Abstract
Separation of metabolic pathways in organelles is critical for eukaryotic life. Accordingly, the number, morphology and function of organelles have to be maintained through processes linked with membrane remodeling events. Despite their acknowledged significance and intense study many questions remain about the molecular mechanisms by which organellar membranes proliferate. Here, using the example of peroxisome proliferation, we give an overview of how proteins elongate membranes. Subsequent membrane fission is achieved by dynamin-related proteins shared with mitochondria. We discuss basic criteria that membranes have to fulfill for these fission factors to complete the scission. Because peroxisome elongation is always associated with unequal distribution of matrix and membrane proteins, we propose peroxisomal division to be non-stochastic and asymmetric. We further show that these organelles need not be functional to carry on membrane elongation and present the most recent findings concerning members of the Pex11 protein family as membrane elongation factors. These factors, beside known proteins such as BAR-domain proteins, represent another family of proteins containing an amphipathic α-helix with membrane bending activity.
Keywords: amphipathic α-helix, DRP1/DLP1, FIS1, membrane remodeling, peroxisome proliferation, Pex11
Introduction
Eukaryotic life relies on the arrangement of specialized intracellular microenvironments, the organelles, with several advantages including an increase in efficiency of metabolic activities. To ensure such functionality, processes exist that control the number, size and shape of organelles as well as their positioning during cell cycle progression. The molecular mechanisms triggering these events depend on specialized proteins, such as anchoring factors for the cytoskeleton, motor proteins or membrane shaping factors.
The above-mentioned processes share a common aspect: they require membrane remodeling and thus proteins that have the ability to shape the organelle. Proteins exist that affect membrane curvature, their specialized domain bends the phospholipid bilayer, thereby stabilizing the charged concave surface of the membrane. In the absence of such morphogenic factors, the endoplasmic reticulum (ER) would be misshaped, mitochondria or peroxisomes would be unable to divide and vesicular trafficking, endocytosis or neuronal function would not be possible.
Evidently, the field of membrane remodeling is very broad and we are unable to cover it entirely in only few pages. Therefore, we point at excellent overviews on endocytosis and vesicular trafficking involving factors such as BAR proteins (1–7). Here, we focus on processes that ensure proper maintenance of peroxisomes for cellular homeostasis. We elaborate particularly on proteins involved in the elongation of the peroxisomal membrane.
The peroxisome, a dynamically shaped organelle
Peroxisomes integrate into the organellar system in all eukaryotic organisms to perform a variety of tasks mostly associated with lipid metabolism, e.g., β-oxidation in S. cerevisiae, methanol oxidation in Y. lipolytica as well as α- and β-oxidation of very long chain fatty acids or plasmalogen synthesis in mammals, and detoxification of reactive oxygen species (ROS) (8–11). A role for peroxisomes in ageing and inflammation response has also been suggested (12–14). Consequently, the absence of functional peroxisomes causes severe diseases eventually leading to early death, e.g., Zellweger spectrum diseases such as the Zellweger syndrome, neonatal adrenoleukodystrophy or infantile Refsum disease (15–17). Similarly, yeast mutant cells lacking peroxisomes are unable to grow on media containing fatty acids as the sole carbon source, but they can easily ferment if the culture medium is supplemented with sugars such as glucose (18, 19).
To perform their wide-ranging tasks, peroxisomes are adaptable organelles. Indeed, they exchange material with the endoplasmic reticulum (ER) and the mitochondria (20–24). They also adjust their size, shape, number and even their protein content according to the organism, the tissue or the environmental conditions (8, 25). To ensure such high versatility the maintenance of the peroxisomal compartment must be precisely regulated. Regulatory steps include the selective degradation of superfluous or elderly peroxisomes via micro and macropexophagy, a mechanism conserved throughout kingdoms (26, 27). In addition, tight regulation of peroxisome inheritance during cell division was shown to occur in yeast through the function of specialized proteins controlling peroxisome positioning in the mother cell or in the bud (28). Furthermore, when their function is required peroxisomes can proliferate. Their propagation is either constitutive during cell cycle progression or inducible upon environmental pressure, e.g., growth of yeasts on fatty acids; fibrate supply for rodents or UV-light, high-levels of ROS, and xenobiotics in mammals (8, 11, 29, 30).
Biogenesis of peroxisomes, a need for membrane proliferation
What is the origin of the peroxisomal compartment? The peroxisome field has been highly studied and debated over the last decades but the mechanistics of peroxisome biogenesis and proliferation still requires investigation. However, owing to the characterization of mutant cells, the use of GFP- or photoactivatable GFP-fused proteins in vivo, it is now clear that two main routes lead to peroxisome formation: (i) de novo biogenesis from the ER and (ii) growth and division from existing peroxisomes (19, 31–39).
Studies either report on de novo biogenesis or on growth and division, yet focusing on only one side of peroxisome proliferation. However, the two pathways leading to formation of peroxisomes might not be controlled by completely independent mechanisms. How could the growth and division model possibly work without membrane recruitment? Although a role for the ER in the import of peroxisomal membrane proteins has been suggested (40, 41), little is known on how peroxisomes exchange material with the ER or acquire their membrane lipids.
Generally, most proteins involved in peroxisome biogenesis and proliferation belong to the group of PEX genes-encoded peroxins, most of which act as part of the peroxisomal matrix protein import machinery (42). Only a subset of peroxins, to which the Pex11-protein family belongs, controls the size, shape and number of peroxisomes. Conceptually, peroxisome proliferation can be divided into five steps: (i) organellar polarization, (ii) membrane protrusion, (iii) membrane elongation, (iv) protein import and (v) membrane scission (43). While the Pex11 proteins have been suggested to control the first steps (44), the actual peroxisomal membrane scission is performed by factors also known to operate in mitochondrial fission (45–47).
The Pex11 protein was first identified in the yeast S. cerevisiae. Deletion of the PEX11 gene led to the occurrence of fewer and enlarged peroxisomes and upon overexpression of Pex11p, the cells contained more and smaller peroxisomes than wild type cells (48). Homologues of ScPex11p are known in most eukaryotic organisms and these usually contain more than one Pex11 protein (44, 49–60). Depending on the species, up to three proteins of the Pex11 family were identified in yeasts, e.g., Pex11p, Pex25p and Pex27p in S. cerevisiae; Pex11p, Pex11Cp and Pex25p in H. polymorpha; Pex11p and Pex11Cp in Y. lipolytica; and Pex11p in P. pastoris (61). Plants typically contain five Pex11 proteins, PEX11a to -e, whereas mammals harbor three, namely PEX11α, PEX11β and PEX11γ. Noteworthy, PEX11α, PEX11β and PEX11γ are related to ScPex11p only, and no homolog has been identified for ScPex25p or ScPex27p in mammals, so far.
Dynamin-related proteins are involved in mitochondrial and peroxisomal fission
The role of Pex11 proteins in peroxisome proliferation was strengthened by the results of several studies notably showing that human PEX11β was able to interact with hFis1, a component of the peroxisomal fission machinery (43, 62). In human, the peroxisomal fission apparatus consists of hFis1, a tail-anchored recruitment factor, and the dynamin-related protein DRP1/DLP1, the actual scission factor (62–67). Recently, a new protein, Mff (mitochondrial fission factor), has been identified that acts in both, mitochondrial and peroxisomal fission processes (68). Furthermore, in knockdown studies Mff RNAi seemed to have a stronger effect than hFis1 RNAi. Similar to DRP1 knockdown, they induced tubulation of peroxisomes suggesting that Mff is an important player in the process of peroxisome proliferation (68, 69).
Similarly, in plant proteins of the dynamin family, DRP3A/B and DRP5, were identified as proliferation factors and shown to be accountable for peroxisome fission (70–73). Again, these are recruited to the peroxisomal membrane by FIS1 proteins, homologues of the mammalian hFis1 (70, 72, 74). In biomolecular fluorescence complementation assays with split YFP, FIS1B interacted with all five plant Pex11 proteins (70).
In yeasts, the dynamin-like protein Dnm1p was identified as the peroxisomal membrane scission factor. Dnm1p is recruited by Fis1p through adaptor proteins, either Mdv1p or its paralogue Caf4p (45, 75–79). In addition, a second and apparently independent pathway was identified relying on the function of another dynamin-related protein, Vps1p, as fission factor (80–82). In contrast to plant or human, no interaction between the scission factors and Pex11 proteins could be established in yeast, so far. Yet, a very recent study in S. cerevisiae reported the characterization of peroxin 34, a peroxisomal membrane protein (83). Its interaction with the Pex11 proteins as well as with Fis1p was illustrated in yeast-two-hybrid assays establishing the first link between Pex11 proteins and the fission machinery in yeast. Noteworthy, Pex34p seems to only exist in yeasts and no homolog could be identified in higher eukaryotes (83).
Together with the fission machinery, the cytoskeleton also plays a crucial role in organellar maintenance. Indeed, peroxisomes attach to the cytoskeleton and move along cytoskeletal tracks i.e., microtubules in human (84, 85) or actin in plant (86) and yeast (87). Additionally, it has been shown that organellar fission depends on the cytoskeleton as exemplified by Dnm1p-dependent scission of mitochondria in S. pombe (88). In human cells, functional microtubules and dynein motors were shown to be essential for peroxisome biogenesis (89).
Interactions of Pex11 proteins with fission factors give some insight into a molecular mechanism for peroxisomal proliferation; however, many questions remain unanswered. How does the peroxisomal membrane arrange for scission? What are the factors involved in the membrane remodeling process? Do proteins of the Pex11 family organize this whole process and why do most organisms contain more than one Pex11 protein? In the following sections we integrate the most recent findings that tackle these questions.
Mechanistic aspects of peroxisome division, Pex11 steering membrane elongation
Although several modes of proliferation are possible for organelles, the peroxisome relies on an apparently simple growth and division process. A simplified model depicts a single round-shaped peroxisome starting to elongate (Figure 1A). Once a critical size is reached, the membrane tightens and constricts until scission occurs through the action of the fission machinery. This leads to stochastic distribution of lipids and proteins between the two newly formed organelles. Evidently, this model is questionable: how does a typically round-shaped organelle start to elongate and what are the factors that squeeze the membrane and generate the constriction? In a more realistic model, extension of the peroxisome would be controlled in a concerted manner such that both, membrane elongation and assembly of the fission machinery take place at the site of membrane protrusion. Then, scission would occur across the axe of elongation generating a new daughter organelle (Figure 1B). Here, two alternatives can be foreseen, namely (i) non-polarized elongation equally dividing the peroxisomal matrix content or (ii) polarized elongation of the membrane followed by protein import at the site of membrane outgrowth. In such a model the peroxisome does not require a constriction factor per se since the thin membrane protrusion already fulfills the criteria for scission i.e., suitable membrane diameter to adapt the fission factors. Nevertheless, in both models proposed the membrane must elongate and factors are required to initiate its outgrowth. The findings that the Pex11 proteins interact with the fission machinery in plant and mammal suggest that they act as recruitment factor for the fission apparatus. But, this does not explain how the peroxisomal membrane arranges for fission.
Figure 1.
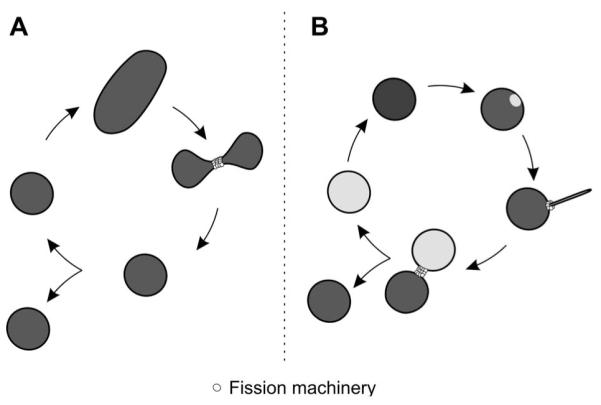
Stochastic versus asymmetric peroxisome proliferation.
(A) In a simplified model for peroxisome proliferation, a peroxisome grows and elongates and, upon a critical size, the membrane is constricted and divides through fission. Herein, the inheritance of membranes and proteins is stochastic. (B) In asymmetric proliferation, the peroxisome becomes polarized and its membrane elongates at a specific site reorganizing membrane proteins. The fission machinery assembles at the site of membrane protrusion and import of new matrix proteins assembles a daughter peroxisome which separates from the mother organelle through membrane scission.
Assessment of the information known about the fission machinery, especially proteins of the dynamin family, might allow for mechanistic assumptions. Dynamin proteins, including DRP1, are self-assembling and self-activating large GTPases. They typically carry three distinct domains, an N-terminal GTPase domain, a middle domain and a GTPase effector domain (GED) at their C-terminus (90). These three domains arrange into an evolutionary conserved structure: the middle domain and the GED region form a neck and a trunk, respectively, whereas the GTPase domain lies on the top. All dynamin-related proteins dimerize along their GTPase domain, further stabilized by their GED region (91, 92). This dimerization step seems to correlate with nucleotide binding and was proposed to arrange the catalytic machinery for GTP hydrolysis (93, 94). Recent structural data however, suggest that the dynamin dimers build spirals around the membrane in its GDP-bound form, which implies that GTP hydrolysis is not the trigger for membrane fission (95). The exact structure of the dynamin spiral is still a matter of discussion. Nonetheless, it creates such high curvature and instability in the membrane that the sudden breakdown of the spiral through GDP dissociation is ultimately resolved by membrane fission (96, 97). Electron microscopy analyses showed that Dnm1p-spirals are exactly fitting mitochondrial constriction sites exhibiting a diameter of about 110 nm. In vitro, high non-physiological levels of Dnm1p were able to elongate liposomes (1 μm in diameter) to 110 nm wide tubules (98). Elegant experiments making use of giant unilamellar vesicles (GUVs) demonstrated that dynamin polymerization requires high membrane curvature. The authors demonstrated that adsorption of dynamin monomers to the bare tubes did not significantly affect curvature of the membrane, however, clusters of dynamins occurred by pulling tubes from these GUVs thereby decreasing the tube radius (99). In agreement, at physiological concentrations, dynamin proteins were shown to only assemble and function on already curved membranes (100, 101). In fact, BAR-domain proteins were reported to prepare the membrane and target the function of dynamin such as amphiphysin in the scission of clathrin coated vesicles (101). No BAR-domain protein has been identified that acts on the peroxisomal membrane.
The conformation of dynamin proteins appears to be regulated through GTP hydrolysis performed by the intrinsic GED region thought to function as internal GTPase-activating protein (GAP). However, Lee et al. reported a role for phospholipase D as external GAP for dynamin increasing its GTPase activity in a more effective manner than the inherent GED. The molecular mechanism appears similar to that of other GAPs based on the positioning of an arginine finger (102). Interestingly, Erdmann and colleagues showed in S. cerevisiae that Lpx1p, a phospholipase, is targeted to peroxisomes (103). Although this enzyme was suggested to have a metabolic function, the authors report drastic changes in peroxisome morphology including membrane invaginations and formation of intra-peroxisomal vesicles in mutant cells lacking LPX1. It is thus tempting to speculate that besides its metabolic activities Lpx1p influences the remodeling of the peroxisomal membrane during proliferation.
Several studies connected the function of Pex11 proteins not only to the recruitment of the fission machinery, but also to peroxisomal membrane remodeling, elongation, prior to fission (52, 104, 105). In previous studies we showed that overexpression of Pex11 proteins from yeast, plant and human resulted in elongation and thereafter clustering of peroxisomes in human cells (43). Peroxisome clustering had already been reported for HsPEX11γ only (60). Close analysis of the peroxisomal clusters in 3D-reconstitutions and electron microscopy revealed that these are composed of individual, elongated peroxisomes that intertwined in a superstructure that we called juxtaposed elongated peroxisomes (JEP). Fluorescence recovery after photobleaching (FRAP) experiments demonstrated that the membranes of the individual peroxisomes in JEPs did not share components (43). Furthermore, we observed an evident separation of matrix and membrane proteins, with the matrix proteins accumulating at one or both extremities of the tubular peroxisomes (Figure 2). In parallel, Schrader and coworkers described the formation of tubular peroxisome accumulations after overexpression of PEX11β tagged with YFP at its extreme C-terminus (106). The authors also state the separation between matrix proteins in the tubular peroxisomes and report the differential localization of some peroxisomal membrane proteins. Interestingly, the early peroxisome biogenesis factors, PEX3, PEX16 and PEX19, were rather found on the stretched and elongated part of the peroxisome, whereas other membrane proteins, e.g., PMP70, PMP22 localized to the globular part. A very recent study in H. polymorpha on differential localization of various peroxisomal membrane proteins during membrane elongation showed that the spatiotemporal dynamic of membrane proteins ultimately depends on Pex11p function (107).
Figure 2.
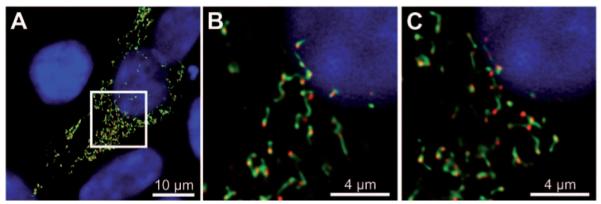
Pex11 induces peroxisome elongation.
(A) Maximum intensity projection of a confocal microscopic image showing the effect of ectopic expression of EGFP–HsPEX11β (green channel) on peroxisomes in human embryonic kidney cells (HEK293T). The elongated peroxisomal membrane shows segregation of the matrix marker, mCherry–Px (red channel). (B, C) Single z-layers from the insert region indicated in panel (A).
Asymmetric division of peroxisomes – segregation of the matrix protein content
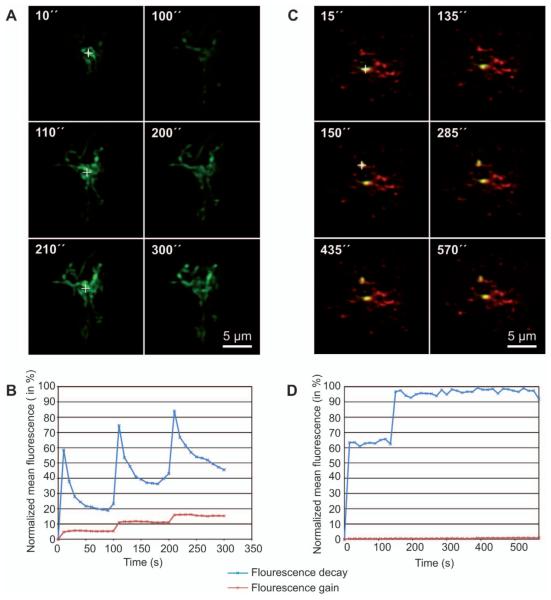
The finding that upon Pex11 overexpression matrix proteins were unequally distributed alongside JEP cast some doubts about the current view that peroxisome division is stochastic. The observation could be merely due to a dilution effect with low amounts of matrix proteins in the elongated structures being below the detection limit in fluorescence microscopy. Alternatively, during the process of membrane protrusion matrix proteins could be sequestered leading to their exclusion from the thin tubular elongation. To differentiate between the two possibilities we measured repetitive fluorescence decay after photoactivation (rFDAP) of photoactivatable-GFP targeted either to mitochondria (paGFP-Mito) or to peroxisomes (paGFP-Px). A small region in mitochondria was photoactivated and the GFP signal was monitored in living mammalian cells. Mitochondria constantly fuse and divide giving them a network-like appearance. Hence, the paGFP-Mito signal could quickly diffuse through the mitochondrial network (Figure 3A, B). In contrast, in cells co-expressing mRFP-HsPEX11β and paGFP-Px the activated GFP signal did not decline with time suggesting that paGFP-Px remained static and sequestered at one side of the elongated peroxisomal membrane (Figure 3C, D). In agreement with this observation, using the HALO-tag, Delille et al. demonstrated that the matrix content in the globular part of the elongated peroxisomes was present before membrane elongation occurred (106). In summary, under the effect of PEX11 peroxisomes elongate in a polarized fashion leaving their matrix content trapped at its original position although we cannot exclude that limited diffusion of small amounts of matrix content occurs during the elongation process. Hence, elongation of the peroxisomal membrane seems to create a matrix protein gradient, thereby segregating the ‘old’ matrix from the ‘new’ membrane. Segregation of matrix proteins during peroxisome elongation could ensure that old and possibly damaged proteins do not populate the new organelle. New matrix proteins would then target to the tip of the new membrane thereby inflating the new peroxisome and modeling the membrane constriction required for fission.
Figure 3.
Peroxisomal matrix proteins are kept back during peroxisome elongation.
HEK293T cells expressing either the mitochondrial matrix marker, paGFP-mito (A, B), or the peroxisomal matrix marker, paGFP-Px and mRFP-HsPEX11γ (C, D) were analyzed 48 h after transfection. Diffusion of matrix proteins was analyzed in repetitive fluorescence decay after photoactivation (rFDAP) experiments. paGFP was activated in a small area and fluorescence was monitored for decay along with measurement of fluorescence gain in the rest of the cell. (A) For mitochondria, repetitive activation of a single area (white crosses) led to rapid diffusion of the paGFP signal throughout the mitochondrial network. (B) Quantification of (A) showing fluorescence decay in the activated region (blue line) and gain of fluorescence in the non-activated region (red line). (C) paGFP was activated in JEPs caused by overexpression of mRFP-HsPEX11γ. Since no decay was measured in the activated region (white cross 15″), a second area was activated (white cross 150″). (D) Quantification clearly shows that no signal was lost during acquisition and no diffusion took place. Image acquisition parameters: LSM DuoScan (ApoChromat 63×1.4; settings: paGFP (489 nm, MBS490, BP 500–525), mRFP (532 nm, MBS 535, BP 560–675); activation: 405 nm).
The observations by Delille et al. upon expression of a PEX11β-YFP suggest that the chimera inhibits peroxisomal fission while allowing their elongation. We showed that PEX11-driven peroxisomal elongation and even JEPs could be dissolved by providing high amounts of hFis1 to the cells (43). Interestingly, overexpression of the dynamin protein, DRP1, led to the appearance of elongated peroxisomes or to an increase in JEP size in cells expressing PEX11 proteins rather than to fission. These findings place hFis1 as limiting factor in the process of peroxisomal fission and highlight the importance of PEX11 as recruitment factor.
Recent experiments on mitochondrial fission described that Mff, another tail-anchored protein, is the ultimate recruiter of hFis1 for membrane fission (69). Although this latter study focused on mitochondrial fission, it had been shown earlier that Mff also played a role in peroxisome proliferation (68). Indeed, mammalian cells transfected with Mff RNAi presented peroxisomes that were more elongated than peroxisomes in cells depleted for hFis1. In consequence, assuming that the interplay between Mff, hFis1 and DRP1 is comparable in mitochondrial and peroxisomal fission, hFis1 might rather modulate DRP1 function than act as recruitment factor. In the light of these new observations it would be intriguing to test whether Pex11 proteins interact with Mff. Interaction with hFis1 only would suggest that Pex11 proteins act as membrane elongation factors, which stimulate the fission machinery. But, interaction with both, Mff and hFis1, would strengthen the role of Pex11 proteins in powering fission of the peroxisomal membrane.
Pex11 proteins elongate membranes in vitro
All these findings strongly point at the involvement of the Pex11 proteins in the membrane elongation event. A first hint about the molecular function of Pex11p was presented by Opalinski et al. (2010). The authors report the presence of an amphipathic α-helix at the N-terminus of several Pex11 proteins from yeast to mammal (108). Incubation of peptides containing the Pex11 amphipathic region with small unilamellar vesicles (SUVs) clearly showed an ability to restructure membranes. The initially round SUVs elongated and formed tubules in the presence of the Pex11 peptides. Similar results were obtained using the purified first 95 amino acids of P. chrysogenum Pex11p. The size and shape of the elongated SUVs could be altered by introducing bulky tryptophan residues in the amphipathic peptide. Changes in the peptide composition, such as introduction of negative charges or proline residues, annihilated the effect on membrane elongation. In vivo, expression of a mutated Pex11p protein lacking this alpha-helix was unable to protrude the peroxisomal membrane suggesting a mechanistic role for this helical structure in membrane elongation.
Amphipathic helices have been reported in a variety of proteins, well-known examples being the BAR proteins (1, 109–112). It has been suggested that two types of amphipathic helices exist namely, curvature sensors or inducers (113). Upon insertion of the helix into one leaflet of the lipid bilayer, the space requirement of this leaflet increases with respect to the other, which leads to membrane bending (Figure 4). This insertion requires the lipids to be pushed aside. If the energy cost is compensated by the presence of positively charged amino acids, it favors interaction between the charged head groups of the lipids and the polar face of the amphipathic helix, the helix can actively curve the membrane (Figure 4A). Alternatively, the helix contains mainly negatively charged residues, which hinder its insertion into a flat membrane. Hence, such helices are unable to induce membrane curvature and require a membrane already curved to insert. These amphipathic helices are membrane curvature sensors (Figure 4B). Evidently, this mechanism depends on the nature of the membrane including its lipid composition and local enrichment in specific lipids. Indeed, the often neglected physical properties of membrane lipids might determine the limits in which proteins can act (114). A well-studied example of curvature sensors is the ArfGAP1 lipid packing sensor (ALPS) motif, which contains numerous serine and threonine residues that favor its adsorption onto membranes with strong positive curvature (113). Curvature inducers are for instance the BAR domain proteins. The N-BAR domain e.g., in endophilin adopts a banana-wedge shape that bends the membrane to give it a curved form. Interestingly, mathematic modeling suggests that induction of membrane curvature relies on the sole property of the amphipathic helix and not on the entire N–BAR domain (115).
Figure 4.
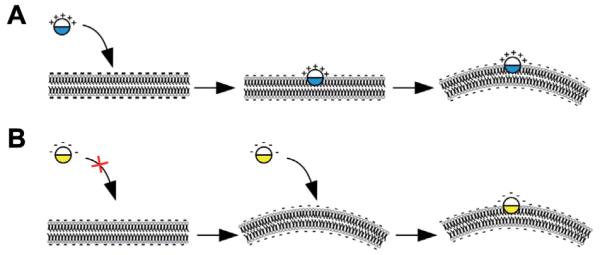
Amphipathic helices as membrane curvature sensors or inducers.
(A) A positively charged amphipathic helix leads to membrane bending upon insertion into one leaflet of the lipid bilayer. The energy cost for helix insertion can be compensated through electrostatic interactions. (B) If the amphipathic helix displays a negatively charged surface, it cannot deform the membrane, and acts as membrane curvature sensor.
Consequently, amphipathic helices play pivotal roles in a plethora of intracellular processes and their presence in Pex11 proteins seems to be crucial for proliferation of the peroxisomal membrane. The generation of high curvature in the peroxisomal membrane could explain the redistribution of peroxisomal membrane proteins along the peroxisome tubules. Recent quantitative fluorescence microscopy analyses showed that membrane curvature as such can account for redistribution of integral or membrane anchored proteins (116). In the context of peroxisome proliferation such reorganization could lead to (i) attraction of the fission machinery and (ii) redistribution of membrane proteins including the import machinery to ensure efficient transport of matrix proteins into the newly formed peroxisome. Because the polar face of the Pex11 amphipathic helix contains lysine and arginine residues, it seems to rather induce membrane curvature. However, membrane curvature still needs to be tightly regulated. No polarized outgrowth would occur if all Pex11 amphipathic helices equally distributed and inserted into the peroxisomal membrane. Therefore, spatiotemporally confined protrusion has to be established to ensure elongation of the peroxisomal membrane. Thus, a strict control is required for Pex11 protein positioning on the membrane or for molecular interactions. This could arise through post-translational modifications. A study in the yeast S. cerevisiae showed that Pex11p is modified through phosphorylation. Cells expressing a phospho-mimicry mutant of Pex11p displayed more and smaller (S→D, ‘phosphorylated’) or less and bigger (S→A, ‘non-phosphorylated’) peroxisomes than wild type cells (117).
Several Pex11 proteins interact to orchestrate peroxisome proliferation
The interplay of the various Pex11 proteins in organisms that contain more than one Pex11 protein remains to be elucidated. Earlier studies showed the homodimerization properties of several Pex11 proteins including the human PEX11β and ScPex11p (62, 118). In addition to ScPex11p homodimerization, yeast-2-hybrid analyses showed homo-dimerization of ScPex25p and ScPex27p, respectively, but no hetero-oligomerization (56). In human cells, all three Pex11 proteins homo-oligomerized and both, PEX11α and PEX11β were shown to interact with PEX11γ. Co-immunoprecipitation experiments also revealed that the three proteins interacted with the fission machinery (43). In vitro binding assays demonstrated a direct interaction between HsPEX11β and hFis1 (62). Importantly, all these experiments were performed using digitonin, a mild detergent that preserves lipid environment, and the addition of Triton X-100 abolished interactions. This implies the requirement of membrane lipids for interactions. The orientation of several Pex11 proteins has been studied based on differential cell permeabilization with digitonin or protease accessibility of their extreme termini (50, 52, 60, 105, 119) however, their exact topology in the membrane remains to be elucidated. Hence, such information would be important to comprehend the mutual influence of Pex11 proteins and the fission machinery.
It is still unclear whether all Pex11 proteins are equally important for peroxisome proliferation. In yeast, the absence of Pex11p resulted in reduced growth of the cells on oleic acid (48) and in the abscence of Pex11p, Pex25p and Pex27p cells were unable to grow on oleate-containing medium. Interestingly, Pex25p alone was able to rescue the oleate non-utilizing phenotype of the pex11Δpex25Δpex27Δ mutant cells (56). In mammal, while PEX11α expression is inducible, PEX11β is constitutively present in the cell (49, 50, 120, 121). Knockout mouse models showed that in the absence of PEX11β mice developed pathologies similar to those of Zellweger patients and the number of peroxisomes per cell was significantly decreased (121, 122). Deletion of PEX11α did not have a phenotype neither did it worsen the condition in PEX11α–/–/β–/– mice (120). These data suggest that in mammal, two routes exist for peroxisome proliferation, one inducible and one constitutive, driven by either PEX11α or PEX11β, respectively. Both ways might require the function of PEX11γ. Homodimerization, interaction with PEX11γ or both could allow for recruitment of the fission machinery. Analysis of PEX11β suggested that its C-terminus was required to interact with hFis1 (62). Proteineaceous interactions were proposed to depend on one of the tetratricopeptide repeat (TPR) regions of hFis1 (123). Peptide-scan analyses demonstrated that proline-rich peptides efficiently bind hFis1, specifically in the TPR region (124). Interestingly, plant and human Pex11 proteins contain proline-rich regions, among which some resemble a Fis1 binding site. In contrast, none of the S. cerevisiae Pex11 family member contains such motif suggesting that in this species Pex11 proteins might not directly recruit Fis1 to peroxisomes.
Oligomerization of Pex11 proteins could regulate their activity. In S. cerevisiae, dimerization of Pex11p was suggested to act as molecular switch. Considering that ScPex11p was localized to the inner surface of the peroxisomal membrane it could easily be influenced by the peroxisomal redox state. Hence, redox-sensitive dimerization of Pex11p could represent a signal for proliferation (118). A redox-sensitive dimerization of Pex11 proteins has not been reported in mammalian cells. However, a recent study investigated the mammalian peroxisomes redox-balance using a redox-sensitive variant of EGFP and an artificial light-triggered ROS-induction protein. The authors demonstrate that although peroxisomes resist to an oxidative stress produced elsewhere in the cell, the intraperoxisomal redox status is strongly affected by the environmental growth conditions. Interestingly, the redox state of peroxisomes did not correlate with their age (125).
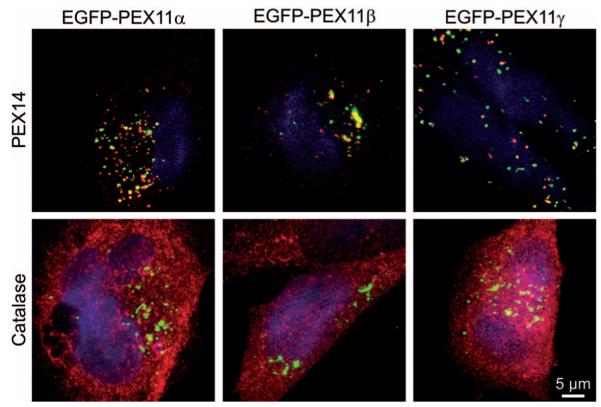
To address whether the peroxisomal matrix content exerts an influence on the function of Pex11 proteins we assessed whether PEX11 could act on empty peroxisomal membranes (remnants) in cells expressing a mutated PEX5, a receptor for peroxisomal matrix proteins (126). Most peroxisome remnants elongated and formed JEPs upon overexpression of either of the human Pex11 proteins (Figure 5). This observation points to an independent mode of regulation for peroxisome function and proliferation. Nevertheless, the expression of some Pex11 proteins is tightly regulated, which allows for coordination of the proliferation machinery and the metabolic state of peroxisomes. Alternatively, matrix proteins could affect the properties of the peroxisomal membrane thereby modulating proliferation of the organelle as already suggested for the peroxisomal enzyme acyl-CoA oxidase in Yarrowia lipolytica (127).
Figure 5.
Pex11 membrane elongation factors do not require peroxisomal matrix content to function.
Analysis of mutant fibroblast cells with mutated PEX5 containing empty peroxisomal membranes for the effect of ectopic expression of GFP-tagged human Pex11 proteins. Pex11 proteins elongated the peroxisomal membrane in the absence of matrix content as demonstrated by immunofluorescent stainings for the peroxisomal membrane protein PEX14 (red channel, upper panel) and the matrix protein catalase (red channel, lower panel). Images represent maximum intensity projection of confocal images acquired on a LSM510META [objective 100×1.45; settings: EGFP (488 nm, MBS 488, BP 500–525), AlexaFluor594 (561 nm, MBS 561, LP 585)].
Perspectives
Recent reports placed the Pex11 proteins as key actors in the process of peroxisomal membrane remodeling. These proteins elongate the peroxisomal membrane. It will be important to test how their positioning selects the site of membrane protrusion, and how they interact with the fission machinery to coordinate membrane scission. Future experiments will be required to determine whether Pex11 proteins represent a new family of amphipathic alpha-helix-containing proteins with membrane bending activities.
Furthermore, evidence exists that peroxisome elongation is polarized. Asymmetric division of the matrix protein content during membrane elongation might allow for import of new material at the site of membrane growth. We propose this mechanism to ensure selective retention instead of dilution of old matrix content. Whether the selective degradation of peroxisomes via pexophagy is specifically targeted to old organelles is an attractive question.
Although the distribution of matrix proteins seems to be highly regulated, the action of the Pex11 proteins does not depend on the functionality or maturity of the peroxisomes. As shown in our experiments, overexpressed Pex11 acts on the membrane obviously without requiring feedback from the matrix. It remains to be elucidated whether the function of the Pex11 proteins is directly or indirectly influenced by the metabolic state of the cell.
The Pex11 interactome was shown to require the integrity of the peroxisomal membrane. Thus, understanding the membrane topology of Pex11 proteins is important in order to gain insight in its role as membrane elongation factor. Eventually, structural studies will deliver the missing elements to understand how these proteins act at the molecular level.
In conclusion, the two pathways leading to peroxisome formation, de novo biogenesis and growth and division, are presumably connected at the stage of membrane uptake. Consequently, with Pex11 proteins as membrane shaping factors, it would not be surprising that some of these proteins also contribute to de novo peroxisome biogenesis from the ER. Interestingly, a very recent study on the identification of peroxisome biogenesis factors in the yeast H. polymorpha revealed the importance of Pex25p for the reintroduction of peroxisomes in mutant cells lacking these organelles (128). Noteworthy, an interaction between the rat PEX11 and Arf1/coatomer has been reported and coatomer inhibition in temperature sensitive CHO-mutant cells correlated with the occurrence of tubular peroxisomes (119).
Alterations in peroxisomal metabolism and peroxisome proliferation cause neurodegenerative diseases and might also represent a trigger for cellular ageing. Understanding how peroxisomes proliferate and, more specifically, generate membrane protrusion to facilitate scission, will have a major impact on understanding the dynamics of biological membranes. The concept of organelle polarization and asymmetric membrane growth and division might engage the re-investigation of the proliferation of other organellar membranes.
Highlights.
Most organisms contain more than one Pex11 protein and all Pex11 proteins act on the peroxisomal membrane
Pex11 proteins are regulated at transcriptional, and post-translational levels through modifications as well as homo- and heterodimerization
Pex11 proteins influence the shape of the peroxisomal membrane
Pex11 proteins coordinate the fission machinery shared between peroxisomes and mitochondria
Pex11 proteins contain an amphipathic alpha-helix suggested to bend the peroxisomal membrane
Pex11 proteins act as membrane elongation factors regardless of whether peroxisomes are functional
Asymmetric inheritance of peroxisomal matrix proteins during peroxisome proliferation might lead to rejuvenation of the peroxisome pool in the cell
Acknowledgments
We are grateful to Ronald Wanders (University of Amsterdam, The Netherlands) and Ralf Erdmann (Ruhr–University Bochum, Germany) for the PEX5 deficient fibroblast cell line. The authors wish to thank Josef Gotzmann (MFPL, Austria) for technical assistance. This work was funded by a grant from the Austrian Science Fund (FWF) P-20803 to C.B. C.B. is supported by the Elise-Richter-Program (V39-B09) of the Austrian Science Fund (FWF, http://www.fwf.ac.at/) and the Austrian Federal Ministry for Science and Research (BMWF, http://www.bmwf.gv.at/).
Biography

Johannes Koch studied Chemistry and received his Diploma from the University of Vienna in 2008. Then, he studied for a PhD in Biochemistry at the Max F. Perutz Laboratories (Vienna, Austria) under the supervision of Dr. Cécile Brocard. In 2010, he received an EMBO short-term fellowship for a collaborative work in the laboratory of Prof. Jeffrey Gerst at the Weizmann Institute of Science (Rehovot, Israel). He is currently finalizing his PhD thesis.

Cécile Brocard studied Biochemistry and Molecular Cell Biology and received her Diploma from the University of Burgundy (Dijon, France), and moved to the University of Vienna (Austria) where she was awarded a PhD in Biochemistry in 1998. Following post-doctoral training in Cell Biology from the University of Western Ontario (London, Canada), supported by a CIHR fellowship and several positions as Biochemist and Molecular Cell Biologist in Austria, she was appointed guest professor in Biochemistry at the University of Vienna. She was awarded an Elise-Richter grant from the Austrian Science Fund (FWF) and the Austrian Federal Ministry of Science and Research (BM-WF) to establish an independent research group at the Max F. Perutz Laboratories from the University of Vienna and the Medical University of Vienna. In 2010 she received her “Habilitation” in Biochemistry and Cell Biology awarded from the University of Vienna.
Footnotes
The authors declare having no financial or other conflicts of interest regarding the work presented in this publication.
References
- 1.Frost A, Unger VM, De Camilli P. The BAR domain superfamily: membrane-molding macromolecules. Cell. 2009;137:191–6. doi: 10.1016/j.cell.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dawson JC, Legg JA, Machesky LM. Bar domain proteins: a role in tubulation, scission and actin assembly in clathrin-mediated endocytosis. Trends Cell Biol. 2006;16:493–8. doi: 10.1016/j.tcb.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 3.Hurley JH, Boura E, Carlson LA, Rozycki B. Membrane budding. Cell. 2010;143:875–87. doi: 10.1016/j.cell.2010.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lundmark R, Carlsson SR. Driving membrane curvature in clathrin-dependent and clathrin-independent endocytosis. Semin Cell Dev Biol. 2010;21:363–70. doi: 10.1016/j.semcdb.2009.11.014. [DOI] [PubMed] [Google Scholar]
- 5.Kozlov MM, McMahon HT, Chernomordik LV. Protein-driven membrane stresses in fusion and fission. Trends Biochem Sci. 2010;35:699–706. doi: 10.1016/j.tibs.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prinz WA, Hinshaw JE. Membrane-bending proteins. Crit Rev Biochem Mol. 2009;44:278–91. doi: 10.1080/10409230903183472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006;124:573–86. doi: 10.1016/j.cell.2005.11.047. [DOI] [PubMed] [Google Scholar]
- 8.van den Bosch H, Schutgens RB, Wanders RJ, Tager JM. Biochemistry of peroxisomes. Annu Rev Biochem. 1992;61:157–97. doi: 10.1146/annurev.bi.61.070192.001105. [DOI] [PubMed] [Google Scholar]
- 9.Angermuller S, Islinger M, Volkl A. Peroxisomes and reactive oxygen species, a lasting challenge. Histochem Cell Biol. 2009;131:459–63. doi: 10.1007/s00418-009-0563-7. [DOI] [PubMed] [Google Scholar]
- 10.Antonenkov V, Grunau S, Ohlmeier S, Hiltunen JK. Peroxisomes are oxidative organelles. Antioxid Redox Signal. 2009;13:525–37. doi: 10.1089/ars.2009.2996. [DOI] [PubMed] [Google Scholar]
- 11.Bonekamp NA, Volkl A, Fahimi HD, Schrader M. Reactive oxygen species and peroxisomes: struggling for balance. Biofactors. 2009;35:346–55. doi: 10.1002/biof.48. [DOI] [PubMed] [Google Scholar]
- 12.Dixit E, Boulant S, Zhang Y, Lee AS, Odendall C, Shum B, Hacohen N, Chen ZJ, Whelan SP, Fransen M, Nibert ML, Superti-Furga G, Kagan JC. Peroxisomes are signaling platforms for antiviral innate immunity. Cell. 2010;141:668–81. doi: 10.1016/j.cell.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koepke JI, Nakrieko KA, Wood CS, Boucher KK, Terlecky LJ, Walton PA, Terlecky SR. Restoration of peroxisomal catalase import in a model of human cellular aging. Traffic. 2008;8:1590–600. doi: 10.1111/j.1600-0854.2007.00633.x. [DOI] [PubMed] [Google Scholar]
- 14.Titorenko VI, Terlecky SR. Peroxisome metabolism and cellular aging. Traffic. 2011;12:252–9. doi: 10.1111/j.1600-0854.2010.01144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fidaleo M. Peroxisomes and peroxisomal disorders: the main facts. Exp Toxicol Pathol. 2010;62:615–25. doi: 10.1016/j.etp.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 16.Wanders RJ, Waterham HR. Peroxisomal disorders I: biochemistry and genetics of peroxisome biogenesis disorders. Clin Genet. 2005;67:107–33. doi: 10.1111/j.1399-0004.2004.00329.x. [DOI] [PubMed] [Google Scholar]
- 17.Steinberg SJ, Dodt G, Raymond GV, Braverman NE, Moser AB, Moser HW. Peroxisome biogenesis disorders. Biochimica et Biophysica Acta. 2006;1763:1733–48. doi: 10.1016/j.bbamcr.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 18.Veenhuis M, Mateblowski M, Kunau WH, Harder W. Proliferation of microbodies in Saccharomyces cerevisiae. Yeast. 1987;3:77–84. doi: 10.1002/yea.320030204. [DOI] [PubMed] [Google Scholar]
- 19.Erdmann R, Veenhuis M, Mertens D, Kunau WH. Isolation of peroxisome-deficient mutants of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1989;86:5419–23. doi: 10.1073/pnas.86.14.5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raychaudhuri S, Prinz WA. Nonvesicular phospholipid transfer between peroxisomes and the endoplasmic reticulum. Proc Natl Acad Sci USA. 2008;105:15785–90. doi: 10.1073/pnas.0808321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lam SK, Yoda N, Schekman R. A vesicle carrier that mediates peroxisome protein traffic from the endoplasmic reticulum. Proc Natl Acad Sci USA. 2010;107:21523–8. doi: 10.1073/pnas.1013397107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perry RJ, Mast FD, Rachubinski RA. Endoplasmic reticulum-associated secretory proteins Sec20p, Sec39p, and Dsl1p are involved in peroxisome biogenesis. Eukaryot Cell. 2009;8:830–43. doi: 10.1128/EC.00024-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neuspiel M, Schauss AC, Braschi E, Zunino R, Rippstein P, Rachubinski RA, Andrade-Navarro MA, McBride HM. Cargo-selected transport from the mitochondria to peroxisomes is mediated by vesicular carriers. Curr Biol. 2008;18:102–8. doi: 10.1016/j.cub.2007.12.038. [DOI] [PubMed] [Google Scholar]
- 24.Braschi E, Goyon V, Zunino R, Mohanty A, Xu L, McBride HM. Vps35 mediates vesicle transport between the mitochondria and peroxisomes. Curr Biol. 2010;20:1310–5. doi: 10.1016/j.cub.2010.05.066. [DOI] [PubMed] [Google Scholar]
- 25.Mast FD, Fagarasanu A, Knoblach B, Rachubinski RA. Peroxisome biogenesis: something old, something new, something borrowed. Physiology (Bethesda) 2010;25:347–56. doi: 10.1152/physiol.00025.2010. [DOI] [PubMed] [Google Scholar]
- 26.Oku M, Sakai Y. Peroxisomes as dynamic organelles: autophagic degradation. FEBS J. 2010;277:3289–94. doi: 10.1111/j.1742-4658.2010.07741.x. [DOI] [PubMed] [Google Scholar]
- 27.Manjithaya R, Nazarko TY, Farre JC, Subramani S. Molecular mechanism and physiological role of pexophagy. FEBS Letters. 2010;584:1367–73. doi: 10.1016/j.febslet.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fagarasanu A, Mast FD, Knoblach B, Rachubinski RA. Molecular mechanisms of organelle inheritance: lessons from peroxisomes in yeast. Nat Rev Mol Cell Biol. 2010;11:644–54. doi: 10.1038/nrm2960. [DOI] [PubMed] [Google Scholar]
- 29.Gurvitz A, Rottensteiner H. The biochemistry of oleate induction: transcriptional upregulation and peroxisome proliferation. Biochimica et Biophysica Acta. 2006;1763:1392–402. doi: 10.1016/j.bbamcr.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 30.Seo KW, Kim KB, Kim YJ, Choi JY, Lee KT, Choi KS. Comparison of oxidative stress and changes of xenobiotic metabolizing enzymes induced by phthalates in rats. Food Chem Toxicol. 2004;42:107–14. doi: 10.1016/j.fct.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 31.Titorenko VI, Smith JJ, Szilard RK, Rachubinski RA. Peroxisome biogenesis in the yeast Yarrowia lipolytica. Cell Biochem Biophys. 2000;32:21–6. doi: 10.1385/cbb:32:1-3:21. [DOI] [PubMed] [Google Scholar]
- 32.Fujiki Y, Okumoto K, Kinoshita N, Ghaedi K. Lessons from peroxisome-deficient Chinese hamster ovary (CHO) cell mutants. Biochimica et Biophysica Acta. 2006;1763:1374–81. doi: 10.1016/j.bbamcr.2006.09.012. [DOI] [PubMed] [Google Scholar]
- 33.Geuze HJ, Murk JL, Stroobants AK, Griffith JM, Kleijmeer MJ, Koster AJ, Verkleij AJ, Distel B, Tabak HF. Involvement of the endoplasmic reticulum in peroxisome formation. Mol Biol Cell. 2003;14:2900–7. doi: 10.1091/mbc.E02-11-0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoepfner D, Schildknegt D, Braakman I, Philippsen P, Tabak HF. Contribution of the endoplasmic reticulum to peroxisome formation. Cell. 2005;122:85–95. doi: 10.1016/j.cell.2005.04.025. [DOI] [PubMed] [Google Scholar]
- 35.Kim PK, Mullen RT, Schumann U, Lippincott-Schwartz J. The origin and maintenance of mammalian peroxisomes involves a de novo PEX16-dependent pathway from the ER. J Cell Biol. 2006;173:521–32. doi: 10.1083/jcb.200601036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tabak HF, Murk JL, Braakman I, Geuze HJ. Peroxisomes start their life in the endoplasmic reticulum. Traffic. 2003;4:512–8. doi: 10.1034/j.1600-0854.2003.00110.x. [DOI] [PubMed] [Google Scholar]
- 37.Tam YY, Fagarasanu A, Fagarasanu M, Rachubinski RA. Pex3p initiates the formation of a preperoxisomal compartment from a subdomain of the endoplasmic reticulum in Saccharomyces cerevisiae. J Biol Chem. 2005;280:34933–9. doi: 10.1074/jbc.M506208200. [DOI] [PubMed] [Google Scholar]
- 38.Toro A, Arredondo C, Córdova G, Araya C, Palacios JL, Venegas A, Morita M, Imanaka T, Santos MJ. Evaluation of the role of the endoplasmic reticulum-Golgi transit in the biogenesis of peroxisomal membrane proteins in wild type and peroxisome biogenesis mutant CHO cells. Biol Res. 2007;40:231–49. doi: 10.4067/s0716-97602007000200014. [DOI] [PubMed] [Google Scholar]
- 39.Toro AA, Araya CA, Córdova GJ, Arredondo CA, Cárdenas HG, Moreno RE, Venegas A, Koenig CS, Cancino J, Gonzalez A, Santos MJ. Pex3p-dependent peroxisomal biogenesis initiates in the endoplasmic reticulum of human fibroblasts. J Cell Biochem. 2009;107:1083–96. doi: 10.1002/jcb.22210. [DOI] [PubMed] [Google Scholar]
- 40.van der Zand A, Braakman I, Tabak HF. Peroxisomal Membrane Proteins Insert into the Endoplasmic Reticulum. Mol Biol Cell. 2010;21:2057–65. doi: 10.1091/mbc.E10-02-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma C, Subramani S. Peroxisome matrix and membrane protein biogenesis. IUBMB Life. 2009;61:713–22. doi: 10.1002/iub.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Distel B, Erdmann R, Gould SJ, Blobel G, Crane DI, Cregg JM, Dodt G, Fujiki Y, Goodman JM, Just WW, Kiel JA, Kunau WH, Lazarow PB, Mannaerts GP, Moser HW, Osumi T, Rachubinski RA, Roscher A, Subramani S, Tabak HF, Tsukamoto T, Valle D, van der Klei I, van Veldhoven PP, Veenhuis M. A unified nomenclature for peroxisome biogenesis factors. J Cell Biol. 1996;135:1–3. doi: 10.1083/jcb.135.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koch J, Pranjic K, Huber A, Ellinger A, Hartig A, Kragler F, Brocard C. PEX11 family members are membrane elongation factors that coordinate peroxisome proliferation and maintenance. J Cell Sci. 2010;123:3389–400. doi: 10.1242/jcs.064907. [DOI] [PubMed] [Google Scholar]
- 44.Marshall PA, Krimkevich YI, Lark RH, Dyer JM, Veenhuis M, Goodman JM. Pmp27 promotes peroxisomal proliferation. J Cell Biol. 1995;129:345–55. doi: 10.1083/jcb.129.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mozdy AD, McCaffery JM, Shaw JM. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J Cell Biol. 2000;151:367–80. doi: 10.1083/jcb.151.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.James DI, Parone PA, Mattenberger Y, Martinou JC. hFis1, a novel component of the mammalian mitochondrial fission machinery. J Biol Chem. 2003;278:36373–9. doi: 10.1074/jbc.M303758200. [DOI] [PubMed] [Google Scholar]
- 47.Yoon Y, Krueger EW, Oswald BJ, McNiven MA. The mitochondrial protein hFis1 regulates mitochondrial fission in mammalian cells through an interaction with the dynamin-like protein DLP1. Mol Cell Biol. 2003;23:5409–20. doi: 10.1128/MCB.23.15.5409-5420.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Erdmann R, Blobel G. Giant peroxisomes in oleic acid-induced Saccharomyces cerevisiae lacking the peroxisomal membrane protein Pmp27p. J Cell Biol. 1995;128:509–23. doi: 10.1083/jcb.128.4.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abe I, Fujiki Y. cDNA cloning and characterization of a constitutively expressed isoform of the human peroxin Pex11p. Biochem Bioph Res Co. 1998;252:529–33. doi: 10.1006/bbrc.1998.9684. [DOI] [PubMed] [Google Scholar]
- 50.Abe I, Okumoto K, Tamura S, Fujiki Y. Clofibrate-inducible, 28–kDa peroxisomal integral membrane protein is encoded by PEX11. FEBS Letters. 1998;431:468–72. doi: 10.1016/s0014-5793(98)00815-1. [DOI] [PubMed] [Google Scholar]
- 51.Krikken AM, Veenhuis M, van der Klei IJ. Hansenula polymorpha pex11 cells are affected in peroxisome retention. FEBS J. 2009;276:1429–39. doi: 10.1111/j.1742-4658.2009.06883.x. [DOI] [PubMed] [Google Scholar]
- 52.Lingard MJ, Trelease RN. Five Arabidopsis peroxin 11 homologs individually promote peroxisome elongation, duplication or aggregation. J Cell Sci. 2006;119:1961–72. doi: 10.1242/jcs.02904. [DOI] [PubMed] [Google Scholar]
- 53.Nayidu NK, Wang L, Xie W, Zhang C, Fan C, Lian X, Zhang Q, Xiong L. Comprehensive sequence and expression profile analysis of PEX11 gene family in rice. Gene. 2008;412:59–70. doi: 10.1016/j.gene.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 54.Orth T, Reumann S, Zhang X, Fan J, Wenzel D, Quan S, Hu J. The PEROXIN11 protein family controls peroxisome proliferation in Arabidopsis. Plant Cell. 2007;19:333–50. doi: 10.1105/tpc.106.045831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rottensteiner H, Hartig A, Hamilton B, Ruis H, Erdmann R, Gurvitz A. Saccharomyces cerevisiae Pip2p-Oaf1p regulates PEX25 transcription through an adenine-less ORE. Eur J Biochem. 2003;270:2013–22. doi: 10.1046/j.1432-1033.2003.03575.x. [DOI] [PubMed] [Google Scholar]
- 56.Rottensteiner H, Stein K, Sonnenhol E, Erdmann R. Conserved function of pex11p and the novel pex25p and pex27p in peroxisome biogenesis. Mol Biol Cell. 2003;14:4316–28. doi: 10.1091/mbc.E03-03-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sakai Y, Marshall PA, Saiganji A, Takabe K, Saiki H, Kato N, Goodman JM. The Candida boidinii peroxisomal membrane protein Pmp30 has a role in peroxisomal proliferation and is functionally homologous to Pmp27 from Saccharomyces cerevisiae. J Bacteriol. 1995;177:6773–81. doi: 10.1128/jb.177.23.6773-6781.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith JJ, Marelli M, Christmas RH, Vizeacoumar FJ, Dilworth DJ, Ideker T, Galitski T, Dimitrov K, Rachubinski RA, Aitchison JD. Transcriptome profiling to identify genes involved in peroxisome assembly and function. J Cell Biol. 2002;158:259–71. doi: 10.1083/jcb.200204059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tam YY, Torres-Guzman JC, Vizeacoumar FJ, Smith JJ, Marelli M, Aitchison JD, Rachubinski RA. Pex11-related proteins in peroxisome dynamics: a role for the novel peroxin Pex27p in controlling peroxisome size and number in Saccharomyces cerevisiae. Mol Biol Cell. 2003;14:4089–102. doi: 10.1091/mbc.E03-03-0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tanaka A, Okumoto K, Fujiki Y. cDNA cloning and characterization of the third isoform of human peroxin Pex11p. Biochem Bioph Res Co. 2003;300:819–23. doi: 10.1016/s0006-291x(02)02936-4. [DOI] [PubMed] [Google Scholar]
- 61.Kiel JA, Veenhuis M, van der Klei IJ. PEX genes in fungal genomes: common, rare or redundant. Traffic. 2006;7:1291–303. doi: 10.1111/j.1600-0854.2006.00479.x. [DOI] [PubMed] [Google Scholar]
- 62.Kobayashi S, Tanaka A, Fujiki Y. Fis1, DLP1, and Pex11p coordinately regulate peroxisome morphogenesis. Exp Cell Res. 2007;313:1675–86. doi: 10.1016/j.yexcr.2007.02.028. [DOI] [PubMed] [Google Scholar]
- 63.Delille HK, Schrader M. Targeting of hFis1 to peroxisomes is mediated by Pex19p. J Biol Chem. 2008;283:31107–15. doi: 10.1074/jbc.M803332200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Koch A, Schneider G, Luers GH, Schrader M. Peroxisome elongation and constriction but not fission can occur independently of dynamin-like protein 1. J Cell Sci. 2004;117:3995–4006. doi: 10.1242/jcs.01268. [DOI] [PubMed] [Google Scholar]
- 65.Koch A, Thiemann M, Grabenbauer M, Yoon Y, McNiven MA, Schrader M. Dynamin–like protein 1 is involved in peroxisomal fission. J Biol Chem. 2003;278:8597–605. doi: 10.1074/jbc.M211761200. [DOI] [PubMed] [Google Scholar]
- 66.Koch A, Yoon Y, Bonekamp NA, McNiven MA, Schrader M. A role for Fis1 in both mitochondrial and peroxisomal fission in mammalian cells. Mol Biol Cell. 2005;16:5077–86. doi: 10.1091/mbc.E05-02-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li X, Gould SJ. The dynamin-like GTPase DLP1 is essential for peroxisome division and is recruited to peroxisomes in part by PEX11. J Biol Chem. 2003;278:17012–20. doi: 10.1074/jbc.M212031200. [DOI] [PubMed] [Google Scholar]
- 68.Gandre-Babbe S, van der Bliek AM. The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol Biol Cell. 2008;19:2402–12. doi: 10.1091/mbc.E07-12-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Otera H, Wang C, Cleland MM, Setoguchi K, Yokota S, Youle RJ, Mihara K. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J Cell Biol. 2010;191:1141–58. doi: 10.1083/jcb.201007152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lingard MJ, Gidda SK, Bingham S, Rothstein SJ, Mullen RT, Trelease RN. Arabidopsis PEROXIN11c-e, FISSION1b, and DYNAMIN-RELATED PROTEIN3A cooperate in cell cycle-associated replication of peroxisomes. Plant Cell. 2008;20:1567–85. doi: 10.1105/tpc.107.057679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mano S, Nakamori C, Kondo M, Hayashi M, Nishimura M. An Arabidopsis dynamin-related protein, DRP3A, controls both peroxisomal and mitochondrial division. Plant J. 2004;38:487–98. doi: 10.1111/j.1365-313X.2004.02063.x. [DOI] [PubMed] [Google Scholar]
- 72.Zhang X, Hu J. Two small protein families, DYNA-MIN–RELATED PROTEIN3 and FISSION1, are required for peroxisome fission in Arabidopsis. Plant J. 2009;57:146–59. doi: 10.1111/j.1365-313X.2008.03677.x. [DOI] [PubMed] [Google Scholar]
- 73.Zhang X, Hu J. The Arabidopsis chloroplast division protein DYNAMIN-RELATED PROTEIN5B also mediates peroxisome division. Plant Cell. 2010;22:431–42. doi: 10.1105/tpc.109.071324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang XC, Hu JP. FISSION1A and FISSION1B proteins mediate the fission of peroxisomes and mitochondria in Arabidopsis. Mol Plant. 2008;1:1036–47. doi: 10.1093/mp/ssn056. [DOI] [PubMed] [Google Scholar]
- 75.Koirala S, Bui HT, Schubert HL, Eckert DM, Hill CP, Kay MS, Shaw JM. Molecular architecture of a dynamin adaptor: implications for assembly of mitochondrial fission complexes. J Cell Biol. 2010;191:1127–39. doi: 10.1083/jcb.201005046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Motley AM, Ward GP, Hettema EH. Dnm1p-dependent peroxisome fission requires Caf4p, Mdv1p and Fis1p. J Cell Sci. 2008;121:1633–40. doi: 10.1242/jcs.026344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nagotu S, Krikken AM, Otzen M, Kiel JA, Veenhuis M, van der Klei IJ. Peroxisome fission in Hansenula polymorpha requires Mdv1 and Fis1, two proteins also involved in mitochondrial fission. Traffic. 2008;9:1471–84. doi: 10.1111/j.1600-0854.2008.00772.x. [DOI] [PubMed] [Google Scholar]
- 78.Nagotu S, Saraya R, Otzen M, Veenhuis M, van der Klei IJ. Peroxisome proliferation in Hansenula polymorpha requires Dnm1p which mediates fission but not de novo formation. Biochimica et Biophysica Acta. 2008;1783:760–9. doi: 10.1016/j.bbamcr.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 79.Schauss AC, Bewersdorf J, Jakobs S. Fis1p and Caf4p, but not Mdv1p, determine the polar localization of Dnm1p clusters on the mitochondrial surface. J Cell Sci. 2006;119:3098–106. doi: 10.1242/jcs.03026. [DOI] [PubMed] [Google Scholar]
- 80.Hoepfner D, van den Berg M, Philippsen P, Tabak HF, Hettema EH. A role for Vps1p, actin, and the Myo2p motor in peroxi-some abundance and inheritance in Saccharomyces cerevisiae. J Cell Biol. 2001;155:979–90. doi: 10.1083/jcb.200107028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jourdain I, Sontam D, Johnson C, Dillies C, Hyams JS. Dynamin-dependent biogenesis, cell cycle regulation and mitochondrial association of peroxisomes in fission yeast. Traffic. 2008;9:353–65. doi: 10.1111/j.1600-0854.2007.00685.x. [DOI] [PubMed] [Google Scholar]
- 82.Kuravi K, Nagotu S, Krikken AM, Sjollema K, Deckers M, Erdmann R, Veenhuis M, van der Klei IJ. Dynamin-related proteins Vps1p and Dnm1p control peroxisome abundance in Saccharomyces cerevisiae. J Cell Sci. 2006;119:3994–4001. doi: 10.1242/jcs.03166. [DOI] [PubMed] [Google Scholar]
- 83.Tower RJ, Fagarasanu A, Aitchison JD, Rachubinski RA. The peroxin Pex34p functions with the Pex11 family of peroxisomal divisional proteins to regulate the peroxisome population in yeast. Mol Biol Cell. 2011;22:1727–1738. doi: 10.1091/mbc.E11-01-0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nguyen T, Bjorkman J, Paton BC, Crane DI. Failure of microtubule-mediated peroxisome division and trafficking in disorders with reduced peroxisome abundance. J Cell Sci. 2006;119:636–45. doi: 10.1242/jcs.02776. [DOI] [PubMed] [Google Scholar]
- 85.Wiemer EA, Wenzel T, Deerinck TJ, Ellisman MH, Subramani S. Visualization of the peroxisomal compartment in living mammalian cells: dynamic behavior and association with microtubules. J Cell Biol. 1997;136:71–80. doi: 10.1083/jcb.136.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mathur J, Mathur N, Hulskamp M. Simultaneous visualization of peroxisomes and cytoskeletal elements reveals actin and not microtubule-based peroxisome motility in plants. Plant Physiol. 2002;128:1031–45. doi: 10.1104/pp.011018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fagarasanu A, Fagarasanu M, Eitzen GA, Aitchison JD, Rachubinski RA. The peroxisomal membrane protein Inp2p is the peroxisome-specific receptor for the myosin V motor Myo2p of Saccharomyces cerevisiae. Dev Cell. 2006;10:587–600. doi: 10.1016/j.devcel.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 88.Jourdain I, Gachet Y, Hyams JS. The dynamin related protein Dnm1 fragments mitochondria in a microtubule-dependent manner during the fission yeast cell cycle. Cell Motil Cytoskeleton. 2009;66:509–23. doi: 10.1002/cm.20351. [DOI] [PubMed] [Google Scholar]
- 89.Brocard CB, Boucher KK, Jedeszko C, Kim PK, Walton PA. Requirement for microtubules and dynein motors in the earliest stages of peroxisome biogenesis. Traffic. 2005;6:386–95. doi: 10.1111/j.1600-0854.2005.00283.x. [DOI] [PubMed] [Google Scholar]
- 90.Low HH, Lowe J. Dynamin architecture – from monomer to polymer. Curr Opin Struc Biol. 2010;20:791–8. doi: 10.1016/j.sbi.2010.09.011. [DOI] [PubMed] [Google Scholar]
- 91.Gao S, von der Malsburg A, Paeschke S, Behlke J, Haller O, Kochs G, Daumke O. Structural basis of oligomerization in the stalk region of dynamin-like MxA. Nature. 2010;465:502–6. doi: 10.1038/nature08972. [DOI] [PubMed] [Google Scholar]
- 92.Low HH, Sachse C, Amos LA, Lowe J. Structure of a bacterial dynamin-like protein lipid tube provides a mechanism for assembly and membrane curving. Cell. 2009;139:1342–52. doi: 10.1016/j.cell.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chappie JS, Acharya S, Leonard M, Schmid SL, Dyda F. G domain dimerization controls dynamin’s assembly–stimulated GTPase activity. Nature. 2010;465:435–40. doi: 10.1038/nature09032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ghosh A, Praefcke GJ, Renault L, Wittinghofer A, Herrmann C. How guanylate-binding proteins achieve assembly – stimulated processive cleavage of GTP to GMP. Nature. 2006;440:101–4. doi: 10.1038/nature04510. [DOI] [PubMed] [Google Scholar]
- 95.Low HH, Lowe J. A bacterial dynamin-like protein. Nature. 2006;444:766–9. doi: 10.1038/nature05312. [DOI] [PubMed] [Google Scholar]
- 96.Bashkirov PV, Akimov SA, Evseev AI, Schmid SL, Zimmerberg J, Frolov VA. GTPase cycle of dynamin is coupled to membrane squeeze and release, leading to spontaneous fission. Cell. 2008;135:1276–86. doi: 10.1016/j.cell.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pucadyil TJ, Schmid SL. Real-time visualization of dynamin-catalyzed membrane fission and vesicle release. Cell. 2008;135:1263–75. doi: 10.1016/j.cell.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ingerman E, Perkins EM, Marino M, Mears JA, McCaffery JM, Hinshaw JE, Nunnari J. Dnm1 forms spirals that are structurally tailored to fit mitochondria. J Cell Biol. 2005;170:1021–7. doi: 10.1083/jcb.200506078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Roux A, Koster G, Lenz M, Sorre B, Manneville JB, Nassoy P, Bassereau P. Membrane curvature controls dynamin polymerization. Proc Natl Acad Sci USA. 2010;107:4141–6. doi: 10.1073/pnas.0913734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ramachandran R. Vesicle scission: dynamin. Semin Cell Dev Biol. 2011;22:10–7. doi: 10.1016/j.semcdb.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 101.Yoshida Y, Kinuta M, Abe T, Liang S, Araki K, Cremona O, Di Paolo G, Moriyama Y, Yasuda T, De Camilli P, Takei K. The stimulatory action of amphiphysin on dynamin function is dependent on lipid bilayer curvature. EMBO J. 2004;23:3483–91. doi: 10.1038/sj.emboj.7600355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lee CS, Kim IS, Park JB, Lee MN, Lee HY, Suh PG, Ryu SH. The phox homology domain of phospholipase D activates dynamin GTPase activity and accelerates EGFR endocytosis. Nat Cell Biol. 2006;8:477–84. doi: 10.1038/ncb1401. [DOI] [PubMed] [Google Scholar]
- 103.Thoms S, Debelyy MO, Nau K, Meyer HE, Erdmann R. Lpx1p is a peroxisomal lipase required for normal peroxisome morphology. FEBS J. 2008;275:504–14. doi: 10.1111/j.1742-4658.2007.06217.x. [DOI] [PubMed] [Google Scholar]
- 104.Lay D, Gorgas K, Just WW. Peroxisome biogenesis: where Arf and coatomer might be involved. Biochim Biophys Acta. 2006;1763:1678–87. doi: 10.1016/j.bbamcr.2006.08.036. [DOI] [PubMed] [Google Scholar]
- 105.Schrader M, Reuber BE, Morrell JC, Jimenez-Sanchez G, Obie C, Stroh TA, Valle D, Schroer TA, Gould SJ. Expression of PEX11beta mediates peroxisome proliferation in the absence of extracellular stimuli. J Biol Chem. 1998;273:29607–14. doi: 10.1074/jbc.273.45.29607. [DOI] [PubMed] [Google Scholar]
- 106.Delille HK, Agricola B, Guimaraes SC, Borta H, Luers GH, Fransen M, Schrader M. Pex11pβ-mediated growth and division of mammalian peroxisomes follows a maturation pathway. J Cell Sci. 2010;123:2750–62. doi: 10.1242/jcs.062109. [DOI] [PubMed] [Google Scholar]
- 107.Cepinska MN, Veenhuis M, van der Klei IJ, Nagotu S. Peroxisome Fission is Associated with Reorganization of Specific Membrane Proteins. Traffic. 2011;12:925–37. doi: 10.1111/j.1600-0854.2011.01198.x. [DOI] [PubMed] [Google Scholar]
- 108.Opalinski L, Kiel JA, Williams C, Veenhuis M, van der Klei IJ. Membrane curvature during peroxisome fission requires Pex11. EMBO J. 2010;30:5–16. doi: 10.1038/emboj.2010.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Campelo F, Fabrikant G, McMahon HT, Kozlov MM. Modeling membrane shaping by proteins: focus on EHD2 and N-BAR domains. FEBS Lett. 2010;584:1830–9. doi: 10.1016/j.febslet.2009.10.023. [DOI] [PubMed] [Google Scholar]
- 110.Masuda M, Mochizuki N. Structural characteristics of BAR domain superfamily to sculpt the membrane. Semin Cell Dev Biol. 2010;21:391–8. doi: 10.1016/j.semcdb.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 111.Suetsugu S. The proposed functions of membrane curvatures mediated by the BAR domain superfamily proteins. J Biochem. 2010;148:1–12. doi: 10.1093/jb/mvq049. [DOI] [PubMed] [Google Scholar]
- 112.Suetsugu S, Toyooka K, Senju Y. Subcellular membrane curvature mediated by the BAR domain superfamily proteins. Semin Cell Dev Biol. 2010;21:340–9. doi: 10.1016/j.semcdb.2009.12.002. 2010. [DOI] [PubMed] [Google Scholar]
- 113.Drin G, Casella JF, Gautier R, Boehmer T, Schwartz TU, Antonny B. A general amphipathic α-helical motif for sensing membrane curvature. Nat Struct Mol Biol. 2007;14:138–46. doi: 10.1038/nsmb1194. [DOI] [PubMed] [Google Scholar]
- 114.van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9:112–24. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Campelo F, McMahon HT, Kozlov MM. The hydrophobic insertion mechanism of membrane curvature generation by proteins. Biophys J. 2008;95:2325–39. doi: 10.1529/biophysj.108.133173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hatzakis NS, Bhatia VK, Larsen J, Madsen KL, Bolinger P-Y, Kunding AH, Castillo J, Gether U, Hedegård P, Stamou D. How curved membranes recruit amphipathic helices and protein anchoring motifs. Nat Chem Biol. 2009;5:835–41. doi: 10.1038/nchembio.213. [DOI] [PubMed] [Google Scholar]
- 117.Knoblach B, Rachubinski RA. Phosphorylation-dependent activation of peroxisome proliferator protein PEX11 controls peroxisome abundance. J Biol Chem. 2010;285:6670–80. doi: 10.1074/jbc.M109.094805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Marshall PA, Dyer JM, Quick ME, Goodman JM. Redoxsensitive homodimerization of Pex11p: a proposed mechanism to regulate peroxisomal division. J Cell Biol. 1996;135:123–37. doi: 10.1083/jcb.135.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Passreiter M, Anton M, Lay D, Frank R, Harter C, Wieland FT, Gorgas K, Just WW. Peroxisome biogenesis: involvement of ARF and coatomer. J Cell Biol. 1998;141:373–83. doi: 10.1083/jcb.141.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Li X, Baumgart E, Dong GX, Morrell JC, Jimenez-Sanchez G, Valle D, Smith KD, Gould SJ. PEX11α is required for peroxisome proliferation in response to 4-phenylbutyrate but is dispensable for peroxisome proliferator-activated receptor α-mediated peroxisome proliferation. Mol Cell Biol. 2002;22:8226–40. doi: 10.1128/MCB.22.23.8226-8240.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Li X, Gould SJ. PEX11 promotes peroxisome division independently of peroxisome metabolism. J Cell Biol. 2002;156:643–51. doi: 10.1083/jcb.200112028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Li X, Baumgart E, Morrell JC, Jimenez-Sanchez G, Valle D, Gould SJ. PEX11 β deficiency is lethal and impairs neuronal migration but does not abrogate peroxisome function. Mol Cell Biol. 2002;22:4358–65. doi: 10.1128/MCB.22.12.4358-4365.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yu T, Fox RJ, Burwell LS, Yoon Y. Regulation of mitochondrial fission and apoptosis by the mitochondrial outer membrane protein hFis1. J Cell Sci. 2005;118:4141–51. doi: 10.1242/jcs.02537. [DOI] [PubMed] [Google Scholar]
- 124.Serasinghe MN, Seneviratne AM, Smrcka AV, Yoon Y. Identification and characterization of unique proline-rich peptides binding to the mitochondrial fission protein hFis1. J Biol Chem. 2010;285:620–30. doi: 10.1074/jbc.M109.027508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ivashchenko O, Van Veldhoven PP, Brees C, Ho YS, Terlecky SR, Fransen M. Intraperoxisomal redox balance in mammalian cells: oxidative stress and interorganellar cross-talk. Mol Biol Cell. 2011;22:1440–51. doi: 10.1091/mbc.E10-11-0919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Slawecki ML, Dodt G, Steinberg S, Moser AB, Moser HW, Gould SJ. Identification of three distinct peroxisomal protein import defects in patients with peroxisome biogenesis disorders. J Cell Sci. 1995;108:1817–29. doi: 10.1242/jcs.108.5.1817. [DOI] [PubMed] [Google Scholar]
- 127.Guo T, Gregg C, Boukh-Viner T, Kyryakov P, Goldberg A, Bourque S, Banu F, Haile S, Milijevic S, San KH, Solomon J, Wong V, Titorenko VI. A signal from inside the peroxisome initiates its division by promoting the remodeling of the peroxisomal membrane. J Cell Biol. 2007;177:289–303. doi: 10.1083/jcb.200609072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Saraya R, Krikken AM, Veenhuis M, van der Klei IJ. Peroxisome reintroduction in Hansenula polymorpha requires Pex25 and Rho1. J Cell Biol. 2011;193:885–900. doi: 10.1083/jcb.201012083. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]