Abstract
The rapid increase in the prevalence of antibiotic-resistant pathogens is a global problem that has challenged our ability to treat serious infections. Currently, clinical decisions on treatment are often based on in vitro susceptibility data. The role of the immune system in combating bacterial infections is unequivocal, but it is not well captured quantitatively. In this study, the impact of neutrophils on bacterial clearance was quantitatively assessed in a murine pneumonia model. In vitro time-growth studies were performed to determine the growth rate constants of Acinetobacter baumannii ATCC BAA 747 and Pseudomonas aeruginosa PAO1. The absolute neutrophil count in mice resulting from different cyclophosphamide preparatory regimens was determined. The dynamic change of bacterial (A. baumannii BAA 747) burden in mice with graded immunosuppression over 24 h was captured by a mathematical model. The fit to the data was satisfactory (r2 = 0.945). The best-fit maximal kill rate (Kk) of the bacterial population by neutrophils was 1.743 h−1, the number of neutrophils necessary for 50% maximal killing was 190.8/μl, and the maximal population size was 1.8 × 109 CFU/g, respectively. Using these model parameter estimates, the model predictions were subsequently validated by the bacterial burden change of P. aeruginosa PAO1 at 24 h. A simple mathematical model was proposed to quantify the contribution of neutrophils to bacterial clearance and predict the bacterial growth/suppression in animals. Our results provide a novel framework to link in vitro and in vivo information and may be used to improve clinical treatment of bacterial infections.
INTRODUCTION
The rapid increase in the prevalence of antibiotic-resistant pathogens is a global problem that has challenged our ability to treat serious infections. To prevent further spread of multidrug resistance and returning to the preantibiotic era, it is imperative that the current approach to treatment be improved. Presently, clinical decisions on treatment are often based on in vitro susceptibility data (MICs), which only provide information about antibacterial activity at a single time point and potentially miss information on the changes in bacterial population dynamics over time. Drug selections solely based on MIC values may not always correlate to patient outcomes (11). One major reason for the discrepancy may be due to in vitro investigations neglecting the presence of the immune system in patients, which is well known to be significant.
The immune system (especially neutrophils) plays an important role in combating bacterial infections. This is best exemplified by patients with neutropenia or after transplantation, who have significantly higher mortality than patients without neutropenia (8, 10, 13). However, the effect of the immune system is not well accounted for when data obtained from in vitro investigations are used to provide guidance for treatment. The overall antimicrobial effect in a patient is the sum of the antibacterial activity from drug therapy and the effect from the immune system. The functional contribution from the latter is often overlooked, and the quantitative contribution of the neutrophils to bacterial clearance is not well established.
Acinetobacter baumannii and Pseudomonas aeruginosa are important Gram-negative pathogens associated with serious nosocomial infections (5, 12). Multidrug resistance in both species has been increasing over the past decades and correlates with significant morbidity and mortality (3, 6). They have been implicated in numerous global outbreaks and pose serious challenges to clinicians (4, 7, 14). In this study, we investigated the impact of neutrophils on the clearance of A. baumannii in an animal infection model. A simple mathematical model was developed to characterize bacterial behavior over time under various degrees of immunosuppression. The model predictions of bacterial behavior were subsequently validated using another important pathogen, P. aeruginosa. Our quantitative results may provide useful information to improve clinical treatment of bacterial infections.
MATERIALS AND METHODS
Microorganisms.
A wild-type A. baumannii ATCC BAA 747 strain (American Type Culture Collection, Rockville, MD) and a laboratory strain of P. aeruginosa PAO1 were used in the study. The bacteria were stored at −70°C in Protect (Key Scientific Products, Round Rock, TX) storage vials. Fresh isolates were subcultured twice on 5% blood agar plates (Hardy Diagnostics, Santa Maria, CA) for 24 h at 35°C prior to each experiment.
In vitro time-growth studies.
The in vitro growth rates of A. baumannii BAA 747 and P. aeruginosa PAO1 were determined in cation-adjusted Mueller-Hinton broth (Ca-MHB) (BBL, Sparks, MD). Briefly, 1 or 2 medium-sized fresh colonies were inoculated in Ca-MHB until reaching log-phase growth. The bacterial suspension was then diluted based on absorbance at 630 nm and transferred to a 50-ml flask containing 20 ml of Ca-MHB. To be consistent with subsequent animal experiments, the baseline inocula of A. baumannii BAA 747 and P. aeruginosa PAO1 used were approximately 1 × 107 and 1 × 103 CFU/ml, respectively. The experiments were conducted for 24 h in a shaker water bath at 35°C. Serial samples (baseline and 1, 2, 3, 4, 6, 8, and 24 h) were obtained in triplicate, and bacterial burden was determined by quantitative culture on Mueller-Hinton agar (MHA) plates (BBL) after 10× serial dilutions. Colony counts were enumerated after incubation at 35°C in a humidified incubator for 24 h. The theoretical lower limit of detection was 100 CFU/ml.
Modeling of in vitro growth.
The exponential growth of the bacterial population over 24 h was analyzed using a mathematical model (18). Details of the model are shown in Fig. 1. Based on the best-fit model, the bacterial growth rate constants (Kg) were determined with the ADAPT II program (1).
Fig. 1.
In vivo bacterial growth dynamics model.
Animals.
Female Swiss-Webster mice weighing 21 to 25 g were used (Harlan Laboratories, Indianapolis, IN). The animals were housed in ventilated microisolator cages to decrease the risk of infection from extraneous pathogens. The mice were allowed to eat and drink ad libitum. The experimental protocol was approved by the Institutional Animal Care and Use Committee of the University of Houston.
Determination of ANC.
A total of 16 mice were randomly divided into 4 groups. One group was used for baseline absolute neutrophil count (ANC) determination, and the other 3 groups were given different cyclophosphamide preparatory regimens to produce graded levels of immunosuppression in the animals. Each cyclophosphamide regimen contained two doses administered intraperitoneally; the first dose (50, 100, or 200 mg/kg of body weight) was given on day −4, followed by the second dose (50, 50, or 150 mg/kg) on day −1. For each treatment group, blood (approximately 40 μl) was drawn on day zero via the tail vein and was collected into BD Microtainer tubes with EDTA (BD, Franklin Lakes, NJ). The blood samples were transported in ice and were analyzed within 2 h of collection using a HemaVet 950FS multispecies blood analyzer (Drew Scientific, Inc., Oxford, CT). The total number of white blood cells, the number of neutrophils, and the percentage of neutrophils were determined following the instructions of the manufacturer.
In vivo growth experiments.
To examine the impact of neutrophils on bacterial clearance, a total of 72 mice were randomly divided into 4 groups (18 mice in each group). Each group was given a cyclophosphamide regimen as described above. The mice were anesthetized by a single intraperitoneal injection of 1.25% 2,2,2-tribromoethanol (Sigma-Aldrich, St. Louis, MO) at a dosage of 25 mg/kg. A. baumannii BAA 747 was grown to log-phase growth in Ca-MHB; the suspension was washed once and concentrated in sterile saline based on absorbance at 630 nm. The bacteria were inoculated into the trachea of anesthetized mice under laryngoscopic guidance. The inoculum (approximately 1 × 106 CFU in 10 μl) used was guided by previous investigations mimicking a clinical course of infection but with no excessive mortality by 24 h. In each group, 3 mice were sacrificed by CO2 asphyxiation at baseline to ascertain the infective inoculum, and 3 mice each were sacrificed at 4, 8, 12, 20, and 24 h after infection. The lungs from each mouse were aseptically collected for quantitative culture. Prior to being cultured, lungs were homogenized in 10 ml of sterile saline. The homogenates were centrifuged (4°C at 4,000 × g for 15 min), decanted, and reconstituted with sterile saline at 10 times the original volume. The samples were subsequently serially diluted (10×) and quantitatively plated on MHA plates. The reliable lower limit of detection was 1,000 CFU/g.
In vivo growth dynamics model.
The change of bacterial burden in lung tissues over time was described using the mathematical model shown in Fig. 1. The rate of change of bacteria over time was expressed as the difference between the intrinsic bacterial growth rate and the kill rate provided by the neutrophils. Previous investigations demonstrated that the growth of A. baumannii BAA 747 was not considerably affected by nutrient depletion, so the Kg value derived from the in vitro time-growth experiments was adopted (data not shown). Additional model parameters were used to account for other relevant physiological phenomena, such as contact inhibition (using a maximum population size) and nonlinear (sigmoidal) kill rate to account for saturable killing. The killing profiles in all animal groups were comodeled with the ADAPT II program (1).
Prospective validation of mathematical model.
The above in vivo growth experiment was repeated using P. aeruginosa PAO1. An inoculum of approximately 1 × 103 CFU (in 10 μl) was used. Four mice were sacrificed at baseline to ascertain the infective inoculum, and 4 mice in each group were sacrificed at 24 h as described above.
RESULTS
In vitro time-growth studies.
The best-fit growth rate constant (Kg) of A. baumannii BAA 747 in full-strength Ca-MHB was 1.463 ± 0.191 h−1 (mean ± standard deviation). Previous experiments demonstrated that the growth courses in various strengths (full-strength and 0.1 strength) of broth were similar, indicating that the growth of A. baumannii BAA 747 was not considerably affected by nutrient depletion (data not shown). In contrast, the growth rate constants (Kg) of P. aeruginosa PAO1 in full-strength and 0.1-strength Ca-MHB were found to be 1.509 ± 0.014 h−1 and 1.047 ± 0.015 h−1, respectively.
Determination of ANC.
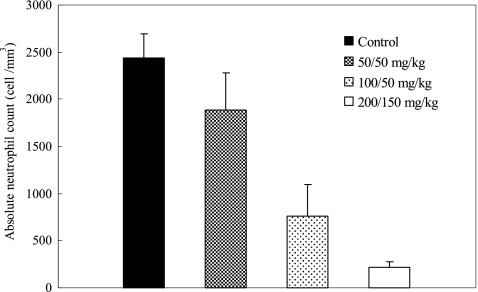
The ANC values of animals given cyclophosphamide are shown in Fig. 2. Compared to the neutrophil count in the control animals, a graded neutropenia was successfully induced by the cyclophosphamide regimens. The neutrophil count was reduced approximately 90% after the administration of 200 mg/kg and 150 mg/kg of cyclophosphamide. For the other two treatment groups, the percentages of ANC reduction were approximately 70% and 20%, respectively.
Fig. 2.
The absolute neutrophil count (ANC) observed in mice with different cyclophosphamide preparatory regimens. Data are shown as means ± standard deviations. The mice were given the indicated cyclophosphamide doses on day −4 and −1.
In vivo growth experiments.
A consistent baseline tissue burden (ranged from 7.07 to 7.32 log CFU/g) was achieved. The dynamic change of bacterial burden in lung tissues over time is displayed in Fig. 3. A divergence of bacterial burden profiles was observed after 8 h. At 24 h, the net change of bacterial burden from the baseline ranged from −1.08 log CFU/g to 1.72 log CFU/g. The bacterial burden observed was negatively correlated with the ANC achieved in the animals. A net growth in bacterial burden was observed in the animal groups given 200/150 and 100/50 mg/kg of cyclophosphamide (1.72 and 0.56 log CFU/g, respectively). On the other hand, net reductions of 0.52 and 1.08 log CFU/g were seen in animals given the 50/50 mg/kg regimen and the control animals, respectively.
Fig. 3.
Time course of pulmonary bacterial burden of A. baumannii BAA 747 in mice administered different cyclophosphamide regimens (0/0, 50/50, 100/50, and 200/150 mg/kg of bodyweight). Data are shown as means ± standard deviations.
Mathematical modeling of in vivo data.
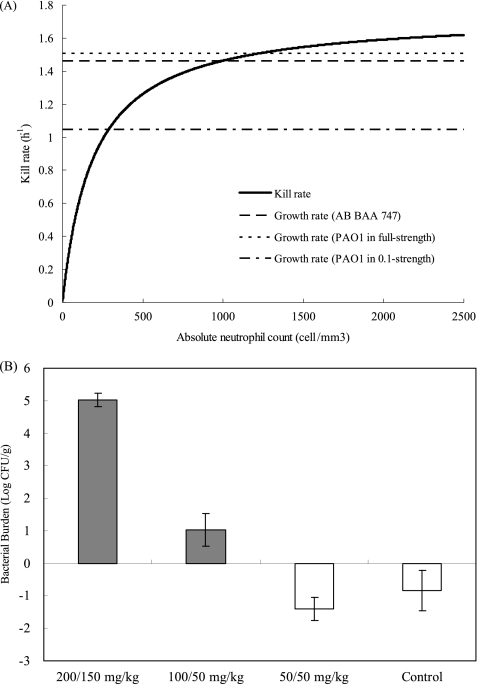
Overall, the model fit to the data was satisfactory (r2 = 0.945) based on the best-fit parameter estimates (Fig. 4). The best-fit maximal kill rate by neutrophils (Kk) was 1.743 h−1. In addition, the number of neutrophils necessary for 50% maximal killing was 190.8/μl and the maximal population size was 1.82 × 109 CFU/g, respectively. The best-fit relationship between the killing rate and ANC is shown in Fig. 5A.
Fig. 4.
Performance of the best-fit model.
Fig. 5.
Prediction and validation of bacterial growth/suppression. (A) Comparison of the kill rate by neutrophils and bacterial growth rates. The solid curve depicts kill rate changes with absolute neutrophil count. The dotted horizontal lines represent growth rates of A. baumannii (AB) BAA 747 and P. aeruginosa PAO1 in different strengths of broth. (B) Observed pulmonary bacterial (P. aeruginosa PAO1) burden changes from baseline at 24 h. The shaded bars indicate conditions under which bacterial growth was predicted, while the open bars indicate conditions under which bacterial suppression was predicted.
Prediction and validation of mathematical model.
A baseline tissue burden ranging from 3.10 to 3.51 log CFU/g was achieved. The behavior of P. aeruginosa PAO1 with similar cyclophosphamide preparatory regimens was predicted using the best-fit parameter estimates. The best-fit kill rate was compared to the growth rates of the bacteria, as shown in Fig. 5A. Qualitatively, P. aeruginosa PAO1 was expected to be suppressed in 2 out of 4 experimental groups (the control group and the dosing group given 50/50 mg/kg cyclophosphamide), using the Kg value derived from experiments using full-strength Ca-MHB. Alternatively, it was predicted that 1 out of 4 experiment groups (the dosing group given 200/150 mg/kg of cyclophosphamide) would not be suppressed, using the Kg value derived from 0.1-strength Ca-MHB experiments. The validation experiments revealed that bacterial growth was suppressed in only 2 dosing groups (Fig. 5B), attesting to the predicting performance of our mathematical model.
DISCUSSION
The important role of the immune system (neutrophils) in combating bacterial infections is indisputable. Several preclinical studies have demonstrated that depletion of neutrophils would result in a serious deficiency in bacterial clearance (15, 16, 19). In addition, retrospective cohort clinical studies have demonstrated that severe neutropenia or immunosuppression was associated with higher mortality in patients (8, 17).
The quantitative impact of the immune system on bacterial clearance is not well established. Recently, an investigation delineated the impact of granulocytes on bacterial clearance in a mouse thigh infection model (2). To the best of our knowledge, this is the only recent published study investigating the quantitative impact of granulocytes. In that study, different inocula were injected into the thighs of immunocompetent mice, and the bacterial burden changes were observed over 24 h. A mathematical model was used to elucidate the impact of bacterial burden on bacterial clearance by granulocytes. The authors concluded that bacterial kill by granulocytes was saturable and a higher reliance on chemotherapy would be necessary to drive clearance of a high bacterial burden.
In this study, we attempted to capture the quantitative impact of neutrophils in bacterial infections. Instead of just relying on an early and late observation, we observed the growth course of bacteria serially over 24 h to more closely track the dynamic change of bacterial burden. Also, escalating cyclophosphamide regimens were used to produce graded levels of immunosuppression in the animals. This would allow us to assess the quantitative relationship between neutrophil counts and bacterial killing with more precision. Finally, in addition to fitting the experimental data, the mathematical model also provided a reasonable prediction of the bacterial growth/suppression of another bacterium under similar experimental conditions.
Several assumptions were made in the model. One assumption was that the in vivo bacterial growth rate was identical to that derived from our in vitro time-growth investigations. Conceptually, it was regarded as the in vivo maximal growth rate in the absence of neutrophils. The growth rate constant of A. baumannii BAA 747 was derived from experiments using full-strength Ca-MHB, which was similar to that obtained using 0.1-strength Ca-MHB previously. However, nutrient depletion appeared to influence the growth of P. aeruginosa PAO1 to a greater extent. We were unsure which growth rate constant would be more realistic under in vivo conditions. Nonetheless, observations in the validation experiments seemed more reliable using the value from full-strength Ca-MHB. Second, the ANC data on day zero were the sole data used to represent the impact of neutrophils. Although neutrophil recruitment to the infection site is expected following bacterial inoculation, we did not expect that the neutrophil count would increase dramatically within a short period of time. Thus, our assumption was deemed reasonable for the time frame of the experiments (24 h). Finally, we assumed that neutrophils were the only immune component responsible for bacterial clearance and there was no substantial residual killing from other immune components, such as antibodies or the complement system. Residual killing was previously considered during the model development stage, with the use of additional model parameter(s). However, these hierarchically more complex mathematical models did not improve the predictions of the validation experiments and were thus abandoned.
The model predictions were reasonable in predicting bacterial behavior in vivo in a qualitative sense. There were several limitations in this study, which could have affected the predicting ability of the model. Observations of ANC on the day of infection could only be made with an ANC of >200 cells/μl (approximately 90% neutrophil reduction from baseline). As such, it was difficult for us to fully define and assess the necessity to incorporate residual killing from immune components other than neutrophils. We briefly investigated a cyclophosphamide dosing regimen higher than 200/150 mg/kg (the highest reported in this study). However, it was found to be associated with excessive mortality (likely due to drug toxicity), and thus, it was not pursued further (data not shown). Another limitation of the study was the bacterial inocula used. In contrast to the previous investigation (2), we did not evaluate the impact of neutrophils on the bacterial clearance for different inocula. Finally, only two representative Gram-negative bacteria were examined. Further studies with other clinically important pathogens (e.g., Staphylococcus aureus and Streptococcus pneumoniae) could further validate the utility of the proposed mathematical model.
In conclusion, our quantitative results provided new insights into the impact of neutrophils on bacterial clearance. The mathematical model established in the study may be used as a foundation to investigate the impact of other immune components on various bacterial species. Furthermore, the beneficial effect of neutrophils in combination with antimicrobials should be investigated.
ACKNOWLEDGMENTS
We thank Alice Chen and Allison Rosen for technical assistance in blood cell profiling.
The study was partially supported by the National Science Foundation (grant CBET-0730454) and the Ministry of Science and Technology of China (grant 2008ZX09312-010).
Footnotes
Published ahead of print on 1 August 2011.
REFERENCES
- 1. D' Argenio D. Z., Schumitzky A. 1997. ADAPT II user's guide: pharmacokinetic/pharmacodynamic systems analysis software. Biomedical simulations resource, University of Southern California, Los Angeles, CA [Google Scholar]
- 2. Drusano G. L., Fregeau C., Liu W., Brown D. L., Louie A. 2010. Impact of burden on granulocyte clearance of bacteria in a mouse thigh infection model. Antimicrob. Agents Chemother. 54:4368–4372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gaynes R., Edwards J. R. 2005. Overview of nosocomial infections caused by gram-negative bacilli. Clin. Infect. Dis. 41:848–854 [DOI] [PubMed] [Google Scholar]
- 4. Giamarellou H., Antoniadou A., Kanellakopoulou K. 2008. Acinetobacter baumannii: a universal threat to public health? Int. J. Antimicrob. Agents 32:106–119 [DOI] [PubMed] [Google Scholar]
- 5. Joly-Guillou M. L. 2005. Clinical impact and pathogenicity of Acinetobacter. Clin. Microbiol. Infect. 11:868–873 [DOI] [PubMed] [Google Scholar]
- 6. Kwa A. L., et al. 2007. The impact of multidrug resistance on the outcomes of critically ill patients with Gram-negative bacterial pneumonia. Diagn. Microbiol. Infect. Dis. 58:99–104 [DOI] [PubMed] [Google Scholar]
- 7. Landman D., et al. 2002. Citywide clonal outbreak of multiresistant Acinetobacter baumannii and Pseudomonas aeruginosa in Brooklyn, NY: the preantibiotic era has returned. Arch. Intern. Med. 162:1515–1520 [DOI] [PubMed] [Google Scholar]
- 8. Lin M. Y., Weinstein R. A., Hota B. 2008. Delay of active antimicrobial therapy and mortality among patients with bacteremia: impact of severe neutropenia. Antimicrob. Agents Chemother. 52:3188–3194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Reference deleted.
- 10. Lyman G. H., et al. 2010. Risk of mortality in patients with cancer who experience febrile neutropenia. Cancer 116:5555–5563 [DOI] [PubMed] [Google Scholar]
- 11. Mouton J. W., Jacobs N., Tiddens H., Horrevorts A. M. 2005. Pharmacodynamics of tobramycin in patients with cystic fibrosis. Diagn. Microbiol. Infect. Dis. 52:123–127 [DOI] [PubMed] [Google Scholar]
- 12. Obritsch M. D., Fish D. N., MacLaren R., Jung R. 2005. Nosocomial infections due to multidrug-resistant Pseudomonas aeruginosa: epidemiology and treatment options. Pharmacotherapy 25:1353–1364 [DOI] [PubMed] [Google Scholar]
- 13. Offner F., Schoch G., Fisher L. D., Torok-Storb B., Martin P. J. 1996. Mortality hazard functions as related to neutropenia at different times after marrow transplantation. Blood 88:4058–4062 [PubMed] [Google Scholar]
- 14. Perez F., et al. 2007. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 51:3471–3484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Robertson C. M., et al. 2008. Neutrophil depletion causes a fatal defect in murine pulmonary Staphylococcus aureus clearance. J. Surg. Res. 150:278–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sun L., et al. 2009. Effect of IL-10 on neutrophil recruitment and survival after Pseudomonas aeruginosa challenge. Am. J. Respir. Cell Mol. Biol. 41:76–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tam V. H., et al. 2010. Impact of multidrug-resistant Pseudomonas aeruginosa bacteremia on patient outcomes. Antimicrob. Agents Chemother. 54:3717–3722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tam V. H., Schilling A. N., Nikolaou M. 2005. Modelling time-kill studies to discern the pharmacodynamics of meropenem. J. Antimicrob. Chemother. 55:699–706 [DOI] [PubMed] [Google Scholar]
- 19. van Faassen H., et al. 2007. Neutrophils play an important role in host resistance to respiratory infection with Acinetobacter baumannii in mice. Infect. Immun. 75:5597–5608 [DOI] [PMC free article] [PubMed] [Google Scholar]