SUMMARY
While chromosomal translocations are common pathogenetic events in cancer, mechanisms that promote them are poorly understood. To elucidate translocation mechanisms in mammalian cells, we developed high throughput, genome-wide translocation sequencing (HTGTS). We employed HTGTS to identify tens of thousands of independent translocation junctions involving fixed I-SceI meganuclease-generated DNA double strand breaks (DSBs) within the c-myc oncogene or IgH locus of B lymphocytes induced for Activation Induced-cytidine Deaminase (AID)-dependent IgH class-switching. DSBs translocated very widely across the genome, but were preferentially targeted to transcribed chromosomal regions and also to numerous AID-dependent and AID-independent hotspots, with the latter being comprised mainly of cryptic genomic I-SceI targets. Comparison of translocation junctions with genome-wide nuclear run-ons revealed a marked association between transcription start sites and translocation targeting. The majority of translocation junctions were formed via end-joining with short micro-homologies. We discuss implications of our findings for diverse fields including gene therapy and cancer genomics.
INTRODUCTION
Recurrent oncogenic translocations are common in hematopoietic malignancies including lymphomas (Kuppers and Dalla-Favera, 2001) and also occur frequently in solid tumors such as prostate and lung cancers (Shaffer and Pandolfi, 2006). DNA double-strand breaks (DSBs) are common intermediates of these genomic aberrations (Stratton et al., 2009). DSBs are generated by normal metabolic processes, by genotoxic agents including some cancer therapeutics, and by V(D)J and immunoglobulin (Ig) heavy (H) chain (IgH) class switch recombination (CSR) in lymphocytes (Zhang et al., 2010). Highly conserved pathways repair DSBs to preserve genome integrity (Lieber, 2010). Nevertheless, repair can fail, resulting in unresolved DSBs and translocations. Recurrent translocations in tumors usually arise as low frequency events that are selected during oncogenesis. However, other factors influence the appearance of recurrent translocations including chromosomal location of oncogenes (Gostissa et al., 2009). Chromosomal environment likely affects translocation frequency by influencing mechanistic factors, including DSB frequency at translocation targets, factors that contribute to juxtaposition of broken loci for joining, and mechanisms that circumvent repair functions that promote intra-chromosomal DSB joining (Zhang et al., 2010).
IgH CSR is initiated by DSBs that result from transcription-targeted AID-cytidine deamination activity within IgH switch (S) regions that lie just 5′ of various sets of CH exons. DSBs within the donor Sμ region and a downstream acceptor S region are fused via end-joining to complete CSR and allow expression of a different antibody class (Chaudhuri et al., 2007). Clonal translocations in human and mouse B cell lymphomas often involve IgH S regions and an oncogene, such as c-myc (Kuppers and Dalla-Favera, 2001; Gostissa et al., 2011). In this regard, AID-generated IgH S region DSBs directly participate in translocations to c-myc and other genes (Franco et al., 2006; Ramiro et al., 2006; Wang et al., 2009). Through its role in somatic hypermutation (SHM) of IgH and Ig light (IgL) variable region exons, AID theoretically might generate lower frequency DSBs in Ig loci that serve as translocation intermediates (Liu and Schatz, 2009). In addition, AID mutates many non-Ig genes in activated B cells at far lower levels than Ig genes (Liu et al., 2008), such off-target AID activity also may contribute to translocations of non-Ig genes (Robbiani et al., 2008). Indeed, AID even has been suggested to initiate lesions leading to translocations in non-lymphoid cancers, including prostate cancer (Lin et al., 2009). However, potential roles of AID in generating DSBs genome-wide has not been addressed. In this regard, other sources of translocation-initiating DSBs could include intrinsic factors, such as oxidative metabolism, replication stress, and chromosome fragile sites, or extrinsic factors such as ionizing radiation or chemotherapeutics (Zhang et al., 2010).
DSBs lead to damage response foci formation over 100kb or larger flanking regions, promoting DSB joining and suppressing translocations (Zhang et al., 2010; Nussenzweig and Nussenzweig, 2010). IgH class-switching in activated B cells can be mediated by yeast I-SceI endonuclease-generated DSBs without AID or S regions, suggesting general mechanisms promote efficient intra-chromosomal DSB joining over at least 100 kb (Zarrin et al., 2007). In somatic cells, classical non-homologous end-joining (C-NHEJ) repairs many DSBs (Zhang et al., 2010). C-NHEJ suppresses translocations by preferentially joining DSBs intra-chromosomally (Ferguson et al., 2000). Deficiency for C-NHEJ leads to frequent translocations, demonstrating that other pathways fuse DSBs into translocations (Zhang et al., 2010). Correspondingly, an alternative end-joining pathway (A-EJ), that prefers ends with short micro-homologies (MHs), supports CSR in the absence of C-NHEJ (Yan et al., 2007) and joins CSR DSBs to other DSBs to generate translocations (Zhang et al., 2010). Indeed, C-NHEJ suppresses p53-deficient lymphomas with recurrent IgH/c-myc translocations catalyzed by A-EJ (Zhu et al., 2002). Various evidence suggests A-EJ may be translocation prone (e.g. Simsek and Jasin, 2010).
The mammalian nucleus is occupied by non-randomly positioned genes and chromosomes (Meaburn et al., 2007). Fusion of DSBs to generate translocations requires physical proximity; thus, spatial disposition of chromosomes might impact translocation patterns (Zhang et al., 2010). Cytogenetic studies revealed that certain loci involved in oncogenic translocations are spatially proximal (Meaburn et al., 2007). Studies of recurrent translocations in mouse B cell lymphomas suggested that aspects of particular chromosomal regions, as opposed to broader territories, might promote proximity and influence translocation frequency (Wang et al., 2009). Non-random position of genes and chromosomes in the nucleus led to two general models for translocation initiation. “Contact-first” poses translocations to be restricted to proximally-positioned chromosomal regions, while “breakage-first” poses that distant DSBs can be juxtaposed (Meaburn et al., 2007). In depth evaluation of how chromosomal organization influences translocations requires a genome-wide approach.
To elucidate translocation mechanisms, we have developed approaches that identify genome-wide translocations arising from a specific cellular DSB. Thereby, we have isolated large numbers of translocations from primary B cells activated for CSR, to provide a genome-wide analysis of the relationship between translocations and particular classes of DSBs, transcription, chromosome domains, and other factors.
RESULTS
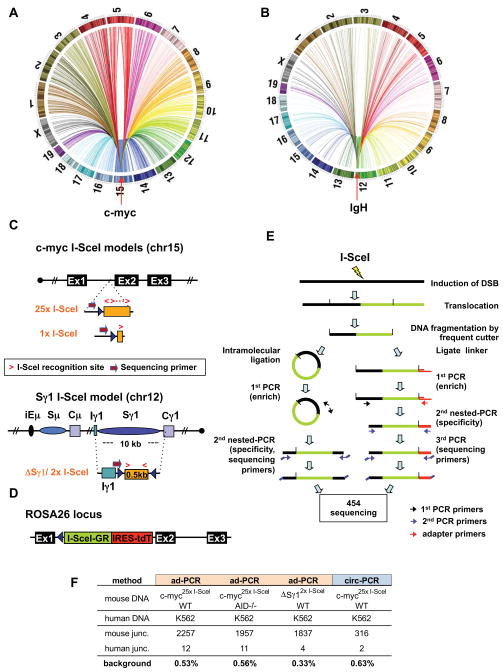
Development of High Throughput Genomic Translocation Sequencing (HTGTS)
We developed HTGTS to isolate junctions between a chromosomal DSB introduced at a fixed site and other sequences genome-wide. Such junctions, other than those involving breaksite resection, mostly should result from end-joining of introduced DSBs to other genomic DSBs. Thus, HTGTS will identify other genomic DSBs capable of joining to the test DSBs. With HTGTS, we isolated from primary mouse B cells junctions that fused IgH or c-myc DSBs to sequences distributed widely across the genome (Fig. 1A,B). We chose c-myc and IgH as targets because they participate in recurrent oncogenic translocations in B cell lymphomas. To generate c-myc- or IgH-specific DSBs, we employed an 18bp canonical I-SceI meganuclease target sequence, which is absent in mouse genomes (Jasin, 1996). One c-myc target was a cassette with 25 tandem I-SceI sites, to increase cutting efficiency, within c-myc intron 1 on chromosome (chr)15 (termed c-myc25xI-SceI; Fig. 1C; Wang et al., 2009). For comparison, we employed an allele with a single I-SceI site in the same position (termed c-myc1xI-SceI) (Figs. 1C; S1A–C). For IgH, we employed an allele with two I-SceI sites in place of endogenous Sγ1 (termed ΔSγ12xI-SceI) on chr12 (Zarrin et al., 2007). As a cellular model, we used primary splenic B cells activated in culture with αCD40 plus IL4 to induce AID, transcription, DSBs and CSR at Sγ1 (IgG1) and Sε (IgE), during days 2–4 of activation. At 24 hours, we infected B cells with I-SceI-expressing retrovirus to induce DSBs at I-SceI targets (Zarrin et al., 2007). Cells were processed at day 4 to minimize doublings and potential cellular selection. As high-titer retroviral infection can impair C-NHEJ (Wang et al., 2009), we also assayed B cells that express from their Rosa26 locus an I-SceI-glucocorticoid receptor fusion protein (I-SceI-GR) that can be activated via triamcinolone acetonide (TA) (Figs. 1D; S1D–F). The c-myc2`I-SceI cassette was frequently cut in TA-treated c-myc25xI-SceI/ROSAI-SceI-GR B cells (Fig. S1G).
Figure 1. High Throughput Genomic Translocation Sequencing.
(A and B) Circos plots of genome-wide translocation landscape of representative c-myc (A) or IgH (B) HTGTS libraries. Chromosome ideograms comprise the circumference. Individual translocations are represented as arcs originating from specific I-SceI breaks and terminating at partner site. (C) Top: a cassette containing either 25 or one I-SceI target(s) was inserted into intron 1 of c-myc (see Fig. S1A–C). Bottom: a cassette composed of a 0.5 kb spacer flanked by I-SceI target replaced the IgH Sγ1 region. Relative orientation of I-SceI sites is indicated by red arrows. Position of primers for generation and sequencing HTGTS libraries is shown. (D) An expression cassette for I-SceI fused to a glucocorticoid receptor (I-SceI-GR) was targeted into Rosa26 (see Fig. S1D–G). The red fluorescent protein Tomato (tdT) is co-expressed via an IRES. (E) Schematic representation of HTGTS methods; left: circularization-PCR, right: adapter-PCR. See text for details. (F) Background for HTGTS approaches, calculated as percent of artifactual human:mouse hybrid junctions when human DNA was mixed 1:1 with mouse DNA from indicated samples.
We employed two HTGTS methods. For the adapter-PCR approach (Fig. 1E, Siebert et al., 1995), genomic DNA was fragmented with a frequently cutting restriction enzyme, ligated to an asymmetric adapter, and further digested to block amplification of germline or unrearranged target alleles. We then performed nested-PCR with adapter- and locus-specific primers. Depending on the locus-specific PCR primers, one or the other side of the I-SceI DSB provides the “bait” translocation partner (Fig. 1C), with the “prey” provided by DSBs at other genomic sites. As a second approach, we employed circularization-PCR (Fig. 1E; Mahowald et al., 2009), in which enzymatically fragmented DNA was intra-molecularly ligated, digested with blocking enzymes, and nested-PCR performed with locus-specific primers. Following sequencing of PCR products, we aligned HTGTS junctions to reference genomes and scripted filters to remove artifacts from aligned databases. We experimentally controlled for potential background by generating HTGTS libraries from mixtures of human DNA and mouse DNA from activated I-SceI-infected c-myc25xI-SceI or ΔSγ12xI-SceI B cells; junctions fusing mouse and human sequences were less than 1% of the total (Fig. 1F). We identified nearly 150,000 independent junctions from numerous libraries from different mice (Supp. Table 1). Resulting genome-wide junction maps are shown either as colored dot plots of overall distribution of translocation numbers in selected size bins (useful for visualizing hotspots) or bar plots that compress hotspots and illustrate translocation site density. HTGTS yields an average of 1 unique junction/5 ng of DNA, corresponding to about 1 junction/1,000 genomes. Major findings were reproduced with both HTGTS methods (e.g. Fig. S2A). Moreover, while the largest portion of data was obtained with c-myc25xI-SceI alleles cut via retroviral I-SceI, major findings were reproduced via HTGTS from the c-myc25xI-SceI allele cleaved by I-SceI-GR and the c-myc1XI-SceI allele cleaved by retroviral I-SceI (Fig. S2C,D).
Analysis of Genome-wide Translocations from c-myc DSBs
For HTGTS of c-myc25xI-SceI or c-myc1xI-SceI alleles, we used primers about 200bp centromeric to the cassette (Fig. 1C) to detect junctions involving broken ends (BEs) on the 5′ side of c-myc I-SceI DSBs (“5′c-myc-I-SceI BEs”). Based on convention, prey sequences joined to 5′c-myc-I-SceI BEs are in (+) orientation if read from the junction in centromere to telomere direction and in (−) orientation if read in the opposite direction (Fig. S3A–D). Joins in which 5′c-myc-I-SceI BEs are fused to resected 3′c-myc-I-SceI BEs would be (+) (Fig. S3A). Intra-chromosomal joins to DSBs centromeric or telomeric to 5′c-myc-I-SceI BEs would be (+) or (−) depending on the side of the second DSB to which they were joined, with potential outcomes including deletions, inversions, and extra-chromosomal circles (Fig. S3B,C). Junctions to DSBs on different chromosomes could be (+) or (−) and derivative chromosomes centric or dicentric (Fig. S3D). Analyses of over 100,000 independent junctions from 5′c-myc-I-SceI BEs from WT and AID−/− backgrounds revealed prey to be distributed widely throughout the genome with similar general distribution patterns (Fig. 2; Fig. S2B, E,F). Other than 200kb downstream of the bait DSB, intra-chromosomal and inter-chromosomal junctions were evenly distributed into (+) and (−) orientation (Fig. 2; Fig. S3I). This finding implies that extra-chromosomal circles and acentric fragments are represented similarly to other translocation classes, suggesting little impact of cellular selection on junction distribution. The junctions of 5′c-myc-I-SceI BE from c-myc25xI-SceI, c-myc1xI-SceI and c-myc25xI-SceI/ROSAI-SceI-GR models were all consistent with end-joining and most (75–90%) had short junctional MHs (Table S1).
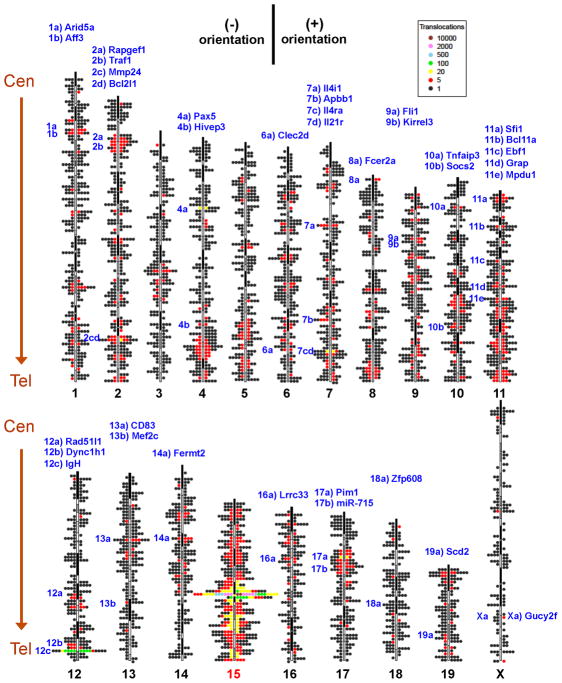
Figure 2. Genome-wide distribution and orientation of translocations from c-myc DSBs.
Genome-wide map of translocations originating from the c-myc25x I-SceI cassette (chr15) in αCD40/IL4-activated and I-SceI-infected B cells. Single junctions are represented by dots located at corresponding chromosomal position. The dot scale is 2 Mb. Clusters of translocations are indicated with color codes, as shown in legend. (+) and (−) orientation junctions (see Fig. S3) are plotted on righ and left side of each ideogram, respectively. Hotspots (see Fig. 4A), are listed in blue on top, with notation on the left side of chromosomes to indicate position. Data are from HTGTS libraries from 7 different mice. Centromere (Cen) and telomere (Tel) positions are indicated. See also Fig. S2.
WT and AID−/− HTGTS maps for 5′c-myc-I-SceI BEs had other common features. First, the majority of junctions (75%) arose from joining 5′c-myc-I-SceI BEs to sequences within 10 kb, with most lying 3′ of the breaksite (Figs. 3A; S4A). The density of joins remained relatively high within a region 200kb telomeric to the breaksite (Figs. 3A; S4A). Notably, most junctions within this 200kb region, but not beyond, were in the (+) orientation, consistent with joining to resected 3′c-myc-I-SceI BEs (Figs. 3A; S4A). About 15% of junctions occurred within the region 100kb centromeric to the breaksite. As these could not have resulted from resection (due to primer removal), they may reflect the known propensity for joining intra-chromosomal DSBs separated at such distances (Zarrin et al., 2007). Compared with other chromosomes, chr15 had a markedly high density of translocations along its 50Mb telomeric portion and also a high density along its centromeric portion (Fig. 2). Many chromosomes had smaller regions of relatively high or low translocation density, with such overall patterns conserved between WT and AID−/− backgrounds (Figs.2; S2A–F). Finally, although the majority of hotspots were WT-specific, a number were shared between WT and AID−/− backgrounds (see below).
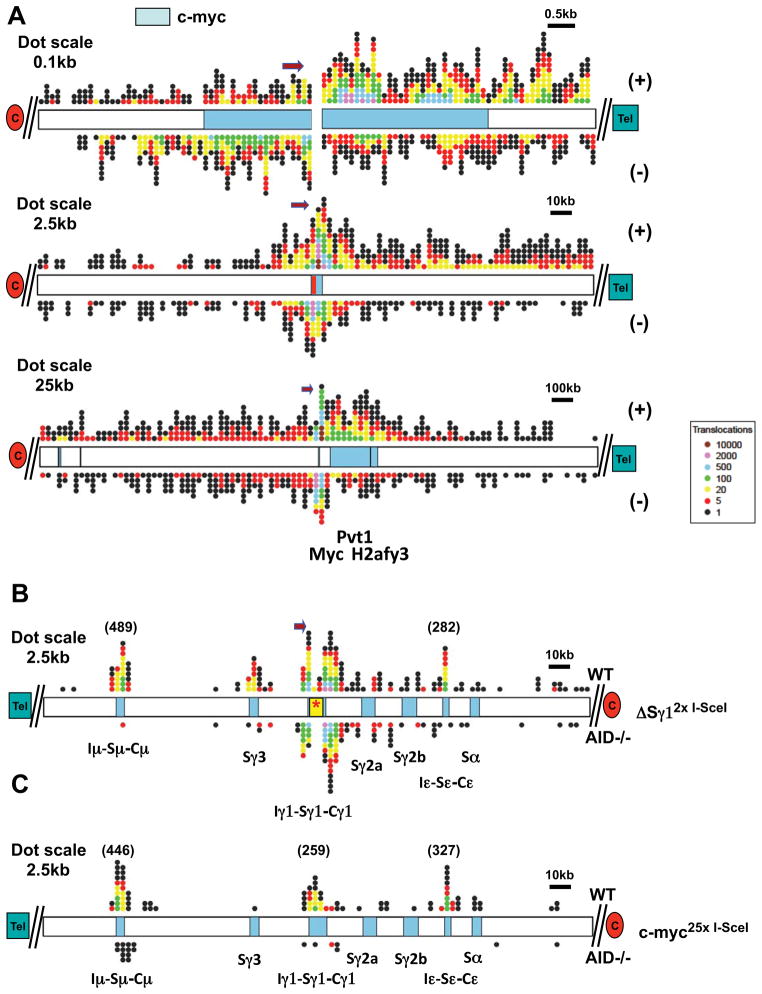
Figure 3. Distribution of IgH and c-myc breakpoint-proximal junctions.
(A) Distribution of junctions around chr15 breaksite in the pooled c-myc25xI-SceI HTGTS library. Top: 10 kb around breaksite (represented as a split). Middle: 250 kb around breaksite (represented by red bar); Bottom: 2.5 Mb around breaksite. (+) and (−)-oriented junctions are plotted on top and bottom of chromosome diagrams, respectively. (B,C) Distribution of translocation junctions at IgH in the pooled ΔSγ12xI-SceI (B) or c-myc25xI-SceI (C) HTGTS libraries. Translocations in WT (top) and AID−/− (bottom) B cells are shown. Positions of S regions within the 250 kb IgH CH region are indicated. Color codes are as in Fig. 2. Dot size, position of centromere (red oval) and telomere (green rectangle), and orientation of the sequencing primer are indicated. See also Fig. S4.
Analysis of HTGTS Libraries from IgH DSBs
For HTGTS of the ΔSγ12xI-SceI alleles, we used primers about 200 bp telomeric to the I-SceI cassette (Fig. 1C), allowing detection of junctions involving BEs on the 5′ side of Sγ1 I-SceI DSBs (“5′Sγ1-I-SceI BEs”). Intra- and inter-chromosomal joins involving 5′Sγ1-I-SceI BEs result in (+) or (−) junctions with the range of potential chromosomal outcomes including deletions, inversions, extra-chromosomal circles and acentrics (Fig. S3E–H). We isolated and analyzed approximately 9,000 and 8,000 5′Sγ1-I-SceI BE junctions from WT and AID−/− libraries, respectively (Fig. S2G,H). Reminiscent of the 5′c-myc-I-SceI junctions, about 75% of these junctions were within 10 kb of the breaksite, with a larger proportion on the 3′ side and predominantly in the (−) orientation, consistent with joining to resected 3′-I-SceI BEs (Fig. S4B–D). Outside the breaksite region, the general 5′Sγ1-I-SceI BE translocation patterns resembled those observed for 5′c-myc-I-SceI BEs, with both (+) and (−) translocations occurring on all chromosomes (Figs. S3J; S2G). While we analyzed more limited numbers of 5′Sγ1-I-SceI BE junctions (Table S2 and Fig. S2G,H), the broader telomeric region of chr12 had a notably large number of hits and, within this region, there were IgH hotspots in WT but not AID−/− libraries (Fig. 3B).
Sμ and Sε are major targets of AID-initiated DSBs in B cells activated with αCD40/IL4. Correspondingly, substantial numbers of 5′Sγ1-SceI BE junctions from WT, but not AID−/−, B cells joined to either Sμ or to Sε, which, respectively, lie approximately 100kb upstream and downstream of the ΔSγ12x I-SceI cassette (Fig. 3B; Fig. S4B–D). These findings support the notion that DSBs separated by 100–200 kb can be joined at high frequency by general repair mechanisms (Zarrin et al., 2007). We also observed frequent junctions from WT libraries specifically within Sγ3, which lies about 20 kb upstream of the breaksite, a finding of interest as joining Sγ3 to donor Sμ DSBs during CSR in αCD40/IL4 activated B cells occurs at low levels (see below). Notably, in WT, but not in AID−/− libraries, we found numerous junctions within Sγ1 (Fig. S4D), which is also targeted by AID in αCD40/IL4-activated B cells. As Sγ1 is present only on the non-targeted chr12 homolog due to the ΔSγ12xI-SceI replacement, these findings demonstrate robust translocation of 5′Sγ1-ISceI BEs to AID-dependent Sγ1 DSBs on the homologous chromosome, consistent with trans-CSR (Reynaud et al., 2005). Finally, while AID deficiency greatly reduced junctions into S regions, we observed a focal cluster of five 5′Sγ1-I-SceI BE junctions in or near Sμ in AID−/− ΔSγ12xI-SceI libraries (Fig. 3B; Fig. S4C).
Most c-myc translocation hotspots are targeted by AID
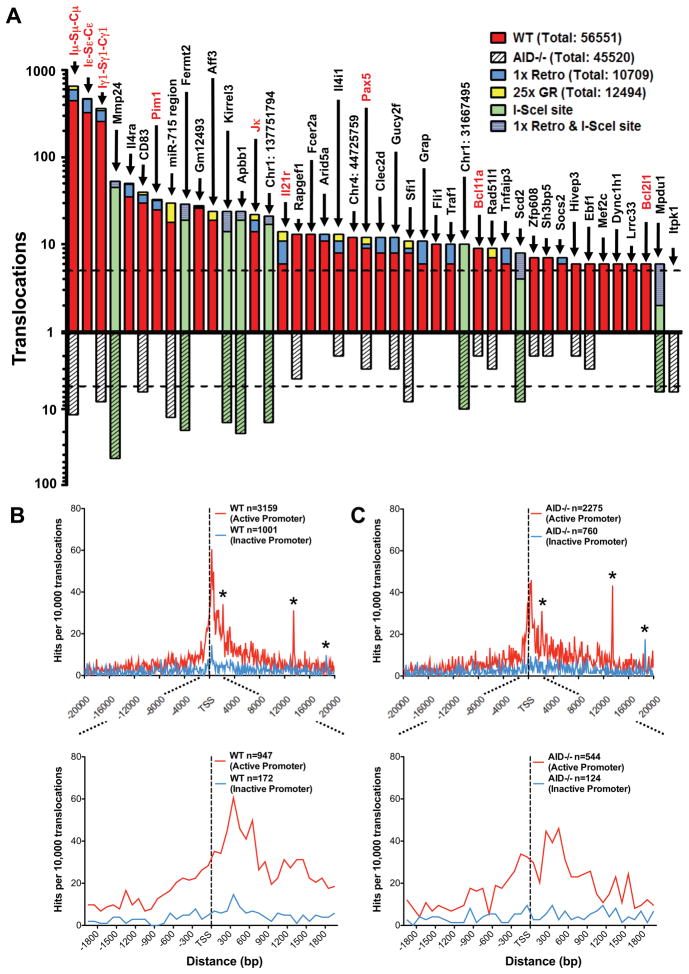
To identify 5′c-myc-I-SceI BE translocation hotspots in an unbiased manner, we separated the genome into 250 kb bins and identified bins containing a statistically significant enrichment of translocations (Suppl. Experimental Procedures). This approach identified 55 hotspots in WT libraries and 15 in AID−/− libraries (Table S3; Fig. 4A). Among the 43 most significant hotspots, 39 were in genes and 4 were in intergenic regions. Of these 43 hotspots, 21 were present at significantly greater levels in WT versus AID−/− backgrounds, and, therefore, classified as AID-dependent; while 9 more were enriched (from 3 to 6 fold) in the WT background and were potentially AID-dependent (Table S3; Fig. 4A). The other 13 were equally represented between WT and AID−/− backgrounds (Table S3; Fig. 4A). Of these 13, two exist in multiple copies (Sfi1 and miR-715), which may have contributed to their classification as hotspots (Quinlan et al., 2010; Ira Hall, personal communication); and 5 reached hotspot significance in only one of the two backgrounds (Table S3; Fig. 4A).
Figure 4. Identification of specific and general translocation hotspots.
(A) Graph representing translocation numbers in frequently hit genes and non-annotated chromosomal regions. Only hotspots with more than 5 hits are shown and are ordered based on frequency of translocations in the pooled c-myc25xI-SceI/WT HTGTS library (top bars). Respective frequencies of translocations in the pooled c-myc25xI-SceI/AID−/− HTGTS library are displayed underneath (bottom bars). Green bars represent frequent hits involving cryptic I-SceI sites. Blue and yellow portions of top bars represent translocations found in c-myc1xI-SceI and c-myc25xI-SceI/ROSAI-SceI-GR libraries, respectively. Genes translocated in human and mouse lymphoma or leukemia are in red. The dashed line represents the cutoff for significance over random occurrence for each of the two groups (see Table S3). (B and C) Genome-wide distribution of translocations relative to TSSs. Junctions from c-myc25xI-SceI/WT (B) or c-myc25xI-SceI/AID−/− (C) libraries (excluding 2 Mb around chr15 breaksite and IgH S regions) are assigned a distance to the nearest TSS and separated into “active” and “inactive” promoters as determined by GRO-seq. Translocation junctions are binned at 100 bp intervals. n represents the number of junctions within 20 kb (upper panels) or 2 kb (lower panels) of TSS. Asterisks indicate cryptic genomic I-SceI sites. See also Fig. S5.
The Sμ, Sγ1 and Sε regions, which are targeted for CSR DSBs by αCD40/IL4 treatment, were by far the strongest AID hotspots for 5′c-myc-I-SceI BEs, with other non-IgH AID-dependent hotspots ranging from 1% to 10% of Sμ levels (Fig. 4A). Translocation specificity to these three S regions, which together comprise less than 20 kb, was striking; there were only a few junctions in the remainder of the CH locus, which includes 4 other S regions not substantially activated by αCD40/IL4 (Fig. 3C). Notably, there was only one 5′c-myc-I-SceI BE junction with Sγ3, even though Sγ3 was a marked hotspot for 5′Sγ1-I-SceI BEs. In this regard, while AID-dependent DSBs in Sγ3 likely are much less frequent than in Sμ, Sγ1 and Sε under αCD40/IL4 stimulation conditions, Sγ3 DSBs may be favored targets of 5′Sγ1-I-SceI BEs because of linear proximity. Finally, translocations occurred in Sμ and Sγ1 in AID−/− B cells at much lower levels than in WT, but frequently enough to qualify them as AID-independent hot-spots (Fig. 4A).
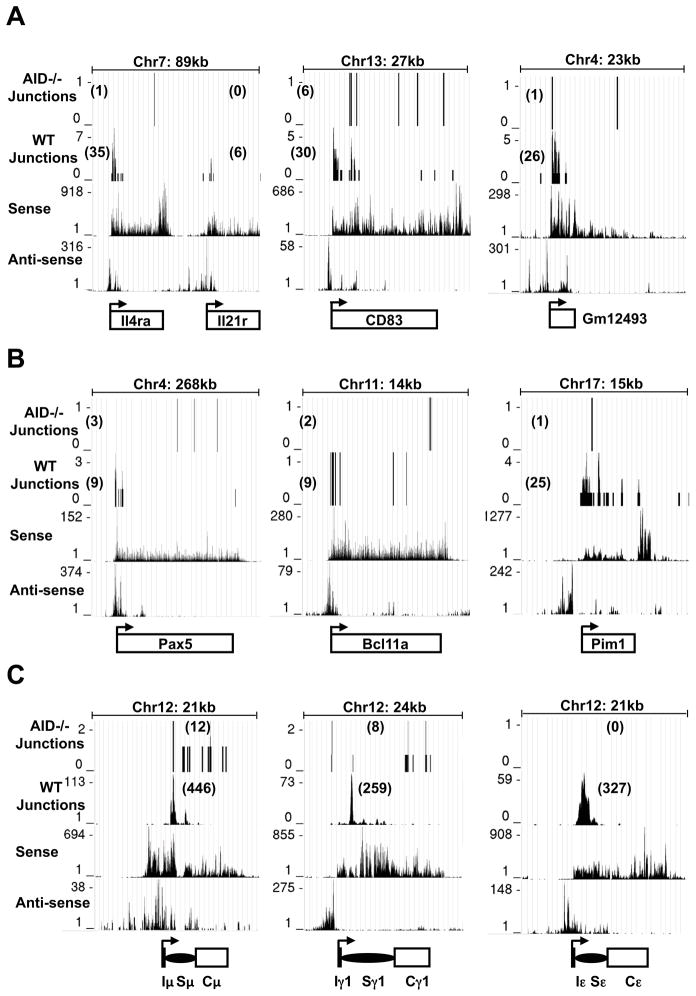
Several top AID SHM or binding targets in activated B cells (Liu, et al 2008; Yamane, et al. 2011) were translocation hotspots for 5′c-myc-I-SceI BEs, including our top 3 non-IgH hotspots (Il4ra, CD83, and Pim1) and probable AID-dependent translocation targets (e.g. Pax5 and Rapgef1) (Fig. 4A; Table S3). We also identified other AID-dependent translocation hotspots including the Aff3, Il21r, and Socs2 genes, and a non-annotated intergenic transcript on chr4 (gm12493, Fig. 4A; Table S3). We confirmed the ability of such hotspots to translocate to the c-myc25x I-SceI cassette by direct PCR (Table S4). We conclude that AID not only binds and mutates numerous non-Ig target genes but also acts on them to cause DSBs and translocations.
Translocations Genome-wide Frequently Occur Near Active Transcription Start Sites
To quantify transcription genome-wide, we applied unbiased global run-on sequencing (GRO-seq; Core et al., 2008) to αCD40/IL4-activated, I-SceI-infected B cells. GRO-seq measures elongating Pol II activity and distinguishes transcription on both strands. For all analyses, we excluded junctions within 1 Mb of the c-myc breaksite to avoid biases from this dominant class of junctions. To analyze remaining junctions from WT and AID−/− backgrounds, we determined nearest transcription start sites (TSSs) and divided translocations based on whether or not the TSS had promoter proximal activity based on GRO-seq (Supp. Exp. Procedures). Strikingly, both WT and AID−/− junctions, when dominant IgH translocations were excluded, showed a distinct peak that reached a maximum about 300–600bp on the sense side of the active TSSs and spanned from about 600bp on the anti-sense side to about 1kb on the sense side (Fig. 4B,C). Translocation hotspot genes, including Il4ra, CD83, Gm12493, Pim1, as well as potential hotspots including Pax5 and Bcl11a, had a substantial proportion of their translocations within 1–2kb regions starting 200–400bp in the sense direction from their bidirectional TSSs (Fig. 5A,B). In one striking example of TSS-proximal translocation targeting, there were distinct translocation peaks downstream of the TSSs of Il4ra and Il21r, which lies just 20 kb downstream; yet, there were no detected translocations into the 3′ portion of Il4ra even though it was highly transcribed (Fig. 5A). While lower level translocations into some AID-hotspot genes in AID−/− mice had less correlation with TSS proximity(Fig. 5A,B); the overall correlation of translocations and active TSS appeared similar in WT and AID−/− mice (Figs. 4B,C; S5A,B). Together, our findings indicate a relationship between active TSSs and AID-dependent and independent translocations genome-wide. In this context, we did not find a marked TSS correlation for translocations into non-transcribed genes (Fig. 4B,C).
Figure 5. Translocations Preferentially Translocate Near TSSs.
WT and AID−/− c-myc25xI-SceI HTGTS libraries were analyzed. In each panel, translocation junctions are in the first and second rows (WT and AID−/− as indicated). The third and fourth rows represent sense and antisense nascent RNA signals from GRO-seq. The IgH μ, γ1, ε genes are shown in (C), the next most-frequently hit hotspots in (A) and three selected oncogene hotspots in (B). The transcriptional start site (arrow) is at the bottom of each panel. The size of each genomic region and number of junctions in each are shown.
When the dominant IgH hotspots were included in the translocation/transcription analyses, the translocation peak shifted from about 300–600bp to about 1.5 kb downstream of the TSS in the sense direction (compare Fig. 4B,C to S5C,D). In B cells, transcription through Sμ initiates from the V(D)J exon and Iμ exon promoters upstream of Sμ. B cell activation with αCD40/IL4 stimulates CSR between Sμ and Sγ1 or Sε by inducing AID and by activating Iγ1 and Iε promoters upstream of Sγ1 and Sε. Indeed, most translocations into germline CH genes in WT αCD40/IL4-activated B cell were tightly clustered 1–2 kb downstream in the 5′ portion of Sμ, Sγ1 and Sε, consistent with transcription robustly targeting AID to S regions (Fig. 5C). Finally, AID-independent IgH translocations were scattered more broadly through S and C regions, suggesting that DSBs that initiate them arise by a different, AID-independent mechanism of S region instability (Fig. 5C).
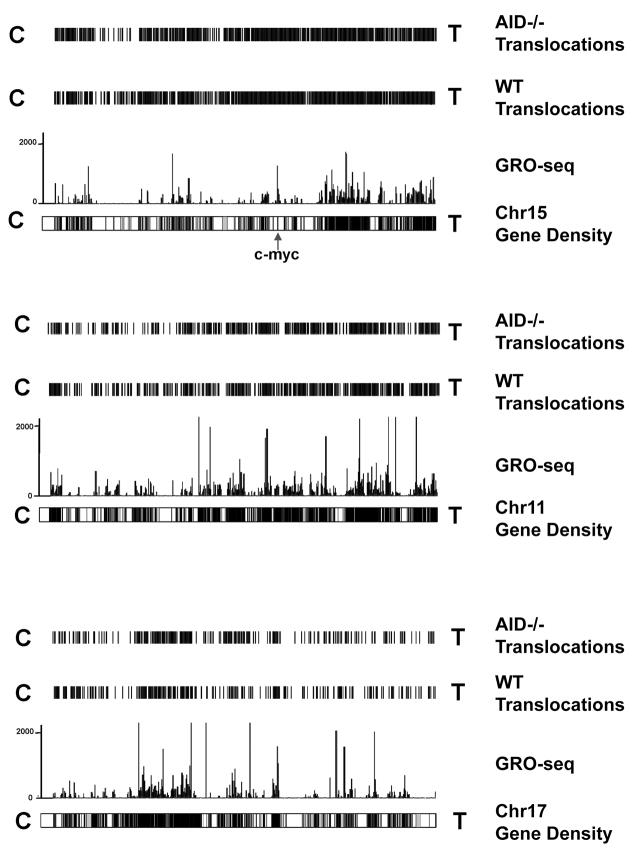
For 5′c-myc-I-SceI BEs (outside the breaksite region), 55% of translocations were within genes, whereas genes account for only 36% of the genome (Table S5). Therefore, we asked whether translocations from 5′c-myc-I-SceI BEs varied with gene density. For this purpose, we compared translocation densities to available gene density maps and to our GRO-seq transcription maps of all genes (Fig. 6; Figs. S6,S7). Strikingly, translocation distribution was highly correlated with gene density and transcription level. In general, chromosomal regions with highest transcriptional activity had highest translocation density. In contrast, regions with very low or undetectable transcription generally were very low in translocations (Fig. 6; S6; S7). Notably, we found no obvious regions with high overall transcription and low translocation levels, supporting a direct relationship between active transcription and translocation targeting genome-wide. In this context, we observed several robust AID-independent hotspot peaks that were relatively distant to the TSS and/or occurred in non-active genes (Fig. 4B,C, asterisks); these hotspots were generated by I-SceI activity at cryptic endogenous I-SceI sites as discussed next.
Figure 6. Translocations cluster to transcribed regions.
Translocation density maps from pooled c-myc25xI-SceI/WT and c-myc25xI-SceI/AID−/− HTGTS libraries are aligned with combined sense and antisense nascent RNA signals for chr 15, 11, and 17 using the UCSC genome browser. Chromosome gene densities are displayed below GRO-seq traces. Chromosomal orientation from left to right is centromere (C) to Telomere (T). See also Figs. S6, S7.
HTGTS Libraries Reveal Numerous Cryptic Genomic I-SceI target sites
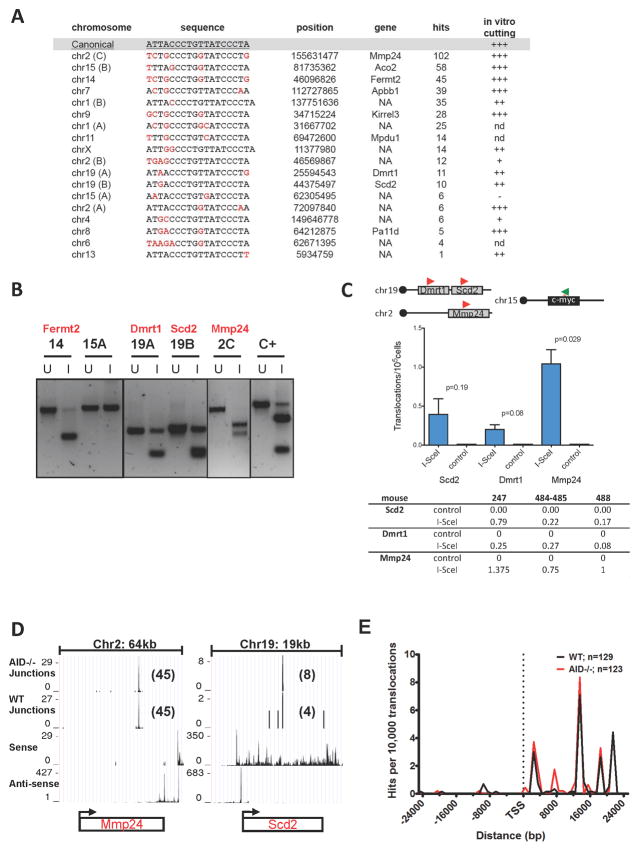
Eleven AID-independent translocation targets for 5′c-myc-I-SceI BEs were in genes and 2 were in intergenic regions (Table S3). Eight of these hotspot regions, in which junctions were tightly clustered, contained potential I-SceI-related sites, many of which were very near (within 50 bp) or actually contributed to translocation junctions. These putative cryptic I-SceI sites had from 1 to 5 divergent nucleotides with respect to the canonical 18 bp target site (Fig. 7A). We scanned the mouse genome for potential cryptic I-SceI sites that diverged up to 3 positions and identified 10 additional sites within 400 bp of one or more 5′c-myc-I-SceI BE translocation junctions (Fig. 7A). In vitro I-SceI digestion of PCR-amplified genomic fragments demonstrated that all 8 putative I-SceI targets at hotspots, and six of seven tested additional putative I-SceI targets, were bona fide I-SceI substrates (Fig. 7A,B). We performed direct translocation PCRs with three selected cryptic I-SceI sites and confirmed I-SceI-dependent translocation to the c-myc25x I-SceI cassette (Fig. 7C). Finally, GRO-seq analyses showed that 5 of 8 cryptic I-SceI translocation hotspots were in transcriptionally silent areas and that two I-SceI generated hotspots in transcribed genes were distant from the TSS (Fig. 4B,C, asterisks; Figs. 7D,E), highlighting the distinction between the I-SceI-generated hotspots and most other genomic translocation hotspots.
Figure 7. Identification of cryptic I-SceI sites in the mouse genome by HTGTS.
(A) Cryptic I-SceI site translocation targets. The canonical I-SceI recognition sequence is on top; nucleotides divergent from the consensus are in red. Chromosomal position and gene location of each cryptic site are indicated. “Hits” represent total number of unique junctions in a 4 kb region centered around each site in the pool of all HTGTS libraries (see also Table S6). In vitro cutting efficiency, evaluated as in Suppl. Exp. Procedures, is indicated. NA, intergenic or not annotated; nd, not determined. (B) In vitro cutting of PCR products encompassing indicated cryptic I-SceI sites. C+, positive control: PCR fragment containing a canonical I-SceI site. U, uncut; I, I-SceI-digested. (C) PCR to detect translocations between c-myc25xI-SceI and cryptic I-SceI sites in Scd2, Dmrt1 and Mmp24 genes. (Top) Position of primers used for PCR amplification. (Middle) Average frequency of translocations ±SEM. (Bottom) Number of translocations/105 cells from three independent c-myc25xI-SceI WT mice. (D) Transcription in genes containing I-SceI sites determined by GRO-seq. Translocation junctions are in the first (AID−/−) and second (WT) rows; sense and antisense nascent RNA signals are in the third and fourth rows. (E). Distance of cryptic I-SceI hotspots from the nearest TSS in pooled HTGTS libraries from WT and AID−/− c-myc25xI-SceI B cells.
DISCUSSION
With HTGTS, we have identified the genome-wide translocations that emanate from DSBs introduced into c-myc or IgH in activated B cells. A substantial percentage of these translocations (80–90%) join introduced DSBs to sequences on the same chromosome proximal to the join, likely reflecting the strong preference for C-NHEJ to join DSBs intra-chromosomally (Ferguson et al., 2000; Zarrin et al., 2007; Mahowald et al., 2009). The remaining 10–20% translocate broadly across all chromosomes, with translocation density correlating with transcribed gene density. Translocations are most often near TSSs within individual genes. Despite c-myc and IgH DSBs translocating broadly, there are translocation hotspots, with the majority being generated by cellular AID activity and most of the rest by ectopically expressed I-SceI activity at cryptic genomic I-SceI target sequences. Notably, targeted DSBs join at similar levels to both (+) and (−) orientations of hotspot sequences, arguing against a role for cellular selection in their appearance. This finding also suggests that both sides of hotspot DSBs have similar opportunity to translocate to a DSB on another chromosome.
The majority of HTGTS junctions from the c-myc I-SceI DSBs are mediated by end-joining and contain short MHs, reminiscent of joins in cancer genomes (Stratton et al., 2009) and consistent with roles for either (or both) C-NHEJ or A-EJ (Zhang et al., 2010). Recurrency of translocations in cancer genomes is a characteristic used to consider them as potential oncogenic “drivers”. Our HTGTS studies establish that many recurrent translocations form in the absence of selection and, thus, are caused by factors intrinsic to the translocation mechanism (Wang et al., 2009; Lin et al., 2009). HTGTS also provides a method to discover recurrent genomic DSBs, as evidenced by ability of HTGTS to find known DSBs, such as AID-initiated DSBs in S regions, and previously unrecognized genomic I-SceI targets. HTGTS should be readily applicable for genome-wide screens for translocations and recurrent DSBs in a wide range of cell types.
AID has a dominant role in targeting recurrent translocations genome-wide
Prior studies demonstrated that AID binds to and mutates non-Ig genes (Pasqualucci et al, 2001; Liu et al., 2008; Yamane et al., 2011). We find that AID also induces DSBs and translocations in non-Ig genes with the peak of translocation junctions spanning the region of the TSS. Thus, processes closely associated with transcription and, potentially, transcriptional initiation may attract AID activity to these non-Ig gene targets, consistent with ectopically expressed AID mutating yeast promoter regions (Gomez-Gonzalez and Aguilera, 2007). IgH translocation junctions mostly fall 1.5–2 kb downstream of the activated I region TSSs within S regions, which are known to be specialized AID targets. Thus, transcription through S regions attracts and focuses AID activity, at least in part via pausing mechanisms and by generating appropriate DNA substrates, such as R-loops, for this single-strand DNA-specific cytidine deaminase (Yu et al., 2003; Pavri and Nussenzweig, 2011; Chaudhuri et al., 2007). Notably, S regions still qualified as translocation hotspots for 5′ c-myc-I-SceI BEs in AID−/− B cells, supporting suggestions that these regions, perhaps via transcription, may be intrinsically prone to DSBs (Dudley et al., 2002; Kovalchuk et al., 2007; Unniraman et al., 2004). Given the differential targeting of CSR and SHM (Liu and Schatz, 2009), application of HTGTS to germinal center (GC) B cells, in which AID initiates SHM within variable region exons, may reveal novel AID genomic targets not observed in B cells activated for IgH CSR in culture, potentially including genes that could contribute to GC B cell lymphoma (Kuppers and Dalla-Favera, 2001).
A General Role for Transcription and Transcription Initiation in Targeting Translocations
We find a remarkable genome-wide correlation between transcription and translocations even in AID−/− cells, with a peak of translocation junctions lying near active TSSs. In this context, while the majority of junctions were located in the sense transcriptional direction, junctions also occurred at increased levels close to the TSS on the anti-sense side (e.g. Fig. 4B,C; Fig. 5), correlating with focal anti-sense transcription in the immediate vicinity of active promoters (Core et al., 2008; Fig. 5). Notably, we observed a number of regions genome-wide that were quite low in or devoid of translocations and transcription, but few, if any, that were low in translocations but high in transcription (Fig. 6). On the other hand, we found that transcription is not required for high frequency translocations, since many I-SceI-dependent hotspots are in non-transcribed regions. Together, our observations are consistent with transcription mechanistically promoting translocations by promoting DSBs. Thus, our findings strongly support the long-standing notion of a mechanistic link between transcription, DSBs, and genomic instability (Aguilera, 2002; Haffner et al., 2011; Li and Manley, 2006).
Potential Influences of Genome Organization on Translocations
The high level of translocations of 5′c-myc-I-SceI BEs to other sequences along much the length of chr15, while generally correlated with transcription, likely may be further promoted by high relative proximity of many intra-chromosomal regions (Lieberman-Aiden et al., 2009). Proximity might also contribute to the apparently increased frequency of 5′c-myc-I-SceI BEs to certain regions of various chromosomes (e.g. Fig. 2). In this regard, the relative frequency of chr15 5′c-myc-I-SceI BE translocations to the Sμ and Sε regions on chr12 were only 5 and 7 fold less, respectively, than levels of intra-IgH 5′Sγ1-ISceI BE joins to Sμ and Sε (Fig. 3,C). Thus, even though DSBs are rare in c-myc, their translocation to IgH when they do occur is driven at a high rate by other mechanistic aspects, most likely proximal position (Wang et al., 2009). However, we also note that sequences lying in regions across all chromosomes translocate to DSBs in c-myc on chr15 and IgH in chr12, suggesting the possibility that, in some cases, DSBs might move into proximity before joining, perhaps during the cell cycle or via other mechanisms (e.g. Dimitrova et al., 2008).
HTGTS Reveals An Unexpectedly Large Number of Genomic I-SceI targets
Our HTGTS studies revealed eighteen cryptic genomic I-SceI sites as translocation targets. There could potentially be more cryptic I-SceI sites; to find the full spectrum, bait sequences may need to be introduced into a variety of chromosomal locations to neutralize position effects. Beyond I-SceI, the HTGTS approach could readily be extended through the use Zinc finger nucleases (Handel and Cathomen, 2011), meganucleases (Arnould et al, 2011), or TALENS (Christian et al., 2010) designed to cleave specific endogenous sites, thereby, obviating the need to introduce a cutting site and greatly facilitating the process. The above three classes of endonucleases are being developed for targeted gene correction of human mutations in stem cells for gene therapy. One major concern with such nucleases is relative activity on the specific target versus off-target activity, with the latter being difficult to assess. HTGTS provides a means for identifying off-target DSBs generated by such enzymes, for assessing ability of such off-target DSBs to translocate, and for identifying the sequences to which they translocate.
EXPERIMENTAL PROCEDURES
Mouse strains utilized
ΔSγ12xI-SceI, c-myc25xI-SceI and AID−/− mice were described (Zarrin et al., 2007; Wang et al., 2009; Muramatsu et al., 2000). c-myc1xI-SceI mice were generated similarly to c-myc25xI-SceI mice (see Suppl. Exp. Procedures). ROSAI-SceI-GR mice were generated by targeting an I-SceI-GR/IRES-tdTomato expression cassette into Rosa26 (Suppl. Experimental Procedures). All mice used were heterozygous for modified alleles containing I-SceI cassettes. The Institutional Animal Care and Use Committee of Children’s Hospital, Boston approved all animal work.
Splenic B cell Purification, Activation in Culture and Retroviral Infection
All procedures were performed as previously described (Wang et al., 2009). c-myc25xI-SceI/ROSAI-SceI-GR B cells were cultured in medium containing charcoal-stripped serum and I-SceI-GR was activated with 10 μM triamcinolone acetate (TA, Sigma).
Generation of HTGTS libraries
Genomic DNA was digested with HaeIII for c-myc25xI-SceI samples or MspI for ΔSγ12xI-SceI samples. For adapter-PCR libraries, an asymmetric adapter was ligated to cleaved genomic DNA. Ligation products were incubated with restriction enzymes chosen to reduce background from germline and unrearranged targeted alleles. Three rounds of nested-PCR were performed with adapter- and locus-specific primers. For circularization-PCR libraries, HaeIII- or MspI-digested genomic DNA was incubated at 1.6 ng/μl to favor intramolecular ligation and samples treated with blocking enzymes as above. Two rounds of nested-PCR were performed with primers specific for sequences upstream of the I-SceI cassette. Libraries were sequenced by Roche-454. See Suppl. Exp. Procedures for details.
Data analysis
Alignment and Filtering
Sequences were aligned to the mouse reference genome (NCBI37/mm9) with the BLAT program. Custom filters were used to purge PCR repeats and multiple types of artifacts including those caused by in vitro ligation and PCR mis-priming. Hotspot Identification: Translocations from WT or AID−/− libraries minus those on chr15 or the IgH locus were pooled. The adjusted genome was then divided into 250 kb bins and bins containing ≥ 5 hits constituted a hotspot (details in Suppl. Exp. Procedures).
In vitro testing of putative cryptic I-SceI sites
A genomic region encompassing each candidate I-SceI site was PCR-amplified and 500 ng of purified products were incubated with 5 units of I-SceI for 3 hours. Reactions were separated on agarose gel and relative intensity of uncut and I-SceI-digested bands calculated with the FluorchemSP program (Alpha Innotech) (see Suppl. Exp. Procedures).
PCR detection of translocations between c-myc and cryptic I-SceI sites
Translocation junctions between c-myc and cryptic I-SceI targets were PCR-amplified according to the standard protocol (Wang et al., 2009). Primers and PCR conditions are detailed in Suppl. Exp. Procedures.
GRO-seq
Nuclei were isolated from day 4 αCD40/IL4-stimulated and I-SceI-infected c-myc25xI-SceI B cells as described (Giallourakis et al., 2010). GRO-seq libraries were prepared from 5×106 cells from two independent mice using a described protocol (Core et al., 2008). Both libraries were sequenced on the Hi-Seq 2000 platform with single-end reads and analyzed as described (see Suppl. Exp. Procedures). After filtering and alignment, we obtained 34,212,717 reads for library 1 and 15,913,244 reads for library 2. As results between libraries were highly correlated, we show results only from replicate 1.
Supplementary Material
Acknowledgments
We thank Barry Sleckman for providing unpublished information about circular PCR translocation cloning of RAG-generated DSBs. This work was supported by NIH grant 5P01CA92625 and a Leukemia and Lymphoma Society of America (LLS) SCOR grant to FA, grants from AIRC and grant FP7 ERC-2009-StG (Proposal No. 242965 “Lunely”) to RC, an NIH KO8 grant AI070837 to CG, and a V Foundation Scholar award to MG. YZ was supported by CRI postdoctoral fellowship and RF by NIH training grant 5T32CA070083-13. FA is and Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aguilera A. The connection between transcription and genomic instability. The EMBO journal. 2002;21:195–201. doi: 10.1093/emboj/21.3.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnould S, Delenda C, Grizot S, Desseaux C, Paques F, Silva GH, Smith J. The I-CreI meganuclease and its engineered derivatives: applications from cell modification to gene therapy. Protein Eng Des Sel. 2011;24:27–31. doi: 10.1093/protein/gzq083. [DOI] [PubMed] [Google Scholar]
- Chaudhuri J, Basu U, Zarrin A, Yan C, Franco S, Perlot T, Vuong B, Wang J, Phan RT, Datta A, et al. Evolution of the immunoglobulin heavy chain class switch recombination mechanism. Adv Immunol. 2007;94:157–214. doi: 10.1016/S0065-2776(06)94006-1. [DOI] [PubMed] [Google Scholar]
- Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova N, Chen YC, Spector DL, de Lange T. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature. 2008;456:524–528. doi: 10.1038/nature07433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley DD, Manis JP, Zarrin AA, Kaylor L, Tian M, Alt FW. Internal IgH class switch region deletions are position-independent and enhanced by AID expression. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:9984–9989. doi: 10.1073/pnas.152333499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson DO, Sekiguchi JM, Chang S, Frank KM, Gao Y, DePinho RA, Alt FW. The nonhomologous end-joining pathway of DNA repair is required for genomic stability and the suppression of translocations. Proc Natl Acad Sci USA. 2000;97:6630–6633. doi: 10.1073/pnas.110152897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco S, Gostissa M, Zha S, Lombard DB, Murphy MM, Zarrin AA, Yan C, Tepsuporn S, Morales JC, Adams MM, et al. H2AX prevents DNA breaks from progressing to chromosome breaks and translocations. Molecular Cell. 2006;21:201–214. doi: 10.1016/j.molcel.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Giallourakis CC, Franklin A, Guo C, Cheng HL, Yoon HS, Gallagher M, Perlot T, Andzelm M, Murphy AJ, Macdonald LE, et al. Elements between the IgH variable (V) and diversity (D) clusters influence antisense transcription and lineage-specific V(D)J recombination. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:22207–22212. doi: 10.1073/pnas.1015954107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Gonzalez B, Aguilera A. Activation-induced cytidine deaminase action is strongly stimulated by mutations of the THO complex. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:8409–8414. doi: 10.1073/pnas.0702836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gostissa M, Alt FW, Chiarle R. Mechanisms that Promote and Suppress Chromosomal Translocations in Lymphocytes. Annu Rev Immunol. 2011 doi: 10.1146/annurev-immunol-031210-101329. [DOI] [PubMed] [Google Scholar]
- Gostissa M, Ranganath S, Bianco JM, Alt FW. Chromosomal location targets different MYC family gene members for oncogenic translocations. Proc Natl Acad Sci U S A. 2009;106:2265–2270. doi: 10.1073/pnas.0812763106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffner M, De Marzo AM, Meeker AK, Nelson WG, Yegnasubramanian S. Transcription-induced DNA double strand breaks: both an oncogenic force and potential therapeutic target? Clinical cancer research : an official. Journal of the American Association for Cancer Research. 2011 doi: 10.1158/1078-0432.CCR-10-2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handel EM, Cathomen T. Zinc-finger nuclease based genome surgery: it’s all about specificity. Current gene therapy. 2011;11:28–37. doi: 10.2174/156652311794520120. [DOI] [PubMed] [Google Scholar]
- Jasin M. Genetic manipulation of genomes with rare-cutting endonucleases. Trends Genet. 1996;12:224–228. doi: 10.1016/0168-9525(96)10019-6. [DOI] [PubMed] [Google Scholar]
- Kovalchuk AL, duBois W, Mushinski E, McNeil NE, Hirt C, Qi CF, Li Z, Janz S, Honjo T, Muramatsu M, et al. AID-deficient Bcl-xL transgenic mice develop delayed atypical plasma cell tumors with unusual Ig/Myc chromosomal rearrangements. The Journal of experimental medicine. 2007;204:2989–3001. doi: 10.1084/jem.20070882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuppers R, Dalla-Favera R. Mechanisms of chromosomal translocations in B cell lymphomas. Oncogene. 2001;20:5580–5594. doi: 10.1038/sj.onc.1204640. [DOI] [PubMed] [Google Scholar]
- Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annual review of biochemistry. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Manley JL. Cotranscriptional processes and their influence on genome stability. Genes Dev. 2006;20:1838–1847. doi: 10.1101/gad.1438306. [DOI] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Yang L, Tanasa B, Hutt K, Ju BG, Ohgi K, Zhang J, Rose DW, Fu XD, Glass CK, et al. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell. 2009;139:1069–1083. doi: 10.1016/j.cell.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Duke JL, Richter DJ, Vinuesa CG, Goodnow CC, Kleinstein SH, Schatz DG. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 2008;451:841–845. doi: 10.1038/nature06547. [DOI] [PubMed] [Google Scholar]
- Liu M, Schatz DG. Balancing AID and DNA repair during somatic hypermutation. Trends in immunology. 2009;30:173–181. doi: 10.1016/j.it.2009.01.007. [DOI] [PubMed] [Google Scholar]
- Mahowald GK, Baron JM, Mahowald MA, Kulkarni S, Bredemeyer AL, Bassing CH, Sleckman BP. Aberrantly resolved RAG-mediated DNA breaks in Atm-deficient lymphocytes target chromosomal breakpoints in cis. Proc Natl Acad Sci USA. 2009;106:18339–18344. doi: 10.1073/pnas.0902545106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaburn KJ, Misteli T, Soutoglou E. Spatial genome organization in the formation of chromosomal translocations. Seminars in cancer biology. 2007;17:80–90. doi: 10.1016/j.semcancer.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- Nussenzweig A, Nussenzweig MC. Origin of chromosomal translocations in lymphoid cancer. Cell. 2010;141:27–38. doi: 10.1016/j.cell.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualucci L, Neumeister P, Goossens T, Nanjangud G, Chaganti RS, Kuppers R, Dalla-Favera R. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature. 2001;412:341–346. doi: 10.1038/35085588. [DOI] [PubMed] [Google Scholar]
- Pavri R, Nussenzweig MC. AID targeting in Antibody Diversity. Adv Immunol. 2011 doi: 10.1016/B978-0-12-387663-8.00005-3. In press. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Clark RA, Sokolova S, Leibowitz ML, Zhang Y, Hurles ME, Mell JC, Hall IM. Genome-wide mapping and assembly of structural variant breakpoints in the mouse genome. Genome research. 2010;20:623–635. doi: 10.1101/gr.102970.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramiro AR, Jankovic M, Callen E, Difilippantonio S, Chen HT, Mcbride KM, Eisenreich TR, Chen J, Dickins RA, Lowe SW, et al. Role of genomic instability and p53 in AID-induced c-myc-Igh translocations. Nature. 2006;440:105–109. doi: 10.1038/nature04495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynaud S, Delpy L, Fleury L, Dougier HL, Sirac C, Cogne M. Interallelic class switch recombination contributes significantly to class switching in mouse B cells. Journal of immunology. 2005;174:6176–6183. doi: 10.4049/jimmunol.174.10.6176. [DOI] [PubMed] [Google Scholar]
- Robbiani DF, Bothmer A, Callen E, Reina-San-Martin B, Dorsett Y, Difilippantonio S, Bolland DJ, Chen HT, Corcoran AE, Nussenzweig A, et al. AID Is Required for the Chromosomal Breaks in c-myc that Lead to c-myc/IgH Translocations. Cell. 2008;135:1028–1038. doi: 10.1016/j.cell.2008.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer DR, Pandolfi PP. Breaking the rules of cancer. Nature medicine. 2006;12:14–15. doi: 10.1038/nm0106-14. [DOI] [PubMed] [Google Scholar]
- Siebert PD, Chenchik A, Kellogg DE, Lukyanov KA, Lukyanov SA. An improved PCR method for walking in uncloned genomic DNA. Nucleic acids research. 1995;23:1087–1088. doi: 10.1093/nar/23.6.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek D, Jasin M. Alternative end-joining is suppressed by the canonical NHEJ component Xrcc4-ligase IV during chromosomal translocation formation. Nature structural & molecular biology. 2010;17:410–416. doi: 10.1038/nsmb.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unniraman S, Zhou S, Schatz DG. Identification of an AID-independent pathway for chromosomal translocations between the Igh switch region and Myc. Nature immunology. 2004;5:1117–1123. doi: 10.1038/ni1127. [DOI] [PubMed] [Google Scholar]
- Wang JH, Gostissa M, Yan CT, Goff P, Hickernell T, Hansen E, Difilippantonio S, Wesemann DR, Zarrin AA, Rajewsky K, et al. Mechanisms promoting translocations in editing and switching peripheral B cells. Nature. 2009;460:231–236. doi: 10.1038/nature08159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane A, Resch W, Kuo N, Kuchen S, Li Z, Sun HW, Robbiani DF, McBride K, Nussenzweig MC, Casellas R. Deep-sequencing identification of the genomic targets of the cytidine deaminase AID and its cofactor RPA in B lymphocytes. Nature immunology. 2011;12:62–69. doi: 10.1038/ni.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan CT, Boboila C, Souza EK, Franco S, Hickernell TR, Murphy M, Gumaste S, Geyer M, Zarrin AA, Manis JP, et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- Zarrin AA, Del Vecchio C, Tseng E, Gleason M, Zarin P, Tian M, Alt FW. Antibody class switching mediated by yeast endonuclease-generated DNA breaks. Science. 2007;315:377–381. doi: 10.1126/science.1136386. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Gostissa M, Hildebrand DG, Becker MS, Boboila C, Chiarle R, Lewis S, Alt FW. The role of mechanistic factors in promoting chromosomal translocations found in lymphoid and other cancers. Adv Immunol. 2010;106:93–133. doi: 10.1016/S0065-2776(10)06004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Mills KD, Ferguson DO, Lee C, Manis J, Fleming J, Gao Y, Morton CC, Alt FW. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell. 2002;109:811–821. doi: 10.1016/s0092-8674(02)00770-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.