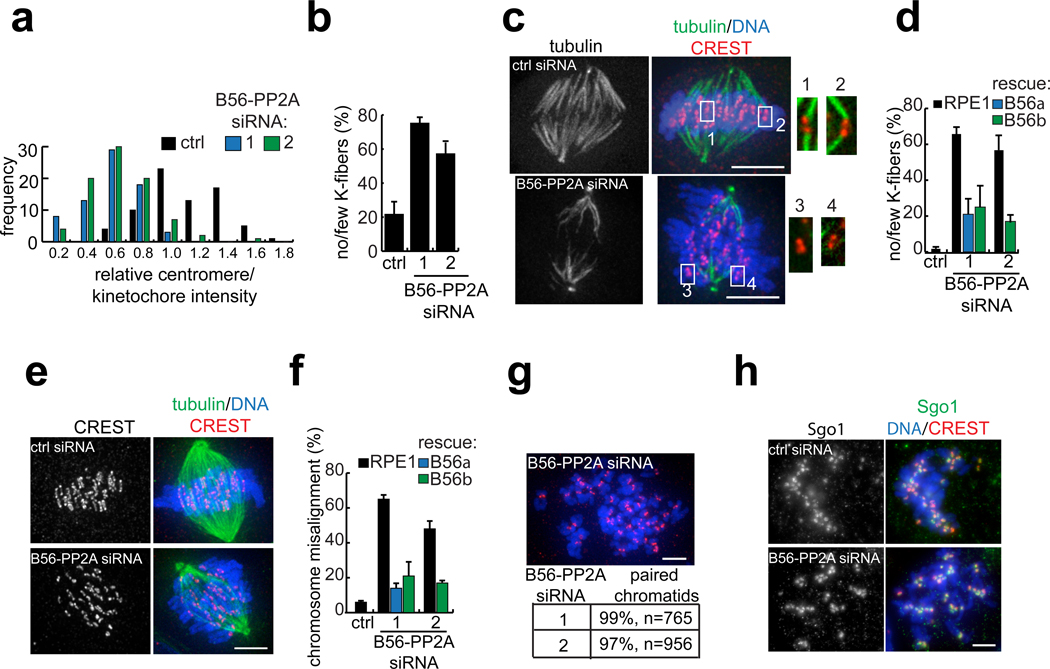
In mammals, one challenge in assigning PP2A functions during the cell cycle is the potential for redundancy in regulatory subunit function. Thus, while PP2A is essential for mitosis12, 14, knockdown of individual regulatory subunits in human cells has not been observed to perturb chromosome segregation22. Redundancy may be particularly relevant to the B56 family, which share a pseudo HEAT repeat structure with ~ 80% sequence identity23, 24. Furthermore, depletion of the scaffold or chemical inhibition of the catalytic subunit cannot inform on specific regulatory subunits. Therefore, we chose to deplete B56α-ε proteins using RNAi. We used two non-overlapping pools, composed of one siRNA targeting each B56 subunit, and analyzed the extent of depletion by two methods. First, by western blot analysis, each pool partially reduced protein levels of endogenous B56α, β, and δ (Fig. S3a). We confirmed that GFP-fusions of B56γ and ε were depleted, as we were unable to detect the endogenous proteins using available antibodies (Fig. S3b). Second, we confirmed that B56-PP2A siRNA cells had reduced levels of GFP-scaffold at centromeres/kinetochores (Fig. 3a). As expected12, 14, nocodazole-treated B56-PP2A siRNA cells accumulated in mitosis (Fig. S4a), indicating an intact spindle checkpoint.
Figure 3.
B56-PP2A is required for stable kinetochore-microtubule attachments and chromosome alignment. (a) Analysis of GFP-scaffold levels at centromeres/kinetochores after B56-PP2A siRNA. An RPE1 cell line expressing GFP-scaffold was transfected with control or either of two B56-PP2A siRNA pools (1, 2) and treated with nocodazole (3.2 µM, 60 min) before processing for immunofluorescence. The GFP signal at centromeres/kinetochores was measured, processed, and normalized to the average value in cells treated with control siRNA. An intensity distribution histogram is plotted from one experiment. B56-PP2A siRNA reduced scaffold targeting to 0.52 ± 0.05 (pool 1) or 0.55 ± 0.05 (pool 2) relative to control cells (mean ± s.e.m, 4 experiments, >50 centromeres/kinetochores from 5 cells per condition, per time). (b–d) K-fiber defects in B56-PP2A siRNA cells. (b) The frequency of pre-anaphase mitotic cells with few or no K-fibers is shown. (c–d) Rescue of siRNA phenotype by stable over-expression of siRNA-resistant B56α or B56β. Cells were arrested in mitosis with nocodazole (0.32 µM, 2.5 h) and released into MG132 (10 µM, 40 min) before cold-treatment and fixation. (c) Cold-stable microtubules in a control and B56-PP2A (pool 2) siRNA treated cell. Insets show 2x enlargement of the boxed regions. (d) The frequency of K-fiber defects is shown. (e–f) Chromosome alignment defects in B56-PP2A siRNA cells. Control or B56-PP2A siRNA-treated cells were arrested with MG132 (10 µM, 60 min). (e) Example of chromosome alignment defects in B56-PP2A (pool 2) treated cells versus control cells. (f) The frequency of mitotic cells with misaligned chromosomes is shown. (g) Cohesion is preserved in B56-PP2A siRNA cells. Chromosome spreads were prepared from nocodazole-arrested cells (3.2 µM, 4 h) treated with either of two B56-PP2A siRNA pools. The fraction of paired chromatids from two experiments is shown. (h) Chromosome spreads were prepared from control and B56-PP2A (pool 2) siRNA treated RPE1 cells arrested as in (g). Equivalently scaled Sgo1 images and an overlay with DNA and kinetochores are shown. Images show maximum intensity projections with tubulin or Sgo1 (green), DNA (blue) and kinetochores (CREST, red). Scale bars, 5 µm. Bars show mean ± s.e.m. (n=3 experiments, >80 cells per condition per time).
To examine whether B56-PP2A siRNA treatment impairs chromosome-microtubule attachment, we made use of the fact that K-fibers are preferentially stable during a brief incubation at 4 °C, while other spindle microtubules depolymerize25. First, we scored the presence of K-fibers in any mitotic cell that had not yet entered anaphase. B56-PP2A siRNA increased the fraction of mitotic cells that contained few or no K-fibers (Fig. 3b), indicating that B56-PP2A is required for proper chromosome-spindle interactions. Second, we used transient nocodazole arrest to accumulate mitotic cells lacking microtubule attachments and then released cells into media containing proteasome inhibitor for 40 minutes, sufficient time for control cells to generate K-fibers (Fig. 3c). In contrast, B56-PP2A siRNA cells typically had numerous kinetochores lacking K-fibers (Fig. 3c) and most contained few or no K-fibers (Fig. 3d). To confirm the specificity of our phenotype, we generated stable cell lines over-expressing siRNA-resistant B56α or B56β (Fig. S3c). When these cell lines were transfected with B56-PP2A siRNA, K-fibers persisted (Fig. 3d).
To examine whether B56-PP2A siRNA impairs alignment of chromosomes at the spindle equator, we analyzed chromosome alignment in metaphase-arrested cells. Consistent with defects in chromosome-spindle attachments, individual B56-PP2A siRNA cells had numerous misaligned chromosomes (Fig. 3e). Overall, B56-PP2A siRNA resulted in an eight-fold increase in the fraction of cells with misaligned chromosomes, and over-expression of an siRNA-resistant B56 regulatory subunit rescued this defect (Fig. 3f). We conclude that B56-PP2A is required to establish stable kinetochore-microtubule attachments and align chromosomes at the spindle equator.
Recent reports indicate that PP2A plays an important role in regulating sister-chromatid cohesion12–14. B56 regulatory subunits have been linked to this function via their association with Sgo112–14, a centromeric protein that maintains cohesion and regulates K-fiber stability26–28. To examine if cohesion is lost in our experiments, we isolated chromosomes from B56-PP2A siRNA cells and found that at least 97% of chromatids had paired kinetochores (Fig. 3g), indicating that centromeric cohesion is preserved. Additionally, we measured inter-kinetochore distances in metaphase-arrested cells, which are expected to increase if cohesion at the centromere is compromised. We found, however, that inter-kinetochore distances in B56-PP2A siRNA cells were equal to or less than in control cells, consistent with intact cohesion (Fig. S5). Finally, we confirmed that centromere targeting of Sgo1 is preserved in B56-PP2A siRNA cells (Fig. 3h and S4b–c). Together, these data indicate that perturbations in centromeric cohesion and/or Sgo1 targeting cannot account for the defects in kinetochore-microtubule attachment observed in B56-PP2A siRNA cells.
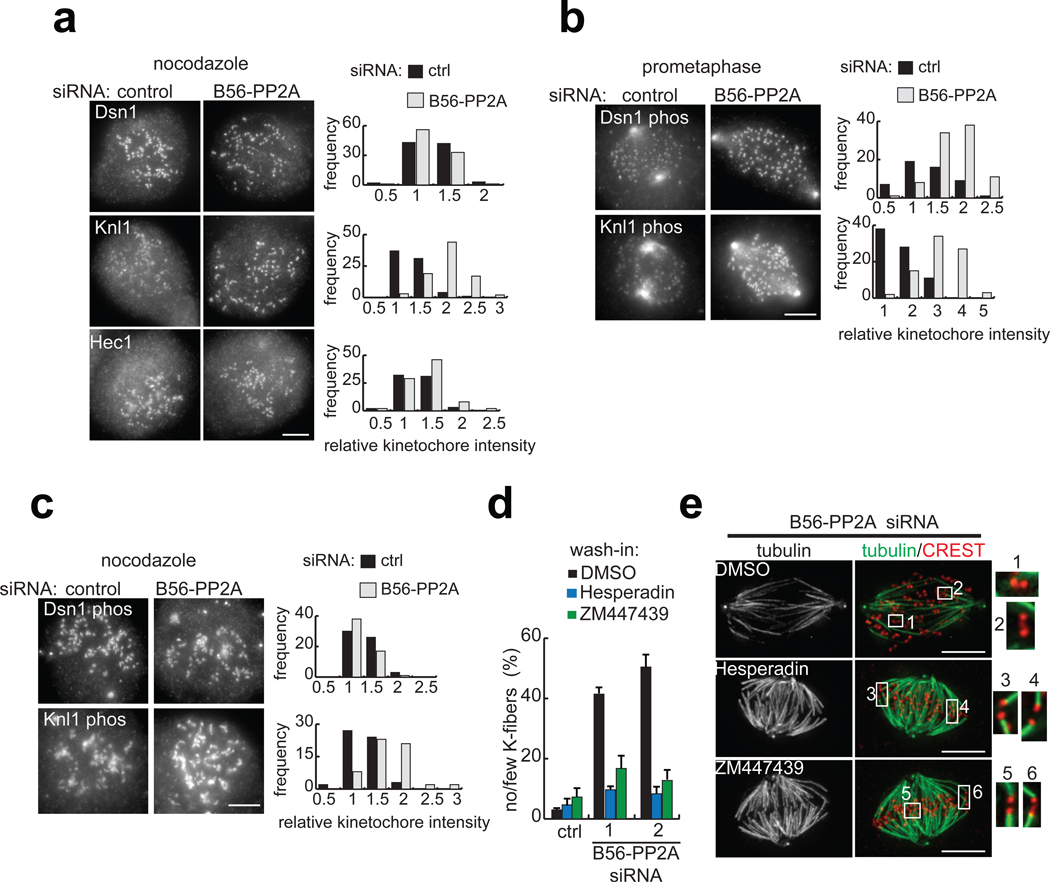
Defects in K-fiber stability in B56-PP2A siRNA cells could be due to an imbalance in substrate phosphorylation and/or failure to recruit proteins that bind kinetochores to microtubules. To test the latter possibility, we examined the kinetochore targeting of three proteins in the KMN network (Dsn1, Knl1, and Hec1), the core microtubule binding complex at the kinetochore29. It has been shown that Dsn1 and Hec1 levels at kinetochores are not sensitive to microtubule binding, while Knl1 recruitment increases 27% in nocodazole-arrested cells compared to metaphase cells6. Therefore, to exclude effects of microtubule sensitivity, we included nocodazole in our analyses. Under these conditions, Dsn1 and Hec1 levels were unchanged by B56-PP2A siRNA treatment, while Knl1 levels were modestly increased (1.4-fold), indicating that overall KMN network targeting is preserved in B56-PP2A siRNA cells (Fig. 4a).
Figure 4.
B56-PP2A depletion increases the phosphorylation of Aurora B substrates and Aurora inhibition suppresses the B56-PP2A siRNA phenotype. (a–c) RPE1 cells were transfected with control or B56-PP2A siRNA (pool 2) and (a, c) incubated with nocodazole (3.2 µM, 60 min) before fixation, or (b) fixed, and stained using indicated antibodies. Images are maximum intensity projections with equivalent scaling. The signal at kinetochores was measured, processed, and normalized to the average value in control siRNA cells. Histograms show intensity distributions from one experiment. (a) KMN network targeting is preserved in B56-PP2A siRNA cells. In B56-PP2A siRNA cells, the mean kinetochore staining intensities were calculated for Dsn1 (0.85 ± 0.11), Knl1 (1.41 ± 0.23), and Hec1 (1.05 ± 0.08) relative to control cells (n=2–3 experiments, >75 kinetochores from 5 cells per condition, per time). (b–c) Analysis of Aurora B substrate phosphorylation. (b) In prometaphase cells, kinetochores with an inter-kinetochore stretch of 1.2 to 1.5 µM were analyzed. In B56-PP2A siRNA cells, the mean phosho-Dsn1 and phospho-Knl1 intensity was 1.79 ± 0.32 and 2.26 ± 0.07, respectively (n=2 experiments, >50 kinetochores from 5 cells per condition, per time). (c) In nocodazole-treated B56-PP2A siRNA cells, the mean intensity of phospho-Dsn1 and phospho-Knl1 was 1.03 ± 0.13 and 1.21 ± 0.15, respectively, relative to control cells (3 experiments, >60 kinetochores from 5 cells per condition per experiment). (d–e) RPE1 cells treated with control or either of two pools of B56-PP2A siRNA (1, 2) were incubated in MG132 (10 µm, 15 min), followed by addition of Hesperadin (50 nM) or ZM447439 (1 µM) or control solvent (DMSO) for 45 min. (d) The frequency of mitotic cells with few or no cold-stable K-fibers is plotted (n=3 experiments, >80 cells per condition per time). (e) Maximum intensity projection of tubulin (green) and an overlay with kinetochores (CREST, red) in B56-PP2A siRNA (pool 2) cells treated with indicated inhibitor. Insets are 2x enlargement of the optical sections spanning the boxed regions. Scale bars, 5 µm. Mean ± s.e.m provided.
To test whether phosphorylation at kinetochores is increased in B56-PP2A siRNA cells, we analyzed substrates of Aurora B, a key regulator of microtubule attachment stability30. We chose two KMN network substrates (Ser100 on Dsn1 and Ser24 on Knl1), whose phosphorylation decreases microtubule binding affinity6. In prometaphase cells, phosphorylation of both substrates was increased in B56-PP2A siRNA cells compared to controls (Fig. 4b). However, because modification of these substrates is sensitive to microtubule attachment status, as is Knl1 recruitment6 (and because B56-PP2A siRNA destabilizes K-fibers), it was necessary to compare phosphorylation levels on kinetochores with similar inter-kinetochore spacing, a read-out for microtubule attachments. This analysis revealed that on kinetochores under comparable microtubule-dependent pulling forces (defined as a 1.2 to 1.5 µm inter-kinetochore stretch), B56-PP2A siRNA increased the mean phosphorylation of Dsn1 and Knl1 by 1.8- and 2.3-fold, respectively (Fig. 4b). After accounting for changes in Knl1 targeting (Fig. 4a), the net increase in phospho-Knl1 is 1.6-fold. Together, these analyses suggest that B56-PP2A limits the phosphorylation of these Aurora B substrates. In nocodazole, B56-PP2A siRNA did not increase Dsn1 phosphorylation and the slight increase in Knl1 phosphorylation could be attributed to higher levels of Knl1 at the kinetochore (Fig. 4c). It is noteworthy that phosphorylation of Dsn1 and Knl1 at these sites increases 7- and 2-fold respectively in nocodazole-treated compared to prometaphase cells6. Thus, phosphorylation at these sites may approach 100% in nocodazole, in which case loss of B56-PP2A would not further increase phosphorylation.
To test whether suppressing Aurora B can rescue the B56-PP2A siRNA phenotype, cells were arrested in mitosis, followed by wash-in of chemical inhibitors of Aurora31, 32 (Hesperadin or ZM447439) or control solvent. As expected, control cells had cold-stable K-fibers, whereas many B56-PP2A siRNA cells did not (Fig. 4d). Strikingly, Aurora inhibition was sufficient to restore K-fibers in B56-PP2A siRNA treated cells (Fig. 4d) and both inhibitors restored K-fibers to nearly all kinetochores (Fig. 4e). Together, these data suggest that the defects in kinetochore-microtubule interactions in B56-PP2A siRNA cells are due, at least in part, to increased phosphorylation of Aurora B substrates.
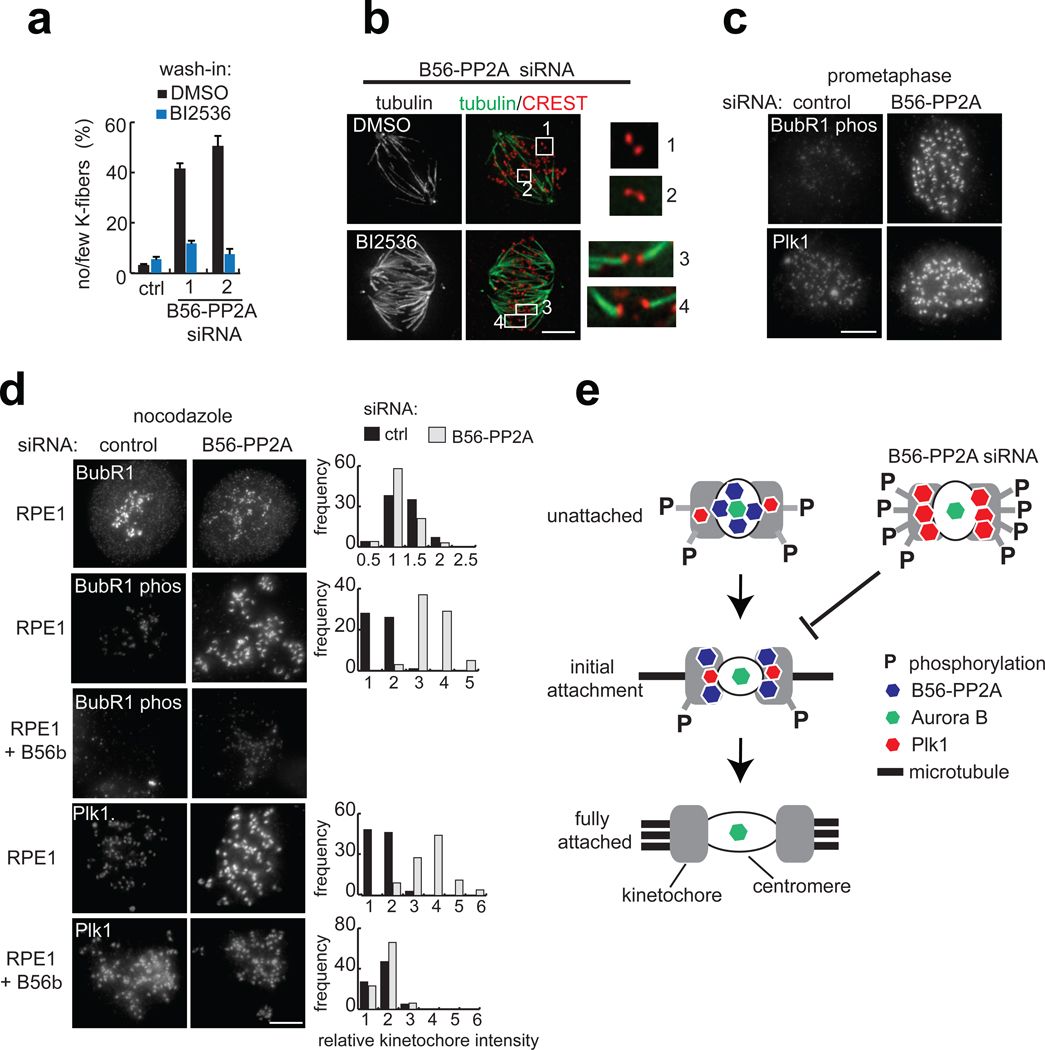
To examine whether B56-PP2A attenuates the signaling of kinetochore kinases other than Aurora B, we examined Plk1. Remarkably, wash-in of the Plk1 inhibitor BI253633 also restored K-fibers in B56-PP2A siRNA cells (Fig. 5a, b). Consistent with this, phosphorylation of a Plk1 kinetochore substrate, Ser 676 on BubR134 was increased in prometaphase cells following B56-PP2A siRNA (Fig. 5c). Furthermore, even in the absence of attachments, B56-PP2A siRNA treatment resulted in a three-fold increase in phospho-BubR1 staining, without affecting BubR1 protein levels at kinetochores (Fig. 5d), indicating that B56-PP2A modulates the phosphorylation level of a Plk1 substrate in addition to Aurora B substrates. BubR1 phosphorylation was undetectable in a cell line stably over-expressing siRNA-resistant B56β (Fig. 5d), consistent with this site as a potential B56-PP2A substrate. Taken together, these analyses reveal that reduction of PP2A at the centromere increases the phosphorylation of multiple kinetochore proteins.
Figure 5.
B56-PP2A regulates Plk1 substrate phosphorylation and Plk1 targeting to the kinetochore, and Plk1 inhibition suppresses the B56-PP2A siRNA phenotype. (a–b) RPE1 cells transfected with control or either of two B56-PP2A siRNA pools (1, 2) were incubated in MG132 (10 µM, 15 min), followed by addition of BI2536 (40 nM) or DMSO for 45 min. (a) The frequency of mitotic cells with few or absent cold-stable K-fibers is plotted (n=3 experiments, >80 cells per condition per time). (b) Maximum intensity projection of tubulin (green) and an overlay with kinetochores (CREST, red) in B56-PP2A siRNA (pool 2) cells treated with DMSO or BI2536 (40 nM). Insets are 3x enlargement of the optical sections spanning the boxed centromeres. (c) RPE1 cells transfected with control or B56-PP2A siRNA (pool2) were fixed and stained. Maximum intensity projections with equivalent scaling are shown. (d) RPE1 cells or a cell line expressing siRNA-resistant B56β were transfected with control or B56-PP2A (pool2) siRNA and incubated with nocodazole (3.2 µM, 60 min) before processing for immunofluorescence. The signal at kinetochores was measured, processed, and normalized to the average value in control siRNA cells. Histograms show intensity distributions from one experiment. In B56-PP2A siRNA cells, mean kinetochore intensity for BubR1 (0.84 ± 0.24), phospho-BubR1 (3.54 ± 1.41), and Plk1 (2.57 ± 0.32) were calculated relative to control cells (n=3–6 experiments, >60 kinetochores from five cells per condition, per time). In the cell line over-expressing siRNA resistant B56β, Plk1 targeting was similar after B56-PP2A siRNA (1.08 ± 0.12) or control siRNA (0.97 ± 0.11), relative to RPE1 cells treated with control siRNA (n=3 experiments, >60 kinetochores from five cells per condition, per time). Scale bars, 5 µm. Mean ± s.e.m provided. (e) Schematic shows a model for how B56-PP2A localization to centromeres (white circle) and kinetochores (grey) may promote microtubule binding.
Considering that Plk1 targeting depends on docking to phospho-epitopes35, we reasoned that B56-PP2A siRNA treatment might increase Plk1 recruitment to kinetochores. Consistent with this, B56-PP2A siRNA increased Plk1 kinetochore targeting in prometaphase cells (Fig. 5c). However, because Plk1 recruitment to kinetochores is sensitive to microtubule attachment status2, we examined Plk1 targeting in nocodazole-treated cells. Strikingly, B56-PP2A siRNA resulted in a 2.5-fold increase in Plk1 kinetochore targeting (Fig. 5d). This result was confirmed with a second pool of B56-PP2A siRNA (data not shown). Critically, over-expression of siRNA-resistant B56β rescued this defect (Fig. 5d). We conclude that, in addition to regulating the phosphorylation of kinase substrates, B56-PP2A also controls targeting of Plk1.
At the start of mitosis, all chromosomes are unattached to the spindle. Cytological studies indicate that initial kinetochore-microtubule interactions during prometaphase consist of one or a few microtubules, with microtubule occupancy increasing to ~ 25 microtubules per kinetochore on aligned chromosomes by metaphase36, 37. Several kinases, including Aurora B and Plk12, 8, are enriched at unattached kinetochores, suggesting high substrate phosphorylation. For Aurora B, a vast body of data involving phospho-mimetic substrates, kinase mis-targeting, and kinase inhibition suggests that high substrate phosphorylation at the kinetochore would lead to unstable attachments, both by disrupting kinetochore-microtubule contacts and by increasing the turnover of kinetochore-bound microtubules30. This creates a paradox for how K-fibers form during prometaphase. Additionally, it has been shown that phosphorylation can ‘fine-tune’ the strength of kinetochore-microtubule interactions6, but contributions from PP2A, or any other phosphatase, during the capture of microtubules by kinetochores remained unclear.
Our data reveal that B56-PP2A is an essential regulator of chromosome-spindle attachments. At the start of mitosis, B56-PP2A is enriched on unattached chromosomes (Fig. 5e, top) where it counteracts kinases, reducing phosphorylation to levels that stabilize kinetochore-microtubule binding (Fig. 5e, middle). Notably, even a modest reduction in B56-PP2A alters the phosphorylation landscape at kinetochores, preventing the stabilization of K-fibers. As microtubules contact the kinetochore, B56-PP2A is removed, as are a subset of kinases (e.g. Plk12). Kinetochore accessibility of the remaining kinase, Aurora B, is reduced when chromosomes are bi-oriented and proper inter- and intra-kinetochore tension is established8 (Fig. 5e, bottom). Interestingly, the re-distribution of B56-PP2A from the centromere towards the kinetochore on chromatids that have come under tension (Fig. 5e, middle) may ensure timely dephosphorylation of Aurora B and Plk1 substrates on kinetochores that have bi-oriented. While this function has been attributed to PP115, 16, recent work in yeast reveals the essential function of kinetochore PP1 is to silence the spindle checkpoint 38.
Misregulation of PP2A is considered a pre-requisite for malignancy in human cells, but less is understood about which phospho-signaling networks are associated with tumorigenesis9. Interestingly, point mutations in the scaffold that disrupt binding to the B56 regulatory subunits have been identified in lung and breast carcinomas39. Our data suggest these mutations will increase the frequency of whole chromosome gain or loss, the most common form of chromosomal instability in human tumors40, through disruption of kinetochore-microtubule interactions.
Materials and Methods
Cell Culture, transfection, and inhibitor treatments
Cells were grown at 37 °C in a humidified atmosphere with 5% CO2 in DMEM (HeLa, 293-ampho) or 1:1 DMEM:F12 media (RPE1) from Invitrogen supplemented with 10% fetal bovine serum (Atlanta Biologicals), 1X penicillin-streptomcyin and non-essential amino acids (Invitrogen), and 2 mM L-glutamine (Invitrogen). Cells used for live imaging or immunofluorescence were grown on no. 1.5 glass coverslips (Fisher Scientific) coated with Poly-D-lysine (Sigma). 293-ampho cells were transfected according to a calcium phosphate protocol to generate retroviruses. Virus-containing media was supplemented with 4 µg/mL polybrene (Sigma) and applied to target cells, followed by selection with puromycin (Sigma). Nocodazole (Sigma), MG132 (Boston Biochem), ZM447439 (Tocris Bioscience), BI2536 (Selleck Biochemicals) and Hesperadin (synthesized in the Kapoor lab) were dissolved in DMSO. For siRNA treatments, 1.7 × 105 RPE1 cells were transfected with 150 pmol siRNA and 7.5 µL Lipofectamine RNAi Max (Invitrogen) following the manufacturer’s reverse transfection protocol and immediately plated onto no 1.5 glass coverslips. In all experiments, cells were fixed or lysed 42–46 h post-transfection (Fisher Scientific). An siRNA targeting mCherry (5’-GCUCCAAGGCCUACGUGAAUU-3’) was used for control transfections.
To generate B56 family siRNA pools, two individual siRNAs from Dharmacon smart pools for each B56 gene determined to be effective in reducing protein levels of endogenous and/or GFP-tagged B56 genes were chosen. siRNAs were mixed at equimolar ratios, with the exception of B56ε siRNA, which was included at 1.5-fold relative to the other siRNAs. siRNAs used in the B56 family pools are: B56α: GCUCAAAGAUGCCACUUCA (pool 1), and UGAAUGAACUGGUUGAGUA (pool 2); B56β: CGCAUGAUCUCAGUGAAUA (pool 1), and GAACAAUGAGUAUAUCCUA (pool 2); B56γ: GGAUUUGCCUUACCACUAA (pool 1), and GGAAGAUGAACCAACGUUA (pool 2); B56δ: UCCAUGGACUGAUCUAUAA (pool 1), and UGACUGAGCCGGUAAUUGU (pool 2); B56ε: UUAAUGAACUGGUGGACUA (pool 1), and GCACAGCUGGCAUAUUGUA (pool 2).
Plasmid Construction
Open reading frames for PP2A subunits were purchased from OpenBiosystems and cloned into pDONR201 using Gateway technology (Invitrogen) and sequenced prior to recombination into GFP-tagged retroviral vectors. N- and C-terminal GFP fusion retroviral destination vectors were cloned from a parent vector, pMSCVpuro (Clontech) compatible with Gateway cloning (a gift from Wade Harper, Harvard Medical School). A strep-eGFP cassette was inserted before or after the Gateway cassette, to yield GFP-tagged destination vectors. For RPE1 stable cell lines expressing N-terminally tagged PP2A scaffold, the destination vector described above was sub-cloned to include an FKBP tag before the strep tag. These vectors were used in recombination reactions with entry clones containing PP2A regulatory subunits, and the open reading frames were sequence-verified.
Immunological methods
For immunofluorescence of BubR1, cells on coverslips were fixed in methanol at −20 C for 10 min. For all other antibodies, cells on coverslips were pre-extracted for 40 seconds at 37 C in PEM buffer (100 mM PIPES, 10 mM EGTA, 1 mM MgCl2 pH to 6.9 by KOH) with 0.5% Triton-X and 4 M glycerol and then fixed for 5 min at 37 C in form fix (3.7% formaldehyde and 0.2% TritonX-100 in PEM buffer). For analysis of cold-stable microtubules, cells were incubated for 10 min at 4 C in L-15 media (Invitrogen) with 10% FBS, and then fixed in form fix for 10 min at 22 C. For the preparation of chromosome spreads, cells were collected by trypsinization, pelleted in media, resuspended in 0.075M KCl for 20 min at 22 C and then spun onto poly-lysine coated glass coverslips at 2,000 r.p.m. for 2 minutes in a Shandon Cytospin 3. All subsequent immunofluorescence incubations were performed at 22 C. Cells were blocked in 2% Donkey serum (Jackson Immunoresearch). The following antibodies were used for immunofluorescence: anti-B56α (BD-Transduction Laboratories, #610615, used at 1:250), anti-Plk1 (Santa Cruz # sc-17783 used at 1:200), anti-GFP (raised against full-length GFP and affinity purified and used at 1 µg/mL), anti-phospho Ser 100 Dsn1 and phospho Ser 24 Knl1 (gifts from Iain Cheeseman1 used at 1:1,000), anti- phospho Ser 676 BubR1 (a gift from Sabine Elowe2 used at 1:1,000), anti-BubR1 (Millipore #MAB3612, used at 1:500), anti-Dsn1 and anti-Knl1 (gifts from Arshad Desai 3 used at 1:1,000), anti-Sgo1 antibodies (a gift from Hongtao Yu4 used at 1:1,000 and Abcam, ab58023, used at 1:200). FITC-conjugated mouse anti-tubulin monoclonal (Sigma # F2168) was used at 1:3,000, and human CREST anti-serum was used at 1:80,000 (a gift from W. Brinkley, Baylor College of Medicine). Secondary antibodies raised in donkey (Jackson Immunoresearch) were used at 2 µg/mL and DNA was stained with Hoechst 33342 (Sigma). Coverslips were mounted in 0.5 % p-phenylenediamine (Sigma) in 20 mM Tris, pH 8.8, 90% glycerol and sealed with nail polish. For western blot analysis, the following antibodies were used: anti-B56α (Bethyl Laboratories, #A300-967A used at 1:3,000), anti-B56β (raised against a peptide corresponding to amino acids 477–497 of human B56β and affinity purified and used at 0.2 µg/mL), anti-B56δ (Santa Cruz, #sc-81605, used at 1:500) or GFP (used at 0.1 µg/mL. HRP-conjugated secondary antibodies raised in donkey were purchased from Jackson Immunoresearch and used at 0.02 µg/mL.
Imaging, data acquisition, and processing
For live imaging, cells grown on 22 × 22 mm poly-lysine coverslips were mounted in a custom Rose chamber in 10% fetal bovine serum in L-15 media (Invitrogen) without phenol-red and maintained at 35–37 C. Confocal GFP fluorescence images were acquired using a Nikon TE2000 microscope (Morrell Instruments), with a 100X objective (PlanApo, 1.4 NA), equipped with a z-motor. 0.4 µm z-stacks were taken through the entire cell with a PerkinElmer Wallac UltraView confocal head, 488 nm excitation filters, and an argon ion laser (Solamere Inc.). A single DIC image was taken prior to each z-stack. Images were acquired with an EMCCD Photometric Cascade 512B camera (Roper Scientific) with 2×2 pixel binning using Metamorph software (MDS Analytical Technologies). Image J software (NIH) was used to crop images, adjust contrast, and create maximum intensity projections.
Quantifications of mitotic index, chromosome mis-alignment, and the presence of cold-stable microtubules were scored visually on a Zeiss Axioplan2 microscope (Carl Zeiss MicroImaging, Inc.), with a 40X objective (Plan Neo, NA 0.75).
Images of fixed cells were acquired as z-stacks with 0.2–0.3 µm spacing using a 100x, 1.35 NA objective on a DeltaVision Image Restoration Microscope (Applied Precision Instruments and Olympus). Immunofluorescence images in Figs. 1, and 3 were processed by iterative constrained deconvolution (SoftWoRx, Applied Precision Instruments) and corrected for chromatic aberration. Maximal intensity projections of the entire cell or of select optical sections (insets) spanning individual centromeres/kinetochores were converted to tiff files and linescans were generated using Image J. For immunofluorescence images in Figs. 2, 4, and 5, where intensities are compared, the acquired images, without deconvolution, were used to generate a maximum projection of the cell. Images for a given antibody staining were scaled identically, and this scale was then applied when converting to a tiff file. Images were cropped and adjusted for contrast equivalently in Image J. For quantifications of kinetochore pairing, randomly selected chromosome spreads were imaged and individual kinetochores were scored.
For quantification of antibody staining intensities at individual kinetochores, five to eight cells or chromosome spreads were randomly selected and acquired on a Deltavision Restoration microscope as described above. Images were scaled identically when converted to tiff files. Quantification of intensities at individual kinetochores was performed essentially as described in5. Briefly, using Metamorph software, integrated staining intensity was quantified from a region of interest drawn manually around a kinetochore. To subtract local background fluorescence, this region of interest was dilated by 6 pixels in Metamorph to generate an ‘outer’ region. The signal intensity of the ‘outer’ region was subtracted from the ‘inner’ kinetochore region, after scaling values for differences in area, as described5. Finally, the background-subtracted, kinetochore-staining intensity was divided by the area of the region of interest.
Supplementary Material
Figure S1. Live-cell localization analysis of PP2A subunits during mitosis. (a) Localization of the GFP-tagged scaffold during mitosis in HeLa cells. DIC images show chromosomes. Centrosome (arrow) and centromere (arrowhead) localizations are indicated. (b–e) Targeting of PP2A regulatory subunits during mitosis. For each regulatory subunit, cell lines stably expressing N- and C-terminal GFP fusions were analyzed, but for clarity only one cell line is presented. Fluorescence images GFP distribution at different stages of mitosis are presented alongside DIC images showing chromosomes. Regulatory subunits are listed by gene, with alternate names in parentheses. (b) The B56α, β, γ, δ, and ε subunits are enriched at mitotic centromeres/kinetochores. Arrows indicate central spindle targeting at anaphase. (c) GFP-PPP2R3B concentrated at puncta prior to anaphase. (d) GFP-PPP2R3C targeted to centrosomes (arrows). (e) PP2A regulatory subunits classified as cytosolic during cell division. Regulatory subunits were imaged in HeLa cell lines, except for B56γ and B56δ, which were images in RPE1 cell lines. All images are maximum intensity confocal projections except (d) where optical sections spanning the centrosomes are presented. Scale bars 5 µm.
Figure S2. Localization of endogenous B56α during mitosis. RPE1 cells were fixed and stained for kinetochores (CREST, red in overlay), B56α (green), and DNA (blue). Identically scaled maximum intensity projections are shown and an overlay is shown. (b) 3x enlargement of the optical sections spanning the boxed centromeres in (a) with DNA omitted. Plotted is the intensity profile of the CREST (red) and B56α (green) signal measured along a line (white) drawn across the centromere. Scale bar 5 µm.
Figure S3. Western blot analysis of B56 regulatory subunit depletion by siRNA and regulatory subunit over-expression in siRNA-resistant cell lines. (a) siRNA depletion of B56 regulatory subunits. Representative western blots for B56α, β, and δ from the siRNA experiments presented in Figs. 3-5. (b) Western blot confirming that siRNAs targeting B56γ and ε used in the indicated B56-PP2A siRNA pools depleted an over-expressed GFP fusion protein. The PP2A catalytic subunit is included as a loading control. (c) Representative western blot showing over-expression of HA-FLAG tagged, siRNA-resistant B56α and β in RPE1 cell lines. Varying amounts of cell lysate from the siRNA-resistant cell lines were run alongside RPE1 cell lysate. Full-length blots are presented in Fig. S6.
Figure S4. B56-PP2A siRNA cells arrest in mitosis and maintain Sgo1’s centromeric targeting. RPE1 cells were treated with the indicated siRNAs and arrested in mitosis with nocodazole (3.2 µM, 4h). (a) Cells were fixed and stained for DNA. The mitotic index is plotted (n=2 experiments, >800 cells counted per condition, per time). (b, c) Chromosome spreads were prepared and stained for kinetochores, and Sgo1 using two different antibodies. (b) Plotted is the frequency of chromosome spreads where centromeres were positive for Sgo1 (n=3 experiments, >80 cells counted per condition, per time. Fig. 3h shows a representative image. (c) The Sgo1 signal at kinetochores was measured, processed, and normalized to the average value in control cells. Histograms show intensity distributions from one experiment. Sgo1 staining intensities in B56-PP2A siRNA treated cells 1.07 ± 0.34 (pool 1) or 1.40 ± 0.15 (pool 2) were calculated relative to control cells (n=2 experiments, >50 centromeres analyzed per condition, per experiment). Mean ± s.e.m presented.
Figure S5. Measurement of inter-kinetochore distance in B56-PP2A siRNA cells. RPE1 cells were treated with the indicated siRNAs and arrested in mitosis with nocodazole (3.2 µM, 60 min) or MG132 (10 µM, 60 min) prior to fixation. Plotted is the distribution of inter-kinetochore distances from at least 60 kinetochores from five randomly selected cells. The mean inter-kinetochore distance (d) for each condition is indicated ± s.e.m. Similar results were obtained in two independent experiments.
Figure S6. Full scans of electrophoretic data from Fig. S3. Boxes indicate edges of blot (black) or regions relevant to Fig. S3b–c (red).
Acknowledgements
We thank Iain Cheeseman, Arshad Desai, Sabine Elowe, and Hongtao Yu for antibodies and Wade Harper for plasmids. E.A.F. was supported by a Damon Runyon Cancer Research postdoctoral fellowship (DRG 1936-07) and the Charles H. Revson Foundation. This work was supported by NIH GM65933 (T.M.K.).
Footnotes
The authors declare no competing financial interests.
Author Contributions
E.A.F. and T.M.K. designed the experiments and wrote the paper. E.A.F. carried out essentially all the experiments. M.M. contributed to the live cell imaging.
References
- 1.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 2.Ahonen LJ, et al. Polo-like kinase 1 creates the tension-sensing 3F3/2 phosphoepitope and modulates the association of spindle-checkpoint proteins at kinetochores. Curr Biol. 2005;15:1078–1089. doi: 10.1016/j.cub.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 3.Cheeseman IM, et al. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 2002;111:163–172. doi: 10.1016/s0092-8674(02)00973-x. [DOI] [PubMed] [Google Scholar]
- 4.Lampson MA, Renduchitala K, Khodjakov A, Kapoor TM. Correcting improper chromosome-spindle attachments during cell division. Nat Cell Biol. 2004;6:232–237. doi: 10.1038/ncb1102. [DOI] [PubMed] [Google Scholar]
- 5.Pinsky BA, Kung C, Shokat KM, Biggins S. The Ipl1-Aurora protein kinase activates the spindle checkpoint by creating unattached kinetochores. Nat Cell Biol. 2006;8:78–83. doi: 10.1038/ncb1341. [DOI] [PubMed] [Google Scholar]
- 6.Welburn JP, et al. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol Cell. 2010;38:383–392. doi: 10.1016/j.molcel.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wong OK, Fang G. Plx1 is the 3F3/2 kinase responsible for targeting spindle checkpoint proteins to kinetochores. J Cell Biol. 2005;170:709–719. doi: 10.1083/jcb.200502163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu D, Vader G, Vromans MJ, Lampson MA, Lens SM. Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science. 2009;323:1350–1353. doi: 10.1126/science.1167000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Westermarck J, Hahn WC. Multiple pathways regulated by the tumor suppressor PP2A in transformation. Trends Mol Med. 2008;14:152–160. doi: 10.1016/j.molmed.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 10.Ubersax JA, Ferrell JE., Jr. Mechanisms of specificity in protein phosphorylation. Nat Rev Mol Cell Biol. 2007;8:530–541. doi: 10.1038/nrm2203. [DOI] [PubMed] [Google Scholar]
- 11.Trinkle-Mulcahy L, et al. Time-lapse imaging reveals dynamic relocalization of PP1gamma throughout the mammalian cell cycle. Mol Biol Cell. 2003;14:107–117. doi: 10.1091/mbc.E02-07-0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kitajima TS, et al. Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature. 2006;441:46–52. doi: 10.1038/nature04663. [DOI] [PubMed] [Google Scholar]
- 13.Riedel CG, et al. Protein phosphatase 2A protects centromeric sister chromatid cohesion during meiosis I. Nature. 2006;441:53–61. doi: 10.1038/nature04664. [DOI] [PubMed] [Google Scholar]
- 14.Tang Z, et al. PP2A is required for centromeric localization of Sgo1 and proper chromosome segregation. Dev Cell. 2006;10:575–585. doi: 10.1016/j.devcel.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 15.Liu D, et al. Regulated targeting of protein phosphatase 1 to the outer kinetochore by KNL1 opposes Aurora B kinase. J Cell Biol. 2010;188:809–820. doi: 10.1083/jcb.201001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Posch M, et al. Sds22 regulates aurora B activity and microtubule-kinetochore interactions at mitosis. J Cell Biol. 2010;191:61–74. doi: 10.1083/jcb.200912046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Janssens V, Goris J. Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem J. 2001;353:417–439. doi: 10.1042/0264-6021:3530417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baharians Z, Schonthal AH. Autoregulation of protein phosphatase type 2A expression. J Biol Chem. 1998;273:19019–19024. doi: 10.1074/jbc.273.30.19019. [DOI] [PubMed] [Google Scholar]
- 19.Hutchins JR, et al. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science. 2010;328:593–599. doi: 10.1126/science.1181348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffman DB, Pearson CG, Yen TJ, Howell BJ, Salmon ED. Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol Biol Cell. 2001;12:1995–2009. doi: 10.1091/mbc.12.7.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waters JC, Chen RH, Murray AW, Salmon ED. Localization of Mad2 to kinetochores depends on microtubule attachment, not tension. J Cell Biol. 1998;141:1181–1191. doi: 10.1083/jcb.141.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neumann B, et al. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature. 2010;464:721–727. doi: 10.1038/nature08869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cho US, Xu W. Crystal structure of a protein phosphatase 2A heterotrimeric holoenzyme. Nature. 2007;445:53–57. doi: 10.1038/nature05351. [DOI] [PubMed] [Google Scholar]
- 24.Xu Y, et al. Structure of the protein phosphatase 2A holoenzyme. Cell. 2006;127:1239–1251. doi: 10.1016/j.cell.2006.11.033. [DOI] [PubMed] [Google Scholar]
- 25.Rieder CL. The structure of the cold-stable kinetochore fiber in metaphase PtK1 cells. Chromosoma. 1981;84:145–158. doi: 10.1007/BF00293368. [DOI] [PubMed] [Google Scholar]
- 26.Kitajima TS, Kawashima SA, Watanabe Y. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature. 2004;427:510–517. doi: 10.1038/nature02312. [DOI] [PubMed] [Google Scholar]
- 27.Rabitsch KP, et al. Two fission yeast homologs of Drosophila Mei-S332 are required for chromosome segregation during meiosis I and II. Curr Biol. 2004;14:287–301. doi: 10.1016/j.cub.2004.01.051. [DOI] [PubMed] [Google Scholar]
- 28.Salic A, Waters JC, Mitchison TJ. Vertebrate shugoshin links sister centromere cohesion and kinetochore microtubule stability in mitosis. Cell. 2004;118:567–578. doi: 10.1016/j.cell.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 29.Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 30.Lampson MA, Cheeseman IM. Sensing centromere tension: Aurora B and the regulation of kinetochore function. Trends Cell Biol. 2010 doi: 10.1016/j.tcb.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ditchfield C, et al. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J Cell Biol. 2003;161:267–280. doi: 10.1083/jcb.200208091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hauf S, et al. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J Cell Biol. 2003;161:281–294. doi: 10.1083/jcb.200208092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lenart P, et al. The small-molecule inhibitor BI 2536 reveals novel insights into mitotic roles of polo-like kinase 1. Curr Biol. 2007;17:304–315. doi: 10.1016/j.cub.2006.12.046. [DOI] [PubMed] [Google Scholar]
- 34.Elowe S, Hummer S, Uldschmid A, Li X, Nigg EA. Tension-sensitive Plk1 phosphorylation on BubR1 regulates the stability of kinetochore microtubule interactions. Genes Dev. 2007;21:2205–2219. doi: 10.1101/gad.436007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elia AE, Cantley LC, Yaffe MB. Proteomic screen finds pSer/pThr-binding domain localizing Plk1 to mitotic substrates. Science. 2003;299:1228–1231. doi: 10.1126/science.1079079. [DOI] [PubMed] [Google Scholar]
- 36.Rieder CL, Alexander SP. Kinetochores are transported poleward along a single astral microtubule during chromosome attachment to the spindle in newt lung cells. J Cell Biol. 1990;110:81–95. doi: 10.1083/jcb.110.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McEwen BF, Heagle AB, Cassels GO, Buttle KF, Rieder CL. Kinetochore fiber maturation in PtK1 cells and its implications for the mechanisms of chromosome congression and anaphase onset. J Cell Biol. 1997;137:1567–1580. doi: 10.1083/jcb.137.7.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosenberg JS, Cross FR, Funabiki H. KNL1/Spc105 Recruits PP1 to Silence the Spindle Assembly Checkpoint. Curr Biol. 2011;21:942–947. doi: 10.1016/j.cub.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruediger R, Pham HT, Walter G. Disruption of protein phosphatase 2A subunit interaction in human cancers with mutations in the A alpha subunit gene. Oncogene. 2001;20:10–15. doi: 10.1038/sj.onc.1204059. [DOI] [PubMed] [Google Scholar]
- 40.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
Materials and Methods References
- 1.Welburn JP, et al. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol Cell. 2010;38:383–392. doi: 10.1016/j.molcel.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elowe S, Hummer S, Uldschmid A, Li X, Nigg EA. Tension-sensitive Plk1 phosphorylation on BubR1 regulates the stability of kinetochore microtubule interactions. Genes Dev. 2007;21:2205–2219. doi: 10.1101/gad.436007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kline SL, Cheeseman IM, Hori T, Fukagawa T, Desai A. The human Mis12 complex is required for kinetochore assembly and proper chromosome segregation. J Cell Biol. 2006;173:9–17. doi: 10.1083/jcb.200509158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tang Z, et al. PP2A is required for centromeric localization of Sgo1 and proper chromosome segregation. Dev Cell. 2006;10:575–585. doi: 10.1016/j.devcel.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 5.Hoffman DB, Pearson CG, Yen TJ, Howell BJ, Salmon ED. Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol Biol Cell. 2001;12:1995–2009. doi: 10.1091/mbc.12.7.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Live-cell localization analysis of PP2A subunits during mitosis. (a) Localization of the GFP-tagged scaffold during mitosis in HeLa cells. DIC images show chromosomes. Centrosome (arrow) and centromere (arrowhead) localizations are indicated. (b–e) Targeting of PP2A regulatory subunits during mitosis. For each regulatory subunit, cell lines stably expressing N- and C-terminal GFP fusions were analyzed, but for clarity only one cell line is presented. Fluorescence images GFP distribution at different stages of mitosis are presented alongside DIC images showing chromosomes. Regulatory subunits are listed by gene, with alternate names in parentheses. (b) The B56α, β, γ, δ, and ε subunits are enriched at mitotic centromeres/kinetochores. Arrows indicate central spindle targeting at anaphase. (c) GFP-PPP2R3B concentrated at puncta prior to anaphase. (d) GFP-PPP2R3C targeted to centrosomes (arrows). (e) PP2A regulatory subunits classified as cytosolic during cell division. Regulatory subunits were imaged in HeLa cell lines, except for B56γ and B56δ, which were images in RPE1 cell lines. All images are maximum intensity confocal projections except (d) where optical sections spanning the centrosomes are presented. Scale bars 5 µm.
Figure S2. Localization of endogenous B56α during mitosis. RPE1 cells were fixed and stained for kinetochores (CREST, red in overlay), B56α (green), and DNA (blue). Identically scaled maximum intensity projections are shown and an overlay is shown. (b) 3x enlargement of the optical sections spanning the boxed centromeres in (a) with DNA omitted. Plotted is the intensity profile of the CREST (red) and B56α (green) signal measured along a line (white) drawn across the centromere. Scale bar 5 µm.
Figure S3. Western blot analysis of B56 regulatory subunit depletion by siRNA and regulatory subunit over-expression in siRNA-resistant cell lines. (a) siRNA depletion of B56 regulatory subunits. Representative western blots for B56α, β, and δ from the siRNA experiments presented in Figs. 3-5. (b) Western blot confirming that siRNAs targeting B56γ and ε used in the indicated B56-PP2A siRNA pools depleted an over-expressed GFP fusion protein. The PP2A catalytic subunit is included as a loading control. (c) Representative western blot showing over-expression of HA-FLAG tagged, siRNA-resistant B56α and β in RPE1 cell lines. Varying amounts of cell lysate from the siRNA-resistant cell lines were run alongside RPE1 cell lysate. Full-length blots are presented in Fig. S6.
Figure S4. B56-PP2A siRNA cells arrest in mitosis and maintain Sgo1’s centromeric targeting. RPE1 cells were treated with the indicated siRNAs and arrested in mitosis with nocodazole (3.2 µM, 4h). (a) Cells were fixed and stained for DNA. The mitotic index is plotted (n=2 experiments, >800 cells counted per condition, per time). (b, c) Chromosome spreads were prepared and stained for kinetochores, and Sgo1 using two different antibodies. (b) Plotted is the frequency of chromosome spreads where centromeres were positive for Sgo1 (n=3 experiments, >80 cells counted per condition, per time. Fig. 3h shows a representative image. (c) The Sgo1 signal at kinetochores was measured, processed, and normalized to the average value in control cells. Histograms show intensity distributions from one experiment. Sgo1 staining intensities in B56-PP2A siRNA treated cells 1.07 ± 0.34 (pool 1) or 1.40 ± 0.15 (pool 2) were calculated relative to control cells (n=2 experiments, >50 centromeres analyzed per condition, per experiment). Mean ± s.e.m presented.
Figure S5. Measurement of inter-kinetochore distance in B56-PP2A siRNA cells. RPE1 cells were treated with the indicated siRNAs and arrested in mitosis with nocodazole (3.2 µM, 60 min) or MG132 (10 µM, 60 min) prior to fixation. Plotted is the distribution of inter-kinetochore distances from at least 60 kinetochores from five randomly selected cells. The mean inter-kinetochore distance (d) for each condition is indicated ± s.e.m. Similar results were obtained in two independent experiments.
Figure S6. Full scans of electrophoretic data from Fig. S3. Boxes indicate edges of blot (black) or regions relevant to Fig. S3b–c (red).