Abstract
Two daptomycin (DAP) regimens were evaluated in a pharmacokinetic/pharmacodynamic (PK/PD) model, and the mutants recovered were examined for changes in phenotypic characteristics. Three Enterococcus faecium strains (vancomycin-resistant Enterococcus [VRE] ATCC 51559, VRE 12311, and VRE SF 12047) were utilized in a 7-day, 1-compartment in vitro PK/PD model. The simulated dosing regimens were DAP at 6 mg/kg/day (free Cmax [fCmax] = 7.9 μg/ml, half-life [t1/2] = 8 h) and DAP at 10 mg/kg/day (fCmax = 13.17 μg/ml, t1/2 = 8 h). Samples were plated daily on Mueller-Hinton agar containing DAP at 16 μg/ml and 50 mg/liter Ca2+ to assess the emergence of DAP resistance. For each strain, the mutant with the highest DAP MIC was then evaluated for changes in relative surface charge, cell wall thickness, and cytoplasmic membrane depolarization induced by DAP. The initial DAP MICs were 4 μg/ml for all 3 strains. A dose-dependent response and regrowth were observed for DAP 6 mg/kg/day and DAP 10 mg/kg/day against all 3 strains. Mutants of VRE ATCC 51559 (MIC = 128 and 64 μg/ml) and VRE 12311 (MIC = 256 and 32 μg/ml) were recovered from the DAP 6 mg and DAP 10 mg regimen, respectively. For VRE SF 12047, a mutant (MIC = 64 μg/ml) was recovered from the DAP 6 mg model. All mutants displayed an increase in relative surface charge compared to those of their respective parent strains. The DAP-resistant mutants displayed a 43 to 58% increase in cell wall thickness (P < 0.0001), while DAP membrane depolarization decreased by 53 to 65% compared to that of the susceptible strains. VRE with DAP resistance displayed increased surface charge, increased cell wall thickness, and decreased depolarization induced by DAP, consistent with previous observations in Staphylococcus aureus with reduced DAP susceptibility. Further characterization of DAP-resistant VRE is warranted.
INTRODUCTION
Daptomycin is a cyclic lipopeptide antibiotic that has bactericidal activity against a wide variety of Gram-positive bacteria, including methicillin-resistant Staphylococcus aureus and vancomycin-resistant enterococci (VRE) (4). Although clinical strains of S. aureus and enterococci displaying nonsusceptibility and resistance to daptomycin have been reported, the mechanism of this phenotype has not been clearly elucidated (2, 4, 16, 18, 20–22, 26, 31, 33, 34). The majority of work in this area has concentrated on S. aureus. Mutations and/or changes in the expression of several genes (mprF, yycG, rpoB, rpoC, and dltABCD) and alterations within the cell membrane and cell wall (fluidity, membrane potential, membrane proteins, and the amount and distribution of phospholipids) have been discovered but are not universally found in nonsusceptible strains (17, 22, 29, 37, 38).
Recent studies investigating S. aureus strains developing daptomycin nonsusceptibility during daptomycin therapy have revealed a decrease of cell membrane depolarization secondary to daptomycin exposure (20, 22). In addition, daptomycin nonsusceptibility has been associated with vancomycin-intermediate S. aureus (VISA) and an increase in cell wall thickness (6, 11). The mechanism of reduced daptomycin susceptibility is hypothesized to be secondary to an affinity trapping of antibiotic molecules in the thickened cell wall (11). Daptomycin-nonsusceptible S. aureus strains have been shown to have diminished daptomycin and cationic peptide binding to the cytoplasmic membrane secondary to an increase in positive cell surface charge (20, 22, 37).
Mutations in the mprF gene have been commonly found in postexposure clinical isolates that have developed nonsusceptibility to daptomycin (37, 38). The MprF protein is responsible for modifying net surface charge by lysinylation of the membrane phosphatidylglycerol (PG) to generate lysyl-PG (LPG) and translocation of the positively charged phospholipids to the outer leaflet of the cytoplasmic membrane (14, 29). Recently, Yang et al. have identified increased expression of mprF and dltABCD in daptomycin-nonsusceptible strains of S. aureus (29, 37, 38). The operon dltABCD contributes to the net positive surface charge by the d-alanylation of wall teichoic acids (37). A shift in the cytoplasmic membrane surface charge toward a more positive state is thought to impede binding via repulsion of the daptomycin-Ca2+ complex, the active form that simulates a cationic antimicrobial peptide (20, 37).
In stark contrast, only three studies have examined genetic changes in enterococci resistant to daptomycin (1, 5, 30). Two gene clusters (EF2694 to EF2701 and EF1751 to EF1753) associated with cell membrane proteins and the phage shock protein C were found to be upregulated in daptomycin-resistant Enterococcus faecalis (5). Examination of a clinical isogenic pair of E. faecalis isolates found alterations in genes associated with phospholipid metabolism and a cell envelope stress response system (1). It has previously been demonstrated that deletion of dltA in Enterococcus faecium leads to increased susceptibility to cationic peptides; therefore, it seems logical that an increase in the expression of the dltABCD operon would lead to decreased daptomycin susceptibility (15). E. faecalis possesses the ability to increase its cell wall thickness as a mechanism to develop resistance to monolaurin, a fatty acid ester with antibacterial activity via action on the membrane (13). Therefore, verification of the changes in the cytoplasmic membrane, cell wall, and cationic peptide binding are important to understand daptomycin resistance in enterococci.
The purpose of the current study was to evaluate two daptomycin dosing regimens (6 and 10 mg/kg/day) in a 1-compartment pharmacokinetic/pharmacodynamic (PK/PD) model and examine recovered mutants for phenotypic characteristics and mechanisms of daptomycin resistance (7, 25).
(This study was presented as a poster presentation at the 21st European Congress of Clinical Microbiology and Infectious Diseases/27th International Congress of Chemotherapy [ECCMID/ICC], Milan, Italy, 2011.)
MATERIALS AND METHODS
Bacterial strains.
The following vancomycin-resistant E. faecium strains (vanA positive) were examined: VRE SF 12047, VRE 12311, and American Type Culture Collection (ATCC) 51559 (23).
Antimicrobials.
Daptomycin and vancomycin (Sigma-Aldrich Co., St. Louis, MO) were commercially purchased. Stock solutions were freshly prepared daily according to Clinical and Laboratory Standards Institute (CLSI) recommendations (8).
Media.
Mueller-Hinton broth II (Difco, Detroit, MI) with 25 mg/liter of calcium and 12.5 mg/liter magnesium (MHB) was used for vancomycin susceptibility testing. Due to its dependence on calcium for antimicrobial activity, MHB II was supplemented to 50 mg/liter of calcium (50 SMHB) for susceptibility testing and 1-compartment in vitro PK/PD models containing daptomycin. Colony counts were determined using brain heart infusion agar (BHIA; Difco, Detroit, MI) plates.
Susceptibility testing.
MICs were determined in duplicate by broth microdilution at ∼106 CFU/ml in MHB as specified above, according to CLSI guidelines (8).
One-compartment in vitro PK/PD model.
An in vitro PK/PD model consisting of a 250-ml 1-compartment glass chamber with multiple ports for the removal of medium, delivery of antibiotics, and collection of bacterial and antimicrobial samples was utilized. The apparatus was prefilled with medium, and antibiotics were administered as boluses over 1 min into the central compartment via an injection port. All model simulations were conducted over 168 h and were performed in duplicate to ensure reproducibility. A starting inoculum of approximately 8 log10 CFU/ml was used. Each model was placed in a 37°C water bath for the duration of the experiment, with a magnetic stir bar to produce continuous mixing of the medium. Daptomycin was added to models to achieve exposures reflecting approximate free (f) daptomycin concentrations based on its protein binding (91%) (3). Regimens consisted of daptomycin at 6 mg/kg every 24 h (estimated fCmax, 7.9 μg/ml; half-life [t1/2] = 8 h) and daptomycin 10 mg/kg every 24 h (estimated fCmax, 13.17 μg/ml; t1/2 = 8 h) (3). A peristaltic pump (Masterflex; Cole-Parmer Instrument Company, Chicago, IL) was used to continually replace antibiotic-containing medium with fresh 50 SMHB at a rate simulating the t1/2 h of daptomycin.
Pharmacodynamic samples from each model (1 ml) were collected at 0, 4, 8, 24, 28, 32, 48, 56, 72, 96, 120, 144, and 168 h and serially diluted in cold normal saline (0.9% NaCl). Bacterial counts were determined by plating 100-μl aliquots of each diluted sample on BHI, using an automated spiral dispenser (Whitley automatic spiral plater; Don Whitley Scientific Limited, West Yorkshire, England). Plated samples were incubated at 37°C for 24 h, and colony counts (log10 CFU/ml) were determined using a laser colony counter (ProtoCOL version 2.05.02; Synbiosis, Cambridge, England). The limit of detection for this method of colony count determination is 2.0 log10 CFU/ml. For all samples, antimicrobial carryover was accounted for by serial dilution of the plated samples. If the anticipated dilution was near the MIC, then vacuum filtration was also used. When vacuum filtration was used, samples were washed through a 0.45-μm filter with normal saline to remove the antimicrobial agent. Filters were then plated onto BHI and incubated at 37°C for 24 h. The model time-kill curves were determined by plotting mean colony counts (log10 CFU/ml) from each model versus time. Antibacterial activity was defined by the following reductions in the starting inoculum: bactericidal, 3 log10 (99.9% kill); bacteriostatic, <3 log10 CFU/ml; or inactivity, no reduction. The time to achieve a 99.9% bacterial load reduction was determined by linear regression (if r2 was ≥0.95) or visual inspection of log10 CFU/ml.
Emergence of daptomycin resistance.
Mutants with increased daptomycin MIC values were recovered from the model in the following manner. Samples (100 μl) from the 24, 48, 72, 96, 120, 144, and 168 h time points were plated on Mueller-Hinton agar (Difco, Detroit, MI) supplemented with 50 mg/liter Ca2+ and containing daptomycin at 16 μg/ml. Plates were visually inspected for growth after 24 and 48 h of incubation at 37°C in both aerobic and anaerobic conditions. MIC testing was performed on the overall population at 0, 96 (midpoint), and 168 h. MIC testing was performed on the mutants (if available) at the first time point where regrowth occurred, at 96 h (midpoint), and at 168 h.
PFGE.
Pulsed-field gel electrophoresis (PFGE) of SmaI DNA digests was performed to verify relatedness of parent strains and derivative mutants selected in the PK/PD models. Gels were run at 6 V/cm, 14°C on a 0.8% agarose gel with pulse times of 5 to 35 s for 19 h using a CHEF-DRIII apparatus (Bio-Rad Laboratories, Richmond, CA), as previously described (12). Strain relatedness was determined by visual inspection of the gel using the criteria of Tenover et al. (36).
Cytochrome c binding.
The relative surface charge was determined following a modified protocol of the method described by Kraus et al. (24). Overnight cultures (5 ml) in brain heart infusion (BHI; Difco, MI) were centrifuged at 4,000 RPM for 10 min. The pellet was washed twice with MOPS (morpholinepropanesulfonic acid) buffer (20 mm, pH = 7.0) and then resuspended in 1 ml of MOPS. A volume of 0.1 ml of the bacterial suspension was serially diluted in normal saline and spiral plated onto BHI plates in order to assess the inoculum size of each sample. Plates were incubated at 37°C for 18 to 24 h prior to reading. A volume of 0.1 ml of cytochrome c from bovine heart (5 mg/ml; Sigma-Aldrich Co., St. Louis, MO) was added to the remaining 0.9 ml of the bacterial suspension in MOPS. The mixture was incubated at room temperature for 30 min, and then cells were pelleted by centrifugation at 13,000 rpm for 5 min. The absorbance of the supernatant was measured at 530 nm using a spectrophotometer (SpectraMax M5; Molecular Devices, Sunnyvale, CA), and the concentration of cytochrome c present was quantified using a standard curve derived from the regression line, adjusted to 109 CFU/ml.
TEM.
Cell wall thickness was determined by transmission electron microscopy (TEM) as described previously (10). Ultrathin sections were evaluated at a magnification of 48,000× with a JEOL 100CX electron microscope, and images were captured with a MegaView III side-mounted digital camera. The resulting images were analyzed using ImageJ 1.39t software. Cell wall thickness was determined for at least 25 cells per sample using four separate quadrants of each cell for a total of ≥100 measurements per sample.
Determination of cytoplasmic membrane depolarization.
The ability of daptomycin to depolarize the cytoplasmic membrane was determined using a modified protocol of the method using the membrane potential-sensitive fluorescent dye DiSC3 (35). Enterococci were grown in 50 SMHB to the early exponential phase (optical density at 600 nm [OD600] = 0.25). Cells were pelleted, washed twice with HEPES buffer (pH 7.2, 50 mg/liter Ca2+), and then resuspended in HEPES buffer at an OD600 of 0.2. A volume of 0.04 ml of DiSC3 (0.1 mg/ml) was added to 2 ml of the bacterial suspension in HEPES in a polystyrene fluorometer cuvette (Fisher Scientific) containing a stir bar, and the cuvette was placed in the heated (37°C) chamber of a Fluoromax-3 spectrofluorometer (Horiba Jobin Yvon). The dye was allowed to load the cell for 10 min, and then 100 mM KCl was added. Nisin and daptomycin were added 2 min later. The parameters were 622 nm (excitation) and 670 nm (emission). Measurements were collected for 60 min after addition of drugs. The drug regimens consisted of a control (HEPES), nisin (25 μg/ml), and daptomycin (8 μg/ml). The membrane potential-dissipating activity of daptomycin at 60 min was calculated using the following formula: % membrane depolarization = 100 × [(Fd − Fc)/(Fn − Fc)], where Fd is the fluorescence measured with daptomycin, Fc is the fluorescence measured with the buffer, and Fn is the fluorescence measured with nisin. The results presented are the means of 2 independent experiments.
Statistical analysis.
Paired continuous data were evaluated with a paired t test or sign test (if distribution was not normal). A P value of ≤0.05 was considered significant.
RESULTS
Susceptibility testing.
Vancomycin and daptomycin MICs for study isolates are displayed in Table 1.
Table 1.
Susceptibility testing and transmission electron microscopy results for Enterococcus faecium strains and their mutants
| Straina | MIC (μg/ml) |
Cell wall thickness ± SD (nm) | |
|---|---|---|---|
| Vancomycin | Daptomycin | ||
| VRE SF 12047 | >256 | 4 | 30.3 ± 5.5 |
| VRESF 12047 M | >256 | 64 | 43.5 ± 9.2 |
| VRE 12311 | >256 | 4 | 22.2 ± 4.5 |
| VRE 12311 M | >256 | 256 | 32.4 ± 5.2 |
| VRE ATCC 51559 | >256 | 2-4 | 24.9 ± 2.6 |
| VRE ATCC 51559 M | >256 | 128 | 39.5 ± 6.2 |
M, mutant recovered from pharmacodynamic sample at 168 h in model simulating daptomycin at 6 mg/kg every 24 h.
One-compartment in vitro PK/PD model.
The results of the 7-day, 1-compartment models are displayed in Table 2. Daptomycin simulated regimens of 6 mg/kg/day and 10 mg/kg/day produced dose-dependent responses in VRE SF 12047 and VRE 12311. Daptomycin simulated regimens of 6 mg/kg/day produced mutants with elevated daptomycin MIC values for all strains, while 10 mg/kg/day produced mutants for VRE 12311 and VRE ATCC 51559. Regrowth occurred with both dosing regimens in all strains.
Table 2.
Daptomycin activity and changes in MIC values for the 1-compartment in vitro pharmacokinetic/pharmacodynamic model
| Strain | Daptomycin dose | Maximum kill (log10 CFU/ml) | Time of 1st regrowth (h) | Inoculum at 168 h (log10 CFU/ml) | MIC (μg/ml)a |
||||
|---|---|---|---|---|---|---|---|---|---|
| Regrowth (Mut) | 96 h |
168 h |
|||||||
| Pop | Mut | Pop | Mut | ||||||
| VRE SF 12047 | 6 mg/kg | −3.73 | 72 | 8.65 | NA | 16 | NA | 16 | 32–64 |
| 10 mg/kg | −6.96 | 96 | 4.32 | NA | 4 | NA | 4 | NA | |
| VRE ATCC 51559 | 6 mg/kg | −3.22 | 24 | 7.86 | 16 | 16–32 | 64–128 | 16 | 64–128 |
| 10 mg/kg | −3.27 | 24 | 7.9 | 32 | 16–32 | 32 | 16 | 32–64 | |
| VRE 12311 | 6 mg/kg | −2.7 | 24 | 7.94 | 16–32 | 16–32 | 64 | 16 | 128–256 |
| 10 mg/kg | −2.47 | 48 | 7.44 | 32 | 8 | 32 | 16 | 32 | |
Mut, recovered mutant; Pop, overall bacterial population; NA, not applicable.
Emergence of derivative mutants with increased daptomycin MICs.
For VRE SF 12047, only one mutant (MIC 16 to 32 μg/ml) was recovered at 168 h during exposure to the daptomycin simulated regimen of 6 mg/kg/day. For both VRE 12311 and VRE ATCC 51559, mutants were recovered from both simulated regimens at the time of first regrowth and all time points tested thereafter. The mutants recovered from the DAP 6 mg/kg/day 168 h sample were chosen as the isogenic pair to be tested secondary to having the highest MIC values.
PFGE.
Pulsed-field gel electrophoresis (PFGE) confirmed the strain relatedness of the three strains utilized for the in vitro PK/PD model and the respective mutants recovered from the 168 h PD sample for the daptomycin 6 mg/kg simulation.
Cytochrome c binding.
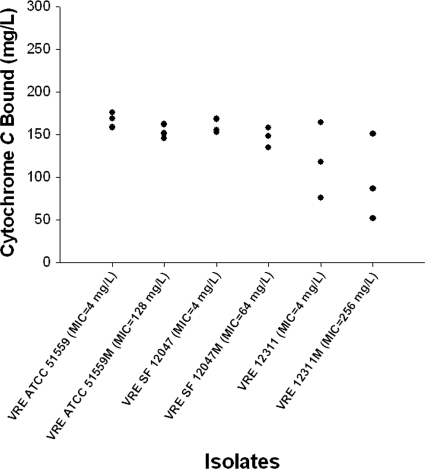
The relative cell surface charges as determined by cytochrome c binding experiments are displayed in Fig. 1. For the isogenic pairs, all E. faecium mutants with elevated daptomycin MIC values had a decrease in cytochrome c binding, suggesting an increase in cell surface positivity compared to that of the daptomycin-susceptible parent strain.
Fig. 1.
Binding of cationic cytochrome c by isogenic Enterococcus faecium strains and their mutants from the 1-compartment model.
TEM.
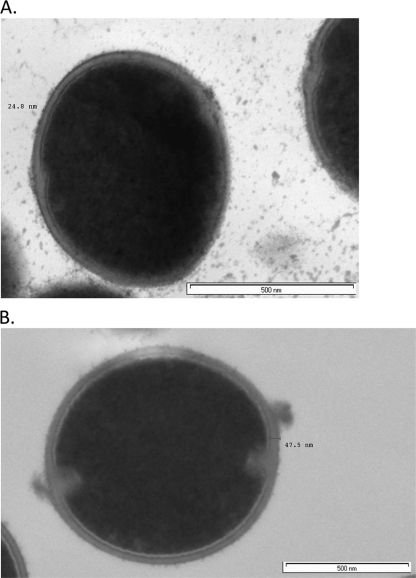
The TEM results for cell wall thickness are displayed in Table 1 and Fig. 2. For the isogenic pairs, the E. faecium mutants with elevated daptomycin MIC values displayed significant increases in mean cell wall thickness, increasing from 30.3 nm to 43.5 nm (43% increase), from 22.2 nm to 32.4 nm (46% increase), and from 24.9 nm to 39.5 nm (58% increase) for VRE SF 12047, VRE 12311, and VRE ATCC 51559, respectively (P < 0.0001 for all comparisons).
Fig. 2.
Cell wall thickness for single cell of VRE ATCC 51559 (A) and VRE ATCC 51559 M (B).
Determination of cytoplasmic membrane depolarization.
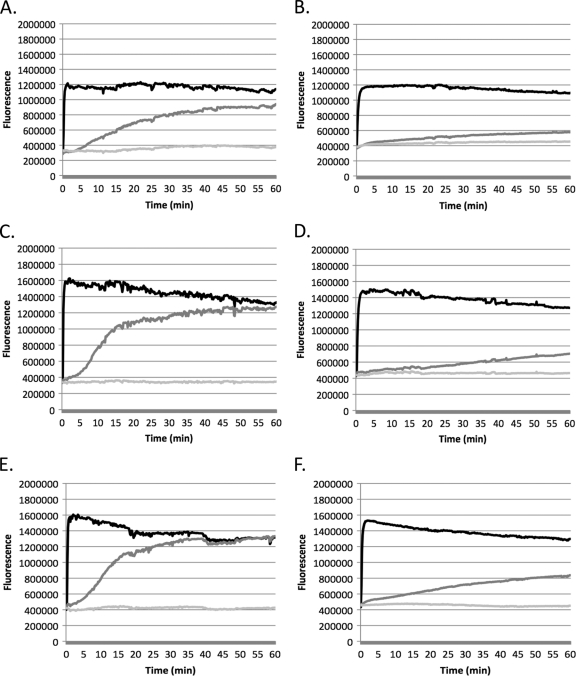
The effect of daptomycin on the cytoplasmic membrane in E. faecium isolates was evaluated using the membrane potential-sensitive fluorescent dye DiSC3. Dye was added into the medium containing bacteria and allowed to penetrate and accumulate inside the cells, and the fluorescence measured during the assay represents the amount of dye present in the medium. If the bacterial membrane is depolarized in the presence of drug, dye will escape from inside the cells out to the medium, causing an increase in measurable fluorescence.
All daptomycin-resistant mutants demonstrated a decrease in depolarization due to daptomycin compared to that of the daptomycin-susceptible parent strains (Fig. 3). For VRE 12311 and the VRE 12311 mutant, daptomycin-induced depolarization decreased significantly, from 93.03% ± 9.86% (mean ± standard deviation) to 27.65% ± 4.95% (P = 0.017). VRE ATCC 51559 and its mutant showed a similar decrease (75.95% ± 2.64% versus 22.29% ± 11.45%, P = 0.06). The daptomycin depolarization was 98.52% ± 5.29% for VRE SF 12047 and 45.89% ± 4.42% for the VRE SF 12047 mutant (P = 0.008), illustrating a >50% loss in daptomycin's depolarization ability for this VRE strain.
Fig. 3.
Cytoplasmic membrane depolarization of VRE ATCC 51559 (A) and the VRE ATCC 51559 mutant (B), VRE 12311 (C) and the VRE 12311 mutant (D), and VRE SF 12047 (E) and the VRE SF 12047 mutant (F). Black lines show results with nisin, dark-gray lines show results with daptomycin, and light-gray lines show results for control.
DISCUSSION
To our knowledge, this is the first published paper to discuss phenotypic changes and potential mechanisms associated with daptomycin-resistant E. faecium strains, the predominant species responsible for nosocomial infections caused by VRE. It is also the first study to utilize multiple isogenic pairs developed in vitro. The three strains utilized in the in vitro PK/PD models, VRE SF 12047, VRE 12311, and VRE ATCC 51559, varied in their susceptibility to daptomycin activity and in their propensity to develop daptomycin-resistant mutants during therapy, therefore allowing the recovery of mutants with MICs ranging from 32 to 256 μg/ml. Our study examined the activity of daptomycin and the development of resistance in a high-inoculum and high-MIC setting. Daptomycin displayed the most potent and sustained activity against VRE SF 12047. The propensity of this strain to develop resistance (only one mutant recovered at 168 h under daptomycin pressure of 6 mg/kg/day) was strikingly less than that of the other two strains tested. VRE ATCC 51559 and VRE 12311 developed resistance to daptomycin at 24/24 h and 24/48 h under exposure to daptomycin at 6 and 10 mg/kg/day, respectively. The variability among strains highlights the need for further research to determine the optimal daptomycin dose for enterococcal infections and to prevent the emergence of resistance.
Comparison of the isogenic in vitro-derived pairs found a small increase in cell surface positivity (decrease in cytochrome c binding) in the daptomycin-resistant mutants. An increase in the positive cell surface charge of E. faecium strains relative to that of the daptomycin-susceptible parent strains may be associated with daptomycin resistance. Preliminary data from one study examining a clinical E. faecalis isogenic pair also found a decrease in cytochrome c binding in the daptomycin-resistant strain (MIC = 16 μg/ml) compared to that of the daptomycin-susceptible (MIC = 1 μg/ml) parent (27). As stated previously, daptomycin is bound to calcium in its active state, forming a positively charged complex that simulates a cationic peptide antibiotic. It is hypothesized that, in S. aureus, a change in the cytoplasmic membrane surface charge from negative to more positive contributes to the increase in daptomycin MIC values by charge repulsion of the positively charged daptomycin-Ca2+ complex (20, 37). This increase in membrane surface charge in S. aureus with increased daptomycin MIC values has been demonstrated in clinical, in vitro, and genetically manipulated isolates via decreased binding of cytochrome c or other cationic peptide antibiotics (14, 20, 24, 29, 37). It would appear that enterococci also possess the ability to alter cytoplasmic membrane surface charge and this contributes to daptomycin resistance. Further analysis, including the use of more sensitive assays, is required to confirm these findings in clinical E. faecium strains and explore the mechanisms by which enterococci are able to alter cytoplasmic membrane surface charge.
With regard to cell wall thickness, comparison of the in vitro-derived isogenic pairs revealed a significant increase for all the daptomycin-resistant E. faecium mutants compared to the cell wall thickness of the parent strain. This finding suggests that thickening of the cell wall is one mechanism by which E. faecium may increase its resistance to daptomycin under prolonged exposure. Previous studies have shown that enterococci possess the ability to thicken cell walls in an effort to survive under selective pressure. Comparison of E. faecalis AR01/DGVS and DGRM2, a monolaurin-resistant mutant, found an increase in cell wall thickness and contraction of the cytoplasm in DGRM2 compared to these characteristics in AR01/DGVS in the presence of monolaurin (13). Preliminary work examining the same clinical E. faecalis isogenic pair (daptomycin MIC values of 1 and 16 μg/ml) mentioned previously found both an increase in cell wall thickness and localized protrusions in the cell envelope in the mutant strain (28). Increases in daptomycin MIC values in S. aureus have been associated with increases in cell wall thickness and VISA strains (6, 11, 29). In S. aureus, thickening of the cell wall is hypothesized to contribute to decreased susceptibility to daptomycin via an affinity trapping mechanism within the cell wall (11). Based on the data for the in vitro-derived isogenic pairs, it would appear that a thickened cell wall in E. faecium contributes to daptomycin resistance.
Daptomycin acts like a cationic antimicrobial peptide in its active calcium-bound form and exerts bactericidal activity in S. aureus through the insertion of a lipophilic acyl tail into the cytoplasmic membrane, leading to potassium efflux, destruction of the ion concentration gradient, membrane depolarization, and probably, more generalized disruption of the cell membrane (9, 19). While the mechanism of action of daptomycin has not been specifically studied in Enterococcus spp., it is logical that it would act via similar mechanisms in these two Gram-positive organisms. Clinical S. aureus strains with decreased susceptibility to daptomycin have been found to have decreases in membrane depolarization from daptomycin compared with their respective isogenic susceptible strains (20, 22). The in vitro-derived E. faecium mutants in this study displayed similar characteristics, with decreased depolarization due to daptomycin compared to that in their susceptible isogenic parent strains. In addition, preliminary data examining the clinical E. faecalis isogenic pair also found decreased membrane depolarization by daptomycin in the resistant strain (MIC = 16 μg/ml) compared to that in the susceptible parent (MIC = 1 μg/ml) (27).
This is the first study to examine phenotypic characteristics associated with daptomycin-resistant E. faecium strains and is strengthened by the number of in vitro-derived strains examined. Even with its novelty, positive findings, and larger number of strains, this study does have limitations. Since this study utilized an in vitro model to simulate clinical daptomycin exposure, it is logical that the mechanisms by which the examined E. faecium strains developed resistance to daptomycin will more closely mimic in vivo conditions than other laboratory methods, such as serial passage. It is still possible, however, that daptomycin-resistant E. faecium strains developed in vivo may differ from the in vitro-developed strains here, because E. faecium isolates in the human body are also exposed to cationic peptides that are part of the immune system. The lack of white cells in the model compared to the immune system may also alter the rate of resistance selection in vitro compared to the rate in vivo. We also did not examine any genetic changes associated with the emergence of daptomycin-resistant strains.
Clearly, more research is needed to continue to explore the mechanisms by which E. faecium strains change their cell wall and cytoplasmic membrane, leading to the development of daptomycin resistance. In S. aureus, changes in surface charge are secondary to alterations in phospholipid asymmetry (translocation of the positively charged phospholipids to the outer leaflet of the cytoplasmic membrane) and increased alanylation of cell wall teichoic acid (14, 20, 29, 37). Research into the phosphatidylglycerol content of the cytoplasmic membrane of various species of bacteria has found differences in amino acid substrate specificity (32). Enterococcus faecium TX1330 possesses a multispecific amino acid aminoacylphosphatidylglycerol synthase, allowing it to synthesize greater amounts of phosphatidylglycerols and perform elaborate remodeling of these lipids within the cell membrane in response to environmental stress (32). Therefore, while changes in phospholipids may also be responsible for the daptomycin resistance in enterococci, the exact mechanisms may vary between the species. Additional work examining the mechanisms, including the dose exposure, leading to the development of daptomycin resistance in enterococci is needed.
ACKNOWLEDGMENT
We are grateful to Marcus J. Zervos and his team for providing isolates (VRE SF 12047 and VRE 12311) and for confirmation of vanA in VRE 12311.
Footnotes
Published ahead of print on 25 July 2011.
REFERENCES
- 1. Arias C. A., et al. 2010. Whole-genome analysis of daptomycin-susceptible Enterococcus faecalis and its daptomycin-resistant derivative that arose during therapy, poster 245. Infect. Dis. Soc. Am. 48th Annu. Meet., Vancouver, BC, Canada, 21 to 24 October 2010 [Google Scholar]
- 2. Arias C. A., et al. 2007. Failure of daptomycin monotherapy for endocarditis caused by an Enterococcus faecium strain with vancomycin-resistant and vancomycin-susceptible subpopulations and evidence of in vivo loss of the vanA gene cluster. Clin. Infect. Dis. 45:1343–1346 [DOI] [PubMed] [Google Scholar]
- 3. Benvenuto M., Benziger D. P., Yankelev S., Vigliani G. 2006. Pharmacokinetics and tolerability of daptomycin at doses up to 12 milligrams per kilogram of body weight once daily in healthy volunteers. Antimicrob. Agents Chemother. 50:3245–3249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boucher H. W., Sakoulas G. 2007. Perspectives on daptomycin resistance, with emphasis on resistance in Staphylococcus aureus. Clin. Infect. Dis. 45:601–608 [DOI] [PubMed] [Google Scholar]
- 5. Bourgogne A., Garcia D., Quinn J. P., Murray B. E., Arias C. A. 2009. Transcriptome analysis of daptomycin-resistant Enterococcus faecalis, poster C1-614. Abstr. 49th Intersci. Conf. Antimicrob. Agents Chemother., San Francisco, CA, 12 to 15 September 2009 [Google Scholar]
- 6. Camargo I. L., Neoh H. M., Cui L., Hiramatsu K. 2008. Serial daptomycin selection generates daptomycin-nonsusceptible Staphylococcus aureus strains with a heterogeneous vancomycin-intermediate phenotype. Antimicrob. Agents Chemother. 52:4289–4299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cha R., Grucz R. G., Jr., Rybak M. J. 2003. Daptomycin dose-effect relationship against resistant gram-positive organisms. Antimicrob. Agents Chemother. 47:1598–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Clinical and Laboratory Standards Institute 2008. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 8th ed CLSI document M7-A8. CLSI, Wayne, PA [Google Scholar]
- 9. Cotroneo N., Harris R., Perlmutter N., Beveridge T., Silverman J. A. 2008. Daptomycin exerts bactericidal activity without lysis of Staphylococcus aureus. Antimicrob. Agents Chemother. 52:2223–2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cui L., et al. 2003. Cell wall thickening is a common feature of vancomycin resistance in Staphylococcus aureus. J. Clin. Microbiol. 41:5–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cui L., Tominaga E., Neoh H. M., Hiramatsu K. 2006. Correlation between reduced daptomycin susceptibility and vancomycin resistance in vancomycin-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 50:1079–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Donabedian S., Chow J. W., Shlaes D. M., Green M., Zervos M. J. 1995. DNA hybridization and contour-clamped homogeneous electric field electrophoresis for identification of enterococci to the species level. J. Clin. Microbiol. 33:141–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dufour M., et al. 2007. Characterization of monolaurin resistance in Enterococcus faecalis. Appl. Environ. Microbiol. 73:5507–5515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ernst C. M., et al. 2009. The bacterial defensin resistance protein MprF consists of separable domains for lipid lysinylation and antimicrobial peptide repulsion. PLoS Pathog. 5:e1000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fabretti F., et al. 2006. Alanine esters of enterococcal lipoteichoic acid play a role in biofilm formation and resistance to antimicrobial peptides. Infect. Immun. 74:4164–4171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fowler V. G., Jr, et al. 2006. Daptomycin versus standard therapy for bacteremia and endocarditis caused by Staphylococcus aureus. N. Engl. J. Med. 355:653–665 [DOI] [PubMed] [Google Scholar]
- 17. Friedman L., Alder J. D., Silverman J. A. 2006. Genetic changes that correlate with reduced susceptibility to daptomycin in Staphylococcus aureus. Antimicrob. Agents Chemother. 50:2137–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gqada Z., et al. 2008. Clinical rationale for treatment of endocarditis caused by methicillin-susceptible Staphylococcus aureus developing nonsusceptibility to daptomycin. J. Clin. Microbiol. 46:2471–2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hobbs J. K., Miller K., O'Neill A. J., Chopra I. 2008. Consequences of daptomycin-mediated membrane damage in Staphylococcus aureus. J. Antimicrob. Chemother. 62:1003–1008 [DOI] [PubMed] [Google Scholar]
- 20. Jones T., et al. 2008. Failures in clinical treatment of Staphylococcus aureus infection with daptomycin are associated with alterations in surface charge, membrane phospholipid asymmetry, and drug binding. Antimicrob. Agents Chemother. 52:269–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Julian K., et al. 2007. Characterization of a daptomycin-nonsusceptible vancomycin-intermediate Staphylococcus aureus strain in a patient with endocarditis. Antimicrob. Agents Chemother. 51:3445–3448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kaatz G. W., Lundstrom T. S., Seo S. M. 2006. Mechanisms of daptomycin resistance in Staphylococcus aureus. Int. J. Antimicrob. Agents 28:280–287 [DOI] [PubMed] [Google Scholar]
- 23. Khan S. A., Sung K., Layton S., Nawaz M. S. 2008. Heteroresistance to vancomycin and novel point mutations in Tn1546 of Enterococcus faecium ATCC 51559. Int. J. Antimicrob. Agents 31:27–36 [DOI] [PubMed] [Google Scholar]
- 24. Kraus D., et al. 2008. The GraRS regulatory system controls Staphylococcus aureus susceptibility to antimicrobial host defenses. BMC Microbiol. 8:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. LaPlante K. L., Rybak M. J., Leuthner K. D., Chin J. N. 2006. Impact of Enterococcus faecalis on the bactericidal activities of arbekacin, daptomycin, linezolid, and tigecycline against methicillin-resistant Staphylococcus aureus in a mixed-pathogen pharmacodynamic model. Antimicrob. Agents Chemother. 50:1298–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lewis J. S., II, et al. 2005. Emergence of daptomycin resistance in Enterococcus faecium during daptomycin therapy. Antimicrob. Agents Chemother. 49:1664–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McGrath D., et al. 2010. Resistance to daptomycin in vancomycin-resistant Enterococcus faecalis is associated with alterations of cell membrane (CM) potential and surface charge, poster C1-078. Abstr. 50th Intersci. Conf. Antimicrob. Agents Chemother., Boston, MA, 12 to 15 September 2010 [Google Scholar]
- 28. McGrath D., et al. 2010. Transmission electron microscopy analysis of daptomycin-susceptible Enterococcus faecalis and its daptomycin-resistant derivative that arose during therapy, poster C1-079. Abstr. 50th Intersci. Conf. Antimicrob. Agents Chemother., Boston, MA, 12 to 15 September 2010 [Google Scholar]
- 29. Mishra N. N., et al. 2009. Analysis of cell membrane characteristics of in vitro-selected daptomycin-resistant strains of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 53:2312–2318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Montero C. I., Stock F., Murray P. R. 2008. Mechanisms of resistance to daptomycin in Enterococcus faecium. Antimicrob. Agents Chemother. 52:1167–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Munoz-Price L. S., Lolans K., Quinn J. P. 2005. Emergence of resistance to daptomycin during treatment of vancomycin-resistant Enterococcus faecalis infection. Clin. Infect. Dis. 41:565–566 [DOI] [PubMed] [Google Scholar]
- 32. Roy H., Ibba M. 2009. Broad range amino acid specificity of RNA-dependent lipid remodeling by multiple peptide resistance factors. J. Biol. Chem. 284:29677–29683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sakoulas G., et al. 2008. Evaluation of endocarditis caused by methicillin-susceptible Staphylococcus aureus developing nonsusceptibility to daptomycin. J. Clin. Microbiol. 46:220–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sharma M., Riederer K., Chase P., Khatib R. 2008. High rate of decreasing daptomycin susceptibility during the treatment of persistent Staphylococcus aureus bacteremia. Eur. J. Clin. Microbiol. Infect. Dis. 27:433–437 [DOI] [PubMed] [Google Scholar]
- 35. Silverman J. A., Perlmutter N. G., Shapiro H. M. 2003. Correlation of daptomycin bactericidal activity and membrane depolarization in Staphylococcus aureus. Antimicrob. Agents Chemother. 47:2538–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tenover F. C., et al. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yang S. J., et al. 2009. Enhanced expression of dltABCD is associated with the development of daptomycin nonsusceptibility in a clinical endocarditis isolate of Staphylococcus aureus. J. Infect. Dis. 200:1916–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yang S. J., et al. 2009. Regulation of mprF in daptomycin-nonsusceptible Staphylococcus aureus strains. Antimicrob. Agents Chemother. 53:2636–2637 [DOI] [PMC free article] [PubMed] [Google Scholar]