Abstract
The HIR complex, which is comprised of the four proteins Hir1, Hir2, Hir3 and Hpc2, was first characterized as a repressor of three of the four histone gene loci in Saccharomyces cerevisiae. Using a bioinformatical approach, previous studies have identified a region of Hpc2 that is conserved in Schizosaccharomyces pombe and humans. Using a similar approach, we identified two additional domains, CDI and CDII, of the Hpc2 protein that are conserved amongst yeast species related to S. cerevisiae. We showed that the N terminal CDI domain (spanning amino acids 63–79) is dispensable for HIR complex assembly, but plays an essential role in the repression of the histone genes by recruiting the HIR complex to the HIR-dependent histone gene loci. The second conserved domain, CDII (spanning amino acids 452–480), is required for the stability of the Hpc2 protein itself as well as for the assembly of the HIR complex. In addition, we report a novel separation-of-function mutation within CDI of Hpc2, which causes derepression of the histone genes but does not confer other reported hir/hpc-phenotypes (such as Spt phenotypes, heterochromatin silencing defects and repression of cryptic promoters). This is the first direct demonstration that a separation-of-function mutation exists within the HIR complex.
Keywords: histone gene, HIR complex, HPC2, transcription, chromatin, yeast
1. Introduction
The eukaryotic genome is extensively packaged into a nucleoprotein complex called chromatin. The structural and regulatory unit of eukaryotic chromatin is the nucleosome, which consists of ~146 bp of DNA wrapped 1.65 turns around the core histone octamer [1]. Chromatin is therefore the substrate for all of the fundamental cellular processes that require DNA, including replication, transcription, recombination and repair.
A crucial step in the initial formation of chromatin occurs in S phase where newly replicated DNA is assembled with core histone octamers into nucleosomes. This step requires a tight coordination between histone synthesis and DNA replication since an altered level of core histones is detrimental for the cell, affecting processes such as DNA replication, chromosome segregation, transcription, heterochromatin silencing, leading to genomic instability, cell cycle perturbation and aging [2–5]. This is evident in the budding yeast Saccharomyces cerevisiae, where aberrant expression of the core histone genes outside of S phase, as well as altered histone gene dosage, result in alteration of chromatin structure [3, 5]. Therefore, transcription of the core histone genes is tightly regulated, occurring in late G1 and S phases concomitantly with DNA replication, while being repressed during the rest of the cell cycle [4]. The S. cerevisiae genome contains two copies of each of the four core histone genes which are organized by a pair of dimer partners into four loci: HTA1-HTB1 and HTA2-HTB2 encoding H2A and H2B; HHT1-HHF1 and HHT2-HHF2 encoding H3 and H4 [4]. The histone genes are positively and negatively regulated during the cell cycle by a number of factors acting through cis-acting elements present in their promoter [4].
Several genes involved in the repression of three of the four histone gene pairs, HTA1-HTB1, HHT1-HHF1 and HHT2-HHF2, were identified by genetic screens in S. cerevisiae. These include the four histone regulatory (HIR) and histone promoter control (HPC) genes, HIR1, HIR2, HIR3 and HPC2, which are required for the repression of these histone gene pairs outside of S phase and in response to hydroxyurea (HU), which causes stalling of DNA replication forks [6, 7]. The HIR-mediated transcriptional repression of these three histone pairs is dependent on a cis-acting element, known as the negative regulatory element (named NEG or CCR), which is present in their promoters but absent from the promoter of the HIR-independent HTA2-HTB2 loci [6, 8]. We have previously shown that these four HIR/HPC genes encode the proteins that stably associate to form the HIR complex (HIRA in S. pombe and humans) [9, 10]. The HIR complex is a histone chaperone that can assemble nucleosomes on to DNA in a replication-independent manner [9, 10]. In addition, the histone chaperone Asf1 was found to co-purify with the HIR complex and is also required for regulation of the HIR-dependent histone genes [9, 11]. The HIR complex stably binds to DNA and nucleosomes without any known sequence specificity [10]. Therefore, the HIR complex is postulated to be recruited to the negative (NEG) site of the three HIR-dependent histone gene pairs by a yet unidentified sequence-specific DNA binding factor [12–14]. The exact mechanism by which the HIR complex represses histone gene transcription is unknown but is likely to involve the modulation of chromatin by creating a repressive structure outside of S phase of the cell cycle. A recent study showed that a repressive chromatin structure at the histone gene promoter is established by the interplay of the HIR complex along with histone chaperones Asf1 and Rtt106 [15]. In late G1 and early S phase, cell cycle signals are suggested to be involved in the relief of HIR-mediated repression along with the HIR-dependent recruitment of the SWI/SNF complex to activate histone gene transcription [16].
In addition to the regulation of histone gene transcription, the HIR complex has been shown to be required for heterochromatic gene silencing [17], suppression of cryptic promoters [18, 19], kinetochore formation [17, 20] and maintenance of genomic integrity through its role in the assembly and maintenance of heterochromatin [21, 22]. Indeed, the HIR complex is part of a nucleosome assembly pathway that functionally overlaps with chromatin assembly factor I (CAF-I), which are together required for the assembly and maintenance of heterochromatin [17, 20, 23]. Combined mutations in either of the three genes that encode CAF-1 (CAC1, CAC2 or CAC3) in combination with mutations in any of the HIR genes result in a synergistic decrease in silencing at telomeres and the mating-type loci as well as an increase in retrotransposition of the Ty1 and Ty2 elements [17, 23]. HIR mutants have been found to suppress the deleterious effect of Δ element insertion in HIS4 and LYS2 in S. cerevisiae, evident by Spt− (suppressor of Ty element) phenotypes exhibited by hir/hpc null mutants [24]. The HIR complex has been shown to be important for repression of spurious transcription originating from cryptic promoters within coding regions suggesting that the HIR complex plays a role in the maintenance of a proper chromatin structure in transcribed genes [18, 19]. The role and importance of HIR-mediated nucleosome assembly in maintaining the protective functions of chromatin is also highlighted in fission yeast by the increased sensitivity of HIRA mutants to agents that causes DNA double-strand breaks [21].
The importance of the HIR complex is seen by its evolutionary conservation, as it is found in several species, including yeasts, Arabidopsis, Drosophila, Xenopus, chicken, fish, mice and humans [25–32]. In the fission yeast Schizosaccharomyces pombe, the HIRA complex is composed of Hip1, Slm9, Hip3 and Hip4 which are homologues of Hir1, Hir2, Hir3 and Hpc2, respectively [33–35]. The human HIRA protein is the homologue of Hir1 and Hir2 and is required for histone H3.3 deposition at active genes independently of DNA replication [25, 36–38]. HIRA is found associated with multiple proteins, including Cabin1 and Ubinuclein 1 (UBN1) which have been recently identified as homologues of Hir3 and Hpc2, respectively [39, 40]. HIRA is a candidate gene for the DiGeorge developmental syndrome [41, 42], and is important for heart formation [43], regulation of angiogenesis [44] and essential for development as mice lacking HIRA die at around embryonic day 10–11 [45].
In this study, we focused on further characterizing the molecular function of the Hpc2 protein within the HIR complex. Besides Hir1 and Hir2, the molecular function of the two other subunits, Hir3 and Hpc2, has not been fully investigated. Recently, studies on the human and S. pombe homologs of Hpc2 (UBN1 and Hip4, respectively) have shown that Hpc2 is a well conserved protein that plays an important role in the HIR complex not only in S. cerevisiae but in other species as well [33, 39, 40]. Here, using a bioinformatical approach, we have shown the existence of two previously unreported conserved regions, CDI and CDII, among Hpc2 fungi homologues. We found that the CDI domain, located within the Hpc2 N-terminal end, is important for the HIR-mediated repression of the histone genes through its essential role in recruiting the HIR complex to the histone gene promoter. Interestingly, we have shown that HPC2 mutants that affect histone gene expression without disrupting the HIR complex do not display other hir/hpc-phenotypes such as the Spt− phenotype, expression of cryptic TATA and heterochromatin silencing defects. This is the first direct demonstration that the hir/hpc-dependent derepression of the histone genes is not responsible for other hir/hpc-phenotypes, and this strongly supports previous evidence that implicates a direct role of the nucleosome assembly activity of the HIR complex in these other HIR-dependent pathways.
2. Materials and methods
2.1. Yeast strains, medium and growth assay
All strains used in this study are listed in Table 1. Yeast strains were grown in rich YPD medium (1% yeast extract, 2% peptone and 2% dextrose) or in minimal SD medium (0.67% yeast nitrogen base, 2% dextrose and the appropriate drop-out mixture of amino acids and bases) and were genetically manipulated by standard techniques [46]. YPP940, used for the Spt phenotype assay, was derived from PKY918 (gift from Dr. Paul Kaufman) by deleting HPC2 using HIS3MX6 cassette. YPP1200, used for the telomeric silencing assay, was derived from the original strain, PKY619 (gift from Dr. Paul Kaufman), by deleting HPC2 using KANMX6 cassette.
TABLE 1. Saccharomyces cerevisiae strains.
| Strain | Genotype | Reference |
|---|---|---|
| YPP461 | MATa his3Δ1 leu2Δ0 ura3Δ0 hpc2::KanMX6 HIR2-TAP::His3MX6 HIR3-13Myc::LEU2 | This work |
| YPP463 | MATa his3Δ1 leu2Δ0 ura3Δ0 hpc2::KanMX6 HIR1-TAP::His3MX6 HIR2-13Myc::LEU2 | This work |
| YPP492 | MATa his3Δ1 leu2Δ0 ura3Δ0 hpc2::KanMX6 HIR3-TAP::His3MX6 HIR1-13Myc::LEU2 | This work |
| YPP940 | MATa ura3-1 trp1-1 his3-11,15 leu2-3,112 can1-100 ade2-1 lys2-128δ hpc2::His3MX6 | This work |
| YPP996 | MATa ura3-1 trp1-1 his3-11,15 leu2-3,112 can1-100 ade2-1 hpc2::KanMX6 HIR2-TAP::His3MX6 | This work |
| YPP1213 | MATa ura3-1 trp1-1 his3-11,15 leu2-3,112 can1-100, ade2-1 hpc2::KanMX6, Hir1-TAP::His3MX6, | This work |
| YPP1200 | MATa ura3-1 trp1-1 his3-11,15 leu2-3,112 can1-100 ade2-1 ADE2-VR URA3-VIIL cac1Δ::hisG hpc2::KanMX6 | This work |
| L1133 | MATα KanMx-Gal1pr-FLO8-HIS3 his3Δ200/Δ1 leu2Δ0/Δ1 lys2-128δ ura3-52/Δ0 hpc2Δ::KanMx | F. Winston |
Telomeric silencing assays using the URA3-VIIL reporter (YPP1200) were performed as previously described [47]. Semi quantitative estimates of telomeric silencing were obtained by spotting 10 μl and 5 μl of 3-fold serial dilutions of log phase cells (adjusted to a starting OD of 2.0) on complete synthetic media with or without 0.3g/L 5-FOA, respectively. The cell viability assay was performed as previously described [17] to assess the proportion of viable cells in a population. Cells were platted on complete synthetic media with or without 1g/L 5-FOA. After 5 days of incubation at 30°C, the numbers of FOA-resistant colonies per viable cells plated were determined and normalized to the wild type strain. The results shown are averages and standard errors were calculated from two independent experiments.
2.2. Plasmids
Escherichia coli strain DH5α was used for cloning and plasmid amplification. Plasmids for HPC2 wild-type and mutants were constructed by sequential cloning of a PCR amplified product from the native promoter (500bp upstream of +1 ATG) and coding sequences in YCp:URA3 (pRS416) and YIp:TRP1 (pRS304) vectors with three HA-tag sequences followed by an ADH1 terminator sequence. All plasmids were sequenced to ensure that no errors were introduced during the PCR or sub cloning. Details of constructs used are available upon request.
2.3. Co-immunoprecipitation assays and Western blot analysis
Yeast whole cell extract were prepared in lysis buffer (40 mM HEPES-KOH pH 7.5, 350 mM NaCl, 10% Glycerol, 0.1% Tween-20, 1μg/ml pepstatin, 2μg/ml leupeptin, 0.5 mM DTT and 1 mM PMSF) and diluted to 150mM NaCl. One milligram protein extract was immunoprecipitated overnight at 4°C with 15 μl of IgG Sepharose beads (GE healthcare Life Science, product # 17096901) that binds to the Protein A tag of the TAP tagged subunit. The beads were washed three times and samples were prepared for Western blot. The immunoprecipitates were then resolved on 8 or 10% SDS-PAGE and blotted to PVDF membrane (Thermo Scientific, product # 88518). Western hybridization was then carried out using antibodies against HA (Roche product # 12013819001), c-Myc (Roche product # 11814150001) to check for the presence of the indicated tagged HIR subunit. Rabbit PAP (Peroxidase-Anti-Peroxidase) antibody (Sigma-Aldrich, product # P1291) was used for detection of the TAP tagged subunits.
2.4. Chromatin Immunoprecipitation assays
Chromatin immunoprecipitation was performed as described previously [48] with minor changes. Briefly, 100 ml of cells were grown in YPD media to an OD600 of 0.7 and treated with formaldehyde (1% v/v) for 20 minutes at room temperature, followed by a 10 minutes treatment with glycine (250 mM final). Chromatin lysates were prepared by glass bead disruption and sheared using a Bioruptor XL from Diagenode. One milligram of chromatin lysate was incubated overnight with 2ul of rabbit anti-Protein A antibody (Sigma Aldrich, product # P3775) to immunoprecipitate the TAP tagged subunit. The immune complexes were recovered by incubation with 20 μl of Protein A Sepharose beads (GE healthcare Life Science, product # 17528001) for 1 hr. The beads were washed and DNA was eluted. After reversing the crosslinks at 65°C overnight, the immunoprecipitated and input DNA were analyzed by quantitative PCR, using MyiQ (BioRad) and Power SYBR PCR Master Mix (Applied Biosystems, product # 4367659). An intergenic region of chromosome V was used as a non-target control. Primer sequences are available upon request. The amplified immunoprecipitated DNA was normalized to DNA amplified from input samples. The results shown are averages and standard errors were calculated from at least three independent experiments.
2.5. RT-qPCR analysis
Cells of the indicated strains were collected using log-phase culture (OD600 ~0.6) and total RNA was extracted using E.Z.N.A. Total RNA Kit I (Omega, product # R6834-02). cDNA was synthesized from 400ng of total RNA using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, product # 4368814). cDNA samples were then analyzed by quantitative PCR, using MyiQ (BioRad) and Power SYBR PCR Master Mix (Applied Biosystems, product # 4367659). Primer sequences are available upon request. Specific gene expression levels were normalized to ACT1 level. The results shown are averages and standard errors were calculated from at least three independent experiments.
3. Results
3.1. Phylogenetic analysis of Hpc2 between distantly related yeast species reveals three conserved domains
Multiple sequence alignment across proteins that are evolutionary related reveals regions of functional significance, as these regions are more conserved during evolution than the rest of the protein sequence [49–51]. In order to characterize the molecular role of the Hpc2 subunit within the HIR complex, we first looked at the sequence conservation of Hpc2 between related budding yeast species - Saccharomyces cerevisiae, Kluyveromyces lactis, Saccharomyces kluyveri (Lachancea kluyveri), and Saccharomyces castellii. The four Hpc2 protein sequences were aligned using the ‘Vector NTI AlignX’ software (Fig. 1). The overall sequence identity of the S. cerevisiae Hpc2 protein compared to the Hpc2 orthologues in S. castellii, Kluyveromyces lactis and S. Kluyveri is 23.5%, 27.9% and 30.9%, respectively. The Hpc2 alignment highlighted three distinct, highly conserved regions, CDI, CDII, and CDIII (Fig. 1). The first conserved domain (CDI) of Hpc2 corresponds to an N-terminal sequence spanning from amino acids 63 to 79 in S. cerevisiae. The second conserved domain (CDII) corresponds to a C-terminal sequence spanning amino acids 460 to 478. The third domain (CDIII) (also named HRD (Hpc2 Related Domain) or HUN (for HPC2-Ubinuclein-1) [39, 40]) also corresponds to a C-terminal sequence spanning amino acids 554 to 610. This bioinformatical analysis clearly identifies two new evolutionary conserved regions of Hpc2, CDI and CDII, and confirms the conservation of the CDIII/HRD domain amongst yeast as well as higher eukaryotes as shown previously [39, 40]. The Hpc2 CDIII/HRD domain has been previously shown to be important for the interaction of Ubinuclein-1 (UBN1) with HIRA in humans, and Hpc2 with Hir1 in yeast [40]. The conservation of Hpc2 CDI and CDII domains, in closely related yeast species, strongly suggest that they might play a significant biological role in the function of the HIR complex.
Figure 1. Identification of three highly conserved regions in Hpc2 yeast homologues.
Sequence alignment of Hpc2 from the yeast species Saccharomyces cerevisiae, Kluyveromyces lactis, Saccharomyces kluyveri, and Saccharomyces castellii is shown. The four Hpc2 protein sequences were aligned using the ‘Vector NTI AlignX’ software. The sequences are color coded according to similarities: Identical, red text on yellow background; Weakly similar, dark green text on a white background; Block of Similarity, black on a light green background; Conservative, dark blue on a light blue background; Non-similar, black on a white background. The three conserved domains CDI (63 to 79 aa), CDII (460 to 478 aa) and CDIII (554 to 610 aa) are underlined.
3.2. The conserved C-terminal CDII and CDIII (HRD) domains of Hpc2 are required for its assembly with the HIR complex while its N-terminal conserved region is dispensable for HIR complex formation
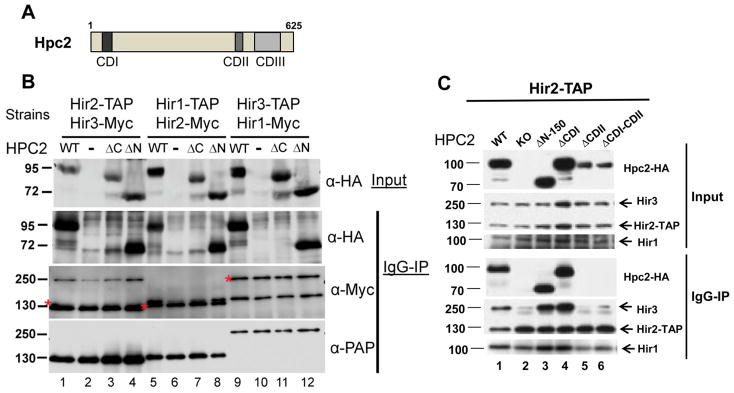
In order to address the biological relevance of the two novel conserved regions that we identified by sequence alignments (Fig. 1), we made a series of truncation and internal deletion mutants of Hpc2 (Fig. 2A) and first asked whether these conserved regions play a role in the interaction of Hpc2 with the rest of the HIR complex. We used whole cell extract from three different hpc2 deleted strains bearing different combinations of endogenously tagged HIR complex subunits with TAP and Myc epitopes. Into each of these strains, we transformed the indicated HPC2 plasmids and carried out immunoprecipitation using IgG beads, followed by analysis by Western blotting (Fig. 2B and 2C). Co-immunoprecipitations of wild-type (WT) Hpc2-HA with all three HIR complex TAP tagged subunits are shown as a positive control (Fig. 2B, lanes 1, 5, and 9, and Fig. 2C lane 1). The Hpc2ΔN-HA (ΔN) mutant, which is deleted of the first 150 amino acids, encompassing the CDI domain, co-precipitated with Hir2-, Hir1- and Hir3-TAP tagged proteins, similarly to WT Hpc2, as indicated by the α-HA signal present in the IgG-IP panel (Fig. 2B, lanes 4, 8, and 12). In agreement with this result, deletion of the CDI domain alone (Hpc2ΔCDI) did not affect the association of Hpc2 with the rest of the HIR complex (Fig. 2C, lane 4). Interestingly, we found that the Hpc2ΔC-HA (ΔC) mutant, which is truncated after the 548th amino acid, which removes the CDIII (HRD) domain, failed to co-precipitate with all three Hir-TAP subunits (Fig. 2B, lanes 3, 7, and 11) suggesting that this conserved region is important for the association of Hpc2 with the rest of the HIR complex. This result corroborates the previous report from Banumathy and colleagues [40] showing that the Hpc2 HRD domain was important for Hpc2 binding to Hir1 subunit by in vitro GST pull-down assays. Here, we confirm that this interaction is important for the in vivo assembly of Hpc2 with the HIR complex. Moreover, we found that the Hpc2 mutants deleted of the novel CDII domain (ΔCDII, and ΔCDI-CDII) also failed to co-precipitate with Hir2-TAP subunit as shown in Fig. 2C (lanes 5 and 6), suggesting that in addition to the CDIII/HRD domain, the CDII region also plays an important role for Hpc2 assembly with the HIR complex. In addition, the CDII domain seems to be important for the stability of Hpc2 as Hpc2 mutants lacking CDII domain are consistently less abundant in whole cell extract than wild-type Hpc2 (Fig. 2C compare lanes 5 and 6 to 1, 3 and 4). It is interesting to note that the association of the Hpc2 subunit to the Hir2-TAP subunit seems to be required for the stable association of the Hir3 subunit to the rest of the complex as shown by the reduced level of Hir3 proteins co-precipitated in a strain deleted of HPC2 (KO) (Fig. 2C, compare lane 2 to lane1) or containing Hpc2 mutant forms lacking the CDII region (Fig. 2C, compare lane 5 and 6 to lane 1). Therefore, these results presented here, in combination with data by Banumathy and colleagues [40], suggest that the Hpc2 CDII and CDIII/HRD conserved domains are both required for the interaction of Hpc2 with the rest of the HIR complex, while the CDI region does not seem to play any structural role in the assembly of the HIR complex.
Figure 2. The Hpc2 C-terminal region is essential for HIR complex assembly.
(A) Schematic of the wild-type Hpc2 protein with three conserved domain CDI, CDII and CDIII. (B) Whole cell extracts was made from the indicated strains, YPP461, YPP463, and YPP492, containing different TAP tag and Myc tag HIR subunits and deleted of the endogenous HPC2, as indicated, and transformed with the indicated HPC2-HA tag YCp:URA3 plasmid: WT, wild-type HPC2; −, empty plasmid; ΔC, HPC2ΔC-548 mutant; ΔN, HPC2ΔN-150 mutant. Co-immunoprecipitation assays were performed using IgG-sepharose beads and analyzed by Western blotting with anti-HA (to probe for Hpc2-HA), anti-Myc (to probe for the indicated Myc tag proteins), and peroxidase anti-peroxidase (α-PAP) (to probe for indicated TAP tag subunit) antibodies. The asterisks correspond to the TAP tagged proteins signal revealed by the HRP-couple secondary antibody used for the anti-Myc Western blot. (C) Same as (B) except that YPP996 (HIR2-TAP hpc2::KANMX6) strain was used and transformed with indicated YIp:TRP1 plasmid encoding the indicated forms of HPC2 wild-type (WT) and mutants.
3.3. The Hpc2 N-terminal conserved region CDI is important for histone gene repression
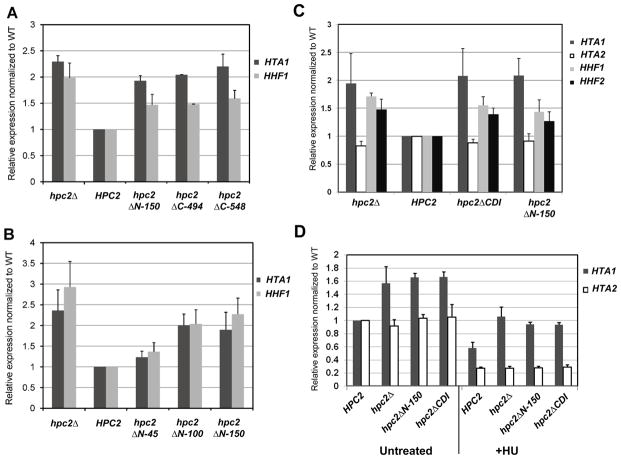
In yeast, the HIR complex regulates the transcription of three of the four histone gene loci during the cell cycle and represses their transcription outside of S phase and in response to hydroxyurea (HU), which causes stalling of DNA replication forks [8, 14, 52]. Deletion of any of the four HIR complex subunit encoding genes leads to an overall increase in the level of expression of the histone genes in an asynchronous population resulting from a specific loss of repression outside of S phase [6, 7]. To better understand the role of the newly identified Hpc2 conserved regions within the HIR complex, we asked whether these conserved regions play a role in the regulation of histone gene transcription. We monitored the level of the histone gene expression by RT-qPCR in strains carrying various mutant forms of Hpc2 as indicated in Figure 3. As shown in Figure 3A, Hpc2 C-terminal truncated mutants (hpc2ΔC-494 and hpc2ΔC-548) displayed a higher expression level of histones HTA1 and HHF1, similar to the derepression observed in an hpc2Δ mutant when compared to the wild type (HPC2). This result is consistent with our co-immunoprecipitation data showing that the Hpc2 C-terminal truncated mutants failed to assemble into the HIR complex (Fig. 2B and 2C). Therefore, these mutants mimic the hpc2Δ mutant phenotype. Furthermore, we found that the hpc2ΔN-150 mutant displayed a similar derepression of HTA1 and HHF1 comparable to the hpc2ΔC and hpc2Δ mutants (Fig. 3A). Since the absence of the N-terminal domain of Hpc2 does not disrupt the HIR complex (Fig. 2), this strongly suggests that the Hpc2 N-terminal region plays a direct role in the regulation of histone gene transcription. To narrow down the region of interest within the first 150 amino acids, we first tested a series of shorter N-terminal truncation mutants (Fig. 3B). The hpc2ΔN-100 mutant showed a similar derepression of HTA1 and HHF1 as the hpc2ΔN-150 mutant, while the hpc2ΔN-45 mutant displayed an almost wild-type level (Fig. 3B), suggesting that the sequences between amino acid 45 and 100 are important for HIR-mediated histone gene repression. This region of Hpc2 contains the newly identified CDI conserved domain (63–79 aa) (Fig. 1). Therefore, we asked whether the hpc2ΔCDI would fail to correctly regulate histone gene transcription. As can be seen in Figure 3C, the Hpc2 mutant lacking the CDI domain (hpc2ΔCDI) failed to properly regulate the histone genes. hpc2ΔCDI exhibited an elevated rate of transcript levels for HTA1, HHF1, and HHF2 that was similar to the rates observed in the hpc2Δ and hpc2ΔN-150 mutants when compared to wild-type (Fig. 3C). This suggests that the CDI domain is important for histone gene repression.
Figure 3. The Hpc2 N-terminal conserved domain is essential for histone gene repression.
(A) N-terminal- and C-terminal Hpc2 truncations cause derepression of the histone genes similarly to hpc2Δ. YPP996 strain was transformed with the indicated YCp:URA3 plasmid and grown in SD Ura- media, total RNA was extracted and HTA1 and HHF1 mRNA expression were tested by RT-qPCR. (B) The Hpc2 region between amino acid 100 and 150 is responsible for the depression of the histone genes. The strain and method are the same as in (A). (C) The Hpc2 CDI conserved domain is responsible for the derepression of the histone genes observed in an Hpc2 N-terminal truncated mutant. YPP996 strain was transformed with the indicated YIp:TRP1 plasmid and, after selection, were grown in YPD. RNA extraction and RT-qPCR were carried out as in (A), to check the level of HTA1, HTA2, HHF1, and HHF2. (D) Hpc2 CDI domain is necessary for repression of histone genes upon HU (hydroxyurea) treatment. The same strains were used as in (C). Cell were grown to log phase and collected from untreated culture or subjected to HU treatment (200mM) for 45min. RT-qPCR was used to monitor the level of HTA1 and HTA2 expression. HTA2 is used as a HIR-independent histone gene control. All the RT-qPCR were normalized to ACT1 expression level.
One of the key hir- phenotypes is manifested by the failure to repress HIR-dependent histone gene transcription upon treatment by hydroxyurea (HU) [6, 24]. Therefore, we asked whether the Hpc2 CDI region is involved in histone gene repression in response to HU treatment. As shown in Fig. 3D, the hpc2ΔN-150 and hpc2ΔCDI mutants fail to repress transcription of the HTA1 gene upon HU treatment at a similar level as the hpc2Δ mutant when compared to the WT strain (Fig. 3D). The level of HTA2 mRNA is shown as an internal positive control as the HTA2-HTB2 locus responds to HU treatment independently of the HIR complex. Here, we have identified that the CDI conserved region located at the N-terminal end of Hpc2 plays an essential role in the transcriptional regulation of the HIR-dependent histone genes as the hpc2ΔCDI mutant displays a hir/hpc null phenotype. These results, taken together with the co-immunoprecipitation assays, strongly suggest that the Hpc2 CDI region plays a direct role in histone gene transcriptional regulation and repression since the lack of the CDI domain does not disrupt the HIR complex.
3.4. The Hpc2 N-terminal conserved CDI region is essential for localization of the HIR complex to the HIR-dependent histone gene loci
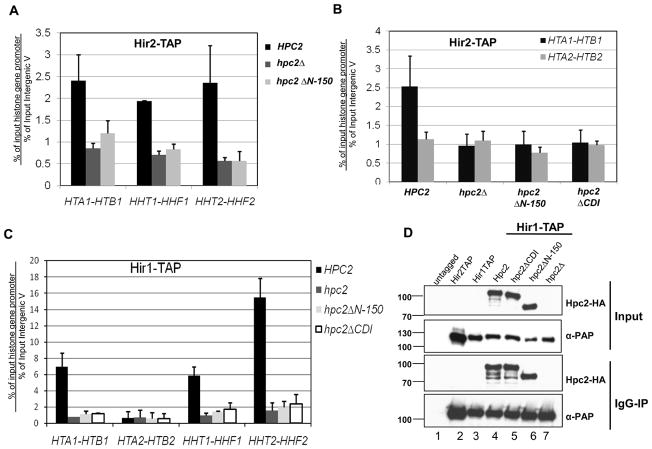
Having found that the N-terminus of Hpc2 and its CDI domain play an important role in HIR-mediated repression of the histone genes without disrupting the HIR complex, we sought to understand how this region was involved in controlling transcription of the HIR-dependent histone genes. We hypothesized that the Hpc2 N-terminal region and its CDI domain are involved in the localization of the HIR complex to the histone gene loci. Indeed, localization of the HIR complex to the histone gene loci is essential for HIR-mediated histone gene repression ([9, 15], and Prochasson unpublished observations). To address this question, we used chromatin immunoprecipitation (ChIP) assays to monitor whether the Hpc2 N-terminal region is necessary for localization of the HIR complex to the histone gene promoter. Consistent with previous results [9, 15], we show that Hir2-TAP specifically localized to all three HIR-dependent histone gene loci in a wild-type background (HPC2) while deletion of HPC2 (hpc2Δ) completely abrogates its recruitment (Fig. 4A). Strikingly, deletion of the N-terminal domain of Hpc2 (hpc2ΔN-150) prevented the recruitment of Hir2-TAP to all three histone gene loci similar to hpc2Δ (Fig. 4A). We then looked at the effect of the specific deletion of the CDI domain, and found that the hpc2ΔCDI mutant also prevented the localization of Hir2-TAP to the HTA1-HTB1 histone gene loci in a similar manner to hpc2ΔN-150 and hpc2Δ (Fig. 4B). We also carried out ChIP using the Hir1-TAP subunit which specifically localized to all three HIR-dependent histone gene loci in a wild-type background (HPC2) (Fig. 4C). Similarly to the results obtained with the Hir2-TAP subunit, deletion of the N-terminal domain of Hpc2 (hpc2ΔN-150) or the CDI domain (hpc2ΔCDI) prevented the recruitment of Hir1-TAP to all three HIR-dependent histone gene loci to the same extent as hpc2Δ (Fig. 4C). Co-immunoprecipitation assays were carried out and confirmed the correct expression and assembly of the Hpc2 wild-type (Hpc2) and mutant proteins (Hpc2ΔCDI and Hpc2ΔN-150) with the Hir1-TAP subunit (Fig. 4D, lane 4, 5 and 6). All together, these results show that the N-terminal region of Hpc2, as well as its CDI domain, is necessary for the localization of the HIR complex to the histone genes, thus explaining the hir- phenotype observed on the regulation of the histone gene transcription (Fig. 3).
Figure 4. The Hpc2 N-terminal conserved CDI domain is necessary for HIR recruitment at the HIR-dependent histone genes.
(A) The Hpc2 N-terminal region is necessary for the recruitment of the HIR complex to the HIR-dependent histone gene loci. Cells from yeast strain YPP996 (HIR2-TAP) transformed with indicated YCp:URA3 plasmid were analyzed by chromatin immunoprecipitation (ChIP) for the association of Hir2-TAP to the indicated histone gene loci, as described in the Material and Methods. Specific primer pairs to the promoter of the indicated histone gene loci were used (primers sequence available upon request). The fold enrichment is calculated as the ratio of percent IP of the indicated region to percent IP of a non-transcribed control region (Inter V). The values shown represent the average and standard errors from three independent experiments. (B) Hpc2 CDI is necessary for association of the HIR complex to HTA1-HTB1 loci. ChIP assay done as in (A) except that YPP996 strain was transformed with the indicated YIp:TRP1 plasmid. (C) The Hpc2 CDI domain is necessary for the association of Hir1-TAP to the HIR-dependent histone gene loci. Cells from yeast strain YPP1213 (HIR1-TAP) transformed with indicated YIp:TRP1 plasmid were analyzed by ChIP for the association of Hir1-TAP to the indicated histone gene loci. The values shown represent the average and standard deviation from two independent experiments. (D) Whole cell extracts were made from the strains used in (C) and other indicated strains. Co-immunoprecipitation assays were performed using IgG-sepharose beads and analyzed by Western hybridization with anti-HA and peroxidase anti-peroxidase (α-PAP) antibodies to verify the correct expression and interaction of the indicated Hpc2 wild-type and mutant proteins with Hir1-TAP tagged subunit.
3.5. Although the Hpc2 N-terminal region affects histone gene transcription, it is not responsible for other hir/hpc-phenotypes
Deletion of HIR/HPC genes have been shown to display Spt− (suppressor of Ty) phenotypes [7, 24], cryptic transcription [18, 19], and synergistically reduce heterochromatin silencing when combined with deletion mutants of the chromatin assembly complex (CAF-1), as well as showing temperature sensitivity, suggesting that the global aspect of chromatin structure could be affected [17]. These studies pointed toward a direct role of the nucleosome assembly activity of the HIR complex in regulating these various pathways. However, increased histone levels and alteration of the core histone stoichiometry have been reported to cause similar defects in yeast such as Spt− phenotype and loss of heterochromatin silencing when combined with cac mutants [17, 24, 53, 54]. Although hir/hpc-mutants are well known to deregulate histone gene levels and stoichiometry [6, 7], the question of the direct or indirect role played by the HIR complex in these various pathways has never been clearly answered. As we found that the Hpc2 N-terminal deletion mutant derepresses histone gene transcription similarly to the hpc2Δ mutant (Fig. 3), but without disrupting the HIR complex (Fig. 2), we could ask whether the hir/hpc-dependent histone gene deregulation is responsible for the aforementioned hir/hpc-phenotypes.
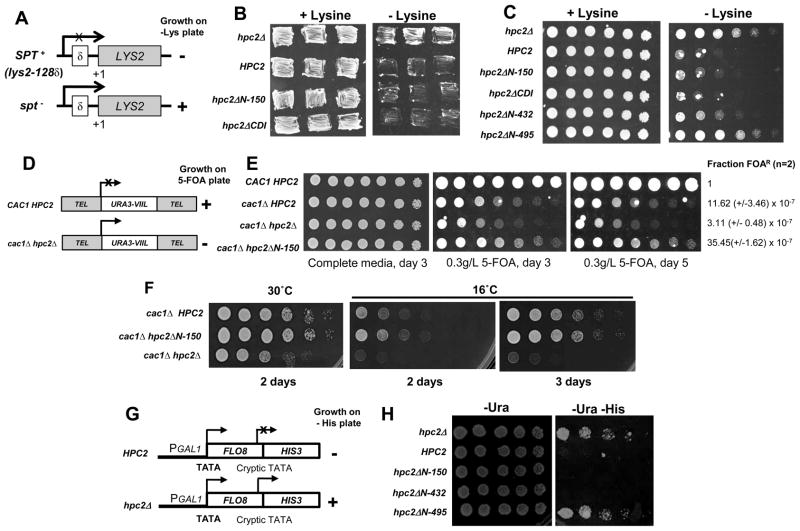
We first looked at the Spt phenotype of the N-terminal and CDI deletion mutants of Hpc2, as the hir/hpc null mutants have been shown to display an Spt− phenotype [7, 24]. Mutations in SPT genes suppress Ty and δ insertion mutations by restoring functional transcription to the adjacent genes [55–57] (Fig. 5A). In a wild-type (HPC2) background the δ sequence at lys2-128δ inserted within the 5′ end of the LYS2 coding region alters its transcription causing lysine auxotrophy (SPT+ phenotype) as seen by the absence of growth on solid media lacking lysine (−lys), while a hpc2Δ mutant strain displays an Spt− phenotype as shown by prototrophy to lysine (Fig. 5B and C, compare hpc2Δ and HPC2). Interestingly, a strain carrying the hpc2ΔN-150 or hpc2ΔCDI mutant failed to grow in absence of lysine, thus displaying a SPT+ phenotype as seen in the wild-type HPC2 strain (Fig. 5B and C). Additionally, we found that an Hpc2 mutant deleted of the first 431 amino acids (hpc2ΔN-432) also displayed an SPT+ phenotype, as shown by auxotrophy to lysine similar to the hpc2ΔN-150 mutant and the WT control (Fig. 5C). In contrast, an Hpc2 mutant deleted of the first 494 amino acids (hpc2ΔN-495), which additionally removes the CDII domain, displays a strong Spt− phenotype similar to hpc2Δ mutant (Fig. 5C). This is consistent with the requirement of the CDII domain for the association of Hpc2 with the rest of the HIR complex (Fig. 2). Together, these results strongly suggest that the deregulation of the histone genes in hir/hpc null mutants is not responsible for their Spt-phenotype since the hpc2ΔN-150 and hpc2ΔCDI mutants display an SPT+ phenotype.
Figure 5. The hir/hpc dependent deregulation of the histone genes is not responsible for the other hir/hpc-associated phenotypes.
(A) Schematic of the Spt assay phenotype. Mutations in SPT gene (Spt−) suppresses the transcriptional defect of the LYS2 gene in the SPT+ strain caused by insertion of the Ty element. The Spt phenotype is measured by growth on media lacking lysine. (B) The Hpc2 mutant deleted of the CDI domain presents an SPT+ phenotype similar to Hpc2 WT. Cells of yeast strain YPP940 transformed with the indicated YIp:TRP1 plasmid were patched onto synthetic complete media with or without Lysine to assess the Spt phenotype. (C) The first 432 amino acids of Hpc2 are dispensable for the SPT+ phenotype. Fourfold serial dilutions of cells from the YPP940 strain transformed with the indicated YIp:TRP1 plasmid were spotted onto synthetic complete media with or without lysine plates and grown for 5 days at 23°C. (D) Schematic of the telomeric silencing loci (URA3-VIIL). The URA3 gene is inserted next to the chromosome VIIL telomere [58]. Under a wild-type background, URA3 is silenced and so cells cannot metabolize 5-FOA into a lethally toxic metabolite and thus will survive. However mutations affecting telomeric silencing leads to URA3 expression resulting into toxicity and cell death in presence of 5-FOA. (E) Telomeric silencing defects of the hir/hpcΔ mutant when combined to cac1Δ are not due to hir/hpc-dependent histone gene derepression. Threefold serial dilution of cells of the yeast strain YPP1200 (URA3-VIIL) transformed with the indicated YIp:TRP1 plasmid were spotted onto synthetic complete media with or without 5-FOA and grown at 30°C. As previously reported by Sharp and colleagues [47], the cac1Δ mutant alone displays a severe loss of telomeric silencing, which is further exacerbated upon deletion of HIR/HPC genes. Therefore, we had to decrease the amount of 5-FOA to reduce the toxicity of the cac1Δ mutant alone and to observe enhanced sensitivity due to the hir/hpc-dependent loss of telomeric silencing. The cac1Δ HPC2 and cac1Δ hpc2ΔN-150 have a reduced sensitivity to 5-FOA compare to the cac1Δ hpc2Δ double mutant as shown by their better growth. The fraction of viable cells resistant to FOA (FOAR) was normalized to the value obtained for the wild-type (CAC1 HPC2). The averages of data from two experiments carried out with duplicate plating are shown (F). The hpc2ΔN-150 mutant does not display any synthetic temperature sensitive growth defect when combined with cac1Δ mutant. Cells of strains used in (D) were spotted on YPD media and incubated at 30°C or at 16°C for the indicated number of days. (G) Schematic of the pGAL1::FLO8-HIS3 reporter gene (adapted from [19]). (F) hir/hpc-dependent histone gene derepression is not responsible for the HIR-dependent inhibition of transcription initiation from the cryptic promoter of the pGAL1::FLO8-HIS3 reporter gene. The yeast strain L1133 containing the pGAL1::FLO8::HIS3 reporter, described in [19], was transformed with the indicated YCp:URA3 plasmid. Fourfold serial dilution of cells were spotted onto SD -Ura or SD -Ura -His to assess for the derepression of the FLO8 cryptic promoter allowing expression of HIS3 and growth in absence of histidine. Similarly to (C), most of Hpc2 is dispensable for the HIR-mediated repression of the FLO8 cryptic promoter. Only further N-terminal truncation which removed the CDII domain (hpc2ΔN-495), necessary for Hpc2 to interact with the rest of the HIR complex, displays a growth on -His media similar to hpc2Δ showing a loss of repression of the cryptic promoter.
In S. cerevisiae, telomere-proximal genes are subject to position-dependent but gene-independent silencing, a phenomenon termed as the ‘telomeric position effect’ (TPE) [58]. TPE can be quantified by placing the URA3 gene adjacent to a telomere (URA3-VIIL). The fraction of cells in a population that are resistant to the drug 5-FOA, a metabolic poison for Ura+ cells, represents the level of silencing of the URA3 gene (Fig. 5D). In cacΔ mutants, TPE is greatly reduced but not completely abolished [17], while in hirΔ mutants alone there is no effect on TPE [17]. However, the cacΔ hirΔ double mutants show a stronger reduction of the TPE compared to cacΔ single mutant [17]. However, alterations in histone H3 and H4 levels or in core histone stoichiometry also reduced silencing in cacΔ mutants but not in wild-type cells. This suggests that the HIR genes might contribute to silencing indirectly through deregulation of histone gene transcription [17]. To address the direct role of the HIR complex, we asked whether the histone gene derepression observed in the hpc2ΔN-150 mutant in combination with cac1Δ would affect TPE or not (Fig. 5E). We generated a cac1Δ hpc2Δ double mutant which displays the same reduction of TPE as the previously published cac1Δ hir1Δ double mutants [17] (data not shown). Because of the limited differences in TPE between single cac1Δ and double cac1Δ hir/hpcΔ mutants in presence of 1g/l of 5-FOA [17], we used a lower concentration of 5-FOA to increase the sensitivity of the assay to monitor more subtle effect on URA3 silencing. As can be seen in Figure 5E, the cac1Δ HPC2 and cac1Δ hpc2ΔN-150 mutants have a similar reduced sensitivity to 5-FOA as shown by the slightly better growth compared to the cac1Δ hpc2Δ double mutant. In addition, we measured the fraction of FOA-resistant (Fraction FOAR) colonies by doing a cell viability assay and found that cac1Δ HPC2 and cac1Δ hpc2ΔN-150 are 4 to 10 fold higher relative to cac1Δ hpc2Δ (Fig 5E), thus reflecting a more silenced URA3 gene. These results show that the hpc2ΔN-150 mutant does not enhance the loss of silencing, while it derepresses the histone genes similarly to hirΔ/hpcΔ mutants (Fig. 3), strongly suggesting that the HIR complex has a direct role in telomeric position effect. Interestingly, cac1Δ hpc2ΔN-150 shows a slightly better TPE than cac1Δ HPC2, suggesting that the overexpression of the histone genes slightly enhances silencing.
Kaufman and colleagues also reported that cacΔ hirΔ double mutants grow more slowly than wild-type or single mutant cells at 30°C and this phenotype is exacerbated at an even lower temperature (16°C) [17]. They speculate that the synthetic growth phenotypes do not solely result from changes in histone gene expression due to hirΔ mutants, however they could not rule out subtle effects due to changes in histone levels [17]. Here, equipped with the hpc2ΔN-150 mutant, we could rule out the implication of hir/hpc-dependent changes in histone levels in the synthetic growth defect of cacΔ hirΔ double mutants. As can be seen in Figure 5F, the cac1Δ hpc2ΔN-150 double mutant grows as well as the cac1Δ mutant alone (cac1Δ HPC2) at 30°C and at 16°C while the growth of cac1Δ hpc2Δ double mutants was reduced at 30°C and this phenotype was severely exacerbated at 16°C, shown by the strong reduction of growth. These results clearly indicate that the synthetic growth defects observed between cac1Δ mutant and hpc2Δ mutants is not due to the hpc2Δ-dependent histone gene deregulation but more likely from a direct role of the HIR complex on global aspect of chromatin structure has previously suggested [17].
Finally, we looked at the cryptic initiation, since the HIR/HPC genes are required for repression of cryptic promoters [18, 19] and asked whether histone gene deregulation could play a role. To test if hpc2ΔN-150 allows cryptic initiation, we used the previously described reporter gene for FLO8 cryptic initiation in which the 3′ coding region of FLO8 has been replaced with the HIS3 coding region such that HIS3 is expressed only when the FLO8 cryptic promoter is active (Fig. 5G) [18, 19]. A strain deleted of HPC2 (hpc2Δ) resulted in growth in the absence of histidine (His−) reflecting the activation of the cryptic promoter within FLO8, which allows expression of the HIS3 gene (Fig. 5H). In contrast, similarly to the wild-type (HPC2), the hpc2ΔN-150 mutant failed to grow on plates lacking histidine showing that the hir/hpc-dependent deregulation of the histone genes is not responsible for cryptic initiation observed in hir/hpcΔ mutants (Fig. 5H). Consistent with the Spt− phenotype (Fig. 5C), we found that the hpc2ΔN-432 mutant properly repressed cryptic transcription, as shown by the lack of growth on His-plate (Fig. 5H), while the hpc2ΔN-495 mutant allowed cryptic initiation as shown by growth similar to the hpc2Δ strain. These results again strongly suggest that the derepression of the histone genes observed in hir/hpc null mutants is not responsible for the hir/hpc-effect on the expression of spurious transcripts originating from cryptic promoters. This therefore strongly supports the direct role of the HIR complex in nucleosome re-assembly during transcriptional elongation, as previously suggested [19].
4. Discussion
Based on the presence of the conserved HRD (CDIII) domain, sophisticated bioinformatical analyses have identified orthologues of the S. cerevisiae protein Hpc2 in S. pombe (Hip4) and in humans (UBN1 and UBN2) [39, 40]. Interestingly, aside from this HRD domain, no other significant homology was seen in the rest of the Hpc2 protein, which is indicative of the degree of sequence divergence that has arisen during evolution. In this study, we report two new highly conserved Hpc2 regions, CDI and CDII, among yeast species that are related to S. cerevisiae (Fig. 1). We show that each domain plays a significant biological role in the functions of the HIR complex. The CDI domain is important for repression of the HIR-dependent histone genes (Fig. 3) and is necessary for the recruitment of the HIR complex to these loci (Fig. 4), while CDII is important for the stability of the Hpc2 protein itself as well as for the binding of Hpc2 to the rest of the HIR complex (Fig. 2). It is interesting to note that the human and S. pombe HIR complex homologues are also involved in the regulation of the histone genes as well as transcriptional repression [34, 59, 60]. Since the overall function of the HIR complex is conserved, it was surprising that neither of these two domains, CDI and CDII, was identified in other Hpc2 orthologues, raising the question regarding their degree of conservation in higher eukaryotes. We can speculate that these sequences have become too divergent during evolution, so that they can no longer be identified using bioinformatic analysis. Alternatively, we cannot rule out the possibility that one of the subunits of HIRA (in higher eukaryotes) has evolved in such a way that it carries out the functions carried out by the S. cerevisiae Hpc2 CDI and CDII domains. As we have shown the importance of the CDI domain in the localization of the HIR complex to the histone genes and in their regulation, we can hypothesize that either a domain present in the orthologue HIRA complex, or perhaps one of its associated proteins, assumes the biological function of the Hpc2 CDI domain. Further bioinformatical analysis and molecular characterizations of the S. cerevisiae Hpc2 orthologues and/or HIRA subunits will be necessary to unravel this interesting question regarding the evolution of histone gene regulation by HIR complexes.
The mechanism of recruitment of the HIR complex to the HIR-dependent histone gene loci has remained elusive since the first identification of the HIR/HPC genes and their role in the regulation of histone gene transcription. While it was initially reported that the Hir/Hpc proteins did not possess intrinsic DNA binding activity [36, 61], we previously showed that the HIR complex binds non-specifically to DNA [10]. Hence, the HIR complex is postulated to be recruited to the negative site of the histone gene promoters by an uncharacterized sequence-specific DNA binding factor [13]. Here, we found that the Hpc2 CDI domain plays an essential role in the recruitment of the HIR complex as deletion of the CDI domain from Hpc2 completely abrogates HIR recruitment to the histone genes (Fig. 4) resulting in their derepression (Fig. 3) but without disrupting the HIR complex (Fig. 2) nor displaying other hir/hpc-phenotypes (Fig. 5). We also showed that the Hpc2 CDI domain is dispensable for the non-specific DNA binding activity of the HIR complex (data not shown). All together, this strongly suggests that the Hpc2 CDI domain could bind to this yet uncharacterized sequence-specific DNA binding factor that is hypothesized to bind to the negative site of the HIR-dependent histone gene promoters, hence mediating the recruitment of the HIR complex to these loci. Therefore, future experiments aiming at identifying specific proteins interacting with Hpc2 CDI domain is likely to lead to the identification of the negative site (NEG/CCR) specific DNA binding protein which tethers the HIR complex to the histone genes.
In S. cerevisiae, increased histone levels and alteration of the core histone stoichiometry have been reported to cause similar phenotypes as deletion of the HIR/HPC genes, such as Spt− phenotype and loss of heterochromatin silencing when combined with cacΔ mutants [17, 53, 54]. Since one of the key phenotypes of the hir/hpc null mutants is the de-repression of HTA1-HTB1, HHT1-HHF1, and HHT2-HHF2 histone genes, it has always been difficult to discriminate between the direct effects of the HIR complex and its indirect effects due to the deregulation of the histone genes on those various hir/hpc- phenotypes. However, several studies pointed out the direct implications of the HIR complex as forced overexpression of the histone genes could not recapitulate the full extent of the hir/hpc-phenotypes observed in position-dependent gene silencing, for example, when combined with cacΔ mutants [17]. In the present study, we identified two HPC2 mutants, hpc2ΔN-150 and hpc2ΔCDI, which fully recapitulate the hir/hpc-dependent derepression of the histone genes but without displaying other hir/hpc-phenotypes (e.g. Spt− phenotype, loss of heterochromatin silencing and suppression of cryptic promoters (Fig. 5)). These are the first mutants that show a separation-of-function of the HIR complex activity between its role in the regulation of histone genes and its role in the maintenance of chromatin structure. Therefore, our results strongly support previous studies that suggested a direct involvement of the nucleosome assembly activity of the HIR complex in regulating and maintaining a proper chromatin structure. We can then speculate that the HIR nucleosome assembly activity [10, 17] is directly involved in regulating the chromatin structure that is affected in hir/hpc mutants phenotypes. However, as previously suggested, it is likely that the HIR nucleosome assembly activity is also required for repression of histone gene transcription to maintain a repressive chromatin structure at their promoters [15]. By using these specific Hpc2 mutants (hpc2ΔN-150 and hpc2ΔCDI) as controls, we will be able to potentially discriminate between the nucleosome assembly and histone repression roles of the HIR complex. Interestingly, our data strongly suggests that although the Hpc2 CDI domain is involved in the recruitment of the HIR complex at the histone genes, it does not play any role in targeting the HIR complex to regions that are involved in causing the Spt− phenotype, heterochromatin silencing and the repression of cryptic promoters. This raises the question of how the HIR complex is brought to these various places in the genome to maintain proper chromatin integrity, either by recognizing directly a loose chromatin structure or by interacting directly with other factors involved in these pathways, such as the transcription elongation machinery. Further studies of the HIR complex will address these exciting questions and extend our understanding of its role in the maintenance of repressive chromatin structures required for proper cell homeostasis.
Research Highlights.
New functional conserved Hpc2 domains CDI and CDII among fungi.
Hpc2 N-terminal CDI essential for HIR-dependent histone gene repression.
Hpc2 CDI domain needed for HIR complex localization to histone gene loci.
Histone gene derepression in hpc2ΔCDI is not responsible for other hir/hpc phenotypes.
First report of separation-of-function mutation within the HIR complex.
Acknowledgments
This publication was made possible by NIH grant number P20 RR016475 from the INBRE Program of the National Center for Research Resources. This work was supported by a Leukemia and Lymphoma Society Special Fellowship to P.P. and by the Kansas Bioscience Authority (KBA). We thank Aimee Nienstedt for her technical help in the lab. We thank Dr Paul Kaufman for the generous gift of the PKY619 and PKY918 strains, and Dr Fred Winston for the strain carrying the FLO8-HIS3 reported gene (L1133).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Feser J, Truong D, Das C, Carson JJ, Kieft J, Harkness T, Tyler JK. Elevated histone expression promotes life span extension. Mol Cell. 2010;39:724–735. doi: 10.1016/j.molcel.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gunjan A, Paik J, Verreault A. Regulation of histone synthesis and nucleosome assembly. Biochimie. 2005;87:625–635. doi: 10.1016/j.biochi.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 4.Osley MA. The regulation of histone synthesis in the cell cycle. Annu Rev Biochem. 1991;60:827–861. doi: 10.1146/annurev.bi.60.070191.004143. [DOI] [PubMed] [Google Scholar]
- 5.Singh RK, Liang D, Gajjalaiahvari UR, Kabbaj MH, Paik J, Gunjan A. Excess histone levels mediate cytotoxicity via multiple mechanisms. Cell Cycle. 2010;9:4236–4244. doi: 10.4161/cc.9.20.13636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Osley MA, Lycan D. Trans-acting regulatory mutations that alter transcription of Saccharomyces cerevisiae histone genes. Mol Cell Biol. 1987;7:4204–4210. doi: 10.1128/mcb.7.12.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu H, Kim UJ, Schuster T, Grunstein M. Identification of a new set of cell cycle-regulatory genes that regulate S-phase transcription of histone genes in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:5249–5259. doi: 10.1128/mcb.12.11.5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Osley MA, Gould J, Kim S, Kane MY, Hereford L. Identification of sequences in a yeast histone promoter involved in periodic transcription. Cell. 1986;45:537–544. doi: 10.1016/0092-8674(86)90285-0. [DOI] [PubMed] [Google Scholar]
- 9.Green EM, Antczak AJ, Bailey AO, Franco AA, Wu KJ, Yates JR, 3rd, Kaufman PD. Replication-independent histone deposition by the HIR complex and Asf1. Curr Biol. 2005;15:2044–2049. doi: 10.1016/j.cub.2005.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prochasson P, Florens L, Swanson SK, Washburn MP, Workman JL. The HIR corepressor complex binds to nucleosomes generating a distinct protein/DNA complex resistant to remodeling by SWI/SNF. Genes Dev. 2005;19:2534–2539. doi: 10.1101/gad.1341105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sutton A, Bucaria J, Osley MA, Sternglanz R. Yeast ASF1 protein is required for cell cycle regulation of histone gene transcription. Genetics. 2001;158:587–596. doi: 10.1093/genetics/158.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeSilva H, Lee K, Osley MA. Functional dissection of yeast Hir1p, a WD repeat-containing transcriptional corepressor. Genetics. 1998;148:657–667. doi: 10.1093/genetics/148.2.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moran L. PhD Thesis. New York: Cornell University; 1994. Factors involved in temporal and autogenous control of histone synthesis in Saccharomyces cerevisiae. [Google Scholar]
- 14.Spector MS, Raff A, DeSilva H, Lee K, Osley MA. Hir1p and Hir2p function as transcriptional corepressors to regulate histone gene transcription in the Saccharomyces cerevisiae cell cycle. Mol Cell Biol. 1997;17:545–552. doi: 10.1128/mcb.17.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fillingham J, Kainth P, Lambert JP, van Bakel H, Tsui K, Pena-Castillo L, Nislow C, Figeys D, Hughes TR, Greenblatt J, Andrews BJ. Two-color cell array screen reveals interdependent roles for histone chaperones and a chromatin boundary regulator in histone gene repression. Mol Cell. 2009;35:340–351. doi: 10.1016/j.molcel.2009.06.023. [DOI] [PubMed] [Google Scholar]
- 16.Dimova D. PhD Thesis. New York: Cornell University; 2000. Antagonizing transcriptional repression of a cell cycle regulated yeast promoter. [Google Scholar]
- 17.Kaufman PD, Cohen JL, Osley MA. Hir proteins are required for position-dependent gene silencing in Saccharomyces cerevisiae in the absence of chromatin assembly factor I. Mol Cell Biol. 1998;18:4793–4806. doi: 10.1128/mcb.18.8.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheung V, Chua G, Batada NN, Landry CR, Michnick SW, Hughes TR, Winston F. Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol. 2008;6:e277. doi: 10.1371/journal.pbio.0060277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nourani A, Robert F, Winston F. Evidence that Spt2/Sin1, an HMG-like factor, plays roles in transcription elongation, chromatin structure, and genome stability in Saccharomyces cerevisiae. Mol Cell Biol. 2006;26:1496–1509. doi: 10.1128/MCB.26.4.1496-1509.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharp JA, Franco AA, Osley MA, Kaufman PD. Chromatin assembly factor I and Hir proteins contribute to building functional kinetochores in S. cerevisiae. Genes Dev. 2002;16:85–100. doi: 10.1101/gad.925302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anderson HE, Wardle J, Korkut SV, Murton HE, Lopez-Maury L, Bahler J, Whitehall SK. The fission yeast HIRA histone chaperone is required for promoter silencing and the suppression of cryptic antisense transcripts. Mol Cell Biol. 2009;29:5158–5167. doi: 10.1128/MCB.00698-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Formosa T, Ruone S, Adams MD, Olsen AE, Eriksson P, Yu Y, Rhoades AR, Kaufman PD, Stillman DJ. Defects in SPT16 or POB3 (yFACT) in Saccharomyces cerevisiae cause dependence on the Hir/Hpc pathway: polymerase passage may degrade chromatin structure. Genetics. 2002;162:1557–1571. doi: 10.1093/genetics/162.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qian Z, Huang H, Hong JY, Burck CL, Johnston SD, Berman J, Carol A, Liebman SW. Yeast Ty1 retrotransposition is stimulated by a synergistic interaction between mutations in chromatin assembly factor I and histone regulatory proteins. Mol Cell Biol. 1998;18:4783–4792. doi: 10.1128/mcb.18.8.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sherwood PW, Osley MA. Histone regulatory (hir) mutations suppress delta insertion alleles in Saccharomyces cerevisiae. Genetics. 1991;128:729–738. doi: 10.1093/genetics/128.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell. 2002;9:1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- 26.Kirov N, Shtilbans A, Rushlow C. Isolation and characterization of a new gene encoding a member of the HIRA family of proteins from Drosophila melanogaster. Gene. 1998;212:323–332. doi: 10.1016/s0378-1119(98)00143-7. [DOI] [PubMed] [Google Scholar]
- 27.Llevadot R, Marques G, Pritchard M, Estivill X, Ferrus A, Scambler P. Cloning, chromosome mapping and expression analysis of the HIRA gene from Drosophila melanogaster. Biochem Biophys Res Commun. 1998;249:486–491. doi: 10.1006/bbrc.1998.9165. [DOI] [PubMed] [Google Scholar]
- 28.Phelps-Durr TL, Thomas J, Vahab P, Timmermans MC. Maize rough sheath2 and its Arabidopsis orthologue ASYMMETRIC LEAVES1 interact with HIRA, a predicted histone chaperone, to maintain knox gene silencing and determinacy during organogenesis. Plant Cell. 2005;17:2886–2898. doi: 10.1105/tpc.105.035477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roberts C, Daw SC, Halford S, Scambler PJ. Cloning and developmental expression analysis of chick Hira (Chira), a candidate gene for DiGeorge syndrome. Hum Mol Genet. 1997;6:237–245. doi: 10.1093/hmg/6.2.237. [DOI] [PubMed] [Google Scholar]
- 30.Scamps C, Lorain S, Lamour V, Lipinski M. The HIR protein family: isolation and characterization of a complete murine cDNA. Biochim Biophys Acta. 1996;1306:5–8. doi: 10.1016/0167-4781(96)00010-3. [DOI] [PubMed] [Google Scholar]
- 31.Wilming LG, Snoeren CA, van Rijswijk A, Grosveld F, Meijers C. The murine homologue of HIRA, a DiGeorge syndrome candidate gene, is expressed in embryonic structures affected in human CATCH22 patients. Hum Mol Genet. 1997;6:247–258. doi: 10.1093/hmg/6.2.247. [DOI] [PubMed] [Google Scholar]
- 32.Zhao ZK, Li W, Wang MY, Zhou L, Wang JL, Wang YF. The role of HIRA and maternal histones in sperm nucleus decondensation in the gibel carp and color crucian carp. Mol Reprod Dev. 2011;78:139–147. doi: 10.1002/mrd.21278. [DOI] [PubMed] [Google Scholar]
- 33.Anderson HE, Kagansky A, Wardle J, Rappsilber J, Allshire RC, Whitehall SK. Silencing mediated by the Schizosaccharomyces pombe HIRA complex is dependent upon the Hpc2-like protein, Hip4. PLoS One. 2010;5:e13488. doi: 10.1371/journal.pone.0013488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blackwell C, Martin KA, Greenall A, Pidoux A, Allshire RC, Whitehall SK. The Schizosaccharomyces pombe HIRA-like protein Hip1 is required for the periodic expression of histone genes and contributes to the function of complex centromeres. Mol Cell Biol. 2004;24:4309–4320. doi: 10.1128/MCB.24.10.4309-4320.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Greenall A, Williams ES, Martin KA, Palmer JM, Gray J, Liu C, Whitehall SK. Hip3 interacts with the HIRA proteins Hip1 and Slm9 and is required for transcriptional silencing and accurate chromosome segregation. J Biol Chem. 2006;281:8732–8739. doi: 10.1074/jbc.M512170200. [DOI] [PubMed] [Google Scholar]
- 36.Lamour V, Lecluse Y, Desmaze C, Spector M, Bodescot M, Aurias A, Osley MA, Lipinski M. A human homolog of the S. cerevisiae HIR1 and HIR2 transcriptional repressors cloned from the DiGeorge syndrome critical region. Hum Mol Genet. 1995;4:791–799. doi: 10.1093/hmg/4.5.791. [DOI] [PubMed] [Google Scholar]
- 37.Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 38.Ray-Gallet D, Quivy JP, Scamps C, Martini EM, Lipinski M, Almouzni G. HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol Cell. 2002;9:1091–1100. doi: 10.1016/s1097-2765(02)00526-9. [DOI] [PubMed] [Google Scholar]
- 39.Balaji S, Iyer LM, Aravind L. HPC2 and ubinuclein define a novel family of histone chaperones conserved throughout eukaryotes. Mol Biosyst. 2009;5:269–275. doi: 10.1039/b816424j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Banumathy G, Somaiah N, Zhang R, Tang Y, Hoffmann J, Andrake M, Ceulemans H, Schultz D, Marmorstein R, Adams PD. Human UBN1 is an ortholog of yeast Hpc2p and has an essential role in the HIRA/ASF1a chromatin-remodeling pathway in senescent cells. Mol Cell Biol. 2009;29:758–770. doi: 10.1128/MCB.01047-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Halford S, Wadey R, Roberts C, Daw SC, Whiting JA, O’Donnell H, Dunham I, Bentley D, Lindsay E, Baldini A, Scambler PJ. Isolation of a putative transcriptional regulator from the region of 22q11 deleted in DiGeorge syndrome, Shprintzen syndrome and familial congenital heart disease. Hum Mol Genet. 1993;2:2099–2107. doi: 10.1093/hmg/2.12.2099. [DOI] [PubMed] [Google Scholar]
- 42.Llevadot R, Scambler P, Estivill X, Pritchard M. Genomic organization of TUPLE1/HIRA: a gene implicated in DiGeorge syndrome. Mamm Genome. 1996;7:911–914. doi: 10.1007/s003359900268. [DOI] [PubMed] [Google Scholar]
- 43.Farrell MJ, Stadt H, Wallis KT, Scambler P, Hixon RL, Wolfe R, Leatherbury L, Kirby ML. HIRA, a DiGeorge syndrome candidate gene, is required for cardiac outflow tract septation. Circ Res. 1999;84:127–135. doi: 10.1161/01.res.84.2.127. [DOI] [PubMed] [Google Scholar]
- 44.Dutta D, Ray S, Home P, Saha B, Wang S, Sheibani N, Tawfik O, Cheng N, Paul S. Regulation of angiogenesis by histone chaperone HIRA-mediated incorporation of lysine 56-acetylated histone H3.3 at chromatin domains of endothelial genes. J Biol Chem. 2010;285:41567–41577. doi: 10.1074/jbc.M110.190025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roberts C, Sutherland HF, Farmer H, Kimber W, Halford S, Carey A, Brickman JM, Wynshaw-Boris A, Scambler PJ. Targeted mutagenesis of the Hira gene results in gastrulation defects and patterning abnormalities of mesoendodermal derivatives prior to early embryonic lethality. Mol Cell Biol. 2002;22:2318–2328. doi: 10.1128/MCB.22.7.2318-2328.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaiser C, Michaelis S, Mitchell A. Methods in yeast genetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y.: 1994. [Google Scholar]
- 47.Sharp JA, Fouts ET, Krawitz DC, Kaufman PD. Yeast histone deposition protein Asf1p requires Hir proteins and PCNA for heterochromatic silencing. Curr Biol. 2001;11:463–473. doi: 10.1016/s0960-9822(01)00140-3. [DOI] [PubMed] [Google Scholar]
- 48.Sharma VM, Li B, Reese JC. SWI/SNF-dependent chromatin remodeling of RNR3 requires TAF(II)s and the general transcription machinery. Genes Dev. 2003;17:502–515. doi: 10.1101/gad.1039503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cliften P, Sudarsanam P, Desikan A, Fulton L, Fulton B, Majors J, Waterston R, Cohen BA, Johnston M. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science. 2003;301:71–76. doi: 10.1126/science.1084337. [DOI] [PubMed] [Google Scholar]
- 50.La D, Sutch B, Livesay DR. Predicting protein functional sites with phylogenetic motifs. Proteins. 2005;58:309–320. doi: 10.1002/prot.20321. [DOI] [PubMed] [Google Scholar]
- 51.Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Freeman KB, Karns LR, Lutz KA, Smith MM. Histone H3 transcription in Saccharomyces cerevisiae is controlled by multiple cell cycle activation sites and a constitutive negative regulatory element. Mol Cell Biol. 1992;12:5455–5463. doi: 10.1128/mcb.12.12.5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Clark-Adams CD, Norris D, Osley MA, Fassler JS, Winston F. Changes in histone gene dosage alter transcription in yeast. Genes Dev. 1988;2:150–159. doi: 10.1101/gad.2.2.150. [DOI] [PubMed] [Google Scholar]
- 54.Dollard C, Ricupero-Hovasse SL, Natsoulis G, Boeke JD, Winston F. SPT10 and SPT21 are required for transcription of particular histone genes in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:5223–5228. doi: 10.1128/mcb.14.8.5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fassler JS, Winston F. Isolation and analysis of a novel class of suppressor of Ty insertion mutations in Saccharomyces cerevisiae. Genetics. 1988;118:203–212. doi: 10.1093/genetics/118.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Silverman SJ, Fink GR. Effects of Ty insertions on HIS4 transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:1246–1251. doi: 10.1128/mcb.4.7.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Winston F, Durbin KJ, Fink GR. The SPT3 gene is required for normal transcription of Ty elements in S. cerevisiae. Cell. 1984;39:675–682. doi: 10.1016/0092-8674(84)90474-4. [DOI] [PubMed] [Google Scholar]
- 58.Gottschling DE, Aparicio OM, Billington BL, Zakian VA. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- 59.Hall C, Nelson DM, Ye X, Baker K, DeCaprio JA, Seeholzer S, Lipinski M, Adams PD. HIRA, the human homologue of yeast Hir1p and Hir2p, is a novel cyclin-cdk2 substrate whose expression blocks S-phase progression. Mol Cell Biol. 2001;21:1854–1865. doi: 10.1128/MCB.21.5.1854-1865.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nelson DM, Ye X, Hall C, Santos H, Ma T, Kao GD, Yen TJ, Harper JW, Adams PD. Coupling of DNA synthesis and histone synthesis in S phase independent of cyclin/cdk2 activity. Mol Cell Biol. 2002;22:7459–7472. doi: 10.1128/MCB.22.21.7459-7472.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sherwood PW, Tsang SV, Osley MA. Characterization of HIR1 and HIR2, two genes required for regulation of histone gene transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:28–38. doi: 10.1128/mcb.13.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]