Abstract
MicroRNAs (miRs) have emerged as important gene silencers affecting many target mRNAs. Here, we report the identification of 244 miRs that target the 3′-untranslated regions of different cholinesterase transcripts: 116 for butyrylcholinesterase (BChE), 47 for the synaptic acetylcholinesterase (AChE-S) splice variant, and 81 for the normally rare splice variant AChE-R. Of these, 11 and 6 miRs target both AChE-S and AChE-R, and AChE-R and BChE transcripts, respectively. BChE and AChE-S showed no overlapping miRs, attesting to their distinct modes of miR regulation. Generally, miRs can suppress a number of targets; thereby controlling an entire battery of functions. To evaluate the importance of the cholinesterase-targeted miRs in other specific biological processes we searched for their other experimentally validated target transcripts and analyzed the gene ontology enriched biological processes these transcripts are involved in. Interestingly, a number of the resulting categories are also related to cholinesterases. They include, for BChE, response to glucocorticoid stimulus, and for AChE, response to wounding and two child terms of neuron development: regulation of axonogenesis and regulation of dendrite morphogenesis. Importantly, all of the AChE-targeting miRs found to be related to these selected processes were directed against the normally rare AChE-R splice variant, with three of them, including the neurogenesis regulator miR-132, also directed against AChE-S. Our findings point at the AChE-R splice variant as particularly susceptible to miR regulation, highlight those biological functions of cholinesterases that are likely to be subject to miR post-transcriptional control, demonstrate the selectivity of miRs in regulating specific biological processes, and open new venues for targeted interference with these specific processes.
Keywords: AChE, BChE, microRNA
Introduction
MicroRNAs (miRs) are small RNA molecules which target many mRNA transcripts, leading to their post-transcriptional silencing (Bartel, 2009). Many mRNAs can be silenced by multiple miRs and miRs often target more than one mRNA participating in a particular biological function (Bartel, 2009). Together, this suggests that the miR networks affecting specific mRNA transcripts may provide useful information on the biological roles in which these transcripts are involved. Cholinesterases are involved in many biological functions (Massoulie, 2002). However, miR-132 is the only miR so far that has been experimentally validated as targeting AChE, with consequences on inflammatory responses (Shaked et al., 2009). To delineate additional miRs which might regulate cholinesterase functions, we explored the 3′-untranslated regions (3′-UTR) of human cholinesterase transcripts (acetyl- and butyrylcholinesterase, AChE, BChE; Soreq and Seidman, 2001).
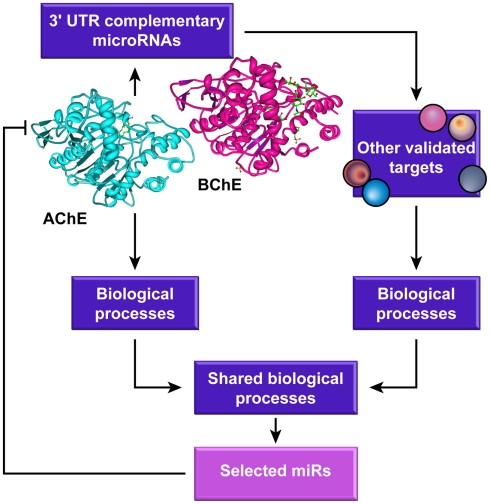
Given that several of the proteins involved in a specific function are often repressed by the same miR (Girardot et al., 2010), changes in a particular miR might down-regulate the entire process. Hence, we surmised that those functions that are shared by cholinesterases and the other targets of the cholinesterase-complementary miRs would be more susceptible for being affected by miR control than other processes. That concept is schematically presented as a workflow in Figure 1.
Figure 1.
The study’s flow chart. MicroRNAs complementary to the 3′-UTR domains of AChE and BChE transcripts were identified using several algorithms and other validated targets for those miRs were searched for and analyzed for common biological processes in which both these miR targets and cholinesterases are involved.
Materials and Methods
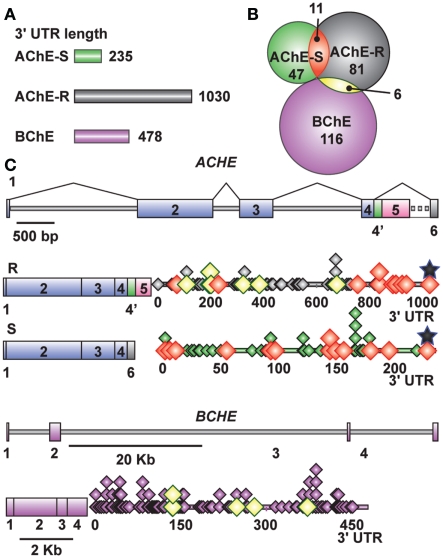
MicroRNA candidates were identified on each of the 3′-UTR sequences of AChE and BChE, which are 235, 1030, and 478 nucleotides long for BChE, the major “synaptic” AChE-S variant and the stress-inducible AChE-R variant, respectively (Figure 2A). We used the PicTar1, miRanda2, miRbase3, and microCosm4 algorithms to identify these transcript-specific miRs. All predictions ensured a threshold P-value < 0.05, and analysis specifications allowed both evolutionarily conserved and non-conserved miRs, which enabled us to include primate-targeting miRs as well.
Figure 2.
Cholinesterase-targeted miRs show distinct 3′UTR distributions and partial overlaps. (A) The length of the studied 3′UTR domains. (B) Each 3′-UTR is targeted by many different miRs, part of which shared by BChE/AChE-R and AChE-R/AChE-S. (C) Gene and transcript compositions (exons shown as boxes, introns – as lines) and miR distribution patterns on the 3′-UTR domain (not to scale). Overlaps are color coded. MiR (diamonds) localizations are marked. Black stars show miR-132 position.
Validation of miR-target interactions generally involved a 3′UTR luciferase assay. In some cases, it was complemented by protein blots, real-time RT-qPCR, microarrays, transgenic technology, β-galactosidase, or GFP-tagged targets. See, for example the Shaked et al. (2009) report for several of the latter technologies used to explore the miR-132 target AChE, and (Hansen et al., 2010) for the “classical” 3′-UTR and transgenic approaches, in exploring p250GAP which is also a miR-132 target.
To search for gene ontology (GO) categories which are also relevant for the other mRNA targets of cholinesterase-related miRs, we used the DAVID functional annotation clustering tool5. For each of the miRs identified as targeting one of the cholinesterases we searched for other experimentally validated targets; and we then used the lists of the other validated targets as gene lists for the DAVID search. Each list was normalized to the entire human genome, which served as a background.
Results
We identified 116, 81, and 47 miRs (24, 8, and 20 miRs/100 nucleotides) that are complementary to the 3′-UTR domains of the BChE, AChE-R, and AChE-S transcripts, respectively. Of these, 6 miRs target both BChE and AChE-R whereas 11 miRs are common to both AChE-R and AChE-S, but BChE and AChE-S do not share any miR (Figure 2B). Positions of the identified miRs are presented in Figure 2C, with miR-132 targeting a similar seed domain localized at the very 3′-end of the 3′-UTR in both the AChE-S and AChE-R transcripts. Of the cholinesterase-targeting miRs, seven had multiple binding sites to the target AChE-S, nine to AChE-R, and seven to the BChE transcript, suggesting that they have a higher prospect for being functional (John et al., 2004). Compatible with the different conceptual principles on which each of the algorithms employed is based, only 8.6, 17, and 13.7% (7/81), (8/47), (16/116) of the miRs identified as targeting AChE-R, AChE-S, and BChE, respectively, were predicted by more than one of the algorithms. For AChE-R, these are hsa-miR-28-5p, −423-3p, −484, −483-5p, −663, −582-3p, −380*. For AChE-S, hsa-miR-194, −939, −658, −608,-615-5p, −423-5p −920, and let-7f-2* and for BChE, hsa-miR-203, −218, −221, −222, −181a, −181b, −181c, −181d, −494, −200b, −200c, −576-3p, −16-2*, −625, −195*, −889.
These cholinesterase-targeting miRs and their other validated non-cholinesterase targets are listed in Tables 1– 3 with the corresponding functions attributed to these other targets. The relevant citations appear in Tables A1– A4 in Appendix. Of note, numerous cholinesterase-targeting miRs have no experimentally validated targets at this time, yet others have more than one validated target and associate with more than one biological function. Examples include miR-124 which targets both the AChE-S and IQGAP1-(Furuta et al., 2010), a GTPase activating protein which promotes neurite outgrowth (Table 1). Additionally miR-152 and miR-148a, which target AChE-R, also target the calmodulin regulating kinase CaMKIIα (Liu et al., 2010; Table 2). Lastly, the BChE-targeting cluster of miRs-222 and −221 also target the neuronal early immediate protein c-fos (Ichimura et al., 2010; Table 3).
Table 1.
Additional targets of AChE-S targeting microRNAs.
| miR ID | Validated targets | ||
|---|---|---|---|
| hsa-miR-491-5p | Bcl-X(L; cell death) | ||
| hsa-miR-605 | Mdm2 (ubiquitination) | ||
| hsa-miR-608 | CD44 (cell–cell/cell–matrix interaction) | CDC42 (cell division) | |
| hsa-miR-124 | Glucocorticoid receptor | LAMC1 (laminin γ1) | IQGAP1(neurite outgrowth; Furuta et al., 2010) |
| NeuroD1 (neurogenic differentiation 1) | BAF53a (chromatin remodeling) | C14orf24 (chromosome 14 ORF 24) | |
| Mtpn (myotrophin) | PTBP1 (splicing) | CDK6 (cyclin-dependent kinase 6) | |
| Mapk14 (mitogen activated protein kinase 14) | PTBP2 (splicing) | SOX9 (glial cell specification) | |
| CDK2 (cyclin-dependent kinase 2) | C/EBPα (transcription) | Lhx2 (transcription) | |
| MCP1 (monocyte chemoattractant protein 1) | FOXA2 (transcription) | EfnB1(projecting axons) | |
| Itgb1 (integrin 1) | VIM (cytoskeleton; Furuta et al., 2010) | NR3C2 (Mineralocorticoid and glucocorticoid receptor) | |
| SCP1 (synaptonemal filaments) | SMYD3 (transcription; Furuta et al., 2010) | ||
| hsa-let-7g | C-Myc (transcription) | Collagen alpha2 (COL1A2) | Bcl-xL (cell death) |
| hsa-miR-196a | HOX-B7 (transcription) | SPRR2C (small proline-rich protein 2C) | Annexin A1 (exocytosis) |
| S100A9 (calcium-binding protein A9) | KRT5 (keratin 5) | HOXC8 (transcription) | |
| hsa-miR-542-3p | Survivin | ||
| hsa-miR-525-5p | VPAC1 (vasoactive intestinal peptide receptor 1) | ||
Table 3.
Additional targets of BChE-targeting microRNAs.
| miR ID | Validated target | ||
|---|---|---|---|
| hsa-miR-203 | SOCS-3 (cytokine signaling) | Lef1 (lymphoid enhancer-binding factor) | p63 (transcription) |
| ABL1 (cell growth) | Barx1 (transcription) | CKAP2 (cytoskeleton associated protein 2) | |
| LASP1 (cytoskeletal activities) | BIRC5 (regulator of mitosis) | WASF1 (signal transmission) | |
| ASAP1 (membrane trafficking) | RUNX2 (runt-related transcription factor 2) | ||
| hsa-miR-340 | MITF (microphthalmia-associated transcription factor) | ||
| hsa-miR-218 | IKK-β (NFκB activation) | ROBO1 (roundabout, axon guidance receptor, homolog 1) | BIRC5 (mitosis) |
| GJA1 (gap junction protein, α1) | ROBO2 (roundabout, axon guidance receptor homolog 2) | GLCE (glucuronic acid epimerase) | |
| PXN (paxillin, cytoskeletal protein) | |||
| hsa-miR-221 | ERα (estrogen receptor α) | ICAM-1(leukocyte adhesion protein) | p27 (cell cycle) |
| p57 (cyclin-dependent kinase inhibitor 1C) | DNA damage-inducible transcript 4 (DDIT4) | TIMP3 (TIMP metallopeptidase inhibitor 3) | |
| PTEN (tumor suppressor) | PUMA (apoptosis) | C-fos (cell proliferation; Ichimura et al., 2010) | |
| Bmf (apoptosis) | Mdm2 (ubiquitination) | CDKN1B (cyclin-dependent kinase inhibitor 1B) | |
| hsa-miR-222 | ERα (estrogen receptor α) | p27 (cell cycle) | PTEN (tumor suppressor) |
| STAT5A (transcription) | p57 (cyclin-dependent kinase inhibitor 1C) | TIMP3 (TIMP metallopeptidase inhibitor 3) | |
| Bim (apoptosis) | ETS-1 (transcription) | PUMA (apoptosis) | |
| PPP2R2A (protein phosphatase 2A subunit B) | C-fos (cell proliferation; Ichimura et al., 2010) | ICAM-1(leukocyte adhesion protein) | |
| MMP1 (cleaves collagens) | SOD2 (superoxide dismutase 2) | ||
| hsa-miR-181a | SIRT1 (apoptosis) | Ataxia telangiectasia mutated (ATM; cell cycle) | Hox-A11 (transcription) |
| p27(cell cycle) | PLAG1 (transcription) | BCL2 (B-cell CLL/lymphoma 2; apoptosis) | |
| Bim (apoptosis) | Tcl1 (cell proliferation) | OPN (osteopontin) | |
| hsa-miR-181b | AID (RNA-editing) | PLAG1 (transcription) | BCL2 (B-cell CLL/lymphoma 2; apoptosis) |
| TIMP3 (TIMP metallopeptidase inhibitor 3) | Ataxia telangiectasia mutated (ATM; cell cycle) | SIRT1 (apoptosis) | |
| ZNF37A (transcriptional regulation) | ZNF83 (transcriptional regulation) | ZNF182 (transcriptional regulation) | |
| Mcl-1 (myeloid cell leukemia-1; apoptosis) | |||
| hsa-miR-181c | IL2 (immune response) | BCL2 (B-cell CLL/lymphoma 2; apoptosis) | NOTCH4 (transcriptional activator) |
| KRAS (GTPase activity) | |||
| hsa-miR-181d | BCL2 (B-cell CLL/lymphoma 2; apoptosis) | ||
| hsa-miR-494 | CaMKIIδ (CNS kinase) | ROCK-1 (apoptosis) | LIF [leukemia inhibitory factor (cholinergic differentiation factor)] |
| PTEN (phosphatase and tensin homolog) | TEL-AML1 (hematopoiesis) | FGFR2 (fibroblast growth factor receptor 2) | |
| hsa-miR-129-5p | CAMTA1 (calmodulin binding transcription activator 1) | EIF2C3 (eukaryotic translation initiation factor 2C, 3) | GALNT1 (oligosaccharide biosynthesis) |
| SOX4 (transcriptional activator) | |||
| hsa-miR-30d | Galphai2 (G protein, α inhibiting activity polypeptide 2) | ||
| hsa-miR-30c | Runx1 (runt-related transcription factor 1) | CTGF (connective tissue growth factor) | |
| hsa-miR-30a | SOD2 (superoxide dismutase 2) | BDNF (brain-derived neurotrophic factor) | Beclin 1 (autophagy) |
| Xlim1/Lhx1 (transcription factor) | |||
| hsa-miR-30e | Ubc9 (ubiquitin-conjugating enzyme E2I) | B-Myb (transcription factor) | |
| hsa-miR-320a | (Hsp20 heat-shock protein 20) | AQP1 (aquaporin 1) | AQP4 (aquaporin 4) |
| TfR-1; CD71 (development of erythrocytes and the nervous system) | Mcl-1 (myeloid cell leukemia sequence 1; apoptosis) | ||
| hsa-miR-140-5p | Smad3 (transcription) | HDAC4 (histone deacetylase 4) | |
| hsa-miR-519c-3p | HIF-1α (hypoxia-inducible factor 1α) | ABCG2 (exclusion of xenobiotics from the brain) | |
| hsa-miR-489 | PTPN11 (signal transduction) | ||
| hsa-miR-584 | NXA1 (exocytosis) | ROCK-1 (actin assembly) | |
Table 2.
Additional targets of microRNAs targeting AChE-R.
| miR ID | Validated targets | ||
|---|---|---|---|
| Hsa-miR-708 | MPL (thrombopoietin receptor; Girardot et al., 2010) | ||
| Hsa-miR-28-5p | MPL (thrombopoietin receptor; Girardot et al., 2010) | OTUB1 (immune system transcription; Girardot et al., 2010) | |
| N4BP1 (NEDD4 binding protein 1; Girardot et al., 2010) | TEX261 (apoptosis; Girardot et al., 2010) | MAPK1 (megakaryocyte differentiation; Girardot et al., 2010) | |
| hsa-miR-503 | ANLN (actin-binding protein anillin) | ATF6 (activating transcription factor 6) | CHEK1 (cell cycle) |
| EIF2C1 (argonaute1) | KIF23 (mitotic kinesin-like protein 1) | WEE1 (mitosis regulator) | |
| CCNE1 (cyclin E1) | CDC25A (cell cycle) | ||
| CCND1 (cyclin D1) | CDC14A (CDC14 cell division cycle 14 homolog A) | ||
| hsa-miR-148a | CaMKIIα (CNS kinase; Liu et al., 2010) | MLC1 (megalencephalic leukoencephalopathy with subcortical cysts 1) | MSK1 (histone phosphorylase) |
| DNMT1 (DNA methyltransferase 1) | DNMT3B (CpG island methylation) | MITF (microphthalmia-associated transcription factor) | |
| CCKBR (modulates anxiety and neuroleptic activity) | EPAS1 (endothelial PAS domain-containing protein 1) | HLA-G (asthma susceptibility) | |
| POMC (pro-opiomelanocortin) | CAND1 (ubiquitin ligase regulation) | PXR (pregnane X receptor) | |
| hsa-miR-152 | CaMKIIα (CNS kinase; Liu et al., 2010) | DNMT1 (DNA methyltransferase 1) | |
| hsa-miR-125b | TNFα (tumor necrosis factor α) | ERBB2 (erythroblastic leukemia viral oncogene homolog 2) | BMPR1B (bone morphogenic receptor type 1B) |
| IRF4 (interferon regulatory factor 4) | ERBB3 (erythroblastic leukemia viral oncogene homolog 3) | E2F3 (cell cycle) | |
| Blimp1 (zinc finger protein) | TEF (thyrotroph embryonic factor) | Bcl2 modifying factor (apoptosis) | |
| Vdr (vitamin D receptor) | MUC1 (adhesion) | Bak1 (pro-apoptotic Bcl2 antagonist killer 1) | |
| CYP24A1 (cytochrome P450 family 24A) | p53 (tumor suppressor) | SMO (smoothened receptors) | |
| IGF2 (insulin-like growth factor 2) | Suv39h1 (histone methyltransferase) | Stat3 (Transcription factor, binds to IL-6) | |
| LIN28 (translational enhancer) | NMDA receptor subunit NR2A | ATM (ataxia telangiectasia mutated) | |
| hsa-miR-125a-5p | LIN28 (translational enhancer) | T-TrkC (neurotrophic tyrosine kinase receptor 3) | HuR (cell growth) |
| p53 (tumor suppressor) | KLF13 (transcription factor) | AT-rich interactive domain 3B (transcription) | |
| PDPN 9 (actin organization) | Bak1 (pro-apoptotic Bcl2 antagonist killer 1) | ||
| N-ras (oncogene) | MEK3 (phosphorylation of MAP kinase) | ||
| hsa-miR-214 | SrGAP1(neuronal migration) | Ezh2 (stem cell identity) | N-ras (oncogene) |
| JNK1 (MAPK8) | PTEN (tumor suppressor) | MEK3 (phosphorylation) | |
| hsa-miR-199a-5p | Hif-1α (Hypoxia-inducible factor 1) | IKKβ (NFκB activation) | DDR1 (discoidin domain receptor 1) |
| Sirt1 (apoptosis) | |||
| hsa-miR-31 | ICAM-1 (leukocyte adhesion protein) | Fgf13 (fibroblast growth factor 13) | Dkk-1 (canonical Wnt signaling) |
| DACT-3 (epigenetic regulator of Wnt) | E-selectin (inflammation) | p16Ink4a (cell cycle) | |
| LATS2 (tumor suppression) | PPP2R2A (signal transduction) | Krt16 (keratin 16) | |
| Krt17 (keratin 17) | Dlx3 (development of ventral forebrain) | E2F6 (cell cycle) | |
| TIAM1 (T-cell lymphoma invasion and metastasis 1) | Fzd3 (accumulation of β-catenin) | Integrin α (fibronectin) | |
| M-RIP (regulation of actin) | MMP16 (blood vessels matrix remodeling) | RDX (actin filaments binding to plasma membrane) | |
| RhoA (signal transduction) | SATB2 (upper-layer neurons initiation) | PROX1 (CNS development) | |
| WAVE3 (signal transmission) | |||
| hsa-miR-185 | Six1 (limb development) | ||
| hsa-miR-193b | Estrogen receptor α Mcl-1 (myeloid cell leukemia sequence 1) | ETS-1 (oncogene) uPA (urokinase-type plasminogen activator) | CCND1 (cyclin D1) |
| hsa-miR-7 | Alpha-synuclein (SNCA) | SFRS1 (splicing) | ERF (cell proliferation) |
| LSH (lymphoid-specific helicase) | DAP (cell death-associated protein) | MRP1 (human multidrug resistance-associated protein 1) | |
| Associated cdc42 kinase 1 | Yan (cell differentiation) | EGFR (epidermal growth factor receptor) | |
| CD98 (sodium transport) | Pak1 (p21-activated kinase 1) | IGF1R (insulin-like growth factor 1 receptor) | |
| hsa-miR-483-5p | Socs-3 (cytokine signaling) | BBC3/PUMA (apoptosis) | |
| hsa-miR-663 | TGFβ1 (proliferation) | JunB (jun B proto-oncogene) | JunD (jun D proto-oncogene) |
| hsa-miR-765 | TRK3 (neurotrophic tyrosine kinase) | ||
| hsa-miR-146b-3p | IRAK1 (IL1 receptor-associated kinase 1) | EGFR (epidermal growth factor receptor) | MMP16 (degrades extracellular matrix) |
We focused our survey on those functions of those miRs for which experimental validation is available. Table 4 presents these miRs which are shared for AChE-R and AChE-S or AChE-R and BChE and some of their additional targets, highlighting the multitude of miR targets with predicted regulatory functions (e.g., the chromatin modulator zinc finger proteins ZEB1 and ZEB2 targeted by miR-200b, miR-200c, and miR-429 that are also directed to both AChE-R and AChE-S; Gregory et al., 2008). Likewise, the AChE-S-targeted miR-132 (Shaked et al., 2009; Soreq and Wolf, 2011) also targets the GTPase regulator p250GAP involved in neurite extension (Vo et al., 2005; Hansen et al., 2010; Table 4).
Table 4.
Additional targets of ChE-targeting miRs (common to more than one ChE).
| miR ID | Validated target common to ACHE-R and AChE-S | ||
|---|---|---|---|
| hsa-miR-186 | Pro-apoptotic P2 × 7 purinergic receptor | AKAP12 (tumor suppressor) | |
| hsa-miR-199b-5p | Dyrk1a (brain development) | HES1 (transcriptional repressor) | SET (apoptosis) |
| hsa-miR-429 | ZEB1 (transcriptional repression of IL2; Gregory et al., 2008) | ZEB2 (SIP1; zinc finger protein; Gregory et al., 2008) | PLCgamma1(apoptosis) |
| RERE (apoptosis) | |||
| hsa-miR-200b | ZEB1 (transcriptional repression of IL2; Gregory et al., 2008) | ZEB2 (SIP1; zinc finger protein; Gregory et al., 2008) | PLCgamma1(apoptosis) |
| Serca2 (sarco/endoplasmic reticulum Ca2+ ATPase) | Suz12 (chromatin silencing) | Ets-1 (transcriptions factor) | |
| OREBP (osmotic response element) | Cyclin D1 | ||
| hsa-miR-200c | ZEB1 (transcriptional repression of IL2; Gregory et al., 2008) | ZEB2 (SIP1; zinc finger protein; Gregory et al., 2008) | PLCgamma1(apoptosis) |
| VEGF (angiogenesis) | TUBB3 (neurogenesis and axon guidance) | TRPS1 (transcription factor) | |
| KLF13 (transcription factor) | MBNL2 (muscleblind-like protein 2) | FAP1 (apoptosis) | |
| miR ID | Validated targets common to ACHE-R and BChE | ||
| hsa-miR-24 | SOD1 (superoxide dismutase 1) | ALK4 (transducer of activin) | Notch1 (Bergmann glia differentiation) |
| MKK4 (survival signal in T cells) | E2F2 (cell cycle) | H2AX (histone-formation) | |
| FAF1 (apoptosis) | HNF4α (cell proliferation) | FURIN (processing of TGFβ1) | |
| DHFR (dihydrofolate reductase) | DND1(miRNA-mediated gene suppression) | ||
| hsa-miR-212 | MeCP2 (interaction with histone deacetylase) PED (apoptosis) | MYC (transcription) | Rb1(tumor suppressor) |
| hsa-miR-132 | AChE-S (Shaked et al., 2009) | P250GAP (neuron-associated GTPase; Vo et al., 2005) | Per1 (circadian clock) |
| SirT1 (apoptosis) | MeCP2 (modification of eukaryotic genomes) | p300 (chromatin remodeling) | |
| Jarid1a (histone demethylase) | Btg2 (cell cycle) | Paip2a (translation regulation) | |
| p120RasGAP (angiogenesis) | |||
| hsa-miR-198 | Cyclin T1 | ||
| hsa-miR-194 | Rac1 (GTP-binding protein) | Per family (circadian) | EP300 (transcriptional co-activator) |
| MDM2 (p53 negative regulator) | |||
The process-regulation hypothesis of miR function predicts the existence of biological functions in which both cholinesterases, and those other targets which share miRs with cholinesterases, would be involved. To challenge this hypothesis, we first identified the GO categories in which AChE and BChE are involved, and found 24 and 11 biological processes for these two proteins, respectively. Twenty-three, 13, and 18 enriched biological processes emerged as shared processes for the other validated targets of AChE-R, AChE-S, and BChE-targeting miRs, respectively (P-value threshold < 0.05).
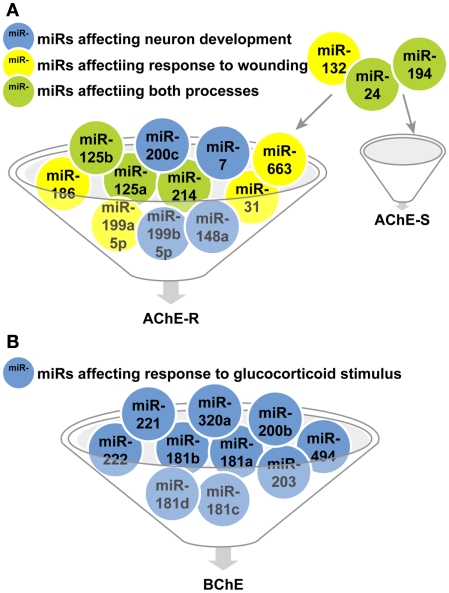
Out of over 20 ontology categories attributed to AChE, only two are shared with the categories attributed to the other validated targets of the cholinesterase-targeting miRs. These are: Response to wounding (GO: 0009611; 68 transcripts) and Neuron development (GO: 0048666), and specifically its AChE-relevant child terms Regulation of axonogenesis (GO: 0050770; 78 transcripts) and regulation of dendrite morphogenesis (GO: 0048814; 27 transcripts). Surprisingly, all 10 miRs that regulate Response to wounding and Neuron development selectively target the normally rare, stress-responsive AChE-R transcript, (miR-186, −125b, −200c, −199a-5p, −199b-5p, −125a, −214, −7, −663, −31, and −148a) whereas only three of these miRs also target the prevalent AChE-S mRNA (miR-194, −24, and −132). For BChE, we found only one shared category out of 11 relevant ontology groups: Response to glucocorticoid stimulus (GO: 0051384; 119 transcripts), and no overlap with the AChE-relevant categories (Figures 3A,B).
Figure 3.
MiR regulators of biological processes shared by cholinesterases and validated targets of these miRs. (A) miRs targeting transcripts participating in the AChE-S and AChE-R relevant response to wounding (yellow)and neuron development processes(blue) or both categories(green). (B) miRs targeting transcripts participating in the BChE-relevant response to glucocorticoid stimulus category.
Discussion
Using a variety of available algorithms, we found a plethora of cholinesterase-targeted miRs. Some of these were already validated as functionally capable of silencing other mRNA transcripts. A study of the functionally relevant biological processes in which these other targets are involved revealed a highly focused overlap with only few of the biological processes in which cholinesterases participate. Given that miRs regulate targets which share biological processes, cholinesterases appear to be primarily subject to miR regulation when involved in neuronal development, response to wounding, and glucocorticoid stimulus; and specific cholinergic processes are regulated by miRs targeting both AChE and other targets participating in the same biological process.
Several limitations should be considered in the context of this study. First, the currently available search algorithms for miR candidates appear to differ substantially, which casts a shadow on the veracity of such identification. Second, research bias has focused much of the efforts in the miR field toward cancer research, whereas neuroscience-focused miRs were relatively neglected. Therefore, we might have overlooked important miRs simply because they have not yet been validated experimentally. This being said, that many of the biological functions in which cholinesterases are involved show no relevant cholinesterase-targeting miR sequences suggests other modes of regulation of cholinesterase levels for most of these functions [e.g., transcriptional (Hill and Treisman, 1995), epigenetic (Allshire and Karpen, 2008), or post-translational processes (Fukushima et al., 2009)]. Alternatively, or in addition, miRs might exist which control these functions, but have no role in cancer biology and are therefore not yet characterized. MiR regulation of cholinesterase functions will therefore need to be re-inspected in the near future.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to E. R. Bennett, Jerusalem, for critical evaluation of this manuscript. This work was supported by the Legacy Heritage Biomedical Science Partnership Program of the Israel Science Foundation (Grant No. 1876/08, to Hermona Soreq).
Appendix
Table A1.
Additional targets of AChE-S targeting microRNAs.
| miR ID | Validated targets | ||
|---|---|---|---|
| hsa-miR-491-5p | Bcl-X (L; cell death; Nakano et al., 2010) | ||
| hsa-miR-605 | Mdm2 (ubiquitination; Xiao et al., 2011) | ||
| hsa-miR-608 | CD44 (cell–cell/cell–matrix interaction; Jeyapalan et al., 2011) | CDC42 (cell division; Jeyapalan et al., 2011) | |
| hsa-miR-124 | Glucocorticoid receptor (Vreugdenhil et al., 2009) | LAMC1 (laminin γ1; Cao et al., 2007) | IQGAP1(neurite outgrowth; Furuta et al., 2010) |
| NeuroD1 (neurogenic differentiation 1; Liu et al., 2011) | BAF53a (chromatin remodeling; Yoo et al., 2009) | C14orf24 (chromosome 14 open reading frame 24; Ko et al., 2009) | |
| Mtpn (myotrophin; Krek et al., 2005) | PTBP1 (splicing; Makeyev et al., 2007) | CDK6 (cyclin-dependent kinase 6; Pierson et al., 2008) | |
| Mapk14 (mitogen activated protein kinase 14; Krek et al., 2005) | PTBP2 (splicing; Makeyev et al., 2007) | SOX9 (glial cell specification; Cheng et al., 2009) | |
| CDK2 (cyclin-dependent kinase 2; Nakamachi et al., 2009) | C/EBPα (transcription factor; Hackanson et al., 2008) | Lhx2 (transcription; Qiu et al., 2009) | |
| MCP- (1 monocyte chemoattractant protein 1; Nakamachi et al., 2009) | FOXA2 (transcription factor; Baroukh et al., 2007) | EfnB1(projecting axons; Arvanitis et al., 2010) | |
| Itgb1 (integrin 1; Cao et al., 2007) | VIM (cytoskeleton; Furuta et al., 2010) | NR3C2 (mineralocorticoid and glucocorticoid receptor; Sober et al., 2010) | |
| SCP1 (synaptonemal filaments; Cao et al., 2007) | SMYD3 (transcription; Furuta et al., 2010) | ||
| hsa-let-7g | C-Myc (transcription factor; Lan et al., 2011) | Collagen alpha2 (COL1A2; Ji et al., 2010) | Bcl-xL (cell death; Shimizu et al., 2010) |
| hsa-miR-196a | HOX-B7 (transcription factor; Braig et al., 2010) | SPRR2C (small proline-rich protein 2C; Maru et al., 2009) | Annexin A1 (exocytosis; Luthra et al., 2008) |
| S100A9 (calcium-binding protein A9; Maru et al., 2009) | KRT5 (keratin 5; Maru et al., 2009) | HOXC8 (transcription factor; Kim et al., 2009a) | |
| hsa-miR-542-3p | Survivin (Yoon et al., 2010) | ||
| hsa-miR-525-5p | VPAC1 (vasoactive intestinal peptide receptor 1; Cocco et al., 2010) | ||
miRs without validated targets: hsa-miR-920, −506, −27b *, −541, −92a-2 *, −658, −423-5p, −615-5p, −25 *, −4688, −4776-3p, −668, −3613-5p, −4700-5p, −718, let-7f-2 *, −455-3p, −633, −554, −524-3p, −638, −525-3p, −611, let-7e *, −4283, −4329, −4278, −4300, −3184, −149 *.
Table A2.
Additional targets of microRNAs targeting AChE-R.
| miR ID | Validated targets | ||
|---|---|---|---|
| hsa-miR-708 | MPL (thrombopoietin receptor; Girardot et al., 2010) | ||
| hsa-miR-28-5p | MPL (thrombopoietin receptor; Girardot et al., 2010) | OTUB1 (immune system transcription; Girardot et al., 2010) | |
| N4BP1 (NEDD4 binding protein 1; Girardot et al., 2010) | TEX261 (apoptosis; Girardot et al., 2010) | MAPK1 (megakaryocyte differentiation; Girardot et al., 2010) | |
| hsa-miR-503 | ANLN (actin-binding protein anillin; Forrest et al., 2010) | ATF6 (activating transcription factor 6; Forrest et al., 2010) | CHEK1 (checkpoint mediated cell cycle arrest; Forrest et al., 2010) |
| EIF2C1 (argonaute1; Forrest et al., 2010) | KIF23 (mitotic kinesin-like protein 1; Forrest et al., 2010) | WEE1 (mitosis regulator; Forrest et al., 2010) | |
| CCNE1 (cyclin E1; Forrest et al., 2010) | CDC25A (cell cycle progression; Forrest et al., 2010) | ||
| CCND1 (cyclin D1; Forrest et al., 2010) | CDC14A (CDC14 cell division cycle 14 homolog A; Forrest et al., 2010) | ||
| hsa-miR-148a | CaMKIIα (CNS kinase; Liu et al., 2010e) | MLC1 (megalencephalic leukoencephalopathy with subcortical cysts 1; Geisler et al., 2011) | MSK1 (histone phosphorylase; Fujita et al., 2010) |
| DNMT1 (DNA methyltransferase 1; Pan et al., 2010) | DNMT3B (CpG island methylation; Duursma et al., 2008) | MITF (microphthalmia-associated transcription factor; Haflidadottir et al., 2010) | |
| CCKBR (modulates anxiety, analgesia, arousal, and neuroleptic activity; Muinos-Gimeno et al., 2011) | EPAS1 (endothelial PAS domain-containing protein 1; Giraud-Triboult et al., 2011) | HLA-G (asthma susceptibility; Tan et al., 2007) | |
| POMC (pro-opiomelanocortin; Muinos-Gimeno et al., 2011) | CAND1 (ubiquitin ligase regulation; Murata et al., 2010) | PXR (pregnane X receptor; Takagi et al., 2008) | |
| hsa-miR-152 | CaMKIIα (CNS kinase; Liu et al., 2010e) | DNMT1 (DNA methyltransferase 1; Braconi et al., 2010a) | |
| hsa-miR-125b | TNFα (tumor necrosis factor α; Tili et al., 2007) | ERBB2 (erythroblastic leukemia viral oncogene homolog 2; Scott et al., 2007) | BMPR1B (bone morphogenic receptor type 1B; Saetrom et al., 2009) |
| IRF4 (interferon regulatory factor 4; Malumbres et al., 2009) | ERBB3 erythroblastic leukemia viral oncogene homolog 3; Scott et al., 2007) | E2F3 (cell cycle control; Huang et al., 2011a) | |
| Blimp1 (zinc finger protein; Zhang et al., 2011b) | TEF (thyrotroph embryonic factor; Gutierrez et al., 2011) | Bcl2 modifying factor (apoptosis; Xia et al., 2009b) | |
| Vdr (vitamin D receptor; Zhang et al., 2011b) | MUC1 (adhesion; Rajabi et al., 2010) | Bak1 (pro-apoptotic Bcl2 antagonist killer 1; Zhou et al., 2010) | |
| CYP24A1 (cytochrome P450, family 24A, polypeptide 1; Komagata et al., 2009) | p53 (tumor suppressor; Le et al., 2009) | SMO (smoothened receptors; Ferretti et al., 2008) | |
| IGF2 (insulin-like growth factor 2; Ge et al., 2011) | Suv39h1 (histone methyltransferase; Villeneuve et al., 2010) | Stat3 (transcription factor binds to IL-6; Surdziel et al., 2011) | |
| LIN28 (translational enhancer; Zhong et al., 2010) | NMDA receptor subunit NR2A (Edbauer et al., 2010) | ATM (ataxia telangiectasia mutated; Smirnov and Cheung, 2008) | |
| hsa-miR-125a-5p | LIN28 (translational enhancer; Wu and Belasco, 2005) | T-TrkC (neurotrophic tyrosine kinase receptor 3; Ferretti et al., 2009) | HuR (cell growth; Guo et al., 2009) |
| p53 (tumor suppressor; Zhang et al., 2009) | KLF13 (transcription factor; Zhao et al., 2010) | AT-rich interactive domain 3B (transcription factor; Cowden Dahl et al., 2009) | |
| PDPN 9 (actin organization; Cortez et al., 2010) | Bak1 (pro-apoptotic Bcl2 antagonist killer 1; Guo et al., 2010) | ||
| N-ras (oncogene; Juan et al., 2009) | MEK3 (phosphorylation of MAP kinase; Li et al., 2011) | ||
| hsa-miR-214 | SrGAP1(neuronal migration; Zhang et al., 2011a) | Ezh2 (stem cell identity; Juan et al., 2009) | N-ras (oncogene; Liu et al., 2010b) |
| JNK1 (MAPK8; Yang et al., 2009) | PTEN (tumor suppressor; Yang et al., 2009) | MEK3 (phosphorylation; Yang et al., 2009) | |
| hsa-miR-199a-5p | Hif-1α (hypoxia-inducible factor 1; Rane et al., 2009) | Sirt1 (apoptosis; Rane et al., 2009) | DDR1 (discoidin domain receptor 1; Shen et al., 2010) |
| IKKβ (NFκB activation; Chen et al., 2008) | |||
| hsa-miR-31 | ICAM-1 (leukocyte adhesion protein; Suarez et al., 2010) | Fgf13 (fibroblast growth factor 13; Mardaryev et al., 2010) | Dkk-1 (canonical Wnt signaling; Xi et al., 2010) |
| DACT-3 (epigenetic regulator of Wnt; Xi et al., 2010) | E-selectin (inflammation; Suarez et al., 2010) | p16Ink4a (cell cycle; Malhas et al., 2010) | |
| LATS2 (tumor suppression; Liu et al., 2010c) | PPP2R2A (signal transduction; Liu et al., 2010c) | Krt16 (keratin 16; Mardaryev et al., 2010) | |
| Krt17 (keratin 17; Mardaryev et al., 2010) | Dlx3 (development of ventral forebrain; Mardaryev et al., 2010) | E2F6 (cell cycle; Bhatnagar et al., 2010) | |
| TIAM1 (T-cell lymphoma invasion and metastasis 1; Cottonham et al., 2010) | Fzd3 (accumulation of β-catenin; Valastyan et al., 2009) | Integrin α (fibronectin; Valastyan et al., 2009) | |
| M-RIP (regulation of actin; Valastyan et al., 2009) | MMP16 (blood vessels matrix remodeling; Valastyan et al., 2009) | RDX (actin filaments binding to plasma membrane; Valastyan et al., 2009) | |
| RhoA (signal transduction; Valastyan et al., 2009) | SATB2 (upper-layer neurons initiation; Aprelikova et al., 2010) | PROX1 (CNS development; Pedrioli et al., 2010) | |
| WAVE3 (signal transmission; Sossey-Alaoui et al., 2010) | |||
| hsa-miR-185 | Six1 (limb development; Imam et al., 2010) | ||
| hsa-miR-193b | Estrogen receptor α (Leivonen et al., 2009) | ETS-1 (oncogene; Xu et al., 2010a) | CCND1 (cyclin D1; Xu et al., 2010a) |
| Mcl-1 (myeloid cell leukemia sequence 1; Braconi et al., 2010b) | uPA (urokinase-type plasminogen activator; Li et al., 2009b) | ||
| hsa-miR-7 | Alpha-synuclein (SNCA; Junn et al., 2009) | SFRS1 (Wu et al., 2010b) | ERF (cell proliferation; Chou et al., 2010) |
| LSH (lymphoid-specific helicase; Ilnytskyy et al., 2008) | DAP (cell death-associated protein; Yu et al., 2009) | MRP1 (human multidrug resistance-associated protein 1; Pogribny et al., 2010) | |
| Associated cdc42 kinase 1 (Saydam et al., 2011) | Yan (cell differentiation; Li and Carthew, 2005) | EGFR (epidermal growth factor receptor; Kefas et al., 2008) | |
| CD98 (sodium transport; Nguyen et al., 2010) | Pak1 (p21-activated kinase 1; Reddy et al., 2008) | IGF1R (insulin-like growth factor 1 receptor; Jiang et al., 2010) | |
| hsa-miR-483-5p | Socs-3 (cytokine signaling; Ma et al., 2011) | BBC3/PUMA (apoptosis; Veronese et al., 2010) | |
| hsa-miR-663 | TGFβ1 (proliferation; Tili et al., 2010b) | JunB (jun B proto-oncogene; Tili et al., 2010a) | JunD (jun D proto-oncogene; Tili et al., 2010a) |
| hsa-miR- 765 | TRK3 (neurotrophic tyrosine kinase; Guidi et al., 2010) | ||
| hsa-miR- 146b-3p | IRAK1 (interleukin-1 receptor-associated kinase 1; Taganov et al., 2006) | EGFR (epidermal growth factor receptor; Shao et al., 2011) | MMP16 (degrades extracellular matrix; Xia et al., 2009a) |
miRs without validated targets that are predicted to target AChE-R: hsa-miR-590-3p, −148b, −193a-3p, −182*, −4298, −4644, −4739, −1224-3p, −4769-5p, –582-3p, −380*, −1825, −892b, −1275, −3155, −765, −3119, −3139, −563, −92b*, −1321, −4283, −1228*, −4323, −4319, −761, −767-5p, −224*, −522, −4271, −1226*, −3179, −92a-1*, −3202, −20b*, −4303, −4306, −3065-5p, −4297, −4329, −3148, −3163, −22*, −4302, −513a-5p, −542-5p, −377*, −1908, −92a-2*, −608, −625.
Table A3.
Additional targets of BChE-targeting microRNAs.
| miR ID | Validated target | ||
|---|---|---|---|
| hsa-miR-203 | SOCS-3 (cytokine signaling; Wei et al., 2010) | Lef1 (lymphoid enhancer-binding factor; Thatcher et al., 2008) | p63 (transcriptional activator or repressor; Yi et al., 2008) |
| ABL1 (cell growth; Bueno et al., 2008) | Barx1 (transcription factor; Kim et al., 2011) | CKAP2 (cytoskeleton associated protein 2; Viticchie et al., 2011) | |
| LASP1 (cytoskeletal activities; Viticchie et al., 2011) | BIRC5 (regulator of mitosis; Viticchie et al., 2011) | WASF1 (signal transmission; Viticchie et al., 2011) | |
| ASAP1 (membrane trafficking; Viticchie et al., 2011) | RUNX2 (runt-related transcription factor 2; Viticchie et al., 2011) | ||
| hsa-miR-340 | MITF (microphthalmia-associated transcription factor; Goswami et al., 2010) | ||
| hsa-miR-218 | IKK-β (cytokine-activated intracellular signaling pathway; Song et al., 2010) | ROBO1 (roundabout, axon guidance receptor, homolog 1; Alajez et al., 2011) | BIRC5 (regulator of mitosis; Alajez et al., 2011) |
| GJA1 (gap junction protein, α1; Alajez et al., 2011) | ROBO2 (roundabout, axon guidance receptor homolog 2; Alajez et al., 2011) | GLCE (glucuronic acid epimerase; Small et al., 2010) | |
| PXN (paxillin, cytoskeletal protein; Wu et al., 2010a) | |||
| hsa-miR-221 | ERα (estrogen receptor α; Zhao et al., 2008) | ICAM-1 (leukocyte adhesion protein; Hu et al., 2010) | p27 (cell cycle; Garofalo et al., 2008) |
| p57 (cyclin-dependent kinase inhibitor 1C; Kim et al., 2009b) | DNA damage-inducible transcript 4 (DDIT4; Pineau et al., 2010) | TIMP3 (TIMP metallopeptidase inhibitor 3; Garofalo et al., 2009) | |
| PTEN (tumor suppressor; Garofalo et al., 2009) | PUMA (apoptosis; Zhang et al., 2010) | C-fos (cell proliferation; Ichimura et al., 2010) | |
| Bmf (apoptosis; Gramantieri et al., 2009) | Mdm2 (ubiquitination; Kim et al., 2010) | CDKN1B (cyclin-dependent kinase inhibitor 1B; Kotani et al., 2009) | |
| hsa-miR-222 | ERα (estrogen receptor α; Zhao et al., 2008) | p27 (cell cycle; Garofalo et al., 2008) | PTEN (tumor suppressor; Garofalo et al., 2009) |
| STAT5A (signal transducer and activator of transcription 5; Dentelli et al., 2010) | p57 (cyclin-dependent kinase inhibitor 1C; Kim et al., 2009b) | TIMP3 (TIMP metallopeptidase inhibitor 3; Garofalo et al., 2009) | |
| Bim (apoptosis; Terasawa et al., 2009) | ETS-1 (transcription factor; Zhu et al., 2011) | PUMA (apoptosis; Zhang et al., 2010) | |
| PPP2R2A (protein phosphatase 2A subunit B; Wong et al., 2010) | C-fos (cell proliferation; Ichimura et al., 2010) | ICAM-1 (Ueda et al., 2009) | |
| MMP1 (cleaves collagens; Liu et al., 2009) | SOD2 (superoxide dismutase 2; Liu et al., 2009) | ||
| hsa-miR-181a | SIRT1 (apoptosis, muscle differentiation; Saunders et al., 2010) | Ataxia telangiectasia mutated (ATM; cell cycle; Wang et al., 2011) | Hox-A11 (transcription factor; Naguibneva et al., 2006) |
| p27(cell cycle; Cuesta et al., 2009) | PLAG1 (transcription factor; Pallasch et al., 2009) | BCL2 (B-cell CLL/lymphoma 2; apoptosis; Zhu et al., 2010) | |
| Bim (apoptosis; Lwin et al., 2010) | Tcl1 (cell proliferation; Pekarsky et al., 2006) | OPN (osteopontin; Bhattacharya et al., 2010) | |
| hsa-miR-181b | AID (activation-induced cytidine deaminase; RNA-editing; De Yebenes et al., 2008) | PLAG1 (transcription factor; Pallasch et al., 2009) | BCL2 (B-cell CLL/lymphoma 2; apoptosis; Zhu et al., 2010) |
| TIMP3 (TIMP metallopeptidase inhibitor 3; Wang et al., 2010a) | Ataxia telangiectasia mutated (ATM; cell cycle; Wang et al., 2011) | SIRT1 (apoptosis, muscle differentiation; Saunders et al., 2010) | |
| ZNF37A (transcriptional regulation; Huang et al., 2010) | ZNF83 (transcriptional regulation; Huang et al., 2010) | ZNF182 (transcriptional regulation; Huang et al., 2010) | |
| Mcl-1 (myeloid cell leukemia-1; apoptosis; Zimmerman et al., 2010) | |||
| hsa-miR-181c | IL2 (immune response; Xue et al., 2011) | BCL2 (B-cell CLL/lymphoma 2; apoptosis; Zhu et al., 2010) | NOTCH4 (transcriptional activator complex; Hashimoto et al., 2010) |
| KRAS (GTPase activity; Hashimoto et al., 2010) | |||
| hsa-miR-181d | BCL2 (B-cell CLL/lymphoma 2; Zhu et al., 2010) | ||
| hsa-miR-494 | CaMKIIδ (CNS kinase; Wang et al., 2010b) | ROCK-1 (apoptosis; Wang et al., 2010b) | LIF (leukemia inhibitory factor (cholinergic differentiation factor); Wang et al., 2010b) |
| PTEN (phosphatase and tensin homolog; Wang et al., 2010b) | TEL-AML1 (hematopoiesis; Diakos et al., 2010) | FGFR2 (fibroblast growth factor receptor 2; Wang et al., 2010b) | |
| hsa-miR-129-5p | CAMTA1 (calmodulin binding transcription activator 1; Liao et al., 2008) | EIF2C3 (eukaryotic translation initiation factor 2C, 3; Liao et al., 2008) | GALNT1 (oligosaccharide biosynthesis; Dyrskjot et al., 2009) |
| SOX4 (transcriptional activator; Dyrskjot et al., 2009) | |||
| hsa-miR-30d | Galphai2 (G protein, α inhibiting activity polypeptide 2; Yao et al., 2010) | ||
| hsa-miR-30c | Runx1 (runt-related transcription factor 1; Ben-Ami et al., 2009) | CTGF (connective tissue growth factor; Duisters et al., 2009) | |
| hsa-miR-30a | SOD2 (superoxide dismutase 2; Xia et al., 2006) | BDNF (brain-derived neurotrophic factor; Mellios et al., 2008) | Beclin 1 (autophagy; Zhu et al., 2009) |
| Xlim1/Lhx1 (transcription factor; Agrawal et al., 2009) | |||
| hsa-miR-30e | Ubc9 (ubiquitin-conjugating enzyme E2I; Wu et al., 2009) | B-Myb (transcription factor; Martinez et al., 2011) | |
| hsa-miR-320a | (Hsp20 heat-shock protein 20; Ren et al., 2009) | AQP1 (aquaporin 1; Sepramaniam et al., 2010) | AQP4 (aquaporin 4; Sepramaniam et al., 2010) |
| The transferrin receptor 1(TfR-1; CD71; development of erythrocytes and the nervous system; Schaar et al., 2009) | Mcl-1 (myeloid cell leukemia sequence 1; apoptosis; Chen et al., 2009) | ||
| hsa-miR-140-5p | Smad3 (transcriptional modulator; Pais et al., 2010) | HDAC4 (histone deacetylase 4; Tuddenham et al., 2006) | |
| hsa-miR-519c-3p | HIF-1α (hypoxia-inducible factor 1α; Cha et al., 2010) | ABCG2 (exclusion of xenobiotics from the brain; To et al., 2008) | |
| hsa-miR-489 | PTPN11 (signal transduction; Kikkawa et al., 2010) | ||
| hsa-miR-584 | NXA1 (exocytosis; Luthra et al., 2008) | ROCK-1 (actin assembly; Ueno et al., 2011) | |
miRs without validated targets: hsa-miR-147b, −532-5p, −508-3p, −889, −325, −573, −195*, −567, −193b*, −625, −16-2*, −576-3p, −190b, −518e*, −518f*, −518d-5p, −147, −320d, −320c, −320b, −875-5p, −758, −30b, −1279, −3145, −1183, −664, −4261, −4262, −1237, −1972, −3146, let-7a-2*, let-7g*, −1911*, −2052, −15a*, −3148, −555, −656, −636, −3182, −513a-3p, −501-3p, −502-3p, −579, −4316, −4312, −1294, −142-5p, −3128, −30a*, −30d*, −30e*, −4268, −3137, −20b*, −651, −32*, −362-5p, −500b, −501-5p, −1976, −449c*, −1224-5p, −302a*, −1248, −99b*, −99a*, −369-3p, −1256, −629, −187*, −514b-3p, −378*, −1305, −331-5p, −1200, −4272, −4260, −493*, −582-5p, −4255, −3133, −4273, −19a*, −19b-1*, −19b-2*, −4271, −15b*, −1826.
Table A4.
Additional targets of ChE-targeting miRs (common to more than one ChE).
| miR ID | Validated target common to ACHE-R and AChE-S | ||
|---|---|---|---|
| Hsa-miR-186 | Pro-apoptotic P2 × 7 purinergic receptor(Zhou et al., 2008) | AKAP12 (tumor suppressor; Goeppert et al., 2010) | |
| Hsa-miR-199b-5p | Dyrk1a (brain development; Da Costa Martins et al., 2010) | HES1 (transcriptional repressor; Garzia et al., 2009) | SET (apoptosis; Chao et al., 2010) |
| Hsa-miR-429 | ZEB1 (transcriptional repression of IL2; Gregory et al., 2008) | ZEB2 (SIP1; zinc finger protein; Gregory et al., 2008) | PLCgamma1(apoptosis; Uhlmann et al., 2010) |
| RERE (apoptosis; Karres et al., 2007) | |||
| Hsa-miR-200b | ZEB1 (transcriptional repression of IL2; Gregory et al., 2008) | ZEB2 (SIP1; zinc finger protein; Gregory et al., 2008) | PLCgamma1(apoptosis; Uhlmann et al., 2010) |
| Serca2 (sarco/endoplasmic reticulum Ca2+-ATPase; Salomonis et al., 2010) | Suz12 (chromatin silencing; Iliopoulos et al., 2010) | Ets-1 (transcriptions factor; Chan et al., 2011) | |
| OREBP (osmotic response element; Huang et al., 2011b) | Cyclin D1 (Xia et al., 2010) | ||
| Hsa-miR-200c | ZEB1 (transcriptional repression of IL2; Gregory et al., 2008) | ZEB2 (SIP1; zinc finger protein; Gregory et al., 2008) | PLCgamma1(apoptosis; Uhlmann et al., 2010) |
| VEGF (angiogenesis; Liu et al., 2010a) | TUBB3 (neurogenesis and axon guidance; Cochrane et al., 2009) | TRPS1 (transcription factor; Li et al., 2009a) | |
| KLF13 (transcription factor; Li et al., 2009a) | MBNL2 (muscleblind-like protein 2; Li et al., 2009a) | FAP1 (apoptosis; Schickel et al., 2010) | |
| miR ID | Validated targets common to ACHE-R and BChE | ||
| Hsa-miR-24 | SOD1 (superoxide dismutase 1; Papaioannou et al., 2011) | ALK4 (transducer of activin; Wang et al., 2008) | Notch1 (Bergmann glia differentiation; Fukuda et al., 2005) |
| MKK4 (survival signal in T cells; Marasa et al., 2009) | E2F2 (cell cycle; Lal et al., 2009a) | H2AX (histone-formation; Lal et al., 2009b) | |
| FAF1 (apoptosis; Qin et al., 2010) | HNF4α (cell proliferation; Takagi et al., 2010) | FURIN (processing of TGFβ1; Luna et al., 2011) | |
| DHFR (dihydrofolate reductase; Mishra et al., 2009) | DND1(miRNA-mediated gene suppression; Liu et al., 2010d) | ||
| Hsa-miR-212 | MeCP2 (interaction with histone deacetylase; Im et al., 2010) | MYC (transcription; Xu et al., 2010b) | Rb1(tumor suppressor; Park et al., 2011) |
| PED (apoptosis; Incoronato et al., 2010) | |||
| Hsa-miR-132 | AChE-S (Shaked et al., 2009) | P250GAP (neuron-associated GTPase; Vo et al., 2005) | Per1 (circadian clock; Cheng et al., 2007) |
| SirT1 (apoptosis; Strum et al., 2009) | MeCP2 (modification of eukaryotic genomes; Klein et al., 2007) | p300 (chromatin remodeling; Lagos et al., 2010) | |
| Jarid1a (histone demethylase; Alvarez-Saavedra et al., 2011) | Btg2 (cell cycle; Alvarez-Saavedra et al., 2011) | Paip2a (translation regulation; Alvarez-Saavedra et al., 2011) | |
| p120RasGAP (angiogenesis; Anand et al., 2010) | |||
| Hsa-miR-198 | Cyclin T1(Xu et al., 2010b) | ||
| Hsa-miR-194 | Rac1 (GTP-binding protein; Venugopal et al., 2010) | Per family (circadian; Nagel et al., 2009) | EP300 (transcriptional co-activator; Mees et al., 2010) |
| MDM2 (p53 negative regulator; Pichiorri et al., 2010) | |||
miRs without validated targets: hsa-miR-423-3p, −484, −4728-3p, −939, −484, −4728-3p.
Footnotes
References
- Allshire R. C., Karpen G. H. (2008). Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat. Rev. Genet. 9, 923–937 10.1038/nrg2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima N., Furuta D., Hidaka Y., Moriyama R., Tsujiuchi T. (2009). Post-translational modifications of tubulin in the nervous system. J. Neurochem. 109, 683–693 10.1111/j.1471-4159.2009.06013.x [DOI] [PubMed] [Google Scholar]
- Furuta M., Kozaki K. I., Tanaka S., Arii S., Imoto I., Inazawa J. (2010). miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis 31, 766–776 10.1093/carcin/bgp250 [DOI] [PubMed] [Google Scholar]
- Girardot M., Pecquet C., Boukour S., Knoops L., Ferrant A., Vainchenker W., Giraudier S., Constantinescu S. N. (2010). miR-28 is a thrombopoietin receptor targeting microRNA detected in a fraction of myeloproliferative neoplasm patient platelets. Blood 116, 437–445 10.1182/blood-2008-06-165985 [DOI] [PubMed] [Google Scholar]
- Gregory P. A., Bert A. G., Paterson E. L., Barry S. C., Tsykin A., Farshid G., Vadas M. A., Khew-Goodall Y., Goodall G. J. (2008). The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 10, 593–601 10.1038/ncb1722 [DOI] [PubMed] [Google Scholar]
- Hansen K. F., Sakamoto K., Wayman G. A., Impey S., Obrietan K. (2010). Transgenic miR132 alters neuronal spine density and impairs novel object recognition memory. PLoS ONE 5, e15497. 10.1371/journal.pone.0015497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C. S., Treisman R. (1995). Transcriptional regulation by extracellular signals: mechanisms and specificity. Cell 80, 199–211 10.1016/0092-8674(95)90403-4 [DOI] [PubMed] [Google Scholar]
- Ichimura A., Ruike Y., Terasawa K., Shimizu K., Tsujimoto G. (2010). MicroRNA-34a inhibits cell proliferation by repressing mitogen-activated protein kinase kinase 1 during megakaryocytic differentiation of K562 cells. Mol. Pharmacol. 77, 1016–1024 10.1124/mol.109.063321 [DOI] [PubMed] [Google Scholar]
- John B., Enright A. J., Aravin A., Tuschl T., Sander C., Marks D. S. (2004). Human MicroRNA targets. PLoS Biol. 2, e363. 10.1371/journal.pbio.0020363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Zhan Z., Xu L., Ma F., Li D., Guo Z., Li N., Cao X. (2010). MicroRNA-148/152 impair innate response and antigen presentation of TLR-triggered dendritic cells by targeting CaMKII alpha. J. Immunol. 185, 7244–7251 10.4049/jimmunol.1000953 [DOI] [PubMed] [Google Scholar]
- Massoulie J. (2002). The origin of the molecular diversity and functional anchoring of cholinesterases. Neurosignals 11, 130–143 10.1159/000065054 [DOI] [PubMed] [Google Scholar]
- Shaked I., Meerson A., Wolf Y., Avni R., Greenberg D., Gilboa-Geffen A., Soreq H. (2009). MicroRNA-132 potentiates cholinergic anti-inflammatory signaling by targeting acetylcholinesterase. Immunity 31, 965–973 10.1016/j.immuni.2009.09.019 [DOI] [PubMed] [Google Scholar]
- Soreq H., Seidman S. (2001). Acetylcholinesterase – new roles for an old actor. Nat. Rev. Neurosci. 2, 294–302 10.1038/35067100 [DOI] [PubMed] [Google Scholar]
- Soreq H., Wolf Y. (2011). NeurimmiRs: micro-RNAs in the neuroimmune interface. Trends Mol. Med. [Epub ahead of print]. 10.1016/j.molmed.2011.06.009 [DOI] [PubMed] [Google Scholar]
- Vo N., Klein M. E., Varlamova O., Keller D. M., Yamamoto T., Goodman R. H., Impey S. (2005). A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc. Natl. Acad. Sci. U.S.A. 102, 16426–16431 10.1073/pnas.0508448102 [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- Agrawal R., Tran U., Wessely O. (2009). The miR-30 miRNA family regulates Xenopus pronephros development and targets the transcription factor Xlim1/Lhx1. Development 136, 3927–3936 10.1242/dev.037432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alajez N. M., Lenarduzzi M., Ito E., Hui A. B., Shi W., Bruce J., Yue S., Huang S. H., Xu W., Waldron J., O’sullivan B., Liu F. F. (2011). MiR-218 suppresses nasopharyngeal cancer progression through downregulation of survivin and the SLIT2-ROBO1 pathway. Cancer Res. 71, 2381–2391 10.1158/0008-5472.CAN-10-2754 [DOI] [PubMed] [Google Scholar]
- Alvarez-Saavedra M., Antoun G., Yanagiya A., Oliva-Hernandez R., Cornejo-Palma D., Perez-Iratxeta C., Sonenberg N., Cheng H. Y. (2011). miRNA-132 orchestrates chromatin remodeling and translational control of the circadian clock. Hum. Mol. Genet. 20, 731–751 10.1093/hmg/ddq519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand S., Majeti B. K., Acevedo L. M., Murphy E. A., Mukthavaram R., Scheppke L., Huang M., Shields D. J., Lindquist J. N., Lapinski P. E., King P. D., Weis S. M., Cheresh D. A. (2010). MicroRNA-132-mediated loss of p120RasGAP activates the endothelium to facilitate pathological angiogenesis. Nat. Med. 16, 909–914 10.1038/nm.2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aprelikova O., Yu X., Palla J., Wei B. R., John S., Yi M., Stephens R., Simpson R. M., Risinger J. I., Jazaeri A., Niederhuber J. (2010). The role of miR-31 and its target gene SATB2 in cancer-associated fibroblasts. Cell Cycle 9, 4387–4398 10.4161/cc.9.21.13674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvanitis D. N., Jungas T., Behar A., Davy A. (2010). Ephrin-B1 reverse signaling controls a posttranscriptional feedback mechanism via miR-124. Mol. Cell. Biol. 30, 2508–2517 10.1128/MCB.01620-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroukh N., Ravier M. A., Loder M. K., Hill E. V., Bounacer A., Scharfmann R., Rutter G. A., Van Obberghen E. (2007). MicroRNA-124a regulates Foxa2 expression and intracellular signaling in pancreatic beta-cell lines. J. Biol. Chem. 282, 19575–19588 10.1074/jbc.M611841200 [DOI] [PubMed] [Google Scholar]
- Ben-Ami O., Pencovich N., Lotem J., Levanon D., Groner Y. (2009). A regulatory interplay between miR-27a and Runx1 during megakaryopoiesis. Proc. Natl. Acad. Sci. U.S.A. 106, 238–243 10.1073/pnas.0811466106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatnagar N., Li X., Padi S. K., Zhang Q., Tang M. S., Guo B. (2010). Downregulation of miR-205 and miR-31 confers resistance to chemotherapy-induced apoptosis in prostate cancer cells. Cell Death Dis. 1, e105. 10.1038/cddis.2010.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya S. D., Garrison J., Guo H., Mi Z., Markovic J., Kim V. M., Kuo P. C. (2010). Micro-RNA-181a regulates osteopontin-dependent metastatic function in hepatocellular cancer cell lines. Surgery 148, 291–297 10.1016/j.surg.2010.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braconi C., Huang N., Patel T. (2010a). MicroRNA-dependent regulation of DNA methyltransferase-1 and tumor suppressor gene expression by interleukin-6 in human malignant cholangiocytes. Hepatology 51, 881–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braconi C., Valeri N., Gasparini P., Huang N., Taccioli C., Nuovo G., Suzuki T., Croce C. M., Patel T. (2010b). Hepatitis C virus proteins modulate microRNA expression and chemosensitivity in malignant hepatocytes. Clin. Cancer Res. 16, 957–966 10.1158/1078-0432.CCR-09-2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braig S., Mueller D. W., Rothhammer T., Bosserhoff A. K. (2010). MicroRNA miR-196a is a central regulator of HOX-B7 and BMP4 expression in malignant melanoma. Cell. Mol. Life Sci. 67, 3535–3548 10.1007/s00018-010-0394-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueno M. J., Perez De Castro I., Gomez De Cedron M., Santos J., Calin G. A., Cigudosa J. C., Croce C. M., Fernandez-Piqueras J., Malumbres M. (2008). Genetic and epigenetic silencing of microRNA-203 enhances ABL1 and BCR-ABL1 oncogene expression. Cancer Cell 13, 496–506 10.1016/j.ccr.2008.04.018 [DOI] [PubMed] [Google Scholar]
- Cao X., Pfaff S. L., Gage F. H. (2007). A functional study of miR-124 in the developing neural tube. Genes Dev. 21, 531–536 10.1101/gad.1519207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha S. T., Chen P. S., Johansson G., Chu C. Y., Wang M. Y., Jeng Y. M., Yu S. L., Chen J. S., Chang K. J., Jee S. H., Tan C. T., Lin M. T., Kuo M. L. (2010). MicroRNA-519c suppresses hypoxia-inducible factor-1alpha expression and tumor angiogenesis. Cancer Res. 70, 2675–2685 10.1158/1538-7445.AM10-2383 [DOI] [PubMed] [Google Scholar]
- Chan Y. C., Khanna S., Roy S., Sen C. K. (2011). miR-200b targets Ets-1 and is down-regulated by hypoxia to induce angiogenic response of endothelial cells. J. Biol. Chem. 286, 2047–2056 10.1074/jbc.C110.212621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao A., Tsai C. L., Wei P. C., Hsueh S., Chao A. S., Wang C. J., Tsai C. N., Lee Y. S., Wang T. H., Lai C. H. (2010). Decreased expression of microRNA-199b increases protein levels of SET (protein phosphatase 2A inhibitor) in human choriocarcinoma. Cancer Lett. 291, 99–107 10.1016/j.canlet.2009.10.005 [DOI] [PubMed] [Google Scholar]
- Chen L., Yan H. X., Yang W., Hu L., Yu L. X., Liu Q., Li L., Huang D. D., Ding J., Shen F., Zhou W. P., Wu M. C., Wang H. Y. (2009). The role of microRNA expression pattern in human intrahepatic cholangiocarcinoma. J. Hepatol. 50, 358–369 10.1016/S0168-8278(09)60364-0 [DOI] [PubMed] [Google Scholar]
- Chen R., Alvero A. B., Silasi D. A., Kelly M. G., Fest S., Visintin I., Leiser A., Schwartz P. E., Rutherford T., Mor G. (2008). Regulation of IKKbeta by miR-199a affects NF-kappaB activity in ovarian cancer cells. Oncogene 27, 4712–4723 10.1038/onc.2008.288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H. Y., Papp J. W., Varlamova O., Dziema H., Russell B., Curfman J. P., Nakazawa T., Shimizu K., Okamura H., Impey S., Obrietan K. (2007). microRNA modulation of circadian-clock period and entrainment. Neuron 54, 813–829 10.1016/j.neuron.2007.04.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L. C., Pastrana E., Tavazoie M., Doetsch F. (2009). miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat. Neurosci. 12, 399–408 10.1038/nn.2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou Y. T., Lin H. H., Lien Y. C., Wang Y. H., Hong C. F., Kao Y. R., Lin S. C., Chang Y. C., Lin S. Y., Chen S. J., Chen H. C., Yeh S. D., Wu C. W. (2010). EGFR promotes lung tumorigenesis by activating miR-7 through a Ras/ERK/Myc pathway that targets the Ets2 transcriptional repressor ERF. Cancer Res. 70, 8822–8831 10.1158/1538-7445.AM10-5398 [DOI] [PubMed] [Google Scholar]
- Cocco E., Paladini F., Macino G., Fulci V., Fiorillo M. T., Sorrentino R. (2010). The expression of vasoactive intestinal peptide receptor 1 is negatively modulated by microRNA 525-5p. PLoS ONE 5, e12067. 10.1371/journal.pone.0012067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochrane D. R., Spoelstra N. S., Howe E. N., Nordeen S. K., Richer J. K. (2009). MicroRNA-200c mitigates invasiveness and restores sensitivity to microtubule-targeting chemotherapeutic agents. Mol. Cancer Ther. 8, 1055–1066 10.1158/1535-7163.MCT-08-1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez M. A., Nicoloso M. S., Shimizu M., Rossi S., Gopisetty G., Molina J. R., Carlotti C., Jr., Tirapelli D., Neder L., Brassesco M. S., Scrideli C. A., Tone L. G., Georgescu M. M., Zhang W., Puduvalli V., Calin G. A. (2010). miR-29b and miR-125a regulate podoplanin and suppress invasion in glioblastoma. Genes Chromosomes Cancer 49, 981–990 10.1002/gcc.20808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottonham C. L., Kaneko S., Xu L. (2010). miR-21 and miR-31 converge on TIAM1 to regulate migration and invasion of colon carcinoma cells. J. Biol. Chem. 285, 35293–35302 10.1074/jbc.M110.160069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowden Dahl K. D., Dahl R., Kruichak J. N., Hudson L. G. (2009). The epidermal growth factor receptor responsive miR-125a represses mesenchymal morphology in ovarian cancer cells. Neoplasia 11, 1208–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuesta R., Martinez-Sanchez A., Gebauer F. (2009). miR-181a regulates cap-dependent translation of p27(kip1) mRNA in myeloid cells. Mol. Cell. Biol. 29, 2841–2851 10.1128/MCB.01971-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Costa Martins P. A., Salic K., Gladka M. M., Armand A. S., Leptidis S., El Azzouzi H., Hansen A., Coenen-De Roo C. J., Bierhuizen M. F., Van Der Nagel R., Van Kuik J., De Weger R., De Bruin A., Condorelli G., Arbones M. L., Eschenhagen T., De Windt L. J. (2010). MicroRNA-199b targets the nuclear kinase Dyrk1a in an auto-amplification loop promoting calcineurin/NFAT signalling. Nat. Cell Biol. 12, 1220–1227 10.1038/ncb2126 [DOI] [PubMed] [Google Scholar]
- De Yebenes V. G., Belver L., Pisano D. G., Gonzalez S., Villasante A., Croce C., He L., Ramiro A. R. (2008). miR-181b negatively regulates activation-induced cytidine deaminase in B cells. J. Exp. Med. 205, 2199–2206 10.1084/jem.20080579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dentelli P., Rosso A., Orso F., Olgasi C., Taverna D., Brizzi M. F. (2010). microRNA-222 controls neovascularization by regulating signal transducer and activator of transcription 5A expression. Arterioscler. Thromb. Vasc. Biol. 30, 1562–1568 10.1161/ATVBAHA.110.206201 [DOI] [PubMed] [Google Scholar]
- Diakos C., Zhong S., Xiao Y., Zhou M., Vasconcelos G. M., Krapf G., Yeh R. F., Zheng S., Kang M., Wiencke J. K., Pombo-De-Oliveira M. S., Panzer-Grumayer R., Wiemels J. L. (2010). TEL-AML1 regulation of survivin and apoptosis via miRNA-494 and miRNA-320a. Blood 116, 4885–4893 10.1182/blood-2009-02-206706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duisters R. F., Tijsen A. J., Schroen B., Leenders J. J., Lentink V., Van Der Made I., Herias V., Van Leeuwen R. E., Schellings M. W., Barenbrug P., Maessen J. G., Heymans S., Pinto Y. M., Creemers E. E. (2009). miR-133 and miR-30 regulate connective tissue growth factor: implications for a role of microRNAs in myocardial matrix remodeling. Circ. Res. 104, 170–178 10.1161/CIRCRESAHA.108.182535 [DOI] [PubMed] [Google Scholar]
- Duursma A. M., Kedde M., Schrier M., Le Sage C., Agami R. (2008). miR-148 targets human DNMT3b protein coding region. RNA 14, 872–877 10.1261/rna.972008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyrskjot L., Ostenfeld M. S., Bramsen J. B., Silahtaroglu A. N., Lamy P., Ramanathan R., Fristrup N., Jensen J. L., Andersen C. L., Zieger K., Kauppinen S., Ulhoi B. P., Kjems J., Borre M., Orntoft T. F. (2009). Genomic profiling of microRNAs in bladder cancer: miR-129 is associated with poor outcome and promotes cell death in vitro. Cancer Res. 69, 4851–4860 10.1158/0008-5472.CAN-08-4043 [DOI] [PubMed] [Google Scholar]
- Edbauer D., Neilson J. R., Foster K. A., Wang C. F., Seeburg D. P., Batterton M. N., Tada T., Dolan B. M., Sharp P. A., Sheng M. (2010). Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron 65, 373–384 10.1016/j.neuron.2010.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti E., De Smaele E., Miele E., Laneve P., Po A., Pelloni M., Paganelli A., Di Marcotullio L., Caffarelli E., Screpanti I., Bozzoni I., Gulino A. (2008). Concerted microRNA control of hedgehog signalling in cerebellar neuronal progenitor and tumour cells. EMBO J. 27, 2616–2627 10.1038/emboj.2008.172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti E., De Smaele E., Po A., Di Marcotullio L., Tosi E., Espinola M. S., Di Rocco C., Riccardi R., Giangaspero F., Farcomeni A., Nofroni I., Laneve P., Gioia U., Caffarelli E., Bozzoni I., Screpanti I., Gulino A. (2009). MicroRNA profiling in human medulloblastoma. Int. J. Cancer 124, 568–577 10.1002/ijc.23948 [DOI] [PubMed] [Google Scholar]
- Forrest A. R., Kanamori-Katayama M., Tomaru Y., Lassmann T., Ninomiya N., Takahashi Y., De Hoon M. J., Kubosaki A., Kaiho A., Suzuki M., Yasuda J., Kawai J., Hayashizaki Y., Hume D. A., Suzuki H. (2010). Induction of microRNAs, mir-155, mir-222, mir-424 and mir-503, promotes monocytic differentiation through combinatorial regulation. Leukemia 24, 460–466 10.1038/leu.2009.246 [DOI] [PubMed] [Google Scholar]
- Fujita Y., Kojima K., Ohhashi R., Hamada N., Nozawa Y., Kitamoto A., Sato A., Kondo S., Kojima T., Deguchi T., Ito M. (2010). MiR-148a attenuates paclitaxel resistance of hormone-refractory, drug-resistant prostate cancer PC3 cells by regulating MSK1 expression. J. Biol. Chem. 285, 19076–19084 10.1074/jbc.M109.079525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda Y., Kawasaki H., Taira K. (2005). Exploration of human miRNA target genes in neuronal differentiation. Nucleic Acids Symp. Ser. (Oxf.) 49, 341–342 10.1093/nass/49.1.341 [DOI] [PubMed] [Google Scholar]
- Garofalo M., Di Leva G., Romano G., Nuovo G., Suh S. S., Ngankeu A., Taccioli C., Pichiorri F., Alder H., Secchiero P., Gasparini P., Gonelli A., Costinean S., Acunzo M., Condorelli G., Croce C. M. (2009). miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell 16, 498–509 10.1016/j.ccr.2009.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Garofalo M., Quintavalle C., Di Leva G., Zanca C., Romano G., Taccioli C., Liu C. G., Croce C. M., Condorelli G. (2008). MicroRNA signatures of TRAIL resistance in human non-small cell lung cancer. Oncogene 27, 3845–3855 10.1038/onc.2008.6 [DOI] [PubMed] [Google Scholar]
- Garzia L., Andolfo I., Cusanelli E., Marino N., Petrosino G., De Martino D., Esposito V., Galeone A., Navas L., Esposito S., Gargiulo S., Fattet S., Donofrio V., Cinalli G., Brunetti A., Vecchio L. D., Northcott P. A., Delattre O., Taylor M. D., Iolascon A., Zollo M. (2009). MicroRNA-199b-5p impairs cancer stem cells through negative regulation of HES1 in medulloblastoma. PLoS ONE 4, e4998. 10.1371/journal.pone.0004998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y., Sun Y., Chen J. (2011). IGF-II is regulated by microRNA-125b in skeletal myogenesis. J. Cell Biol. 192, 69–81 10.1083/jcb.201007165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler A., Jungmann A., Kurreck J., Poller W., Katus H. A., Vetter R., Fechner H., Muller O. J. (2011). microRNA122-regulated transgene expression increases specificity of cardiac gene transfer upon intravenous delivery of AAV9 vectors. Gene Ther. 18, 199–209 10.1038/gt.2010.141 [DOI] [PubMed] [Google Scholar]
- Giraud-Triboult K., Rochon-Beaucourt C., Nissan X., Champon B., Aubert S., Pietu G. (2011). Combined mRNA and microRNA profiling reveals that miR-148a and miR-20b control human mesenchymal stem cell phenotype via EPAS1. Physiol. Genomics 43, 77–86 10.1152/physiolgenomics.00077.2010 [DOI] [PubMed] [Google Scholar]
- Goeppert B., Schmezer P., Dutruel C., Oakes C., Renner M., Breinig M., Warth A., Vogel M. N., Mittelbronn M., Mehrabi A., Gdynia G., Penzel R., Longerich T., Breuhahn K., Popanda O., Plass C., Schirmacher P., Kern M. A. (2010). Down-regulation of tumor suppressor A kinase anchor protein 12 in human hepatocarcinogenesis by epigenetic mechanisms. Hepatology 52, 2023–2033 10.1002/hep.23939 [DOI] [PubMed] [Google Scholar]
- Goswami S., Tarapore R. S., Teslaa J. J., Grinblat Y., Setaluri V., Spiegelman V. S. (2010). MicroRNA-340-mediated degradation of microphthalmia-associated transcription factor mRNA is inhibited by the coding region determinant-binding protein. J. Biol. Chem. 285, 20532–20540 10.1074/jbc.M110.109298 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gramantieri L., Fornari F., Ferracin M., Veronese A., Sabbioni S., Calin G. A., Grazi G. L., Croce C. M., Bolondi L., Negrini M. (2009). MicroRNA-221 targets Bmf in hepatocellular carcinoma and correlates with tumor multifocality. Clin. Cancer Res. 15, 5073–5081 10.1158/1078-0432.CCR-09-0092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidi M., Muinos-Gimeno M., Kagerbauer B., Marti E., Estivill X., Espinosa-Parrilla Y. (2010). Overexpression of miR-128 specifically inhibits the truncated isoform of NTRK3 and upregulates BCL2 in SH-SY5Y neuroblastoma cells. BMC Mol. Biol. 11, 95. 10.1186/1471-2199-11-95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S., Lu J., Schlanger R., Zhang H., Wang J. Y., Fox M. C., Purton L. E., Fleming H. H., Cobb B., Merkenschlager M., Golub T. R., Scadden D. T. (2010). MicroRNA miR-125a controls hematopoietic stem cell number. Proc. Natl. Acad. Sci. U.S.A. 107, 14229–14234 10.1073/pnas.1008946107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Wu Y., Hartley R. S. (2009). MicroRNA-125a represses cell growth by targeting HuR in breast cancer. RNA Biol. 6, 575–583 10.4161/rna.6.5.10079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez O., Berciano M. T., Lafarga M., Fernandez-Luna J. L. (2011). A novel pathway of TEF regulation mediated by microRNA-125b contributes to the control of actin distribution and cell shape in fibroblasts. PLoS ONE 6, e17169. 10.1371/journal.pone.0017169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackanson B., Bennett K. L., Brena R. M., Jiang J., Claus R., Chen S. S., Blagitko-Dorfs N., Maharry K., Whitman S. P., Schmittgen T. D., Lubbert M., Marcucci G., Bloomfield C. D., Plass C. (2008). Epigenetic modification of CCAAT/enhancer binding protein alpha expression in acute myeloid leukemia. Cancer Res. 68, 3142–3151 10.1158/0008-5472.CAN-07-6859 [DOI] [PubMed] [Google Scholar]
- Haflidadottir B. S., Bergsteinsdottir K., Praetorius C., Steingrimsson E. (2010). miR-148 regulates Mitf in melanoma cells. PLoS ONE 5, e11574. 10.1371/journal.pone.0011574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y., Akiyama Y., Otsubo T., Shimada S., Yuasa Y. (2010). Involvement of epigenetically silenced microRNA-181c in gastric carcinogenesis. Carcinogenesis 31, 777–784 10.1093/carcin/bgq013 [DOI] [PubMed] [Google Scholar]
- Hu G., Gong A. Y., Liu J., Zhou R., Deng C., Chen X. M. (2010). miR-221 suppresses ICAM-1 translation and regulates interferon-gamma-induced ICAM-1 expression in human cholangiocytes. Am. J. Physiol. Gastrointest. Liver Physiol. 298, G542–G550 10.1152/ajpgi.00234.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Luo J., Cai Q., Pan Q., Zeng H., Guo Z., Dong W., Huang J., Lin T. (2011a). MicroRNA-125b suppresses the development of bladder cancer by targeting E2F3. Int. J. Cancer 128, 1758–1769 10.1002/ijc.25633 [DOI] [PubMed] [Google Scholar]
- Huang W., Liu H., Wang T., Zhang T., Kuang J., Luo Y., Chung S. S., Yuan L., Yang J. Y. (2011b). Tonicity-responsive microRNAs contribute to the maximal induction of osmoregulatory transcription factor OREBP in response to high-NaCl hypertonicity. Nucleic Acids Res. 39, 475–485 10.1093/nar/gkq1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Wu S., Ding J., Lin J., Wei L., Gu J., He X. (2010). MicroRNA-181a modulates gene expression of zinc finger family members by directly targeting their coding regions. Nucleic Acids Res. 38, 7211–7218 10.1093/nar/gkq164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliopoulos D., Lindahl-Allen M., Polytarchou C., Hirsch H. A., Tsichlis P. N., Struhl K. (2010). Loss of miR-200 inhibition of Suz12 leads to polycomb-mediated repression required for the formation and maintenance of cancer stem cells. Mol. Cell 39, 761–772 10.1016/j.molcel.2010.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilnytskyy Y., Zemp F. J., Koturbash I., Kovalchuk O. (2008). Altered microRNA expression patterns in irradiated hematopoietic tissues suggest a sex-specific protective mechanism. Biochem. Biophys. Res. Commun. 377, 41–45 10.1016/j.bbrc.2008.09.080 [DOI] [PubMed] [Google Scholar]
- Im H. I., Hollander J. A., Bali P., Kenny P. J. (2010). MeCP2 controls BDNF expression and cocaine intake through homeostatic interactions with microRNA-212. Nat. Neurosci. 13, 1120–1127 10.1038/nn.2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imam J. S., Buddavarapu K., Lee-Chang J. S., Ganapathy S., Camosy C., Chen Y., Rao M. K. (2010). MicroRNA-185 suppresses tumor growth and progression by targeting the Six1 oncogene in human cancers. Oncogene 29, 4971–4979 10.1038/onc.2010.233 [DOI] [PubMed] [Google Scholar]
- Incoronato M., Garofalo M., Urso L., Romano G., Quintavalle C., Zanca C., Iaboni M., Nuovo G., Croce C. M., Condorelli G. (2010). miR-212 increases tumor necrosis factor-related apoptosis-inducing ligand sensitivity in non-small cell lung cancer by targeting the antiapoptotic protein PED. Cancer Res. 70, 3638–3646 10.1158/1538-7445.AM10-3638 [DOI] [PubMed] [Google Scholar]
- Jeyapalan Z., Deng Z., Shatseva T., Fang L., He C., Yang B. B. (2011). Expression of CD44 3′-untranslated region regulates endogenous microRNA functions in tumorigenesis and angiogenesis. Nucleic Acids Res. 39, 3026–3041 10.1093/nar/gkq1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji J., Zhao L., Budhu A., Forgues M., Jia H. L., Qin L. X., Ye Q. H., Yu J., Shi X., Tang Z. Y., Wang X. W. (2010). Let-7g targets collagen type I alpha2 and inhibits cell migration in hepatocellular carcinoma. J. Hepatol. 52, 690–697 10.1016/j.jhep.2009.12.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Liu X., Chen Z., Jin Y., Heidbreder C. E., Kolokythas A., Wang A., Dai Y., Zhou X. (2010). MicroRNA-7 targets IGF1R (insulin-like growth factor 1 receptor) in tongue squamous cell carcinoma cells. Biochem. J. 432, 199–205 10.1042/BJ20100505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan A. H., Kumar R. M., Marx J. G., Young R. A., Sartorelli V. (2009). Mir-214-dependent regulation of the polycomb protein Ezh2 in skeletal muscle and embryonic stem cells. Mol. Cell 36, 61–74 10.1016/j.molcel.2009.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junn E., Lee K. W., Jeong B. S., Chan T. W., Im J. Y., Mouradian M. M. (2009). Repression of alpha-synuclein expression and toxicity by microRNA-7. Proc. Natl. Acad. Sci. U.S.A. 106, 13052–13057 10.1073/pnas.0906277106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karres J. S., Hilgers V., Carrera I., Treisman J., Cohen S. M. (2007). The conserved microRNA miR-8 tunes atrophin levels to prevent neurodegeneration in Drosophila. Cell 131, 136–145 10.1016/j.cell.2007.09.020 [DOI] [PubMed] [Google Scholar]
- Kefas B., Godlewski J., Comeau L., Li Y., Abounader R., Hawkinson M., Lee J., Fine H., Chiocca E. A., Lawler S., Purow B. (2008). microRNA-7 inhibits the epidermal growth factor receptor and the Akt pathway and is down-regulated in glioblastoma. Cancer Res. 68, 3566–3572 10.1158/0008-5472.CAN-07-6639 [DOI] [PubMed] [Google Scholar]
- Kikkawa N., Hanazawa T., Fujimura L., Nohata N., Suzuki H., Chazono H., Sakurai D., Horiguchi S., Okamoto Y., Seki N. (2010). miR-489 is a tumour-suppressive miRNA target PTPN11 in hypopharyngeal squamous cell carcinoma (HSCC). Br. J. Cancer 103, 877–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B. M., Woo J., Kanellopoulou C., Shivdasani R. A. (2011). Regulation of mouse stomach development and Barx1 expression by specific microRNAs. Development 138, 1081–1086 10.1242/dev.066233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Song J., Jin E. J. (2010). MicroRNA-221 regulates chondrogenic differentiation through promoting proteosomal degradation of slug by targeting Mdm2. J. Biol. Chem. 285, 26900–26907 10.1074/jbc.M109.078204 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kim Y. J., Bae S. W., Yu S. S., Bae Y. C., Jung J. S. (2009a). miR-196a regulates proliferation and osteogenic differentiation in mesenchymal stem cells derived from human adipose tissue. J. Bone Miner. Res. 24, 816–825 10.1359/jbmr.081230 [DOI] [PubMed] [Google Scholar]
- Kim Y. K., Yu J., Han T. S., Park S. Y., Namkoong B., Kim D. H., Hur K., Yoo M. W., Lee H. J., Yang H. K., Kim V. N. (2009b). Functional links between clustered microRNAs: suppression of cell-cycle inhibitors by microRNA clusters in gastric cancer. Nucleic Acids Res. 37, 1672–1681 10.1093/nar/gkn813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein M. E., Lioy D. T., Ma L., Impey S., Mandel G., Goodman R. H. (2007). Homeostatic regulation of MeCP2 expression by a CREB-induced microRNA. Nat. Neurosci. 10, 1513–1514 10.1038/nn2010 [DOI] [PubMed] [Google Scholar]
- Ko H. Y., Lee D. S., Kim S. (2009). Noninvasive imaging of microRNA124a-mediated repression of the chromosome 14 ORF 24 gene during neurogenesis. FEBS J. 276, 4854–4865 10.1111/j.1742-4658.2009.07185.x [DOI] [PubMed] [Google Scholar]
- Komagata S., Nakajima M., Takagi S., Mohri T., Taniya T., Yokoi T. (2009). Human CYP24 catalyzing the inactivation of calcitriol is post-transcriptionally regulated by miR-125b. Mol. Pharmacol. 76, 702–709 10.1124/mol.109.056986 [DOI] [PubMed] [Google Scholar]
- Kotani A., Ha D., Hsieh J., Rao P. K., Schotte D., Den Boer M. L., Armstrong S. A., Lodish H. F. (2009). miR-128b is a potent glucocorticoid sensitizer in MLL-AF4 acute lymphocytic leukemia cells and exerts cooperative effects with miR-221. Blood 114, 4169–4178 10.1182/blood-2008-12-191619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek A., Grun D., Poy M. N., Wolf R., Rosenberg L., Epstein E. J., Macmenamin P., Da Piedade I., Gunsalus K. C., Stoffel M., Rajewsky N. (2005). Combinatorial microRNA target predictions. Nat. Genet. 37, 495–500 10.1038/ng1536 [DOI] [PubMed] [Google Scholar]
- Lagos D., Pollara G., Henderson S., Gratrix F., Fabani M., Milne R. S., Gotch F., Boshoff C. (2010). miR-132 regulates antiviral innate immunity through suppression of the p300 transcriptional co-activator. Nat. Cell Biol. 12, 513–519 10.1038/ncb2054 [DOI] [PubMed] [Google Scholar]
- Lal A., Navarro F., Maher C. A., Maliszewski L. E., Yan N., O’day E., Chowdhury D., Dykxhoorn D. M., Tsai P., Hofmann O., Becker K. G., Gorospe M., Hide W., Lieberman J. (2009a). miR-24 inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3′UTR microRNA recognition elements. Mol. Cell 35, 610–625 10.1016/j.molcel.2009.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal A., Pan Y., Navarro F., Dykxhoorn D. M., Moreau L., Meire E., Bentwich Z., Lieberman J., Chowdhury D. (2009b). miR-24-mediated downregulation of H2AX suppresses DNA repair in terminally differentiated blood cells. Nat. Struct. Mol. Biol. 16, 492–498 10.1038/nsmb.1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan F. F., Wang H., Chen Y. C., Chan C. Y., Ng S. S., Li K., Xie D., He M. L., Lin M. C., Kung H. F. (2011). Hsa-let-7g inhibits proliferation of hepatocellular carcinoma cells by downregulation of c-Myc and upregulation of p16(INK4A). Int. J. Cancer 128, 319–331 10.1002/ijc.25336 [DOI] [PubMed] [Google Scholar]
- Le M. T., Teh C., Shyh-Chang N., Xie H., Zhou B., Korzh V., Lodish H. F., Lim B. (2009). MicroRNA-125b is a novel negative regulator of p53. Genes Dev. 23, 862–876 10.1101/gad.1767609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivonen S. K., Makela R., Ostling P., Kohonen P., Haapa-Paananen S., Kleivi K., Enerly E., Aakula A., Hellstrom K., Sahlberg N., Kristensen V. N., Borresen-Dale A. L., Saviranta P., Perala M., Kallioniemi O. (2009). Protein lysate microarray analysis to identify microRNAs regulating estrogen receptor signaling in breast cancer cell lines. Oncogene 28, 3926–3936 10.1038/onc.2009.241 [DOI] [PubMed] [Google Scholar]
- Li L. M., Hou D. X., Guo Y. L., Yang J. W., Liu Y., Zhang C. Y., Zen K. (2011). Role of microRNA-214-targeting phosphatase and tensin homolog in advanced glycation end product-induced apoptosis delay in monocytes. J. Immunol. 186, 2552–2560 10.4049/jimmunol.1190019 [DOI] [PubMed] [Google Scholar]
- Li S. S., Yu S. L., Kao L. P., Tsai Z. Y., Singh S., Chen B. Z., Ho B. C., Liu Y. H., Yang P. C. (2009a). Target identification of microRNAs expressed highly in human embryonic stem cells. J. Cell. Biochem. 106, 1020–1030 10.1002/jcb.22084 [DOI] [PubMed] [Google Scholar]
- Li X. F., Yan P. J., Shao Z. M. (2009b). Downregulation of miR-193b contributes to enhance urokinase-type plasminogen activator (uPA) expression and tumor progression and invasion in human breast cancer. Oncogene 28, 3937–3948 10.1038/onc.2009.171 [DOI] [PubMed] [Google Scholar]
- Li X., Carthew R. W. (2005). A microRNA mediates EGF receptor signaling and promotes photoreceptor differentiation in the Drosophila eye. Cell 123, 1267–1277 10.1016/j.cell.2005.10.040 [DOI] [PubMed] [Google Scholar]
- Liao R., Sun J., Zhang L., Lou G., Chen M., Zhou D., Chen Z., Zhang S. (2008). MicroRNAs play a role in the development of human hematopoietic stem cells. J. Cell. Biochem. 104, 805–817 10.1002/jcb.21730 [DOI] [PubMed] [Google Scholar]
- Liu H., Brannon A. R., Reddy A. R., Alexe G., Seiler M. W., Arreola A., Oza J. H., Yao M., Juan D., Liou L. S., Ganesan S., Levine A. J., Rathmell W. K., Bhanot G. V. (2010a). Identifying mRNA targets of microRNA dysregulated in cancer: with application to clear cell renal cell carcinoma. BMC Syst. Biol. 4, 51. 10.1186/1752-0509-4-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Luo X. J., Xiong A. W., Zhang Z. D., Yue S., Zhu M. S., Cheng S. Y. (2010b). MicroRNA-214 promotes myogenic differentiation by facilitating exit from mitosis via down-regulation of proto-oncogene N-ras. J. Biol. Chem. 285, 26599–26607 10.1074/jbc.M109.098558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Sempere L. F., Ouyang H., Memoli V. A., Andrew A. S., Luo Y., Demidenko E., Korc M., Shi W., Preis M., Dragnev K. H., Li H., Direnzo J., Bak M., Freemantle S. J., Kauppinen S., Dmitrovsky E. (2010c). MicroRNA-31 functions as an oncogenic microRNA in mouse and human lung cancer cells by repressing specific tumor suppressors. J. Clin. Invest. 120, 1298–1309 10.1172/JCI39566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Wang A., Heidbreder C. E., Jiang L., Yu J., Kolokythas A., Huang L., Dai Y., Zhou X. (2010d). MicroRNA-24 targeting RNA-binding protein DND1 in tongue squamous cell carcinoma. FEBS Lett. 584, 4115–4120 10.1016/j.febslet.2010.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Zhan Z., Xu L., Ma F., Li D., Guo Z., Li N., Cao X. (2010e). MicroRNA-148/152 impair innate response and antigen presentation of TLR-triggered dendritic cells by targeting CaMKIIalpha. J. Immunol. 185, 7244–7251 10.4049/jimmunol.1000953 [DOI] [PubMed] [Google Scholar]
- Liu K., Liu Y., Mo W., Qiu R., Wang X., Wu J. Y., He R. (2011). MiR-124 regulates early neurogenesis in the optic vesicle and forebrain, targeting NeuroD1. Nucleic Acids Res. 39, 2869–2879 10.1093/nar/gkq956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Yu J., Jiang L., Wang A., Shi F., Ye H., Zhou X. (2009). MicroRNA-222 regulates cell invasion by targeting matrix metalloproteinase 1 (MMP1) and manganese superoxide dismutase 2 (SOD2) in tongue squamous cell carcinoma cell lines. Cancer Genomics Proteomics 6, 131–139 [PMC free article] [PubMed] [Google Scholar]
- Luna C., Li G., Qiu J., Epstein D. L., Gonzalez P. (2011). MicroRNA-24 regulates the processing of latent TGFbeta1 during cyclic mechanical stress in human trabecular meshwork cells through direct targeting of FURIN. J. Cell. Physiol. 226, 1407–1414 10.1002/jcp.22476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthra R., Singh R. R., Luthra M. G., Li Y. X., Hannah C., Romans A. M., Barkoh B. A., Chen S. S., Ensor J., Maru D. M., Broaddus R. R., Rashid A., Albarracin C. T. (2008). MicroRNA-196a targets annexin A1: a microRNA-mediated mechanism of annexin A1 downregulation in cancers. Oncogene 27, 6667–6678 10.1038/onc.2008.256 [DOI] [PubMed] [Google Scholar]
- Lwin T., Lin J., Choi Y. S., Zhang X., Moscinski L. C., Wright K. L., Sotomayor E. M., Dalton W. S., Tao J. (2010). Follicular dendritic cell-dependent drug resistance of non-Hodgkin lymphoma involves cell adhesion-mediated Bim down-regulation through induction of microRNA-181a. Blood 116, 5228–5236 10.1182/blood-2010-03-275925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma N., Wang X., Qiao Y., Li F., Hui Y., Zou C., Jin J., Lv G., Peng Y., Wang L., Huang H., Zhou L., Zheng X., Gao X. (2011). Coexpression of an intronic microRNA and its host gene reveals a potential role for miR-483-5p as an IGF2 partner. Mol. Cell. Endocrinol. 333, 96–101 10.1016/j.mce.2010.11.027 [DOI] [PubMed] [Google Scholar]
- Makeyev E. V., Zhang J., Carrasco M. A., Maniatis T. (2007). The microRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol. Cell 27, 435–448 10.1016/j.molcel.2007.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhas A., Saunders N. J., Vaux D. J. (2010). The nuclear envelope can control gene expression and cell cycle progression via miRNA regulation. Cell Cycle 9, 531–539 10.4161/cc.9.3.10511 [DOI] [PubMed] [Google Scholar]
- Malumbres R., Sarosiek K. A., Cubedo E., Ruiz J. W., Jiang X., Gascoyne R. D., Tibshirani R., Lossos I. S. (2009). Differentiation stage-specific expression of microRNAs in B lymphocytes and diffuse large B-cell lymphomas. Blood 113, 3754–3764 10.1182/blood-2008-10-184077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marasa B. S., Srikantan S., Masuda K., Abdelmohsen K., Kuwano Y., Yang X., Martindale J. L., Rinker-Schaeffer C. W., Gorospe M. (2009). Increased MKK4 abundance with replicative senescence is linked to the joint reduction of multiple microRNAs. Sci. Signal. 2, ra69. 10.1126/scisignal.2000442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardaryev A. N., Ahmed M. I., Vlahov N. V., Fessing M. Y., Gill J. H., Sharov A. A., Botchkareva N. V. (2010). Micro-RNA-31 controls hair cycle-associated changes in gene expression programs of the skin and hair follicle. FASEB J. 24, 3869–3881 10.1096/fj.10-160663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez I., Cazalla D., Almstead L. L., Steitz J. A., Dimaio D. (2011). miR-29 and miR-30 regulate B-Myb expression during cellular senescence. Proc. Natl. Acad. Sci. U.S.A. 108, 522–527 10.1073/pnas.1017346108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maru D. M., Singh R. R., Hannah C., Albarracin C. T., Li Y. X., Abraham R., Romans A. M., Yao H., Luthra M. G., Anandasabapathy S., Swisher S. G., Hofstetter W. L., Rashid A., Luthra R. (2009). MicroRNA-196a is a potential marker of progression during Barrett’s metaplasia-dysplasia-invasive adenocarcinoma sequence in esophagus. Am. J. Pathol. 174, 1940–1948 10.2353/ajpath.2009.080718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mees S. T., Mardin W. A., Wendel C., Baeumer N., Willscher E., Senninger N., Schleicher C., Colombo-Benkmann M., Haier J. (2010). EP300 – a miRNA-regulated metastasis suppressor gene in ductal adenocarcinomas of the pancreas. Int. J. Cancer 126, 114–124 10.1002/ijc.24695 [DOI] [PubMed] [Google Scholar]
- Mellios N., Huang H. S., Grigorenko A., Rogaev E., Akbarian S. (2008). A set of differentially expressed miRNAs, including miR-30a-5p, act as post-transcriptional inhibitors of BDNF in prefrontal cortex. Hum. Mol. Genet. 17, 3030–3042 10.1093/hmg/ddn201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra P. J., Song B., Wang Y., Humeniuk R., Banerjee D., Merlino G., Ju J., Bertino J. R. (2009). MiR-24 tumor suppressor activity is regulated independent of p53 and through a target site polymorphism. PLoS ONE 4, e8445. 10.1371/journal.pone.0008445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muinos-Gimeno M., Espinosa-Parrilla Y., Guidi M., Kagerbauer B., Sipila T., Maron E., Pettai K., Kananen L., Navines R., Martin-Santos R., Gratacos M., Metspalu A., Hovatta I., Estivill X. (2011). Human microRNAs miR-22, miR-138-2, miR-148a, and miR-488 are associated with panic disorder and regulate several anxiety candidate genes and related pathways. Biol. Psychiatry 69, 526–533 10.1016/j.biopsych.2010.10.010 [DOI] [PubMed] [Google Scholar]
- Murata T., Takayama K., Katayama S., Urano T., Horie-Inoue K., Ikeda K., Takahashi S., Kawazu C., Hasegawa A., Ouchi Y., Homma Y., Hayashizaki Y., Inoue S. (2010). miR-148a is an androgen-responsive microRNA that promotes LNCaP prostate cell growth by repressing its target CAND1 expression. Prostate Cancer Prostatic Dis. 13, 356–361 10.1038/pcan.2010.32 [DOI] [PubMed] [Google Scholar]
- Nagel R., Clijsters L., Agami R. (2009). The miRNA-192/194 cluster regulates the period gene family and the circadian clock. FEBS J. 276, 5447–5455 10.1111/j.1742-4658.2009.07229.x [DOI] [PubMed] [Google Scholar]
- Naguibneva I., Ameyar-Zazoua M., Polesskaya A., Ait-Si-Ali S., Groisman R., Souidi M., Cuvellier S., Harel-Bellan A. (2006). The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nat. Cell Biol. 8, 278–284 10.1038/ncb1373 [DOI] [PubMed] [Google Scholar]
- Nakamachi Y., Kawano S., Takenokuchi M., Nishimura K., Sakai Y., Chin T., Saura R., Kurosaka M., Kumagai S. (2009). MicroRNA-124a is a key regulator of proliferation and monocyte chemoattractant protein 1 secretion in fibroblast-like synoviocytes from patients with rheumatoid arthritis. Arthritis Rheum. (Munch.) 60, 1294–1304 10.1002/art.24475 [DOI] [PubMed] [Google Scholar]
- Nakano H., Miyazawa T., Kinoshita K., Yamada Y., Yoshida T. (2010). Functional screening identifies a microRNA, miR-491 that induces apoptosis by targeting Bcl-X(L) in colorectal cancer cells. Int. J. Cancer 127, 1072–1080 10.1002/ijc.25143 [DOI] [PubMed] [Google Scholar]
- Nguyen H. T., Dalmasso G., Yan Y., Laroui H., Dahan S., Mayer L., Sitaraman S. V., Merlin D. (2010). MicroRNA-7 modulates CD98 expression during intestinal epithelial cell differentiation. J. Biol. Chem. 285, 1479–1489 10.1074/jbc.M110.111757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pais H., Nicolas F. E., Soond S. M., Swingler T. E., Clark I. M., Chantry A., Moulton V., Dalmay T. (2010). Analyzing mRNA expression identifies Smad3 as a microRNA-140 target regulated only at protein level. RNA 16, 489–494 10.1261/rna.1701210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallasch C. P., Patz M., Park Y. J., Hagist S., Eggle D., Claus R., Debey-Pascher S., Schulz A., Frenzel L. P., Claasen J., Kutsch N., Krause G., Mayr C., Rosenwald A., Plass C., Schultze J. L., Hallek M., Wendtner C. M. (2009). miRNA deregulation by epigenetic silencing disrupts suppression of the oncogene PLAG1 in chronic lymphocytic leukemia. Blood 114, 3255–3264 10.1182/blood-2009-06-229898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan W., Zhu S., Yuan M., Cui H., Wang L., Luo X., Li J., Zhou H., Tang Y., Shen N. (2010). MicroRNA-21 and microRNA-148a contribute to DNA hypomethylation in lupus CD4+ T cells by directly and indirectly targeting DNA methyltransferase 1. J. Immunol. 184, 6773–6781 10.4049/jimmunol.0904060 [DOI] [PubMed] [Google Scholar]
- Papaioannou M. D., Lagarrigue M., Vejnar C. E., Rolland A. D., Kuhne F., Aubry F., Schaad O., Fort A., Descombes P., Neerman-Arbez M., Guillou F., Zdobnov E. M., Pineau C., Nef S. (2011). Loss of Dicer in Sertoli cells has a major impact on the testicular proteome of mice. Mol. Cell. Proteomics 10, M900587MCP900200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. K., Henry J. C., Jiang J., Esau C., Gusev Y., Lerner M. R., Postier R. G., Brackett D. J., Schmittgen T. D. (2011). miR-132 and miR-212 are increased in pancreatic cancer and target the retinoblastoma tumor suppressor. Biochem. Biophys. Res. Commun. 406, 518–523 10.1016/j.bbrc.2011.02.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrioli D. M., Karpanen T., Dabouras V., Jurisic G., Van De Hoek G., Shin J. W., Marino D., Kalin R. E., Leidel S., Cinelli P., Schulte-Merker S., Brandli A. W., Detmar M. (2010). miR-31 functions as a negative regulator of lymphatic vascular lineage-specific differentiation in vitro and vascular development in vivo. Mol. Cell. Biol. 30, 3620–3634 10.1128/MCB.00185-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekarsky Y., Santanam U., Cimmino A., Palamarchuk A., Efanov A., Maximov V., Volinia S., Alder H., Liu C. G., Rassenti L., Calin G. A., Hagan J. P., Kipps T., Croce C. M. (2006). Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Res. 66, 11590–11593 10.1158/0008-5472.CAN-06-3613 [DOI] [PubMed] [Google Scholar]
- Pichiorri F., Suh S. S., Rocci A., De Luca L., Taccioli C., Santhanam R., Zhou W., Benson D. M., Jr., Hofmainster C., Alder H., Garofalo M., Di Leva G., Volinia S., Lin H. J., Perrotti D., Kuehl M., Aqeilan R. I., Palumbo A., Croce C. M. (2010). Downregulation of p53-inducible microRNAs 192, 194, and 215 impairs the p53/MDM2 autoregulatory loop in multiple myeloma development. Cancer Cell 18, 367–381 10.1016/j.ccr.2010.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Pierson J., Hostager B., Fan R., Vibhakar R. (2008). Regulation of cyclin dependent kinase 6 by microRNA 124 in medulloblastoma. J. Neurooncol. 90, 1–7 10.1007/s11060-008-9624-3 [DOI] [PubMed] [Google Scholar]
- Pineau P., Volinia S., Mcjunkin K., Marchio A., Battiston C., Terris B., Mazzaferro V., Lowe S. W., Croce C. M., Dejean A. (2010). miR-221 overexpression contributes to liver tumorigenesis. Proc. Natl. Acad. Sci. U.S.A. 107, 264–269 10.1073/pnas.0907904107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogribny I. P., Filkowski J. N., Tryndyak V. P., Golubov A., Shpyleva S. I., Kovalchuk O. (2010). Alterations of microRNAs and their targets are associated with acquired resistance of MCF-7 breast cancer cells to cisplatin. Int. J. Cancer 127, 1785–1794 10.1002/ijc.25191 [DOI] [PubMed] [Google Scholar]
- Qin W., Shi Y., Zhao B., Yao C., Jin L., Ma J., Jin Y. (2010). miR-24 regulates apoptosis by targeting the open reading frame (ORF) region of FAF1 in cancer cells. PLoS ONE 5, e9429. 10.1371/journal.pone.0009429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu R., Liu K., Liu Y., Mo W., Flynt A. S., Patton J. G., Kar A., Wu J. Y., He R. (2009). The role of miR-124a in early development of the Xenopus eye. Mech. Dev. 126, 804–816 10.1016/j.mod.2009.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajabi H., Jin C., Ahmad R., Mcclary C., Joshi M. D., Kufe D. (2010). Mucin 1 oncoprotein expression is suppressed by the mir-125b oncomir. Genes Cancer 1, 62–68 10.1177/1947601909357933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rane S., He M., Sayed D., Vashistha H., Malhotra A., Sadoshima J., Vatner D. E., Vatner S. F., Abdellatif M. (2009). Downregulation of miR-199a derepresses hypoxia-inducible factor-1alpha and sirtuin 1 and recapitulates hypoxia preconditioning in cardiac myocytes. Circ. Res. 104, 879–886 10.1161/CIRCRESAHA.108.193102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy S. D., Ohshiro K., Rayala S. K., Kumar R. (2008). MicroRNA-7, a homeobox D10 target, inhibits p21-activated kinase 1 and regulates its functions. Cancer Res. 68, 8195–8200 10.1158/0008-5472.CAN-08-2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X. P., Wu J., Wang X., Sartor M. A., Qian J., Jones K., Nicolaou P., Pritchard T. J., Fan G. C. (2009). MicroRNA-320 is involved in the regulation of cardiac ischemia/reperfusion injury by targeting heat-shock protein 20. Circulation 119, 2357–2366 10.1161/CIRCULATIONAHA.108.814145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saetrom P., Biesinger J., Li S. M., Smith D., Thomas L. F., Majzoub K., Rivas G. E., Alluin J., Rossi J. J., Krontiris T. G., Weitzel J., Daly M. B., Benson A. B., Kirkwood J. M., O’dwyer P. J., Sutphen R., Stewart J. A., Johnson D., Larson G. P. (2009). A risk variant in an miR-125b binding site in BMPR1B is associated with breast cancer pathogenesis. Cancer Res. 69, 7459–7465 10.1158/0008-5472.CAN-09-1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomonis N., Schlieve C. R., Pereira L., Wahlquist C., Colas A., Zambon A. C., Vranizan K., Spindler M. J., Pico A. R., Cline M. S., Clark T. A., Williams A., Blume J. E., Samal E., Mercola M., Merrill B. J., Conklin B. R. (2010). Alternative splicing regulates mouse embryonic stem cell pluripotency and differentiation. Proc. Natl. Acad. Sci. U.S.A. 107, 10514–10519 10.1073/pnas.0912260107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders L. R., Sharma A. D., Tawney J., Nakagawa M., Okita K., Yamanaka S., Willenbring H., Verdin E. (2010). miRNAs regulate SIRT1 expression during mouse embryonic stem cell differentiation and in adult mouse tissues. Aging (Albany NY) 2, 415–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saydam O., Senol O., Wurdinger T., Mizrak A., Ozdener G. B., Stemmer-Rachamimov A. O., Yi M., Stephens R. M., Krichevsky A. M., Saydam N., Brenner G. J., Breakefield X. O. (2011). miRNA-7 attenuation in schwannoma tumors stimulates growth by upregulating three oncogenic signaling pathways. Cancer Res. 71, 852–861 10.1158/0008-5472.CAN-10-1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaar D. G., Medina D. J., Moore D. F., Strair R. K., Ting Y. (2009). miR-320 targets transferrin receptor 1 (CD71) and inhibits cell proliferation. Exp. Hematol. 37, 245–255 10.1016/j.exphem.2008.10.002 [DOI] [PubMed] [Google Scholar]
- Schickel R., Park S. M., Murmann A. E., Peter M. E. (2010). miR-200c regulates induction of apoptosis through CD95 by targeting FAP-1. Mol. Cell 38, 908–915 10.1016/j.molcel.2010.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott G. K., Goga A., Bhaumik D., Berger C. E., Sullivan C. S., Benz C. C. (2007). Coordinate suppression of ERBB2 and ERBB3 by enforced expression of micro-RNA miR-125a or miR-125b. J. Biol. Chem. 282, 1479–1486 10.1074/jbc.M609383200 [DOI] [PubMed] [Google Scholar]
- Sepramaniam S., Armugam A., Lim K. Y., Karolina D. S., Swaminathan P., Tan J. R., Jeyaseelan K. (2010). MicroRNA 320a functions as a novel endogenous modulator of aquaporins 1 and 4 as well as a potential therapeutic target in cerebral ischemia. J. Biol. Chem. 285, 29223–29230 10.1074/jbc.M110.144576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao M., Rossi S., Chelladurai B., Shimizu M., Ntukogu O., Ivan M., Calin G. A., Matei D. (2011). PDGF induced microRNA alterations in cancer cells. Nucleic Acids Res. 39, 4035–4047 10.1093/nar/gkq908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q., Cicinnati V. R., Zhang X., Iacob S., Weber F., Sotiropoulos G. C., Radtke A., Lu M., Paul A., Gerken G., Beckebaum S. (2010). Role of microRNA-199a-5p and discoidin domain receptor 1 in human hepatocellular carcinoma invasion. Mol. Cancer 9, 227. 10.1186/1476-4598-9-227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu S., Takehara T., Hikita H., Kodama T., Miyagi T., Hosui A., Tatsumi T., Ishida H., Noda T., Nagano H., Doki Y., Mori M., Hayashi N. (2010). The let-7 family of microRNAs inhibits Bcl-xL expression and potentiates sorafenib-induced apoptosis in human hepatocellular carcinoma. J. Hepatol. 52, 698–704 10.1016/j.jhep.2009.12.024 [DOI] [PubMed] [Google Scholar]
- Small E. M., Sutherland L. B., Rajagopalan K. N., Wang S., Olson E. N. (2010). MicroRNA-218 regulates vascular patterning by modulation of Slit-Robo signaling. Circ. Res. 107, 1336–1344 10.1161/CIRCRESAHA.110.227926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnov D. A., Cheung V. G. (2008). ATM gene mutations result in both recessive and dominant expression phenotypes of genes and microRNAs. Am. J. Hum. Genet. 83, 243–253 10.1016/j.ajhg.2008.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sober S., Laan M., Annilo T. (2010). MicroRNAs miR-124 and miR-135a are potential regulators of the mineralocorticoid receptor gene (NR3C2) expression. Biochem. Biophys. Res. Commun. 391, 727–732 10.1016/j.bbrc.2009.11.128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L., Huang Q., Chen K., Liu L., Lin C., Dai T., Yu C., Wu Z., Li J. (2010). miR-218 inhibits the invasive ability of glioma cells by direct downregulation of IKK-beta. Biochem. Biophys. Res. Commun. 402, 135–140 10.1016/j.bbrc.2010.10.003 [DOI] [PubMed] [Google Scholar]
- Sossey-Alaoui K., Downs-Kelly E., Das M., Izem L., Tubbs R., Plow E. F. (2010). WAVE3, an actin remodeling protein, is regulated by the metastasis suppressor microRNA, miR-31, during the invasion-metastasis cascade. Int. J. Cancer 129, 1331–1343 10.1002/ijc.25793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strum J. C., Johnson J. H., Ward J., Xie H., Feild J., Hester A., Alford A., Waters K. M. (2009). MicroRNA 132 regulates nutritional stress-induced chemokine production through repression of SirT1. Mol. Endocrinol. 23, 1876–1884 10.1210/me.2009-0117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez Y., Wang C., Manes T. D., Pober J. S. (2010). Cutting edge: TNF-induced microRNAs regulate TNF-induced expression of E-selectin and intercellular adhesion molecule-1 on human endothelial cells: feedback control of inflammation. J. Immunol. 184, 21–25 10.4049/jimmunol.0902369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surdziel E., Cabanski M., Dallmann I., Lyszkiewicz M., Krueger A., Ganser A., Scherr M., Eder M. (2011). Enforced expression of miR-125b affects myelopoiesis by targeting multiple signaling pathways. Blood 117, 4338–4348 10.1182/blood-2010-06-289058 [DOI] [PubMed] [Google Scholar]
- Taganov K. D., Boldin M. P., Chang K. J., Baltimore D. (2006). NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. U.S.A. 103, 12481–12486 10.1073/pnas.0605298103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi S., Nakajima M., Kida K., Yamaura Y., Fukami T., Yokoi T. (2010). MicroRNAs regulate human hepatocyte nuclear factor 4alpha, modulating the expression of metabolic enzymes and cell cycle. J. Biol. Chem. 285, 4415–4422 10.1074/jbc.M109.085431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi S., Nakajima M., Mohri T., Yokoi T. (2008). Post-transcriptional regulation of human pregnane X receptor by micro-RNA affects the expression of cytochrome P450 3A4. J. Biol. Chem. 283, 9674–9680 10.1074/jbc.M709382200 [DOI] [PubMed] [Google Scholar]
- Tan Z., Randall G., Fan J., Camoretti-Mercado B., Brockman-Schneider R., Pan L., Solway J., Gern J. E., Lemanske R. F., Nicolae D., Ober C. (2007). Allele-specific targeting of microRNAs to HLA-G and risk of asthma. Am. J. Hum. Genet. 81, 829–834 10.1086/521200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terasawa K., Ichimura A., Sato F., Shimizu K., Tsujimoto G. (2009). Sustained activation of ERK1/2 by NGF induces microRNA-221 and 222 in PC12 cells. FEBS J. 276, 3269–3276 10.1111/j.1742-4658.2009.06901.x [DOI] [PubMed] [Google Scholar]
- Thatcher E. J., Paydar I., Anderson K. K., Patton J. G. (2008). Regulation of zebrafish fin regeneration by microRNAs. Proc. Natl. Acad. Sci. U.S.A. 105, 18384–18389 10.1073/pnas.0803713105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tili E., Michaille J. J., Adair B., Alder H., Limagne E., Taccioli C., Ferracin M., Delmas D., Latruffe N., Croce C. M. (2010a). Resveratrol decreases the levels of miR-155 by upregulating miR-663, a microRNA targeting JunB and JunD. Carcinogenesis 31, 1561–1566 10.1093/carcin/bgq143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tili E., Michaille J. J., Alder H., Volinia S., Delmas D., Latruffe N., Croce C. M. (2010b). Resveratrol modulates the levels of microRNAs targeting genes encoding tumor-suppressors and effectors of TGFbeta signaling pathway in SW480 cells. Biochem. Pharmacol. 80, 2057–2065 10.1016/j.bcp.2010.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tili E., Michaille J. J., Cimino A., Costinean S., Dumitru C. D., Adair B., Fabbri M., Alder H., Liu C. G., Calin G. A., Croce C. M. (2007). Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J. Immunol. 179, 5082–5089 [DOI] [PubMed] [Google Scholar]
- To K. K., Zhan Z., Litman T., Bates S. E. (2008). Regulation of ABCG2 expression at the 3′ untranslated region of its mRNA through modulation of transcript stability and protein translation by a putative microRNA in the S1 colon cancer cell line. Mol. Cell. Biol. 28, 5147–5161 10.1128/MCB.00331-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuddenham L., Wheeler G., Ntounia-Fousara S., Waters J., Hajihosseini M. K., Clark I., Dalmay T. (2006). The cartilage specific microRNA-140 targets histone deacetylase 4 in mouse cells. FEBS Lett. 580, 4214–4217 10.1016/j.febslet.2006.06.080 [DOI] [PubMed] [Google Scholar]
- Ueda R., Kohanbash G., Sasaki K., Fujita M., Zhu X., Kastenhuber E. R., Mcdonald H. A., Potter D. M., Hamilton R. L., Lotze M. T., Khan S. A., Sobol R. W., Okada H. (2009). Dicer-regulated microRNAs 222 and 339 promote resistance of cancer cells to cytotoxic T-lymphocytes by down-regulation of ICAM-1. Proc. Natl. Acad. Sci. U.S.A. 106, 10746–10751 10.1073/pnas.0811817106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno K., Hirata H., Shahryari V., Chen Y., Zaman M. S., Singh K., Tabatabai Z. L., Hinoda Y., Dahiya R. (2011). Tumour suppressor microRNA-584 directly targets oncogene Rock-1 and decreases invasion ability in human clear cell renal cell carcinoma. Br. J. Cancer 104, 308–315 10.1038/sj.bjc.6606028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann S., Zhang J. D., Schwager A., Mannsperger H., Riazalhosseini Y., Burmester S., Ward A., Korf U., Wiemann S., Sahin O. (2010). miR-200bc/429 cluster targets PLCgamma1 and differentially regulates proliferation and EGF-driven invasion than miR-200a/141 in breast cancer. Oncogene 29, 4297–4306 10.1038/onc.2010.201 [DOI] [PubMed] [Google Scholar]
- Valastyan S., Reinhardt F., Benaich N., Calogrias D., Szasz A. M., Wang Z. C., Brock J. E., Richardson A. L., Weinberg R. A. (2009). A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell 137, 1032–1046 10.1016/j.cell.2009.03.047 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Venugopal S. K., Jiang J., Kim T. H., Li Y., Wang S. S., Torok N. J., Wu J., Zern M. A. (2010). Liver fibrosis causes downregulation of miRNA-150 and miRNA-194 in hepatic stellate cells, and their overexpression causes decreased stellate cell activation. Am. J. Physiol. Gastrointest. Liver Physiol. 298, G101–G106 10.1152/ajpgi.00220.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veronese A., Lupini L., Consiglio J., Visone R., Ferracin M., Fornari F., Zanesi N., Alder H., D’elia G., Gramantieri L., Bolondi L., Lanza G., Querzoli P., Angioni A., Croce C. M., Negrini M. (2010). Oncogenic role of miR-483-3p at the IGF2/483 locus. Cancer Res. 70, 3140–3149 10.1158/0008-5472.CAN-09-4456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve L. M., Kato M., Reddy M. A., Wang M., Lanting L., Natarajan R. (2010). Enhanced levels of microRNA-125b in vascular smooth muscle cells of diabetic db/db mice lead to increased inflammatory gene expression by targeting the histone methyltransferase Suv39h1. Diabetes 59, 2904–2915 10.2337/db10-0208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viticchie G., Lena A. M., Latina A., Formosa A., Gregersen L. H., Lund A. H., Bernardini S., Mauriello A., Miano R., Spagnoli L. G., Knight R. A., Candi E., Melino G. (2011). MiR-203 controls proliferation, migration and invasive potential of prostate cancer cell lines. Cell Cycle 10, 1121–1131 10.4161/cc.10.7.15180 [DOI] [PubMed] [Google Scholar]
- Vreugdenhil E., Verissimo C. S., Mariman R., Kamphorst J. T., Barbosa J. S., Zweers T., Champagne D. L., Schouten T., Meijer O. C., De Kloet E. R., Fitzsimons C. P. (2009). MicroRNA 18 and 124a down-regulate the glucocorticoid receptor: implications for glucocorticoid responsiveness in the brain. Endocrinology 150, 2220–2228 10.1210/en.2008-1335 [DOI] [PubMed] [Google Scholar]
- Wang B., Hsu S. H., Majumder S., Kutay H., Huang W., Jacob S. T., Ghoshal K. (2010a). TGFbeta-mediated upregulation of hepatic miR-181b promotes hepatocarcinogenesis by targeting TIMP3. Oncogene 29, 1787–1797 10.1038/onc.2009.302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zhang X., Ren X. P., Chen J., Liu H., Yang J., Medvedovic M., Hu Z., Fan G. C. (2010b). MicroRNA-494 targeting both proapoptotic and antiapoptotic proteins protects against ischemia/reperfusion-induced cardiac injury. Circulation 122, 1308–1318 10.1161/CIRCULATIONAHA.110.964684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Huang Z., Xue H., Jin C., Ju X. L., Han J. D., Chen Y. G. (2008). MicroRNA miR-24 inhibits erythropoiesis by targeting activin type I receptor ALK4. Blood 111, 588–595 10.1182/blood-2007-07-103119 [DOI] [PubMed] [Google Scholar]
- Wang Y., Yu Y., Tsuyada A., Ren X., Wu X., Stubblefield K., Rankin-Gee E. K., Wang S. E. (2011). Transforming growth factor-beta regulates the sphere-initiating stem cell-like feature in breast cancer through miRNA-181 and ATM. Oncogene 30, 1470–1480 10.1038/onc.2010.531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei T., Orfanidis K., Xu N., Janson P., Stahle M., Pivarcsi A., Sonkoly E. (2010). The expression of microRNA-203 during human skin morphogenesis. Exp. Dermatol. 19, 854–856 10.1111/j.1600-0625.2010.01118.x [DOI] [PubMed] [Google Scholar]
- Wong Q. W., Ching A. K., Chan A. W., Choy K. W., To K. F., Lai P. B., Wong N. (2010). MiR-222 overexpression confers cell migratory advantages in hepatocellular carcinoma through enhancing AKT signaling. Clin. Cancer Res. 16, 867–875 10.1158/1078-0432.CCR-09-1840 [DOI] [PubMed] [Google Scholar]
- Wu D. W., Cheng Y. W., Wang J., Chen C. Y., Lee H. (2010a). Paxillin predicts survival and relapse in non-small cell lung cancer by microRNA-218 targeting. Cancer Res. 70, 10392–10401 10.1158/0008-5472.CAN-10-1656 [DOI] [PubMed] [Google Scholar]
- Wu H., Sun S., Tu K., Gao Y., Xie B., Krainer A. R., Zhu J. (2010b). A splicing-independent function of SF2/ASF in microRNA processing. Mol. Cell 38, 67–77 10.1016/j.molcel.2010.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhu S., Ding Y., Beck W. T., Mo Y. Y. (2009). MicroRNA-mediated regulation of Ubc9 expression in cancer cells. Clin. Cancer Res. 15, 1550–1557 10.1158/1078-0432.CCR-08-1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Belasco J. G. (2005). Micro-RNA regulation of the mammalian lin-28 gene during neuronal differentiation of embryonal carcinoma cells. Mol. Cell. Biol. 25, 9198–9208 10.1128/MCB.25.22.9741-9752.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi S., Yang M., Tao Y., Xu H., Shan J., Inchauste S., Zhang M., Mercedes L., Hong J. A., Rao M., Schrump D. S. (2010). Cigarette smoke induces C/EBP-beta-mediated activation of miR-31 in normal human respiratory epithelia and lung cancer cells. PLoS ONE 5, e13764. 10.1371/journal.pone.0013764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H., Qi Y., Ng S. S., Chen X., Li D., Chen S., Ge R., Jiang S., Li G., Chen Y., He M. L., Kung H. F., Lai L., Lin M. C. (2009a). microRNA-146b inhibits glioma cell migration and invasion by targeting MMPs. Brain Res. 1269, 158–165 10.1016/j.brainres.2009.02.037 [DOI] [PubMed] [Google Scholar]
- Xia H. F., He T. Z., Liu C. M., Cui Y., Song P. P., Jin X. H., Ma X. (2009b). MiR-125b expression affects the proliferation and apoptosis of human glioma cells by targeting Bmf. Cell. Physiol. Biochem. 23, 347–358 10.1159/000218181 [DOI] [PubMed] [Google Scholar]
- Xia W., Li J., Chen L., Huang B., Li S., Yang G., Ding H., Wang F., Liu N., Zhao Q., Fang T., Song T., Wang T., Shao N. (2010). MicroRNA-200b regulates cyclin D1 expression and promotes S-phase entry by targeting RND3 in HeLa cells. Mol. Cell. Biochem. 344, 261–266 10.1007/s11010-010-0550-2 [DOI] [PubMed] [Google Scholar]
- Xia X. G., Zhou H., Samper E., Melov S., Xu Z. (2006). Pol II-expressed shRNA knocks down Sod2 gene expression and causes phenotypes of the gene knockout in mice. PLoS Genet. 2, e10. 10.1371/journal.pgen.0020010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J., Lin H., Luo X., Wang Z. (2011). miR-605 joins p53 network to form a p53:miR-605:Mdm2 positive feedback loop in response to stress. EMBO J. 30, 524–532 10.1038/emboj.2010.347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Liu S., Fu H., Li S., Tie Y., Zhu J., Xing R., Jin Y., Sun Z., Zheng X. (2010a). MicroRNA-193b regulates proliferation, migration and invasion in human hepatocellular carcinoma cells. Eur. J. Cancer 46, 2828–2836 10.1016/j.ejca.2010.06.017 [DOI] [PubMed] [Google Scholar]
- Xu L., Wang F., Xu X. F., Mo W. H., Xia Y. J., Wan R., Wang X. P., Guo C. Y. (2010b). Down-regulation of miR-212 expression by DNA hypermethylation in human gastric cancer cells. Med. Oncol. 10.1007/s12032-010-9691-0 [DOI] [PubMed] [Google Scholar]
- Xue Q., Guo Z. Y., Li W., Wen W. H., Meng Y. L., Jia L. T., Wang J., Yao L. B., Jin B. Q., Wang T., Yang A. G. (2011). Human activated CD4(+) T lymphocytes increase IL-2 expression by downregulating microRNA-181c. Mol. Immunol. 48, 592–599 10.1016/j.molimm.2010.10.021 [DOI] [PubMed] [Google Scholar]
- Yang Z., Chen S., Luan X., Li Y., Liu M., Li X., Liu T., Tang H. (2009). MicroRNA-214 is aberrantly expressed in cervical cancers and inhibits the growth of HeLa cells. IUBMB Life 61, 1075–1082 10.1002/iub.252 [DOI] [PubMed] [Google Scholar]
- Yao J., Liang L., Huang S., Ding J., Tan N., Zhao Y., Yan M., Ge C., Zhang Z., Chen T., Wan D., Yao M., Li J., Gu J., He X. (2010). MicroRNA-30d promotes tumor invasion and metastasis by targeting Galphai2 in hepatocellular carcinoma. Hepatology 51, 846–856 [DOI] [PubMed] [Google Scholar]
- Yi R., Poy M. N., Stoffel M., Fuchs E. (2008). A skin microRNA promotes differentiation by repressing “stemness.” Nature 452, 225–229 10.1038/nature06642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo A. S., Staahl B. T., Chen L., Crabtree G. R. (2009). MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature 460, 642–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S., Choi Y. C., Lee S., Jeong Y., Yoon J., Baek K. (2010). Induction of growth arrest by miR-542-3p that targets survivin. FEBS Lett. 584, 4048–4052 10.1016/j.febslet.2010.08.025 [DOI] [PubMed] [Google Scholar]
- Yu J. Y., Reynolds S. H., Hatfield S. D., Shcherbata H. R., Fischer K. A., Ward E. J., Long D., Ding Y., Ruohola-Baker H. (2009). Dicer-1-dependent Dacapo suppression acts downstream of insulin receptor in regulating cell division of Drosophila germline stem cells. Development 136, 1497–1507 10.1242/dev.022087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C. Z., Zhang J. X., Zhang A. L., Shi Z. D., Han L., Jia Z. F., Yang W. D., Wang G. X., Jiang T., You Y. P., Pu P. Y., Cheng J. Q., Kang C. S. (2010). MiR-221 and miR-222 target PUMA to induce cell survival in glioblastoma. Mol. Cancer 9, 229. 10.1186/1476-4598-9-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H. Y., Zheng S. J., Zhao J. H., Zhao W., Zheng L. F., Zhao D., Li J. M., Zhang X. F., Chen Z. B., Yi X. N. (2011a). MicroRNAs 144, 145, and 214 are down-regulated in primary neurons responding to sciatic nerve transection. Brain Res. 1383, 62–70 10.1016/j.brainres.2011.01.067 [DOI] [PubMed] [Google Scholar]
- Zhang L., Stokes N., Polak L., Fuchs E. (2011b). Specific MicroRNAs are preferentially expressed by skin stem cells to balance self-renewal and early lineage commitment. Cell Stem Cell 8, 294–308 10.1016/j.stem.2010.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Gao J. S., Tang X., Tucker L. D., Quesenberry P., Rigoutsos I., Ramratnam B. (2009). MicroRNA 125a and its regulation of the p53 tumor suppressor gene. FEBS Lett. 583, 3725–3730 10.1016/j.febslet.2009.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J. J., Lin J., Yang H., Kong W., He L., Ma X., Coppola D., Cheng J. Q. (2008). MicroRNA-221/222 negatively regulates estrogen receptor alpha and is associated with tamoxifen resistance in breast cancer. J. Biol. Chem. 283, 31079–31086 10.1074/jbc.M803963200 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhao X., Tang Y., Qu B., Cui H., Wang S., Wang L., Luo X., Huang X., Li J., Chen S., Shen N. (2010). MicroRNA-125a contributes to elevated inflammatory chemokine RANTES levels via targeting KLF13 in systemic lupus erythematosus. Arthritis Rheum. (Munch.) 62, 3425–3435 10.1002/art.27632 [DOI] [PubMed] [Google Scholar]
- Zhong X., Li N., Liang S., Huang Q., Coukos G., Zhang L. (2010). Identification of microRNAs regulating reprogramming factor LIN28 in embryonic stem cells and cancer cells. J. Biol. Chem. 285, 41961–41971 10.1074/jbc.M109.060467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Qi X., Potashkin J. A., Abdul-Karim F. W., Gorodeski G. I. (2008). MicroRNAs miR-186 and miR-150 down-regulate expression of the pro-apoptotic purinergic P2X7 receptor by activation of instability sites at the 3′-untranslated region of the gene that decrease steady-state levels of the transcript. J. Biol. Chem. 283, 28274–28286 10.1074/jbc.M710232200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Liu Z., Zhao Y., Ding Y., Liu H., Xi Y., Xiong W., Li G., Lu J., Fodstad O., Riker A. I., Tan M. (2010). MicroRNA-125b confers the resistance of breast cancer cells to paclitaxel through suppression of pro-apoptotic Bcl-2 antagonist killer 1 (Bak1) expression. J. Biol. Chem. 285, 21496–21507 10.1074/jbc.M110.133280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Wu H., Liu X., Li B., Chen Y., Ren X., Liu C. G., Yang J. M. (2009). Regulation of autophagy by a beclin 1-targeted microRNA, miR-30a, in cancer cells. Autophagy 5, 816–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Chen S., Liu X., Lin L., Huang X., Guo Z., Liu J., Wang Y., Yuan W., Qin Y. (2011). Endothelial enriched microRNAs regulate angiotensin II-induced endothelial inflammation and migration. Atherosclerosis 215, 286–293 10.1016/j.atherosclerosis.2010.12.024 [DOI] [PubMed] [Google Scholar]
- Zhu W., Shan X., Wang T., Shu Y., Liu P. (2010). miR-181b modulates multidrug resistance by targeting BCL2 in human cancer cell lines. Int. J. Cancer 127, 2520–2529 10.1002/ijc.25260 [DOI] [PubMed] [Google Scholar]
- Zimmerman E. I., Dollins C. M., Crawford M., Grant S., Nana-Sinkam S. P., Richards K. L., Hammond S. M., Graves L. M. (2010). Lyn kinase-dependent regulation of miR181 and myeloid cell leukemia-1 expression: implications for drug resistance in myelogenous leukemia. Mol. Pharmacol. 78, 811–817 10.1124/mol.110.066258 [DOI] [PMC free article] [PubMed] [Google Scholar]