Abstract
Oxidative stress in cells and tissues leads to the formation of an assortment of lipid electrophiles, such as the quantitatively important 4-hydroxy-2-trans-nonenal (HNE). Although this cytotoxic aldehyde is atherogenic the mechanisms involved are unclear. We hypothesize that elevated HNE levels can directly inactivate esterase and lipase activities in macrophages via protein adduction, thus generating a biochemical lesion that accelerates foam cell formation and subsequent atherosclerosis. In the present study we examined the effects of HNE treatment on esterase and lipase activities in human THP1 monocytes/macrophages at various physiological scales (i.e., pure recombinant enzymes, cell lysate, and intact living cells). The hydrolytic activities of bacterial and human carboxylesterase enzymes (pnbCE and CES1, respectively) were inactivated by HNE in vitro in a time- and concentration-dependent manner. In addition, so were the hydrolytic activities of THP1 cell lysates and intact THP1 monocytes and macrophages. A single lysine residue (Lys105) in recombinant CES1 was modified by HNE via a Michael addition reaction, whereas the lone reduced cysteine residue (Cys389) was found unmodified. The lipolytic activity of cell lysates and intact cells was more sensitive to the inhibitory effects of HNE than the esterolytic activity. Moreover, immunoblotting analysis using HNE antibodies confirmed that several cellular proteins were adducted by HNE following treatment of intact THP1 monocytes, albeit at relatively high HNE concentrations (>50 µM). Unexpectedly, in contrast to CES1, the treatment of a recombinant human CES2 with HNE enhanced its enzymatic activity ~3-fold compared to untreated enzyme. In addition, THP1 monocytes/macrophages can efficiently metabolize HNE, and glutathione conjugation of HNE is responsible for ~43% of its catabolism. The functional importance of HNE-mediated inactivation of cellular hydrolytic enzymes with respect to atherogenesis remains obscure, although this study has taken a first step toward addressing this important issue by examining the potential of HNE to inhibit this biochemical activity in a human monocyte/macrophage cell line.
Keywords: carboxylesterases, 4-hydroxynonenal, THP1 monocytes, THP1 macrophages
1. Introduction
Oxidative stress in cells and tissues occurs when the levels of reactive oxygen species (ROS) and reactive nitrogen species (RNS) exceed amounts that the body can detoxify. This imbalance can lead to lipid peroxidation, a free radical chain reaction by which ROS/RNS oxidize lipids in the cell membrane, thus leading to the formation of an assortment of reactive aldehydes, including 4-hydroxy-2-trans-nonenal (HNE), an α,β-unsaturated aldehyde that is a diffusible lipid electrophile [1]. Esterified ω6-polyunsaturated fatty acids, such as arachidonic acid and linoleic acid, are particularly prone to peroxidation reactions. HNE is one of the most abundant reactive aldehydes found in cell membranes and it is also a toxic by-product of oxidized low-density lipoproteins (oxLDLs) [2–4]; local HNE concentrations have been estimated to be in the millimolar range in cell membranes [5, 6] and oxidized low-density lipoproteins [7]. Although HNE is a reactive molecule, it can modify cellular targets far from the site of its origination, including proteins and DNA [8]. Reaction of proteins with HNE typically occurs via initial Michael addition to histidine, cysteine, or lysine, which subsequently cyclize to yield hemiacetals. Schiff base formation between lysine and HNE is also observed, but is less frequent than Michael addition [1]. HNE can chemically modify a large number of proteins, thereby altering their structure and function [9], and it also significantly modulates the transcriptional program in cells [1]. Conjugation of HNE with the tripeptide glutathione (GSH) is a major pathway of detoxification and GST A4-4 is the predominant isoform of glutathione transferase responsible for this reaction [10]. Interestingly, a recent study indicated that GSTA4 deficient mice were prone to obesity due to their inability to detoxify endogenous HNE, thus resulting in dysregulated lipid metabolism and weight gain [11]. Furthermore, because it is associated with oxLDL, HNE has gained increased attention and has been linked to the development of atherosclerosis [12–14]. For example, HNE has been shown to induce the scavenger receptor CD36 in macrophages and to promote foam cell formation [15]. Further, apoB-100 protein within oxLDL particles is covalently modified by HNE, which may contribute to the atherogenic properties of these lipoproteins [16]. Although this cytotoxic aldehyde is known to be pro-atherogenic the mechanisms that account for its pathogenicity remain enigmatic [15, 17].
Atherosclerosis is in part a manifestation of the inflammatory response of macrophages and lymphocytes to pathogenic lipoproteins [18, 19]. In particular, numerous studies have shown that monocyte-derived macrophages play a major role in atherogenesis by promoting the formation of atherosclerotic lesions via uptake of cholesterol-loaded oxLDL and subsequent foam cell formation [19–23]. The importance of macrophages in this pathology was underscored by Smith et al. [22] who showed that macrophage-deficient hypercholesterolemic mice are drastically resistant to atherosclerosis. Although these findings help explain the histological formation of lesions, they do not sufficiently elucidate the molecular pathways that may promote or inhibit the atherogenic processes. Foam cell formation results from an imbalance of cholesterol influx and efflux, which is in part caused by the dysregulation of hydrolytic enzymes [24]. Hydrolytic enzymes are classified on the basis of the substrates that they hydrolyze; for example, esterases and lipases hydrolyze water soluble and water insoluble esters, respectively. Accordingly, in the present study we examined the effects of the lipid electrophile HNE on esterase and lipase activities in intact human THP1 monocytes/macrophages [25] and on recombinant carboxylesterase (CES) 1 enzyme, which is the most abundant serine hydrolase expressed in this cell line [26] and is proposed to regulate the levels of cholesterol and lipid glyceryl esters in macrophages [24, 27]. We examined the effects of HNE on esterase and lipase activities at various physiological scales (i.e. pure recombinant protein, cell lysate, intact cells) and the extent of HNE-protein adduct formation. In addition, we examined the catabolism of HNE by THP1 monocytes/macrophages and whether this reactive aldehyde could induce CES1 protein levels. Our working hypothesis for these studies is that elevated HNE levels inactivate esterase and lipase activities in macrophages, thus generating a biochemical lesion that contributes to foam cell formation and subsequent atherosclerosis.
2. Materials and methods
2.1. Materials
4-Hydroxynonenal, 4-oxononenal, and the lactate dehydrogenase (LDH) cytotoxicity kit were from Cayman Chemical (Ann Arbor, MI). Human THP1 monocytes, RPMI-1640 medium, gentamicin sulfate solution (50mg/ml), and Hanks' balanced salt solution without calcium, magnesium and phenol red were purchased from the American Type Culture Collection (ATCC Manassas, VA). Fetal bovine serum (FBS) was purchased from Invitrogen (Carlsbad, CA). Trypan Blue solution (0.4%), β-mercaptoethanol, 4-methylumbelliferyl oleate (4-MUBO), p-nitrophenyl valerate (pNPV), monoclonal antibodies against β-actin, phorbol 12-myristate 13-acetate (PMA), and all components of the buffers were purchased from Sigma (St. Louis, MO). The activity-based serine hydrolase probe, fluorophosphonate-biotin (FP-biotin) was from Toronto Research Chemicals (North York, Ontario). HPLC grade solvents were from Burdick and Jackson. Polyvinylidine fluoride (PVDF), goat anti-rabbit IgG secondary antibody conjugated to horse radish peroxidase (HRP), and goat anti-mouse IgG secondary antibody conjugated to HRP were purchased from Bio-Rad (Hercules, CA). SuperSignal WestPico Chemiluminescent and BCA protein reagents were purchased from Thermo Scientific (Rockford, IL). Anti-HNE antibodies were purchased from Oxis (Foster City, CA). Recombinant bacterial pnbCE protein was expressed in Escherichia coli and purified as previously described [28]. Recombinant human CES1 (isoform C) and CES2 proteins were expressed in baculovirus-infected Spodoptera frugiperda cells and purified [29].
2.2. Culture conditions
THP1 monocytes were grown in suspension in RPMI-1640 medium supplemented with 10% FBS, 0.05 mM β-mercaptoethanol, and 50 µg gentamicin/ml (which is defined as complete growth medium) at 37°C and 5% CO2. The cells were grown at a density between 0.2 × 106 and 1 × 106 cells/ml, as recommended by ATCC. THP1 monocytes were differentiated into macrophages by incubating in complete growth medium containing 100 nM PMA for 24–48 h at 37°C and 5% CO2.
2.3. Preparation of cell lysates
THP1 monocyte and macrophage cell lysates were prepared in 100 mM Tris-HCl (pH 7.4) buffer as previously described [27]. Membrane and cytosolic fractions were obtained by ultracentrifugation of monocyte lysate (100,000×g, 60 min, 4°C). Protein concentrations of cell lysates and subcellular fractions were determined using the BCA reagent according to the manufacturer's instructions.
2.4. Treatment of recombinant carboxylesterases with HNE
Recombinant pnbCE or CES1 protein was diluted to 50 µg/ml (0.83 µM) in 100 µL of 50 mM Tris-HCl (pH 7.4) containing various concentrations of HNE (0, 0.1, 0.5, and 3.2 mM) and incubated at 37° C. HNE stock solutions were in ethanol; therefore, ethanol was used for the vehicle control reactions (<1% v/v). At various time points (pnbCE: 0, 90, 150, 210, and 270 min; CES1: 0, 1, 2, 3, 4, 24, 48, and 96 h), a 10 µL aliquot from each reaction was removed and diluted in 50 mM Tris-HCl (pH 7.4) buffer to measure the residual esterase activity using pNPV as substrate [30]. The final concentrations of enzyme and substrate in the esterase assay were 0.17 µg/ml (~300-fold dilution) and 500 µM, respectively, in a total volume of 300 µl. Preliminary incubations of HNE and recombinant enzyme were done in either 50 mM potassium phosphate (pH 7.4) or 50 mM Tris-HCl (pH 7.4) buffer; no difference in the extent of HNE-mediated inhibition of carboxylesterase activity was observed when the two buffers were compared, indicating that Tris buffer was acceptable to use for these experiments. An additional reaction contained CES1 protein and 4-oxononenal (1 mM), which was incubated for 1h at 37°C prior to assaying its esterase activity. Furthermore, recombinant pnbCE and CES1 proteins were treated with HNE (3.2 mM final concentration) or ethanol vehicle for 5 h, followed by analysis by SDS-PAGE and western blotting for the presence of HNE-protein adducts (see below).
In a separate experiment, CES1 and CES2 proteins (0.033 µM, 0.2 µg) were treated with 1 mM HNE or ethanol vehicle in 100 µl of 50 mM Tris-HCl (pH 7.4) for 15 min at 37°C. Because the ethanol vehicle that HNE is dissolved in can significantly inhibit CES2 activity (data not shown), 6 µl aliquots of stock HNE solution (100 nmol) or ethanol (vehicle control) were added to separate microfuge tubes and the ethanol was evaporated in a vacuum centrifuge. One-hundred µl of a freshly prepared dilute enzyme/buffer solution was then immediately added to each dried tube and incubated for 15 min (37°C) at 600 rpm. The treated enzymes were then desalted using Zeba spin desalting columns (7kDa MWCO, Thermo) and assayed for carboxylesterase activity using pNPV.
2.5.1. Covalent modification of CES1 and mass spectrometry
Recombinant CES1 protein (1 µM, 5 µg) was treated with HNE (100 or 1000 µM) or vehicle (ethanol) in 50 mM Hepes (pH 7.4). The final amount of ethanol was 1.6% v/v. The reactions proceeded for 2h (37°C) at which point HNE-protein adducts were stabilized by addition of 10 µl of 100 mM NaBH4 (dissolved in 1M NaOH) and further incubated for 30 min at 37°C. The samples were desalted using 3kDa MWCO microfuge filters (Millipore) and buffer exchanged 3× with 10 mM potassium phosphate (pH 7.0). The final volume of samples after desalting/buffer exchange was ~30 µl. Samples were subjected to electrophoresis on a 10% SDS-PAGE gel. The proteins were visualized with Coomassie blue, excised, and trypsin digested as described [31]. After digestion, the samples were dried in a vacuum centrifuge and the tryptic peptides solubilized in 50 µl of water containing 2% acetonitrile/0.1% formic acid. Peptides were analyzed by LC-MS/MS by using a 75-µm i.d. × 15 cm reverse phase column (fused-silica C18 column, Thermo) controlled by an Ultimate 3000 nanoflow HPLC (Dionex). Peptides were eluted using a 50-minute gradient from 2%–95% solvent B (95% acetonitrile, 0.1% formic acid) at a flow rate of 0.5 µl/min, and further introduced into an OrbiTrap Velos mass spectrometer (Thermo Fisher). The OrbiTrap mass spectrometer was operated in data-dependent mode, automatically switching between MS and MS/MS. Full scan MS spectra (300–2,000 amu) were acquired with a resolution of 60,000 and analyzed by FTMS analyzer. The five most intense ions were selected for collision-induced fragmentation in the OrbiTrap at normalized collision energy of 35% and activation time of 10 ms. The acquired data were analyzed by Proteome Discoverer 1.1 (Thermo Fisher) using SEQUEST algorithm and a human IPI database as well as a reversed decoy database. Searches were done using a precursor and fragment tolerance of 0.5 Da and the following dynamic modifications were included: HNE+Delta:H(2) (+158.131 Da) on cysteine, histidine and lysine; oxidation on methionine; and carbamidomethylation on cysteine. The results were filtered using normalized XCorr values for different charge states [32] and were accepted as valid identifications only if the precursor ion mass accuracy was below 10 ppm and the XCorr values were >1.7, >2, and >3 for singly, doubly and triply charged peptides, respectively. More-over, for mapping of the post-translational modifications, each spectrum was manually inspected and accepted only if a series of at least 4 continuous y or b ions was observed.
2.6. Treatment of THP1 cell lysates with HNE
Cell lysates of monocytes or macrophages were diluted to 500 µg protein/ml in 50 mM Tris-HCl (pH 7.4) (for esterase assay) or 10 mM Tris-HCl (pH 7.4)/150 mM NaCl/0.01% Triton X-100 (for lipase assay) containing different concentrations of HNE (0, 0.1, 0.5, and 3.2 mM) and incubated at 37°C (ethanol vehicle, <1% v/v). At different time points (0, 1, 2, 3, 4, and 5 h), a 50 µl aliquot (25 µg protein) from each reaction was taken to measure the residual esterase activity of the lysate. The final concentration of protein in the esterase assay was 16.7 µg/ml (~30-fold dilution). For the lipase assay, a 150 µl aliquot from each reaction was diluted in lipase assay buffer (10 mM Tris-HCl, 150 mM NaCl, 0.01% Triton X-100, pH 7.4) to yield a final protein concentration of 83.3 µg/ml (~6-fold dilution).
2.7. Treatment of intact THP1 cells with HNE
THP1 monocytes (3 × 106 cells) were placed into T-25 flasks in 5 mL of complete growth medium. HNE from the ethanol stock solution was added to the flasks to give the desired final concentration of compound. The final amount of ethanol in the medium in all flasks was always 1% (v/v). The treated cells were incubated at 37°C in 95% air/5% CO2 for either 2 h or 17 h. Following the HNE exposure, cell viability was assessed using Trypan Blue exclusion. Cells were then harvested by centrifugation (100 × g) and washed three times with PBS to remove excess HNE. Cell pellets were then re-suspended in 100 mM Tris-HCl (pH 7.4) and lysed by sonication. The resulting cell lysates were used in the hydrolysis assays described below.
Alternatively, monocytes were seeded into wells of a 96-well plate in complete growth medium supplemented with PMA (100 nM) in order to differentiate cells into macrophages (24 h). After washing with PBS, the cells received serum-free RPMI culture medium supplemented with HNE (indicated concentrations shown in figures) or ethanol vehicle. The macrophages were treated with HNE for 2 h (37°C, 5% CO2), at which point the medium was removed and the cells washed with PBS. The LDH assay was used to determine cell viability of macrophages treated with HNE (LDH kit from Cayman Chemicals). Positive controls (100% cell death) consisted of macrophages treated with RPMI supplemented with 0.1% Triton X-100 (2 h, 37°C, 5% CO2) and negative controls consisted of macrophages treated only with RPMI (no supplements) (2 h, 37°C, 5% CO2).
2.8. Hydrolytic activity assays
2.8.1. Hydrolysis of p-nitrophenyl valerate (esterase activity)
Esterase assays to measure the activity of recombinant enzyme, THP1 cell lysates or intact cells following treatments with HNE were performed as previously described [30]. Data were expressed as percent of vehicle (ethanol) control.
2.8.2. Hydrolysis of 4-methylumbelliferyl oleate (lipase activity)
Hydrolysis of the fluorogenic substrate 4-MUBO by lysates of HNE-treated THP1 cells was done as previously described [26]. Data were expressed as percent of vehicle (ethanol) control.
2.8.3. Hydrolysis of 2-arachidonoylglycerol (2AG)
Hydrolysis of 2AG and the analysis of its product (arachidonic acid) by LC-MS were done as described previously [27].
2.8.4. Detection of serine hydrolases in THP1 cell lysates using the activity-based protein probe, fluorophosphonate (FP)-biotin
Cytosolic and membrane fractions of THP1 monocytes (1 mg/ml protein in 50 mM Tris-HCl, pH 7.4) were treated with FP-biotin (2 µM, final) and analyzed as previously described [27].
2.8.5. In situ determination of esterase and lipase activities of macrophages
Adherent THP1 macrophages were challenged with HNE for 2 h in a 96-well format, as described above. After 2h, the culture medium was removed and cells were washed with PBS. Three hundred µl of 0.5 mM pNPV, prepared in PBS, was immediately added to each well to determine the in situ esterase activity of the cells [33]. The rate of intracellular hydrolysis of pNPV was determined by monitoring the absorbance (405 nm) caused by release of p-nitrophenol using a plate reader (Molecular Devices). Negative control reactions contained ethanol vehicle instead of HNE, and positive control reactions contained 10 µM paraoxon since it can effectively inhibit CES1 activity in intact THP1 cells [26]. Data are reported as % of control esterase activity (ethanol vehicle). For lipase activity determinations, 150 µl of 50 mM Tris-HCl (pH 7.4) containing 0.1% v/v Triton X-100 was added to the washed cells and the plate was incubated at room temperature for 30 min with gentle shaking to lyse the cells. The 4-MUBO activity of the lysate was then determined by adding 150 µl of 150 µM 4-MUBO, suspended in 10 mM Tris-HCl (pH 7.4) containing 150 mM NaCl and 0.01% Triton X-100, to each well and monitoring the fluorescence caused by the release of 4-methylumbelliferone using λex 355 nm and λem 460 nm.
2.9. Western blot analysis
Cell lysates or recombinant proteins were subjected to SDS-PAGE as described by Laemmli [34]. Following electrophoresis, proteins were transferred to PVDF membranes and probed for HNE protein adducts with mouse anti-HNE antibodies. For western blot analysis of CES1 and β-actin, PVDF membranes were probed with rabbit anti-CES1 and mouse anti-β-actin.
2.10. Glutathione conjugation of HNE
THP1 monocytes (~1×106 cells) were suspended in 50 µl of PBS and treated with 50 µM HNE or vehicle (ethanol) for 5 min at 37°C. The cells were lysed by adding 100 µl of ice-cold methanol containing an internal standard (1 nmol atrazine-mercapturate, synthesized as described in Ross and Filipov [35]) and sonicated. After brief centrifugation the supernatant was analyzed by LC-MS (20 µl injection) using the conditions described below. In addition, lysates of naïve monocytes and macrophages (0.5 mg protein/ml, final) were supplemented with 5 mM GSH and 50 µM HNE in 50 mM Tris-HCl (pH 7.4) buffer, and incubated for 5 min at 37°C. The reactions were terminated with two volumes of ice-cold methanol/acetic acid (90/10 v/v) containing 1 nmol of internal standard (atrazine-mercapturate), followed by LC-UV-MS analysis. For LC-UV-MS, a Thermo MSQ single-quadrupole mass spectrometer was interfaced with a Surveyor LC pump and UV detector (Thermo Fisher Scientific, San Jose, CA). The mobile phases were a blend of solvent A (0.1% v/v acetic acid in water) and solvent B (0.1% v/v acetic acid in acetonitrile). Samples (20 µl) were injected onto a C18 column (2.1 mm × 100 mm, Thermo) equipped with guard column, and analytes were eluted with the following gradient program: 0 min (95% A, 5% B), 2 min (95% A, 5% B), 12 min (10% A, 90% B), 17 min (10% A, 90% B), 20 min (95% A, 5% B), and 25 min (95% A, 5% B). Flow rate was 0.3 mL/min and column eluate was directed into a UV detector (λ 230nm) and mass spectrometer in tandem. Ions were introduced into the MS by electrospray ionization in positive or negative ion modes. The MS single quadrupole was operated in single-ion mode (SIM) to detect the following ions: GS-HNE ([M-H]− m/z 462), atrazine-mercapturate ([M-H] − m/z 341), GS-HNE ([M+H]+ m/z 464), and HNE ([M+H]+ m/z 157). Glutathione transferase (GST) activity of monocytes and macrophages were determined according to the method of Habig [36] using 1-chloro-2,4-dinitrobenzene as substrate.
2.11 Statistical analysis
Significant differences were determined by Student’s t-test or ANOVA using Tukey or Dunnett’s post-hoc tests (SigmaStat). Significant differences are indicated by letters or asterisks in the figures (p<0.05).
3. Results and Discussion
3.1 Inhibition of recombinant carboxylesterase activity by HNE
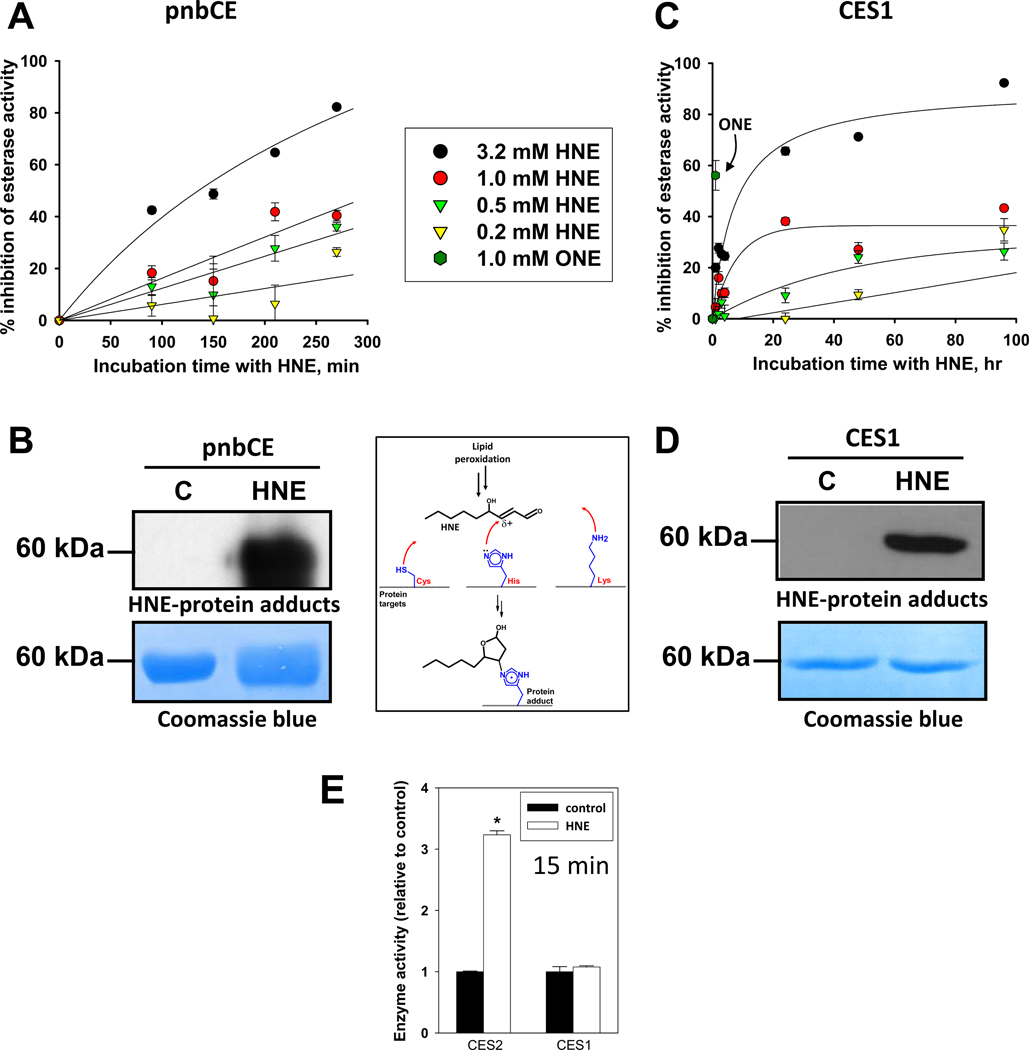
Bacterial pnbCE enzyme is a model CES with high sequence and structural homology to the mammalian CES [28]. Treatment of recombinant pnbCE with increasing concentrations of HNE caused the esterase activity of the enzyme to be inhibited in a time- and HNE concentration-dependent manner (Fig. 1A). Nearly 90% of the pnbCE esterase activity was inhibited following treatment with 3.2 mM HNE at 4.5 hours. Western blot analysis using antibodies that recognize HNE adducts demonstrated that the pnbCE protein was covalently modified by HNE when treated with 3.2 mM HNE, whereas the vehicle control-treated protein was not (Fig. 1B). Recombinant human CES1 enzyme was also sensitive to covalent modification and inhibition by HNE, although longer incubation times were necessary to achieve the same degree of inhibition seen with pnbCE (Fig. 1C,D). Prolonged incubation times at 37°C up to 96 hours were feasible with CES1 protein due to its remarkable stability, whereas pnbCE was much less stable during extended incubations (data not shown). The stability of CES1 may be attributed to the fact that it is a glycosylated protein, whereas the bacterial protein is not. In addition, treatment of CES1 with 4-oxononenal (ONE; 1 mM, 1 h), another reactive aldehyde formed during lipid peroxidiation, inhibited the esterase activity of CES1 by nearly 60% (Fig. 1C), which is markedly higher than the level of inhibition when CES1 was treated with 1 mM HNE for the same amount of time. This result is consistent with the greater chemical reactivity of ONE compared to HNE [37]. Fitting HNE inhibition data in Fig. 1C to a single-phase exponential equation, %inhib = %inhibmax (1-e−kobs*t), allowed secondary plots of the apparent first-order rate constant of enzyme inhibition (kobs) versus [HNE] to be produced, which, assuming that the rate of adduction of CES1 by HNE is reflected by the rate of enzyme inhibition, permits an estimate of the second-order rate constant (kinact/Ki) for the covalent reaction of HNE and CES1 to be estimated (i.e., 0.73 h−1mM−1; Supplementary Fig. 1). Additionally, the first-order rate constant (kinact) that describes the maximal rate of CES1 inactivation by HNE was 0.19 h−1, which is in line with the reactivity of human serum albumin toward HNE [38].
Fig. 1.
Inhibition of esterase activity of recombinant carboxylesterases (pnbCE and CES1) following treatment with HNE (A,C) and formation of HNE-protein adducts (B,D). The pnbCE and CES1 recombinant proteins were treated with increasing concentrations of HNE for various times, followed by esterase assay or western blot using anti-HNE antibodies. PVDF membranes were also stained with Coomassie blue to demonstrate equal protein loading. (E) Comparison of CES1 and CES2 activities following treatment with 1 mM HNE for 15 min. ONE, 4-oxononenal.
Human CES1 and CES2 are both members of the carboxylesterase family, but are encoded by separate genes [39]. CES2 exhibits 48% amino acid sequence homology to CES1. The covalent modification of CES2 protein by HNE (1 mM, 15 min, 37°C) caused an unexpected 3.2-fold enhancement in CES2 activity (Fig. 1E). In contrast, CES1 protein treated under the same conditions did not exhibit any difference from control. On the basis of data shown in Fig. 1C, it was expected that CES1 activity would not be significantly affected by 1 mM HNE after just 15 min of incubation; for example, marked inhibition of CES1 was observed only after long incubation times (~24 h). Because prolonged incubation of vehicle-treated CES2 at 37°C (≥1h) caused significant losses in its enzymatic activity (data not shown), a short incubation time (15 min) was chosen in order to compare CES2 with CES1. The result suggests that CES2 activity can be activated by HNE-mediated covalent modification, whereas CES1 activity is reduced but only after prolonged incubation periods. The nature and location of the amino acid modified by HNE in CES2 is currently unknown but is being investigated. Although the concentrations of HNE are relatively high in these experiments, it is known that the levels of HNE in subcellular locales such as in lipid membranes can reach millimolar levels during lipid peroxidation [5],[6]. This may be particularly relevant for interactions of HNE with CES1 because this protein is found in the lumen of the endoplasmic reticulum and is intimately associated with the lipid membrane of this organelle [39].
3.2 Mapping the HNE adduct in recombinant CES1
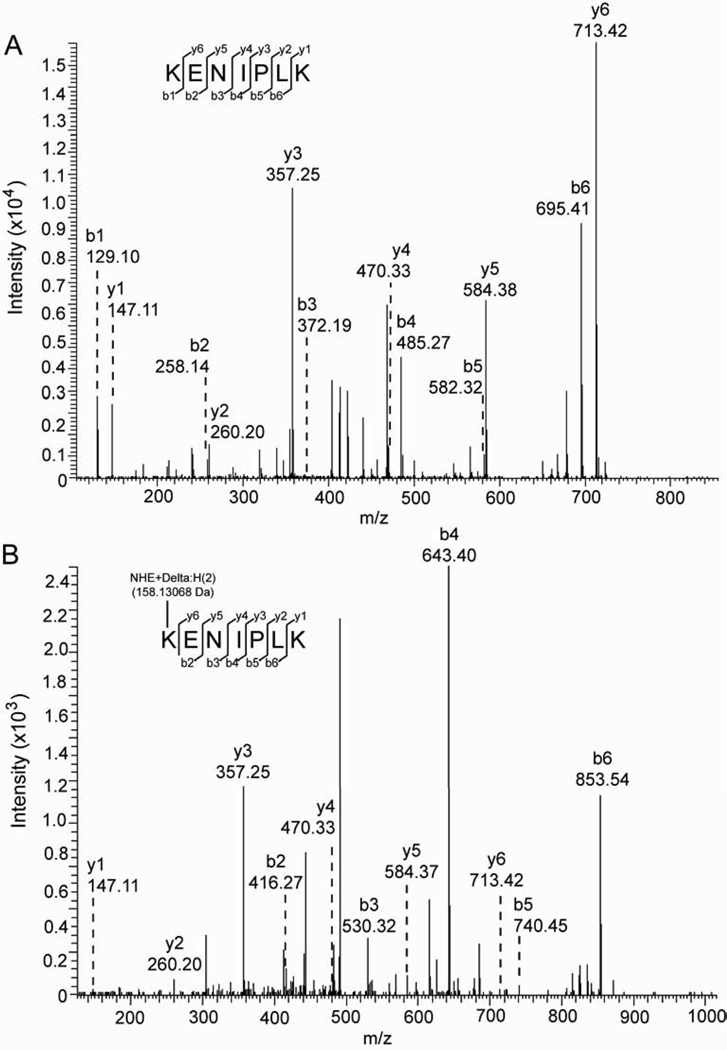
Analysis of tryptic peptides obtained from HNE-modified recombinant CES1 protein by mass spectrometry demonstrated that a single amino acid (Lys105) was modified by the addition of +158 amu, which is the mass addition expected following Michael addition of HNE and subsequent NaBH4-mediated reduction of the resulting hemiacetal [38]. No evidence of Schiff base adducts (+140 amu) was found and the sequence coverage of CES1 determined by mass spectrometry was ~60%. Fig. 2 shows the MS/MS spectra obtained for the +158 amu modified peptide K105(HNE)ENIPLK and the unmodified KENIPLK peptides. The HNE modification on the N-terminal lysine of KENIPLK added a mass of 158.13068 Da to all b fragment ions in the modified peptide (Fig. 2B) when compared to the b ions in the unmodified peptide (Fig. 2A). Because the HNE modification is located on the N-terminus of this peptide, only the N-terminal fragment ions (b ions) were affected, each having a mass altered by +158.13068 (b2, b3, b4, b5 and b6 ions, Fig. 2B). In contrast, the detected C-terminal fragment ions (y ions) have the same mass in both the unmodified and modified KENIPLK peptides. This was expected because the HNE modification would affect only the mass of the y7 ion, which was not detected in this MS/MS experiment. In addition, extracted ion chromatograms of the modified ([M+2H]2+, 500.33) and unmodified ([M+2H]2+, 421.26) KENIPLK peptides are shown in Supplementary Fig. 2, along with quantitative data. Lys105 is a surface exposed residue found in β-sheet 7 within the catalytic domain of CES1; its pKa is estimated to be 10.55 according to analysis of PDB file 3K9B (http://propka.ki.ku.dk/pka). Lys105 is not one of the residues that comprise the catalytic triad of CES1 (i.e., Ser221, Glu354, and His468); therefore, it is uncertain whether Lys105 modification by HNE completely ablates catalytic activity or only partially inhibits it. Interestingly, the lone reduced cysteine residue (Cys389) in CES1 was not modified by HNE, which was somewhat surprising given that Cys residues are more reactive than either His or Lys toward HNE [40]. However, modification by HNE is dependent not only on the nucleophilicity of individual residues, but also on their steric accessibility [38]. The prolonged incubation time needed for inhibition of CES1 (Fig. 1B) suggests that the kinetics of the reaction between HNE and its putative amino acid target (i.e., Lys105) are slow. However, CES1 is a stable protein and expressed at high concentrations in certain cells, such as hepatocytes and monocytes; therefore, it may be continuously exposed to steady-state, basal levels of HNE, which over time may cause a reduction in its catalytic activity. It appears from Fig. 1A that pnbCE is more rapidly inhibited by HNE than CES1, suggesting that multiple amino acids are targeted and that their cumulative modification results in decreased catalytic activity. Alternatively, a critical single amino acid residue may be present in pnbCE that is highly susceptible to covalent modification by HNE.
Fig. 2.
HNE modifies Lys105 residue of CES1. Recombinant CES1 protein was treated with ethanol vehicle (A) or 1 mM HNE (B). The modified proteins were analyzed by SDS-PAGE and Coomassie staining, followed by tryptic digestion of the excised bands and LC-MS/MS analysis by OrbiTrap Velos. The CID fragmentation of the KENIPLK peptide (residues 105–111), as analyzed by Fourier Transform (FT), indicates that HNE modifies Lys105 (B). The detected b- and y- fragment ion series are shown for the unmodified (A; [M+2H]2+, 421.26 Da) and modified (B; [M+2H]2+, 500.33 Da) KENIPLK peptide.
3.3 Inhibition of hydrolytic activity in THP1 cell lysates by HNE
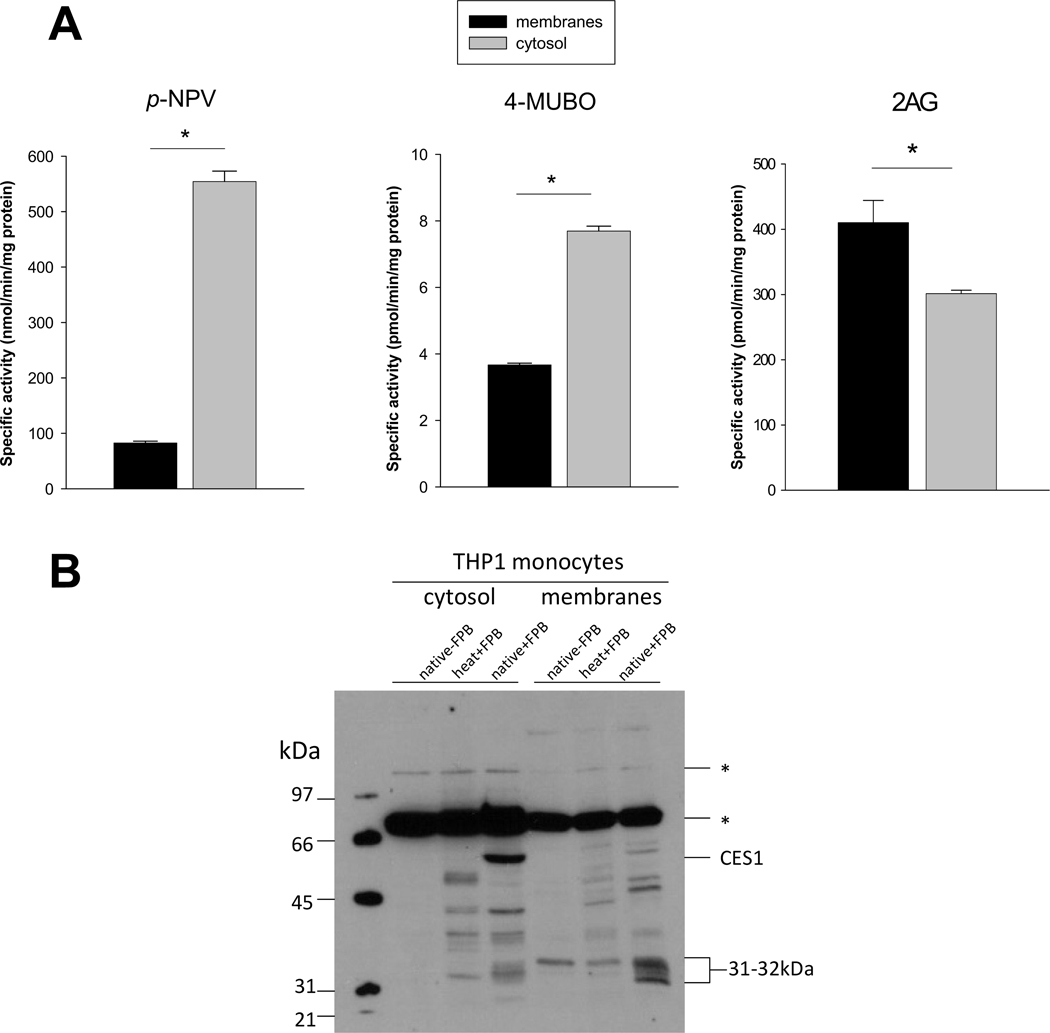
Esterase and lipase activities in THP1 monocyte lysates appear to be due to different sets of enzymes with different subcellular locations (Fig. 3A). For example, esterase activity (pNPV) is 6x higher in the cytosolic fraction compared to membrane fraction, whereas lipase activity levels (4-MUBO or 2AG) are 2× or 0.75× higher in cytosolic versus membrane fractions depending on the substrate used. CES1 and an uncharacterized 31–32 kDa enzyme are the most robust serine hydrolase activity gel bands in the cytosolic and membrane fractions, respectively (Fig. 3B). Each is a candidate enzyme that can hydrolyze the endocannabinoid 2AG in THP1 monocytes/macrophages [27]. Using CES1 specific inhibitors and immunoprecipitation it was previously estimated that CES1 accounted for 40–50% of the 2AG hydrolytic activity in THP1 cells, while the remaining 2AG hydrolytic activity was postulated to be due to the 31–32 kDa serine hydrolase [27]. This is in concordance with the subcellular distribution of 2AG hydrolysis activity in THP1 cells (Fig. 3A) and the subcellular localization of CES1 and the 31–32 kDa enzyme (Fig. 3B).
Fig. 3.
Specific activities of THP1 monocyte cytosolic and membrane fractions toward pNPV, 4MUBO, and 2AG. (A) The esterase (pNPV) and lipase (4MUBO and 2AG) activities of cytosolic and membrane fractions were determined as described in the Materials and Methods. (B) Serine hydrolase activity profile of THP1 monocyte subcellular fractions determined with FP-biotin. heat indicates samples were boiled prior to addition of FP-biotin, native indicates samples were not boiled. Endogenous biotin-containing proteins are indicated by asterisks and are detected in samples that did not receive FP-biotin treatment. *, p<0.05; Student’s t-test.
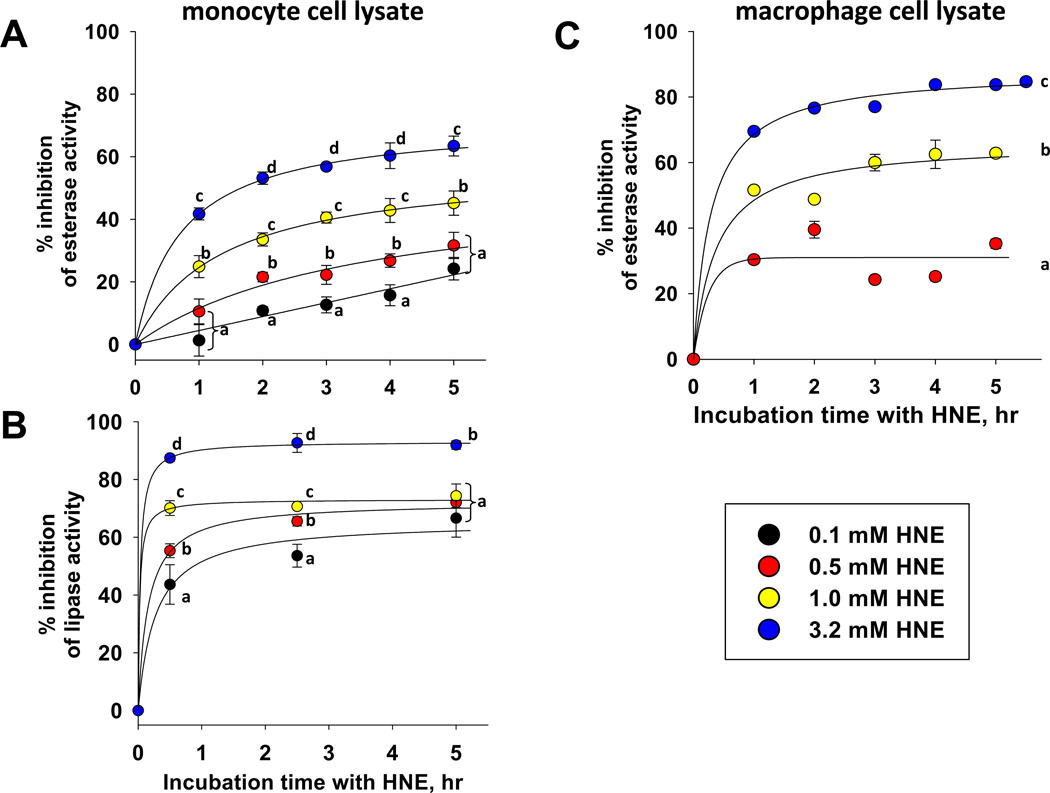
The esterase and lipase activities in THP1 cell lysates were inhibited by HNE (Fig. 4A,B). Lipase activity in THP1 cell lysate appears to be much more sensitive to the inhibitory effects of HNE than the esterase activity, suggesting that two different subsets of enzymes are responsible for these different activities. The enzymes responsible for the lipase activity in THP1 cells (i.e., 4-MUBO hydrolysis) are presently unknown, while 85% of the esterase activity of THP1 monocytes is attributed to CES1 [26]. The lipase activity in lysates was inhibited by 90% within one hour of exposure to the highest concentration of HNE, whereas esterase activity was inhibited by 40% under the same conditions. When THP1 macrophage lysates were treated with increasing amounts of HNE (Fig. 4C), the extent of inhibition of esterase activity appeared to be more pronounced than seen with monocyte lysates (Fig. 4A). The greater sensitivity of esterase activity in macrophage lysates compared to monocytes may be due to differences in subcellular location of esterases in the two cell types.
Fig. 4.
Esterase and lipase activities in THP1 cell lysates are inhibited by HNE. (A) Inhibition of esterase activity in monocyte cell lysate. (B) Inhibition of lipase activity in monocyte cell lysate. (C) Inhibition of esterase activity in macrophage cell lysate. Different alphabetic letters indicate that significant differences were noted for each HNE concentration at each time point (p<0.05; ANOVA, Tukey test); data points denoted by the same alphabetic letter were not significantly different at the specific time point indicated.
3.4 Inhibition of hydrolytic activity in intact THP1 monocytes and macrophages following challenge with HNE
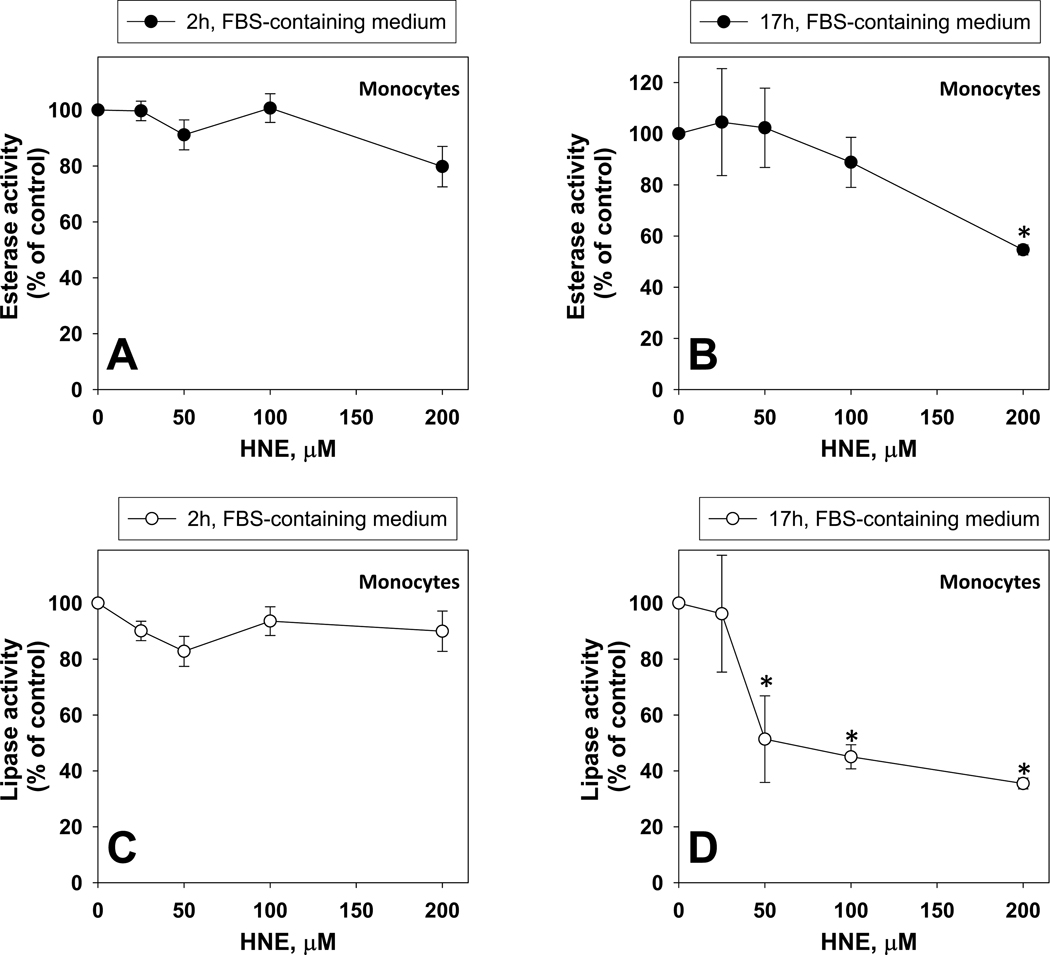
The effects of HNE on esterase and lipase activities following treatment of intact THP1 monocytes are shown in Fig. 5. Treatment durations of two hours in serum-containing medium did not significantly alter the esterase and lipase activities of the monocytes when compared to vehicle controls (Fig. 5A,C). Longer durations of exposure (17 h) could inhibit these activities (Fig. 5B,D), although cytotoxicity was noted in this instance when compared to the 2 h exposures (Supplementary Fig. 3). As seen with HNE-treated cell lysates, the lipase activity was more sensitive to the inhibitory effects of HNE than the esterase activity following treatment of intact cells.
Fig. 5.
Esterase and lipase activities in intact THP1 monocytes challenged with different concentrations of HNE. Inhibition of esterase activity after 2 h (A) and 17 h (B) treatments. Inhibition of lipase activity after 2 h (C) and 17 h (D) treatment. Asterisks indicate that the value is significantly different when compared to the control (0 µM HNE) (p<0.05; ANOVA, Dunnett’s test).
The presence of fetal bovine serum (FBS) in the culture medium may have reduced the inhibitory effect of HNE by lowering the effective concentration of HNE in the culture medium due to adduction reactions between serum albumin protein and HNE. This possibility was verified by treating intact monocytes with increasing amounts of HNE (0–100 µM) for two hours, in the presence or absence of 10% FBS, then measuring the resulting esterase activity. It was found that the potency of HNE-mediated inhibition of esterase activity was enhanced when FBS was absent from the medium [3.8-fold increase when regression slopes of esterase activity vs. HNE concentration were compared; normalized slope values: 1.0/µM HNE with FBS absent and 0.26/µM HNE with FBS present (arbitrary units)], suggesting that more of the reactive aldehyde could penetrate the cells and inhibit intracellular esterases.
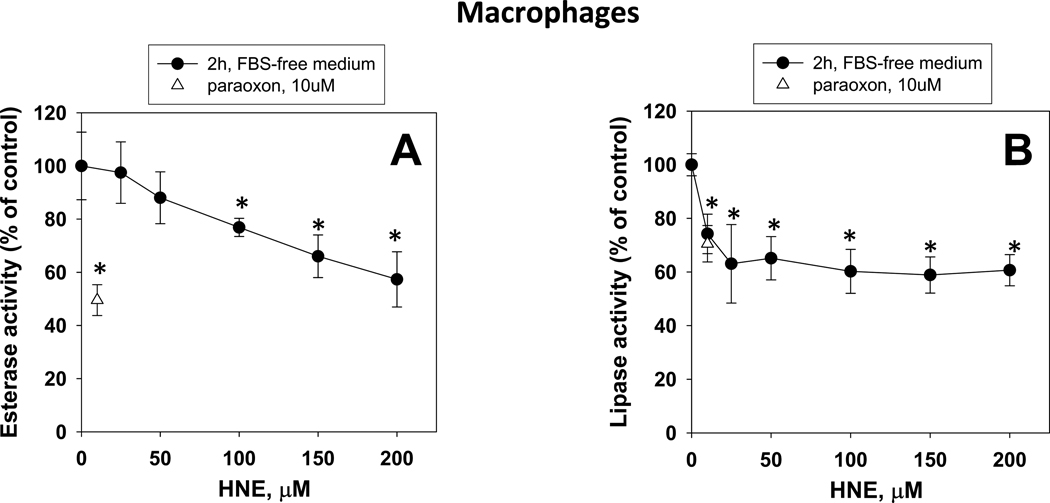
When differentiated THP1 macrophages were treated with increasing amounts of HNE for two hours in the absence of FBS, a concentration-dependent inhibition of esterase and lipase activities was also found (Fig. 6A,B). Maximal inhibition of esterase and lipase activity (~40% for each) was noted at HNE concentrations of 200 µM and 25 µM, respectively. Thus, as with monocytes, the lipase activity in macrophages was more sensitive to the inhibitory effects of HNE than the esterase activity. It should be noted here that cytotoxicity was not observed in the macrophage experiments (Fig. 6) at any of the concentrations of HNE used as assessed by the presence of LDH in the culture medium; short exposures (2 h) to HNE are not cytotoxic to THP1 macrophages, although longer exposures are (17 h; data not shown). Moreover, THP1 macrophages challenged with HNE for short periods of time appear to be more resistant to cytotoxicity than undifferentiated THP1 monocytes, which exhibit significant cytotoxicity when treated with HNE concentrations >50 µM in serum-free medium [41].
Fig. 6.
Inhibition of esterase (A) and lipase (B) activities in THP1 macrophage cells challenged with increasing concentrations of HNE. Asterisks indicate that the value is significantly different when compared to the control (0 µM HNE) (p<0.05; ANOVA, Dunnett’s test).
It should be pointed out that the concentrations of HNE needed to inhibit esterase/lipase activity in THP-1 cells are generally higher in relation to concentrations that have been shown to have other important biological effects on cells with roles in atherosclerosis. For example, Go et al.[13] showed that treatment of cultured endothelial cells with low concentrations of HNE (1–10 µM) enhanced monocyte adhesion to these cells, which is one of the first observable events in atherosclerosis. Yun et al. [15] demonstrated that CD36 expression, a scavenger receptor, on murine J774A.1 macrophages could be robustly induced by HNE at 10 µM. Darley-Usmar et al. [42] showed that when THP-1 macrophages were challenged with 50 µM HNE, the cellular GSH pool was significantly depleted by 1–2 h but had rebounded by 4 h and was markedly higher than the untreated macrophages by 24 h. Therefore, HNE likely affects multiple molecular pathways that contribute to pathophysiological processes, such as atherosclerosis, across a wide range of concentrations.
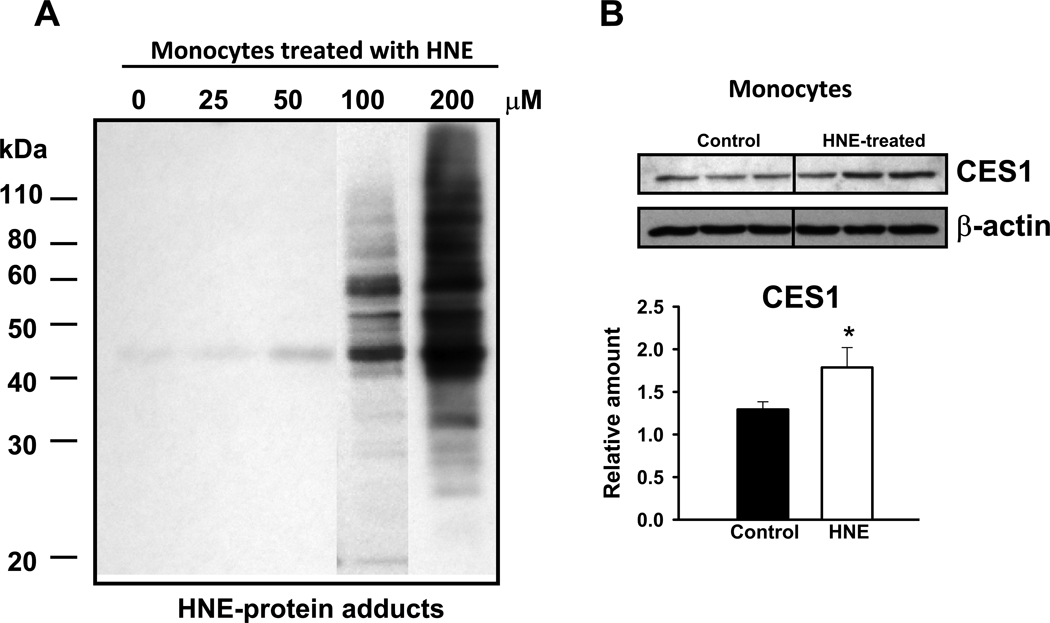
3.5 HNE-mediated damage of THP1 monocyte proteins detected by western blot analysis
It has been previously shown that several cellular proteins and DNA are covalently modified by HNE [43–46], which can alter their cellular activities. Therefore, we examined whether HNE-protein adducts are detectable after treating intact THP1 monocytes with HNE. Indeed, we found that many cellular proteins were covalently modified by HNE and detected by immunoblotting methods using antibodies that recognize HNE adducts, most noticeably after treatment with 100 and 200 µM HNE (Fig. 7A). This result is likely due to saturation of HNE detoxication enzymes such as glutathione transferases and aldehyde dehydrogenase [47]. These findings might explain why the cellular esterase and lipase activities are inactivated following HNE treatments; the enzymes responsible for these activities are likely covalently modified by HNE and their function altered. However, it should be pointed out that a recent proteomic study of THP1 monocytes challenged with this lipid electrophile failed to detect HNE-modified CES1 protein [48]. Nevertheless, in this particular study, the sixteen identified HNE-damaged proteins were primarily cytoskeletal proteins, which are abundant background contaminants that are often detected in proteomic studies, and as such might interfere with mass spectrometry detection of less abundant HNE targets. Therefore, HNE-damaged CES1 protein may have escaped detection in this study and further enrichment procedures will be necessary to obtain in-depth proteomic data.
Fig. 7.
Evidence of HNE-protein adducts and upregulation of CES1 in THP1 monocytes. (A) Formation of HNE-protein adducts in THP1 monocytes treated with different concentrations of HNE. Protein-adducts were detected using an antibody that recognizes these lesions. (B) Expression of CES1 protein is enhanced when THP1 monocytes are challenged with 200 µM HNE. Each lane represents a separate flask of THP1 monocytes challenged with HNE. *, p<0.05; Student’s t-test.
3.6 Induction of CES1 protein in THP1 monocytes following HNE challenge
Next, we investigated whether HNE could modulate the abundance of CES1 protein in THP1 monocytes. Indeed, we found by western blot that the amount of CES1 protein was significantly elevated (1.4-fold) by HNE exposure (200 µM, 17 h) (Fig. 7B) when normalized to β-actin expression. HNE is an electrophilic lipid and is known to activate the transcription factor Nrf2 via covalent modification of its negative regulatory protein Keap 1 [16], thereby inducing the expression of several genes encoding enzymes that detoxify HNE and reactive oxygen species. In the presence of electrophilic agents, Nrf2 is released from Keap1 and translocates from the cytosol into the nucleus where it stimulates the transcription of a host of genes, including CES1 [49]. Thus, while HNE may reduce esterase and lipase activities in THP1 cells by direct chemical adduction of the enzymes responsible for these functions, it is possible that HNE can activate Nrf2 transcriptional activity leading to increased amounts of CES1 protein, partially compensating for deficits in enzyme activity caused by HNE modification.
3.7 Glutathione conjugation of HNE
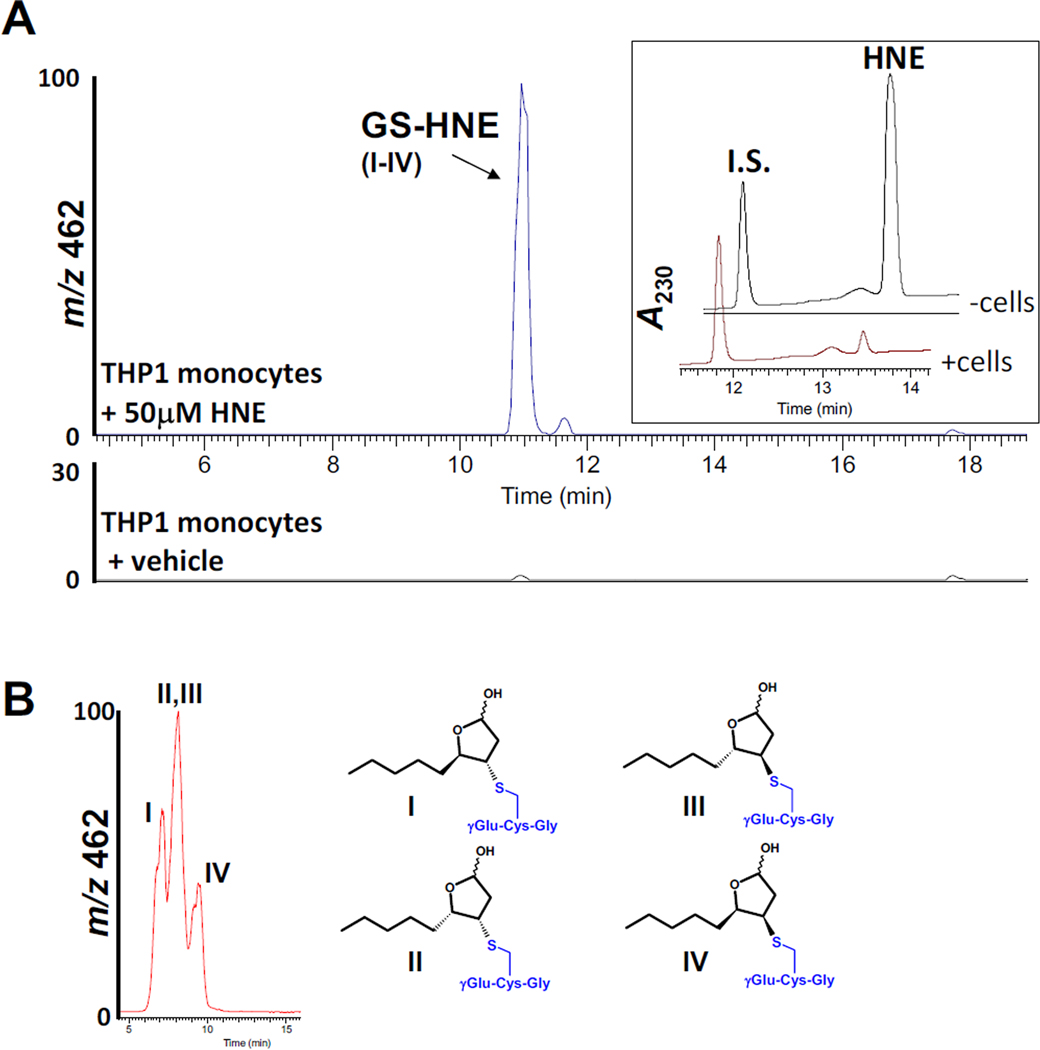
HNE is both non-enzymatically and enzymatically conjugated to GSH, a significant pathway for its detoxification in cells [50]. For instance, it was previously shown that >50% of exogenous HNE was metabolized by GSH conjugation in cultured keratinocytes [51]. When intact THP1 monocytes were challenged for 5 min with 50 µM HNE in the present study, GS-HNE conjugates were readily detected in combined cell/culture medium extracts by LC-MS (Fig. 8A,B). On the basis of LC-UV chromatograms, exogenous HNE was almost completely metabolized (>90%) by the intact monocytes in 5 min (Fig. 8A, inset) and the rate of HNE catabolism was estimated to be 0.46 nmol/min/106 cells, which is similar to the rate seen with isolated human polymorphonuclear leukocytes [47]. The rate of catabolism of HNE (initial concentration, 50 µM) by THP1 monocyte and macrophage lysates ranged from 14 to 16 nmol/min/mg protein, whereas the rates of GS-HNE conjugation for THP1 monocyte and macrophage lysates were 6.5±0.8 and 6.3±0.1 nmol/min/mg protein, respectively. Hence, GSH conjugation of HNE is an important mechanism of its detoxification in both THP1 monocytes and macrophages, accounting for ~43% of the metabolism of HNE. This metabolic rate is comparable to that described by Luckey and Petersen [52], who estimated that GS-HNE conjugation accounted for ~30% of the metabolism of HNE in rat Kupffer cells (macrophage-like cells). The remaining metabolism of HNE in THP1 cells is likely due to oxidative and/or reductive pathways, which are mediated by aldehyde and alcohol dehydrogenases [2].
Fig. 8.
THP1 monocytes produce GS-HNE conjugates when challenged with HNE. (A) THP1 monocytes were treated with HNE (50 µM) for 5 min at 37°C; culture medium (PBS) and cells were mixed with 2 volumes of methanol (containing internal standard) and GS-HNE conjugates were detected by LC-ESI-MS (negative ion mode, m/z 462). Inset, LC-UV chromatograms depicting elution profiles of HNE and internal standard (I.S.) after the cells or culture medium without cells were challenged with 50 µM HNE. (B) The stereochemical configurations of the GS-HNE diastereomers are shown. The diastereomers (I–IV) elute as one peak when analyzed by the gradient method (shown in A); however, separation of diastereomers is partially enabled by using isocratic mobile phase conditions (85% water/15% acetonitrile containing 0.1% v/v acetic acid; flow rate, 0.3 ml/min). Assignment of peaks is based on the study by Balogh et al. [50], which used similar stationary and mobile phases for chromatography. Note that the stereochemistry of the hydroxyl group in the hemiacetal is undefined.
3.8 Conclusions and Significance
Atherosclerosis is a disease having both oxidative and inflammatory components, and HNE is an endogenous molecule found within atherosclerotic lesions that bridges these pathobiological processes [16]. CES1 has been proposed to release cholesterol from cholesterol esters stored in lipid droplets within macrophage foam cells [24]. Furthermore, CES1 appears to regulate the levels of the endocannabinoid, 2-arachidonoylglycerol, and prostaglandin glyceryl esters in THP1 cells [27]. Therefore, the interactions of HNE with hydrolytic enzymes such as CES1 are important to clarify. The present study has taken a first step toward investigating the functional importance of HNE-mediated inactivation of cellular hydrolytic enzymes in THP1 monocytes/macrophages. Although the molecular details remain obscure, it appears that the lipolytic activity of enzymes in THP1 monocytes is more sensitive to inactivation by HNE than the esterolytic activity. Since CES1 appears to be a target of HNE in vitro and its modification has been mapped by mass spectrometry to Lys105 it would be interesting to validate this potential amino acid target in vivo and further define the physiological significance of this modification by testing its effect on CES1’s catalytic activity. Accordingly, the investigation of serine hydrolases in monocytes/macrophages that are targeted by HNE and become functionally inactivated by lipid electrophiles will be important to pursue because of the critical role of this enzyme family in diseases such as atherosclerosis.
Highlights.
Effects of 4-hydroxynonenal (HNE) on esterase and lipase activities was examined.
Hydrolytic activities of pure CES enzymes and intact cells are inhibited by HNE.
Lysine 105 in recombinant CES1 enzyme was covalently modified by HNE.
THP1 cells efficiently metabolize HNE and yield glutathione-HNE conjugates.
Supplementary Material
Acknowledgements
Research support was partially funded by NIH 1R15ES015348 and is gratefully acknowledged. Katelyn L. Hardin was supported by NIH T35RR007071. Funding for M.J.E. and the proteomic work was provided by the Institute for Genomics, Biocomputing & Biotechnology at Mississippi State University (funding number 269181-018100-027000-191400). We thank Dr. Philip M. Potter, St. Jude Children’s Research Hospital, for the generous gift of recombinant human CES proteins.
Abbreviations
- HNE
4-hydroxy-2-trans-nonenal
- CES1
carboxylesterase 1
- CES2
carboxylesterase 2
- oxLDLs
oxidized low-density lipoproteins
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jacobs AT, Marnett LJ. Systems analysis of protein modification and cellular responses induced by electrophile stress. Acc Chem Res. 2010;43:673–683. doi: 10.1021/ar900286y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Esterbauer H, Schaur RJ, Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic Biol Med. 1991;11:81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- 3.Kutuk O, Adli M, Poli G, Basaga H. Resveratrol protects against 4-HNE induced oxidative stress and apoptosis in Swiss 3T3 fibroblasts. Biofactors. 2004;20:1–10. doi: 10.1002/biof.5520200101. [DOI] [PubMed] [Google Scholar]
- 4.Yang J, Wu LJ, Tashiro S, Onodera S, Ikejima T. Nitric oxide activated by p38 and NF-kappaB facilitates apoptosis and cell cycle arrest under oxidative stress in evodiamine-treated human melanoma A375-S2 cells. Free Radic Res. 2008;42:1–11. doi: 10.1080/10715760701762407. [DOI] [PubMed] [Google Scholar]
- 5.Benedetti A, Comporti M, Fulceri R, Esterbauer H. Cytotoxic aldehydes originating from the peroxidation of liver microsomal lipids. Identification of 4,5-dihydroxydecenal. Biochim Biophys Acta. 1984;792:172–181. doi: 10.1016/0005-2760(84)90219-4. [DOI] [PubMed] [Google Scholar]
- 6.Koster JF, Slee RG, Montfoort A, Lang J, Esterbauer H. Comparison of the inactivation of microsomal glucose-6-phosphatase by in situ lipid peroxidation-derived 4-hydroxynonenal and exogenous 4-hydroxynonenal. Free Radic Res Commun. 1986;1:273–287. doi: 10.3109/10715768609051637. [DOI] [PubMed] [Google Scholar]
- 7.Esterbauer H, Jurgens G, Quehenberger O, Koller E. Autoxidation of human low density lipoprotein: loss of polyunsaturated fatty acids and vitamin E and generation of aldehydes. J Lipid Res. 1987;28:495–509. [PubMed] [Google Scholar]
- 8.Catala A. Lipid peroxidation of membrane phospholipids generates hydroxyalkenals and oxidized phospholipids active in physiological and/or pathological conditions. Chem Phys Lipids. 2009;157:1–11. doi: 10.1016/j.chemphyslip.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 9.Fritz KSaP, DR Exploring the Biology of Lipid Peroxidation Derived Protein Carbonylation. Chemical Research in Toxicology. 2011 doi: 10.1021/tx200169n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balogh LM, Atkins WM. Interactions of glutathione transferases with 4-hydroxynonenal. Drug Metab Rev. 2011;43:165–178. doi: 10.3109/03602532.2011.558092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh SP, Niemczyk M, Saini D, Awasthi YC, Zimniak L, Zimniak P. Role of the electrophilic lipid peroxidation product 4-hydroxynonenal in the development and maintenance of obesity in mice. Biochemistry. 2008;47:3900–3911. doi: 10.1021/bi702124u. [DOI] [PubMed] [Google Scholar]
- 12.Leonarduzzi G, Chiarpotto E, Biasi F, Poli G. 4-Hydroxynonenal and cholesterol oxidation products in atherosclerosis. Mol Nutr Food Res. 2005;49:1044–1049. doi: 10.1002/mnfr.200500090. [DOI] [PubMed] [Google Scholar]
- 13.Go YM, Halvey PJ, Hansen JM, Reed M, Pohl J, Jones DP. Reactive aldehyde modification of thioredoxin-1 activates early steps of inflammation and cell adhesion. Am J Pathol. 2007;171:1670–1681. doi: 10.2353/ajpath.2007.070218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miranda CL, Reed RL, Kuiper HC, Alber S, Stevens JF. Ascorbic acid promotes detoxification and elimination of 4-hydroxy-2(E)-nonenal in human monocytic THP-1 cells. Chem Res Toxicol. 2009;22:863–874. doi: 10.1021/tx900042u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yun MR, Im DS, Lee SJ, Park HM, Bae SS, Lee WS, Kim CD. 4-Hydroxynonenal enhances CD36 expression on murine macrophages via p38 MAPK-mediated activation of 5-lipoxygenase. Free Radic Biol Med. 2009;46:692–698. doi: 10.1016/j.freeradbiomed.2008.12.013. [DOI] [PubMed] [Google Scholar]
- 16.Uchida K. 4-Hydroxy-2-nonenal: a product and mediator of oxidative stress. Prog Lipid Res. 2003;42:318–343. doi: 10.1016/s0163-7827(03)00014-6. [DOI] [PubMed] [Google Scholar]
- 17.Esterbauer H. Cytotoxicity and genotoxicity of lipid-oxidation products. Am J Clin Nutr. 1993;57:779S–785S. doi: 10.1093/ajcn/57.5.779S. discussion 785S–786S. [DOI] [PubMed] [Google Scholar]
- 18.Li AC, Glass CK. The macrophage foam cell as a target for therapeutic intervention. Nat Med. 2002;8:1235–1242. doi: 10.1038/nm1102-1235. [DOI] [PubMed] [Google Scholar]
- 19.Woolf N. Pathology of atherosclerosis. Br Med Bull. 1990;46:960–985. doi: 10.1093/oxfordjournals.bmb.a072448. [DOI] [PubMed] [Google Scholar]
- 20.Boring L, Gosling J, Cleary M, Charo IF. Decreased lesion formation in CCR2−/− mice reveals a role for chemokines in the initiation of atherosclerosis. Nature. 1998;394:894–897. doi: 10.1038/29788. [DOI] [PubMed] [Google Scholar]
- 21.Rosenfeld ME, Khoo JC, Miller E, Parthasarathy S, Palinski W, Witztum JL. Macrophage-derived foam cells freshly isolated from rabbit atherosclerotic lesions degrade modified lipoproteins, promote oxidation of low-density lipoproteins, and contain oxidation-specific lipid-protein adducts. J Clin Invest. 1991;87:90–99. doi: 10.1172/JCI115006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith JD, Trogan E, Ginsberg M, Grigaux C, Tian J, Miyata M. Decreased atherosclerosis in mice deficient in both macrophage colony-stimulating factor (op) and apolipoprotein E. Proc Natl Acad Sci U S A. 1995;92:8264–8268. doi: 10.1073/pnas.92.18.8264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takaku M, Wada Y, Jinnouchi K, Takeya M, Takahashi K, Usuda H, Naito M, Kurihara H, Yazaki Y, Kumazawa Y, Okimoto Y, Umetani M, Noguchi N, Niki E, Hamakubo T, Kodama T. An in vitro coculture model of transmigrant monocytes and foam cell formation. Arterioscler Thromb Vasc Biol. 1999;19:2330–2339. doi: 10.1161/01.atv.19.10.2330. [DOI] [PubMed] [Google Scholar]
- 24.Ghosh S, Zhao B, Bie J, Song J. Macrophage cholesteryl ester mobilization and atherosclerosis. Vascul Pharmacol. 2010;52:1–10. doi: 10.1016/j.vph.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Auwerx J. The human leukemia cell line, THP-1: a multifacetted model for the study of monocyte-macrophage differentiation. Experientia. 1991;47:22–31. doi: 10.1007/BF02041244. [DOI] [PubMed] [Google Scholar]
- 26.Crow JA, Middleton BL, Borazjani A, Hatfield MJ, Potter PM, Ross MK. Inhibition of carboxylesterase 1 is associated with cholesteryl ester retention in human THP-1 monocyte/macrophages. Biochim Biophys Acta Mol Cell Biol Lipids. 2008;1781:643–654. doi: 10.1016/j.bbalip.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie S, Borazjani A, Hatfield MJ, Edwards CC, Potter PM, Ross MK. Inactivation of Lipid Glyceryl Ester Metabolism in Human THP1 Monocytes/Macrophages by Activated Organophosphorus Insecticides: Role of Carboxylesterases 1 and 2. Chem Res Toxicol. 2010;23:1890–1904. doi: 10.1021/tx1002194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Streit TM, Borazjani A, Lentz SE, Wierdl M, Potter PM, Gwaltney SR, Ross MK. Evaluation of the 'side door' in carboxylesterase-mediated catalysis and inhibition. Biol Chem. 2008;389:149–162. doi: 10.1515/BC.2008.017. [DOI] [PubMed] [Google Scholar]
- 29.Morton CL, Potter PM. Comparison of Escherichia coli, Saccharomyces cerevisiae, Pichia pastoris, Spodoptera frugiperda, and COS7 cells for recombinant gene expression. Application to a rabbit liver carboxylesterase. Mol Biotechnol. 2000;16:193–202. doi: 10.1385/MB:16:3:193. [DOI] [PubMed] [Google Scholar]
- 30.Ross MK, Borazjani A. Enzymatic Activity of Human Carboxylesterases. Curr Protoc Toxicol. 2007;33:4.24.21–24.24.14. doi: 10.1002/0471140856.tx0424s33. [DOI] [PubMed] [Google Scholar]
- 31.Edelmann MJ, Kramer HB, Altun M, Kessler BM. Post-translational modification of the deubiquitinating enzyme otubain 1 modulates active RhoA levels and susceptibility to Yersinia invasion. FEBS J. 2010;277:2515–2530. doi: 10.1111/j.1742-4658.2010.07665.x. [DOI] [PubMed] [Google Scholar]
- 32.MacCoss MJ, Wu CC, Yates JR., 3rd Probability-based validation of protein identifications using a modified SEQUEST algorithm. Anal Chem. 2002;74:5593–5599. doi: 10.1021/ac025826t. [DOI] [PubMed] [Google Scholar]
- 33.Crow JA, Herring KL, Xie S, Borazjani A, Potter PM, Ross MK. Inhibition of carboxylesterase activity of THP1 monocytes/macrophages and recombinant human carboxylesterase 1 by oxysterols and fatty acids. Biochim Biophys Acta Mol Cell Biol Lipids. 2010;1801:31–41. doi: 10.1016/j.bbalip.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 35.Ross MK, Filipov NM. Determination of atrazine and its metabolites in mouse urine and plasma by LC-MS analysis. Anal Biochem. 2006;351:161–173. doi: 10.1016/j.ab.2006.01.050. [DOI] [PubMed] [Google Scholar]
- 36.Habig WH, Pabst MJ, Jakoby WB. Glutathione S-transferases. The first enzymatic step in mercapturic acid formation. J Biol Chem. 1974;249:7130–7139. [PubMed] [Google Scholar]
- 37.McGrath CE, Tallman KA, Porter NA, Marnett LJ. Structure-activity analysis of diffusible lipid electrophiles associated with phospholipid peroxidation: 4-hydroxynonenal and 4-oxononenal analogues. Chem Res Toxicol. 2011;24:357–370. doi: 10.1021/tx100323m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szapacs ME, Riggins JN, Zimmerman LJ, Liebler DC. Covalent adduction of human serum albumin by 4-hydroxy-2-nonenal: kinetic analysis of competing alkylation reactions. Biochemistry. 2006;45:10521–10528. doi: 10.1021/bi060535q. [DOI] [PubMed] [Google Scholar]
- 39.Ross MK, Crow JA. Human carboxylesterases and their role in xenobiotic and endobiotic metabolism. J Biochem Mol Toxicol. 2007;21:187–196. doi: 10.1002/jbt.20178. [DOI] [PubMed] [Google Scholar]
- 40.Doorn JA, Petersen DR. Covalent modification of amino acid nucleophiles by the lipid peroxidation products 4-hydroxy-2-nonenal and 4-oxo-2-nonenal. Chem Res Toxicol. 2002;15:1445–1450. doi: 10.1021/tx025590o. [DOI] [PubMed] [Google Scholar]
- 41.Foucaud L, Goulaouic S, Bennasroune A, Laval-Gilly P, Brown D, Stone V, Falla J. Oxidative stress induction by nanoparticles in THP-1 cells with 4-HNE production: stress biomarker or oxidative stress signalling molecule? Toxicol In Vitro. 2010;24:1512–1520. doi: 10.1016/j.tiv.2010.07.012. [DOI] [PubMed] [Google Scholar]
- 42.Darley-Usmar VM, Severn A, O'Leary VJ, Rogers M. Treatment of macrophages with oxidized low-density lipoprotein increases their intracellular glutathione content. Biochem J. 1991;278(Pt 2):429–434. doi: 10.1042/bj2780429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bacot S, Bernoud-Hubac N, Baddas N, Chantegrel B, Deshayes C, Doutheau A, Lagarde M, Guichardant M. Covalent binding of hydroxy-alkenals 4-HDDE, 4-HHE, and 4-HNE to ethanolamine phospholipid subclasses. J Lipid Res. 2003;44:917–926. doi: 10.1194/jlr.M200450-JLR200. [DOI] [PubMed] [Google Scholar]
- 44.Bacot S, Bernoud-Hubac N, Chantegrel B, Deshayes C, Doutheau A, Ponsin G, Lagarde M, Guichardant M. Evidence for in situ ethanolamine phospholipid adducts with hydroxy-alkenals. J Lipid Res. 2007;48:816–825. doi: 10.1194/jlr.M600340-JLR200. [DOI] [PubMed] [Google Scholar]
- 45.Lin CC, Su TH, Wang TS. Protein carbonylation in THP-1 cells induced by cigarette smoke extract via a copper-catalyzed pathway. Chem Res Toxicol. 2009;22:1232–1238. doi: 10.1021/tx900008h. [DOI] [PubMed] [Google Scholar]
- 46.Nadkarni DV, Sayre LM. Structural definition of early lysine and histidine adduction chemistry of 4-hydroxynonenal. Chem Res Toxicol. 1995;8:284–291. doi: 10.1021/tx00044a014. [DOI] [PubMed] [Google Scholar]
- 47.Siems W, Crifo C, Capuozzo E, Uchida K, Grune T, Salerno C. Metabolism of 4-hydroxy-2-nonenal in human polymorphonuclear leukocytes. Arch Biochem Biophys. 2010;503:248–252. doi: 10.1016/j.abb.2010.08.018. [DOI] [PubMed] [Google Scholar]
- 48.Chavez J, Chung WG, Miranda CL, Singhal M, Stevens JF, Maier CS. Site-specific protein adducts of 4-hydroxy-2(E)-nonenal in human THP-1 monocytic cells: protein carbonylation is diminished by ascorbic acid. Chem Res Toxicol. 2010;23:37–47. doi: 10.1021/tx9002462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maruichi T, Fukami T, Nakajima M, Yokoi T. Transcriptional regulation of human carboxylesterase 1A1 by nuclear factor-erythroid 2 related factor 2 (Nrf2) Biochem Pharmacol. 2010;79:288–295. doi: 10.1016/j.bcp.2009.08.019. [DOI] [PubMed] [Google Scholar]
- 50.Balogh LM, Roberts AG, Shireman LM, Greene RJ, Atkins WM. The stereochemical course of 4-hydroxy-2-nonenal metabolism by glutathione S-transferases. J Biol Chem. 2008;283:16702–16710. doi: 10.1074/jbc.M801725200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aldini G, Granata P, Orioli M, Santaniello E, Carini M. Detoxification of 4-hydroxynonenal (HNE) in keratinocytes: characterization of conjugated metabolites by liquid chromatography/electrospray ionization tandem mass spectrometry. J Mass Spectrom. 2003;38:1160–1168. doi: 10.1002/jms.533. [DOI] [PubMed] [Google Scholar]
- 52.Luckey SW, Petersen DR. Metabolism of 4-hydroxynonenal by rat Kupffer cells. Arch Biochem Biophys. 2001;389:77–83. doi: 10.1006/abbi.2001.2307. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.