Abstract
Cohesin is a multiprotein ringed complex that is most well known for its role in stabilizing the association of sister chromatids between S phase and M. More recently cohesin was found to be associated with transcriptional insulators, elements that are associated with the organization of chromatin into regulatory domains. The human major histocompatibility complex class II (MHC-II) locuscontains ten intergenic elements, termed MHC-II insulators, which bind the transcriptional insulator protein CCCTC transcription factor (CTCF). MHC-II insulators interact with each other forming a base architecture of discrete loops and potential regulatory domains. When MHC-II genes are expressed, their proximal promoter regulatory regions reorganize to the foci established by the interacting MHC-II insulators. MHC-II insulators also bind cohesin, but the functional role of cohesin in regulating this system is not known. Here we show that the binding of cohesin to MHC-II insulators occurred irrespective of MHC-II expression but was required for optimal expression of the HLA-DR and HLA-DQ genes. In a DNA dependent manner, cohesin subunits interacted with CTCF and the MHC-II specific transcription factors RFX and CIITA. Intriguingly, cohesin subunits were important for DNA looping interactions between the HLA-DRA promoter region and a 5’ MHC-II insulator but were not required for interactions between the MHC-II insulators themselves. This latter observation introduces cohesin as a regulator of MHC-II expression by initiating or stabilizing MHC-II promoter regulatory element interactions with the MHC-II insulator elements; events which are required for maximal MHC-II transcription.
Introduction
The human major histocompatibility complex class II (MHC-II)2 locus located at 6p21.31 contains a cluster of some of the most polymorphic genes of the human genome (1). Within the locus, the three-heterodimeric MHC-II isotypes (HLA-DR, -DQ, and -DP) are encoded by alpha and beta chain genes. These proteins function by presenting antigenic peptides to CD4+ T lymphocytes (2–5). Through the presentation of antigens, MHC-II genes function, control and/or maintain adaptive immune responses (reviewed in (6–8)). Antigenic peptide selection and presentation is aided by the products of the HLA-DM and HLA-DO genes, also encoded within the locus. Several MHC-II evolutionarily defunct pseudogenes and non-MHC-II genes are present within the MHC-II locus as well.
MHC-II genes are tightly regulated in a cell-type dependent manner, being highly expressed in B-lymphocytes, macrophages, dendritic cells, and certain cells within the thymus. Interferon-γ (IFN-γ) is a potent inducer of MHC-II gene expression in diverse cell types (9), and potentially can allow these cells to present antigens. Irrespective of transcription, the factors RFX (regulatory factor X), CREB (cAMP response element binding protein), and NF-Y (nuclear factor-Y) bind constitutively to promoter proximal conserved sequence elements upstream of each MHC-II gene, termed the X1, X2 and Y boxes, respectively (Reviewed in (6, 10, 11)). When transcription is activated, the MHC class II transactivator, CIITA, is recruited to the X-Y box boundfactors. CIITA functions as a coregulatory adapter between the above DNA-bound transcription factors, chromatin modification/remodeling factors, and the general transcription machinery (12). CIITA expression is cell-type specific and the limiting transcription factor for this system. Critically, CIITA expression is regulated and can be induced in most cell types by IFN-γ (13), a process that leads to MHC-II gene expression.
The CCCTC-binding factor, CTCF, demarcates and insulates regions of regulatory activity within the genome by its nature of action either as an enhancer blocker (14) or barrier preventing heterochromatin propagation toward active genes (15). All known insulator elements in vertebrates are associated with CTCF (16–18). Ten CTCF-binding sites were identified within the human MHC-II region (19). Knockdown of CTCF resulted in decreased expression of all CIITA-regulated MHC-II genes but not the constitutively (non-CIITA regulated) expressed genes within the locus (19, 20). A set of self-interactions between the CTCF-bound elements was observed that was independent of MHC-II gene transcription, occurring in the absence or presence of CIITA. These interactions form the basal architecture of chromatin loops across the locus. An additional set of interactions between the CTCF sites and the proximal promoter regions of MHC-II genes was observed but only when CIITA was present. The CIITA-dependent interactions were also dependent on CTCF and placed these CTCF-bound regions as key elements required for the regulation of MHC-II transcription and antigen presentation (19). We termed these CTCF-bound regions “MHC-II insulators” to describe the property that they regulate MHC-II gene expression and can interact with MHC-II proximal promoters in a CIITA-dependent manner.
The cohesin complex is composed of Smc1, Smc3, Rad21/Scc1, and Scc3/SA1 proteins (21). Cohesin is best known for its role in maintaining the pairing of sister chromatids following S phase of the cell cycle (22–24) and in repair of double-stranded DNA breaks (25–29). However, over the last few years, the function of this multisubunit, ringed complex has diversified considerably (30–34). Genome-wide ChIP-seq and ChIP-chip analyses found that the majority of the CTCF binding sites (35, 36) are also bound/associated with the cohesin complex (37–39). Transient knockdown of a cohesin subunit resulted in a loss of CTCF bound enhancer-blocking activity (40). This finding and the fact that cohesin was expressed and bound to chromatin in interphase nuclei suggest that cohesin may play a functional role with CTCF transcriptional insulators. More recent ChIP-seq studies found that cohesin subunits were associated with sites that also bound the mediator coactivator complex of RNA polymerase II, suggesting that cohesin also played a role in the transcriptional regulation of some genes (41). We recently showed that the cohesin subunits Rad21 and Smc3 co-localized with the MHC-II insulator sites identified within the MHC-II locus (19); however, the role of these cohesin subunits in this system was not investigated.
In this study, the role of cohesin in regulating human MHC-II genes was examined using the C1 MHC-II insulator CTCF binding element, which lies 24 kb upstream of the HLA-DRA transcription start site and is the most 5’ of the MHC-II insulators (Figure 1). In the transcriptionally inactive state, C1 interacts with the MHC-II insulator XL9, which is located 3’ to the HLA-DRB1 gene (19). This interaction forms a large chromatin loop that encompasses the HLA-DR subregion. In the transcriptionally active state, C1 interacts directly with the HLA-DRA gene and is the predominant MHC-II insulator controlling HLA-DRA gene expression (19). We show here that C1 is required for maximal HLA-DRA expression; encodes enhancer blocking activity; and that siRNA depletion of cohesin subunits correlates with a loss of the enhancer blocking of the C1 element and with the expression of HLA-DRA and other MHC-II genes. Immunoprecipitation assays showed that cohesin is in a complex with CTCF, RFX, and CIITA, but that such interactions were dependent on the presence of DNA in the extract. Cohesin subunit siRNA depletion experiments showed that cohesin binding to C1 was dependent on CTCF. Cohesin depletion also demonstrated that the interactions between C1 and the HLA-DRA proximal promoter region were dependent on cohesin subunits; but surprisingly, interactions between adjacent MHC-II insulators were not. These data therefore describe the novel role for cohesin in regulating MHC-II expression and separating cohesin function from that of CTCF.
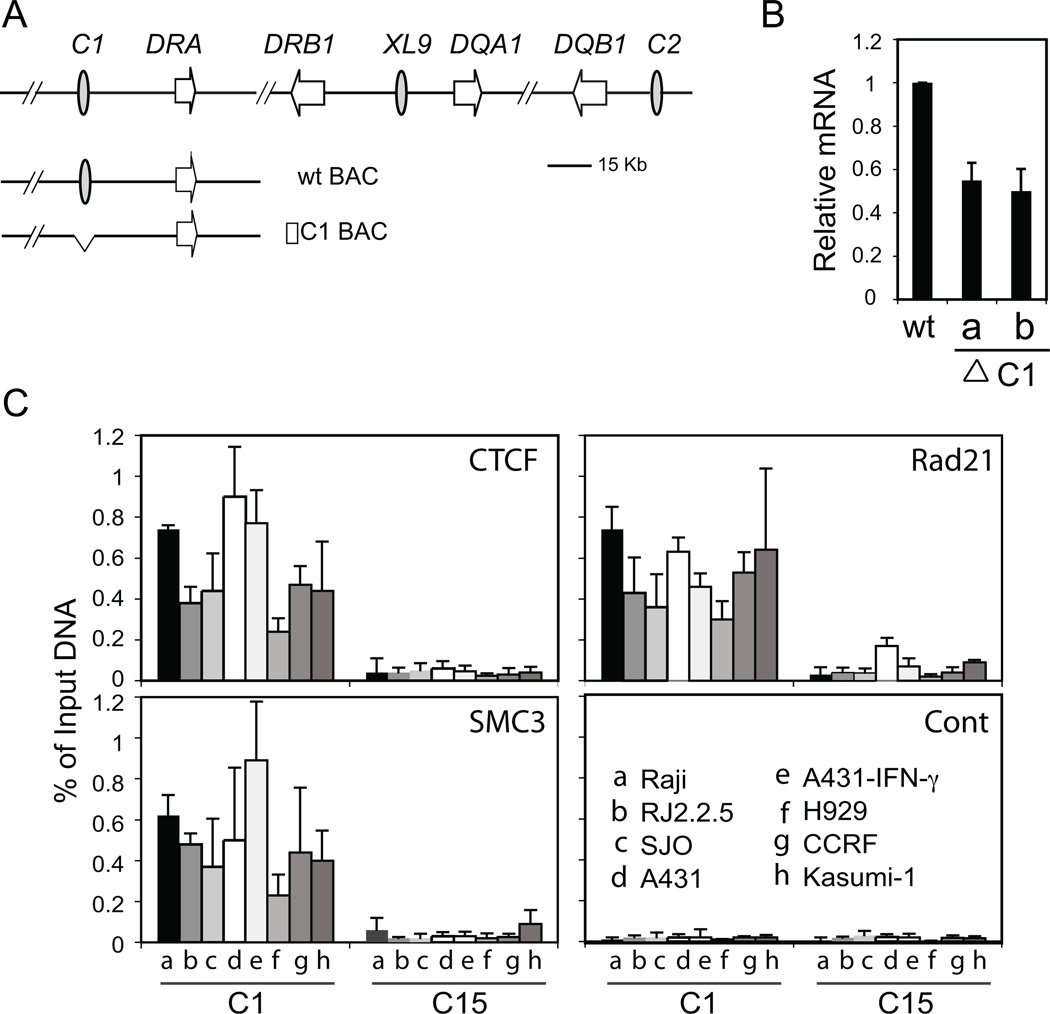
Figure 1. Optimal HLA-DRA gene expression is dependent on the MHC-II insulator C1, which is constitutively occupied by CTCF and cohesin.
A) A relative schematic of the HLA-DR, HLA-DQ region with the MHC-II insulators C1, XL9, and C2 is shown. Wild-type and mutant human BAC clones derived from PR11-974L24, containing the C1-HLA-DRA region are represented. The exact DNA sequence coordinates of PR11-974L24 from the UCSC Genome Browser are Chr6:32,341,463 to 32,456,230. B) Wild-type and two independently derived ΔC1 deleted mutant BACs (ΔC1a and ΔC1b) were transiently transfected into the murine B cell line (A20) by nucleofection. 48 h posttransfection, RNA was purified and analyzed for the presence of HLA-DRA gene transcripts by real time RT-PCR using a primer set specific for the human HLA-DRA gene. Results were normalized with respect to the level of GAPDH mRNA expression and the average plotted as relative expression. C) Quantitative, real-time PCR based ChIP assays were performed using chromatin prepared from the indicated cells to determine the in vivo occupancy of CTCF, Rad21, and SMC3 at C1. A431 cells (−/+IFN-γ) were also used in this ChIP assay. Antisera specific to CTCF, Rad21, and SMC3, and a non-specific IgG control antiserum were used as indicated. Primers used to amplify the C1 and negative control C15 (19) region are provided in Supplemental Figure 1. These results represent an average of at least three independent experiments.
Materials and Methods
Cell culture
The Burkitt’s lymphoma B-cell line Raji (42) expresses MHC-II genes and was obtained from the American Type Culture Collection (ATCC). The cell line RJ2.2.5 (provided by R. Accola, University of Insubria, Varese, Italy) is a derivative of the Raji line that does not express MHC-II genes due to mutations within the CIITAlocus (43, 44). NCI-H929 cells (CRL-9068; ATCC), a plasmacytoma-derived cell line does not express CIITA and MHC-II genes. These cells were grown in RPMI 1640 supplemented with 5% fetal bovine serum (FBS) (Hyclone, Inc., Logan, UT), 5% bovine calf serum (Hyclone), 100 U/ml penicillin/streptomycin, 4.5 g/L glucose, 1.0 mM sodium pyruvate, and 10 mM HEPES. The B-cell line SJO, derived from a patient with bare lymphocyte syndrome, is MHC-II deficient due to mutations in RFX5 (45). SJO cells were grown in the above media supplemented with FBS at 15%. A431 is an epithelial cell line that is negative for MHC-II expression. A431 cells were grown with or without Interferon-γ (500U/ml; PeproTech Inc., NJ; catalog number 300-02) in DMEM with the above supplements and 10% FBS. The CCRF-CEM cells human T cell lymphoblastic leukemia cell line and Kasumi-1 cells, a human acute myeloid leukemia cell line (provided by Dr. L. R. Gooding, Emory University) were also used. These lines were grown in RPMI media as above supplemented with 10 and 20% FBS, respectively. Murine A20 B cells are MHC-II positive and were grown in RPMI 1640 medium with 2 mM L-glutamine and 10% FBS.
Bacterial artificial chromosome modification and plasmid constructions
The bacterial artificial chromosome (BAC) vector containing a section of MHC-II locus that included C1 and HLA-DRA (PR11-974L24, Figure 1) was purchased from BACPAC resources Center at Children’s Hospital Oakland Research Institute (Oakland, CA). This BAC was manipulated to delete C1 using a recA-mediated modification system protocol as described (46). A recombination shuttle vector (pLD53.SCA-E-B) was obtained from Dr. N. Heniz (The Rockefeller University, NY). DNA sequences surrounding C1, representing the homologous targeting left and right arms, respectively (Chr6: 32,488,773-32,489,204 and Chr6: 32,489,725-32,490,117 on UCSC Genome Browser on Human Mar. 2006 Assembly) were introduced into the restriction sites of AscI and PacI of the shuttle vector, resulting in pLD53SCA-E-B-C1. The pLD53SCA-E-B-L-C1 was transformed into competent E. coli bacteria harboring the BAC DNA by electroporation (Bio-Rad Gene Pulser) to allow for in vivo homologous recombination events. Bacteria were selected and grown in 1 ml Luria broth medium containing ampicillin (50 µg/ml) and chloramphenicol (15 µg/ml) overnight at 37°C. The culture was diluted 1,000 fold and incubated at 37°C for another 16 h in the above selection medium. Bacteria were further diluted and spread onto Luria broth plates containing the above antibiotics and incubated at 37°C overnight. Colonies were analyzed for integration of the shuttle vector into the BAC by PCR. Bacteria containing co-integrated BAC/Shuttle vector were grown in Luria broth media supplemented with chloramphenicol (15 µl/l) for 1 h at 37°C and spread onto plates containing chloramphenicol and 7% sucrose for resolution/excision of the recombined targeting vector sequences. Following this step, bacterial colonies were screened by PCR for excision of the targeting vector sequences, and BAC DNAs were analyzed by restriction enzyme digestion and Southern blotting to ensure that the recombination, integration, and resolution processes occurred precisely. Two independent co-integrated BACs were carried through the process, producing ΔC1a and ΔC1b that had no other detectable deletions but those in C1.
The minimal promoter fireflyluciferase reporter plasmid, pGL3-promoter plasmid (Promega, Inc.) was used as a base vector in transient transfection transcriptional reporter assays to determine enhancer activity. The 520 bp encompassing C1 was cloned into pGL3-promoter using primers forward 5’GCATTCTGTAGTATCATCTCTATGG and reverse 5’CTCCAGTCAGTCAGCAAACC to amplify the sequence. The 5’HS4 chicken β-globin insulator (17) and a fragment of λ DNA pGL3-promoter vectors were described previously (47).
To assay the ability of C1 region to function as an enhancer blocker, the pGL3-promoter vector containing SV40 promoter upstream of the luciferase gene was used as the base plasmid vector (Promega, Inc.). pGL3–5′HS4-SV40, derived from pGL3-promoter, was described previously (47) and contains the 5’HS4 insulator upstream of the SV40 enhancer. C1 was placed between the SV40 enhancer and the SV40 promoter in pGL3-5’HS4-SV40 in a manner similar to that described for XL9 previously (47). The other enhancer blocking vectors were also described previously (47). In some constructions, the MHC-II insulator XL9 replaced HS4. The XL9 sequence was described previously (47, 48). Primers used to amplify this sequence were XL9: 5’-TGCTTCCTTTCAGTGTCCAAGTG and 5’-GGCCAGCCACACAGAGTTAGGGC.
Transient Transfection assays
The human wild-type and modified BAC DNAs (ΔC1a and b) were transfected into the murine B cell lineA20 by nucleofection using an Amaxa Nucleoportator with transfection Kit V according to the manufacturer’s protocols (Lonza, Walkersville, MD). In each transfection, ~2 × 106 cells and 4 µg DNA were used. Following 48 h post-transfection, total RNA was isolated using RNA easy kits (Qiagen, Inc.) and the transcripts of human HLA-DRA gene, which was encoded in the human BAC was analyzed by real time RT-PCR as described (46). Transcripts were normalized to the levels of GAPDH mRNA. The data were averaged from three independent transfections.
For the luciferase reporter transient transfection assays, a constitutively expressing Renilla luciferase expression vector (pRLTK) was co-transfected into Raji cells along with the test or control fireflyluciferase vectors described above. All transfections were carried out by nucleofection as above. After 48 h, cell lysates were prepared and luciferase activity for both firefly and Renilla was determined with a luminometer as instructed in the manufacturer's protocol (Dual-luciferase reporter assay system; Promega, Inc.). The value of measured activity of fireflyluciferase for each construct was normalized to the activity of the co-transfected Renilla luciferase reporter. All assays were carried out at least three times, and the data are presented as the average with the standard error of the mean.
SiRNA treatment
SMART pool siRNAs purchased (Dharmacon, Thermo Scientific, Inc.) specific for CTCF, Rad21, and SMC3 were transfected into 4 × 106 cells using a nucleofection apparatus and transfection reagents (kit V) from Amaxa Biosystems to knockdown the expression of these genes as previously described (19). As a negative control, siRNA against GFP was also transfected. Transfected cells were grown for three days and then assessed by either western blotting or qRT-PCR to determine the efficiency of the knockdown and for use in the assays described.
Coimmunoprecipitations and Western blots
Coimmunoprecipitations were carried out essentially as described (20). 1.5 × 107 M-280 sheep anti–rabbit and anti–mouse antibody coupled magnetic beads (Invitrogen) were coupled overnight to 3 µg of the appropriate antibody for each immunoprecipitation. Antibody-bound beads were mixed with 200 µg of nuclear extract, which was prepared as previously described (20), and the volume was adjusted to 300 µl by the addition of lysis buffer (50 mM Tris [pH 8], 150 mM NaCl, and 1% NP-40) plus protease inhibitors. Immunoprecipitation reactions were continued overnight at 4°C with gentle rotation. Immune complex-bound beads were washed four times with the lysis buffer. Washed beads were re-suspended in SDS-PAGE sample buffer. Precipitates were eluted from the beads in a boiling water bath for 5 min, separated by 6 or 7.5% SDS-PAGE, and analyzed by Western blotting. Western blots were performed according to standard protocols with Immobilon-Ptransfer membranes (Millipore, Inc.) coupled with Enhanced Chemiluminescence detection kits (GE Healthcare). Anti-CTCF and β-actin were purchased from Millipore, Inc. and Chemicon, Inc., respectively.
In some reactions, ethidium bromide (50 µg/ml) was added to the nuclear extract and incubated on ice for 30 min as described (20). After incubation, the nuclear extract was cleared by centrifugation and used in the coimmunoprecipitation assay described above. For some immunoprecipitation reactions, samples were treated with DNase I as described previously (20). For these, the coimmunoprecipitation reaction was performed first, and then 30 U/ml DNase I (Roche Applied Science, Inc.) was added for a 5-min incubation at room temperature (20). The protein-bound beads were washed with lysis buffer and analyzed as above.
Chromatin immunoprecipitation (ChIP) assay
ChIP assays were performed as described previously (20, 49). Here, cells were crosslinked in 1% formaldehyde for 10 minutes and a chromatin lysate was prepared and sonicated to generate fragments averaging 500 bp in length. For immunoprecipitations, 5–10 µg of anti-CTCF (Upstate, Cat# 06-917), anti-cohesin (anti-Rad21, Upstate, Cat# 05-908), (anti-SMC3, Abcam, Cat# ab8263), anti-RFX5 (50), anti-CIITA (49), or anti-T-cell receptor (non-specific control) antibodies were used. The immunoprecipitated DNA was quantitated by real-time PCR using a 5-point genomic DNA standard curve and an I-cycler (Biorad Laboratories, Inc.). Quantitative PCR reactions contained 5% DMSO, 1XSYBR green (Bio Whittaker Molecular Applications), 0.04% gelatin, 0.3% Tween 20, 50 mM KCl, and 20 mM Tris [pH 8.3], 3 mM MgCl2, 0.2 mM dNTP, and 100 nM of each primer. Sequences for all primers used in the ChIP real-time PCR assays are listed in Supplemental Figure 1. All ChIP experiments were performed at least three times from independent preparations of chromatin. The data were averaged and plotted with respect to the input chromatin. Under the above conditions of crosslinking we do not detect RFX5 or CIITA at the MHC-II insulators, nor do we detect CTCF or cohesin at MHC-II proximal regulatory regions.
Quantitative 3C assay
A modified 3C assay protocol was employed as described previously (19, 20). In these 3C assays, 1×107 cells were used and crosslinked with formaldehyde to a final concentration of 1% and incubated for 10 minutes at room temperature. Prepared nuclei were digested with EcoRI. Following heat inactivation of the enzyme, the samples were diluted into ligation buffer at a ratio of ~40:1, and then ligated overnight with T4 DNA ligase at 16°C. The 3C DNA was purified and quantified by real-time PCR using a five point standard curve as described previously (19). Standard curve templates for the 3C products were generated in vitro by restriction enzyme cleaving and religating a BAC containing the region being studied. The BACsRP11-974L24, RP11-257P24, and RP11-54H13, which cover the MHC-II region from C1 through C2, were used to generate all 3C product templates and standard curves. All primer combinations listed in the Supplemental Figure 1 were tested previously (19) in the 3C assay to determine whether they could efficiently amplify a single product on EcoRI cleaved/religated BAC DNA. All primers had a greater than 90% PCR efficiency and produce a single amplicon on cleaved/religated BAC DNA. All sites used were previously shown to be equally cleavable by EcoRI in these assays (19). Data are presented as relative crosslinked frequency and represent an average derived from three independent biological replicates. Relative crosslink frequency was calculated as the qPCR value for the 3C sample × 100, divided by the qPCR value for that potential 3C junction represented in a pool of all potential 3C combinations generated using EcoRI cleaved and religated BAC DNA to the regions assayed.
Statistical Analyses
All quantitative experiments were reproduced at least three times from independent biological samples. The data are presented as an average with standard error of the mean. The Student’s t test was used to determine statistical significance between samples and the results of such tests are indicated in the figures or legends.
Results
C1 is required for maximal expression of the HLA-DRA gene
Ten strong CTCF binding regions were identified in the human MHC-II locus (19). C1 represents the MHC-II insulator that is closest to and interacts with the HLA-DRA gene and was chosen here as the model element to study the role of cohesin (Figure 1A). One critical feature of MHC-II insulators that was not previously determined was whether deletion of the insulator region would affect MHC-II gene expression. Although a targeted disruption of the gene in human cells is not practical, the manipulation of a human BAC construct, followed by transfection into murine cells can be used to demonstrate a role for a cis-acting element such as C1 (46). The RP11-974L24 BAC encodes C1 and the HLA-DRA gene (Figure 1A). BAC recombineering, which uses targeted homologous recombination vectors to alter the BAC DNA, was used to create a 520 bp deletion encompassing the C1 MHC-II insulator. Two independent recombineered clones, termed ΔC1a and ΔC1b were isolated with identical mutations. Restriction enzyme digestion, PCR, and Southern blot analyses (data not shown) demonstrated that the mutant BAC DNAs contained only the C1 deletion. Wild-type and ΔC1 mutant BAC DNAs were transfected into the murine B-cell line A20, and 48 h post transfection, RNA was isolated and analyzed. Real-time RT-PCR for HLA-DRA gene expression showed that it was expressed from the wild-type BAC. In contrast, the two mutant ΔC1 BACs showed about 50% reduction in the level of HLA-DRA mRNA (Figure 1B). These results suggest that as a cis-element, C1 is required for maximal expression of HLA-DRA.
Cohesin subunits associate with C1 irrespective of MHC-II gene expression
Using ChIP with Raji cells, the cohesin subunits Rad21 and Smc3 were found to be associated with each of the ten MHC-II insulators, including C1 (19). To determine if the binding of cohesin subunits was correlated with MHC-II gene expression, the binding of Rad21 and Smc3 to C1 in MHC-II positive and negative cells was examined by ChIP. Chromatin was prepared and assayed from several MHC-II negative cell lines: RJ2.2.5 (CIITA-deficient, MHC-II−, B cell line (43, 44)), SJO (RFX5-deficient, MHC-II−, B cell line (45)), A431 (MHC-II− epithelial cell line), H929 (MHC-II− plasma cell line (CRL-9068; ATCC), CCRF (MHC-II− T cell leukemic cell line (51)), and Kasumi-1 (MHC-II− acute myeloid leukemic cell line (52)). In addition to Raji cells, which are constitutive for MHC class II, chromatin was also isolated and examined from A431 epithelial cells treated with IFN-γ to induce CIITA and MHC-II gene expression (49). Although there is some variability in the strength of the signal observed, the results showed that C1 significantly bound CTCF, Rad21, and Smc3 in all cell types/lines examined (Figure 1C). The levels of binding were comparable to the other MHC-II insulators as previously reported for Raji cells (19). Serving as a CTCF-negative site for these ChIP assays, C15 exhibited 6-18-fold lower levels of binding of CTCF and the cohesin subunits depending on the cell type. Student’s t-test analyses showed that binding of CTCF, SMC3, or Rad21 was not significantly different in A431 cells treated with or without IFN-γ (Figure 1C). Thus, irrespective of MHC-II gene expression, CTCF and cohesin are associated with the C1 CTCF site in multiple cell types.
Rad21/Cohesin is required for full expression of the MHC-II genes
The binding of cohesin to the MHC-II insulator sites suggests that cohesin may also have an effect on MHC-II gene expression. To test this hypothesis, Rad21 and SMC3-specific SMART pool siRNA oligonucleotides (Dharmacon, Inc.) were used to deplete transiently Rad21 or SMC3 from Raji cells. This treatment did not affect the overall growth or viability of these cells during the 48 hours of the assay (data not shown). Immunoblotting showed that each of the factors was reduced to about 20–30% of the untreated or irrelevant siRNA treated cells (Figure 2A). Importantly, the levels of Smc3, another component of cohesin, and β-actin were unchanged during the course of the assay (Figure 2A). RNA purified from the above cell groups was analyzed by real-time RT-PCR. The results showed that CIITA and RFX5 mRNA levels were unchanged irrespective of treatment (Figure 2B). As expected, the steady-state levels of Rad21 and SMC3 mRNA were substantially reduced by their respective and specific siRNA treatments. Following either Rad21 or SMC3 siRNA depletion, the levels of HLA-DRA mRNA were reduced to ~40–50% of the control treated cells (Figure 2B). These results demonstrated that at least two components of the cohesin complex were required for maximal HLA-DRA gene expression.
Figure 2. Knockdown of cohesin specifically reduces the expression of HLA-DR and HLA-DQ genes.
Protein lysates and RNAs were prepared following siRNA knockdown in Raji cells using Dharmacon, Inc., SMART pool siRNA oligonucleotides against Rad21, SMC3, or an siRNA to GFP. A) Western blots using anti-Rad21, -actin, or -SMC3 antibodies demonstrate the efficiency of siRNA knockdown. B) Following siRNA depletion of Rad21, SMC3, or GFP, HLA-DRA, Rad21, RFX5, and CIITA mRNA levels were determined by real-time RT-PCR.C) Expression of HLA-DRB1, HLA-DQA1, and HLA-DQB1 were also measured following siRNA depletion as above. These data were compiled from three biological replicates and are presented as relative mRNA expression with respect to the no treatment lane (nt). Standard error is presented along with student’s t test comparisons between siRNAs against Rad21 and GFP. An asterisk (*) indicates p values <0.05 between samples treated with siRNAs against GFP and Rad21.
To define more broadly whether cohesin played a role in other MHC-II genes, the expression of HLA-DRB1, HLA-DQA1, and HLA-DQB1 was examined (Figure 2C). These genes represent the major human MHC-II isotypes. Like HLA-DRA, the expression of these genes was also reduced significantly when either Rad21 or SMC3 were depleted from the system. These data suggest that cohesin is critical for maximal expression of MHC-II genes.
Cohesin is important for the function of C1
One experimental property initially observed for the MHC-II insulator XL9 was its ability to block the activity of an upstream enhancer from acting on a downstream promoter. Such a property may exist for C1 and may be dependent on cohesin for this function. To assess this characteristic, the function of C1 as an element was defined. The 520bp region encompassing C1 was cloned 5’ to a minimal promoter luciferase reporter gene and assayed for enhancer activity following transient transfection into Raji cells. Similar to the chicken β-globin HS4 CTCF insulator (17, 20) and a control DNA from lambda phage, C1 contains no enhancer activity (Figure 3A). This result was also similar to that observed for the MHC-II insulator XL9 (47).
Figure 3. The C1 MHC-II insulator encodes enhancer-blocking activity that is dependent on cohesin.
Schematic diagrams of luciferase reporter gene vectors containing the chicken β globin HS4 insulator element (17), C1 element (520 bp), lambda DNA (47), XL9 (747 bp), and the SV40 transcriptional enhancer inserted upstream of the minimal promoter of luciferase reporter gene in pGL3-promoterare shown next to each set of assays. The constructs were transiently transfected along with pRLTK plasmid containing the Renilla luciferase gene as a normalizing control vector into Raji cells. At 48 h posttransfection, cells were harvested and luciferase activity was determined and normalized to the Renilla luciferase expression. A) This series examines the effect of the inserted sequence for enhancer activity. B) This series examines the ability of the inserted sequence to function as an enhancer-blocking element. C) The ability of C1 to block the activity of the SV40 enhancer was tested in a second series of constructs that used the XL9 MHC-II insulator at the 5’ end instead of the 5’HSR insulator. D) This series of experiments assessed the effect of depletion of Rad21 on enhancer blocking activity. SiRNA SMART pool oligonucleotides against Rad21 or siRNA against GFP (control) were co-transfected into Raji cells with the indicated reporter construct. In all graphs, the average of three independent experiments was plotted with their standard error of the mean.
To determine if C1 encoded an enhancer blocking activity, C1 was placed between an SV40 enhancer and a promoter, transfected into cells, and the ability of the enhancer to drive the luciferase reporter gene was assayed. An additional HS4 insulator was placed on the 5’ side of the enhancer to prevent the enhancer from operating from the other side of the circular plasmid DNA. C1 was as effective as the HS4 insulator at blocking the activity of SV40 enhancer in driving transcription of the reporter gene (Figure 3B). The control lambda DNA exhibited no enhancer blocking activity. To ensure that the HS4 insulator provided no additional activities, it was replaced with XL9 in the enhancer blocking constructs. XL9 is an MHC-II insulator that is normally located between the HLA-DRB1 and HLA-DQA1 genes (47, 48). Following transfection and assessment of the activities of these reporters, similar data were collected indicating that the HS4 insulator was not providing additional enhancer blocking activities to the construct (Figure 3C). Thus, C1 has no intrinsic enhancer activity but can block the activity of an enhancer.
To determine if C1’s enhancer blocking activity was dependent on cohesin, the above enhancer-blocking assay was performed in Rad21-depleted cells. The enhancer-blocking constructs were co-transfected with the Rad21 SMART pool siRNA oligonucleotides into Raji cells, and the luciferase activity of the reporter gene was measured at 48h post transfection (Figure 3D). The HS4 chicken β-globin insulator (17) was also included to determine whether cohesin was also required for its activity in B cells. Constructs containing C1 or HS4 elements showed increased levels of luciferase expression in cells in which cohesin was depleted compared to control siRNA treated cells. Rad21 depletion had no statistically significant effect on the SV40 enhancer mediated expression in the control construct, which did not contain an insulator. Thus, Rad21 is required for the enhancer blocking activity of the C1 and HS4 insulators.
Cohesin associates in a complex with CIITA and RFX5
The above experiments suggest a role for cohesin and C1 in regulating HLA-DRA gene expression but do not provide a mechanism by which this may occur. Because cohesin proteins are not found at the MHC-II promoters, but along with CTCF are found at C1 and the other MHC-II insulators, the ability of cohesin subunits to interact with MHC-II proximal promoter region transcription factors (RFX and/or CIITA) was hypothesized as a way in which these regions may interact. To determine the nature of interactions that may occur, coimmunoprecipitation and immunoblotting experiments were conducted in Raji cells, which are wild-type for all MHC-II transcription factors. Two other cell lines, which contain mutations in CIITA (RJ2.2.5, (43, 44)) and RFX5 (SJO (45)), were used to dissect the interactions. As previously shown (20, 53), CIITA and RFX5 interact in the wild-type cells as indicated by the coimmunoprecipitation (Figure 4A). Antibodies to Rad21 and Smc3 immunoprecipitated RFX5 in Raji cells (Figure 4A) but not in cells lacking CIITA (RJ2.2.5) or the control RFX-deficient (SJO) cells. Reciprocally, RFX5 antibodies precipitated Rad21 and SMC3, but only in Raji cells. Antibodies to CIITA immunoprecipitated Rad21 and Smc3 in Raji but not in RJ2.2.5 or SJO cell lysates. These data suggest that both RFX5 and CIITA form either direct or indirect interactions with cohesin subunits. Such interactions could be through a soluble complex or a complex assembled on DNA.
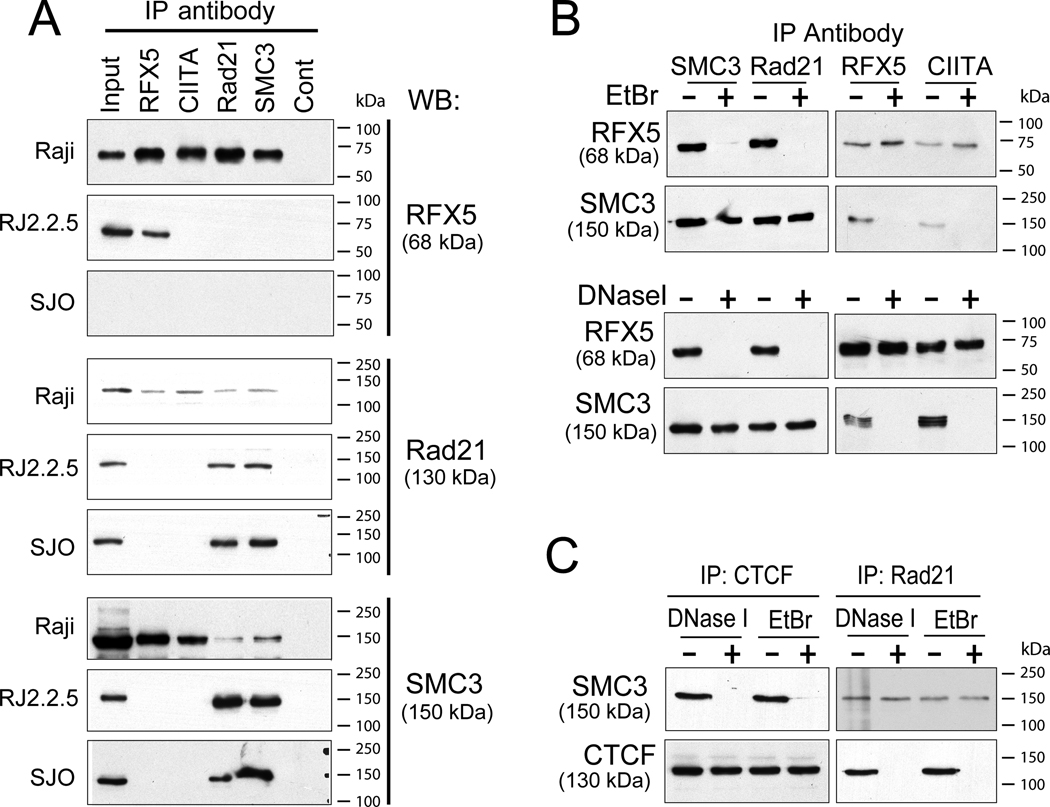
Figure 4. Cohesin is associated in a complex containing CTCF and CIITA.
A) Lysates were prepared from Raji (wild type), RJ2.2.5 (CIITA−, RFX5+), and SJO (CIITA+, RFX5−) cells and immunoprecipitated with the indicated antibodies. The immunoprecipitates were analyzed by western blotting for the presence of RFX5, Rad21, and SMC3 as indicated. No interactions with the nonspecific antiserumto the T cell receptor (Cont) were observed. Input represents 10% of the sample in the immunoprecipitation reaction. B) Coimmunoprecipitation experiments using antisera to Rad21, SMC3, RFX5, and CIITA in the presence of ethidium bromide or treated with DNase I were performed with Raji cell lysates as described in the methods. The precipitations were analyzed by immunoblotting for RFX5 and SMC. C) Immunoprecipitations as performed in B were carried out with antisera to CTCF and Rad21 as indicated and assessed by immunoblotting with antisera to SMC3 and CTCF.
To determine whether the coimmunoprecipitations were dependent on the presence of DNA, DNaseI or ethidium bromide, which prevents protein/DNA interactions (54), were added to the immunoprecipitation reactions. In the presence of ethidium bromide or DNaseI, co-immunoprecipitates between SMC3 and RFX5 or CIITA were no longer detected (Figure 4B). Similarly, both ethidium bromide and DNaseI disrupted Rad21 coimmunoprecipitations with RFX5. Importantly, the interactions between CIITA and RFX5 were not affected, nor were the interactions between SMC3 and Rad21.
Cohesin interactions with CTCF require DNA
The above experiments question whether cohesin and CTCF interactions may also require DNA. Thus, Raji nuclear lysates were generated and coimmunoprecipitations in the presence and absence of either DNaseI or ethidium bromide were performed using anti-CTCF or -Rad21 antibodies (Figure 4C). Whereas coimmunoprecipitation interactions between Smc3 and Rad21 were not altered by the presence of DNaseI or ethidium bromide, these compounds disrupted the ability of Smc3 or Rad21 to interact with CTCF and form coimmunoprecipitates. These data argue that a DNA component is required for cohesin to interact with CTCF, RFX5, and/or CIITA. From our previous work, the association of CTCF with RFX5 or CIITA was not dependent on the presence of DNA (20).
CTCF is required to recruit cohesin to C1
CTCF and cohesin are colocalized at the C1 MHC-II insulator. However, we do not know if one of these factors is required for the other to associate with the MHC-II insulator. To examine the dependency of one factor for the other in associating with the C1 MHC-II insulator, a set of ChIP assays were performed following RNAi depletion of CTCF, SMC3, and Rad21. Depletion of CTCF resulted in the loss of CTCF binding (>80%) at C1 and a >50–60 % reduction in the association of SMC3 and Rad21 (Figure 5). In sharp contrast, depletion of SMC3 and Rad21 did not alter the binding of CTCF to C1 (Figure 5 top left panel). SMC3 and Rad21 depletion by RNAi did however result in a reduction in their own association, as well as each other from C1. These observations suggest that CTCF is required to recruit cohesin to C1, and that CTCF binding/stability is independent of cohesin. The results also suggest that an intact cohesin complex is important for recruitment or stability of cohesin interactions at these sites.
Figure 5. Recruitment of cohesin to C1 is CTCF dependent.
Cross-linked chromatin was prepared from Raji cells following transfection with SMART pool siRNAs against CTCF, SMC3, Rad21 and GFP (nt) as indicated. In the case against siRNA to CTCF, negative HLA-DR cells were isolated as described earlier (19). ChIP assays were performed with antibodies as indicated. Cont represents an IgG control ChIP. The average of three independent ChIP assays are shown with the data plotted with respect to the % of input chromatin in each reaction. Asterisks indicate samples that displayed p values <0.05 as determined by Student’s t tests.
Cohesin is required to form chromatin loops between C1 and the HLA-DRA promoter region
Previously, we reported that CTCF is required for the chromatin interactions between MHC-II insulators and the proximal promoters of MHC-II genes (19). The above data suggest that cohesin could play a role in maintaining the chromatin architecture of the MHC-II locus or in the ability of the MHC-II insulators to interact with MHC-II promoters. To determine whether cohesin is required for similar chromatin interactions, we investigated whether interactions with C1 and XL9 were dependent on the cohesin subunit Rad21. Here, chromatin conformation capture (3C) assays (19, 55) were employed to assess the role that cohesin plays in the shaping of the architecture of the MHC-II region encompassing HLA-DRA. 3C assays utilize formaldehyde crosslinking to fix interactions between regulatory elements. The crosslinked chromatin is subjected to extensive restriction digestion and dilution, and is followed by religation of restriction fragments. Only restriction fragments that are in close spatial proximity to each other (due to the crosslinking) display a high frequency of ligation, which is measured by real-time PCR across the 3C generated novel ligation junction. As shown above (Figure 3 and 5), Rad21 depletion results in a ~70% reduction in Rad21 protein and occupancy at C1 in Raji cells. Using control and Rad21 siRNA-depleted Raji cells, 3C assays were conducted to measure the relative frequency of interactions between C1 and the HLA-DRA proximal regulatory region and several control restriction fragments (Figure 6). Background levels of interactions were detected between C1 and the control restriction fragments (CL1, CL2, and P12). Such background interactions are common with this assay (19, 55, 56). In control cells, the relative crosslink frequency between C1 and the HLA-DRA proximal promoter were robust (Figure 6) and recapitulate previous observations (19). Intriguingly, depletion of Rad21 from the system resulted in a significant reduction in C1/HLA-DRA interactions, suggesting that Rad21 was important for this interaction.
Figure 6. Cohesin controls the chromatin interactions between C1 and the proximal regulatory region of HLA-DRA.
A) A schematic diagram of MHC-II insulators C1, XL9, and C2, the HLA-DRA, HLA-DRB1, HLA-DQB1 genes, the relative position of the EcoR1 sites, and the positions of primer sets used to determine the relative frequency of chromatin interactions in the locus by 3C assay is shown. // indicates gaps in the map that are not shown to reduce the complexity of the illustration and its size. Relative primer positions for C1 and XL9 are colored gray and represent 3C anchor sites. The other primer positions are colored as black. Primer sets are the same as described earlier (19). B) Quantitative 3C assays were performed in untreated Raji cells (cont) or Raji cells depleted for Rad21 by siRNA as described above. The 3C results determining the relative interaction frequencies between the indicated anchor sites and the indicated restriction fragment are plotted as the relative crosslink frequency with standard error. Relative crosslink frequency represents the relative average amount of 3C product (from three independent chromatin preparations) determined from qPCR of the samples compared to a standard curve generated from BAC DNA that was fully digested with EcoRI and religated, times one hundred. Cutting and religating the BAC DNA in this regard provides all possible combinations of EcoRI fragments. The asterisk represents the only Rad21-siRNA treated cell sample that was significantly different from its control as determined by the Student’s t test (p<0.05).
MHC-II insulators can also interact with each other (19, 20) in a manner that is independent of MHC-II gene expression. To determine if cohesin was important for these interactions, 3C was performed using C1, XL9, and C2, the three MHC-II insulators that divide the HLA-DR and DQ subregions (Figure 6). In these 3C reactions, C1 and XL9, and XL9 and C2 interactions were examined along with control fragments CL1, P12, CL4, and CL5. As previously observed (19), XL9 interacted with either C1 or C2 in control cells (Figure 6). Surprisingly, Rad21 siRNA treated cells showed no change in the ability of XL9 to interact with either C1 or C2 (Figure 6). Thus, cohesin depletion by RNAi does not affect the ability of MHC-II insulators to interact with each other.
Discussion
The MHC-II locus is organized architecturally through interactions across the series of CTCF binding sites that we have termed MHC-II insulators. Critically, CTCF was found to be required for the full expression of MHC-II genes, identifying a novel mechanism of MHC-II gene expression (19). In 2008, genome-wide binding studies conducted by several groups found that subunits of cohesin were associated with many but not all CTCF binding sites (38, 39, 57). Cohesin was also found at each of the 10 MHC-II insulators, suggesting that cohesin may play a role in the expression of MHC-II genes (19). Here we chose to determine the role of cohesin on MHC-II gene expression and for the most part focused on one of the MHC-II insulators, C1. Cohesin binding to C1 varied slightly among the different cell lines/types but was always present irrespective of MHC-II gene expression, suggesting that it was part of the base architecture. Importantly, knockdown of cohesin subunits Rad21 or Smc3 showed that the HLA-DR and HLA-DQ genes were both dependent on cohesin for maximal expression. Thus, cohesin acting at MHC-II insulators is a novel and important component of MHC-II transcription regulation. To determine how it functioned, we examined some of the molecular features associated with insulator function. Like XL9 (47), C1 has no intrinsic enhancer activity but does encode an enhancer-blocking activity. This activity was dependent in part on the presence of the cohesin subunit Rad21. A similar observation was made for the chicken β globin insulator (Figure 3 and (39)), suggesting that cohesin may be required for this function at all enhancer-blocking insulators. The ability of MHC-II insulators to block enhancer activity in the artificial system suggests that these elements can do this in vivo and may serve to restrict the MHC-II proximal regulatory elements to single genes, or at least to genes between the MHC-II insulators. With MHC-II insulators surrounding each of the sub-regions (12, 19), including separating the HLA-DM genes from TAP1, PSMB9, and BRD2, this function may indeed be important.
To develop insight into how cohesin functions in this system, coimmunoprecipitation experiments between cohesin and the MHC-II specific transcription factors RFX5 and CIITA were conducted and found that both Rad21 and Smc3 associated with these factors in extracts prepared from MHC-II expressing B cells. The interactions were specific because cells lacking functional RFX5 or CIITA showed no association. The interactions with RFX5 were either not sufficient or direct as no interaction was observed in RJ2.2.5 (CIITA-) cells. This would suggest that the interactions with RFX5 occurred through CIITA when it was bound to the MHC-II promoter regions. Intriguingly, the observed interaction was dependent on the presence of DNA in the extract as treatment with DNaseI or the intercalating agent ethidium bromide, which disrupts protein-DNA interactions, resulted in the loss of the coprecipitates. These data suggest that cohesin is interacting with an MHC-II specific multiprotein/DNA complex or that a specific DNA-dependent protein conformation is necessary. It is assumed that such interactions with DNA are occurring through specific interactions with WXY box DNA that is associated with the proteins in the lysates. However, it is possible that any DNA substrate would serve this purpose. As a point of reference, CTCF-CIITA interactions were not perturbed by the addition of DNaseI or ethidium bromide (20). A surprise was that the CTCF-cohesin interactions were also dependent on the presence of DNA. This observation was not previously reported and supports a novel mechanism of cohesin association with its target substrates. One possible substrate may be looped DNA or a pair of chromatinized DNA helices that are in close proximity, which would occur due to insulator interactions or the pairing of sister chromatids, respectively. Coupled with these findings were experiments to determine the requirements for cohesin binding to MHC-II insulators. Using RNAi to deplete CTCF, Rad21, and Smc3, coupled with ChIP, we were able to determine that CTCF binding was independent of cohesin. This was not the case for either Rad21 or Smc3, which showed reduced binding when CTCF was depleted. Additionally, Rad21 and Smc3 binding was dependent on each other, suggesting that these proteins are binding in a complex with each other.
Because cohesin was important for MHC-II gene expression and interacted at the MHC-II insulators, we initially postulated that siRNA depletion of Rad21 would disrupt the basal architecture associated with the MHC-II insulators. However, the 3C experiments indicated that Rad21 depletion had no affect on the ability of the C1, XL9, and C2 insulators to interact with each other. These insulators separate the HLA-DR and HLA-DQ subregions from each other. The data therefore suggest that the interactions between the MHC-II insulators, which are mediated by CTCF, are potentially sufficient for these interactions to occur. The fact that CTCF can form homodimers would suggest that CTCF dimer formation might be what holds MHC-II insulators together. In contrast, depletion of cohesin subunits disrupted the 3C interactions between the HLA-DRA promoter proximal regulatory and the C1 MHC-II insulator. Thus, the reason for loss of MHC-II gene expression upon cohesin depletion was due to the loss of a chromatin loop between the MHC-II insulator and the promoter. This would suggest that the latter interactions are not stable, allowing for finer gene control; which in this case would be mediated by the level or functional activity of cohesin in the cell. The current data are in agreement with a model predicting that MHC-II insulators serve as focal points for the formation of a functional and efficient transcription complex at MHC-II gene promoters. We have previously shown that the loss of CTCF resulted in a decrease in the presence of transcriptionally active histone modifications at MHC-II gene promoters, suggesting that these interactions are either required for or are a consequence of CTCF-mediated activities and loop formation (20).
While cohesin function in mitosis and DNA repair mechanisms is well established, it is clear that it is involved in many other aspects of genome structure. Kagey et al. recently found cohesin bound to 43,687 sites in human ES cells, but only a fraction of these sites (24,741) overlapped with CTCF occupancy (41). The rest were located mostly at enhancers and core promoter regions of active genes and were associated with the binding of mediator complex subunits. Mediator is a transcriptional coactivator complex that associates with the C-terminal domain repeats of RNA polymerase II (58, 59). 3C experiments showed that these cohesin-bound regions interacted, bridging enhancer regions and core promoters (41). However, one difference observed between the CTCF/cohesin sites and the mediator sites was that the cohesin-loading factor Nipb1 was mostly present at the mediator sites and not at the CTCF sites. Preliminary ChIP experiments did not find the mediator subunit Med12 at MHC-II insulators or core promoters (data not shown). Thus, MHC-II insulator interactions are not similar to cohesin binding regions that use mediator for their function.
In summary, we present a novel role for cohesin in regulating the expression of MHC-II genes by stabilizing interactions between distant CTCF-bound MHC-II insulators and MHC-II promoter proximal regulatory sequences when the genes are actively expressed. It is likely that cohesin carries out this function by encircling the DNAs associated with the loop that is formed between these regions.
Supplementary Material
Acknowledgements
We thank Joshua Lee for generating the BAC deletions and expert technical assistance. We thank Drs. Paul Wade and John Lucchesi and members of the laboratory for helpful discussions and advice with respect to this work.
Footnotes
Disclosures
The authors have no financial conflicts of interests.
This work was supported by a grant from the NIH 5RO1GM47310.
Abbreviations used in this work: 3C, chromatin conformation capture; BAC, bacterial artificial chromosome; ChIP, chromatin immunoprecipitation; CTCF, CCCTC transcription factor; RFX, regulatory factor X; siRNA, silencing RNA.
Literature Cited
- 1.Complete sequence and gene map of a human major histocompatibility complex. The MHC sequencing consortium. Nature. 1999;401:921–923. doi: 10.1038/44853. [DOI] [PubMed] [Google Scholar]
- 2.Sadegh-Nasseri S, Natarajan S, Chou CL, Hartman IZ, Narayan K, Kim A. Conformational heterogeneity of MHC class II induced upon binding to different peptides is a key regulator in antigen presentation and epitope selection. Immunologic research. 2010;47:56–64. doi: 10.1007/s12026-009-8138-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nielsen M, Lund O, Buus S, Lundegaard C. MHC class II epitope predictive algorithms. Immunology. 2010;130:319–328. doi: 10.1111/j.1365-2567.2010.03268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nedjic J, Aichinger M, Mizushima N, Klein L. Macroautophagy, endogenous MHC II loading and T cell selection: the benefits of breaking the rules. Current opinion in immunology. 2009;21:92–97. doi: 10.1016/j.coi.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 5.Rocha N, Neefjes J. MHC class II molecules on the move for successful antigen presentation. The EMBO journal. 2008;27:1–5. doi: 10.1038/sj.emboj.7601945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ting JP, Trowsdale J. Genetic control of MHC class II expression. Cell. 2002;109 Suppl:S21–S33. doi: 10.1016/s0092-8674(02)00696-7. [DOI] [PubMed] [Google Scholar]
- 7.Boss JM. Regulation of transcription of MHC class II genes. Current opinion in immunology. 1997;9:107–113. doi: 10.1016/s0952-7915(97)80166-5. [DOI] [PubMed] [Google Scholar]
- 8.Germain RN, Margulies DH. The biochemistry and cell biology of antigen processing and presentation. Annual review of immunology. 1993;11:403–450. doi: 10.1146/annurev.iy.11.040193.002155. [DOI] [PubMed] [Google Scholar]
- 9.Collins T, Korman AJ, Wake CT, Boss JM, Kappes DJ, Fiers W, Ault KA, Gimbrone MA, Jr, Strominger JL, Pober JS. Immune interferon activates multiple class II major histocompatibility complex genes and the associated invariant chain gene in human endothelial cells and dermal fibroblasts. Proceedings of the National Academy of Sciences of the United States of America. 1984;81:4917–4921. doi: 10.1073/pnas.81.15.4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reith W, Mach B. The bare lymphocyte syndrome and the regulation of MHC expression. Annu Rev Immunol. 2001;19:331–373. doi: 10.1146/annurev.immunol.19.1.331. [DOI] [PubMed] [Google Scholar]
- 11.Boss JM, Jensen PE. Transcriptional regulation of the MHC class II antigen presentation pathway. Current opinion in immunology. 2003;15:105–111. doi: 10.1016/s0952-7915(02)00015-8. [DOI] [PubMed] [Google Scholar]
- 12.Choi NM, Majumder P, Boss JM. Regulation of major histocompatibility complex class II genes. Current opinion in immunology. 2010 doi: 10.1016/j.coi.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steimle V, Siegrist CA, Mottet A, Lisowska-Grospierre B, Mach B. Regulation of MHC class II expression by interferon-gamma mediated by the transactivator gene CIITA. Science. 1994;265:106–109. doi: 10.1126/science.8016643. [DOI] [PubMed] [Google Scholar]
- 14.West AG, Huang S, Gaszner M, Litt MD, Felsenfeld G. Recruitment of histone modifications by USF proteins at a vertebrate barrier element. Mol Cell. 2004;16:453–463. doi: 10.1016/j.molcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 15.Labrador M, Corces VG. Setting the boundaries of chromatin domains and nuclear organization. Cell. 2002;111:151–154. doi: 10.1016/s0092-8674(02)01004-8. [DOI] [PubMed] [Google Scholar]
- 16.Bushey AM, Dorman ER, Corces VG. Chromatin insulators: regulatory mechanisms and epigenetic inheritance. Mol Cell. 2008;32:1–9. doi: 10.1016/j.molcel.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farrell CM, West AG, Felsenfeld G. Conserved CTCF insulator elements flank the mouse and human beta-globin loci. Molecular and cellular biology. 2002;22:3820–3831. doi: 10.1128/MCB.22.11.3820-3831.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Magdinier F, Yusufzai TM, Felsenfeld G. Both CTCF-dependent and -independent insulators are found between the mouse T cell receptor alpha and Dad1 genes. J Biol Chem. 2004;279:25381–25389. doi: 10.1074/jbc.M403121200. [DOI] [PubMed] [Google Scholar]
- 19.Majumder P, Boss JM. CTCF controls the expression and the chromatin architecture of the human major histocompatibility complex class II locus. Molecular and cellular biology. 2010;30 doi: 10.1128/MCB.00327-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Majumder P, Gomez JA, Chadwick BP, Boss JM. The insulator factor CTCF controls MHC class II gene expression and is required for the formation of long-distance chromatin interactions. The Journal of Experimental Medicine. 2008;205:785–798. doi: 10.1084/jem.20071843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peters JM, Tedeschi A, Schmitz J. The cohesin complex and its roles in chromosome biology. Genes & development. 2008;22:3089–3114. doi: 10.1101/gad.1724308. [DOI] [PubMed] [Google Scholar]
- 22.Marston AL, Amon A. Meiosis: cell-cycle controls shuffle and deal. Nat Rev Mol Cell Biol. 2004;5:983–997. doi: 10.1038/nrm1526. [DOI] [PubMed] [Google Scholar]
- 23.Petronczki M, Siomos MF, Nasmyth K. Un menage a quatre: the molecular biology of chromosome segregation in meiosis. Cell. 2003;112:423–440. doi: 10.1016/s0092-8674(03)00083-7. [DOI] [PubMed] [Google Scholar]
- 24.Watanabe Y. Modifying sister chromatid cohesion for meiosis. J Cell Sci. 2004;117:4017–4023. doi: 10.1242/jcs.01352. [DOI] [PubMed] [Google Scholar]
- 25.Strom L, Lindroos HB, Shirahige K, Sjogren C. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol Cell. 2004;16:1003–1015. doi: 10.1016/j.molcel.2004.11.026. [DOI] [PubMed] [Google Scholar]
- 26.Sjogren C, Nasmyth K. Sister chromatid cohesion is required for postreplicative double-strand break repair in Saccharomyces cerevisiae. Curr Biol. 2001;11:991–995. doi: 10.1016/s0960-9822(01)00271-8. [DOI] [PubMed] [Google Scholar]
- 27.Sonoda E, Matsusaka T, Morrison C, Vagnarelli P, Hoshi O, Ushiki T, Nojima K, Fukagawa T, Waizenegger IC, Peters JM, Earnshaw WC, Takeda S. Scc1/Rad21/Mcd1 is required for sister chromatid cohesion and kinetochore function in vertebrate cells. Dev Cell. 2001;1:759–770. doi: 10.1016/s1534-5807(01)00088-0. [DOI] [PubMed] [Google Scholar]
- 28.Kim JS, Krasieva TB, LaMorte V, Taylor AM, Yokomori K. Specific recruitment of human cohesin to laser-induced DNA damage. J Biol Chem. 2002;277:45149–45153. doi: 10.1074/jbc.M209123200. [DOI] [PubMed] [Google Scholar]
- 29.Unal E, Arbel-Eden A, Sattler U, Shroff R, Lichten M, Haber JE, Koshland D. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol Cell. 2004;16:991–1002. doi: 10.1016/j.molcel.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 30.Merkenschlager M. Cohesin: a global player in chromosome biology with local ties to gene regulation. Current opinion in genetics & development. 2010;20:555–561. doi: 10.1016/j.gde.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 31.Mannini L, Menga S, Musio A. The expanding universe of cohesin functions: a new genome stability caretaker involved in human disease and cancer. Human mutation. 2010;31:623–630. doi: 10.1002/humu.21252. [DOI] [PubMed] [Google Scholar]
- 32.Feeney KM, Wasson CW, Parish JL. Cohesin: a regulator of genome integrity and gene expression. The Biochemical journal. 2010;428:147–161. doi: 10.1042/BJ20100151. [DOI] [PubMed] [Google Scholar]
- 33.Wood AJ, Severson AF, Meyer BJ. Condensin and cohesin complexity: the expanding repertoire of functions. Nature reviews. 2010;11:391–404. doi: 10.1038/nrg2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong RW. An update on cohesin function as a 'molecular glue' on chromosomes and spindles. Cell cycle (Georgetown, Tex. 2010;9:1754–1758. doi: 10.4161/cc.9.9.11806. [DOI] [PubMed] [Google Scholar]
- 35.Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 37.Stedman W, Kang H, Lin S, Kissil JL, Bartolomei MS, Lieberman PM. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. The EMBO journal. 2008;27:654–666. doi: 10.1038/emboj.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, Yahata K, Imamoto F, Aburatani H, Nakao M, Imamoto N, Maeshima K, Shirahige K, Peters JM. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 39.Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, Cobb BS, Yokomori K, Dillon N, Aragon L, Fisher AG, Merkenschlager M. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 40.Mishiro T, Ishihara K, Hino S, Tsutsumi S, Aburatani H, Shirahige K, Kinoshita Y, Nakao M. Architectural roles of multiple chromatin insulators at the human apolipoprotein gene cluster. The EMBO journal. 2009;28:1234–1245. doi: 10.1038/emboj.2009.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, Taatjes DJ, Dekker J, Young RA. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Epstein MA, Achong BG, Barr YM, Zajac B, Henle G, Henle W. Morphological and virological investigations on cultured Burkitt tumor lymphoblasts (strain Raji) J Natl Cancer Inst. 1966;37:547–559. [PubMed] [Google Scholar]
- 43.Accolla RS. Human B cell variants immunoselected against a single Ia antigen subset have lost expression of several Ia antigen subsets. The Journal of experimental medicine. 1983;157:1053–1058. doi: 10.1084/jem.157.3.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steimle V, Otten LA, Zufferey M, Mach B. Complementation cloning of an MHC class II transactivator mutated in hereditary MHC class II deficiency (or bare lymphocyte syndrome) Cell. 1993;75:135–146. [PubMed] [Google Scholar]
- 45.Steimle V, Durand B, Barras E, Zufferey M, Hadam MR, Mach B, Reith W. A novel DNA-binding regulatory factor is mutated in primary MHC class II deficiency (bare lymphocyte syndrome) Genes & development. 1995;9:1021–1032. doi: 10.1101/gad.9.9.1021. [DOI] [PubMed] [Google Scholar]
- 46.Yoon H, Boss JM. PU.1 binds to a distal regulatory element that is necessary for B cell-specific expression of CIITA. J Immunol. 2010;184:5018–5028. doi: 10.4049/jimmunol.1000079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Majumder P, Gomez JA, Boss JM. The human major histocompatibility complex class II HLA-DRB1 and HLA-DQA1 genes are separated by a CTCF-binding enhancer-blocking element. J Biol Chem. 2006;281:18435–18443. doi: 10.1074/jbc.M601298200. [DOI] [PubMed] [Google Scholar]
- 48.Gomez JA, Majumder P, Nagarajan UM, Boss JM. X box-like sequences in the MHC class II region maintain regulatory function. J Immunol. 2005;175:1030–1040. doi: 10.4049/jimmunol.175.2.1030. [DOI] [PubMed] [Google Scholar]
- 49.Beresford GW, Boss JM. CIITA coordinates multiple histone acetylation modifications at the HLA-DRA promoter. Nat Immunol. 2001;2:652–657. doi: 10.1038/89810. [DOI] [PubMed] [Google Scholar]
- 50.Moreno CS, Rogers EM, Brown JA, Boss JM. Regulatory factor X, a bare lymphocyte syndrome transcription factor, is a multimeric phosphoprotein complex. J Immunol. 1997;158:5841–5848. [PubMed] [Google Scholar]
- 51.Drexler HG, Gaedicke G, Minowada J. T-leukemia cell lines CCRF-CEM, HPB-ALL, JM and MOLT-4: changes in isoenzyme profiles during induction of differentiation. Blut. 1987;54:79–87. doi: 10.1007/BF00321034. [DOI] [PubMed] [Google Scholar]
- 52.Larizza L, Magnani I, Beghini A. The Kasumi-1 cell line: a t(8;21)-kit mutant model for acute myeloid leukemia. Leukemia & lymphoma. 2005;46:247–255. doi: 10.1080/10428190400007565. [DOI] [PubMed] [Google Scholar]
- 53.DeSandro AM, Nagarajan UM, Boss JM. Associations and interactions between bare lymphocyte syndrome factors. Molecular and cellular biology. 2000;20:6587–6599. doi: 10.1128/mcb.20.17.6587-6599.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lai JS, Herr W. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:6958–6962. doi: 10.1073/pnas.89.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dekker J. The three 'C' s of chromosome conformation capture: controls, controls, controls. Nat Methods. 2006;3:17–21. doi: 10.1038/nmeth823. [DOI] [PubMed] [Google Scholar]
- 56.Oestreich KJ, Cobb RM, Pierce S, Chen J, Ferrier P, Oltz EM. Regulation of TCR beta gene assembly by a promoter/enhancer holocomplex. Immunity. 2006;24:381–391. doi: 10.1016/j.immuni.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 57.Rubio ED, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, Baliga NS, Aebersold R, Ranish JA, Krumm A. CTCF physically links cohesin to chromatin. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:8309–8314. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim YJ, Bjorklund S, Li Y, Sayre MH, Kornberg RD. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 59.Thompson CM, Koleske AJ, Chao DM, Young RA. A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell. 1993;73:1361–1375. doi: 10.1016/0092-8674(93)90362-t. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.