Abstract
The conserved catalytic core of the Tetrahymena group I ribozyme is encircled by peripheral elements. We have carried out a detailed structure-function study of the five long-range tertiary contacts that fasten these distal elements together. Mutational ablation of each of the tertiary contacts destabilizes the folded ribozyme, indicating a role of the peripheral elements in overall stability. Once folded, three of the five tertiary contact mutants exhibit defects in overall catalysis that range from 20- to 100-fold. These and the subsequent results indicate that the structural ring of peripheral elements does not act as a unitary element; rather, individual connections have distinct roles as further revealed by kinetic and thermodynamic dissection of the individual reaction steps. Ablation of P14 or the metal ion core/metal ion core receptor (MC/MCR) destabilizes docking of the substrate-containing P1 helix into tertiary interactions with the ribozyme’s conserved core. In contrast, ablation of the L9/P5 contact weakens binding of the guanosine nucleophile by slowing its association, without affecting P1 docking. The P13 and tetraloop/tetraloop receptor (TL/TLR) mutations had little functional effect and small, local structural changes, as revealed by hydroxyl radical footprinting, whereas the P14, MC/MCR, and L9/P5 mutants show structural changes distal from the mutation site. These changes extended into regions of the catalytic core involved in docking or guanosine binding. Thus, distinct allosteric pathways couple the long-range tertiary contacts to functional sites within the conserved core. This modular functional specialization may represent a fundamental strategy in RNA structure-function interrelationships.
Fundamental chemical and physical principles of biological catalysis have been revealed and reinforced through comparisons of RNA and protein active sites (1, 2). RNA and proteins share active site catalytic strategies, including electrostatic stabilization, facilitation of proton transfer, and positioning of reacting and catalytic groups. These active sites exist within overall structures composed of either RNA or protein, but for RNA in particular, there is limited information on how aspects of the broader structure impact the active site.
Several investigations of regions surrounding the active sites of proteins suggest that mutations of most non-active site regions have modest or negligible functional effects (3–10). For example, 55% of the amino acids in the 164-residue T4 lysozyme and 59% of the amino acids in the 329-residue lac repressor could be mutated to produce functional proteins with activities that were within 30-fold and 10-fold, respectively, of the wild type activities (3, 4). Nevertheless, non-active site regions within a folded protein have been shown to modulate substrate specificity or reactivity in several cases (11–19). For example, changing the substrate specificity of trypsin to that of chymotrypsin required mutation of non-active site loop regions in addition to mutations in the substrate recognition pocket (12); the promiscuous activities of serum paraoxonase were increased ∼100-fold over those of the wild type by mutation of two to four residues located in non-active site loops and helices as well as the substrate-binding pocket (19); and the activity of a catalytic antibody was increased by 100-fold by changing only non-active site residues (18). More broadly, non-active site residues allow allosteric coupling between distal sites and the active site, suggesting that long-range interactions can affect function within the active site (8, 20, 21).
The catalytic contributions of non-active site regions in RNA have been less studied, but structural differences between proteins and RNA suggest that RNA may use non-active site regions differently in catalysis than proteins do. Protein structure appears to be dominated by the formation of a packed hydrophobic core that is bolstered by networks of hydrogen bonding interspersed throughout the structure (22). RNA lacks fully hydrophobic side chains and the side chain diversity of proteins so that packing cannot be as dense and extensive (2). RNA may therefore require different means to fold and adopt highly specific, functional conformations. Visual inspection of the secondary structures of RNA structure reveals rigid helical elements connected by loops and junctions, often in complex topologies (23). Functional sites are often constructed from loops and junctions at the confluence of multiple helical elements within these complex topologies (24–29). These helices, in turn, appear to be positioned by tertiary interactions that are often long-range, bringing together helices that are distal in secondary structure (30–37). These few, punctuated regions of tertiary contact in RNA contrast starkly with the many regions of contact within the tightly packed cores of folded proteins. The subject of this study is the interplay between the aforementioned long-range tertiary interactions and the RNA functional core.
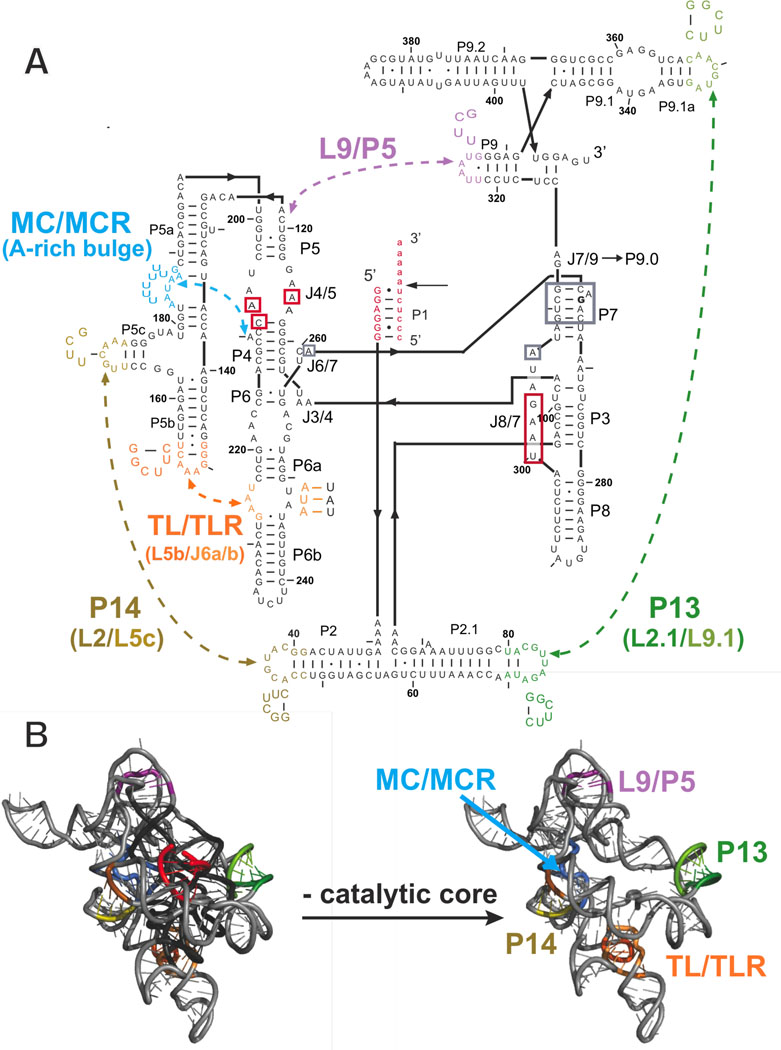
The Tetrahymena group I ribozyme is a powerful system to study RNA catalysis (38). Phylogenetic and x-ray crystallographic data provide a structural view of this group I intron that can guide functional studies (24–27, 39–41). The ribozyme is composed of a catalytic core, conserved in the ∼3000 known group I introns (42), which consists of two helically stacked regions – P5-P4-P6 and P3-P7 (Figure 1A) that interact directly with the substrates (38). The conserved core is encircled by peripheral regions, which contain five long-range tertiary contacts (Figure 1B). These peripheral regions are conserved among a subset of the 1C1 group I introns, the subclass to which the Tetrahymena group I ribozyme belongs (42).
Figure 1.
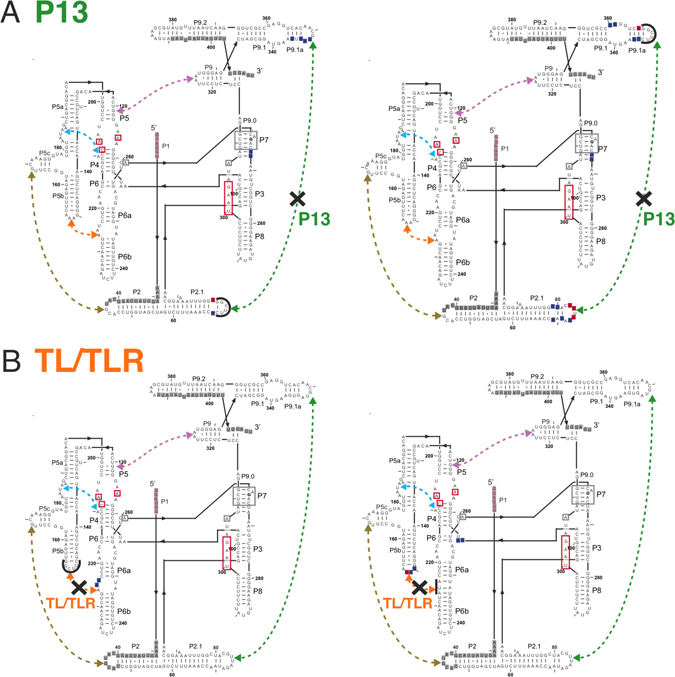
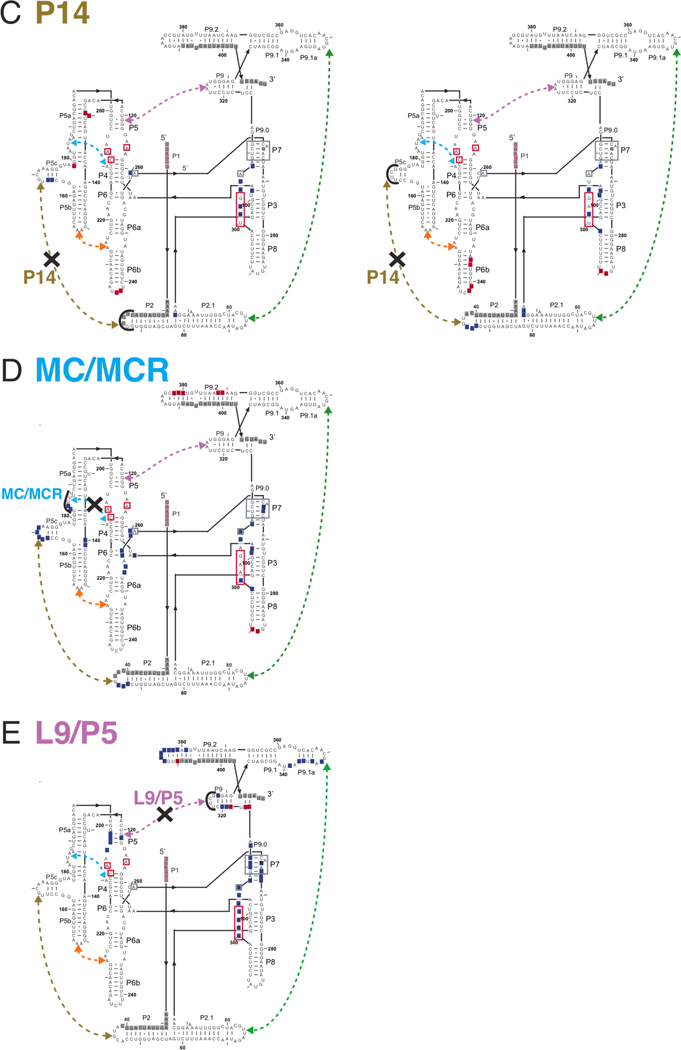
The Tetrahymena group I ribozyme with its five long-range tertiary contacts highlighted. A. The secondary structure of the ribozyme; labels ‘P,’ ‘L,’ and ‘J’ refer to base-paired, loop regions, and junction regions, respectively (43). The five long-range tertiary contacts are indicated by arrows and labeled by their common names or abbreviations of these names (MC/MCR = metal core/metal core receptor; TL/TLR = tetraloop/tetraloop receptor) (40, 44–47). Mutations made to disrupt the long-range tertiary contacts are depicted next to the colored regions, which correspond to the residues that were replaced. For cases in which the tertiary contact is not named by its components, the names of the components that were mutated are shown in parenthesis below the contact name (e.g., L2/L5c for the P14 contact and A-rich bulge for MC/MCR contact) (39, 40). In the case of the P13, P14, and TL/TLR tertiary contacts, each side of the tertiary contact was mutated separately, and each side was monitored separately for structural and functional effects. The P1 duplex contains the oligonucleotide substrate (indicated by lowercase letters) and is shown in red. A black arrow denotes the cleavage site, which corresponds to the 5′-splice site in the normal self-splicing reaction. During the course of the reaction, the P1 duplex forms tertiary interactions with the core of the ribozyme, and regions known to be involved in these interactions are shown in red boxes (38, 48–50). Regions boxed in gray proposed to be involved in stacking interactions with guanosine nucleophile (24). G264 directly hydrogen bonds to the guanosine nucleophile and is bold. B. A three-dimensional phylogenetic model of the ribozyme (40) is shown with and without the conserved catalytic core to highlight the connections between the peripheral elements. Peripheral elements are shown in light gray and encircle the dark gray core; highlighted in color are the residues that make long-range tertiary contacts between the peripheral elements as defined by the phylogenetic model, mutational analysis, and an x-ray crystal structure of a truncated form of this ribozyme (24, 40). In parts A and B and in subsequent figures, the long-range tertiary contacts are color-coded and color-coordinated as follows: P13 (L2.1: dark green, L9.1: olive green); P14 (L2: brown; L5c: tan); MC/MCR (blue); TL/TLR (L5b: red-orange; J6a/b: light orange); L9/P5 (purple).
These long-range tertiary contacts in the Tetrahymena group I ribozyme could contribute to the stability of the folded molecule, to fine-tuning the catalytic conformation of the folded RNA, or to both. To study the contributions of the five long-range tertiary interactions to stability and catalysis, mutants of the individual long-range tertiary contacts within the ribozyme were made. The results reveal effects on overall stability and idiosyncratic effects of the individual tertiary connections on distinct functional steps that arise via allosteric-like communication between the RNA’s periphery and its functional core.
MATERIALS AND METHODS
Materials
Wild type (WT) in vitro transcribed Tetrahymena L-21 ScaI ribozyme was prepared as described previously (51). Ribozyme constructs containing long-range tertiary contact variants were prepared from the plasmid pT7L-21 (51) using the QuikChange protocol (Stratagene) with oligonucleotide primers from Integrated DNA Technologies (Coralville, IA) or the Protein and Nucleic Acid Facility at Stanford (Stanford, CA) that encoded the desired changes. Full nucleotide sequences for all ribozyme genes were confirmed by sequencing. WT and mutant ribozymes were prepared by runoff transcription with the appropriate plasmid and purified on 8% (w/v) polyacrylamide gels. Oligonucleotide substrates were purchased from Dharmacon Inc. (Layfayette, CO) and Integrated DNA Technologies. Standard methods were used in the 5′-32P end-labeling of oligonucleotide substrates for kinetic experiments, and these oligonucleotides were purified by non-denaturing gel prior to use (52). A non-denaturing gel was used to ensure that the common components of a denaturing gel, EDTA and urea, were not present in the purified oligonucleotides and in subsequent kinetic experiments. Oligonucleotides used without end-labeling, UCG, rP, and −1d, rP (Table 1), were purified by anion exchange HPLC using a DNAPac PA-100 column (Dionex, Austin, TX) and desalted by Sep-Pak (Waters, Milford, MA).
Table 1.
Oligonucleotide Substrates Used in this Worka
| Abbreviation | Position | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| −6 | −5 | −4 | −3 | −2 | −1 | +1 | +2 | +3 | +4 | +5 | |
| rSA5 | C | C | C | U | C | U | A | A | A | A | A |
| −1r,dSA5 | dC | dC | dC | dU | dC | U | dA | dA | dA | dA | dA |
| −3m,−1d,rSA5 | C | C | C | mU | C | dU | A | A | A | A | A |
| −1d,rSA5 | C | C | C | U | C | dU | A | A | A | A | A |
| −1d,rSA | C | C | C | U | C | dU | A | ||||
| −5U,−1d,rSA | C | U | C | U | C | dU | A | ||||
| −1d,rP | C | C | C | U | C | dU | |||||
| −3m,−1d,rP | C | C | C | mU | C | dU | |||||
| rP | C | C | C | U | C | U | |||||
Uppercase letters represent RNA bases; m represents a 2′-methoxy moiety, and d represents a 2′-deoxy substitution.
General kinetic methods
All cleavage reactions were single turnover with ribozyme in excess over the 32P-labeled 5′-splice site analog (*S), which was present in trace amounts (<3 nM), and were carried out analogously to prior studies (53–55). Unless otherwise specified, reactions were carried out in 50 mM Na-MOPS, pH 6.9, and 10 mM MgCl2 at 30 °C, conditions in which the WT and mutant ribozymes are essentially fully folded (Figure 4 below). Reaction mixtures containing 50 mM Na-MOPS, pH 6.9, and 10 mM MgCl2 were preincubated at 50 °C for 30 minutes to renature the enzyme. Additional components were then added at room temperature and allowed to equilibrate for at least 5–10 minutes at the reaction temperature before the addition of *S. At least six aliquots were removed at specified times and quenched by 2–4 volumes of 50–100 mM Na-EDTA, pH 8, in 80–90% formamide with 0.02% xylene cyanol and 0.02% bromophenol blue. Radiolabeled oligonucleotide substrates and products were separated by 20% polyacrylamide/7 M urea denaturing gel electrophoresis, and their ratios at each time point were quantified using a PhosphorImager (GE Healthcare) with ImageQuant software. Reactions were typically followed for 3 half-lives or up to 24 hours for slower reactions at 30 °C, and longer at lower temperature. Slow reactions were fit by initial rates, assuming the same endpoints as for reactions that did go to completion. Time courses were fit to a first-order exponential decay and with typical endpoints of 95–98% and R2 values of 0.97–0.998.
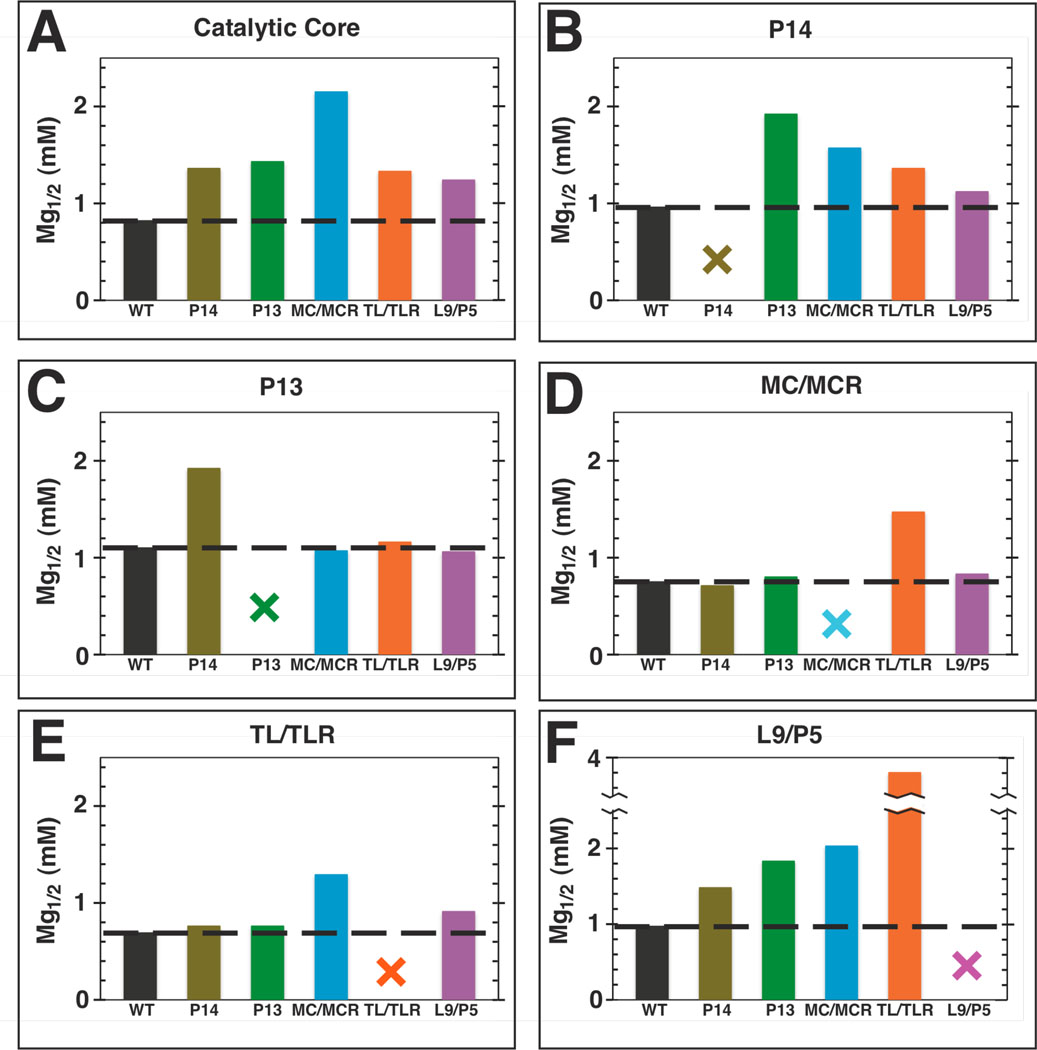
Figure 4.
Mg1/2 values for folding the catalytic core and long-range tertiary contacts shift upon the mutation of a long-range tertiary contact. A–F. Plot labels refer to the region of the ribozyme being monitored for folding over a range of Mg2+ concentrations (Table S3). The x-axis labels represent the long-range tertiary contact mutated. Colors are as described in Figure 1: WT (black); P14 (brown); P13 (green); MC/MCR (blue); TL/TLR (orange); L9/P5 (purple). The black dashed line represents the Mg1/2 value measured for the WT ribozyme, and the colored X’s represent mutation of the contact being queried for folding. Hydroxyl radical footprinting conditions: 100 mM KCl, 10 mM MgCl2, 10 mM K•MOPS, pH 7.
Determination of reactivity over a range of Mg2+ concentrations: kobs
The observed reactivity (kobs) over various Mg2+ concentrations measures the reaction of (E•S)o + UCG → P (Scheme 1 below) at a single concentration of UCG. Oligonucleotide cleavage assays were carried out with E (500 nM) saturating with respect to trace 5′-labeled −1r,dSA5 (Table 1). The concentration of UCG used was 10 µM [ µM for WT], and the mutants have similar or higher values at 10 mM Mg2+ (Table 3 below). In addition, prior studies reveal effects of <3-fold on binding of guanosine and guanosine analogs to the WT ribozyme from increased Mg2+ concentrations up to 100 mM (35, 56–58), and UCG was assumed to be subsaturating across the range of Mg2+ concentrations studied herein. Because of a faster guanosine-independent reaction for the L9/P5 mutant (0.004 min−1 compared to 0.001 min−1 for WT at 10 mM MgCl2, Figure S1 and data not shown) and the guanosine binding deficiency of the mutant [ µM, Table 3], 250 µM UCG was used instead of 10 µM UCG for this mutant. The observation of slower reactions for the WT and L9/P5 ribozymes when UCG was omitted indicated that the observed reactions with UCG present are dominated by the UCG-dependent reaction (Figure S1). To compare the reactivity of the L9/P5 mutant at 250 µM UCG with that of WT at 10 µM UCG, the rate of the L9/P5 mutant was scaled down by a factor of 25 to obtain the ratio of used in Figure 2B below.
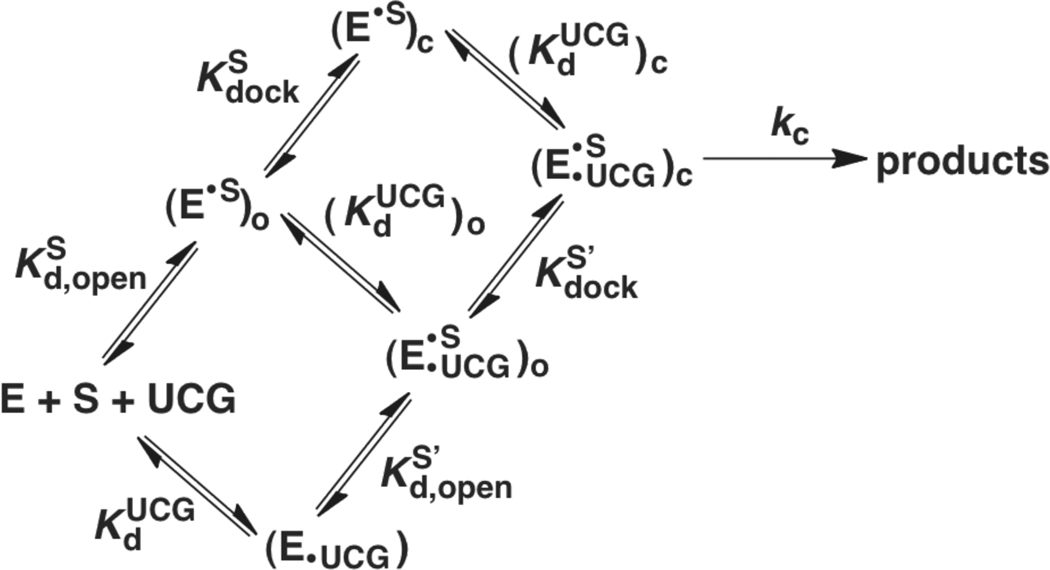
Scheme 1.
Table 3.
Rate and equilibrium constants for individual steps in the Tetrahymena ribozyme reaction for the WT and mutant ribozymes.a
|
(×102 M−1min−1) |
b (µM) |
b (µM) |
Coupling Constantc |
d | kc (min−1) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| −1r,dSA5 | −1r,dSA5 | −1d,rSA | −5U, −1d,rSA | −1d,rSA | −5U, −1d,rSA | −1d,rSA | −5U, −1d,rSA | ||||
| WT | 12 ± 2 | 35 ± 10 | 3.0 ± 0.7 | 3.0 ± 0.2 | 11 ± 4 | 12 ± 4 | 10.0 ± 2.9 | 601 ± 179 | 0.109± 0.008 | 0.142 ± ±0.006 | |
| P14 | L2 | 0.65 ± 0.07 | 25 ± 5 | ≤9 ± 1* | ≤6.1± 1.0* | 4 | 4 | ≤1.2 (2) | 111 ±25 | ≥0.051 | ≥0.079 |
| L5c | 0.67 ± 0.08 | 30 ± 1 | ≤8.1 ± 0.6* | ≤5 ± 1* | 5 | 6 | ≤1.6 (2) | 138 ± 28 | ≥0.059 | ≥0.077 | |
| P13 | L2.1 | 3.3 ± 0.1 | 69 ± 18 | 10 ± 2* | − | 7.2 | − | 6.5 ± 1.3 | 369 | 0.101 | − |
| L9.1 | 3.6 ± 0.3 | 71 ± 22* | 7 ± 1* | − | 10 | − | 6.6 ± 1.1 | 325 | 0.096 | − | |
| MC/MCR | 0.18 ± 0.01 | 50 ± 5 | ≤39 ± 2 | ≤22 ± 1* | 2 | 3 | ≤0.61 (0.5) | 29 ± 4 | ≥0.051 | ≥0.086 | |
| TL/TLR | L5b | 24 ± 5 | 49 ± 7 | 4 ± 1* | − | 11 | − | 14.0 ± 4.0 | 1075 | 0.106 | − |
| J6a/b | 27 ± 8 | 32 ± 7* | 4.1 ± 0.6* | − | 7.8 | − | 9.7 ± 2.6 | 868 | 0.119 | − | |
| L9/P5 | 0.34 ± 0.04 | ≥250 | 35 ± 17 | − | ≥7 | − | 7.6 ± 1.3 | 293 ± 85 | 0.049 ± 0.003 | − | |
Mutants are labeled as in Figure 1. Errors represent standard deviations of two or more measurements unless otherwise noted; if a measurement is not reported with an error, the measurement was only made once or is a limit, as noted. All values were determined as described in Materials and Methods. Values in bold ≥4-fold different from the WT value. Reaction conditions: 30 °C, 10 mM MgCl2, 50 mM Na•MOPS, pH 6.9.
Errors shown for UCG binding curves that are starred (*) are errors from the curve fit; the binding curve itself was generated once.
Coupling constants were determined from the ratio of can be used to determine cooperative binding, or coupling, between the two substrates (eqn 4; see Measurement of UCG affinities in Materials and Methods). For the P14 and MC/MCR mutants, the coupling constants were determined differently, using eqn 4, as described in Measurement of UCG affinities in Materials and Methods.
Numbers in parenthesis for the P14 and MC/MCR mutants are estimates of based on the assumption that the mutations destabilize docking of S and P to the same extent as described in Measurement of docking equilibria in Materials and Methods.
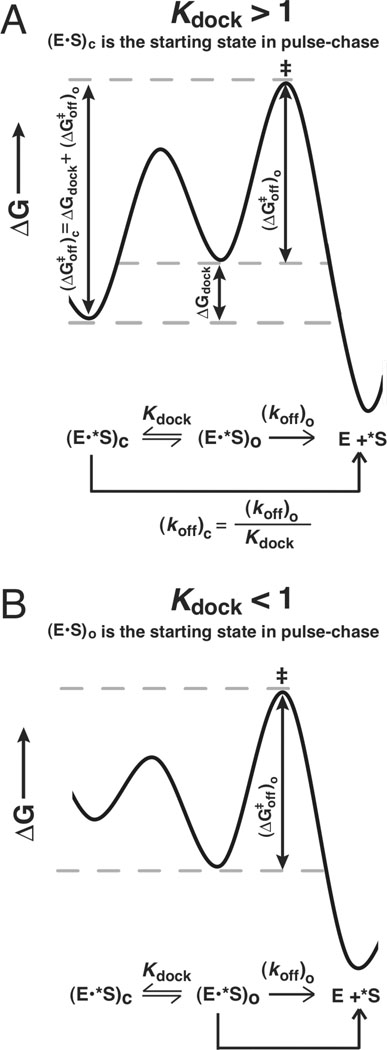
Figure 2.
The dissociation constants for the oligonucleotide substrate from the open and closed complexes can be used to determine the equilibrium constant for docking (Kdock). A. The free energy diagram for a closed complex substrate, which allows measurement of . ΔGdock can be obtained from the difference between and [i.e., ] B. The free energy diagram for an open complex substrate, which allows measurement of The values of ΔG and the corresponding dissociation rate and equilibrium constants can be interconverted via standard equations as described in Measurement of docking equilibria in Materials and Methods.
Measurement of docking equilibria: and
The equilibrium constants for P1 forming tertiary interactions within the core of the ribozyme are termed and for the oligonucleotide substrate (S) and product (P), respectively [Scheme 1 below, Kdock = (E•S)c/(E•S)o]. Previous studies have established that Kdock values can be determined from kinetic measurements (55, 57–62). This determination is possible using the dissociation rate constants (koff) of S (or P) if the closed complex can be significantly populated (i.e., if Kdock > 1).
To derive values of Kdock from dissociation rate constants, the second-order rate constant for the association of S (or P) with E, kon is assumed to be the same for all of the substrates, the stability of the open complex is assumed to be the same for all of the substrates, and the first-order rate constant for the docking of bound S (kdock) is assumed to be faster than (koff)o. These assumptions are supported by the observations that the association of the ribozyme with S involves only base-pairing interactions (59), which are the same for all the ribozymes used, that kon is unaffected using a variety of substrates and ribozymes (55, 57), that the stability of the open complex is unaffected for substrates with and without the 2′-methoxy substitution used herein (Table 1) (60), and that kdock is at least two orders of magnitude faster than (koff)o in the WT ribozyme (61). These assumptions lead to the Gibbs free energy diagrams shown in Figure 2A and 2B for oligonucleotide species that favor the closed and open complexes, respectively.
The difference in free energy between the (E•S)c ground state and the rate-limiting transition state (‡) is shown in Figure 2A and eqn 1, which show that ΔGdock may be obtained from the difference between and . The free energies shown in eqn 1 can be obtained from rate and equilibrium constants using the standard equations for these interconversions: ΔG = −RT lnK and , where R is the ideal gas constant, T is the temperature in Kelvin, h is the Planck constant, k is the Boltzmann constant, K is an equilibrium constant, and k1 is a rate constant. The relationship between Kdock and the off-rates from the open and closed complexes is given by eqn 2, which can be derived from eqn 1.
| (1) |
| (2) |
Thus, if both (koff)o and (koff)c for S (or P) can be measured, a value of Kdock may be obtained. This procedure is possible using S for most of the mutants studied herein by measuring with −3m,−1d,rSA5 and with −1d,rSA5 (Table 1), which predominantly bind the ribozyme in the open and closed complexes, respectively (61). However, for cases in which a mutant ribozyme docks more weakly than the WT ribozyme, an oligonucleotide S that favors the closed complex in the WT could predominantly or significantly populate the open complex in the mutant. In this circumstance, a value of cannot be obtained, and S cannot be used to measure
With the mutants that dock weakly such that the (E•S)c complex may not predominate, the oligonucleotide product (P) can be used to measure docking because P docks more strongly than S does such that the (E•P)c complex may be populated (63, 64). is determined analogously to through measurement of the dissociation rate constants for −3m,−1d,rP and −1d,rP (Table 3 below), which bind the WT ribozyme predominantly in the open and closed complexes, respectively (63, 64). The off-rates of −3m,−1d,rP are within error for all of the mutants, consistent with simple duplex formation as expected for the open complex (Table 2 below). In addition, the −3m,−1d,rP off-rates are slightly faster than those of −3m,−1d,rSA5, as predicted for the loss of 3′-stacking from the A5-tail of −3m,−1d,rSA5 (64, 65). We therefore used docking values obtained with the P analogs for the mutants that did not clearly form the closed complex with S (the P14 and MC/MCR mutants). To estimate values of for the P14 and MC/MCR mutants, we assumed that these mutants also destabilized S and P to the same extent, and we determined docking values of the mutants for S by applying this fold destabilization to the value of for WT (shown in parenthesis in Table 3 below).
Table 2.
Dissociation Rate Constants Used to Determine Kdocka
|
b (min−1) |
(min−1) |
(×10−3 min−1) |
(min−1) |
||
|---|---|---|---|---|---|
| −1d,rSA5 | −3m,−1d,rSA5 | −1d,rP | −3m,−1d,rP | ||
| WT | 0.019 ± 0.004 | 0.19 ± 0.03 | 0.67 ± 0.18 | 0.40 ± 0.05 | |
| P14 | L2 | 0.15 ± 0.07 | 0.25 ± 0.04 | 3.64 ± 0.64 | 0.66 |
| L5c | 0.12 ± 0.05 | 0.23 ± 0.09 | 2.93 ± 0.44 | 0.61 | |
| P13 | L2.1 | 0.029 ± 0.003 | 0.24 ± 0.01 | 1.1 | 0.48 |
| L9.1 | 0.029 ± 0.001 | 0.24 ± 0.04 | 1.2 | 0.59 | |
| MC/MCR | 0.31 ± 0.03 | 0.25 ± 0.18 | 13.7 ± 0.9 | 0.45 | |
| TL/TLR | L5b | 0.013 ± 0.003 | 0.19 ± 0.02 | 0 38 | 0.27 |
| J6a/b | 0.019 ± 0.004 | 0.19 ± 0.02 | 0.47 | 0.30 | |
| L9/P5 | 0.025 ± 0.001 | 0.25 ± 0.08 | 1.38 ± 0.36 | 0.30 | |
Errors are standard deviations of two or more measurements. If errors are not displayed, values served as controls and were measured in a single experiment. Values in bold are ≥4-fold different from the WT value. Reaction conditions: 30 °C, 10 mM MgCl2, 50 mM Na•MOPS, pH 6.9.
Values for the P14 and MC/MCR mutants are apparent because there is a significant population of these mutants in the open complex using −1d,rSA5.
In the experiments used to determine Kdock from dissociation rate constants, modifications −3m and −1d were used in the S and P substrates to predominantly favor the open complex and to slow rate of the chemical step, respectively. The methoxy-substitution at the −3 position (see Table 1) destabilizes docking while maintaining the same duplex stability as a ribose at the site (60, 61, 64). The deoxy-substitution at the −1 position (Table 1) decreases the rate of the chemical step by ∼1000-fold but does not affect docking interactions (55) and is used to decrease the rate of the guanosine-independent reaction of S such that only a small amount of P (which dissociates slower than S) is formed during the assay. The halftimes for the G-independent reaction range from 40 hours to 50 days for the WT and the mutant ribozymes except the L9/P5 mutant, which exhibits a halftime of 2.3 hours (data not shown).
Pulse-chase gel-shift assays were used to determine the dissociation constants from the E•S complexes for the WT and mutant ribozymes (59, 66, 67). Trace amounts of *S were bound to saturating amounts of the ribozyme (10–50 nM) for 10 minutes, sufficient for near-complete binding of *S, and a large excess of unlabeled rP or −1d,rP chase (10–50-fold greater than the ribozyme concentration) was added (Table 1). At specified times, 2 µL aliquots were mixed with loading buffer and carefully loaded onto a native gel that was running in THEM buffer (33 mM Tris, 67 mM HEPES, pH 7.5, 1 mM EDTA, and 10 mM MgCl2) at a low power (15W) and cooled to approximately 7 °C with cooling coils. The loading buffer for the aliquots was 20% glycerol, 80% water, and 0.02% xylene cyanol, and 0.02% bromophenol blue. The fraction of remaining E•*S complex after the chase was plotted over time. Control experiments verified full binding of *S prior to the addition of chase and the efficiency of the chase in preventing *S binding when the chase was added before *S. The dissociation of S oligonucleotides was followed for 1.5–3 half-lives over the course of approximately an hour, and the dissociation of P oligonucleotides was followed for approximately 24 hours, corresponding to 1–3 half-lives. All of the time points for determination of the dissociation rate constants of E•*S or E•*P for a single ribozyme were loaded onto the same gel. To accomplish this for the dissociation of (E•*P)c, which occurs more slowly than the dissociation of (E•*S)c (see Table 2 below), dissociation reactions for the longer time points were initiated earlier than the shorter time point reactions.
Measurement of UCG affinities: and
The equilibrium constants for the dissociation of UCG from the (E•S)o and (E•S)c complexes are termed and , respectively. Oligonucleotide cleavage assays were carried out with E (500 nM) saturating with respect to trace 5′-[32P]-labeled −1r,dSA5 for and with E (50 nM) saturating with respect to trace 5′-[32P]-labeled −1d,rSA for (Table 1). A single A-tail instead of an A5 tail was used for −1d,rSA because previous studies have shown a destabilizing interaction between UCG and the second A of the A5 tail when S analogs are in the closed complex (68). Concentrations of UCG ranged from 0–250 µM, with at least six concentrations used to define each binding curve. The measured values of and for the WT ribozyme are within 3% and 60%, respectively, of previous results (66).
Inhibition of the ribozyme reaction was previously observed at high GUCG concentrations (35). Consistent with these prior results, a small amount of inhibition was observed with UCG concentrations of 1–2 mM, where the binding curves are nearly level (data not shown). This inhibition had a negligible effect at the lower concentrations of UCG that were used to determine the UCG binding curve; for example, for the WT ribozyme and the closed complex substrate −1d,rSA, there was typically a decrease of <20% decrease in the observed reaction rate in the plateau region over the range of 40–250 µM UCG (Figure S2A). Because of this separation between the UCG-dependent and inhibitory phases, the observed inhibition at high [UCG] would be expected to decrease the measured by <40%. Most directly, the UCG concentration dependencies for the WT and seven out of eight mutants were superimposable for binding to the open complex and six of eight for binding to the closed complex, strongly suggesting that binding of UCG is not affected by these mutations and that inhibition by the concentrations of UCG used herein is negligible (Figure S2). In principle, dissociation constants for the mutants with weakened binding could underestimate the extent of weakening, due to a larger inhibitory effect for these mutants, but such an effect would not change any of the conclusions herein.
The rate constant for dissociation of UCG from the ribozyme is faster than the rate constant for the chemical step with the substrates and conditions herein (54, 69); therefore, the affinity of UCG for the ribozyme can be obtained by plotting the observed rate constant (kobs) for single-turnover cleavage of *S as a function of UCG concentration and fitting it to eqn 3.
| (3) |
The value of was obtained using the closed complex substrate −1d,rSA for the P13, TL/TLR, and L9/P5 mutants, which predominantly populate the closed complex and allow a clean determination of . With both and , the coupling constant between S docking and G binding can be determined as described in eqn 4. For the P14 and MC/MCR mutants, the values of with the substrate −1d,rSA are near one (Table 3 below), meaning that these mutants react from a mixture of the open and closed complexes with the −1d,rSA substrate. Observed affinities of UCG for these mutants with bound −1d,rSA are therefore apparent affinities, .
The coupling constant, defined by eqn 4, which was derived from the thermodynamic cycle in Scheme 1, can also be determined from the ratio of to , and the value of can be determined using the measured values of , and , according to eqn 5. Eqn 5 was also derived from Scheme 1 and shows that the observed UCG affinity, , is determined by the degree of coupling [i.e., the ratio ] and is modulated by how much of the enzyme is in the open versus the closed complex when is measured. Using the measured values of and and the estimated value of from Table S1 and Table 3 below, we could solve for the value of (eqn 5) and then use the ratio of this value to to obtain an estimate for the coupling constant (eqn 4). From this approach, the coupling constants were determined to be 4–5 and 2 for the P14 and MC/MCR mutants, respectively (Table S1 & Table 3 below).
| (4) |
| (5) |
The weak docking with the P14 and MC/MCR mutants leads to considerable uncertainty in the value of used in the above estimate of the coupling constant. This uncertainty could be eliminated if the closed complex can be predominantly populated. We therefore turned to the substrate, - 5U,−1d,rSA (Table 1), which docks ∼10-fold more tightly than −1d,rSA to the WT ribozyme [(70); S. Solomatin & D.H., unpublished results] and is predicted to increase docking in the P14 and MC/MCR mutants to greater than 80% (Table S1). The stronger binding of UCG with bound −5U,−1d,rSA than with bound open-complex substrate (−1r,dSA5) or bound −1d,rSA (Table 3 below) for the P14 and MC/MCR mutants indicates that there is coupling between UCG and −5U,−1d,rSA (S−5U) and thus that the closed complex is significantly populated with S−5U for each mutant. Furthermore, UCG binding to the WT ribozyme using S−5U is the same as that for −1d,rSA (abbreviated “S” for the rest of this section), providing evidence that coupled binding to UCG is unaffected by the change in substrate identity to S−5U.
The UCG binding affinities using S−5U were obtained from oligonucleotide cleavage assays with −5U,−1d,rSA as a function of UCG concentration using eqn 3 with 3 µM E, as the −5U,−1d,rSA substrate is >95% bound at 3 µM E by native gel-shift assays for the WT, P14, and MC/MCR mutant ribozymes (data not shown).
As noted above, we estimate that >80% of S−5U is in the closed complex. We therefore used the measured value of with this substrate as a good estimate of the true value of . The ratio of this value to that for UCG binding to the open complex gives coupling constants of 4–6 and 3 for the P14 and MC/MCR mutants, respectively, according to eqn 4. These values are increased by ≤15% when the small fraction of undocked S−5U is accounted for via eqn 5 and are thus the same, within the uncertainty of these measurements.
Measurement of the rate of the chemical step: kc
The first-order rate constant for the reaction of (E•*S•UCG)c→products, obtained with saturating UCG and E saturating with respect to the trace −1d,rSA5 present, is referred to as kc, and prior work strongly suggests that this reaction is limited by the chemical step under the conditions used herein (55, 66). The measured value is an apparent rate constant for ribozymes with large docking deficiencies (the MC/MCR and P14 mutants herein) because these ribozymes may be reacting from a ground state consisting of a mixture of open and closed complexes. These values are therefore reported as and are a lower limit for the actual value of kc. Reported kc and values in Table 3 and Figure 5 below were obtained for the oligonucleotide substrate −1d,rSA (Table 1) from the fits to the binding curves for UCG extrapolated by ≤15% to saturating UCG (eqn 3, where kmax equals kc or ).
Figure 5.
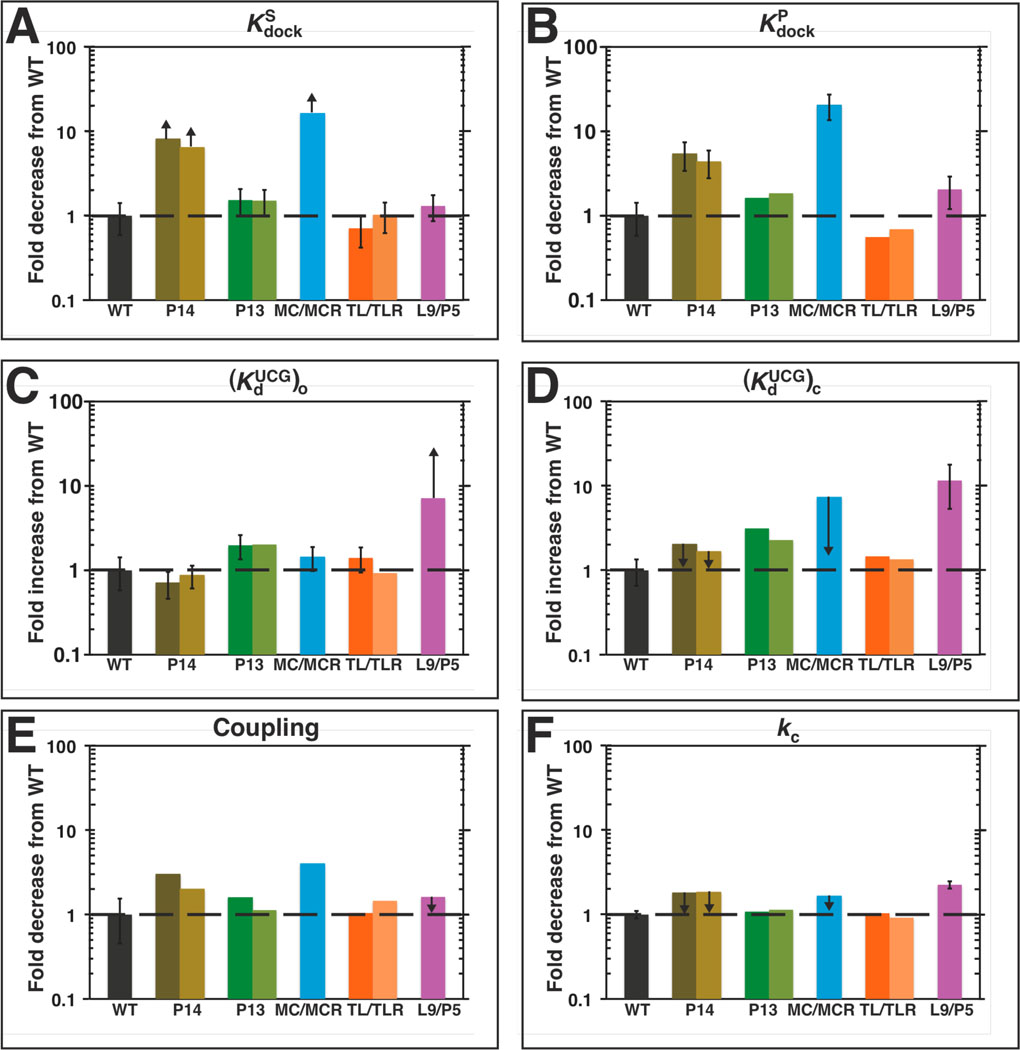
Differential effects on individual reaction steps from different mutations. A–F. Reactions steps are labeled as in Scheme 1 and eqn 4 and determined as described in the Results and Discussion and Materials and Methods. Table 3 contains the kinetic and thermodynamic values for each individual reaction step, which are presented relative to the WT in this figure. The black dashed is drawn to guide the eye at the value of ‘1,’ which represents no difference between the mutant and WT. Colors are as in Figure 1: WT (black); P13 mutants (L2.1: dark green, L9.1: olive green); P14 mutants (L2: brown; L5c: tan); MC/MCR mutant (blue); TL/TLR mutants (L5b: red-orange; J6a/b: light orange); L9/P5 mutant (purple). Reaction conditions: 30 °C, 10 mM MgCl2, 50 mM Na•MOPS, pH 6.9.
For the mutants that dock significantly weaker than the WT (the MC/MCR and P14 mutants), the substrate −5U,−1d,rSA was also used to measure values of . The reported values in Table 3 below are extrapolations from the fits to the binding curves for UCG using eqn 3 as above.
Measurement of the association rate constant of guanosine:
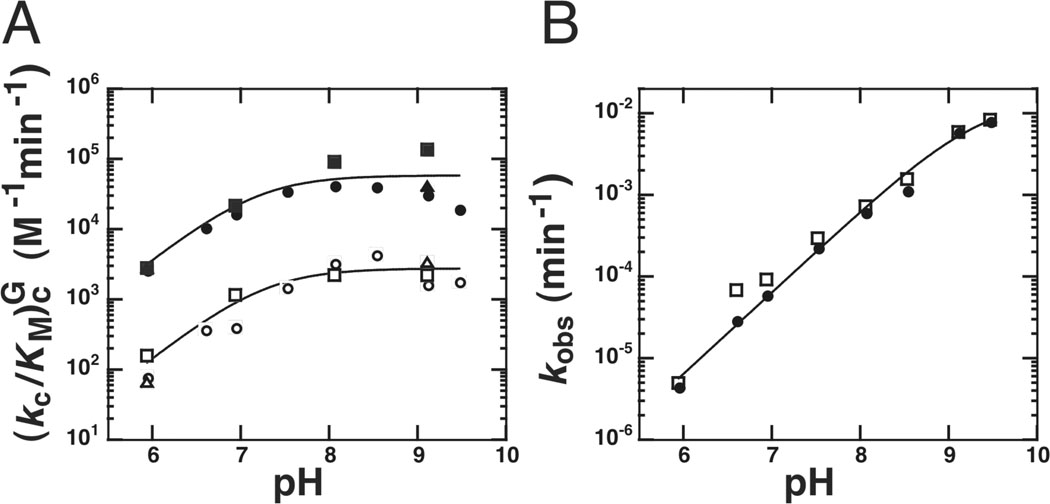
The association rate constant for guanosine binding to the (E•S)c complex is referred to as . For the WT ribozyme, previous data have shown that G binding is rate limiting for above pH 7 using the substrate rSA5 (Table 1) at 4 °C and 30 °C (54). To measure the pH dependence of , the second-order rate constant for the reaction of (E•*S)c + G → products, the following buffers were used: Na-MES, pH 5.9–6.6; Na- MOPS, pH 6.9–7.6; Na-EPPS, pH 8.1–8.5; Na-CHES, pH 9.1–9.5; previous control experiments revealed no buffer-specific effects under analogous conditions (71). Values of were determined with five subsaturating concentrations of G [WT: 0–10 µM, µM; L9/P5 mutant: 50–300 µM, µM], and with E saturating (200 nM) with respect to trace amounts of *S (rSA5). Increasing the concentration of E to 1 µM gave no significant change in the observed values of at low and high pH values, suggesting that S is binding sufficiently fast to not be rate-limiting under these conditions (55, 59). Endpoints were 77–88%, rather than the ∼95% as observed at higher temperatures, likely due to a small population of inactive ribozyme that binds substrate tightly at 5 °C but does not react (56).
For the L9/P5 ribozyme, concentrations below 50 µM G exhibited bi-exponential behavior, attributed to known complications from the G-independent reaction (59, 72). Consequently, concentrations of 50 µM G and above were used to determine the values of for the L9/P5 mutant. Reactions were carried out at 5 °C because the faster G-independent reaction for the L9/P5 mutant renders the L9/P5 mutant reactions too fast to measure manually at 30 °C.
Rate constants were plotted against the concentration of G to yield from the slope. The 0 µM G concentration was not included in the linear fits of for the L9/P5 mutant because it frequently deviated from the line presumably due to the previously reported complications from the G-independent reaction (59, 72) that were magnified for the L9/P5 mutant. Fits of versus pH (eqn 6) gave the apparent , the pH at which G-binding becomes rate-limiting, and the maximum second-order rate constant .
| (6) |
As a control, the pH dependence of the reaction of (E•*S)c + G → P with −1d,rSA5 was measured at a subsaturating concentration of G (10 µM for the WT ribozyme and 300 µM for the L9/P5 mutant) at 5 °C. Here, because a substrate that reacts more slowly in the chemical step is used, G binding is not expected to become rate-limiting and the pH dependence is no longer predicted to level off (55, 66). Ribozyme (50 nM) was saturating with respect to trace amounts of *S (−1d,rSA5). At 5 °C, these reactions were slow and were followed for 24–70 hours. During these long timescale reactions, linear initial rates were observed, indicating that there was no significant activity loss over time. Reactions that did not approach completion after this time were fit by initial rates. The pH dependence of the observed rate constants was fit to eqn 7, which accounts for the deprotonation of the 3′-OH group of the G nucleophile as pH increases. Over a range of pH values from 5.9–9.5, no significant deviation from log-linearity was seen until pH values of 9 or greater are approached (Figure 6B below), consistent with alkaline denaturation of the ribozyme (71). The R2 values for the fits were 0.985 for the WT ribozyme and 0.995 for the L9/P5 mutant. To demonstrate that the measured G-dependent rate was significantly above the G-independent rate, which is increased in the L9/P5 mutant, reaction rates in the absence of G were obtained from pH 5.9–9.5 (Figure S3).
| (7) |
Figure 6.
The weakened guanosine affinity of the L9/P5 mutant arises from slower binding. A. The pH dependence of for the WT ribozyme (closed) and L9/P5 mutant (open), which measures the reaction: E•rSA5 + G → P. Fits to all of the data shown using eqn 6 give maximal values for of 5.9 × 104 and 2.8 × 103 M−1min−1 for the WT and L9/P5 mutant ribozymes, respectively, and apparent pKa values of 7.2 and 7.3 for the WT and L9/P5 mutant ribozymes, respectively. Circles and squares represent independent trials with 200 nM ribozyme, and triangles represent data obtained with 1 µM ribozyme. B. The pH dependence of the reaction of E•−1d,rSA5 + G → P is log-linear for the WT (closed circles) and L9/P5 (open squares) mutant ribozymes. Data were fit to eqn 7 and give pKa values of ≥9.4 and ≥9.3, for the WT (closed) and L9/P5 mutant (open) ribozymes, respectively. The concentration of G in the reaction was subsaturating: 10 µM G (WT) and 300 µM G (L9/P5). The ribozyme concentration was 50 nM and saturating with respect to S. Reaction conditions: 5 °C, 10 mM MgCl2, 50 mM buffer (see Materials and Methods for buffer identities).
Measurement of the overall reaction rate:
The overall reaction rate monitors UCG binding, S docking, and the chemical step and is termed (eqns 10–13 below). The values of were obtained under the conditions described for measuring in Measurement of UCG affinities using the substrate −1r,dSA5. Briefly, the value of was determined by following the reaction of (E•S)o + UCG →P with varied UCG concentrations (0–10 µM and 0–25 µM UCG in repeat experiments; 10–150 µM for the L9/P5 mutant) and obtaining from the linear fit for the initial slope of this dependence.
The overall reaction rate monitors UCG binding, S docking, and the chemical step so that they can be compared to the individual measurements of UCG binding, S docking, and the chemical step, with all of these values evaluated relative to WT, according to eqns 10–13 below. For all of the mutants except the MC/MCR and P14 mutants, the individual rate and equilibrium constants relative to WT (Table S2) used to determine the product of the individual rate and equilibrium constants (eqn 13 below) were those from eqn 11 below, , and kc, and were directly measured herein. For the MC/MCR and P14 mutants, the value of was a limit, and the product of the individual rate and equilibrium constants were determined from those in eqn 12 below: and kc (where , eqn 4). The overall reaction rate was carried out with the substrate −1r,dSA5, whereas some of the individual reaction steps were measured with substrates having a 3′-tail of a single A. The additional residues in the 3′ A5-tail have a small inhibitory effect on coupling of ∼4-fold, giving a coupling constant of ∼3 instead of the 12-fold coupling that is observed herein using the individual rate constants (eqn 4, Table 3 below) (58). To compare the overall reaction rate and the individual rate constants, it was necessary to reduce the 12-fold coupling that was measured with the individual rate constants to the 3-fold coupling observed in . For example, a total loss of coupling or any loss of coupling greater than 3-fold, as measured by the individual rate constants, reflects an effect on of 3-fold, not 12-fold. Since the MC/MCR mutant and L2 side of the P14 mutation are compromised in coupling by estimated amounts of 3- and 4-fold, respectively (Table 3 below), we used a value of 1 for their coupling in eqn 4. Because the L5c side of the P14 mutation is compromised in coupling by 2-fold, we used a coupling value of 1.5 for its coupling in eqn 4 because 1.5 is half of 3, the total coupling in the reaction. Small errors in these values are possible but would not significantly affect the overall trends observed in Figure 7 or the conclusions that are drawn.
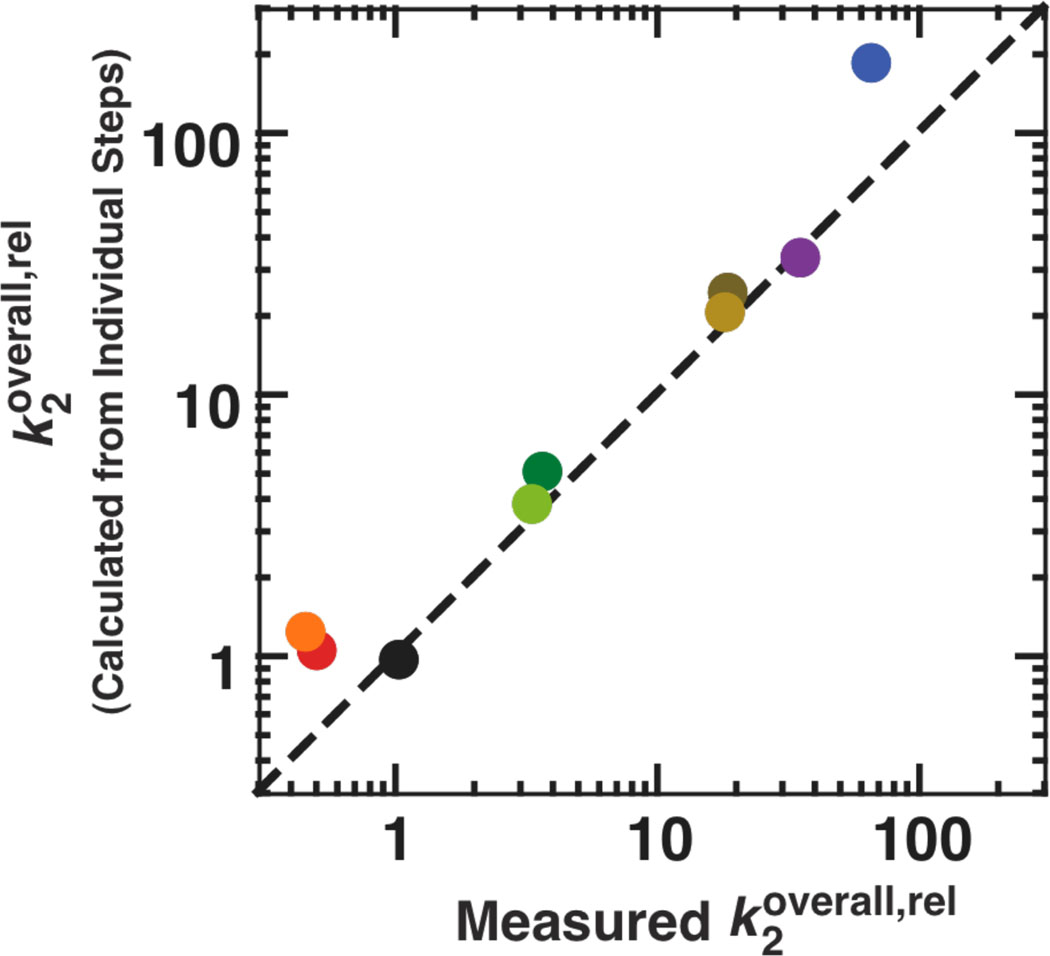
Figure 7.
The individually determined rate and equilibrium constants for the tertiary contact mutants account for the measured overall effect of the mutations. The effects on the individual reaction steps for each mutant are expected to match the effect on the overall reaction of (E•S)o+ UCG → products as measured by , according to eqns 10–13 (see The effects on the individual reaction steps account for the effect on overall reactivity). Here we plot the values of for the mutants relative to the WT, such that values greater than 1 correspond to deleterious effects. The black circle for WT, then, has relative values of 1 by definition, and the dashed line represents a perfect correlation with a slope of 1 between the observed and calculated effects. The values used in calculating the overall rate constant from the individual steps are from Table 3 in the main text and given in Table S2. Colors represent the different mutants and are as defined in Figure 1: P13 mutants (L2.1: dark green, L9.1: olive green); P14 mutants (L2: brown; L5c: tan); MC/MCR mutant (blue); TL/TLR mutants (L5b: red-orange; J6a/b: light orange); L9/P5 mutant (purple). Reaction conditions: 30 °C, 10 mM MgCl2, 50 mM Na•MOPS, pH 6.9.
Hydroxyl radical footprinting with Fe(II)-EDTA
The ribozyme was 32P-labeled at the 5′- or 3′-end using published protocols (73, 74), purified by 8% (w/v) denaturing polyacrylamide gel electrophoresis, and eluted by soaking overnight at 4 °C in water. For the Mg2+ titration experiments (Figure 3 below), each construct was buffer-exchanged into water by gel filtration in P30 Microspin columns (Bio-Rad). Each ribozyme was incubated with 100 mM KCl, 10 mM K-MOPS, pH 7.0, and varying concentrations of MgCl2 at 50 °C for 30 minutes prior to initiating the footprinting reactions. For the residue-by-residue comparisons of the folded structures for WT and the mutant ribozymes (Figure 8 below), each construct was exchanged into 10 mM Na-MOPS, pH 6.9 using a microconcentrator (Millipore). Each ribozyme was preincubated in 50 mM Na-MOPS, pH 6.9, and 10 mM MgCl2 at 50 °C for 30 minutes to renature, or fold, the enzyme. The Mg2+ titrations and residue-by-residue comparisons of the folded ribozymes differ in monovalent ion concentration (110 mM K+ for the Mg2+ titrations versus 20 mM Na+ that is in the 50 mM Na-MOPS buffer for the residue-by-residue comparisons). Comparisons of the WT Mg1/2 values from our Mg2+ titrations with literature Mg1/2 values obtained in lower monovalent ion concentrations (1–10 mM Na+) indicate that Mg1/2 values do not increase upon decreasing the monovalent salt concentration to 20 mM Na+ (75, 76) and provide evidence that the ribozymes in our residue-by-residue comparisons are essentially fully folded in 20 mM Na+.
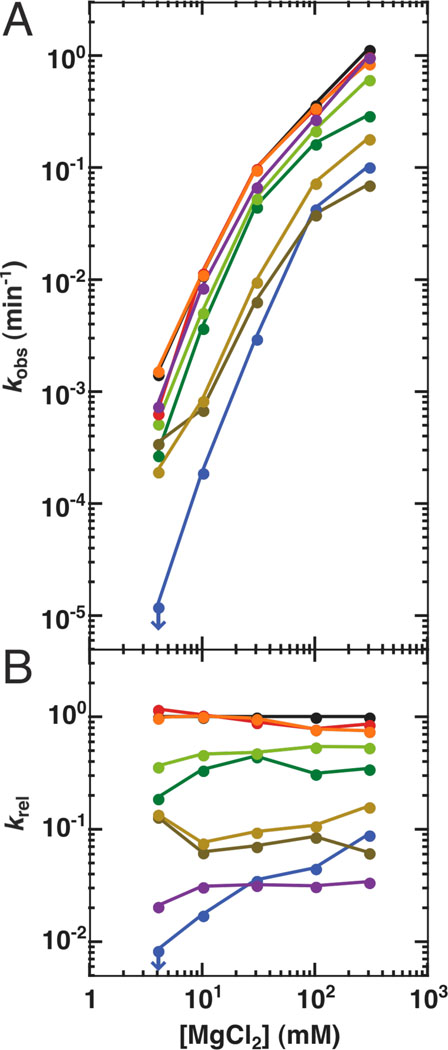
Figure 3.
Mg2+ does not fully rescue the catalytic activity of the long-range tertiary contact mutants. A. The values of kobs for the reaction (E•S)o + UCG → products at 10 µM UCG (250 µM UCG for the L9/P5 mutant) over a range of [Mg2+] (S = −1r,dSA5). Activity of the MC/MCR mutant represents an upper limit for the rate at 4 mM MgCl2 as denoted by the blue arrow and is calculated from the observation of <2% substrate cleavage over 24 hours. B. Rate constants of the reaction relative to the WT ribozyme at each [Mg2+]. The relative rate constant for the L9/P5 mutation is relative to the WT reaction at 10 µM UCG, which has been corrected to account for the different concentrations of UCG used in each reaction (See Determination of the reactivity over a range of Mg2+concentrations in Materials and Methods). Colors are as in Figure 1: WT (black); P13 mutants (L2.1: dark green, L9.1: olive green); P14 mutants (L2: brown; L5c: tan); MC/MCR mutant (blue); TL/TLR mutants (L5b: red-orange; J6a/b: light orange); L9/P5 mutant (purple). Reaction conditions: 30 °C, 50 mM Na•MOPS, pH 6.9.
Figure 8.
Hydroxyl radical footprinting of the tertiary contact mutants in P13 (A), TL/TLR (B), P14 (C), MC/MCR (D), and L9/P5 (E). (A–E): Hydroxyl radical footprinting data (25 °C, 10 mM MgCl2, 50 mM Na•MOPS, pH 6.9) are relative to the WT ribozyme. Blue nucleotides represent regions of the mutant ribozyme with increased cleavage relative WT ribozyme, and red nucleotides represent regions with decreased cleavage. Gray nucleotides represent regions of the mutant ribozyme for which there is no data. Dashed lines represent the long-range tertiary contacts and are colored as in Figure 1 (P13: dark green; P14: brown; MC/MCR: blue; TL/TLR: red-orange; L9/P5: purple). An ‘X’ on the dashed line indicates the tertiary contact ablated by mutation, the regions of mutations are denoted by thick black lines at the site of mutation, and no regions of difference from WT are noted in this region because the lengths of the mutated and WT loops were different. For the P13, P14, and TL/TLR tertiary contact mutants, two footprinting results are shown because each side of the tertiary contact was mutated and probed for structure (e.g., L2.1 or L9.1 was mutated in the case of P13). The mutation to ablate the MC/MCR contact is the same length as the WT contact so that these nucleotides may be directly compared, and cleavage differences from WT within the mutation itself are noted for this mutant. Nucleotides with known functional roles are colored and denoted as in Figure 1. The internal guide sequence, which binds to the oligonucleotide substrate (not shown) to form the P1 duplex, is shown in red. Regions of the ribozyme known to be involved in tertiary interactions with P1 are shown in red boxes. Regions boxed in gray are part of the guanosine binding site. G264 directly hydrogen bonds to the guanosine nucleophile and is bold.
The footprinting reactions were started by the addition of the freshly prepared footprinting reagent to the folded ribozyme. The final concentration of the footprinting reagent in the footprinting reaction was 100 µM Fe(NH4)2(SO4)2, 125 µM Na-EDTA, and 10 mM sodium ascorbate. Reactions were allowed to proceed for 60 minutes (the Mg2+ titration experiments) or 45 minutes (the folded structural comparisons) at 25 °C and were then quenched by the addition of a half volume of 100 mM thiourea in formamide. Cleavage products, a control sample that was untreated with footprinting reagent, and a control sample cleaved by ribonuclease T1 were separated by 8% denaturing polyacrylamide (19:1 acrylamide/bisacrylamide) gel electrophoresis with different running times (typically, two and four hours at 55W for 29.8 × 40.6 cm gel plates) to resolve different regions of the RNA for both the 5′- and 3′-end-labeled RNA, imaged using storage phosphor screens and a PhosphorImager, and quantified using the single-band fitting program SAFA (77).
For the Mg2+ titrations, the 5′- and 3′-ends of each ribozyme were 32P-labeled, and a total of four gels were run for each set of titration curves for each mutant (two gels with 5′-end-labeled and two gels with 3′-end-labeled ribozyme). Each gel contained a single Mg2+ titration (one gel lane per Mg2+ concentration) over which the residues on that gel were compared and normalized. To account for slight loading differences in the amount of radioactivity loaded per lane in the gel, each lane was normalized to so-called invariant residues, as previously described (78). To determine the fraction of a certain region that is folded at each Mg2+ concentration, residues that became protected upon the transition from unfolded to folded were monitored at each Mg2+ for each tertiary contact and for the catalytic core. Each structural region monitored for folding was comprised of a set of multiple residues that are noted in Table S3 (79). The folding of each residue was normalized between unfolded (an arbitrary value of ‘0’) and folded (an arbitrary value of ‘1’). The values between 0 and 1 represent the fraction folded for each set of residues at a single Mg2+ concentration. Each set of residues for a specific tertiary contact or for the catalytic core was averaged together to give a single number representing the fraction folded for that region. These averaged values for each region were plotted versus Mg2+ concentration and fit with an empirical Hill equation (80, 81).
For the folded structural comparisons, seven of eight mutant ribozymes were independently 32P-labeled twice on both the 5′- and 3′-ends and footprinted. The TL/TLR mutants were radiolabeled once on the 5′-end, and the L5b mutant and J6a/b mutant were labeled twice and once on the 3′-ends, respectively. These mutants were functionally and structurally similar to WT so that further radiolabeling and footprinting was deemed unnecessary.
To obtain a single set of footprinting data for a 5′- or 3′-end-labeled RNA, a set of two gels (a 2-hour and 4-hour gel) were run; the analysis of these gels is described briefly below and a detailed sample is given in the supporting information in Analysis of Hydroxyl Radical Footprinting Data for the Folded WT versus Mutant Ribozymes. The analysis described below was somewhat ad hoc, as there is yet no rigorous statistical model to fully account for all of the uncertainties in these footprinting data. We therefore aimed to err on the side of ensuring that the determined structural differences between the mutant and WT ribozymes are real. Typically, each set of gels contained three to five footprinting reactions for three different RNAs (WT and two tertiary contact mutants). For a single RNA, the SAFA output at each nucleotide (the area under the curve in a plot of arbitrary counts versus nucleotide number) for each of the three to five footprinting reactions was normalized between −1 and 1 to the average of the three largest and three smallest values of the SAFA output for each reaction, termed ‘max’ and ‘min’ values (eqn 8, where A represents the curve area). For a given gel, the largest and smallest values of the SAFA output were very similar in magnitude, suggesting that the mutations did not significantly affect the solvent exposure of the majority of the residues (see Figure S5). If bands (or entire lanes) displayed abnormally large values in the SAFA output, they were not used in the normalization or subsequent analysis. Subsequent to normalization, the three to five footprinting reactions for the single RNA were averaged, and these averages for the mutant were compared to the WT average (each with standard deviations). The program Solver (Frontline Systems), which runs within Microsoft Excel, was used to minimize the square of the differences between the 3–5 footprinting traces for a given RNA and also the differences between the averaged traces for the mutant and WT ribozymes. For a single gel, the average footprint for the mutant was subtracted from the average footprint for the WT, and the errors for this difference were propagated. To combine comparisons from multiple gels, these differences from each gel were averaged together, and the errors for these averages were propagated. To determine which residues exhibited altered solvent exposure from the WT, cutoffs that take into account the size of the effect, the number of residues affected, and the error associated with those measurements were applied as described in the Supporting Information (Analysis of Hydroxyl Radical Footprinting Data for the Folded WT versus Mutant Ribozymes).
| (8) |
RESULTS AND DISCUSSION
Do long-range tertiary contacts simply stabilize a folded and active form of the ribozyme?
RNA stability is generally a function of Mg2+ concentration (75, 81, 82), and within the group I ribozymes, there are numerous examples of increased Mg2+ concentration providing a qualitative ‘rescue’ of activity upon deletion of peripheral regions containing long-range tertiary contacts or upon introduction of other potentially destabilizing mutations (40, 83–93). Destabilization from long-range tertiary contact mutations might be rescued to WT levels with additional Mg2+ to stabilize the folded state. On the other hand, long-range tertiary contacts might help the ribozyme maintain an active fold over alternate folds that are less-active; in this case, the long-range contacts may have catalytic roles in substrate binding and positioning and/or transition state stabilization that additional Mg2+ cannot rescue.
To test if the role of long-range tertiary contacts is to stabilize the functional, folded ribozyme over alternatively folded forms, the reactivities of the ribozymes in which one of five long-range tertiary contacts were ablated (Figure 1) were determined over a range of Mg2+ concentrations (Figure 3A). Activities for several of the mutants remained lower than the activity of the wild-type (WT) ribozyme even at the highest Mg2+ concentrations (Figure 3B), suggesting that decreased folding stability cannot account for the full effect of these mutations.
To further and more directly test whether some or all of the rate effects arose from incomplete folding, we carried out structure-mapping studies for each of the mutant ribozymes. Hydroxyl radical protection patterns were monitored as a function of Mg2+ concentration, analogous to previous studies of this ribozyme and of other RNAs (75, 76, 80, 94–98). Formation of the catalytic core, as assessed by the average hydroxyl radical protection of multiple residues over a range of Mg2+ concentrations (Table S3), required higher concentrations of Mg2+ for the mutant ribozymes (Figure 4A), but the same level of protection was attained for the mutants at saturating Mg2+ (data not shown; see also Figure S5). Thus, one biological role of these long-range tertiary interactions is likely to stabilize the folded RNA, allowing it to fold under physiological conditions of ∼0.5–1 mM free Mg2+ (99). Nevertheless, the Mg1/2 values (Figure 4), the Mg2+ concentrations at which each region of RNA is half folded, and folding curves (not shown) indicate that the mutant ribozymes are essentially fully folded by ∼4–10 mM Mg2+, yet rate effects remain for several of the mutants at still higher Mg2+ concentrations (Figure 3B). Thus, the data strongly support additional functional roles of the long-range tertiary contacts in restricting the folded RNA to its most active conformation(s).
An initial model, suggested solely by structural inspection, is that the ring of peripheral elements that surrounds the conserved catalytic core (Figure 1B) acts as a cooperative unit, enforcing the active core conformation. This model predicts that ablation of any of the long-range tertiary contacts would have the same effect on reactivity. The results in Figure 3 provide strong evidence against this model. Whereas the ablation of the same contact via mutation at either interface of the tertiary contact have the same effects, within our experimental error of twofold, mutations of the different tertiary contacts give a much larger range of effects (Figure 3 & Table 3; Figure 7 below).
To better understand the differential effects from ablation of the different long-range tertiary contacts, we dissected these overall effects in terms of individual reaction steps. This effort was guided by the well-established kinetic and thermodynamic framework for the WT ribozyme (Scheme 1 below) (38). Furthermore, to assess the structural differences of the long-range tertiary contact mutants compared to WT ribozyme, we compared hydroxyl radical footprinting protections for the folded WT and mutant ribozymes, the results of which we describe in Structural insights into functional effects of the peripheral mutations below.
Individual long-range tertiary contacts influence distinct reaction steps
The reaction catalyzed by the Tetrahymena group I ribozyme mimics the first phosphoryl transfer reaction of group I intron self-splicing in which CCCUCUpA5, the oligonucleotide substrate (S), transfers its 3′-pA5-tail transferred to the guanosine (GOH) nucleophile to yield the products GpA and CCCUCUOH (P) (eqn 9) (38, 100). Scheme 1 shows the individual steps of the reaction for the guanosine analog UCG, which has nucleotides 5′ to G that base pair and allow it bind more tightly than guanosine itself (68, 101). Both substrates, S (Table 1) and G, bind in multiple steps. S forms a duplex (P1) with the ribozyme by base-pairing (59), forming the “open complex” (subscript ‘o’ in Scheme 1). P1 then docks, forming tertiary interactions with the core of the ribozyme to give the “closed complex” (subscript ‘c’ in Scheme 1) (102, 103). Only one step is shown for G binding because the steps of G binding have not been dissected to the level of individual rate constants. S and G can bind in either order to form the E•S•G ternary complex, and their binding is coupled such that G binds stronger when S is docked and visa versa (69). The chemical step occurs from the ternary complex following the deprotonation 3′-OH group of G (not shown) that attacks S to form the products (38, 66).
| (9) |
Rate and equilibrium constants for the mutants were obtained using methods previously established for the WT ribozyme (54, 58, 59, 66); conditions and details are described in the Materials and Methods. Briefly, reactions were single-turnover with the ribozyme in excess over radiolabeled S1 (*S). Reactions used to measure the binding of G and the guanosine analog UCG, a trinucleotide that binds more tightly than guanosine itself (68, 101), were carried out under conditions in which equilibration is rapid relative to the subsequent chemical reaction, unless specifically noted otherwise (69). Dissociation rate constants for the oligonucleotides S and P were measured using pulse-chase native gel binding assays (66, 67). All equilibrium and rate constants were measured in parallel with those for the WT enzyme to most accurately reveal the kinetic and thermodynamic parameters that are affected by the mutations.
The MC/MCR and P14 mutations affect Kdock
The docking of S was measured to determine whether the tertiary interactions between P1 and the ribozyme’s core were compromised by mutation of the long-range tertiary contacts (Scheme 1). The docking step was isolated by comparing the dissociation rate constants of two S oligonucleotides with and without a 2′-methoxy substitution at the −3 position, −3m,−1d,rSA5 and −1d,rSA5, which favor the open and closed complexes respectively. Briefly, as depicted in Figure 2 and described in Materials and Methods (see Measurement of docking equilibria), ΔGdock can be obtained from the difference between and (eqn 1) so that can be obtained by comparing dissociation rate constants of a substrate that predominantly populates the closed complex and one that predominantly populates the open complex (eqn 2). The 2′-methoxy substitution at the −3 position destabilizes docking by ∼200-fold without affecting the simple duplex stability and thus allows us to probe the stability of the open complex (61, 104), and in the WT ribozyme, −1d,rSA5 predominantly populates the closed complex (61). None of the mutations significantly affect the dissociation rate constant of −3m,−1d,rSA5 [, Table 2)], suggesting, as expected, that the mutations do not affect the stability of the open complex.
To determine whether the mutations affect the stability of the docked complex, we measured for −1d,rSA5, the oligonucleotide that favors the docked, closed state over the open complex by ∼10–30-fold when bound to the WT ribozyme (58, 61, 64). Several of the mutants gave values indistinguishable from the WT value, strongly suggesting that ablation of the P13, TL/TLR, or L9/P5 long-range tertiary contact does not affect docking [, Table 2; , Table 3]. In contrast, the mutants ablating the MC/MCR and P14 long-range tertiary interactions gave increased values of , indicating that these mutations destabilize the docked complex [, Table 2; , Table 3]. The values of for the MC/MCR and P14 mutants are within error of the values for these mutants, suggesting that a significant population of each mutant is in the open complex with the −1d,rSA5 oligonucleotide and that an accurate value of cannot be obtained (see Measurement of docking equilibria in Materials and Methods). Thus, the measured values for with the MC/MCR and the P14 mutants are apparent values and define upper limits for the true values (Table 2; , Figure 5A, Table 3).
To obtain docking values for the MC/MCR and P14 mutants, we turned to oligonucleotides that dock more strongly, as described in the Materials and Methods (see Measurement of docking equilibria). Experiments analogous to those described above for S were performed using product oligonucleotides (−3m,−1d,rP and −1d,rP), as −1d,rP exhibits 20–60-fold greater tertiary stabilization in the closed complex than S does (64). In agreement with values of , the P13, TL/TLR, and L9/P5 mutants have values of that are the same as WT, indicating that these mutations do not affect docking [, Table 2; , Table 3]. For the MC/MCR and P14 mutants, the dissociation rate constants for −1d,rP were smaller than those for −3m,−1d,rP, suggesting that −1d,rP allows both mutants to access the docked, closed state, and thus, allows docking differences from WT to be measured. The values of suggest that the docked complex is destabilized by 20- and 5-fold for the MC/MCR and P14 mutants, respectively [, Table 2; , Figure 5B, Table 3]. To estimate values of for the P14 and MC/MCR mutants, we assumed that these mutants also destabilized docking of S and P to the same extent so that equals , and we determined docking values for the mutants with S by applying this fold destabilization to the value of for WT (shown in parenthesis in Table 3).
The L9/P5 mutant weakens guanosine binding
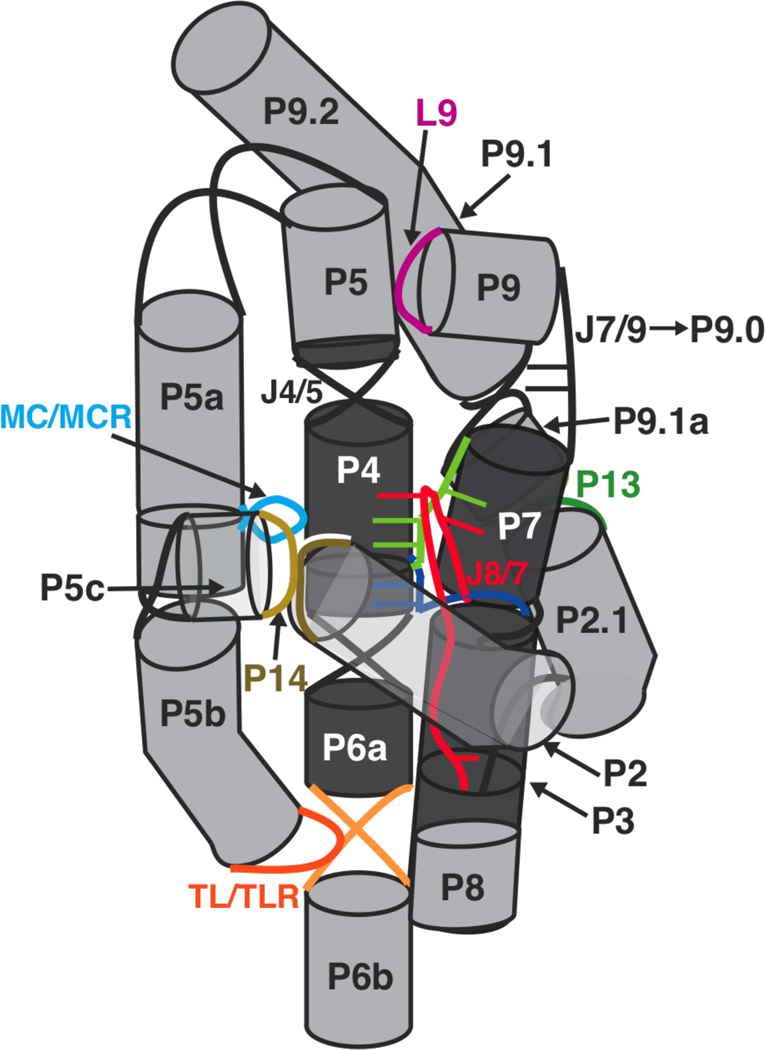
The equilibrium dissociation constant for the binding of guanosine analog UCG to the open complex, was measured by following the dependence of the cleavage of an open complex substrate on UCG concentration. The nucleotides 5′ to the guanosine of the trinucleotide UCG pair with nucleotides G313 and A314 of the ribozyme to form a two base-pair helix P9.0 (Figure 1) (68, 105). P9.0 allows UCG to bind more tightly to the ribozyme than G (68, 101), and this tighter binding, given the limited solubility of G, allows a more accurate determination of guanosine affinities. The values of for the P14, MC/MCR, and TL/TLR mutants were within 2-fold of that of WT; the P13 mutant exhibited slightly weaker UCG binding, by about 2-fold [, Figure 5C, Table 3]. For the L9/P5 mutant, UCG binding was weaker by ≥7-fold [, Figure 5C, Table 3]; only a limit could be obtained due to inhibition at high concentrations of UCG (see Materials and Methods). Thus, only one of the five long-range tertiary contacts affects binding to the guanosine site by more than 2-fold (see also Slower binding causes the weakened guanosine affinity of the L9/P5 mutant below), and this contact is different from the two contacts that affect docking.
Coupling between S and G is reduced by ablation of P14 or MC/MCR
In the WT ribozyme, guanosine binds 4–10-fold more tightly to the E•S closed complex than the open complex (Scheme 1) (66, 69). This coupled binding indicates cooperativity between guanosine binding and S docking (69). To measure the effect of the mutations on coupling, we compared the equilibrium dissociation constant for UCG binding to the closed and open complexes, and , respectively. In some instances, the inability to fully attain the closed complex or the ability to only obtain limits for binding constants necessitated additional analyses as described in Measurement of UCG affinities in Materials and Methods.
The closed complex of the TL/TLR mutants bound UCG the same as WT, as was also observed in the open complex for this mutant, indicating that coupling was unaffected (Figure 5E, Table 3). Coupling was also unaffected for the P13 mutant. This mutant bound UCG 2–3-fold weaker than WT did in both the open complex (Figure 5C) and closed complex (Figure 5D & E; Table 3). The L9/P5 mutant exhibited 12-fold weaker UCG binding to the closed complex than WT did; this value of µM, combined with the lower limit for the binding of UCG to this mutant in the open complex of µM, gives a lower limit of 7-fold coupling, which is within 2-fold of the 10-fold coupling observed for WT herein. Although this mutation could, in principle, increase coupling between guanosine binding and docking, it is likely that the mutation of L9/P5 does not increase coupling to a value greater than that of WT and has at most a <2-fold deleterious effect on coupling.
For the P14 and MC/MCR mutants, the equilibrium constant for UCG binding to the closed complex is an apparent value because the oligonucleotide substrate that favors binding to the WT ribozyme in the closed complex also significantly populates in the open complex for these mutants [i.e., is not much greater than 1 (Table 3, Figure 5A)]. Thus, the measured values of for these mutants are not good measures of and cannot be directly compared with values to provide a measurement of the extent of coupling (eqn 4).
We therefore turned to two different means to determine the coupling constants. First, we corrected for the amount of the E•S complex not in the closed complex according to eqn 5. Second, we used an oligonucleotide S that docks ∼10-fold stronger (S−5U) with the WT ribozyme [(70); S. Solomatin & D.H., unpublished results] to increase the population of the P14 and MC/MCR mutants in the closed complex. These methods gave consistent values for the coupling of 4–6- and 2–3-fold for the P14 and MC/MCR mutants, respectively (Table 3; see Measurement of UCG affinities in Materials and Methods). These values correspond to a 2–3-fold and 4-fold loss of coupling (Figure 5E) for the P14 and MC/MCR mutants, respectively, suggesting that the P14 and MC/MCR mutations are detrimental to both docking and coupling.
The tertiary mutations have no effect on the formation of the P9.0 helix
P9.0 is a two base-pair helix formed between nucleotides G313 and A314 of the ribozyme and the 5′-nucleotides of UCG. This helix formation allows UCG to bind ∼30-fold stronger to the ribozyme than G does (57, 58, 68, 101), and that property has been utilized herein to more accurately measure and compare the guanosine affinities of the long-range tertiary contact mutants. The values of for all of the mutants except L9/P5 are within 2–3-fold of the value for the WT ribozyme, suggesting that P9.0 is formed in these mutants. The weaker binding of UCG to the L9/P5 mutant (≥250 µM: Table 3) could arise from compromised P9.0 formation or from an effect on the guanosine binding site itself.
To determine whether P9.0 formation was compromised, equilibrium binding constants for G and UCG were compared, as equal affinities would suggest the absence of P9.0 formation. An oligonucleotide substrate that favors the closed complex in the WT was used as it increases guanosine affinity, allows saturation of the enzyme to be approached with G and UCG for the weak-binding L9/P5 mutant, and gives more accurate binding constants for the other mutants. The wild type ribozyme bound UCG 30-fold stronger than G, consistent with previous measurements (57, 58, 68, 101), and all of the mutants, including L9/P5, exhibited 20–40-fold stronger UCG binding (Table S4, Figure S4) strongly suggesting that P9.0 is formed in the mutants and not significantly perturbed by deletion of any of the long-range tertiary contacts.
Slower binding results in the weakened guanosine affinity of the L9/P5 mutant
Previous studies have shown that the WT ribozyme binds guanosine with a second-order rate constant of 4×105 M−1min−1, ∼105-fold slower than diffusion (54). The slow binding of guanosine was suggested to stem from a necessary rearrangement of base-triples of the guanosine binding site to accommodate guanosine (38, 54). The L9/P5 tertiary contact is in close proximity to the guanosine binding site (Figure 1), and mutation of this contact weakens guanosine binding by approximately 10-fold (Figure 5D, Table 3). To test whether the weakened binding of guanosine to the L9/P5 mutant was due to still slower G binding, we measured the association rate constant for .
In the WT ribozyme, the association rate constant for G binding is rate-limiting above pH 7 for the reaction of the E•S complex with free when the oligonucleotide substrate used has all-ribose nucleotides that favor the closed complex and a fast chemical step (rSA5 in Table 1) (54). In other words, as the pH increases, the rate of the chemical step increases log-linearly due to deprotonation of the guanosine nucleophile, and the chemical step becomes sufficiently fast such that G binding becomes rate-limiting instead of the chemical step (54). To test if for the L9/P5 mutant is compromised relative to WT, was determined over a range of pH values. The reactions were carried out at 5 °C because the high guanosine-independent reactivity of the L9/P5 mutant rendered the overall reactions too fast for manual measurements at 30 °C (see Measurement of the association rate constant of guanosine in Materials and Methods). The pH-rate profiles for of the WT ribozyme are similar at 4 and 30 °C (54).
Figure 6A shows the pH dependence of for the WT and L9/P5 mutant ribozymes. For both, the pH dependence levels off above pH 7, but the mutant levels off at a maximal rate constant about 20-fold lower than the wild type does, suggesting that slower binding of guanosine has become rate-limiting for the L9/P5 mutant. As a control, the pH dependence of the reaction (E•*S)c+G → P was followed using a subsaturating concentration of G for WT and the L9/P5 mutant and a closed complex substrate, −1d,rSA5, which decreases the rate of the chemical step by ∼1000-fold compared to the all-ribose substrate for the WT (55). The substrate −1d,rSA5 is expected to slow the rate of the chemical step for the L9/P5 mutant as for WT because the effect of L9/P5 mutation on the rate of the chemical step is small (Table 3). The slower chemical step in both the L9/P5 mutant and WT ribozymes should be rate-limiting in the reaction of (E•*S)c + G → P and display a log-linear pH dependence until the pKa (pKa ≥ 10) of the guanosine nucleophile is approached (66). As expected, the pH dependence for both ribozymes remains log-linear with a slope of one until pH values of 9 or greater are approached, consistent with the chemical step being rate limiting over the pH range of 5.9–9 (Figure 6B). The small deviation from linearity in this control reaction around pH 9 is consistent with alkaline denaturation of the ribozyme (71) and does not affect the conclusions drawn at and above pH 7 for the all-ribose substrate.
Thus, the value for the L9/P5 mutant is ∼20-fold slower than that for the WT ribozyme, accounting for all of the observed 12-fold guanosine binding deficit for the L9/P5 mutant compared to WT (Table 3). The decreased rate of association of G with the L9/P5 mutant suggests that the structure of the guanosine binding site in L9/P5 mutant is altered compared to WT ribozyme such that the number of productive guanosine-binding encounters are ∼20-fold fewer than in WT. Hydroxyl radical footprinting data in the sections below also suggest that structural alterations near the guanosine binding site arise from the L9/P5 mutation (see Structure-function relationships provided by hydroxyl radical footprinting and Figure 8E below).
The mutations do not significantly affect reaction of bound substrates
To assess the roles of the long-range tertiary contacts in the chemical step, the rate constant for the cleavage of S from the ternary complex ( kc ) was monitored using saturating UCG concentrations and an oligonucleotide substrate that favors the closed complex. As noted previously for measurements of , the values of kc for the MC/MCR and P14 mutants are apparent values because these ribozymes may be reacting partially from the open complex (, Table 3; Figure 5A). This effect is less severe than in the case of because bound UCG increases the population of the ribozyme that is docked (see Coupling between S and G is reduced by ablation of P14 or MC/MCR in Results). Nevertheless, all long-range tertiary contact mutants have kc or values that are within 2-fold of the WT ribozyme (Table 3, Figure 5F), suggesting that once both S and G are bound, the chemical step proceeds with a similar transition state as in the WT ribozyme. The L9/P5 mutation has a small (2-fold) effect on the rate of the chemical step.
The effects on the individual reaction steps account for the effect on overall reactivity
The overall reaction of eqn 10 starts from the (E•S)o complex and free UCG and monitors UCG binding, S docking, and the chemical step [Scheme 1 & in Table 3]. The thermodynamic cycle in Scheme 1 dictates that the overall reaction can be expressed in terms of either of two reaction pathways shown in eqns 11 and 12, with either UCG binding or S docking occurring first. Thus, the value of can be expressed as a product of the individual rate constants from these schemes, as shown in eqn 13. For a single substrate, this equation must hold, and evaluating it would provide an estimate of the accuracy of the individual measurements. Because we have used different substrates to follow different reaction steps, assessing the congruence of the left and right sides of eqn 13 for WT and the mutants provides an additional check on the assumption that the modifications used to favor or disfavor S docking or slow the chemical step do not have additional, unaccounted for effects outside of those that affect docking. Considerable prior work supports this assumption for the WT ribozyme, and the congruence of the values for the mutant ribozymes relative to the WT (Figure 7) support this assumption for the mutants.
| (10) |
| (11) |
| (12) |
| (13) |
The overall effects of the long-range tertiary contact mutations range from none to 64-fold, with the majority of the effect localized to an individual reaction step for the three mutants that had the largest effects (Figure 5; Tables 3 & S2). To learn more about the origins of these effects, we turned to structural probing of each of the mutants.
Structural insights into functional effects of the peripheral mutations
To determine the structural alterations from mutation of the long-range tertiary contacts, hydroxyl radical protection patterns were obtained for each mutant. Hydroxyl radical cleavage allowed us to monitor changes in solvent accessibility between the mutant and WT ribozymes (106–108), and the results are summarized in Figure 8. As expected, there were changes at and around the site of the tertiary contact mutation in each case (Figures 8). Additional changes for some of the mutants have allowed us to develop models for the communication between the periphery and catalytic core.
Limited and local changes in the P13 and TL/TLR mutants
The P13 and TL/TLR mutants showed limited changes beyond the sites of mutation (Figure 8A & B; Figures S11–S14), consistent with the small functional effects from these mutants (Figure 5, Table 3). A small increase in solvent exposure was observed in P7 for the P13 mutants as was previously seen for a construct deleting the P9.1 and P9.2 helices (107), consistent with the phylogeny model that positions the P9.1a helix over the P7 helix in the folded structure (Figure 9) (40). For the single TL/TLR mutant that had mutations to J6a/b, there was an observed decrease in the protection of J3/4. The affected residues have been suggested to make interdomain interactions that connect P4-P5-P6 to the P3-P7-P8 helical stack (Figure 9) (24, 33, 39).
Figure 9.
Cylinder and line cartoon of the tertiary connectivity within the WT ribozyme. The regions that make up the conserved P4-P5-P6 and P3-P7-P8 helical stacks are colored in dark gray as in Figure 1. The light gray peripheral elements are connected by tertiary interactions that are colored as in Figures 1 and 8: P13 mutants (L2.1: dark green, L9.1: olive green); P14 mutants (L2: brown; L5c: tan); MC/MCR mutant (blue); TL/TLR mutants (L5b: red-orange; J6a/b: light orange); L9/P5 mutant (purple). The MC/MCR and L9/P5 tertiary interactions are denoted by the colored region touching the point of tertiary contact. Helices, long-range tertiary contacts, and selected junctions are numbered and labeled as in Figure 1; single-stranded regions and regions that form non-canonical interactions (loops and junctions) are shown as lines. Junctions within the core of the ribozyme form base-triples that have been suggested to orient the P4-P5-P6 and P3-P7-P8 helical stacks relative to each other. These junctions are colored (J3/4: dark blue, J6/7: light green, J8/7: red), and junction nucleotides that form the base triple with the helix are shown as a thin slab and colored the same color as their corresponding junctions. J7/9 (black) is shown as a line with the two unpaired nucleotides that form P9.0 upon binding of UCG. The base-triples shown in P7 are two “bottom” base-triples guanosine binding site as shown in Figure 10. For clarity, the P1 helix is not shown and the angle between the P4-P5-P6 and P3-P7-P8 helical stacks has been altered from what is seen in the x-ray crystal structure (24).
Larger structural effects from the P14, MC/MCR, and L9/P5 mutations correlate with larger functional effects
The P14, MC/MCR, and L9/P5 mutations, which exhibit larger functional effects of 20–70-fold overall (Table 3 & Table S2), had more extensive structural changes at positions away from the site of tertiary contact mutation (Figure 8C–E; Figures S10, S15–S17). These changes suggest possible routes of allosteric coupling between spatially distinct regions of the RNA. Below we suggest models to relate these structural changes to the observed functional effects.
The L9/P5 mutant
This mutant has a 12-fold decreased affinity for guanosine relative to wild type that is accounted for by slower guanosine binding (Figures 5 & 6; Table 3). The guanosine binding site is located in P7 (Figure 1), and P7 shows changes in hydroxyl radical protections upon mutation of L9/P5 (Figure 8E, Figure S17), suggesting that the L9/P5 long-range tertiary contact is integral to forming the guanosine binding site. For the WT ribozyme, binding of guanosine is five orders of magnitude slower than diffusive binding (54), and this observation has led to a model in which the triple helices above and below the guanosine binding site collapse in the absence of bound guanosine such that guanosine binding can only occur from transient ‘open’ states (Figure 10) (38). The L9/P5 contact might ‘pull on’ or ‘torque’ the P7 helix with its associated base-triples to favor the ‘open state’ that can bind guanosine (Figures 9 & 10). This change in the conformation of P7 in the absence of the L9/P5 contact could also cause a conformational rearrangement of the 3′-end of J8/7, which is connected to P7 (Figure 9) and also shows changes in protection from hydroxyl radicals in the L9/P5 mutant (Figure 8E). Interestingly, crystal structures of Azoarcus and Twort group I introns also show L9 forming a long-range tertiary contact to the P4-P5-P6 helical stack (Figure 11) and analogous P9/L9 motifs are found in most group I introns, suggesting that a tertiary contact involving L9 could be a general strategy for increasing guanosine affinity (39, 42).
Figure 10.
Model in which the ‘open’ state of the guanosine binding site is more favorable when the L9/P5 contact is present. A. The collapsed guanosine binding site is presumably favored for the WT ribozyme (54) and is favored 12-fold more in the L9/P5 mutant (Figure 6A). B. A space-filling model of open and collapsed states of the guanosine binding site. The model for the open state has a pocket for guanosine, but in the model for the collapsed state, the upper triple (purple) collapses into the pocket. While two discrete states are represented, each of the states are likely highly dynamic. As guanosine binding is very slow for the WT ribozyme, it presumably exists in predominantly collapsed states in the absence of G, and this preference is enhanced by the L9/P5 mutation. The open guanosine binding sites are from the x-ray crystal structure of the Tetrahymena group I intron (24) and are shown with the base-triples above (purple) and below (green and red) the guanosine binding site where guanosine (orange) forms a base-triple to the blue base-pair in the P7 helix. The model for the collapsed guanosine binding site shows the upper base-triple (purple) collapsed to sterically block the guanosine binding site. This collapsed site was created from the same x-ray structure as the open guanosine binding site using the program Assemble to alter the position of the upper base-triple (125).
Figure 11.
The L9 tertiary contact to the P4-P5-P6 helical stack is conserved in the Tetrahymena (missing P2-P2.1, P9.1-P9.2), Azoarcus, and Twort group I introns. The L9 tertiary contact (purple) sits above the guanosine binding site (blue). The guanosine nucleophile (orange) hydrogen bonds the site and forms stacking interactions with nucleotides sitting above and below it (Figure 10). The conserved regions that make up the P4-P5-P6 and P3-P7-P8 helical stacks are colored in dark gray as in Figure 1. The light gray peripheral elements are connected by other tertiary interactions that are colored in green. For clarity, the P1 helix and P9.0 are not shown, and nucleotides at the 3′-end of the crystallized ribozymes (24, 26, 27) are not shown (Tetrahymena: A410-C411, Azoarcus: A205, Twort: A248-A249). The x-ray crystal structures for the Tetrahymena (24), Azoarcus (27), and Twort (26) ribozymes are from Protein Data Bank entries 1X8W, 3BO2, and 1Y0Q, respectively.
It is also possible that some or all of the effect on guanosine binding could be transmitted through P9.1a and J9.1/9.1a, as there are also changes in their protection patterns in the L9/P9 mutant. P9.1a and J9.1/9.1a likely sit in close proximity to P7 and may contact it (Figure 9) (40). In addition, there is a small but significant change in P9.1a and P7 protection for both P13 mutants (Figure 8A), and these mutants have a small effect of twofold on guanosine binding (Figure 5C & D; Table 3). An analogous P7 contact to a peripheral helix is possible in the Twort group I intron, which has peripheral helices P7.1 and P7.2 that sit in close proximity to P7 (Figure 11) (26) . While the Azoarcus group I intron lacks an analogous P7 to peripheral element contact, it contains a helix-internal-loop-helix motif (a kink-turn) between P7 and P9 (25, 109, 110). A structured kink-turn motif could allow L9 to induce the ‘open state’ of the P7 helix in lieu of a contact from the P7 peripheral element.
Possible structural origins for docking effects from the P14 and MC/MCR mutants
Both the P14 and MC/MCR mutants affect docking by 5- and 20-fold, respectively (Figure 5B, Table 3). J8/7 and J4/5 directly interact with the docked P1 helix (Figures 1 & 9) (38, 48, 50), and changes in hydroxyl radical footprinting are seen in J8/7, but not J4/5, for both P14 and MC/MCR mutants (Figure 8C & D; Figures S10, S15, S16). In the simplest model, docking is weakened in the P14 and MC/MCR mutants by conformational changes in or reorientation of J8/7.
Alternative models for the MC/MCR mutant effects on docking
The MC/MCR mutant, which gives the larger docking effect, shows changes in the hydroxyl radical protection patterns of J3/4 and J6/7, in addition to the J8/7 changes (Figure 8D, Figure S10). These junction regions form base-triples with P6 (J3/4) and P4 (J6/7 and J8/7) (24, 32, 33, 39, 111, 112) and have been suggested to orient the P4-P5-P6 and P3-P7-P8 helical stacks relative to each other (Figure 9) (31, 39, 111, 112). The metal core (MC) of the MC/MCR contact also contacts P4, where the metal core receptor (MCR) is located (24, 113). Its ablation could alter or disrupt the P4 triples with J8/7 and/or J6/7 (Figure 9), misorient the P4 helix itself or P4-P5-P6 helical stack relative to the P3-P7-P8 helical stack, and thereby misalign tertiary contacts in the P1 docking site, causing the observed 20-fold docking effect. Interestingly, disruption of the J8/7 to P4 base-triple was previously shown to weaken docking by ∼50-fold relative to the WT ribozyme (31).
J6/7 could also be involved in the weaker docking of P1 observed for the MC/MCR mutant. Of the group I introns that contain the same J6/7 nucleotides as the Tetrahymena ribozyme, phylogenetic evidence suggests that ∼80% contain the MC/MCR contact itself, suggesting a potential functional link between J6/7 and the MC/MCR contact (39). A functional link is further suggested in a study in which functional effects from deletion of MC/MCR can be partially rescued by replacement of the base-triples formed by J6/7 and P4 with analogous base-triples from the Azoarcus group I intron, which does not have a MC/MCR contact (91). In summary, either or J6/7 and J8/7 could contribute to the misorientation of P4 itself or the P4-P5-P6 helical stack relative to the P3-P7-P8 helical stack, and hence, the misalignment of P1 docking interactions within the MC/MCR tertiary contact mutant.
Alternative models for how the P14 mutations affect docking
The P14 mutant shows changes in the hydroxyl radical protection patterns in P2.1 and J2.1/3 in addition to J8/7 (Figure 8C; Figures S15 & S16), and J2.1/3 has been suggested from mutational studies to form contacts with the rest of the molecule (48, 114). Changes in J2.1/3 could propagate to the 5′-end of P3, which forms a base-triple with U300 at the 5′-end of J8/7 (Figure 9) (115). If this base-triple were disrupted, U300 could be misoriented, and the docking interaction between U300 and a specific 2′-OH in the P1 duplex (38) could be disrupted and give the observed 5-fold docking effect in the P14 mutants (Figure 5B, Table 3). Mutation of the 2′-OH to a 2′-H in the P1 duplex that interacts with U300 gives a 4-fold effect on docking (64). If the docking effect is propagated through J2.1/3, it might seem that mutation of P13 should also affect docking; however, if P2.1 were held in place by contacts to the rest of the molecule, such as nearby P8 (40), disruption of the P13 contact might not affect the conformation of J2.1/3. Indeed, no change in the J2.1/3 protection is observed for the P13 mutants (Figure 8A).
In perhaps the simplest possible model, disruption of the P14 tertiary contact could also increase the conformational entropy of the undocked P1, through J1/2 that connects P1 and P2 (Figure 1), and hence, increase the entropic cost for docking.
Coupled or independent effects of the P14 and MC/MCR mutations?
For the MC/MCR mutant, the P14 contact unexpectedly exhibited changes in its hydroxyl radical protection pattern (Figure 8D). Alone, this structural data might suggest a coupling between these structural elements, but contrary to this expectation, ablation of P14 did not result in increased cleavage from hydroxyl radicals in the MC/MCR tertiary contact (Figure 8C). Further, a functional double mutant cycle with the P14, MC/MCR, and P14+MC/MCR mutations revealed additive energetic effects (unpublished results), providing further evidence against structural and functional coupling. Thus, the simplest model to account for the hydroxyl radical cleavage changes in P14 upon MC/MCR mutation is a reoriented but still intact P14 contact, possibly effected through interactions of the MC/MCR contact with J5b/5c or L5c itself that were suggested based on x-ray crystallographic data (24, 113).
An L9/P5 effect in J8/7
Another surprising change in protection was in J8/7 of the L9/P5 mutant (Figure 8E, Figure S17). For the MC/MCR and P14 mutants, changes in J8/7 cleavages could be attributed to conformational changes of J8/7 or the P4-P5-P6 helical stack that could lead to the observed weakening of docking (Figure 5E, Table 3). Because the L9/P5 mutant shows no significant change in docking behavior from the WT ribozyme (Figure 5A & B; Table 3), we suggest that the protection change in J8/7, at least for this mutant and possibly the other mutants, may arise from increased solvent exposure of J8/7 and not a conformational rearrangement of J8/7 that might hinder docking.
IMPLICATIONS
We have monitored the contributions of five long-range tertiary contacts of the Tetrahymena group I ribozyme to stability, catalysis, and structure. Each of the long-range tertiary contacts decreases the stability of the RNA fold. Once the RNA is folded, three of the five long-range tertiary contacts have additional roles in catalysis. The P14 and MC/MCR tertiary contacts contribute to the docking step and to coupling between binding of guanosine and the oligonucleotide substrate. In contrast, the L9/P5 tertiary contact modulates the intron’s affinity for guanosine. As there are no direct contacts between these tertiary elements and the substrates, these functional roles must be mediated by structural communication between the periphery of the RNA and functional sites in the catalytic core. The ability of RNA’s tertiary contacts to modulate the structure and function of catalytic sites from afar is analogous to the way allosteric proteins are coupled from one distal site to another through non-active site residues (8, 20, 21, 116).
Our structure mapping results for the tertiary contact mutants, combined with prior structural and mutational data (see Structural insights into functional effects of the peripheral mutations and references therein), provide initial models for the origins of this communication. For example, guanosine binds via the formation of a base triple that is sandwiched between two other base triples within P7 (Figure 10). Interestingly, the isolated guanosine binding site (P7, covalently attached to P9.0 and G) can bind a covalently tethered guanosine in the absence of the rest of the ribozyme (117, 118). Nevertheless, this site lacks many of the interactions present within the folded ribozyme and is unlikely to robustly bind and position exogenous guanosine on its own. Even within the context of the ribozyme, guanosine associates several orders of magnitude slower than diffusion, suggesting that the site must rearrange to allow guanosine to bind (54). It appears that the distal interaction of L9 with the P5 helix aids in opening this binding site for guanosine entry. Thus, remote tertiary contacts in RNA can allosterically communicate with the active site to help fashion local structures and facilitate function.
Long-range tertiary contacts within RNA may also be used to orient structural elements with respect to one another. Docking of the P1 duplex requires the correct positioning of groups located on the two central helical stacks of the catalytic core (25, 26, 31, 39, 111, 112). To position the docking interactions correctly, the MC/MCR tertiary contact may be used to hold the two central helical stacks in the correct orientation with respect to one another. Other functional RNAs, including the hammerhead ribozyme, VS ribozyme, and certain riboswitches, also appear to use long-range tertiary contacts to orient their helices relative to one another (34, 36, 119, 120). The use of long-range tertiary contacts to position helices and modulate function is particularly evident for the hammerhead ribozyme, in which its ligation and cleavage rates can be varied by 102−103-fold by varying its two peripheral helices and the tertiary contact that connects them (34). In the Tetrahymena ribozyme, different long-range tertiary contacts facilitate different reaction steps and thus exhibit distinct functional roles. This observation and the different structural consequences of the individual mutations suggest that the allosteric tertiary elements have modular effects on ribozyme structure and function. Further exploration of long-range RNA interactions will help reveal the properties of the peripheral and core elements that allow their functional communication.
Whereas there are many functional RNAs that contain peripheral elements and long-range tertiary contacts, some ribozymes are notably smaller, exhibiting compact structures with few unpaired residues and highly stable folds (121, 122). For example, the 205-nt Azoarcus group I ribozyme is 200 nucleotides smaller than the Tetrahymena ribozyme and only has two long-range tertiary contacts (121), yet it can perform the same reaction as the Tetrahymena group I ribozyme with similar efficiency and can do so at higher temperatures (123). The ∼90-nt hepatitis delta virus (HDV) ribozyme has a nested double-pseudoknot structure formed from multiple strand crossovers, with no other long-range tertiary interactions (122), yet this fold can maintain activity in 8 M urea (124). It may be that in certain biological milieus additional evolutionary pressures drive the formation of these compact structures, or it may be that RNA has two relatively equivalent solutions to its folding problem, one that engages peripheral elements and one that utilizes more local interaction networks. It will be fascinating to learn more about the strengths and limitations of these strategies used by RNA to attain active structures, how these strategies have been exploited during evolution, and what evolutionary circumstances may favor or disfavor these different strategies and outcomes.
Supplementary Material
ACKNOWLEDGEMENT
We thank Marcello Forconi and Sergey Solomatin for helpful advice and discussions. We thank Jonathan Lassila and other members of the Herschlag lab for comments on the manuscript. We thank Rhiju Das, Nathan Boyd, Vess Diankov, and Manjula Rajendran for technical assistance in the collection and analysis of the Mg1/2 footprinting data. We thank Rishi Porecha for technical assistance with kinetic assays.
Abbreviations
- E
enzyme
- WT
wild type
- MC/MCR
metal core/metal core receptor
- TL/TLR
tetraloop/tetraloop receptor
- S
oligonucleotide substrate
- P
oligonucleotide product
- G
guanosine
Footnotes
This work was supported by a grant from the National Institutes of Health (GM 49243) to D.H.
S and P will be used through out as generic abbreviations for the 5′-splice site analog and 5′-exon analog respectively. The various modifications to these analogs are shown in Table 1.
SUPPORTING INFORMATION AVAILABLE
The docking and coupling constants for the P14 and MC/MCR mutants, rate and equilibrium constants of the tertiary contact mutants relative to the WT for both the overall reaction and individual steps, the nucleotides used to monitor folding different regions of the WT and mutant ribozymes, the G and UCG binding curves for the mutant and WT ribozymes, the pH and Mg2+ dependence of the G-independent reaction for the L9/P5 mutant and WT ribozymes, a detailed procedure for the analysis of the hydroxyl radical footprinting for the folded RNAs, nucleotide-by-nucleotide plots of the differences in hydroxyl radical cleavage between each mutant and the WT ribozyme. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Lilley DM. The origins of RNA catalysis in ribozymes. Trends Biochem Sci. 2003;28:495–501. doi: 10.1016/S0968-0004(03)00191-9. [DOI] [PubMed] [Google Scholar]
- 2.Narlikar GJ, Herschlag D. Mechanistic aspects of enzymatic catalysis: lessons from comparison of RNA and protein enzymes. Annu Rev Biochem. 1997;66:19–59. doi: 10.1146/annurev.biochem.66.1.19. [DOI] [PubMed] [Google Scholar]
- 3.Rennell D, Bouvier SE, Hardy LW, Poteete AR. Systematic mutation of bacteriophage T4 lysozyme. J Mol Biol. 1991;222:67–88. doi: 10.1016/0022-2836(91)90738-r. [DOI] [PubMed] [Google Scholar]
- 4.Markiewicz P, Kleina LG, Cruz C, Ehret S, Miller JH. Genetic studies of the lac repressor. XIV. Analysis of 4000 altered Escherichia coli lac repressors reveals essential and non-essential residues, as well as "spacers" which do not require a specific sequence. J Mol Biol. 1994;240:421–433. doi: 10.1006/jmbi.1994.1458. [DOI] [PubMed] [Google Scholar]
- 5.Matthews BW. Studies on protein stability with T4 lysozyme. Adv Protein Chem. 1995;46:249–278. doi: 10.1016/s0065-3233(08)60337-x. [DOI] [PubMed] [Google Scholar]
- 6.Loeb DD, Swanstrom R, Everitt L, Manchester M, Stamper SE, Hutchison CA., 3rd Complete mutagenesis of the HIV-1 protease. Nature. 1989;340:397–400. doi: 10.1038/340397a0. [DOI] [PubMed] [Google Scholar]
- 7.Lockless SW, Ranganathan R. Evolutionarily conserved pathways of energetic connectivity in protein families. Science. 1999;286:295–299. doi: 10.1126/science.286.5438.295. [DOI] [PubMed] [Google Scholar]
- 8.Ferguson AD, Amezcua CA, Halabi NM, Chelliah Y, Rosen MK, Ranganathan R, Deisenhofer J. Signal transduction pathway of TonB-dependent transporters. Proc Natl Acad Sci U S A. 2007;104:513–518. doi: 10.1073/pnas.0609887104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shulman AI, Larson C, Mangelsdorf DJ, Ranganathan R. Structural determinants of allosteric ligand activation in RXR heterodimers. Cell. 2004;116:417–429. doi: 10.1016/s0092-8674(04)00119-9. [DOI] [PubMed] [Google Scholar]
- 10.Suel GM, Lockless SW, Wall MA, Ranganathan R. Evolutionarily conserved networks of residues mediate allosteric communication in proteins. Nat Struct Biol. 2003;10:59–69. doi: 10.1038/nsb881. [DOI] [PubMed] [Google Scholar]
- 11.Hunt JA, Ahmed M, Fierke CA. Metal binding specificity in carbonic anhydrase is influenced by conserved hydrophobic core residues. Biochemistry. 1999;38:9054–9062. doi: 10.1021/bi9900166. [DOI] [PubMed] [Google Scholar]
- 12.Hedstrom L, Szilagyi L, Rutter WJ. Converting trypsin to chymotrypsin: the role of surface loops. Science. 1992;255:1249–1253. doi: 10.1126/science.1546324. [DOI] [PubMed] [Google Scholar]
- 13.Hedstrom L, Perona JJ, Rutter WJ. Converting trypsin to chymotrypsin: residue 172 is a substrate specificity determinant. Biochemistry. 1994;33:8757–8763. doi: 10.1021/bi00195a017. [DOI] [PubMed] [Google Scholar]
- 14.Perona JJ, Hedstrom L, Rutter WJ, Fletterick RJ. Structural origins of substrate discrimination in trypsin and chymotrypsin. Biochemistry. 1995;34:1489–1499. doi: 10.1021/bi00005a004. [DOI] [PubMed] [Google Scholar]
- 15.Lassila JK, Keeffe JR, Kast P, Mayo SL. Exhaustive mutagenesis of six secondary active-site residues in Escherichia coli chorismate mutase shows the importance of hydrophobic side chains and a helix N-capping position for stability and catalysis. Biochemistry. 2007;46:6883–6891. doi: 10.1021/bi700215x. [DOI] [PubMed] [Google Scholar]
- 16.Ataie NJ, Hoang QQ, Zahniser MP, Tu Y, Milne A, Petsko GA, Ringe D. Zinc coordination geometry and ligand binding affinity: the structural and kinetic analysis of the second-shell serine 228 residue and the methionine 180 residue of the aminopeptidase from Vibrio proteolyticus. Biochemistry. 2008;47:7673–7683. doi: 10.1021/bi702188e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oue S, Okamoto A, Yano T, Kagamiyama H. Redesigning the substrate specificity of an enzyme by cumulative effects of the mutations of non-active site residues. J Biol Chem. 1999;274:2344–2349. doi: 10.1074/jbc.274.4.2344. [DOI] [PubMed] [Google Scholar]
- 18.Patten PA, Gray NS, Yang PL, Marks CB, Wedemayer GJ, Boniface JJ, Stevens RC, Schultz PG. The immunological evolution of catalysis. Science. 1996;271:1086–1091. doi: 10.1126/science.271.5252.1086. [DOI] [PubMed] [Google Scholar]
- 19.Aharoni A, Gaidukov L, Khersonsky O, Mc QGS, Roodveldt C, Tawfik DS. The ‘evolvability’ of promiscuous protein functions. Nat Genet. 2005;37:73–76. doi: 10.1038/ng1482. [DOI] [PubMed] [Google Scholar]
- 20.Dueber JE, Yeh BJ, Bhattacharyya RP, Lim WA. Rewiring cell signaling: the logic and plasticity of eukaryotic protein circuitry. Curr Opin Struct Biol. 2004;14:690–699. doi: 10.1016/j.sbi.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 21.Perutz MF. Regulation of oxygen affinity of hemoglobin: influence of structure of the globin on the heme iron. Annu Rev Biochem. 1979;48:327–386. doi: 10.1146/annurev.bi.48.070179.001551. [DOI] [PubMed] [Google Scholar]
- 22.Dill KA. Dominant forces in protein folding. Biochemistry. 1990;29:7133–7155. doi: 10.1021/bi00483a001. [DOI] [PubMed] [Google Scholar]
- 23.Brion P, Westhof E. Hierarchy and dynamics of RNA folding. Annu Rev Biophys Biomol Struct. 1997;26:113–137. doi: 10.1146/annurev.biophys.26.1.113. [DOI] [PubMed] [Google Scholar]
- 24.Guo F, Gooding AR, Cech TR. Structure of the Tetrahymena ribozyme: base triple sandwich and metal ion at the active site. Mol Cell. 2004;16:351–362. doi: 10.1016/j.molcel.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 25.Adams PL, Stahley MR, Kosek AB, Wang J, Strobel SA. Crystal structure of a self-splicing group I intron with both exons. Nature. 2004;430:45–50. doi: 10.1038/nature02642. [DOI] [PubMed] [Google Scholar]
- 26.Golden BL, Kim H, Chase E. Crystal structure of a phage Twort group I ribozyme-product complex. Nat Struct Mol Biol. 2005;12:82–89. doi: 10.1038/nsmb868. [DOI] [PubMed] [Google Scholar]
- 27.Lipchock SV, Strobel SA. A relaxed active site after exon ligation by the group I intron. Proc Natl Acad Sci U S A. 2008;105:5699–5704. doi: 10.1073/pnas.0712016105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rupert PB, Massey AP, Sigurdsson ST, Ferre-D'Amare AR. Transition state stabilization by a catalytic RNA. Science. 2002;298:1421–1424. doi: 10.1126/science.1076093. [DOI] [PubMed] [Google Scholar]
- 29.Martick M, Scott WG. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell. 2006;126:309–320. doi: 10.1016/j.cell.2006.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lehmann J, Reichel A, Buguin A, Libchaber A. Efficiency of a self-aminoacylating ribozyme: effect of the length and base-composition of its 3' extension. RNA. 2007;13:1191–1197. doi: 10.1261/rna.500907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karbstein K, Tang KH, Herschlag D. A base triple in the Tetrahymena group I core affects the reaction equilibrium via a threshold effect. RNA. 2004;10:1730–1739. doi: 10.1261/rna.7118104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michel F, Ellington AD, Couture S, Szostak JW. Phylogenetic and genetic evidence for base-triples in the catalytic domain of group I introns. Nature. 1990;347:578–580. doi: 10.1038/347578a0. [DOI] [PubMed] [Google Scholar]
- 33.Green R, Szostak JW. In vitro genetic analysis of the hinge region between helical elements P5-P4-P6 and P7-P3-P8 in the sunY group I self-splicing intron. J Mol Biol. 1994;235:140–155. doi: 10.1016/s0022-2836(05)80022-1. [DOI] [PubMed] [Google Scholar]
- 34.Shepotinovskaya IV, Uhlenbeck OC. Catalytic diversity of extended hammerhead ribozymes. Biochemistry. 2008;47:7034–7042. doi: 10.1021/bi7025358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Engelhardt MADE, Knitt DS, Doudna JA, Herschlag D. The P5abc peripheral element facilitates preorganization of the Tetrahymena group I ribozyme for catalysis. Biochemistry. 2000;39:2639–2651. doi: 10.1021/bi992313g. [DOI] [PubMed] [Google Scholar]
- 36.Khvorova A, Lescoute A, Westhof E, Jayasena SD. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat Struct Biol. 2003;10:708–712. doi: 10.1038/nsb959. [DOI] [PubMed] [Google Scholar]
- 37.De la M, Gago S, Flores R. Peripheral regions of natural hammerhead ribozymes greatly increase their self-cleavage activity. EMBO J. 2003;22:5561–5570. doi: 10.1093/emboj/cdg530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hougland JL, Piccirilli JA, Forconi M, Lee J, Herschlag D. How the group I intron works: a case study of RNA structure and function. In: Gesteland RF, Cech TR, Atkins JF, editors. RNA World 3rd Edition. New York: Cold Spring Harbor Laboratory Press; 2005. pp. 133–205. [Google Scholar]
- 39.Michel F, Westhof E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J Mol Biol. 1990;216:585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- 40.Lehnert V, Jaeger L, Michel F, Westhof E. New loop-loop tertiary interactions in self-splicing introns of subgroup IC and ID: a complete 3D model of the Tetrahymena thermophila ribozyme. Chem Biol. 1996;3:993–1009. doi: 10.1016/s1074-5521(96)90166-0. [DOI] [PubMed] [Google Scholar]
- 41.Stahley MR, Strobel SA. Structural evidence for a two-metal-ion mechanism of group I intron splicing. Science. 2005;309:1587–1590. doi: 10.1126/science.1114994. [DOI] [PubMed] [Google Scholar]
- 42.Cannone JJ, Subramanian S, Schnare MN, Collett JR, D’Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Muller KM, Pande N, Shang Z, Yu N, Gutell RR. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burke JM, Belfort M, Cech TR, Davies RW, Schweyen RJ, Shub DA, Szostak JW, Tabak HF. Structural conventions for group I introns. Nucleic Acids Res. 1987;15:7217–7221. doi: 10.1093/nar/15.18.7217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Costa M, Michel F. Frequent use of the same tertiary motif by self-folding RNAs. EMBO J. 1995;14:1276–1285. doi: 10.1002/j.1460-2075.1995.tb07111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Szewczak AA, Kundrot CE, Cech TR, Doudna JA. RNA tertiary structure mediation by adenosine platforms. Science. 1996;273:1696–1699. doi: 10.1126/science.273.5282.1696. [DOI] [PubMed] [Google Scholar]
- 46.Cate JH, Hanna RL, Doudna JA. A magnesium ion core at the heart of a ribozyme domain. Nat Struct Biol. 1997;4:553–558. doi: 10.1038/nsb0797-553. [DOI] [PubMed] [Google Scholar]
- 47.Sattin BD, Zhao W, Travers K, Chu S, Herschlag D. Direct measurement of tertiary contact cooperativity in RNA folding. J Am Chem Soc. 2008;130:6085–6087. doi: 10.1021/ja800919q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ortoleva-Donnelly L, Szewczak AA, Gutell RR, Strobel SA. The chemical basis of adenosine conservation throughout the Tetrahymena ribozyme. RNA. 1998;4:498–519. doi: 10.1017/s1355838298980086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Strauss-Soukup JK, Strobel SA. A chemical phylogeny of group I introns based upon interference mapping of a bacterial ribozyme. J Mol Biol. 2000;302:339–358. doi: 10.1006/jmbi.2000.4056. [DOI] [PubMed] [Google Scholar]
- 50.Szewczak AA, Ortoleva-Donnelly L, Ryder SP, Moncoeur E, Strobel SA. A minor groove RNA triple helix within the catalytic core of a group I intron. Nat Struct Biol. 1998;5:1037–1042. doi: 10.1038/4146. [DOI] [PubMed] [Google Scholar]
- 51.Zaug AJ, Grosshans CA, Cech TR. Sequence-specific endoribonuclease activity of the Tetrahymena ribozyme: enhanced cleavage of certain oligonucleotide substrates that form mismatched ribozyme-substrate complexes. Biochemistry. 1988;27:8924–8931. doi: 10.1021/bi00425a008. [DOI] [PubMed] [Google Scholar]
- 52.Zaug AJ, Cech TR. The Tetrahymena intervening sequence ribonucleic acid enzyme is a phosphotransferase and an acid phosphatase. Biochemistry. 1986;25:4478–4482. doi: 10.1021/bi00364a002. [DOI] [PubMed] [Google Scholar]
- 53.Forconi M, Lee J, Lee JK, Piccirilli JA, Herschlag D. Functional identification of ligands for a catalytic metal ion in group I introns. Biochemistry. 2008;47:6883–6894. doi: 10.1021/bi800519a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karbstein K, Herschlag D. Extraordinarily slow binding of guanosine to the Tetrahymena group I ribozyme: implications for RNA preorganization and function. Proc Natl Acad Sci U S A. 2003;100:2300–2305. doi: 10.1073/pnas.252749799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Herschlag D, Eckstein F, Cech TR. Contributions of 2'-hydroxyl groups of the RNA substrate to binding and catalysis by the Tetrahymena ribozyme. An energetic picture of an active site composed of RNA. Biochemistry. 1993;32:8299–8311. doi: 10.1021/bi00083a034. [DOI] [PubMed] [Google Scholar]
- 56.McConnell TS, Cech TR. A positive entropy change for guanosine binding and for the chemical step in the Tetrahymena ribozyme reaction. Biochemistry. 1995;34:4056–4067. doi: 10.1021/bi00012a024. [DOI] [PubMed] [Google Scholar]
- 57.Forconi M, Piccirilli JA, Herschlag D. Modulation of individual steps in group I intron catalysis by a peripheral metal ion. RNA. 2007;13:1656–1667. doi: 10.1261/rna.632007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Forconi M, Sengupta RN, Piccirilli JA, Herschlag D. A rearrangement of the guanosine-binding site establishes an extended network of functional interactions in the Tetrahymena group I ribozyme active site. Biochemistry. 2010;49:2753–2762. doi: 10.1021/bi902200n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Herschlag D, Cech TR. Catalysis of RNA cleavage by the Tetrahymena thermophila ribozyme. 1. Kinetic description of the reaction of an RNA substrate complementary to the active site. Biochemistry. 1990;29:10159–10171. doi: 10.1021/bi00496a003. [DOI] [PubMed] [Google Scholar]
- 60.Narlikar GJ, Herschlag D. Isolation of a local tertiary folding transition in the context of a globally folded RNA. Nat Struct Biol. 1996;3:701–710. doi: 10.1038/nsb0896-701. [DOI] [PubMed] [Google Scholar]
- 61.Bartley LE, Zhuang X, Das R, Chu S, Herschlag D. Exploration of the transition state for tertiary structure formation between an RNA helix and a large structured RNA. J Mol Biol. 2003;328:1011–1026. doi: 10.1016/s0022-2836(03)00272-9. [DOI] [PubMed] [Google Scholar]
- 62.Bevilacqua PC, Kierzek R, Johnson KA, Turner DH. Dynamics of ribozyme binding of substrate revealed by fluorescence-detected stopped-flow methods. Science. 1992;258:1355–1358. doi: 10.1126/science.1455230. [DOI] [PubMed] [Google Scholar]
- 63.Narlikar GJ, Gopalakrishnan V, McConnell TS, Usman N, Herschlag D. Use of binding energy by an RNA enzyme for catalysis by positioning and substrate destabilization. Proc Natl Acad Sci U S A. 1995;92:3668–3672. doi: 10.1073/pnas.92.9.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Narlikar GJ, Khosla M, Usman N, Herschlag D. Quantitating tertiary binding energies of 2' OH groups on the P1 duplex of the Tetrahymena ribozyme: intrinsic binding energy in an RNA enzyme. Biochemistry. 1997;36:2465–2477. doi: 10.1021/bi9610820. [DOI] [PubMed] [Google Scholar]
- 65.Freier SM, Kierzek R, Caruthers MH, Neilson T, Turner DH. Free energy contributions of G•U and other terminal mismatches to helix stability. Biochemistry. 1986;25:3209–3213. doi: 10.1021/bi00359a019. [DOI] [PubMed] [Google Scholar]
- 66.Karbstein K, Carroll KS, Herschlag D. Probing the Tetrahymena group I ribozyme reaction in both directions. Biochemistry. 2002;41:11171–11183. doi: 10.1021/bi0202631. [DOI] [PubMed] [Google Scholar]
- 67.Mei R, Herschlag D. Mechanistic investigations of a ribozyme derived from the Tetrahymena group I intron: insights into catalysis and the second step of self-splicing. Biochemistry. 1996;35:5796–5809. doi: 10.1021/bi9527653. [DOI] [PubMed] [Google Scholar]
- 68.Russell R, Herschlag D. Specificity from steric restrictions in the guanosine binding pocket of a group I ribozyme. RNA. 1999;5:158–166. doi: 10.1017/s1355838299981839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McConnell TS, Cech TR, Herschlag D. Guanosine binding to the Tetrahymena ribozyme: thermodynamic coupling with oligonucleotide binding. Proc Natl Acad Sci U S A. 1993;90:8362–8366. doi: 10.1073/pnas.90.18.8362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pyle AM, Moran S, Strobel SA, Chapman T, Turner DH, Cech TR. Replacement of the conserved G•U with a G-C pair at the cleavage site of the Tetrahymena ribozyme decreases binding, reactivity, and fidelity. Biochemistry. 1994;33:13856–13863. doi: 10.1021/bi00250a040. [DOI] [PubMed] [Google Scholar]
- 71.Knitt DS, Herschlag D. pH dependencies of the Tetrahymena ribozyme reveal an unconventional origin of an apparent pKa. Biochemistry. 1996;35:1560–1570. doi: 10.1021/bi9521147. [DOI] [PubMed] [Google Scholar]
- 72.Legault P, Herschlag D, Celander DW, Cech TR. Mutations at the guanosine-binding site of the Tetrahymena ribozyme also affect site-specific hydrolysis. Nucleic Acids Res. 1992;20:6613–6619. doi: 10.1093/nar/20.24.6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Donis-Keller H, Maxam AM, Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977;4:2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huang Z, Szostak JW. A simple method for 3'-labeling of RNA. Nucleic Acids Res. 1996;24:4360–4361. doi: 10.1093/nar/24.21.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Celander DW, Cech TR. Visualizing the higher order folding of a catalytic RNA molecule. Science. 1991;251:401–407. doi: 10.1126/science.1989074. [DOI] [PubMed] [Google Scholar]
- 76.Sclavi B, Woodson S, Sullivan M, Chance MR, Brenowitz M. Time-resolved synchrotron X-ray "footprinting", a new approach to the study of nucleic acid structure and function: application to protein-DNA interactions and RNA folding. J Mol Biol. 1997;266:144–159. doi: 10.1006/jmbi.1996.0775. [DOI] [PubMed] [Google Scholar]
- 77.Das R, Laederach A, Pearlman SM, Herschlag D, Altman RB. SAFA: semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA. 2005;11:344–354. doi: 10.1261/rna.7214405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Takamoto K, Chance MR, Brenowitz M. Semi-automated, single-band peak-fitting analysis of hydroxyl radical nucleic acid footprint autoradiograms for the quantitative analysis of transitions. Nucleic Acids Res. 2004;32:E119. doi: 10.1093/nar/gnh117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Uchida T, Takamoto K, He Q, Chance MR, Brenowitz M. Multiple monovalent ion-dependent pathways for the folding of the L-21 Tetrahymena thermophila ribozyme. J Mol Biol. 2003;328:463–478. doi: 10.1016/s0022-2836(03)00247-x. [DOI] [PubMed] [Google Scholar]
- 80.Uchida T, He Q, Ralston CY, Brenowitz M, Chance MR. Linkage of monovalent and divalent ion binding in the folding of the P4-P6 domain of the Tetrahymena ribozyme. Biochemistry. 2002;41:5799–5806. doi: 10.1021/bi020042v. [DOI] [PubMed] [Google Scholar]
- 81.Fang X, Pan T, Sosnick TR. A thermodynamic framework and cooperativity in the tertiary folding of a Mg2+ dependent ribozyme. Biochemistry. 1999;38:16840–16846. doi: 10.1021/bi991700n. [DOI] [PubMed] [Google Scholar]
- 82.Draper DE. A guide to ions and RNA structure. RNA. 2004;10:335–343. doi: 10.1261/rna.5205404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Joyce GF, van der Horst G, Inoue T. Catalytic activity is retained in the Tetrahymena group I intron despite removal of the large extension of element P5. Nucleic Acids Res. 1989;17:7879–7889. doi: 10.1093/nar/17.19.7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.van der Horst G, Christian A, Inoue T. Reconstitution of a group I intron self-splicing reaction with an activator RNA. Proc Natl Acad Sci U S A. 1991;88:184–188. doi: 10.1073/pnas.88.1.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pace U, Szostak JW. Mutations in a semiconserved region of the Tetrahymena intron. FEBS Lett. 1991;280:171–174. doi: 10.1016/0014-5793(91)80230-z. [DOI] [PubMed] [Google Scholar]
- 86.Barfod ET, Cech TR. Deletion of nonconserved helices near the 3' end of the rRNA intron of Tetrahymena thermophila alters self-splicing but not core catalytic activity. Genes Dev. 1988;2:652–663. doi: 10.1101/gad.2.6.652. [DOI] [PubMed] [Google Scholar]
- 87.Caprara MG, Waring RB. Deletion of P9 and stem-loop structures downstream from the catalytic core affects both 5' and 3' splicing activities in a group-I intron. Gene. 1994;143:29–37. doi: 10.1016/0378-1119(94)90600-9. [DOI] [PubMed] [Google Scholar]
- 88.Szostak JW. Enzymatic activity of the conserved core of a group I self-splicing intron. Nature. 1986;322:83–86. doi: 10.1038/322083a0. [DOI] [PubMed] [Google Scholar]
- 89.Doudna JA, Szostak JW. Miniribozymes, small derivatives of the sunY intron, are catalytically active. Mol Cell Biol. 1989;9:5480–5483. doi: 10.1128/mcb.9.12.5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tuschl T, Ng MM, Pieken W, Benseler F, Eckstein F. Importance of exocyclic base functional groups of central core guanosines for hammerhead ribozyme activity. Biochemistry. 1993;32:11658–11668. doi: 10.1021/bi00094a023. [DOI] [PubMed] [Google Scholar]
- 91.Ikawa Y, Yoshimura T, Hara H, Shiraishi H, Inoue T. Two conserved structural components, A-rich bulge and P4 XJ6/7 base-triples, in activating the group I ribozymes. Genes Cells. 2002;7:1205–1215. doi: 10.1046/j.1365-2443.2002.00601.x. [DOI] [PubMed] [Google Scholar]
- 92.Jaeger L, Westhof E, Michel F. Function of P11, a tertiary base pairing in self-splicing introns of subgroup IA. J Mol Biol. 1991;221:1153–1164. doi: 10.1016/0022-2836(91)90925-v. [DOI] [PubMed] [Google Scholar]
- 93.von Ahsen U, Noller HF. Methylation interference experiments identify bases that are essential for distinct catalytic functions of a group I ribozyme. EMBO J. 1993;12:4747–4754. doi: 10.1002/j.1460-2075.1993.tb06163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Travers KJ, Boyd N, Herschlag D. Low specificity of metal ion binding in the metal ion core of a folded RNA. RNA. 2007;13:1205–1213. doi: 10.1261/rna.566007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Doherty EA, Herschlag D, Doudna JA. Assembly of an exceptionally stable RNA tertiary interface in a group I ribozyme. Biochemistry. 1999;38:2982–2990. doi: 10.1021/bi982113p. [DOI] [PubMed] [Google Scholar]
- 96.Das R, Travers KJ, Bai Y, Herschlag D. Determining the Mg2+ stoichiometry for folding an RNA metal ion core. J Am Chem Soc. 2005;127:8272–8273. doi: 10.1021/ja051422h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Buchmueller KL, Webb AE, Richardson DA, Weeks KM. A collapsed non-native RNA folding state. Nat Struct Biol. 2000;7:362–366. doi: 10.1038/75125. [DOI] [PubMed] [Google Scholar]
- 98.Swisher JF, Su LJ, Brenowitz M, Anderson VE, Pyle AM. Productive folding to the native state by a group II intron ribozyme. J Mol Biol. 2002;315:297–310. doi: 10.1006/jmbi.2001.5233. [DOI] [PubMed] [Google Scholar]
- 99.Romani A, Scarpa A. Regulation of cell magnesium. Arch Biochem Biophys. 1992;298:1–12. doi: 10.1016/0003-9861(92)90086-c. [DOI] [PubMed] [Google Scholar]
- 100.Zaug AJ, Cech TR. The intervening sequence RNA of Tetrahymena is an enzyme. Science. 1986;231:470–475. doi: 10.1126/science.3941911. [DOI] [PubMed] [Google Scholar]
- 101.Moran S, Kierzek R, Turner DH. Binding of guanosine and 3′ splice site analogues to a group I ribozyme: interactions with functional groups of guanosine and with additional nucleotides. Biochemistry. 1993;32:5247–5256. doi: 10.1021/bi00070a037. [DOI] [PubMed] [Google Scholar]
- 102.Bevilacqua PC, Turner DH. Comparison of binding of mixed ribose-deoxyribose analogues of CUCU to a ribozyme and to GGAGAA by equilibrium dialysis: evidence for ribozyme specific interactions with 2' OH groups. Biochemistry. 1991;30:10632–10640. doi: 10.1021/bi00108a005. [DOI] [PubMed] [Google Scholar]
- 103.Herschlag D. Evidence for processivity and two-step binding of the RNA substrate from studies of J1/2 mutants of the Tetrahymena ribozyme. Biochemistry. 1992;31:1386–1399. doi: 10.1021/bi00120a015. [DOI] [PubMed] [Google Scholar]
- 104.Narlikar GJ, Bartley LE, Herschlag D. Use of duplex rigidity for stability and specificity in RNA tertiary structure. Biochemistry. 2000;39:6183–6189. doi: 10.1021/bi992858a. [DOI] [PubMed] [Google Scholar]
- 105.Bevilacqua PC, Sugimoto N, Turner DH. A mechanistic framework for the second step of splicing catalyzed by the Tetrahymena ribozyme. Biochemistry. 1996;35:648–658. doi: 10.1021/bi951962z. [DOI] [PubMed] [Google Scholar]
- 106.Russell R, Das R, Suh H, Travers KJ, Laederach A, Engelhardt MA, Herschlag D. The paradoxical behavior of a highly structured misfolded intermediate in RNA folding. J Mol Biol. 2006;363:531–544. doi: 10.1016/j.jmb.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 107.Laggerbauer B, Murphy FL, Cech TR. Two major tertiary folding transitions of the Tetrahymena catalytic RNA. EMBO J. 1994;13:2669–2676. doi: 10.1002/j.1460-2075.1994.tb06557.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Murphy FL, Cech TR. An independently folding domain of RNA tertiary structure within the Tetrahymena ribozyme. Biochemistry. 1993;32:5291–5300. doi: 10.1021/bi00071a003. [DOI] [PubMed] [Google Scholar]
- 109.Antonioli AH, Cochrane JC, Lipchock SV, Strobel SA. Plasticity of the RNA kink turn structural motif. RNA. 2010;16:762–768. doi: 10.1261/rna.1883810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Matsumura S, Ikawa Y, Inoue T. Biochemical characterization of the kink-turn RNA motif. Nucleic Acids Res. 2003;31:5544–5551. doi: 10.1093/nar/gkg760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tanner MA, Anderson EM, Gutell RR, Cech TR. Mutagenesis and comparative sequence analysis of a base triple joining the two domains of group I ribozymes. RNA. 1997;3:1037–1051. [PMC free article] [PubMed] [Google Scholar]
- 112.Tanner MA, Cech TR. Joining the two domains of a group I ribozyme to form the catalytic core. Science. 1997;275:847–849. doi: 10.1126/science.275.5301.847. [DOI] [PubMed] [Google Scholar]
- 113.Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Kundrot CE, Cech TR, Doudna JA. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 114.Downs WD, Cech TR. A tertiary interaction in the Tetrahymena intron contributes to selection of the 5' splice site. Genes Dev. 1994;8:1198–1211. doi: 10.1101/gad.8.10.1198. [DOI] [PubMed] [Google Scholar]
- 115.Szewczak AA, Ortoleva-Donnelly L, Zivarts MV, Oyelere AK, Kazantsev AV, Strobel SA. An important base triple anchors the substrate helix recognition surface within the Tetrahymena ribozyme active site. Proc Natl Acad Sci U S A. 1999;96:11183–11188. doi: 10.1073/pnas.96.20.11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Goodey NM, Benkovic SJ. Allosteric regulation and catalysis emerge via a common route. Nat Chem Biol. 2008;4:474–482. doi: 10.1038/nchembio.98. [DOI] [PubMed] [Google Scholar]
- 117.Watanabe S, Kawai G, Muto Y, Watanabe K, Inoue T, Yokoyama S. An RNA fragment consisting of the P7 and P9.0 stems and the 3'-terminal guanosine of the Tetrahymena group I intron. Nucleic Acids Res. 1996;24:1337–1344. doi: 10.1093/nar/24.7.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kitamura A, Muto Y, Watanabe S, Kim I, Ito T, Nishiya Y, Sakamoto K, Ohtsuki T, Kawai G, Watanabe K, Hosono K, Takaku H, Katoh E, Yamazaki T, Inoue T, Yokoyama S. Solution structure of an RNA fragment with the P7/P9.0 region and the 3'-terminal guanosine of the Tetrahymena group I intron. RNA. 2002;8:440–451. doi: 10.1017/s1355838202026043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.de la Pena M, Dufour D, Gallego J. Three-way RNA junctions with remote tertiary contacts: a recurrent and highly versatile fold. RNA. 2009;15:1949–1964. doi: 10.1261/rna.1889509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Andersen AA, Collins RA. Rearrangement of a stable RNA secondary structure during VS ribozyme catalysis. Mol Cell. 2000;5:469–478. doi: 10.1016/s1097-2765(00)80441-4. [DOI] [PubMed] [Google Scholar]
- 121.Adams PL, Stahley MR, Gill ML, Kosek AB, Wang J, Strobel SA. Crystal structure of a group I intron splicing intermediate. RNA. 2004;10:1867–1887. doi: 10.1261/rna.7140504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Shih IH, Been MD. Catalytic strategies of the hepatitis delta virus ribozymes. Annu Rev Biochem. 2002;71:887–917. doi: 10.1146/annurev.biochem.71.110601.135349. [DOI] [PubMed] [Google Scholar]
- 123.Tanner M, Cech T. Activity and thermostability of the small self-splicing group I intron in the pre-tRNA(lle) of the purple bacterium Azoarcus. RNA. 1996;2:74–83. [PMC free article] [PubMed] [Google Scholar]
- 124.Rosenstein SP, Been MD. Self-cleavage of hepatitis delta virus genomic strand RNA is enhanced under partially denaturing conditions. Biochemistry. 1990;29:8011–8016. doi: 10.1021/bi00487a002. [DOI] [PubMed] [Google Scholar]
- 125.Jossinet F, Ludwig TE, Westhof E. Assemble: an interactive graphical tool to analyze and build RNA architectures at the 2D and 3D levels. Bioinformatics. 2010;26:2057–2059. doi: 10.1093/bioinformatics/btq321. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.