Abstract
Fungal infections are a serious health problem in clinics, especially in the immune-compromised patient. Disease ranges from widespread superficial infections like vulvovaginal infections to life-threatening systemic candidiasis. Especially for systemic mycoses, only a limited arsenal of antifungals is available. The most commonly used classes of antifungal compounds used include azoles, polyenes, and echinocandins. Due to emerging resistance to standard therapy, significant side effects, and high costs for several antifungals, there is a medical need for new antifungals in the clinic and general practice. In order to expand the arsenal of compounds with antifungal activities, we screened a compound library including more than 35,000 individual compounds derived from organic synthesis as well as combinatorial compound collections representing mixtures of compounds for antimycotic activity. In total, more than 100,000 compounds were screened using a new type of activity-selectivity assay, analyzing both the antifungal activity and the compatibility with human cells at the same time. One promising hit, an (S)-2-aminoalkyl benzimidazole derivative, was developed among a series of lead compounds showing potent antifungal activity. (S)-2-(1-Aminoisobutyl)-1-(3-chlorobenzyl) benzimidazole showed the highest antifungal activity and the best compatibility with human cells in several cell culture models and against a number of clinical isolates of several species of pathogenic Candida yeasts. Transcriptional profiling indicates that the newly discovered compound is a potential inhibitor of the ergosterol pathway, in contrast to other benzimidazole derivatives, which target microtubules.
INTRODUCTION
Fungal infections still represent a serious and not-yet-solved health problem, especially in industrialized countries. In Europe, fungal infections account for 17% of infections in intensive care units, and similar numbers are reported from the United States (29, 45). In addition, emerging resistance to almost all commercialized antifungals has been reported (30, 38). Treatment, especially of systemic infections, largely relies on chemotherapy and is accompanied not only by intermediate success rates but also by high costs. Modern antifungals like novel polyene formulations, new azoles, and echinocandins (7, 15, 19, 28, 33) are expensive. In addition, common non-life-threatening superficial infections like recurrent vulvovaginal candidiasis impose significant restrictions to patients, resulting in a reduced quality of life. Due to the eukaryotic nature of fungal pathogens, well-tolerated antifungals are much less frequently identified than antibiotics targeting bacteria. Antifungals in general have been identified using typical screening approaches focusing on growth retardation or killing of the pathogen in an artificial environment (34). This type of screening results in a high number of hits which are often cytotoxic, in general, and therefore only of limited use for clinical development. In addition, the environment might be critical for the effect of a potential antifungal. Several known antifungal compounds have been shown to have different effects depending on the environmental context in which they are employed, possibly due to another set of targets expressed under these conditions (23, 25, 32). Recent studies, especially in Candida albicans, the predominant cause of systemic fungal infections, have shown that interaction with the host results in a significant level of stress response reactions as well as metabolic adaptations which differ from those in the artificial environment generally used in typical drug screening approaches (5, 36). However, primary large-scale screenings for antifungals in a host-like environment have, to our knowledge, not been reported so far, but rather, high-throughput screenings (HTSs) have been conducted in model organisms like Caenorhabditis elegans or Drosophila (26, 27). For viral pathogens, a screening assay mimicking a host-like environment has already been established (21). This assay mimics the smallest unit of a natural infection by incubation of the pathogen in the presence of host cells. The assay therefore is able to monitor both the tolerability of antiviral agents by host cells and their antiviral activity in one step. Instead of measuring growth retardation of the pathogen itself or inhibition of enzymatic functions of the pathogen, the survival of the host cells cultured in the presence of the pathogen and the respective compounds to be tested is measured directly. Thereby, the screening assay covers all accessible potential in vitro targets of both the pathogen and the host simultaneously and is not restricted to a single target.
To identify novel antifungal lead compounds with selective antifungal activity in a host-like environment, we successfully adapted this HTS-compatible assay for screening in the presence of fungal pathogens. In a screen encompassing more than 100,000 small chemical molecules based on heterocyclic scaffolds, several compounds with potential antifungal activity were identified. The antifungal activity of one compound [(S)-2-(1-aminoisobutyl)-1-(3-chlorobenzyl) benzimidazole; EMC120B12] was studied in more detail. In order to identify the molecular structure responsible for the antifungal activity, a structure-activity relationship study was performed by synthesis of related analogues [(S)-2-aminoalkyl benzimidazole derivatives]. The compound with the highest activity (EMC120B12) was tested further for efficiency in complex three-dimensional (3D) tissue models (9) as well as for activity against other pathogenic yeasts and molds, including a series of clinical isolates. The results of the activity-selectivity (AS) assay were compared to those of standard MIC determination of the compounds identified using the European Committee on Antimicrobial Susceptibility Testing (EUCAST) protocol (13). In addition, the transcriptional response of C. albicans revealed the ergosterol pathway to be the most likely target of EMC120B12, whereas no change in tubulin expression, a known target for benzimidazoles, was found. Our results show the feasibility of using simple host-pathogen interaction models for drug screening by identification of a new group of benzimidazole derivatives effective against Candida spp.
MATERIALS AND METHODS
Strains and media.
The Candida strains used in this study were clinical isolate C. albicans SC5314 (14) from several sources (Can14 from the S. Rupp lab and SC5314, KS04-01, and KS04-02 from the K. Schröppel lab); the type strains C. parapsilosis ATCC 22019, C. glabrata ATCC 90030, C. albicans DSMZ 11949, C. albicans ATCC 90028, C. parapsilosis ATCC 90018, C. guilliermondii ATCC 90877, and Issatchenki orientalis ATCC 6258; as well as a collection of 143 clinical isolates (see Table 2) and Aspergillus fumigatus (NRRL6585; U.S. Department of Agriculture, Peoria, IL). Yeast strains were cultured overnight in yeast extract, peptone, dextrose (YPD) medium containing 2% glucose (Difco) from glycerol stock cultures or plated onto YPD agar plates (2% Bacto agar; Difco) for 48 h at 30°C. A. fumigatus NRRL6585 was cultivated using potato-glucose agar.
Table 2.
MICs of 150 strains against EMC120B12 and fluconazole
| Strain no. | Strain (n = 150)a | MIC (μg/ml) |
|
|---|---|---|---|
| EMC120B12 | Fluconazole | ||
| 1 | C. albicans ATCC 90028 | 4 | 2 |
| 2 | C. albicans DSMZ 11949 | 4 | 4 |
| 3 | C. glabrata ATCC 90030 | 16 | 16 |
| 4 | C. guilliermondii ATCC 90877 | 4 | 1 |
| 5 | C. parapsilosis ATCC 22019 | 0.125 | 0.25 |
| 6 | C. parapsilosis ATCC 90018 | 0.125 | 0.125 |
| 7 | Issatchenki orientalis ATCC 6258 | 0.25 | 0.5 |
| 8 | C. neoformans | 4 | 1 |
| 9 | C. albicans 1008* | 32 | 32 |
| 10 | C. albicans 75 | 0.25 | 0.5 |
| 11 | C. albicans 924* | 128 | 128 |
| 12 | C. albicans 931* | 64 | 64 |
| 13 | C. albicans 959* | 128 | 128 |
| 14 | C. albicans 971* | 8 | 32 |
| 15 | C. albicans 974* | 8 | 4 |
| 16 | C. albicans 987* | 16 | 32 |
| 17 | C. albicans 993* | 16 | 16 |
| 18 | C. albicans 994* | 32 | 64 |
| 19 | C. albicans 999* | 64 | 32 |
| 20 | C. albicans AM2001/0007 | 0.5 | 0.25 |
| 21 | C. albicans AN 10481 | 8 | 0.25 |
| 22 | C. albicans AN 10798 | 0.25 | 0.5 |
| 23 | C. albicans AN 10883 | 0.125 | 0.125 |
| 24 | C. albicans AN 11231 | 0.5 | 0.5 |
| 25 | C. albicans AN 11549 | 0.5 | 0.25 |
| 26 | C. albicans AN 13 | 0.125 | 0.125 |
| 27 | C. albicans AN 13244 | 0.125 | 0.125 |
| 28 | C. albicans AN 1699 | 8 | 32 |
| 29 | C. albicans AN 1769 | 0.25 | 0.25 |
| 30 | C. albicans AN 1994 | 128 | 16 |
| 31 | C. albicans AN 2787 | 0.125 | 0.125 |
| 32 | C. albicans AN 2829 | 4 | 64 |
| 33 | C. albicans AN 3156 | 8 | 16 |
| 34 | C. albicans AN 3431 | 0.125 | 0.125 |
| 35 | C. albicans AN 3591 | 0.125 | 0.125 |
| 36 | C. albicans AN 397 | 0.125 | 0.125 |
| 37 | C. albicans AN 4071 | 4 | 16 |
| 38 | C. albicans AN 4462.1 | 2 | 1 |
| 39 | C. albicans AN 4835 | 0.125 | 0.25 |
| 40 | C. albicans AN 562 | 1 | 16 |
| 41 | C. albicans AN 5752.2 | 0.125 | 0.25 |
| 42 | C. albicans AN 5944 | 0.25 | 0.25 |
| 43 | C. albicans AN 5960 | 0.125 | 0.125 |
| 44 | C. albicans AN 6160 | 0.25 | 0.25 |
| 45 | C. albicans AN 62 | 0.125 | 0.125 |
| 46 | C. albicans AN 6896 | 4 | 0.25 |
| 47 | C. albicans AN 7961 | 0.125 | 0.125 |
| 48 | C. albicans AN 8449 | 1 | 0.25 |
| 49 | C. albicans AN 8775 | 0.25 | 0.25 |
| 50 | C. albicans AN 9560 | 1 | 0.25 |
| 51 | C. albicans AN 9645 | 0.25 | 0.25 |
| 52 | C. albicans AN4051 | 2 | 0.5 |
| 53 | C. albicans CA 20 | 0.125 | 0.125 |
| 54 | C. albicans CA 21 | 0.125 | 0.125 |
| 55 | C. albicans Jg 32570 | 128 | 16 |
| 56 | C. albicans MY 2902/2008 | 0.5 | 16 |
| 57 | C. albicans RU IV | 1 | 1 |
| 58 | C. albicans SCS 71865L | 0.125 | 64 |
| 59 | C. albicans VB 1723 | 4 | 0.5 |
| 60 | C. albicans VB 1723 | 0.5 | 0.25 |
| 61 | C. albicans VB 1811 | 0.125 | 0.125 |
| 62 | C. albicans VB 21064 | 0.125 | 0.125 |
| 63 | C. albicans VB 22905 | 0.25 | 0.125 |
| 64 | C. albicans VB 2688 | 0.125 | 0.125 |
| 65 | C. albicans VB 4384 | 2 | 0.5 |
| 66 | C. albicans VB 4606 | 0.125 | 0.125 |
| 67 | C. albicans VB 8567 | 0.125 | 0.125 |
| 68 | C. blankii VB 13063 | 0.5 | 32 |
| 69 | C. glabrata AN 10767 | 0.5 | 8 |
| 70 | C. glabrata AN 12749 | 0.25 | 8 |
| 71 | C. glabrata AN 12862 | 0.25 | 4 |
| 72 | C. glabrata AN 1857 | 1 | 2 |
| 73 | C. glabrata AN 4355 | 16 | 64 |
| 74 | C. glabrata AN 4462.2 | 0.125 | 0.125 |
| 75 | C. glabrata AN 5547 | 0.125 | 4 |
| 76 | C. glabrata AN 8148 | 0.25 | 4 |
| 77 | C. glabrata AN 8626 | 4 | 0.125 |
| 78 | C. glabrata CG 7 | 4 | 2 |
| 79 | C. glabrata VB 3346 | 4 | 64 |
| 80 | C. guilliermondii AN 444 | 0.5 | 4 |
| 81 | C. guilliermondii AN 6494 | 0.25 | 0.5 |
| 82 | C. guilliermondii AN 8832 | 4 | 2 |
| 83 | C. guilliermondii RU II | 4 | 2 |
| 84 | C. krusei 224 | 0.25 | 8 |
| 85 | C. krusei 394 | 4 | 16 |
| 86 | C. krusei 201 | 1 | 32 |
| 87 | C. krusei 222 | 1 | 16 |
| 88 | C. krusei 232 | 2 | 32 |
| 89 | C. krusei 237 | 2 | 32 |
| 90 | C. krusei 241 | 2 | 32 |
| 91 | C. krusei 242 | 1 | 32 |
| 92 | C. krusei 2572 | 2 | 32 |
| 93 | C. krusei 337 | 2 | 32 |
| 94 | C. krusei 364 | 1 | 32 |
| 95 | C. krusei 39.986 | 2 | 32 |
| 96 | C. krusei 639 | 1 | 32 |
| 97 | C. krusei A 1934 | 2 | 32 |
| 98 | C. krusei AN 12026 | 2 | 16 |
| 99 | C. krusei AN 2572 | 1 | 16 |
| 100 | C. krusei AN 2944 | 4 | 32 |
| 101 | C. krusei AN 4557 | 2 | 32 |
| 102 | C. krusei AN 8033 | 8 | 64 |
| 103 | C. krusei AN 8829 | 4 | 32 |
| 104 | C. krusei VB 18175 | 2 | 32 |
| 105 | C. lusitaniae AN 5752.1 | 0.125 | 0.25 |
| 106 | C. lusitaniae AN 6110.2 | 0.125 | 0.25 |
| 107 | C. lusitaniae UR 14911 | 0.5 | 8 |
| 108 | C. nivariensis RU IV | 0.125 | 4 |
| 109 | C. parapsilosis 352 | 2 | 16 |
| 110 | C. parapsilosis 551 | 1 | 0.25 |
| 111 | C. parapsilosis 553 | 0.5 | 0.25 |
| 112 | C. parapsilosis 619 St-R | 1 | 0.5 |
| 113 | C. parapsilosis St-R 623 | 0.5 | 0.25 |
| 114 | C. parapsilosis 1007.2 | 1 | 0.25 |
| 115 | C. parapsilosis 10207 | 1 | 0.5 |
| 116 | C. parapsilosis 10267 | 8 | 2 |
| 117 | C. parapsilosis 4321 | 1 | 0.25 |
| 118 | C. parapsilosis 549 | 2 | 1 |
| 119 | C. parapsilosis 552 | 2 | 0.5 |
| 120 | C. parapsilosis 554 | 2 | 0.25 |
| 121 | C. parapsilosis 640 | 2 | 0.25 |
| 122 | C. parapsilosis AN 1.5845 | 0.5 | 0.5 |
| 123 | C. parapsilosis AN 1.6464 | 0.5 | 0.25 |
| 124 | C. parapsilosis AN 11805 | 0.5 | 0.125 |
| 125 | C. parapsilosis AN 3.7441 | 0.5 | 0.5 |
| 126 | C. parapsilosis AN 3284 | 2 | 0.25 |
| 127 | C. parapsilosis AN 3570 | 0.5 | 0.25 |
| 128 | C. parapsilosis AN 39207 | 0.25 | 0.25 |
| 129 | C. parapsilosis AN 5485 | 0.25 | 1 |
| 130 | C. parapsilosis AN 6218 | 1 | 0.25 |
| 131 | C. parapsilosis AN 6480 | 0.5 | 0.5 |
| 132 | C. parapsilosis AN 7348 | 2 | 0.5 |
| 133 | C. parapsilosis AN 7793 | 2 | 0.5 |
| 134 | C. parapsilosis AN 7903 | 0.125 | 0.25 |
| 135 | C. parapsilosis AV 7675 | 2 | 8 |
| 136 | C. parapsilosis R V II | 4 | 0.5 |
| 137 | C. parapsilosis St-R 613 | 2 | 0.5 |
| 138 | C. parapsilosis UR 13926 | 0.125 | 0.125 |
| 139 | C. parapsilosis UR 2795V | 0.5 | 0.25 |
| 140 | C. parapsilosis UR 5428 | 0.5 | 0.25 |
| 141 | C. parapsilosis VA 9625 | 1 | 0.25 |
| 142 | C. parapsilosis VB 4188 | 1 | 0.5 |
| 143 | C. pelliculosa AN 3525 | 16 | 8 |
| 144 | C. pelliculosa AN 8197 | 16 | 16 |
| 145 | C. tropicalis 550 | 0.5 | 0.25 |
| 146 | C. tropicalis AN 1946 | 128 | 4 |
| 147 | C. tropicalis AN 3241 | 0.25 | 0.5 |
| 148 | C. tropicalis AN 4522.1 | 0.125 | 0.125 |
| 149 | C. tropicalis AN 4572 | 2 | 1 |
| 150 | C. tropicalis UR 15464 | 0.125 | 0.125 |
Strains identified to be resistant to fluconazole with preliminary characterization on resistance mechanisms are identified with asterisks (see Discussion). These strains have been provided by the National Reference Center for Systemic Mycoses, as have strains 10, 84 to 96, and 109 to 121. All other strains were provided by the strain collection of the Interfaculty Institute of Microbiology and Infection Medicine, University of Tübingen.
Epithelial cell line culture.
A431, A549, Caco-2, HeLa, and CHO-K1 cells were grown in 175-cm2 tissue culture flasks (Greiner) and split 1:3 by standard methods just before they reached confluence. All media were obtained from Gibco. All cells were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS), 2 mM l-glutamine, and 0.1 mg gentamicin/ml. The cells were cultivated under standard conditions at 37°C in 5% CO2. All antibiotics were omitted prior to cocultivation with C. albicans.
Compound library and synthesis of analogons.
The compound library used is a proprietary library from EMC Microcollections GmbH. A detailed description of synthesis of the hit compound and its structural compounds is published elsewhere (3).
Screening assay.
The screening assay was based on a screen for antivirals (22) and adapted for C. albicans as follows: 10,000 HeLa cells (human cervix carcinoma cells [ATCC catalog no. CCL-2]) per well were dispensed (50 μl of a solution with 200,000 HeLa cells/ml) in a 96-well plate in the presence or absence of a compound (100 μl of a 40 μM stock solution gives an end concentration of 20 μM) of the respective library infected with 5 to 500 CFU of C. albicans (strain SC5314 from the S. Rupp lab; optimum, 50 CFU; 50 μl, 1,000 CFU/ml). The plates were incubated at 37°C in 5% CO2 in a total volume of 200 μl medium (RPMI 1640, 10% FCS, 1% glutamine) for 5 days. The compounds in general had a purity of >90%. Hits were determined photometrically (Spectra-Photometer V630; Jasco, Gross-Umstadt, Germany). For this purpose, fluorescein generated from fluorescein diacetate by the metabolic activity of live HeLa cells was measured. Metabolic activity resulting in more than 40% fluorescence intensity compared to that for the no-pathogen control (100%) was considered a hit. These compounds were processed further. None of the other compounds were investigated further. The 50% inhibitory concentration (IC50) and the 50% cytotoxic concentration (CC50) were determined using a 2-fold serial dilution of the respective compound (10 different concentrations) and fluorescein diacetate as described above.
Toxicity tests.
To test the toxicity of the hit compounds against mammalian cells, tests were performed with HeLa cells and two additional cell lines, CHO-K1 (Chinese hamster ovary cells [ATCC catalog no. CCL-61]) and A549 (human lung carcinoma cells [ATCC catalog no. CCL-185]), as described above. C. albicans was omitted in these toxicity assays. In addition, viability tests of a 3D tissue model containing a collagen matrix as a supporting scaffold for a confluent layer of A431 cells (ATCC catalog no. CRL-1555) were performed by incubating the model with the hit compound (see below). Viability of cells was confirmed by trypan blue exclusion and histology.
Effectiveness against other fungal pathogens.
In addition to C. albicans, several other yeast and mold strains of virulent fungal species were tested using both the screening assay described above and standard MIC tests, including A. fumigatus, C. glabrata, C. parapsilosis, C. krusei, C. norvegensis, C. guilliermondii, C. nivariensis, C. tropicalis, C. lusitaniae, C. blankii, and Issatchenki orientalis. For the yeast strains, a collection of clinical isolates from diagnostic blood culture, urine, or swab specimens as well as type strains was tested in order to estimate the general potency of the compound identified.
Three-dimensional tissue models.
The three-dimensional epithelial models for measuring the effect of the compound on invasion were generated over a 5-day period as described previously (9, 17) with modifications. An equal volume of 2× DMEM containing 100 mM HEPES was mixed with an equal volume of acidic collagen solution extracted from rat tails. Of this solution, 200 μl was instantly poured into each cell culture insert (diameter, 12 mm); the gels were allowed to solidify for 15 min at 37°C in 5% CO2. Each insert was transferred to a cavity of a 6-well plate and provided basolaterally with 2 ml DMEM. On day 3, the medium was replaced by fresh medium and 2 × 105 A431 cells were placed on top of the gel. Gels were kept at 37°C in 5% CO2 for at least 2 days to secure the formation of a confluent cell layer. The vitality of the epithelium was determined microscopically by using trypan blue. For infection assays, any remaining supernatant on the epithelium was removed to avoid growth of Candida into the medium. Each tissue model was infected with approximately 250 cells for up to 48 h. At the same time, EMC120B12 was added at a concentration of 2 μM (2 μg/ml) to the medium basolaterally supporting the tissue model. The tissue models were fixed after 48 h of incubation at 37°C and processed for histological staining as described previously (17).
Determination of MICs of EMC120B12 for Candida spp. and comparison of EMC120B12 activity to that of a reference antifungal compound.
For the determination of MICs, we applied the standard protocol, the EUCAST reference method EDef 7.1 for antifungal susceptibility testing of yeasts, with modifications (13), to our strain collection, consisting of 143 clinical isolates of C. albicans, C. glabrata, C. parapsilosis, C. neoformans, C. guilliermondii, C. nivariensis, C. tropicalis, C. pelliculosa, C. lusitaniae, and C. blankii, including 17 fluconazole-resistant C. albicans strains (MICs > 4 μg/ml). Furthermore, we used C. albicans DSMZ 11949, C. albicans ATCC 90028, C. glabrata ATCC 90030, C. parapsilosis ATCC 90018, C. parapsilosis ATCC 22019, C. guilliermondii ATCC 90877, and Issatchenki orientalis ATCC 6258 as reference control strains to confirm that the MIC values were within the limits of the EUCAST procedure. In contrast to the classical EUCAST protocol, prior to the test, strains were grown at 25°C for 24 h on solid Sabouraud (Sab) agar plates supplemented with 40 μg/ml gentamicin to avoid any possible bacterial contamination. This has no impact on the MIC in the following tests, as approved by comparison of our modified protocol to the classical EUCAST protocol (data not shown). On each microdilution plate, row 1 was used as a growth control for viability of fungal cells. Rows 2 to 12 contained increasing amounts of the test compound in a 2-fold titration scheme, resulting in a range of final concentrations from 0.125 μg/ml to 128 μg/ml. Fluconazole was used as a control for antifungal activity of the test compounds. Microtiter plates were incubated at 35°C, and the growth of yeast cells was evaluated after 22 ± 2 h by measuring the optical density at 450 nm using a Tecan microtiter plate reader and analyzed with Magellan software. MICs were defined as the lowest drug concentration giving rise to an inhibition of growth of more than 50% of that of the drug-free control. The average MIC (MICav) of EMC120B12 for a subset of test strains was calculated as the geometric mean of the MIC test results of strains included in the subset.
Transcriptome analysis using DNA microarrays. (i) Cultivation of C. albicans SC5314 for DNA microarray experiments.
To reveal the influence of EMC120B12 on the transcriptome of C. albicans, 100 ml RPMI 1640 medium (Invitrogen, Karlsruhe, Germany) with 10% FCS (Gibco Life Technologies GmbH, Karlsruhe, Germany) was inoculated with C. albicans SC5314 from an overnight culture (10 ml YPD medium [1% yeast extract, 2% Bacto peptone, 2% glucose], 30°C) to an optical density of 0.4 and grown for 3 h at 37°C with or without EMC120B12 (IC25, 0.4 μM). Cells were centrifuged (1,200 × g, 3 min), and the supernatant was discarded. Subsequently, cells were resuspended in the remaining medium and dropped into liquid N2 to generate cell beads for RNA isolation. The experiment was performed three times.
(ii) Printing of whole-genome DNA microarrays.
Whole-genome C. albicans SC5314 DNA microarrays were printed on epoxy-coated glass slides (Schott, Jena, Germany) using a Microgrid II robot of Biorobotics (DigiLab Inc., Holliston, MA) by spotting 6,400 oligonucleotides (55- to 70-mers dissolved in spotting buffer, which consisted of 3× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 1.5 M betaine) in duplicate, including 10 bacteriophage lambda oligonucleotide DNA specificity controls. Oligonucleotides (Operon Biotechnologies Custom Array-Ready Longmers; Eurofins MWG Operon, Ebersberg, Germany) showed less than 70% cross hybridization within C. albicans and toward the human genome sequence (BLAST data source, C. albicans assembly 21 [http://www.candidagenome.org/], and Ensembl human transcripts [http://www.ensembl.org/index.html]).
SMP3 pins (Arrayit Corporation, Sunnyvale, CA) were used for spotting. After the spotting process, probes were immobilized on glass slides by incubation at room temperature (RT) for 30 min at high humidity (>80%) and adjacent drying at 80°C for 120 min. DNA microarrays were stored under an argon atmosphere in an exsiccator until use.
For data analysis, a gal file was produced using the TAS Application Suite software, version 2.7.1.18, from Genomic Solutions. Gal files allow the assignment of genes to the corresponding spots on the array. Further details on data analysis are given in “Data analysis” below.
(iii) Blocking of whole-genome DNA microarrays.
To avoid unspecific binding, whole-genome C. albicans SC5314 DNA microarrays were blocked using the following procedure: 0.1% Triton X-100 for 5 min at RT, 1 mM HCl for 4 min at RT, 100 mM KCl for 10 min at RT, double-distilled H2O (ddH2O) for 1 min at RT, blocking solution (Schott, Jena, Germany) for 15 min at 50°C, and subsequent ddH2O for 1 min at RT. Afterwards, slides were dried using compressed N2 and were stored dry in the dark until hybridization.
RNA isolation and precipitation.
Cell beads were destroyed using a mortar and pestle. Grounded cell powder was transferred to a 15-ml Falcon tube filled with 4 ml RLT buffer (RNeasy midikit; Qiagen, Hilden, Germany) and 40 μl β-mercaptoethanol (Merck, Darmstadt, Germany) using an applicator. Prior to use, mortar, pestle, and applicator were cooled with liquid nitrogen. Liquid nitrogen was added during the cell destruction process whenever needed to avoid thawing.
RNA was isolated using an RNeasy midikit (Qiagen, Hilden, Germany) according to the manufacturer's protocol (Yeast II). On-column DNase digestion was performed (RNase-free DNase set; Qiagen, Hilden, Germany). RNA concentration and quality were assessed photometrically (Spectra-Photometer V630; Jasco, Gross-Umstadt, Germany) and by bioanalyzer analysis (RNA 6000 Nano LabChip kit, Agilent Bioanalyzer 2100; Agilent Technologies, CA). The RNA was precipitated using lithium chloride: 500 μl 4 M LiCl solution was added to 500 μl RNA solution, and the mixture was incubated at −20°C overnight. RNA was centrifuged at 13,000 × g and 4°C for 30 min and washed three times with 70% ethanol, and the precipitate was dried for 5 min at 37°C. RNA was redissolved in 50 μl RNase-free ddH2O. Only RNA with a 260 nm/280 nm ratio of 1.8 to 2 as well as a 260 nm/230 nm ratio of 1.8 was used for DNA microarrays. Twenty-five micrograms of total RNA in a maximal volume 19 μl was used for reverse transcription and labeling.
Reverse transcription and labeling.
Identical amounts of concentrated total RNA were reverse transcribed using a LabelStar kit (Qiagen, Hilden, Germany) and labeled with Cy3-dCTP or Cy5-dCTP. All reactions were performed on ice according to the manufacturer's protocol. After reverse transcription, the reference (Cy3 or Cy5 labeled, respectively) and sample (Cy5 or Cy3 labeled, respectively) were combined, purified, and eluted using 30 μl elution buffer.
Experimental design.
In total, three biological replicates were performed. All experiments were performed as dye swaps. Hybridization experiments included an untreated reference sample and a sample of cells treated with EMC120B12.
Hybridization.
Thirty microliters cDNA eluate, 6.5 μl SDS solution (1%), 13 μl 20× SSC, and 15.5 μl RNase-free ddH2O were mixed and denatured at 100°C for 3 min. Hybridization was done overnight (16 to 20 h) at 65°C using LifterSlips hybridization coverslips (25 by 44 mm; VWR, Darmstadt, Germany). After hybridization, the LifterSlips were removed while they were rinsed for 1 min using 2× SSC, 0.2% SDS. Subsequently, DNA microarrays were washed for 10 min in 2× SSC, 0.2% SDS, for 10 min in 2× SSC, and for 10 min in 0.2× SSC. Afterwards, the DNA microarrays were dried with compressed N2 gas and stored protected from light in the dark until scanned.
Scanning process.
The completely dried DNA microarrays were scanned using a Genepix microarray scanner 4300A (resolution, 10 μm; PMT [photomultiplier tube] setting, auto-PMT; Molecular Devices, Sunnyvale, CA). Pictures of both channels were saved as 16-bit TIFF files.
Data analysis.
Raw data were created using GenePix Pro, version 7.0, imaging software (Molecular Devices, Sunnyvale, CA). The data were statistically analyzed using the software R (version 2.10.1), the limma package (version 3.2.1), and the statmod package (version 1.4.2) (31, 41). Limma applies linear models to analyze designed experiments and to identify differentially expressed genes. An empirical Bayes method was chosen to take into account the fact that in the classical t test, small variance values of the gene of interest tend to lead to high t-test statistics and therefore, erroneously, to a higher probability of significant differential expression. The parametric empirical Bayes approach (24) involves a so-called penalty value which is estimated from the mean and standard deviation of the random sample variance. This leads to an adjusted test statistic and therefore to more reliable results. Furthermore, Smyth's empirical Bayes approach (41), which includes the variance of all genes to calculate single t-test statistics, was used to obtain reproducible data. Empirical Bayes and other shrinkage methods are used to borrow information across genes, making the analyses stable even for experiments with a small number of arrays (41–43). The background correction was performed using the normexp (normal exponential model) correction with an offset of 50 (35). The data were normalized by the use of print-tip loess normalization (44). Spots which were classified to be not found by the image analysis software were ignored for fitting of the linear model.
As thousands of hypotheses can be tested with DNA microarrays, the chance to make an α error increases with the number of hypotheses. Therefore, multiple-hypothesis testing was performed (10). The false discovery rate (FDR), which takes into account an expected contingent on errors of genes being identified as differentially expressed, was selected. Multiple-hypothesis testing correction was performed by the method of Benjamini and Hochberg, which controls the FDR (4). The quotient between expected false-positive genes and all differentially expressed genes allows an adjusted P value to be calculated, and this value can be used to make predictions about differential gene expression. Genes were regarded as differentially expressed and followed up when the adjusted P values were 0.05, therefore nominally controlling the expected FDR to less than 0.05, and showed at least 2-fold up- or downregulation.
Of the 6,400 spots present in duplicate on each of the 6 replicate arrays, 3,926 spots showed a signal for all 12 replicate spots and 1,323 spots showed a signal for 11 or 10 of all 12 replicate spots in the Cy3 channel, the Cy5 channel, or both channels. This indicates that 82% of all genes are detectable with very high reproducibility on the arrays, reflecting the high quality of the arrays, and therefore are expressed under one or the other or both conditions used.
Microarray data accession number.
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/ [2, 11]) and are accessible through GEO series accession number GSE21622 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE21622).
RESULTS
Establishing an HTS-compatible activity-selectivity assay for identification of antifungals.
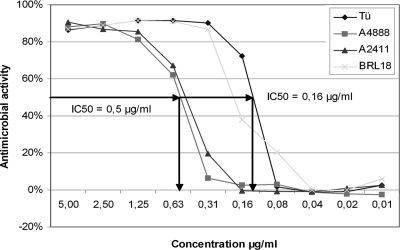
To allow the screening of compound libraries simultaneously for activity against fungal pathogens as well as tolerability by human cells, we adapted an assay originally developed for the identification of antiviral compounds (21). This assay is based on the infection of a human cell line or tissue with the pathogen and the protection of the human cells mediated by compounds of the library. Therefore, in a first step, a human cell line with robust and reproducible growth behavior and susceptibility to C. albicans infections had to be identified. Several cell lines were tested, including Caco-2, A431, A459, and HeLa cells. HeLa cells were chosen for establishing the HTS assay due to their most robust growth behavior and reproducible sensitivity to C. albicans. The other cell lines were considered suitable for confirmatory experiments after the initial screening. In a second step, the optimal multiplicity of infection (MOI) of C. albicans SC5314 on a confluent lawn of HeLa cells in 96-well plates (∼10,000 cells) was determined. For the assay, it is critical to ensure rapid and complete destruction of HeLa cells in the presence of the fungal pathogen but protection of the HeLa cells in the presence of an antifungal compound for the entire incubation period of 5 days. We used the well-established antifungal amphotericin B as a positive control, and it reliably blocked proliferation of the Candida cells employed over the entire course of the experiment (Fig. 1). To monitor the immediate effect of C. albicans on HeLa cells in the presence or absence of the compounds to be tested, the individual wells were microscopically analyzed for growth of C. albicans after 18 h to 24 h of incubation and the state of the HeLa cells (Fig. 2b). After 4 to 5 days, the survival rate of the HeLa cells was determined using fluorescein diacetate. Fluorescein diacetate is a chromogenic substance which is taken up by cells and metabolized to a fluorescent compound which can be detected using multiwell readers. Using this assay, the optimal MOI for C. albicans (SC5314) was determined to be 0.0025 (equivalent to 50 CFU per well) (data not shown). Using this MOI, a dose-response curve was generated using different sources of amphotericin B. This dose-response curve shows that the assay is highly sensitive, reflecting the differences in the drug sources (Fig. 1). The data, furthermore, clearly showed the dose-dependent effect of amphotericin B on C. albicans, and the assay gave values comparable to the standard MIC test values (IC50 in the AS assay, between 0.5 μg/ml and 0.16 μg/ml; MIC determined by CLSI between method, between 0.25 μg/ml and 0.12 μg/ml [46]). These experiments indicated that the assay is suitable for drug screening purposes. Using this setup of the assay, we screened a library of ∼100,000 compounds.
Fig. 1.
Validation of AS HTS assay. Antifungal activity of different sources of amphotericin B in the HeLa cell-based activity-selectivity assay. A 2-fold serial dilution of amphotericin B (10 different concentrations) was used to validate the assay. The four different lines represent four different batches of amphotericin B from different suppliers (Tü, Boehringer/Roche catalog no. 1081497 [no longer available]; A4888 and A2411, Sigma-Aldrich, Taufkirchen, Germany; BRL18, Gibco/Invitrogen catalog no. 15290-018). The IC50 represents the concentration of the compound required for half-maximal metabolization of fluorescein diacetate after 5 days incubation in the presence of C. albicans. One hundred percent metabolization is determined by using controls without C. albicans and without amphotericin B.
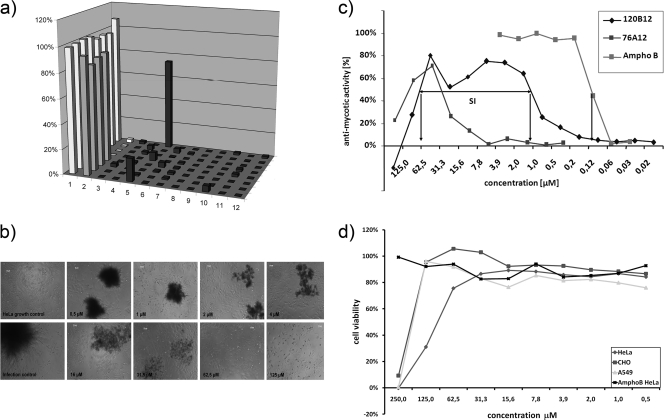
Fig. 2.
(a) Screening assay for the identification of antifungals tolerated by human cells. Typical readout of a screening plate containing 80 wells filled with compounds and 16 control wells. HeLa cells only (wells A1 to H1, growth control, light gray), HeLa cells in the presence of C. albicans (wells A2 to D2, infection control, white), HeLa cells in the presence of amphotericin B (wells E2 to H2, antifungal activity control, gray), and C. albicans (remaining wells, columns 3 to 12, dark gray). (b) Microscopic evaluation of a hit in the dose-response assay. The pictures indicate the microscopic evaluation of the growth control and infection control as well as the titration of an arbitrary hit visualizing the screening process. (c) Photometric readout of the dose-response assay. Determination of dose-response curves of two initial hits, EMC120B12 and 76A12. For the initial batch of EMC120B12, an IC50 of 1.1 μM and a CC50 of 62.5 μM could be determined, resulting in an SI of 57. For 76A12, an IC50 of 32 μM and a CC50 of 125 μM could be determined, resulting in an SI of <10. Amphotericin B serves as a control. Each compound was analyzed four times on the same plate. The assay was performed in duplicate. (d) Inhibitory effects of EMC120B12 on three different mammalian cell lines. EMC120B12 was tested against three cell lines for inhibitory effects in the absence of C. albicans. HeLa cells show the highest sensitivity to EMC120B12 (CC50, ∼62.5), whereas A549 and CHO-K1 show higher resistance (CC50, ∼125 μM).
Screening of a compound library for antifungal compounds.
The compound library available consisted of more than 35,000 heterocyclic compounds with more than 90% purity and 65,000 compounds provided as mixtures of several combinatorial compounds. These individual compounds or compound collections were screened for antifungal activity against C. albicans and tolerability by HeLa cells by simultaneous incubation of the human cell line with C. albicans and the compound samples (∼10 μM) for 4 to 5 days as described in the Materials and Methods section. A cutoff of 40% of total fluorescence intensity per well in comparison to the control wells containing only HeLa cells was chosen to select compounds preventing growth of C. albicans to a significant extent without strongly impairing the vitality of HeLa cells (hits) (Fig. 2a). All primary hits were retested, and if they were positive, resynthesis of the compounds was performed in order to verify the activity of the respective hit. The resynthesized compounds were evaluated in a dose-response assay by both microscopic evaluation (Fig. 2b) and spectrophotometrically (Fig. 2c) in order to determine the IC50 in the setting of the screening assay (by determining the concentration of the compound required to ensure the vitality of 50% of HeLa cells due to growth inhibition of the pathogen after 5 days) and CC50 of the compound (concentration of the compound resulting in a 50% reduction of the metabolic activity of HeLa cells due to the effects of the compound). Hits with a high selectivity (>50) for antifungal activity were selected for further studies (Fig. 2c). The selectivity index (SI) is defined as the ratio between the CC50 and IC50. All hits were derived from the collection of individual compounds. Positively tested compound collections did not result in reproducible hits. The hit rate with regard to the individual compounds was approaching 1:1,000. In this work, we focus on one compound, EMC120B12, an (S)-2-aminoalkyl benzimidazole derivative [(S)-2-(1-aminoisobutyl)-1-(3-chlorobenzyl) benzimidazole]. This compound showed high antifungal activity and low cytotoxicity, resulting in a high selectivity index (CC50/IC50) suitable for further investigations (IC50 = 1.1 μM, CC50 = 62.5 μM, SI = 57) (Fig. 2; Table 1). New synthesis and purification strategies resulted in an even better SI of 130 due to a higher CC50 of 97.5 μM and a slightly lower IC50 of 0.75 μM, indicating the improved removal of potentially inhibitory side products during synthesis. This batch of the compound was used for all further studies.
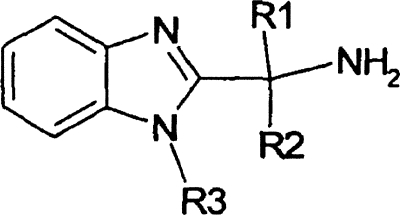
Table 1.
Selection of structural analogues tested modifying R1, R2, and R3
ND, not determined.
Inhibitory effects of EMC120B12 on other cell lines.
Mammalian cells are highly differentiated and may show different susceptibilities to the same chemical compound. In order to compare the sensitivity or resistance of HeLa cells to the hit compound discovered with other cell lines, we explored the cytotoxicity of EMC120B12 to two other mammalian cell lines, CHO-K1 and A549. We determined their CC50s in the absence of fungal pathogens. In both cases the CC50 was even higher than the CC50 on HeLa cells (HeLa cell CC50 = 97 μM, A549 cell CC50 = 125 μM, and CHO-K1 cell CC50 = 125 μM), indicating that HeLa cells may represent rather a more sensitive than resistant cell line with regard to EMC120B12 (Fig. 2d).
Structure-activity relationship of EMC120B12.
To identify the critical structural features of this new molecule, several structural analogues of EMC120B12 [(S)-2-(1-aminoisobutyl)-1-(3-chlorobenzyl) benzimidazole] were synthesized and tested by determining the respective dose-response curves. Especially the phenyl ring (Table 1, R3) and its substituents as well as the methylpropylamine residue (Table 1, R1 and R2) were modified. For all compounds the respective IC50s, CC50s, and SIs were determined in the presence of C. albicans using the activity-selectivity assay. A summary of results for a small subset of the structural analogues of EMC120B12 and their respective activities are given in Table 1. The entire set of compounds tested and their synthesis are in press elsewhere (3). These studies showed that the essential parts of the molecule are the primary amine group and the optically active center of the molecule to which the amine group is bound. The respective enantiomer [(R)-2-(1-aminoisobutyl)-1-(3-chlorobenzyl) benzimidazole] was shown to be not active (Table 1). Also, the position and nature of the substituent on the phenyl ring, among other parameters, are crucial for activity. The highest activity and selectivity were determined for the initial hit, EMC120B12, which in turn was followed up further. Two additional compounds, (S)-2-(1-aminoisobutyl)-1-(3-methylbenzyl) benzimidazole and (S)-2-(1-aminoisobutyl)-1-(3-fluorobenzyl) benzimidazole, showed comparable activity but reduced selectivity, whereas all other structural analogues showed substantially less activity or significant toxicity in the assay used (data not shown).
Using the activity-selectivity assay, we also tested the response of A. fumigatus to EMC120B12 and it structural analogues. However, growth of A. fumigatus was not blocked by EMC120B12, indicating that it is resistant to this type of benzimidazole derivative (data not shown).
Correlation with MIC values according to a standardized EUCAST protocol.
The AS assay described in this work measures the survival rate of the human cells and not necessarily the MIC of the compounds for the fungal pathogen. In order to be able to correlate the results from the AS assay, we performed a standard MIC determination of EMC120B12 using the protocol according to EUCAST. In addition, we compared their activities to the activity of a reference compound with well-established antifungal activity, fluconazole. MICs were determined for a strain collection consisting of seven type strains (C. albicans ATCC 90028, C. albicans DSMZ 11949, C. glabrata ATCC 90030, C. guilliermondii ATCC 90877, C. parapsilosis ATCC 22019, C. parapsilosis ATCC 90018, and Issatchenki orientalis ATCC 6258) as well as 143 clinical isolates of different Candida spp., as described in Table 2, which were mostly isolated from septic patients during testing of blood by culture or patients with urinary tract infections. This included a set of 10 specifically selected C. albicans strains resistant to azoles. We measured a range of MICs of from 0.125 to 128 μg/ml for all strains included in the analysis of EMC120B12, with a median of 1.06 μg/ml. In particular, the MICav of EMC120B12 for the type strains was 1.2 μg/ml, while for the clinical isolates we found a MICav of 1.05 μg/ml. The MICav for all C. albicans clinical isolates tested (61 isolates) was determined to be 1.08 μg/ml. A similar value resulted for fluconazole (fluconazole MICav, 1.10 μg/ml). Looking only at the MICav of the C. albicans clinical isolates not selected for resistance (52 isolates), a MICav for EMC120B12 of 0.87 μg/ml resulted. Interestingly, focusing on the 18 strains with a MICav of >4 for fluconazole, the MICav for EMC120B12 equals 13.5 μg/ml, whereas the MICav for fluconazole equals 33.3 μg/ml, indicating a higher susceptibility of azole-resistant strains to EMC120B12. At the level of non-C. albicans species, C. lusitaniae was the most susceptible species (MICav, 0.25 μg/ml; 3 strains), whereas the rarely isolated C. pelliculosa was the most resistant species in our collection (MICav, 16 μg/ml; 2 strains). Most interestingly, the activity of EMC120B12 is much higher than the activity of fluconazole against C. krusei (EMC120B12 MICav, 1.25 μg/ml; fluconazole MICav, 22.6 μg/ml) and to a lesser extent against C. glabrata (EMC120B12 MICav, 1.06 μg/ml; fluconazole MICav, 4.0 μg/ml), whereas it is in a similar range for C. parapsilosis (EMC120B12 MICav, 0.81 μg/ml, fluconazole MICav, 0.41 μg/ml). Summaries of the MIC distributions of the isolates of C. albicans, C. glabrata, C. parapsilosis, and C. krusei in comparison to the MIC distributions for fluconazole are shown in Tables 2 and 3.
Table 3.
Relative MIC distribution of EMC120B12 and fluconazole
| Species | Compound | % isolates with the following MIC (μg/ml): |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.125 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | ||
| C. albicans (n = 61) | EMC120B12 | 33 | 13 | 8 | 7 | 5 | 10 | 8 | 3 | 3 | 3 | 7 |
| Fluconazole | 30 | 23 | 10 | 3 | 2 | 3 | 0 | 11 | 8 | 7 | 3 | |
| C. glabrata (n = 12) | EMC120B12 | 17 | 25 | 8 | 8 | 0 | 25 | 0 | 17 | 0 | 0 | 0 |
| Fluconazole | 17 | 0 | 0 | 0 | 17 | 25 | 17 | 8 | 0 | 17 | 0 | |
| C. parapsilosis (n = 36) | EMC120B12 | 11 | 6 | 28 | 22 | 28 | 3 | 3 | 0 | 0 | 0 | 0 |
| Fluconazole | 8 | 47 | 31 | 6 | 3 | 0 | 3 | 3 | 0 | 0 | 0 | |
| C. krusei (n = 21) | EMC120B12 | 0 | 5 | 0 | 29 | 48 | 14 | 5 | 0 | 0 | 0 | 0 |
| Fluconazole | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 19 | 71 | 5 | 0 | |
The MIC values obtained using the EUCAST protocol correlated nicely with the results observed in the AS assay. For example, the average MIC of EMC120B12 for the C. albicans isolates was determined to be 1.1 μg/ml (EUCAST). The IC50 in the activity selectivity assay was determined to be 0.75 μM, which corresponds to 0.23 μg/ml (EMC120B12 molecular weight, 313.8), reflecting 50% survival of the human cells in DMEM supplemented with 10% FCS. Significant growth of the pathogen has not been observed under this condition. Thus, the IC values generated in the AS assays are in the same range as the MIC values determined according to the EUCAST (13) or CLSI (6) protocol, which show significant differences themselves.
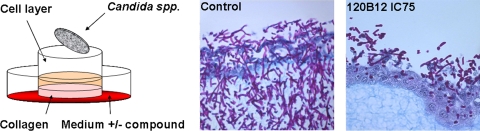
Effectiveness of EMC120B12 in 3D tissue models.
In order to test if EMC120B12 is able to penetrate tissue as well as membranes and therefore will have a high bioavailability, we added the compound at a concentration corresponding to the IC75 (2 μg/ml, according to the AS assay) to a 3D tissue model. The model is composed of a basolateral collagen matrix sitting on a membrane enabling medium supply from the bottom. On top of the collagen, a layer of A431 cells was grown to confluence (9). Several tissue models were infected with C. albicans as described in the Materials and Methods section. At the time point of infection, the medium for the tissue models was replaced by medium containing EMC120B12 at a final concentration of 2 μg/ml, taking into account the volume of the tissue model, or were mock treated. Figure 3 clearly shows the effect of EMC120B12, supplied basolaterally, on the ability of C. albicans to invade into the tissue. Only in the absence of EMC120B12 was the pathogen able to invade into the tissue and to destroy it within 48 h. In the presence of EMC120B12, C. albicans was barely able to proliferate and showed only a minor ability to penetrate into the tissue model. This indicates that EMC120B12 is capable of penetrating through both the collagen and the layer of A431 cells, inhibiting growth of C. albicans at the surface of the tissue model.
Fig. 3.
EMC120B12 is effective in 3D tissue equivalents. (Left) Schematic representation of the general setup of the infection model; (middle and right) representative histological sections of the infection models in the presence and absence of EMC120B12 (IC75) after infection with C. albicans for 48 h.
Global transcription analysis to identify the target of EMC120B12.
In order to identify the potential target of EMC120B12, we performed transcriptional profiling in the presence or absence of the compound as described in the Materials and Methods section. Time course growth studies of C. albicans SC5314 were performed with or without EMC120B12 and controlled by microscopy (1 h, 3 h, 5 h, 24 h; data not shown). Microscopic pictures showed that EMC120B12 had a significant influence on the growth behavior of SC5314 after 3 h of incubation in the presence of 0.4 μM EMC120B12 (IC25), without damaging a major amount of Candida, as judged by microscopic examination. Therefore, this point in time was chosen for transcriptional profiling. The statistical analysis of the data revealed significant differences between the EMC120B12-treated C. albicans SC5314 culture and the untreated reference culture. Genes were regarded as differentially expressed and followed up when their Benjamini-Hochberg (4) adjusted P value from an empirical Bayes moderated t test was ≤0.05 and they showed at least 2-fold up- or downregulation. The calculated average log2 ratios of all considered genes ranged from −2.39 to 3.79. These values corresponded to an up to 5.25-fold downregulation and an up to 13.84-fold upregulation (Table 4).
Table 4.
Differentially expressed genes of EMC120B12-treated C. albicans SC5314a
| Gene group and gene | Function | Fold expression | Adjusted P value |
|---|---|---|---|
| Role in ergosterol biosynthesis | |||
| ERG1 | Squalene epoxidase, catalyzes epoxidation of squalene to 2,3(S)-oxidosqualene in the ergosterol biosynthetic pathway | 4.34 | 0.000108 |
| ERG10 | Protein similar to acetyl-CoA acetyltransferase, role in ergosterol biosynthesis | 2.60 | 0.000948 |
| ERG11 | Lanosterol 14-alpha-demethylase, member of cytochrome P450 family that functions in ergosterol biosynthesis | 8.41 | 0.000000 |
| ERG13 | Protein similar to S. cerevisiae Erg13p, which is involved in ergosterol biosynthesis | 4.91 | 0.000000 |
| ERG2 | C-8 sterol isomerase, enzyme of ergosterol biosynthesis pathway | 2.96 | 0.006752 |
| ERG24 | C-14 sterol reductase, has a role in ergosterol biosynthesis | 3.32 | 0.001618 |
| ERG25 | Putative C-4 methyl sterol oxidase with role in C-4 demethylation of ergosterol biosynthesis intermediates, based on similarity to Saccharomyces cerevisiae Erg25p | 2.64 | 0.000193 |
| ERG251 | Predicted ORF in assemblies 19, 20, and 21; ERG251 appears to be an allele or second copy of the ERG25 gene (39) | 4.66 | 0.000024 |
| ERG26 | C-3 sterol dehydrogenase, catalyzes the second of three steps required to remove two C-4 methyl groups from an intermediate in ergosterol biosynthesis | 2.09 | 0.000224 |
| ERG3 | C-5 sterol desaturase | 6.95 | 0.000036 |
| ERG4 | Protein described to be similar to sterol C-24 reductase | 3.08 | 0.001412 |
| ERG5 | Putative C-22 sterol desaturase, fungal C-22 sterol desaturases are cytochrome P450 enzymes of ergosterol biosynthesis | 3.05 | 0.000069 |
| ERG6 | Delta(24)-sterol C-methyltransferase, converts zymosterol to fecosterol in ergosterol biosynthesis by methylating position C-24 | 11.36 | 0.000028 |
| ERG9 | Putative farnesyl-diphosphate farnesyl transferase (squalene synthase) involved in the sterol biosynthesis pathway | 2.57 | 0.022424 |
| UPC2 | Transcription factor involved in regulation of ergosterol biosynthetic genes and sterol uptake | 3.47 | 0.000159 |
| Described to show changed transcript levels in response to antimycotics | |||
| CHT2 | Chitinase | −3.90 | 0.000036 |
| CRH11 | Putative ortholog of S. cerevisiae Crh1p | 2.02 | 0.006851 |
| CSH1 | Member of aldo- and ketoreductase family, similar to aryl alcohol dehydrogenases | 2.12 | 0.005475 |
| DDR48 | Immunogenic stress-associated protein | 13.84 | 0.000000 |
| FRP1 | Predicted ferric reductase | 2.01 | 0.000820 |
| FTH1 | Protein not essential for viability, similar to S. cerevisiae Fth1p (putative high-affinity iron transporter for intravacuolar stores of iron) | 2.03 | 0.000472 |
| HYR1 | Nonessential, GPI anchored, predicted cell wall protein | 2.72 | 0.003792 |
| NHP6A | Protein described to be a nonhistone chromatin component | −2.04 | 0.001358 |
| orf19.6688 | Decreased transcription is observed upon benomyl treatment or in an azole-resistant strain that overexpresses MDR1 | −3.39 | 0.000147 |
| orf19.7504 | Predicted ORF in assemblies 19, 20, and 21 | 2.46 | 0.007689 |
| orf19.3475 | Described to be a Gag-related protein | −2.20 | 0.046451 |
| orf19.3737 | Predicted ORF in assemblies 19, 20, and 21 | 2.15 | 0.025789 |
| PGA7 | Protein described to be a putative precursor of a hyphal surface antigen | 2.46 | 0.000472 |
| PHR2 | Glycosidase, role in cell wall structure | 4.38 | 0.000116 |
| POL30 | Protein described as proliferating cell nuclear antigen (PCNA) | −2.00 | 0.006851 |
| RTA2 | Putative floppase | 4.18 | 0.000312 |
| RTA3 | Similar to S. cerevisiae Rta1p (role in 7-aminocholesterol resistance) and Rsb1p (flippase) | 3.18 | 0.030754 |
| Other functions | |||
| ARO10 | Aromatic decarboxylase of the Ehrlich fusel oil pathway of aromatic alcohol biosynthesis | −5.25 | 0.000734 |
| CDG1 | Protein described to be similar to cysteine dioxygenases | −2.04 | 0.000281 |
| CHA1 | Protein similar to serine/threonine dehydratases, catabolic | −2.46 | 0.000734 |
| CIP1 | Protein with slight similarity to plant isoflavone reductase | −2.19 | 0.026148 |
| FAV1 | Protein with weak similarity to S. cerevisiae Fus2p | 2.04 | 0.028653 |
| GCV2 | Putative protein of glycine catabolism | −2.18 | 0.010078 |
| HSP31 | Protein repressed during the mating process | −2.83 | 0.016218 |
| HTA2 | Protein described as histone H2A | −2.32 | 0.003792 |
| MAL2 | Alpha-glucosidase that hydrolyzes sucrose | −2.96 | 0.000166 |
| PGA23 | Putative GPI-anchored protein of unknown function | 5.50 | 0.000009 |
| PGA26 | Putative GPI-anchored protein of unknown function | −3.35 | 0.000260 |
| PST3 | Putative flavodoxin | 2.10 | 0.000028 |
| RBT1 | Putative cell wall protein with similarity to Hwp1p | −2.63 | 0.000177 |
| SET3 | Protein similar to S. cerevisiae Set3p, which is an NAD-dependent histone deacetylase | 3.19 | 0.000005 |
| Predicted ORF, protein of unknown function | |||
| MAL31 | Putative protein of unknown function | −2.75 | 0.003792 |
| orf19.2125 | Predicted ORF in assemblies 19, 20, and 21 | 3.12 | 0.000159 |
| orf19.2989 | Predicted ORF in assemblies 19, 20, and 21 | −2.34 | 0.020304 |
| orf19.4476 | Predicted ORF in assemblies 19, 20, and 21 | 2.34 | 0.007689 |
| orf19.5799 | Predicted ORF in assemblies 19, 20, and 21 | 2.59 | 0.000004 |
| orf19.6840 | Predicted ORF in assemblies 19, 20, and 21 | 2.43 | 0.000285 |
| orf19.6852.1 | ORF added to assembly 21 on the basis of comparative genome analysis | 2.09 | 0.019724 |
| orf19.7455 | Predicted ORF in assemblies 19, 20, and 21 | 3.07 | 0.000159 |
| orf19.90 | Predicted ORF in assemblies 19, 20, and 21 | 2.93 | 0.000633 |
Differential expression was determined by comparison to untreated reference samples. Statistical significance was an adjusted P value of ≤0.05; positive values, mRNA level higher than the reference state; negative values, mRNA level lower than the reference state. acetyl-CoA, acetyl coenzyme A; GPI, glycosylphosphatidylinositol.
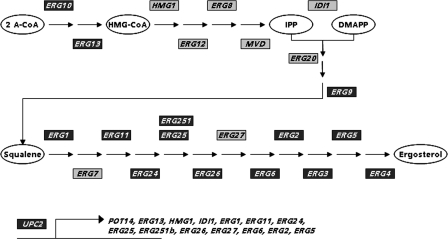
We could detect that in total the transcription of 55 genes changed significantly in the EMC120B12-treated sample compared to the reference sample (Table 4). Thirty-eight genes were upregulated, 17 genes were downregulated, and 15 of the transcripts could be assigned to the ergosterol biosynthesis pathway, each of which was induced (Fig. 4). This includes UPC2, a known regulator of the ergosterol pathway (40). Additionally, 17 genes identified were described to show changed transcript levels in response to antifungals, 9 are predicted open reading frames (ORFs) or encode proteins of unknown function, and 14 could be assigned to other functions.
Fig. 4.
Genes of the ergosterol pathway upregulated in the presence of EMC120B12. All transcripts identified to be upregulated in the presence of EMC120B12 in the schematic representation of the ergosterol pathway of C. albicans are shown in dark gray. In addition, the transcript levels of one of the major regulators of the ergosterol (ERG) pathway, UPC2, were identified as upregulated as well. A-CoA, acetyl coenzyme A; HMG-CoA, 3-hydroxy-3-methyl-glutaryl coenzyme A; IPP, Δ3-isopentenyl pyrophosphate; DMAPP, dimethylallyl pyrophosphate.
Gene ontology (GO) term analysis (http://www.candidagenome.org/cgi-bin/GO/goTermFinder) revealed localization of the gene products mainly in the endoplasmic reticulum (ER; cluster frequency, 29.1%; background frequency, 4.8%; several enzymes of the ergosterol pathway are localized in the ER), the cell surface (cluster frequency, 18.2%; background frequency, 3.1%), the cell wall (cluster frequency, 12.7%; background frequency, 2.3%), and the plasma membrane (cluster frequency, 25.5%; background frequency, 7.6%).
These results indicate that the novel (S)-2-aminoalkyl benzimidazoles identified have a strong impact on membrane and cell wall biosynthesis or structure, similar to what has been shown for the azoles (for a review, see reference 37). Taken together, our results show that we could identify a novel group of benzimidazole derivatives with antifungal activity comparable to that of well-known potent azoles using a screen based on simple host-pathogen interaction models.
DISCUSSION
In this work we adapted a screening assay developed for the identification of antiviral compounds for use in screening for antifungals. Using this novel screening system, we screened a compound library containing pure compounds based on heterocyclic scaffolds as well as mixtures of compounds based on combinatorial chemistry for antifungal activity tolerated by human cells. We identified a new lead structure, from which a group of (S)-2-aminoalkyl benzimidazole derivatives with high antifungal activities was defined. The screening is based on mimicking a minimal site of infection, with the screening assay containing human cells or tissue equivalents as well as the pathogen and the compounds to be tested. The readout is based on the metabolic activity of host cells by using a chromophore which is metabolized in vivo. Using this approach, all potential targets in both the host and the pathogen are addressed at the same time. Therefore, this screening assay is well suited to identify tolerable compounds that have as few side effects on host tissue as possible but that more or less completely block proliferation of the pathogen. This assay has been performed in 96-well plates in an HTS-like manner. Transfer of the assay to a 384-well format is possible, in principle. Screening of more than 35,000 pure compounds gave the best results and several hits (the initial hit rate approached 0.1%), of which the one with the highest activity and best SI value was followed up further. Screening of mixtures of compounds generated as combinatorial libraries in our hands did not result in any reproducible hits. This may be due to a reduced concentration of compounds present in the individual well or the presence of individual toxic compounds within this mix, resulting in a negative result of the assay.
This assay can also be used to titrate the compounds identified for fungistatic or fungicidal effects as well as for growth inhibition or toxicity against the human cells or tissue equivalents employed. Therefore, it was previously termed an activity-selectivity (AS) assay during its development for identification of antivirals (22). We could show that for several cell lines tested, including CHO-K1, A549, and HeLa cells, the metabolic activity of the cell lines is compromised only at high concentrations of the compound, with CC50s (cytotoxic concentrations representing 50% loss of metabolic activity of the cell line) being between 97 μM and 125 μM, depending on the cell line. With an inhibitory concentration representing 50% loss of protection of the cell line from the pathogen (IC50) of 0.75 μM, EMC120B12 shows a high potency against C. albicans (SC5314), resulting in a selectivity index of 130, which is considered appropriate for drug development (22).
To compare the AS assay with conventional MIC tests, we performed MIC assays according to the EUCAST guidelines (13), modified as described in the Materials and Methods section. Furthermore, the MIC determination was performed for a relevant collection of clinical isolates isolated from systemic and urinary tract infections, including a set of specifically selected C. albicans strains resistant to fluconazole (143 strains) plus 7 type strains. Our results showed that EMC120B12 has a similar antifungal activity as fluconazole, a well-known antifungal drug. It acts against a broad spectrum of pathogenic yeasts with an average MICav value of 1.06 μg/ml for all clinical isolates tested and a MICav value of 1.1 μg/ml for all C. albicans isolates analyzed (Table 3). Interestingly, EMC120B12 in general showed a greater antifungal effect than fluconazole against 18 azole-resistant C. albicans strains (MICs > 4 μg/ml) analyzed (Tables 2 and 3). This was also observed for C. glabrata. About 50% of the resistant C. albicans strains (8 strains) showed strong cross-resistance between EMC120B12 and fluconazole, whereas 25% (4 strains) showed no cross-resistance. Azole resistance in C. albicans can be a multifactorial event, including changes in transport activities for multidrug resistance (MDR), changes in regulatory genes or their expression, and changes in genes of the ergosterol biosynthesis pathway. The cross-resistance observed suggests similar resistance mechanisms for both compounds. This is supported by our transcriptional data, suggesting that the ergosterol pathway is the target. For 10 strains specifically selected for their resistance to fluconazole (indicated with asterisks in Table 2), preliminary data for resistance mechanisms are available. For strains 11 to 14, 17, and 18, an enhanced drug efflux pump activity could be observed using rhodamine G accumulation/efflux tests (data not shown). Strains 13 and 14 showed enhanced MDR activity, as determined by testing for susceptibility to brefeldin A (data not shown). For strains 9, 15, and 16, preliminary evidence for mutations in ERG11 exist. The differences between the susceptibilities of these strains to EMC120B12 and fluconazole in these strains are limited, indicating that especially drug efflux mechanisms and alteration of the ergosterol pathway have highly similar effects on both compounds. This, however, clearly has to be investigated in more detail.
Especially, the limited cross-resistance observed with so far uncharacterized resistant C. albicans isolates and non-C. albicans species may also suggest other mechanisms of action for EMC120B12. They still have to be considered until the molecular target is identified directly.
Interestingly, non-C. albicans strains showed a higher susceptibility toward EMC120B12 than fluconazole, as the MICav values were 1.05 μg/ml and 1.95 μg/ml, respectively. Especially for C. krusei, EMC120B12 exhibited much higher antifungal activity than fluconazole, with MICav values of 1.60 μg/ml and 22.63 μg/ml, respectively. MIC80 values of EMC120B12 for C. krusei strains were distributed largely below 2 μg/ml (Table 2). Since C. krusei is notoriously resistant to fluconazole, EMC120B12 might be developed into an alternative drug.
Benzimidazoles are a large group of compounds, some of which have been widely used as antifungal agents in plant protection. However, in medicine they are not used for treatment of fungal infections but rather as antihelminthics (mebendazole, eskazole). Many other derivatives have previously been described to have antifungal activities (12, 39). In addition, an antineoplastic activity of certain benzimidazoles, like nocodazole, has been identified, pointing to a certain level of cytotoxic effects. For several of these compounds, for example, benomyl or nocodazole, their mechanism of action was elucidated. They have been shown to bind to microtubules and inhibit the proper formation of the cytoskeleton, thereby interfering with cellular organization, especially during cell division (8, 16, 18, 20).
The compounds identified in this work have not been described before and are unique through their chiral alkylamine residue, at the 2 position of the benzimidazole ring, which is critical for activity of the compounds. Only the (S)-2-(1-aminoisobutyl)-1-(3-chlorobenzyl) benzimidazole and not the (R) enantiomer showed antifungal activity in the activity-selectivity assay. Interestingly, our analyses indicate that EMC120B12 might not target the microtubules, as described for other benzimidazoles and outlined above. On a transcriptional level, we have no evidence for activities against microtubules, but rather, we have evidence that the ergosterol pathway is the target of EMC120B12. GO term analysis revealed that of the 38 genes upregulated during challenge with EMC120B12, 15 genes could be assigned to ergosterol biosynthesis. GO term analysis, furthermore, revealed an impact of EMC120B12 on gene products localized to the cell surface, the cell wall, and the plasma membrane. These results indicate that the novel (S)-2-aminoalkyl benzimidazoles identified have a strong impact on membrane and cell wall biosynthesis or structure, similar to what has been shown for fluconazole, e.g., by Angiolella et al. (1). They showed by analysis of cell wall protein composition that fluconazole resulted in increased expression of two glucanases as well as an increased amount of polydisperse, highly glycosylated material related to cell wall proteins. Our results also show on a transcriptional level that a set of cell wall proteins is differentially regulated in the presence of EMC120B12. These results indicate, similar to the results by Angiolella et al. (1), a rearrangement and modification of the cell wall structure. These modifications could also be observed for other antifungal compounds and were described by others not to depend on a specific action of the antimycotic but rather might reflect some general indirect metabolic changes provoked by exposure to antimycotics as cell-stressing agents (for a review, see reference 37). Therefore, EMC120B12 might also act only indirectly on cell wall biogenesis.
The results of the MIC tests performed according to EUCAST (13), modified as described in the Materials and Methods section, indicate that the antifungal activity of EMC120B12 is not limited just to the standard reference strain, which was used for the screening procedure. It suggests, rather, that EMC120B12 is active against a broad spectrum of clinically relevant Candida sp. isolates which were represented in our study by a collection of 150 clinical isolates from cultures of blood from septic patients. Most interestingly, non-C. albicans species, especially C. krusei, were much more sensitive to EMC120B12 than fluconazole. Of importance is that the compounds identified have already gone through a first cytotoxicity assay in parallel with screening, which is not the case for most of the primary hit compounds reported in the literature. Therefore, this structure might be an interesting lead for further development toward detection of a new clinically relevant antifungal drug. Further tests will address the potential of EMC120B12 for development as an antimycotic drug using in vivo models of rodents or its application in combination with other antifungals.
ACKNOWLEDGMENTS
We thank the National Reference Center for Systemic Mycoses, especially Oliver Bader, for kindly providing strains and sharing information prior to publication.
This work was supported within the frame of the BioProfile-Program Stuttgart/Neckar-Alb (FKZ 0313709), as well as within the BMBF-Program Basisinnovationen in der genombasierten Infektionsforschung (FKZ 0315221). S.R. is supported by the DFG RU608/4 within the SPP1160.
Footnotes
Published ahead of print on 11 July 2011.
REFERENCES
- 1. Angiolella L., et al. 2002. Identification of major glucan-associated cell wall proteins of Candida albicans and their role in fluconazole resistance. Antimicrob. Agents Chemother. 46:1688–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barrett T., et al. 2009. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 37:D885–D890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bauer J., et al. High-throughput screening-based identification and structure-activity relationship-characterization defined (S)-2-(1-aminoisobutyl)-1-(3-chlorobenzyl) benzimidazole as a highly antimycotic agent, non-toxic to cell lines. J. Med. Chem., in press [DOI] [PubMed] [Google Scholar]
- 4. Benjamini Y., Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57:289–300 [Google Scholar]
- 5. Brown A. J., Odds F. C., Gow N. A. 2007. Infection-related gene expression in Candida albicans. Curr. Opin. Microbiol. 10:307–313 [DOI] [PubMed] [Google Scholar]
- 6. CLSI 2008. Reference method for broth dilution antifungal susceptibility testing of yeasts; approved standard, 3rd ed CLSI document M27-A3, vol. 28 CLSI, Wayne, PA [Google Scholar]
- 7. Cornely O. A., et al. 2007. Posaconazole vs. fluconazole or itraconazole prophylaxis in patients with neutropenia. N. Engl. J. Med. 356:348–359 [DOI] [PubMed] [Google Scholar]
- 8. Davidse L. C. 1986. Benzimidazole fungicides: mechanism of action and biological impact. Annu. Rev. Phytopathol. 24:43–65 [Google Scholar]
- 9. Dieterich C., et al. 2002. In vitro reconstructed human epithelia reveal contributions of Candida albicans EFG1 and CPH1 to adhesion and invasion. Microbiology 148:497–506 [DOI] [PubMed] [Google Scholar]
- 10. Dudoit S., Popper Schaffer J., Boldrick J. C. 2003. Multiple hypothesis testing in microarray experiments. Stat. Sci. 28:71–103 [Google Scholar]
- 11. Edgar R., Domrachev M., Lash A. E. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30:207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Elnima E. I., Zubair M. U., Al-Badr A. A. 1981. Antibacterial and antifungal activities of benzimidazole and benzoxazole derivatives. Antimicrob. Agents Chemother. 19:29–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. EUCAST 2008. Subcommittee on Antifungal Susceptibility Testing (AFST) of the ESCMID European Committee for Antimicrobial Susceptibility Testing (EUCAST). EUCAST definitive document EDef 7.1: method for the determination of broth dilution MICs of antifungal agents for fermentative yeasts. Clin. Microbiol. Infect. 14:398–405 [DOI] [PubMed] [Google Scholar]
- 14. Fonzi W. A., Irwin M. Y. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Francois I. E., Aerts A. M., Cammue B. P., Thevissen K. 2005. Currently used antimycotics: spectrum, mode of action and resistance occurrence. Curr. Drug Targets 6:895–907 [DOI] [PubMed] [Google Scholar]
- 16. Guthrie B. A., Wickner W. 1988. Yeast vacuoles fragment when microtubules are disrupted. J. Cell Biol. 107:115–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hernandez R., Rupp S. 2009. Human epithelial model systems for the study of Candida infections in vitro. Part II. Histologic methods for studying fungal invasion. Methods Mol. Biol. 470:105–123 [DOI] [PubMed] [Google Scholar]
- 18. Hollomon D. W., Butters J. A., Barker H., Hall L. 1998. Fungal beta-tubulin, expressed as a fusion protein, binds benzimidazole and phenylcarbamate fungicides. Antimicrob. Agents Chemother. 42:2171–2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hope W. W., Billaud E. M., Lestner J., Denning D. W. 2008. Therapeutic drug monitoring for triazoles. Curr. Opin. Infect. Dis. 21:580–586 [DOI] [PubMed] [Google Scholar]
- 20. Jacobs C. W., Adams A. E., Szaniszlo P. J., Pringle J. R. 1988. Functions of microtubules in the Saccharomyces cerevisiae cell cycle. J. Cell Biol. 107:1409–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kleymann G., et al. 2002. New helicase-primase inhibitors as drug candidates for the treatment of herpes simplex disease. Nat. Med. 8:392–398 [DOI] [PubMed] [Google Scholar]
- 22. Kleymann G., Werling H. O. 2004. A generally applicable, high-throughput screening-compatible assay to identify, evaluate, and optimize antimicrobial agents for drug therapy. J. Biomol. Screen. 9:578–587 [DOI] [PubMed] [Google Scholar]
- 23. Kojima K., et al. 2004. Fungicide activity through activation of a fungal signalling pathway. Mol. Microbiol. 53:1785–1796 [DOI] [PubMed] [Google Scholar]
- 24. Lönnstedt I., Speed T. P. 2002. Replicated microarray data. Stat. Sin. 12:31–46 [Google Scholar]
- 25. Moosa M. Y., Sobel J. D., Elhalis H., Du W., Akins R. A. 2004. Fungicidal activity of fluconazole against Candida albicans in a synthetic vagina-simulative medium. Antimicrob. Agents Chemother. 48:161–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mylonakis E., Casadevall A., Ausubel F. M. 2007. Exploiting amoeboid and non-vertebrate animal model systems to study the virulence of human pathogenic fungi. PLoS Pathog. 3:e101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Okoli I., et al. 2009. Identification of antifungal compounds active against Candida albicans using an improved high-throughput Caenorhabditis elegans assay. PLoS One 4:e7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pasqualotto A. C., Denning D. W. 2008. New and emerging treatments for fungal infections. J. Antimicrob. Chemother. 61(Suppl. 1):i19–i30 [DOI] [PubMed] [Google Scholar]
- 29. Pfaller M. A., Diekema D. J. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 20:133–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pfaller M. A., et al. 2005. Results from the ARTEMIS DISK Global Antifungal Surveillance Study: a 6.5-year analysis of susceptibilities of Candida and other yeast species to fluconazole and voriconazole by standardized disk diffusion testing. J. Clin. Microbiol. 43:5848–5859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. R Development Core Team 2009, posting date R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 32. Rex J. H., Pfaller M. A., Rinaldi M. G., Polak A., Galgiani J. N. 1993. Antifungal susceptibility testing. Clin. Microbiol. Rev. 6:367–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rex J. H., et al. 2000. Practice guidelines for the treatment of candidiasis. Infectious Diseases Society of America. Clin. Infect. Dis. 30:662–678 [DOI] [PubMed] [Google Scholar]
- 34. Rios J. L., Recio M. C., Villar A. 1988. Screening methods for natural products with antimicrobial activity: a review of the literature. J. Ethnopharmacol. 23:127–149 [DOI] [PubMed] [Google Scholar]
- 35. Ritchie M. E., et al. 2007. A comparison of background correction methods for two-colour microarrays. Bioinformatics 23:2700–2707 [DOI] [PubMed] [Google Scholar]
- 36. Rupp S. 2007. Interactions of the fungal pathogen Candida albicans with the host. Future Microbiol. 2:141–151 [DOI] [PubMed] [Google Scholar]
- 37. Rupp S. 2008. Transcriptomics of the fungal pathogens focusing on Candida albicans human and animal relationships, p. 187–222 In Brakhage A., Zipfel P. (ed.), The mycota, 2nd ed., vol. 7 Human and animal relationships. Springer-Verlag, Berlin, Germany [Google Scholar]
- 38. Sanglard D., Odds F. C. 2002. Resistance of Candida species to antifungal agents: molecular mechanisms and clinical consequences. Lancet Infect. Dis. 2:73–85 [DOI] [PubMed] [Google Scholar]
- 39. Sharma D., et al. 2009. Synthesis, antimicrobial and antiviral activity of substituted benzimidazoles. J. Enzyme Inhib. Med. Chem. 24:1161–1168 [DOI] [PubMed] [Google Scholar]
- 40. Silver P. M., Oliver B. G., White T. C. 2004. Role of Candida albicans transcription factor Upc2p in drug resistance and sterol metabolism. Eukaryot. Cell 3:1391–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Smyth G. K. 2005. Limma: linear models for microarray data, p. 397–420 In Gentleman R., Carrey V., Dudoit S., Irizzary I., Huber W. (ed.), Bioinformatics and computational biology solutions using R and bioconductor. Springer, New York, NY [Google Scholar]
- 42. Smyth G. K. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3:article 3 [DOI] [PubMed] [Google Scholar]
- 43. Smyth G. K., Michaud J., Scott H. S. 2005. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 21:2067–2075 [DOI] [PubMed] [Google Scholar]
- 44. Smyth G. K., Speed T. 2003. Normalization of cDNA microarray data. Methods 31:265–273 [DOI] [PubMed] [Google Scholar]
- 45. Tortorano A. M., et al. 2006. Candidaemia in Europe: epidemiology and resistance. Int. J. Antimicrob. Agents 27:359–366 [DOI] [PubMed] [Google Scholar]
- 46. van Eldere J., Joosten L., Verhaeghe V., Surmont I. 1996. Fluconazole and amphotericin B antifungal susceptibility testing by National Committee for Clinical Laboratory Standards broth macrodilution method compared with E-test and semiautomated broth microdilution test. J. Clin. Microbiol. 34:842–847 [DOI] [PMC free article] [PubMed] [Google Scholar]