Abstract
Background
Prenatal cannabis exposure has been linked to addiction vulnerability, but the neurobiology underlying this risk is unknown.
Methods
Striatal dopamine and opioid-related genes were studied in human fetal subjects exposed to cannabis (as well as cigarettes and alcohol). Cannabis-related gene disturbances observed in the human fetus were subsequently characterized using an animal model of prenatal delta-9-tetrahydrocannabinol (THC; 0.15 mg/kg) exposure.
Results
Prenatal cannabis exposure decreased dopamine receptor D2 (DRD2) mRNA expression in the human ventral striatum (nucleus accumbens; NAc), a key brain reward region. No significant alterations were observed for the other genes in cannabis-exposed subjects. Maternal cigarette use was associated with reduced NAc prodynorphin mRNA expression and alcohol exposure induced broad alterations primarily in the dorsal striatum of most genes. To explore the mechanisms underlying the cannabis-associated disturbances, we exposed pregnant rats to THC and examined the epigenetic regulation of the NAc Drd2 gene in their offspring at postnatal day 2, comparable to the human fetal period studied, and in adulthood. Chromatin immunoprecipitation of the adult NAc revealed increased 2meH3K9 repressive mark and decreased 3meH3K4 and RNA polymerase II at the Drd2 gene locus in the THC-exposed offspring. Decreased Drd2 expression was accompanied by reduced D2R binding sites and increased sensitivity to opiate reward in adulthood.
Conclusions
These data suggest that maternal cannabis use alters developmental regulation of mesolimbic D2R in offspring through epigenetic mechanisms that regulate histone lysine methylation, and the ensuing reduction of D2R may contribute to addiction vulnerability later in life.
Keywords: THC, addiction, development, enkephalin, dynorphin, D1 receptor
Introduction
The prenatal period is sensitive to environmental influences due to dynamic neurobiological events that occur during this stage to ensure proper patterning of the nervous system. These processes are likely to be disrupted by maternal drug use and could have lifelong consequences for their children. In the United States, approximately 4% of pregnant women report using illegal drugs with marijuana (Cannabis sativa) being the illicit drug most commonly abused during pregnancy (1). Cannabis is abused for its psychoactive properties and is prescribed as an antiemetic to treat nausea during pregnancy with the perception that no negative consequences will befall the developing fetus. However, a growing body of evidence suggests that maternal cannabis use can have long-lasting negative consequences on the cannabis-exposed individual including increased risk for developing drug addiction and neuropsychiatric disorders (2-4). Animal studies have also demonstrated that developmental exposure to cannabis-mimetic compounds enhances sensitivity to drugs of abuse, including heroin and morphine in adulthood (5-8). While these studies have been instrumental in establishing a link between prenatal cannabis exposure and addiction risk, little is known regarding which neurobiological systems are vulnerable to early developmental cannabis exposure and how cannabis-induced alterations are maintained into adulthood to contribute to pathological behavior.
The striatal dopamine system has been implicated in the underlying pathogenesis of neuropsychiatric disorders (9). The dorsal striatum (caudate and putamen) is associated with motor control and habit formation, while the ventral striatum (nucleus accumbens; NAc) is linked to goal-directed behavior and reward processing (10). Diverse populations of medium spiny neurons, which are enriched in cannabinoid receptors, constitute the striatal output pathways and are dissociated based on their expression of dopamine receptors and neuropeptides. Dopamine receptor subtype 1 (D1R) is preferentially localized to the striatonigral “direct” pathway that contains the opioid neuropeptide dynorphin, whereas dopamine receptor subtype 2 (D2R) is abundant in striatopallidal “indirect” neurons that also express enkephalin (11). Reduced D2R is a consistent feature observed in adult drug abusers (12) and has raised questions as to whether D2R impairments could also predate adult drug use to increase addiction risk. Based on the significant number of women who smoke marijuana when pregnant and the preclinical studies emphasizing the long-term impact of prenatal cannabis exposure on addiction-related behavior, we hypothesized that in utero cannabis exposure could contribute to D2R impairments characteristic of addiction vulnerability.
Here we studied whether maternal cannabis use alters regulation of DRD2 and related striatal genes in the human fetal brain. Our findings demonstrate that maternal cannabis use decreases NAc DRD2 gene expression in the fetus. Utilizing a THC prenatal rat model, we show that disruption of NAc Drd2 gene regulation persists into adulthood and is maintained by epigenetic alterations, which may contribute to increased opiate sensitivity in adulthood.
Methods and Materials
Human fetal brain material
Fetal brain specimens (18-22 weeks gestation) were previously collected following saline-induced elective abortions under IRB approval at SUNY Downstate Medical Center, Brooklyn, New York (13). Pregnant participants were interviewed to obtain information related to maternal demographic status, substance abuse history and medical history. Fetal brain samples were examined by a pathologist and lightly fixed with 1% paraformaldehyde and frozen in isopentane. Maternal urine and fetal meconium were analyzed for cannabis, opiates, stimulants and their respective metabolites to confirm information obtained by maternal self-report. Samples included in the cannabis-exposed group had positive maternal self-report and/or maternal urine that tested positive for THC, and/or fetal meconium positive for THC. Maternal alcohol and cigarette use was evaluated based on maternal report of average daily alcohol volume and number of cigarettes smoked daily (14). Subjects had no gross brain malformation. Characteristics of the entire fetal population have been published (13) and a subset from this collection (see Table 1) was chosen based on availability of tissue at the level of the NAc which had been cryosectioned in the coronal plane (20 um-thick), slide-mounted and stored at −30°C.
Table 1.
Demographics of the human fetal samples in cannabis-exposed and control groups. Maternal cannabis, cigarette and alcohol use are represented as average daily joint (ADJ), average daily cigarette (ADC), and average daily volume (ADV), respectively. This metric was calculated by converting weekly use into daily use based on the yearly pattern of use [e.g. seven joints per week equals an ADJ score of 0.90 (7 joints/week × 4 weeks/month)/(31 days/month)] (40). For alcohol use, 1 drink was equal to 12 oz. of beer, 4 oz. of wine, or 1.5 oz of liquor. Values are represented as mean ± SEM.
| Control (N = 25) |
Cannabis-exposed (N = 24) |
p-value | |
|---|---|---|---|
| Mother’s Age (years) | 23.84 ± 1.2 | 22.25 ± 0.7 | 0.2724 |
| Mother’s Education (years) | 12.12 ± 0.4 | 11.87 ± 0.4 | 0.6712 |
| Mother’s Race (White/Black/Hispanic) | 2/21/2 | 0/19/5 | - |
| Fetal Sex (male/female) | 13/12 | 13/11 | - |
| Fetal Age (gestation weeks) | 20.36 ± 0.3 | 20.21 ± 0.3 | 0.6961 |
| Postmortem Interval (hours) | 9.58 ± 0.7 | 7.97 ± 0.7 | 0.1248 |
| Maternal Cannabis Use (ADJ) | 0 | 1.24 ± 0.2 | <0.0001 |
| Maternal Cigarette Use (ADC) | 2.16 ± 0.8 | 4.04 ± 0.8 | 0.0983 |
| Maternal Alcohol Use (ADV) | 0.30 ± 0.1 | 0.30 ± 0.1 | 0.9899 |
Drugs
THC (10 mg/ml in ethyl alcohol 95%; NIDA) was evaporated under nitrogen gas and dissolved in saline with 0.3% Tween 80 to a concentration of 1.0 mg/ml. Morphine (Sigma-Aldrich) was dissolved in saline.
Prenatal THC rat model
Adult male (226-250g) and female (151-175g) Long Evans rats (Charles River Laboratories) were housed under a 12h light/dark cycle. Females were surgically implanted with an intravenous (i.v.) jugular catheter and treated post-surgically as previously described (5) with ampicillin (50 mg/kg in Heparin 10U, i.v.) and carprofen (5 mg/kg, s.c.) to prevent infection and manage pain, respectively. Animals were pair mated (2 females: 1 male) for 5 days to ensure that each female went through at least one estrous cycle (15). After mating, females were individually housed and the day of separation was recorded as gestation day 1 (GD1). Females were treated with daily i.v. injections of either THC (0.15 mg/kg) or vehicle (VEH, 0.3 % Tween 80-sterile saline solution) from GD5-PND2. This treatment period corresponds to the neurodevelopmental period examined in our human fetal population (midgestation, ~20 weeks) (16). The dose of THC used in this paradigm is comparable to current estimates of low dose cannabis cigarettes (~16 mg of THC) (17). On PND2, litters were culled 8-10 and pups were fostered such that all pups were raised by vehicle-exposed dams and no dams raised its own offspring. On PND21, pups were weaned and allowed to mature into adulthood. Only males were studied to reduce variability and to be consistent with previous work showing greater alterations in male fetuses with prenatal cannabis exposure (18-19). Two pups/litter were used for different experiments.
Preparation of rat tissue sections for gene expression studies
Adult (PND62) rats were anaesthetized with CO2 and decapitated and neonates (PND2) were sacrificed using live decapitation. After decapitation, brains were frozen in isopentane and stored at −80°C. Coronal cyrosections (20-μm thick) of the striatum were taken and slide-mounted sections stored at −30°C.
In situ hybridization histochemistry (ISHH)
ISHH was used to study DRD1, DRD2, PENK and PDYN in the human fetus (18-19) and Drd2 (NM_012547; sense probe: GAGAAGGCTTTGCAGACCAC, antisense probe: GGATGGATCAGGGAGAGTGA) in the rat. A detailed ISHH procedure has been published previously (20). Briefly, cDNA fragments were obtained from total RNA by reverse transcription-PCR. RNA probes were transcribed in the presence of [35S]-uridine 5′-[α-thio]triphosphate (specific activity 1000–1500 Ci/mmol; New England Nuclear). The labeled probe was applied to the brain sections at a concentration of 2 × 103 cpm/mm2 of the coverslip area. Two adjacent sections/subject were studied at the level of the NAc (anatomical location based on Nissl stain and expression pattern of biochemical marks examined throughout the striatum for each subject). Slides were hybridized overnight at 55°C and apposed to Imaging Plates (Fujifilm) along with 14C-standards (American Radiolabeled Chemicals). Films were developed with FLA-7000 phosphoimaging analyzer (Fujifilm) and images analyzed (MultiGauge software). Relative mRNA expression levels were measured within the human NAc and putamen (dorsal striatum) based on Human Atlas (21) and the rat NAc (+1.60 mm from Bregma) and caudate-putamen (+1.60 mm) in accordance with reference atlas (22). Values from duplicate brain sections for each subject were averaged and expressed as dpm/mg of tissue by reference to the co-exposed standard.
D2 3H – raclopride binding
Adult rat brain sections were incubated in 2nM [3H]raclopride (80Ci/mmol specific activity) in incubation buffer (50mM Tris, 0.1% ascorbic acid, 120nM NACl, 5mM KCl, 2mM CaCl2, 1mM MgCl2, pH 7.4) for 60 mins. Non-specific binding was determined on adjacent brain sections by adding 100mM unlabeled raclopride in the binding buffer. After incubation, the sections were rinsed in cold incubation buffer, rapid dip in cold distilled water and dried. The dried slides were made conductive by an application of metal electric tape (3M, St. Paul, MN) and placed in the β-imager™ 2000 (Biospace Lab, France). Data were collected ~18h and the level of bound radioactivity was directly determined by estimating the number of β-particles obtained from the delineated brain area of interest using the β-vision program (Biospace). The radioligand signal was measured in duplicate, averaged, and expressed as counts/minute/square millimeter (cpm/mm2). Specific binding was determined as the total minus non-specific binding, which for [3H]raclopride was negligible with levels similar to white matter background.
Chromatin immunoprecipitation (ChIP)
NAc was dissected from frozen adult brains of 12 vehicle- and 12 THC-treated rats (2 pups per litter) using a 15-gauge sample punch on a −25°C cold-block . Bilateral tissue from two animals was pooled per sample and fixed with 1% formaldehyde. Samples were sonicated using Bioruptor (Diagenode) in buffer containing 50 mM Hepes pH 7.9, 140 mM NaCl, 1mM EDTA, 1% Triton X-100, 0.1% Na-deoxycholate and 0.1% SDS. 25μg chromatin (measured as DNA at an optical density of 260nm) was immoprecipitated with 9μl α-2meH3K9 antibody (Abcam, catalog #ab1220), 3μl α-3meH3K4 antibody (Active Motif, catalog # 39159), 3μl α-RNA Polymerase II antibody (Covance, 8WG16) and nonspecific mouse IgG (Santa Cruz Biotechnology). Immunocomplexes were collected using protein G Dynabeads (Invitrogen), beads washed, bound fraction eluted, extracted with phenol-chlorophorm, ethanol-precipitated and resuspended in Tris-EDTA buffer. ChIP material was analyzed using SYBR green master mix (Roche) in a LightCycler 480 instrument (Roche) and the default SYBR green PCR program except for annealing temperature (Table S1 in the Supplement). PCR reactions were carried out in triplicate. Cp values from each PCR reaction were obtained using the second derivative maximum method (Roche). The enrichment of a given target sequence precipitated by the antibody was determined as the difference between the amount of target sequence in the immunoprecipitated fraction and the amount of target sequence in the input DNA. Relative quantification and analysis of THC effects were performed using a modified version of the ΔΔCt method using the samples from vehicle-treated animals as the reference group (23). The background immunoprecipitation of chromatin with nonspecific mouse IgG was determined for both groups (Figure S3 in the Supplement).
Morphine place conditioning
PND62 rats with prenatal THC- or vehicle-exposure were tested in an apparatus (ENV-517, MED Associates Inc., Vermont) with two compartments distinct in wall pattern and tactile floor environments. On experiment day 1, rats were allowed to explore both compartments in a 15 minute habituation session. The pre-conditioning test was conducted on day 2 where rats were again allowed to explore both compartments for 15 minutes to assess baseline preference. On days 3, 5 and 7 rats were administered morphine (4 mg/kg s.c.) in their home cages and after 15 minutes were placed in the least preferred compartment for 30 minutes. On days 4, 6 and 8 rats were administered saline in their home cages and after 15 minutes were placed in the other compartment for 30 minutes. On day 9, a post-conditioning test was performed which was identical to the pre-conditioning test. Preference was calculated as the difference in time spent in the drug-paired compartment between the pre-conditioning and post-conditioning tests.
Statistical analysis
For human gene expression studies, data were tested for normality and normalized by natural log transformation if they were not normally distributed. Univariate statistical analyses were used to study the effect of each independent demographic variable (e.g. fetal age, sex, PMI, etc.) on mRNA expression. Variables with a p-value less than 0.10 were included in a multiple regression model and maternal cannabis, alcohol and cigarette were always included in the final model. The dose-dependent effects of prenatal drug exposure on mRNA expression were analyzed by nonparametric correlation (Spearman ranked R). Outliers were excluded by residual analysis. For the animal experiments, statistical comparison between vehicle- and THC-pretreated animals was performed by one-way ANOVA.
Results
Demographics of the human fetal population are summarized in Table 1, which consisted of forty-nine primarily African American subjects with no significant differences between the cannabis and control groups except for the amount of maternal cannabis use.
Maternal cannabis use decreases DRD2 mRNA levels in the NAc of the human fetus
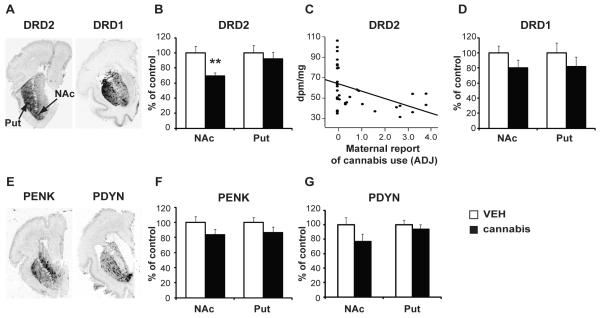
High levels of dopaminergic D1 (DRD1) and D2 (DRD2) receptor mRNA transcripts were detected in the dorsal and ventral striatum (Fig. 1A). Statistical analysis revealed an overall significant alteration of DRD2 mRNA levels [F(3,46)=3.09, p=0.036] with prenatal cannabis exposure accounting for decreased expression detected in the NAc (p=0.003; Fig. 1B). Moreover, NAc DRD2 mRNA levels were negatively correlated with maternal report of cannabis use (r=−0.42, p=0.005; Fig. 1C). No other drug showed a significant association with NAc DRD2 mRNA levels (cigarette p=0.4916, alcohol p=0.604) and DRD2 mRNA expression was not altered in the putamen with cannabis exposure (p=0.736). In contrast to cannabis, maternal alcohol use was associated with a reduction of DRD2 (30%; p=0.0187) and DRD1 (45%; p=0.004) in the dorsal striatum. DRD1 mRNA levels were not significantly altered in the NAc for any drug including cannabis [F(3,35)=1.186, p=0.330; Fig. 1D].
Figure 1.
Dopamine receptor and opioid neuropeptide mRNA levels in human fetal striatum. (A) In situ hybridization autoradiograms showing the distribution pattern of DRD2 and DRD1 mRNA transcripts in the striatum. (B) DRD2 mRNA levels in the NAc and putamen of cannabis-exposed subjects. (C) Correlation of NAc DRD2 mRNA levels with maternal report of cannabis use. (D) DRD1 mRNA levels in the NAc and putamen of cannabis-exposed subjects. (E) Distribution pattern of PENK and PDYN mRNA transcripts in the human fetal striatum. (F) PENK and (G) PDYN mRNA levels in the NAc and putamen of cannabis-exposed subjects. Bar graphs show the cannabis-exposed group (black bars) expressed relative to control (white bars). Date are expressed as mean ± SEM. **, p <0.01 vs control subjects. N=17-24 subjects per group. NAc, nucleus accumbens; Put, putamen; ADJ, average daily joint; dpm/mg, disintegrations per minute per milligram.
Given that the opioid neuropeptides proenkephalin (PENK) and prodynorphin (PDYN) are also highly expressed in the striatum and dissociate striatopallidal (D2R/PENK) and striatonigral (D1R/PDYN) neurons (11), we also analyzed PENK and PDYN mRNA levels in the human fetal striatum (Fig. 1E) to identify any generalized effects of maternal cannabis use. Although PENK mRNA expression tended to be slightly reduced in the cannabis group covaried for age (Fig. 1F), it was not significant since overall PENK alterations in the NAc [F(4,44)=4.76; p=0.003; Figure S1 in the Supplement] and dorsal striatum [F(4,43)=4.720; p=0.003] were strongly influenced by alcohol. Prenatal alcohol exposure was associated with attenuation of the normal developmental upregulation of striatal PENK levels within the NAc (p=0.003; Figure S1 in the Supplement) and putamen (p=0.018). PDYN mRNA levels were not related to maternal cannabis use (p=0.155), but expression in the NAc was significantly associated with cigarette exposure (p=0.007); there was a negative correlation with the amount of reported cigarette use (r=−0.555, p=0.0004; Figure S2 in the Supplement).
Prenatal THC exposure reduces Drd2 mRNA expression in the rat NAc of the offspring that is maintained into adulthood
The observation that prenatal cannabis exposure was related to reduced DRD2 transcripts in the NAc of the human fetal brain prompted us to investigate the specificity of this effect on the developmental regulation of Drd2 expression in the ventral striatum especially given maternal polysubstance use. As such, striatal sections from offspring with prenatal THC exposure were examined for disturbances in Drd2 mRNA levels. There was no significant difference in developmental parameters as a consequence of the THC exposure. Similar to the human cannabis-exposed fetus, Drd2 mRNA expression was decreased by ~40% in the NAc [F(1,15)=16.472, p=0.001], but not the dorsal striatum [F(1,15)=0.846, p=0.373], of the PND2 pup (Fig. 2A). To evaluate whether prenatal THC exposure causes long-term impairments in the expression of the Drd2 gene, a separate cohort of male offspring was studied in adulthood (PND62). Intriguingly, Drd2 mRNA levels continued to be reduced (~30%) at PND62 in the NAc and remained unchanged in the dorsal striatum [F(1,8)=1.12, p=0.322]. The NAc core and shell, important components of motor and reward circuits respectively (24), could be dissociated at PND62, and Drd2 mRNA levels were decreased in both subregions (NAc core: [F(1,8)=9.92, p<0.01]; NAc shell: [F(1,8)=7.09, p<0.05]; Fig. 2B), indicating that THC-induced disruption of mesolimbic Drd2 mRNA levels persisted into adulthood.
Figure 2.
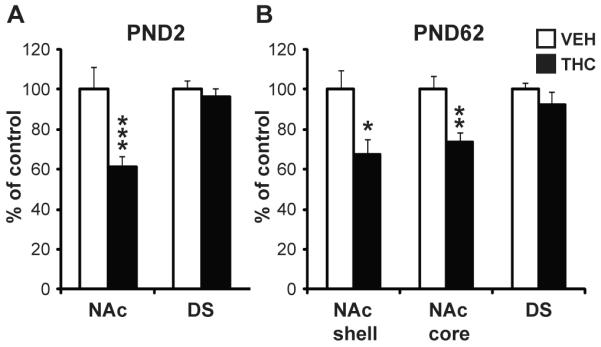
Striatal Drd2 mRNA levels in rats exposed to THC in utero. Drd2 mRNA levels in the striatum of PND2 (A) and PND62 (B) rats with prenatal THC exposure. Bar graphs show the THC-exposed group (black bars) expressed relative to control (white bars). Data are expressed as mean ± SEM. *, p < 0.05; **, p < 0.01; ***, p < 0.001 vs control subjects. N=5-16 rats per group. PND, postnatal day.
Prenatal THC exposure disrupts the long-term epigenetic regulation of Drd2 mRNA expression in the rat NAc
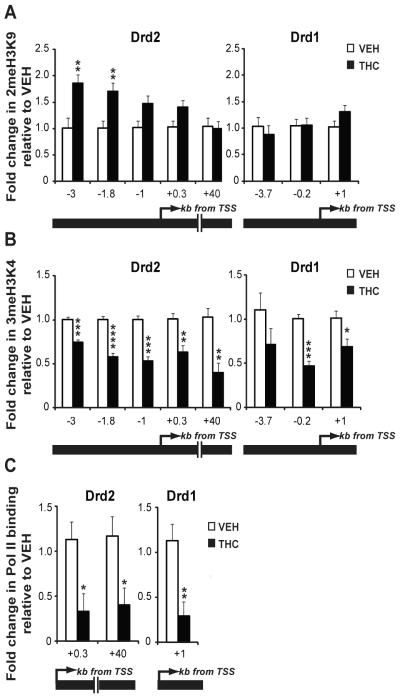
The detection of reduced Drd2 mRNA transcript levels in the NAc of cannabis-exposed human fetuses and in neonatal rats with prenatal THC exposure, together with the persistence of the change into adulthood in rats, indicated that the observed downregulation of Drd2 gene expression may be achieved via epigenetic regulatory processes. One mechanism by which THC could epigenetically modulate the levels of Drd2 mRNA expression is by affecting the post-translational modification of nucleosomal histones in the locus. To investigate this possibility, we performed chromatin immunoprecipitation (ChIP) on extracts isolated from the NAc of adult male rats with prenatal THC exposure and immunopreciptated with antibodies specific for dimethylated lysine 9 (2meH3K9) and trimethylated lysine 4 (3meH3K4) on histone H3. These modifications were chosen because decreased Drd2 expression could be modulated by enhanced transcriptional repression, reduced transcriptional activation or both. It is well accepted that 3meH3K4 is mainly detected in transcriptionally active chromatin regions of the genome (25-26) while dimeH3K9 has been often associated with repression of developmental genes in other experimental paradigms (27-28). After immunoprecipitation, the purified DNA was amplified using primers specific for four evolutionarily conserved genomic regions within 3 kb upstream of the Drd2 transcription start site (TSS) and for one site within the coding sequence. The profile of 2meH3K9 and 3meH3K4 at the Drd2 locus in the NAc of drug-naïve (prenatally vehicle-treated) adult rats revealed higher levels of 3meH3K4 close (+0.3kb) to the TSS, and higher levels of 2meH3K9 upstream of the TSS (Figure S3 in the Supplement).
Interestingly, prenatal THC exposure significantly increased the repressive 2meH3K9 mark between −1.8kb (69% increase vs control) and −3kb (83% increase vs control) upstream of the TSS and decreased 3meH3K4 across the analyzed genomic fragment in the NAc of adult rats exposed to THC in utero (Fig. 3A and B; Figure S4 in the Supplement). In agreement with 3meH3K4 as a mark of transcriptional activity, its reduction was associated with decreased RNA polymerase II (Pol II) at the TSS (+0.3kb) and within the coding region (+40kb) (Fig. 3C). Although no change in 2meH3K9 was observed at the Drd1 gene (Fig. 3A), there was reduced 3meH3K4 (Fig. 3B) and decreased Pol II (Fig. 3C) association at this locus despite the lack of alteration of Drd1 transcripts in response to prenatal THC exposure.
Figure 3.
Dopamine receptor gene regulation in the NAc of adult rats with prenatal THC exposure. (A) Analysis of 2meH3K9 at the Drd2 gene and Drd1 gene. (B) Analysis of 3meH3K4 at the Drd2 gene and Drd1 gene. (C) Analysis of Pol II binding in Drd2 and Drd1 gene. Values are expressed as mean ± SEM. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001vs control subjects. N=5-6 samples (bilateral pooled NAc from 2 rats in each sample)/group. Black bars, THC exposed group; white bars, vehicle-exposed group; kb, kilobases; TSS, transcription start site.
Prenatal THC exposure disrupts D2R function and increases opiate reward sensitivity in adulthood
The relevance of impaired Drd2 gene expression to D2R function was tested by investigating D2R binding using 3H-raclopride in vitro autoradiography. Similar to the alterations observed on gene expression, we found decreased D2R binding sites in the NAc [F(1,8)=6.15, p=0.05], but not in the dorsal striatum [F(1,9)=0.08, p=0.782] of adult rats exposed prenatally to THC (Fig. 4). Since reduced striatal D2R levels are associated with increased addiction vulnerability (29-30), we hypothesized that adult rats with prenatal THC exposure would be more sensitive to the rewarding effects of drugs of abuse since previous studies showed that prenatal THC-exposed adult rats exhibited higher sensitivity to heroin self-administration (5). In order to investigate this possibility, rats were tested in the place condition task with an opiate drug. Adult vehicle animals had no compartmental preference during pre-conditioning (433.75± 11.27s/light and 466.25 ± 11.27s/dark), but THC-exposed animals showed preference to the dark environment (295.57 ± 19.26s/light and 604.43 ± 19.26s/dark). Despite the baseline pre-conditioning aversion, rats exposed to THC in utero showed increased place preference for the initial non-preferred compartment in response to low dose morphine compared to vehicle exposed rats [F(1,10)=5.76, p=0.039] (Fig. 5).
Figure 4.
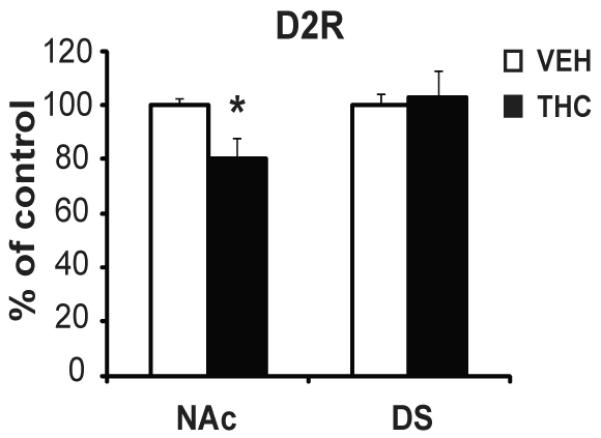
D2R 3H-raclopride binding in the striatum of adult rats with prenatal THC exposure. Bar graphs show the THC-exposed group (black bars) expressed relative to the vehicle group (white bars). Values are expressed as mean ± SEM. *, p < 0.05 vs vehicle group. N=5 rats per group.
Figure 5.
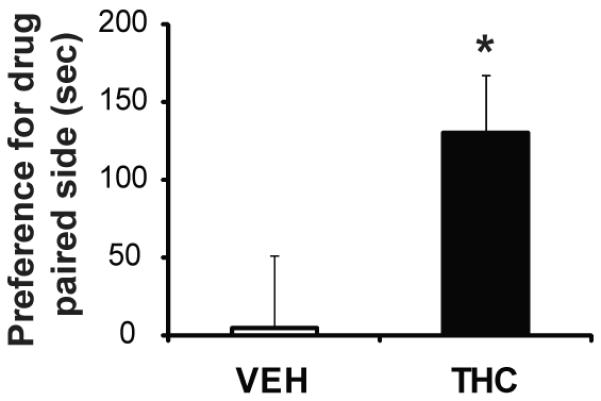
Place conditioning for morphine (4 mg/kg) in adult rats with prenatal THC or vehicle exposure. Data are represented as time spent on drug-paired side on test day. *, p < 0.05 vs vehicle group. N=5-6 rats per group.
Discussion
Our data suggest that the association between prenatal cannabis exposure and addiction vulnerability can be explained, at least in part, by cannabis-induced alterations in the epigenetic regulation of the DRD2 gene in the NAc. Mimicking the human mesolimbic DRD2 expression impairments in a rat prenatal THC exposure model also enabled us to follow the long-term trajectory of disrupted Drd2 regulation, which persisted into adulthood and was associated with increased levels of 2meH3K9. Moreover, prenatal THC-exposed adults showed enhanced opiate reward conditioned place sensitivity that superseded their baseline pre-conditioning aversion that is consistent with increased heroin self-seeking behavior previously documented in animals with similar prenatal treatment (5).
The finding that prenatal cannabis exposure disrupts DRD2 gene expression in the human fetal brain is interesting given that D2R dysregulation has been implicated in addiction risk. Imaging studies of adult subjects with a history of drug abuse have reported reduced levels of D2Rs in the striatum (12), and animal models documented that reduction of NAc Drd2 gene enhances drug intake (31). Human genetic studies have also linked variants of the D2 receptor gene to addiction phenotypes related to alcohol, opiates and psychostimulant abuse (32). While we do not exclude a genetic contribution to the impairment of the DRD2 mRNA expression observed in the cannabis-exposed fetuses, the complementary animal model supports our contention that adult NAc D2R regulation is impacted by prenatal THC exposure.
Studies of the human brain are invariably complex especially given the inability to regulate confounds including multi-drug use and it is expected that alcohol and cigarette could affect DRD2. Examining other striatal genes in addition to DRD2, however, can help to provide important information regarding the specificity of cannabis-related alterations. Similar to DRD2, PENK mRNA is also expressed on striatopallidal neurons in both the ventral and dorsal striatum (11, 33), but striatal PENK expression was strongly influenced by alcohol exposure. The impact of maternal alcohol use on striatal PENK mRNA levels could have masked cannabis-induced effects of PENK mRNA expression considering that reduced NAc PENK mRNA was detected in neonate rats with prenatal THC exposure (5). However, as compared to cannabis, alcohol was associated with a widespread disturbance of most markers studied particularly in the dorsal striatum. The specificity of the molecular observations to cannabis was also supported by the observation that although cigarette smoking is common in cannabis users, only the PDYN mRNA was directly related to the maternal report of cigarette use, in contrast to the DRD2 expression which was significantly correlated to maternal cannabis intake. Thus, it is possible to dissociate specific drug-related effects in the human fetal brain, especially when substantiated by the use of complementary animal models.
Mimicking cannabis-induced human fetal mesolimbic DRD2 gene expression disturbances in the prenatal THC rat model also provided novel information regarding epigenetic disturbances that could maintain Drd2 gene expression impairments into adulthood. Identifying specific epigenetic mechanisms that may play a role in maintaining cannabis-induced alterations in Drd2 gene expression is important since most studies showing long-term epigenetic disturbances in the offspring due to environmental influences has been related to maternal diet-induced changes (34) and no data currently exist on such protracted effects of early developmental drug exposure. The majority of epigenetic studies in the drug abuse literature have focused on cocaine-induced changes in chromatin during adolescence or adulthood, rather than during the prenatal period (26, 28, 35). A particularly interesting observation of our study is the THC-induced increase in 2meH3K9 in a discrete region upstream of the Drd2 TSS. In contrast to 3meH3K4, the 2meH3K9 mark remained unchanged at the Drd1 gene, indicating a developmental regulatory mechanism that might act on specific sequences located between −1.8 and 3 kb upstream of the Drd2 TSS. Developmental regulation of 2meH3K9 has been shown to be important for the appropriate tissue-specific expression of a variety of gene loci not only at promoters but also in broader regulatory regions including enhancers (36-37).
The site-specific increase in 2meH3K9 was accompanied by decreased 3meH3K4 across the analyzed Drd2 genomic fragment. Reduced 3meH3K4 was also detected at the Drd1 gene without significant change in 2meH3K9, suggesting that long-term disruption of 3meH3K4 due to prenatal THC exposure may not on its own be sufficient for altering transcriptional activity. However, the direction of change of both 2meH3K9 (increase) and 3meH3K4 (decrease) at the Drd2 gene is consistent with the observed reduction in Drd2 mRNA expression in adulthood. Antagonistic roles for histone H3 lysine 9 and 4 methylation have been widely documented in gene regulation during brain development (25). For example, it has been shown using in vitro biochemical and in vivo approaches that H3K9 methylation levels can only be increased in the absence of the 3meH3K4 mark due to the specificity of enzymes that modulate H3K9 methylation, the binding of which is blocked by high level 3meH3K4 (38). Such a mechanism would explain the reduced 3meH3K4 levels induced by THC and the recruitment of the enzymes that modulate 2meH3K9 at the upstream sequence element of the Drd2 gene. As the RNA polymerase II transcription machinery is known to physically and functionally interact with chromatin regions containing 3meH3K4 (39), the THC-induced reduction in 3meH3K4 could explain decreased Pol II association with the gene and downregulation of Drd2 mRNA production. Identification of the chromatin modifying enzymes that mediate the long-term effect of prenatal THC exposure will be an important subject of future studies.
In conclusion, these data emphasize the sensitive nature of the prenatal period, during which cannabis exposure can set into motion epigenetic alterations that contribute to long-term disturbances of the D2R in adulthood, thereby laying a foundation for increased vulnerability to addiction and potentially other psychiatric disorders.
Supplementary Material
Acknowledgements
We thank Drs. Howard Minkoff and Diane Ashton for help with access to the VIP clinic and Dr. Virginia Anderson for initial assistance with the fetal morphometrics. We also thank Alexandra Guilliume and Dionne Dunkley for helping with maternal interviews and fetal brain collection and Barbro Berthelsson and Alexandra Tylec for technical assistance with the gene expression experiments. This research was supported by NIH grant DA12030 and DA23214. PC is funded by R01-NS42925.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Financial Disclosures The authors reported no biomedical financial interests or potential conflicts of interest.
References
- 1.SAMHSA . Results from the 2005 National Survey on Drug Use and Health: National Findings. SAMHSA Office of Applied Studies; Rockville, MD: 2006. [Google Scholar]
- 2.Huizink AC, Mulder EJH. Maternal smoking, drinking or cannabis use during pregnancy and neurobehavioral and cognitive functioning in human offspring. NeurosciBiobehavRev. 2006;30:24–41. doi: 10.1016/j.neubiorev.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 3.Porath AJ, Fried PA. Effects of prenatal cigarette and marijuana exposure on drug use among offspring. NeurotoxicolTeratol. 2005;27:267–277. doi: 10.1016/j.ntt.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Leech SL, Larkby CA, Day R, Day NL. Predictors and correlates of high levels of depression and anxiety symptoms among children at age 10. J Am Acad Child Adolesc Psychiatry. 2006;45:223–230. doi: 10.1097/01.chi.0000184930.18552.4d. [DOI] [PubMed] [Google Scholar]
- 5.Spano MS, Ellgren M, Wang X, Hurd YL. Prenatal cannabis exposure increases heroin seeking with allostatic changes in limbic enkephalin systems in adulthood. Biol Psychiatry. 2007;61:554–563. doi: 10.1016/j.biopsych.2006.03.073. [DOI] [PubMed] [Google Scholar]
- 6.Rubio P, de Fonseca F Rodriguez, Martin-Calderon JL, Del Arco I, Bartolome S, Villanua MA, et al. Maternal exposure to low doses of delta9-tetrahydrocannabinol facilitates morphine-induced place conditioning in adult male offspring. Pharmacol Biochem Behav. 1998;61:229–238. doi: 10.1016/s0091-3057(98)00099-9. [DOI] [PubMed] [Google Scholar]
- 7.Singh ME, McGregor IS, Mallet PE. Perinatal exposure to delta(9)-tetrahydrocannabinol alters heroin-induced place conditioning and fos-immunoreactivity. Neuropsychopharmacology. 2006;31:58–69. doi: 10.1038/sj.npp.1300770. [DOI] [PubMed] [Google Scholar]
- 8.Vela G, Martin S, Garcia-Gil L, Crespo JA, Ruiz-Gayo M, Fernandez-Ruiz J Javier, et al. Maternal exposure to delta9-tetrahydrocannabinol facilitates morphine self-administration behavior and changes regional binding to central mu opioid receptors in adult offspring female rats. Brain Res. 1998;807:101. doi: 10.1016/s0006-8993(98)00766-5. [DOI] [PubMed] [Google Scholar]
- 9.Hurd Y, Hall H. Handbook of Chemical Neuroanatomy. 2005. Human Forebrain Dopamine Systems: Characterization of the Normal Brain and in Relation to Psychiatric Disorders; pp. 525–571. [Google Scholar]
- 10.Haber SN. The primate basal ganglia: parallel and integrative networks. J Chem Neuroanat. 2003;26:317–330. doi: 10.1016/j.jchemneu.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 11.Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE, et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Volkow ND, Fowler JS, Wang GJ, Swanson JM. Dopamine in drug abuse and addiction: results from imaging studies and treatment implications. Mol Psychiatry. 2004;9:557–569. doi: 10.1038/sj.mp.4001507. [DOI] [PubMed] [Google Scholar]
- 13.Hurd YL, Wang X, Anderson V, Beck O, Minkoff H, Dow-Edwards D. Marijuana impairs growth in mid-gestation fetuses. Neurotoxicol Teratol. 2005;27:221–229. doi: 10.1016/j.ntt.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 14.Richardson GA, Ryan C, Willford J, Day NL, Goldschmidt L. Prenatal alcohol and marijuana exposure: effects on neuropsychological outcomes at 10 years. NeurotoxicolTeratol. 2002;24:309–320. doi: 10.1016/s0892-0362(02)00193-9. [DOI] [PubMed] [Google Scholar]
- 15.Freeman ME. The ovarian cycle of the rat. In: Knobil E, Neil J, editors. Physiology of reproduction. Raven Press Ltd.; New York: 1988. pp. 1893–1928. [Google Scholar]
- 16.Bayer SA, Altman J, Russo RJ, Zhang X. Timetables of neurogenesis in the human brain based on experimentally determined patterns in the rat. Neurotoxicology. 1993;14:83–144. [PubMed] [Google Scholar]
- 17.Grotenhermen F. Pharmacokinetics and pharmacodynamics of cannabinoids. Clin Pharmacokinet. 2003;42:327–360. doi: 10.2165/00003088-200342040-00003. [DOI] [PubMed] [Google Scholar]
- 18.Wang X, Dow-Edwards D, Anderson V, Minkoff H, Hurd YL. In utero marijuana exposure associated with abnormal amygdala dopamine D2 gene expression in the human fetus. Biol Psychiatry. 2004;56:909–915. doi: 10.1016/j.biopsych.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 19.Wang X, Dow-Edwards D, Anderson V, Minkoff H, Hurd YL. Discrete opioid gene expression impairment in the human fetal brain associated with maternal marijuana use. Pharmacogenomics J. 2006;6:255–264. doi: 10.1038/sj.tpj.6500375. [DOI] [PubMed] [Google Scholar]
- 20.Hurd YL. In situ hybridization with isotopic riboprobes for detection of striatal neuropeptide mRNA expression after dopamine stimulant administration. Methods Mol Med. 2003;79:119–135. doi: 10.1385/1-59259-358-5:119. [DOI] [PubMed] [Google Scholar]
- 21.Feess-Higgins A, Larroche J-C. In: Development of the human foetal brain. An anatomical atlas. INSERM, editor. Masson; Paris: 1987. [Google Scholar]
- 22.Paxinos G, Watson C. The Rat Brain in Stereotaxic Coordinates. 3rd ed Academic Press; New York: 1997. [Google Scholar]
- 23.Chakrabarti SK, James JC, Mirmira RG. Quantitative assessment of gene targeting in vitro and in vivo by the pancreatic transcription factor, Pdx1. Importance of chromatin structure in directing promoter binding. J Biol Chem. 2002;277:13286–13293. doi: 10.1074/jbc.M111857200. [DOI] [PubMed] [Google Scholar]
- 24.Zahm DS. An integrative neuroanatomical perspective on some subcortical substrates of adaptive responding with emphasis on the nucleus accumbens. Neurosci Biobehav Rev. 2000;24:85–105. doi: 10.1016/s0149-7634(99)00065-2. [DOI] [PubMed] [Google Scholar]
- 25.Akbarian S, Huang HS. Epigenetic regulation in human brain-focus on histone lysine methylation. Biol Psychiatry. 2009;65:198–203. doi: 10.1016/j.biopsych.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Black YD, Maclaren FR, Naydenov AV, Carlezon WA, Jr., Baxter MG, Konradi C. Altered attention and prefrontal cortex gene expression in rats after binge-like exposure to cocaine during adolescence. J Neurosci. 2006;26:9656–9665. doi: 10.1523/JNEUROSCI.2391-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horn PJ, Peterson CL. Heterochromatin assembly: a new twist on an old model. Chromosome Res. 2006;14:83–94. doi: 10.1007/s10577-005-1018-1. [DOI] [PubMed] [Google Scholar]
- 28.Maze I, Covington HE, 3rd, Dietz DM, LaPlant Q, Renthal W, Russo SJ, et al. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science. 2010;327:213–216. doi: 10.1126/science.1179438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Volkow ND, Fowler JS, Wang GJ, Swanson JM, Telang F. Dopamine in drug abuse and addiction: results of imaging studies and treatment implications. Arch Neurol. 2007;64:1575–1579. doi: 10.1001/archneur.64.11.1575. [DOI] [PubMed] [Google Scholar]
- 30.Wang GJ, Volkow ND, Fowler JS, Logan J, Abumrad NN, Hitzemann RJ, et al. Dopamine D2 receptor availability in opiate-dependent subjects before and after naloxone-precipitated withdrawal. Neuropsychopharmacology. 1997;16:174–182. doi: 10.1016/S0893-133X(96)00184-4. [DOI] [PubMed] [Google Scholar]
- 31.Thanos PK, Michaelides M, Umegaki H, Volkow ND. D2R DNA transfer into the nucleus accumbens attenuates cocaine self-administration in rats. Synapse. 2008;62:481–486. doi: 10.1002/syn.20523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Le Foll B, Gallo A, Le Strat Y, Lu L, Gorwood P. Genetics of dopamine receptors and drug addiction: a comprehensive review. Behav Pharmacol. 2009;20:1–17. doi: 10.1097/FBP.0b013e3283242f05. [DOI] [PubMed] [Google Scholar]
- 33.Zhou L, Furuta T, Kaneko T. Chemical organization of projection neurons in the rat accumbens nucleus and olfactory tubercle. Neuroscience. 2003;120:783–798. doi: 10.1016/s0306-4522(03)00326-9. [DOI] [PubMed] [Google Scholar]
- 34.Lillycrop KA. Effect of maternal diet on the epigenome: implications for human metabolic disease. Proc Nutr Soc. 2011;70:64–72. doi: 10.1017/S0029665110004027. [DOI] [PubMed] [Google Scholar]
- 35.Laplant Q, Nestler EJ. CRACKing the histone code: Cocaine’s effects on chromatin structure and function. Horm Behav. 2010 doi: 10.1016/j.yhbeh.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hublitz P, Albert M, Peters AH. Mechanisms of transcriptional repression by histone lysine methylation. Int J Dev Biol. 2009;53:335–354. doi: 10.1387/ijdb.082717ph. [DOI] [PubMed] [Google Scholar]
- 37.Rentoft M, Kim K, Cho Y, Lee CH, Kim A. Enhancer requirement for histone methylation linked with gene activation. FEBS J. 2008;275:5994–6001. doi: 10.1111/j.1742-4658.2008.06728.x. [DOI] [PubMed] [Google Scholar]
- 38.Binda O, LeRoy G, Bua DJ, Garcia BA, Gozani O, Richard S. Trimethylation of histone H3 lysine 4 impairs methylation of histone H3 lysine 9: regulation of lysine methyltransferases by physical interaction with their substrates. Epigenetics. 2010;5:767–775. doi: 10.4161/epi.5.8.13278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang P, Lin C, Smith ER, Guo H, Sanderson BW, Wu M, et al. Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1- mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II. Mol Cell Biol. 2009;29:6074–6085. doi: 10.1128/MCB.00924-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Richardson GA, Ryan C, Willford J, Day NL, Goldschmidt L. Prenatal alcohol and marijuana exposure: effects on neuropsychological outcomes at 10 years. Neurotoxicol Teratol. 2002;24:309–320. doi: 10.1016/s0892-0362(02)00193-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.