Abstract
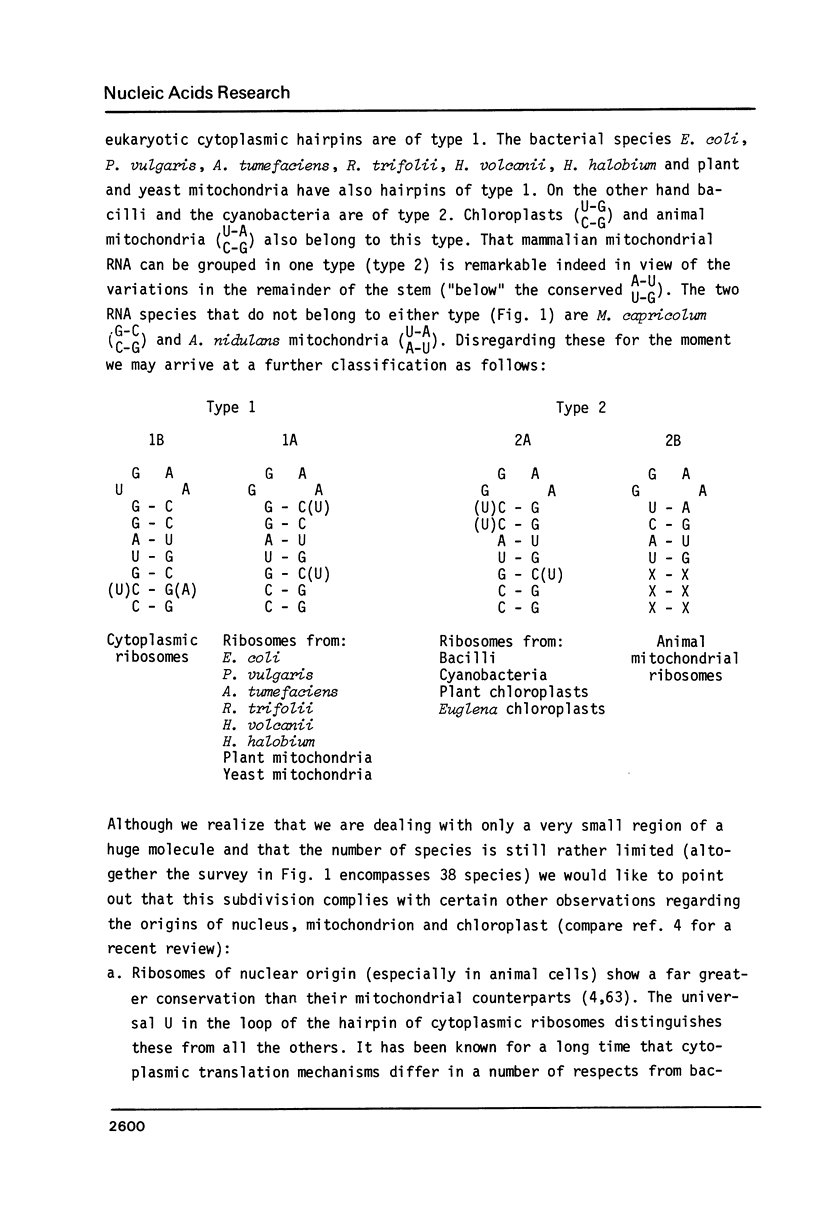
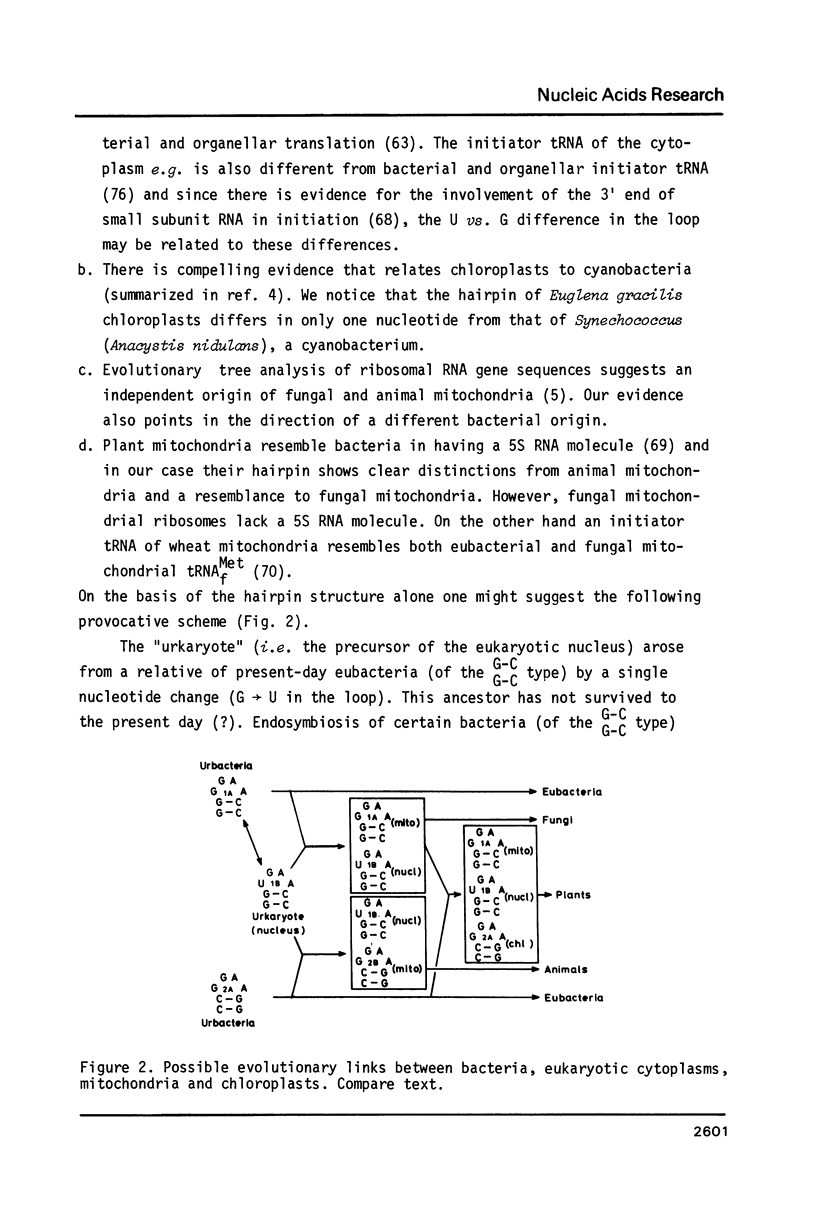
The strongest conserved part of the RNA of small ribosomal subunits is probably located near the 3' end. This paper reviews the primary and secondary structures of some 40 sequenced 3' termini and tries to classify these structures according to common features and differences. The regions under consideration contain at the 5' side an almost universal, supposedly single-stranded stretch of nucleotides with the sequence--AAGUCGUAACAAGGU--. This is followed by a stem-loop structure. The stem always contains 9 basepairs (including U-G pairs) and no mismatches or bulged nucleotides. The loop of the hairpin is either (m2)GGm62Am62A (bacteria, chloroplasts and mitochondria) or UGm62Am62A (cytoplasm). The hairpin is, in most cases, followed at the 3' side by--GGAUCA--. Next to it bacteria and chloroplasts contain the so-called "Shine and Dalgarno" sequence --CCUCC--. The stem region of the hairpin contains a conserved A-U U-G junction. The two basepairs between this junction and the loop are either of type 1 (G-C G-C) or type 2 (C-G C-G). Classification according to type links certain bacteria with mitochondria of yeast and plants and others with chloroplasts and with animal mitochondria.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alberty H., Raba M., Gross H. J. Isolation from rat liver and sequence of a RNA fragment containing 32 nucleotides from position 5 to 36 from the 3' end of ribosomal 18S RNA. Nucleic Acids Res. 1978 Feb;5(2):425–434. doi: 10.1093/nar/5.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S., de Bruijn M. H., Coulson A. R., Eperon I. C., Sanger F., Young I. G. Complete sequence of bovine mitochondrial DNA. Conserved features of the mammalian mitochondrial genome. J Mol Biol. 1982 Apr 25;156(4):683–717. doi: 10.1016/0022-2836(82)90137-1. [DOI] [PubMed] [Google Scholar]
- Azad A. A., Deacon N. J. The 3'-terminal primary structure of five eukaryotic 18S rRNAs determined by the direct chemical method of sequencing. The highly conserved sequences include an invariant region complementary to eukaryotic 5S rRNA. Nucleic Acids Res. 1980 Oct 10;8(19):4365–4376. doi: 10.1093/nar/8.19.4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baan R. A., Hilbers C. W., Van Charldorp R., Van Leerdam E., Van Knippenberg P. H., Bosch L. High-resolution proton magnetic resonance study of the secondary structure of the 3'-terminal 49-nucleotide fragment of 16S rRNA from Escherichia coli. Proc Natl Acad Sci U S A. 1977 Mar;74(3):1028–1031. doi: 10.1073/pnas.74.3.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer R., Dubin D. T. The 3'-terminal sequence of the small subunit ribosomal RNA from hamster mitochondria. Nucleic Acids Res. 1980 Nov 11;8(21):4927–4941. doi: 10.1093/nar/8.21.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borbely G., Simoncsits A. 3'-Terminal conserved loops of 16S rRNAs from the cyanobacterium Synechococcus AN PCC 6301 and maize chloroplast differ only in two bases. Biochem Biophys Res Commun. 1981 Aug 14;101(3):846–852. doi: 10.1016/0006-291x(81)91827-1. [DOI] [PubMed] [Google Scholar]
- Brosius J., Palmer M. L., Kennedy P. J., Noller H. F. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4801–4805. doi: 10.1073/pnas.75.10.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbon P., Ebel J. P., Ehresmann C. The sequence of the ribosomal 16S RNA from Proteus vulgaris. Sequence comparison with E. coli 16S RNA and its use in secondary model building. Nucleic Acids Res. 1981 May 25;9(10):2325–2333. doi: 10.1093/nar/9.10.2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbon P., Ehresmann C., Ehresmann B., Ebel J. P. The complete nucleotide sequence of the ribosomal 16-S RNA from Excherichia coli. Experimental details and cistron heterogeneities. Eur J Biochem. 1979 Oct 15;100(2):399–410. doi: 10.1111/j.1432-1033.1979.tb04183.x. [DOI] [PubMed] [Google Scholar]
- Carbon P., Ehresmann C., Ehresmann B., Ebel J. P. The sequence of Escherichia coli ribosomal 16 S RNA determined by new rapid gel methods. FEBS Lett. 1978 Oct 1;94(1):152–156. doi: 10.1016/0014-5793(78)80926-0. [DOI] [PubMed] [Google Scholar]
- Choi Y. C., Busch H. Modified nucleotides in T1 RNase oligonucleotides of 18S ribosomal RNA of the Novikoff hepatoma. Biochemistry. 1978 Jun 27;17(13):2551–2560. doi: 10.1021/bi00606a015. [DOI] [PubMed] [Google Scholar]
- De Jonge P., Klootwijk J., Planta R. J. Sequence of the 3'-terminal 21 nucleotides of yeast 17S ribosomal RNA. Nucleic Acids Res. 1977 Oct;4(10):3655–3663. doi: 10.1093/nar/4.10.3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dron M., Rahire M., Rochaix J. D. Sequence of the chloroplast 16S rRNA gene and its surrounding regions of Chlamydomonas reinhardii. Nucleic Acids Res. 1982 Dec 11;10(23):7609–7620. doi: 10.1093/nar/10.23.7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubin D. T., HsuChen C. C. The 3'-terminal region of mosquito mitochondrial small ribosomal subunit RNA: sequence and localization of methylated residues. Plasmid. 1983 May;9(3):307–320. doi: 10.1016/0147-619x(83)90008-2. [DOI] [PubMed] [Google Scholar]
- Ehresmann C., Fellner P., Ebel J. -P. The 3'-terminal nucleotide sequence of the 16 S ribosomal RNA from Escherichia coli. FEBS Lett. 1971 Apr 2;13(6):325–328. doi: 10.1016/0014-5793(71)80252-1. [DOI] [PubMed] [Google Scholar]
- Ehresmann C., Stiegler P., Mackie G. A., Zimmermann R. A., Ebel J. P., Fellner P. Primary sequence of the 16S ribosomal RNA of Escherichia coli. Nucleic Acids Res. 1975 Feb;2(2):265–278. doi: 10.1093/nar/2.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eperon I. C., Anderson S., Nierlich D. P. Distinctive sequence of human mitochondrial ribosomal RNA genes. Nature. 1980 Jul 31;286(5772):460–467. doi: 10.1038/286460a0. [DOI] [PubMed] [Google Scholar]
- Fox G. E., Stackebrandt E., Hespell R. B., Gibson J., Maniloff J., Dyer T. A., Wolfe R. S., Balch W. E., Tanner R. S., Magrum L. J. The phylogeny of prokaryotes. Science. 1980 Jul 25;209(4455):457–463. doi: 10.1126/science.6771870. [DOI] [PubMed] [Google Scholar]
- Graf L., Kössel H., Stutz E. Sequencing of 16S--23S spacer in a ribosomal RNA operon of Euglena gracilis chloroplast DNA reveals two tRNA genes. Nature. 1980 Aug 28;286(5776):908–910. doi: 10.1038/286908a0. [DOI] [PubMed] [Google Scholar]
- Gupta R., Lanter J. M., Woese C. R. Sequence of the 16S Ribosomal RNA from Halobacterium volcanii, an Archaebacterium. Science. 1983 Aug 12;221(4611):656–659. doi: 10.1126/science.221.4611.656. [DOI] [PubMed] [Google Scholar]
- Hagenbüchle O., Santer M., Steitz J. A., Mans R. J. Conservation of the primary structure at the 3' end of 18S rRNA from eucaryotic cells. Cell. 1978 Mar;13(3):551–563. doi: 10.1016/0092-8674(78)90328-8. [DOI] [PubMed] [Google Scholar]
- Helser T. L., Davies J. E., Dahlberg J. E. Change in methylation of 16S ribosomal RNA associated with mutation to kasugamycin resistance in Escherichia coli. Nat New Biol. 1971 Sep 1;233(35):12–14. doi: 10.1038/newbio233012a0. [DOI] [PubMed] [Google Scholar]
- Heus H. A., Van Kimmenade J. M., van Knippenberg P. H., Hinz H. J. Calorimetric measurements of the destabilisation of a ribosomal RNA hairpin by dimethylation of two adjacent adenosines. Nucleic Acids Res. 1983 Jan 11;11(1):203–210. doi: 10.1093/nar/11.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heus H. A., van Kimmenade J. M., van Knippenberg P. H., Haasnoot C. A., de Bruin S. H., Hilbers C. W. High-resolution proton magnetic resonance studies of the 3'-terminal colicin fragment of 16 S ribosomal RNA from Escherichia coli. Assignment of iminoproton resonances by nuclear Overhauser effect experiments and the influence of adenine dimethylation on the hairpin conformation. J Mol Biol. 1983 Nov 15;170(4):939–956. doi: 10.1016/s0022-2836(83)80197-1. [DOI] [PubMed] [Google Scholar]
- Kagramanova V. K., Mankin A. S., Baratova L. A., Bogdanov A. A. The 3'-terminal nucleotide sequence of the Halobacterium halobium 16 S rRNA. FEBS Lett. 1982 Jul 19;144(1):177–180. doi: 10.1016/0014-5793(82)80595-4. [DOI] [PubMed] [Google Scholar]
- Kelly J. M., Cox R. A. The nucleotide sequence at the 3'-end of Neurospora crassa 18S-rRNA and studies on the interaction with 5S-rRNA. Nucleic Acids Res. 1982 Nov 11;10(21):6733–6745. doi: 10.1093/nar/10.21.6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klootwijk J., Klein I., Grivell L. A. Minimal post-transcriptional modification of yeast mitochondrial ribosomal RNA. J Mol Biol. 1975 Sep 25;97(3):337–350. doi: 10.1016/s0022-2836(75)80044-1. [DOI] [PubMed] [Google Scholar]
- Kobayashi M., Seki T., Yaginuma K., Koike K. Nucleotide sequences of small ribosomal RNA and adjacent transfer RNA genes in rat mitochondrial DNA. Gene. 1981 Dec;16(1-3):297–307. doi: 10.1016/0378-1119(81)90085-8. [DOI] [PubMed] [Google Scholar]
- Küntzel H., Köchel H. G. Evolution of rRNA and origin of mitochondria. Nature. 1981 Oct 29;293(5835):751–755. doi: 10.1038/293751a0. [DOI] [PubMed] [Google Scholar]
- Leaver C. J., Harmey M. A. Higher-plant mitochondrial ribosomes contain a 5S ribosomal ribonucleic acid component. Biochem J. 1976 Jul 1;157(1):275–277. doi: 10.1042/bj1570275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockard R. E., Connaughton J. F., Kumar A. Nucleotide sequence of the 5'- and 3'- domains for rabbit 18S ribosomal RNA. Nucleic Acids Res. 1982 Jun 11;10(11):3445–3457. doi: 10.1093/nar/10.11.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughney K., Lund E., Dahlberg J. E. tRNA genes are found between 16S and 23S rRNA genes in Bacillus subtilis. Nucleic Acids Res. 1982 Mar 11;10(5):1607–1624. doi: 10.1093/nar/10.5.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller H. F., Woese C. R. Secondary structure of 16S ribosomal RNA. Science. 1981 Apr 24;212(4493):403–411. doi: 10.1126/science.6163215. [DOI] [PubMed] [Google Scholar]
- Orozco E. M., Jr, Rushlow K. E., Dodd J. R., Hallick R. B. Euglena gracilis chloroplast ribosomal RNA transcription units. II. Nucleotide sequence homology between the 16 S--23 S ribosomal RNA spacer and the 16 S ribosomal RNA leader regions. J Biol Chem. 1980 Nov 25;255(22):10997–11003. [PubMed] [Google Scholar]
- Rubtsov P. M., Musakhanov M. M., Zakharyev V. M., Krayev A. S., Skryabin K. G., Bayev A. A. The structure of the yeast ribosomal RNA genes. I. The complete nucleotide sequence of the 18S ribosomal RNA gene from Saccharomyces cerevisiae. Nucleic Acids Res. 1980 Dec 11;8(23):5779–5794. doi: 10.1093/nar/8.23.5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salim M., Maden B. E. Nucleotide sequence of Xenopus laevis 18S ribosomal RNA inferred from gene sequence. Nature. 1981 May 21;291(5812):205–208. doi: 10.1038/291205a0. [DOI] [PubMed] [Google Scholar]
- Samols D. R., Hagenbuchle O., Gage L. P. Homology of the 3' terminal sequences of the 18S rRNA of Bombyx mori and the 16S rRNA of Escherchia coli. Nucleic Acids Res. 1979 Nov 10;7(5):1109–1119. doi: 10.1093/nar/7.5.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnare M. N., Gray M. W. 3'-Terminal sequence of wheat mitochondrial 18S ribosomal RNA: further evidence of a eubacterial evolutionary origin. Nucleic Acids Res. 1982 Jul 10;10(13):3921–3932. doi: 10.1093/nar/10.13.3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnare M. N., Gray M. W. 3'-terminal nucleotide sequence of Crithidia fasciculata small ribosomal subunit RNA. FEBS Lett. 1981 Jun 15;128(2):298–304. doi: 10.1016/0014-5793(81)80103-2. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. Identical 3'-terminal octanucleotide sequence in 18S ribosomal ribonucleic acid from different eukaryotes. A proposed role for this sequence in the recognition of terminator codons. Biochem J. 1974 Sep;141(3):609–615. doi: 10.1042/bj1410609a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sor F., Fukuhara H. Séquence nucléotidique du gène de l'ARN ribosomique 15S mitochondrial de la levure. C R Seances Acad Sci D. 1980 Dec 8;291(12):933–936. [PubMed] [Google Scholar]
- Stiegler P., Carbon P., Ebel J. P., Ehresmann C. A general secondary-structure model for procaryotic and eucaryotic RNAs from the small ribosomal subunits. Eur J Biochem. 1981 Dec;120(3):487–495. doi: 10.1111/j.1432-1033.1981.tb05727.x. [DOI] [PubMed] [Google Scholar]
- Tohdoh N., Sugiura M. The complete nucleotide sequence of 16S ribosomal RNA gene from tobacco chloroplasts. Gene. 1982 Feb;17(2):213–218. doi: 10.1016/0378-1119(82)90074-9. [DOI] [PubMed] [Google Scholar]
- Tomioka N., Sugiura M. The complete nucleotide sequence of a 16S ribosomal RNA gene from a blue-green alga, Anacystis nidulans. Mol Gen Genet. 1983;191(1):46–50. doi: 10.1007/BF00330888. [DOI] [PubMed] [Google Scholar]
- Torczynski R., Bollon A. P., Fuke M. Nucleotide sequence of the 3'-terminal region of rat 18S ribosomal DNA. Mol Gen Genet. 1981;184(3):557–559. doi: 10.1007/BF00352540. [DOI] [PubMed] [Google Scholar]
- Van Buul C. P., Damm J. B., Van Knippenberg P. H. Kasugamycin resistant mutants of Bacillus stearothermophilus lacking the enzyme for the methylation of two adjacent adenosines in 16S ribosomal RNA. Mol Gen Genet. 1983;189(3):475–478. doi: 10.1007/BF00325912. [DOI] [PubMed] [Google Scholar]
- Van Charldorp R., Heus H. A., Van Knippenberg P. H. 16S ribosomal RNA of Escherichia coli contains a N2-methylguanosine at 27 nucleotides from the 3' end. Nucleic Acids Res. 1981 Jun 25;9(12):2717–2725. doi: 10.1093/nar/9.12.2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Charldorp R., Heus H. A., Van Knippenberg P. H. Adenosine dimethylation of 16S ribosomal RNA: effect of the methylgroups on local conformational stability as deduced from electrophoretic mobility of RNA fragments in denaturing polyacrylamide gels. Nucleic Acids Res. 1981 Jan 24;9(2):267–275. doi: 10.1093/nar/9.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Charldorp R., Heus H. A., Van Knippenberg P. H., Joordens J., De Bruin S. H., Hilbers C. W. Destabilization of secondary structure in 16S ribosomal RNA by dimethylation of two adjacent adenosines. Nucleic Acids Res. 1981 Sep 11;9(17):4413–4422. doi: 10.1093/nar/9.17.4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Charldorp R., Van Kimmenade A. M., Van Knippenberg P. H. Sequence and secondary structure of the colicin fragment of Bacillus stearothermophilus 16S ribosomal RNA. Nucleic Acids Res. 1981 Oct 10;9(19):4909–4917. doi: 10.1093/nar/9.19.4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Charldorp R., Van Knippenberg P. H. Sequence, modified nucleotides and secondary structure at the 3'-end of small ribosomal subunit RNA. Nucleic Acids Res. 1982 Feb 25;10(4):1149–1158. doi: 10.1093/nar/10.4.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Charldorp R., Verhoeven J. J., Van Knippenberg P. H., Haasnoot C. A., Hilbers C. W. A carbon-13 nuclear magnetic resonance study of the 3'-terminus of 16S ribosomal RNA of Escherichia coli specifically labeled with carbon-13 in the methylgroups of the m6(2)Am6(2)A sequence. Nucleic Acids Res. 1982 Jul 24;10(14):4237–4245. doi: 10.1093/nar/10.14.4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Etten R. A., Bird J. W., Clayton D. A. Identification of the 3'-ends of the two mouse mitochondrial ribosomal RNAs. The 3'-end of 16 S ribosomal RNA contains nucleotides encoded by the gene for transfer RNALeuUUR. J Biol Chem. 1983 Aug 25;258(16):10104–10110. [PubMed] [Google Scholar]
- Van Etten R. A., Walberg M. W., Clayton D. A. Precise localization and nucleotide sequence of the two mouse mitochondrial rRNA genes and three immediately adjacent novel tRNA genes. Cell. 1980 Nov;22(1 Pt 1):157–170. doi: 10.1016/0092-8674(80)90164-6. [DOI] [PubMed] [Google Scholar]
- Woese C. R. Endosymbionts and mitochondrial origins. J Mol Evol. 1977 Nov 25;10(2):93–96. doi: 10.1007/BF01751802. [DOI] [PubMed] [Google Scholar]
- Young R. A., Steitz J. A. Complementary sequences 1700 nucleotides apart form a ribonuclease III cleavage site in Escherichia coli ribosomal precursor RNA. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3593–3597. doi: 10.1073/pnas.75.8.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Knippenberg P. H., Heus H. A. The conformation of a conserved stem-loop structure in ribosomal RNA. J Biomol Struct Dyn. 1983 Oct;1(2):371–381. doi: 10.1080/07391102.1983.10507448. [DOI] [PubMed] [Google Scholar]



