Abstract
Identifying the types and distributions of organic substrates that support microbial activities around plant roots is essential for a full understanding of plant–microbe interactions and rhizosphere ecology. We have constructed a strain of the soil bacterium Sinorhizobium meliloti containing a gfp gene fused to the melA promoter which is induced on exposure to galactose and galactosides. We used the fusion strain as a biosensor to determine that galactosides are released from the seeds of several different legume species during germination and are also released from roots of alfalfa seedlings growing on artificial medium. Galactoside presence in seed wash and sterile root washes was confirmed by HPLC. Experiments examining microbial growth on α-galactosides in seed wash suggested that α-galactoside utilization could play an important role in supporting growth of S. meliloti near germinating seeds of alfalfa. When inoculated into microcosms containing legumes or grasses, the biosensor allowed us to visualize the localized presence of galactosides on and around roots in unsterilized soil, as well as the grazing of fluorescent bacteria by protozoa. Galactosides were present in patches around zones of lateral root initiation and around roots hairs, but not around root tips. Such biosensors can reveal intriguing aspects of the environment and the physiology of the free-living soil S. meliloti before and during the establishment of nodulation, and they provide a nondestructive, spatially explicit method for examining rhizosphere soil chemical composition.
Keywords: Rhizobium ‖ GFP ‖ protozoa ‖ sugars
Sinorhizobium meliloti is a Gram-negative bacterium that can live as a saprophyte in soil or as a nitrogen-fixing symbiont inside root nodule cells of alfalfa and related legumes (1, 2). The genes uniquely required for free-living rhizobia to nodulate their hosts and fix nitrogen have been targets of recent research, and some of these pathways are now well understood (1–3). Less studied are the activities of nodulating bacteria in the rhizosphere of host plants that are important for host infection. These processes include communication between bacteria (4), competition for access to infection sites on roots (5), gene transfer (6–8), and growth in the rhizosphere (9–11). Although it is generally accepted that growth of large microbial populations in the rhizosphere is supported by organic compounds from roots, identification of these compounds and quantification of their distribution in nonsterile soils has been problematic. To begin examining the roles of particular sugars for the growth of S. meliloti around roots and seeds, we have constructed a strain of S. meliloti in which gfp expression is induced by the presence of galactosides in the soil environment. This biosensor will help identify organic substrates that potentially support microbial growth and activity before and during nodulation. More broadly, such biosensors will provide spatially explicit information (at very small scales) about organic carbon distribution around roots, thus contributing to a deeper understanding of the physiology and ecology of plant–microbial and intermicrobial interactions in the rhizosphere.
The rhizosphere is the volume of soil directly under the influence of plant roots; it is bounded by the root surface on one side and extends millimeters into the soil. It is populated with a high density of bacteria, fungi, protists, and nematodes supported by relatively labile organic compounds, such as sugars, amino acids, organic acids, and polysaccharides. These compounds are actively and passively lost from living and dying root tissues (12, 13). This rhizodeposition allows 5–100 times more organisms per unit volume to be supported in the rhizosphere than in nearby bulk soil, where the microbial community is thought to be carbon limited (14, 15). The amount and spectrum of organic compounds in rhizodeposits vary along the length of individual roots (16), and they change as plants age, undergo physiological changes, or experience altered environmental conditions (17). Ratios of common organic components in rhizodeposits also vary from plant species to species (12, 18). Much, but not all, of this information about rhizodeposits has been gathered from studies with plants grown in sterile culture, and a challenge in rhizosphere research is to extend such observations to nonsterile soils (19). This extension will be particularly useful in microbial ecology, because variations in the composition of rhizodeposits will likely have an impact on the structure and metabolic activities of the associated rhizosphere community (20–22).
Most of the rhizobia cells in the rhizosphere do not directly participate in symbiosis, but they likely derive benefits from the symbiosis by using the rhizodeposits from host plant roots to support growth and division. Persistence of populations of rhizobia in soil depends, in part, on these viable free-living bacteria because nodules are largely filled with highly differentiated bacteroids which do not reproduce. Roots that can host nitrogen-fixing bacteria secrete a spectrum of signal molecules that affect the transcription of nod genes and, hence, are responsible for initiating symbiosis (23, 24). Vitamins, choline, stachydrine, trigonelline, and homoserine have also been shown to be secreted by plant roots and used by different species of rhizobia; rhizopines are thought to be exuded from nodules to provide carbon and nitrogen to rhizobia near the nodule (10, 25–28). However, it is not known whether compounds which are in plentiful supply in root exudates, such as common sugars and organic acids, play an important role in supporting growth of rhizobia in the rhizosphere.
The seeds of many legume species are especially rich in α-galactosides such as raffinose and stachyose (29), and S. meliloti might use these sugars to support growth if they were to be released by germinating seeds or the roots of young plants. We investigated this possibility by examining whether α-galactosides were released during seed germination and in alfalfa root exudates, and by examining whether they could contribute to biomass production in S. meliloti. By using recently characterized genes that are located on the megaplasmid pSymB and that are required for the utilization of α-galactosides by S. meliloti (30, 31), we constructed a GFP-based microbial biosensor which was capable of indicating whether galactosides were present in seed wash or in the rhizosphere of legume and grass species growing in nonsterile soil.
Materials and Methods
Bacterial Strains and Plasmids.
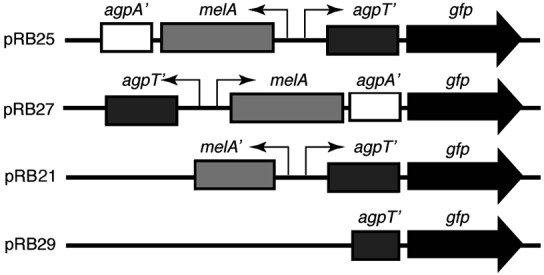
Plasmids pRB25 and pRB27 were constructed by cloning a 2,493-bp NsiI–PstI fragment of the agp operon, which spans the agpT/melA promoters in front of gfp(S65T) (32), in both orientations. The two promoter∷gfp fusions were then inserted into the broad-host-range plasmid pMB393 (33), giving plasmids pRB25 and pRB27. The agpT promoter drives gfp expression in pRB25. The melA promoter drives gfp expression in pRB27 (see Fig. 1). Plasmid pRB21 contains a 1,400-bp BamHI fragment that spans the agpT-melA intergenic region. The agpT promoter drives gfp expression in pRB21. Plasmid pRB29 is a promoterless control plasmid made by removing an 1,157-bp SacII fragment from plasmid pRB21.
Figure 1.
Maps of the gfp fusion plasmids used in this study.
S. meliloti strain RB6 (Rm1021 agpT) was made by cloning an internal fragment of agpT into a suicide vector pMB439 (constructed by M. Barnett, Stanford University) and integrating the resulting plasmid into the chromosome of wild-type R. meliloti strain Rm1021 (34).
Plasmid pDG77 was constructed by placing the gene for the DsRed protein (CLONTECH) downstream of a trp promoter from Salmonella typhimurium. This promoter is constitutively expressed in S. meliloti (33). In addition, a ribosome binding site (RBS) was placed upstream of the DsRed gene by PCR to enhance translation. The upstream primer encoding the RBS was 5′-GGGATCCGGAGGACGCCACCATGAGGTCT-3′; the downstream primer was 5′-GCCCGGGGGCCGCTAAAGGA-3′. The cassette—consisting of the promoter, ribosome binding site, and DsRed gene—was cloned into a stable broad-host-range plasmid pHC41 (35).
Fluorimetry.
Bacteria were grown overnight in M9 minimal medium with succinate as the sole carbon source; succinate ensured that gfp was not expressed. A quantity (1 ml) of cells was pelleted, washed three times with M9 salts to remove residual succinate, and resuspended in 100 μl of M9 salts. This suspension (5 μl) was used to inoculate 2.5 ml of M9 minimal medium with succinate, glucose, arabinose, galactose, lactose, melibiose, or raffinose at 0.4%. Cultures were grown with shaking until bacteria reached stationary phase in all carbon sources, 96 h after inoculation. A quantity (100 μl) of cell culture was collected and used to measure cell density and fluorescence. Cell density was determined by measuring optical density at either 415 nm or 595 nm with the Bio-Rad 550 (Bio-Rad) plate reader. Fluorescence was determined by using a CytoFluor 4000 fluorimeter (PerSeptive Biosystems, Foster City, CA) with excitation at 485 nm and emission at 508 nm. Fluorescence values were corrected for cell density by dividing the fluorescence value of the culture sample by its cell density, giving relative fluorescence. To further test specificity of the biosensor, Biolog (Biolog, Hayward, CA) SFN2 plates containing 95 carbon sources, among them sugars, amino acids, and organic acids, were inoculated with 150 μl of a succinate-grown suspension of Rm1021/pRB27 (pmelA-gfp). After 45 h of incubation at 30°C, relative fluorescence was measured as described above.
Seed Wash Preparation.
Alfalfa (Medicago sativa AS13R), barrel medic (M. truncatula), and white clover (Trifolium repens) seed wash were prepared by soaking 2 g of seeds in 20 ml of water at 4°C for 24 h without shaking for the growth yield experiments, and at room temperature for 1 h with light shaking for the HPLC experiments. The wash was sterilized by filtration through a 0.45-μm syringe filter. Seed washes were stored at 4°C for no more than 48 h before use. Strains Rm1021/pRB27 (pmelA-gfp) and Rm1021/pRB29 (control) were grown to stationary phase in tubes containing 1.25 ml of 2× M9 salts supplemented with 1.25 ml of sterile seed wash from alfalfa, barrel medic, and white clover. GFP fluorescence was measured as described above.
Root Exudate Preparation.
Alfalfa seeds were surface sterilized with ethanol and bleach (33) and sprouted in a Petri dish. Seedlings were transferred to sterile polypropylene centrifuge tubes (contained within larger tubes with loose lids), shoots inside were supported with gauze, and roots grew through the gauze into 4 ml of sterile water. Plants were maintained 7 days in a Sanyo MLR-350 chamber at 26°C for a 14-h light/10-h dark cycle. Each day 1.5 ml of water containing root exudate was collected from one tube and stored at −80°C. Each sample was later evaporated to dryness and redissolved in 100 μl of sterile water.
Sugar Detection.
Sugars in seed wash and root exudate were analyzed by HPLC with a DX-500 system with a CarboPac PA10 column, GP40 gradient pump, and ED40 electrochemical detector using pulse amperimetric detection (Dionex). Eluent was sodium hydroxide at 6 mM for 18 min, then as a linear gradient to 130 mM over the next 16 min, followed by 200 mM for 10 min. Flow rate was 1 ml/min. Sugars in seed wash and root exudate were identified by comparing elution times with those from 5-μl injections of 0.1 mM fructose, galactose, glucose, melibiose, raffinose, stachyose, and sucrose.
Growth Yield Experiments.
Varying amounts of alfalfa seed wash were used to produce 6.25% to 50% (vol/vol) wash in 1× M9 salts. Bacterial strains were grown overnight in M9 minimal medium with succinate as the sole carbon source. A quantity (1 ml) of cells was pelleted, washed three times in 1× M9, and resuspended in 100 μl of M9 salts. This suspension (10 μl) was used to inoculate 10 ml of M9 medium containing seed wash. The 10-ml cultures were grown for 96 h at 30°C with shaking at 250 rpm. Cell density was determined by measuring the optical density of 100 μl of cell culture at 595 nm with the Bio-Rad 550 plate reader.
Plant Growth and Inoculation—Artificial Medium.
Alfalfa plants were grown and inoculated as described (33) with the following changes. Microscope slides were overlaid with 5 ml of basic nodulation medium (BNM; ref. 36) in 1.15% agarose. Bacteria were grown 48 h in succinate M9 minimal medium. A quantity (1 ml) of culture was pelleted, washed three times in BNM, and resuspended in 1 ml of BNM. One-day-old plants grown on BNM were inoculated with 200 μl of bacterial suspension containing about 108 bacteria. Plant roots were monitored by epifluorescence microscopy each day for the following week. On day 6, root sections that were colonized with fluorescent bacteria were excised from the seedling, placed in water, and immediately photographed.
Plant Growth and Inoculation in Nonsterile Soil.
Microcosms were constructed from two 5-cm by 7.5-cm glass microscope slides placed parallel to each other and separated by 6-mm thick neoprene strips. Nylon mesh (500 μm) was placed between the neoprene and one glass side of the microcosm, and the entire assembly was secured along the sides with duct tape. The microcosms were filled with unsterilized, calcareous sandy loam, sieved through a 2-mm sieve, and then mixed 1:2 with fine sand. M. sativa, Macroptillium atropurpureum, Avena sativa, or Bromus hordeadeus seeds were planted at the top of the chambers; the chambers were lightly watered and placed at a 45° angle, nylon-mesh side up. Roots grew downward, collided with the bottom slide of the microcosm, and traveled along it. This method allowed observation of the root system by epifluorescence microscopy. Bacteria were introduced into the microcosms by removing the top slide and dripping a suspension of strains Rm1021/pRB27 (pmel∷gfp) and RM1021/pDG77 (constitutive DsRed expression) mixed in M9 salts onto the mesh. The suspension, containing about 2 × 109 bacteria in total, thoroughly wetted the soil and root systems in the chamber. Production of GFP was monitored over 96 h. At that time, 0.2% solutions of melibiose, sucrose, or glucose were injected into dark areas of the microcosms to test whether the fusion strain was still present and capable of responding to α-galactosides.
Microscopy.
Epifluorescence was viewed on an Eclipse TE300 (Nikon) inverted microscope, and photographs were taken with a Life Science Research (PE Biosystems, Foster City, CA) charge-coupled device camera. GFP fluorescence was imaged by using a filter set with a 465-nm to 495-nm excitation filter and a 515-nm to 555-nm emission filter. DsRed fluorescence was imaged by using a filter set with a 540-nm to 580-nm excitation filter and a 600-nm to 660-nm emission filter. Root autofluorescence was imaged by using a filter set with a 330-nm to 380-nm excitation filter and a 435-nm long pass emission filter. Composite images were colorized and constructed with Adobe photoshop (Adobe Systems, Mountain View, CA).
Confocal microscopy was done with a Zeiss LSM 510 system (Zeiss) using FITC and rhodamine filter sets to collect GFP and DsRed fluorescence, respectively. Reflected laser light was collected to visualize root and soil structure.
Results
Construction of S. Meliloti Reporter Strains.
S. meliloti genes required for the utilization of α-galactosides include the melA gene (encoding an α-galactosidase) and the genes agpA and agpB (encoding part of an α-galactoside transport system; ref. 30). Upstream, agpT encodes an AraC-like transcriptional activator required for induction of melA, agpA, and agpB by galactosides (37). We constructed the reporter strain for galactoside detection by fusing the promoter of the melA gene to gfp in plasmid pRB27 (Fig. 1). Inverting the promoter fragment resulted in the promoter for agpT driving gfp expression in pRB25. Plasmid pRB29 is a promoterless control derived from pRB21. pDG77 provided a constitutive DsRed signal for tracking inoculated bacterial populations.
Induction of gfp Fusions by Sugars.
Strains of Rm1021 containing plasmids pMB393, pRB25, pRB27, and pRB29 were grown in M9 minimal medium containing different carbon sources, and the relative GFP fluorescence was measured in stationary phase cells (Table 1). The control strain Rm1021/pRB29 showed very low levels of fluorescence, roughly equivalent to the autofluorescence of strain Rm1021/pMB393 which contained only the cloning vector. This low fluorescence could be detected by a fluorimeter, but not by epifluorescence microscopy. In contrast, strain Rm1021/pRB27 (pmelA-gfp) fluoresced brightly when grown on galactose, or α-galactosides such as melibiose or raffinose. The β-galactoside lactose also induced the pmelA-gfp reporter, but succinate, glucose and arabinose did not. After 45 h of incubation on Biolog SNF2 plates (Hayward, CA), only cells grown on galactose, lactose, melibiose, raffinose, and lactulose (all galactosides) showed fluorescence values above background (data not shown).
Table 1.
Relative fluorescence of Rm1021 reporter strains grown on various carbon sources
| Carbon source | pMB393 vector alone | pRB27 pmelA-gfp | pRB25 pagpT-gfp | pRB29 ΔpagpT-gfp |
|---|---|---|---|---|
| Succinate | 177 | 210 | 237 | 188 |
| Glucose | 314 | 344 | 455 | 306 |
| Arabinose | 318 | 427 | 429 | 324 |
| Galactose | 263 | 3804 | 492 | 198 |
| Lactose | 304 | 1775 | 529 | 246 |
| Melibiose | 253 | 4961 | 542 | 281 |
| Raffinose | 222 | 3757 | 369 | 235 |
In all media except M9-succinate, strain Rm1021/pRB25 (pagpT-gfp) showed fluorescence levels of 1.5 to 2.0 times greater than those seen in the control strain Rm1021/pRB29. This fact indicates that the gene encoding the transcriptional activator AgpT is expressed constitutively at a low level and is not up-regulated by inducing sugars.
Examination of Seed Wash and Rhizodeposits.
Seed wash from legumes induces the galactoside reporter.
Seed wash induced the pmelA-gfp fusion in strain Rm1021/pRB27 20- to 40-fold above the levels seen in the control strain Rm1021/pRB29 (Fig. 2A). To assure that galactosides were present in seed wash, we analyzed seed wash from alfalfa by HPLC. We found peaks which comigrated with purified galactose, raffinose, and stachyose (Fig. 2B), as well as glucose and sucrose. To ensure that the two peaks from seed wash that comigrated with purified raffinose and stachyose were really α-galactosides, we treated the seed wash with α-galactosidase from Aspergillus niger. After treatment, the putative raffinose (gal-sucrose) and stachyose (gal-gal-sucrose) peaks decreased, and galactose and sucrose peaks concomitantly increased (data not shown) as expected for peaks containing these α-galactosides.
Figure 2.
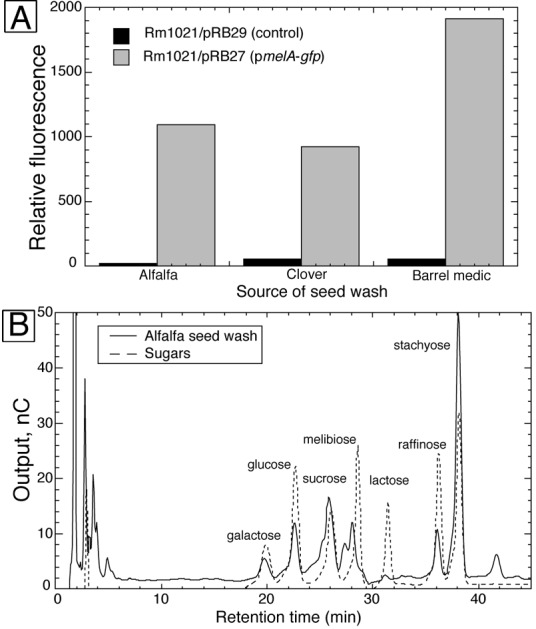
(A) Relative fluorescence of strain Rm1021/pRB29 and strain Rm1021/pRB27 grown in M9 medium with seed wash from three legumes as carbon sources. (B) HPLC chromatogram of alfalfa seed wash overlaid on a chromatogram of a mixture of seven sugars.
α-Galactosides released from alfalfa seeds can contribute to the growth of S.
meliloti.
The ability of α-galactosides from seeds to support the growth of S. meliloti was assessed by comparing the growth yield of strains RB6 (agpT-) and Rm1021 wild type grown in M9 medium containing various concentrations of seed wash. Strain RB6 contains a mutation in the AraC-like transcriptional activator agpT and is unable to use α-galactosides. In these experiments, the yield of strain Rm1021 was 12–30% higher than that of strain RB6 over a range of seed wash concentrations (Fig. 3). As a control, both strains were grown in M9 medium with succinate or sucrose as the carbon source, and the growth yield of strain RB6 was equal to, or slightly greater than, that of strain Rm1021 (data not shown).
Figure 3.
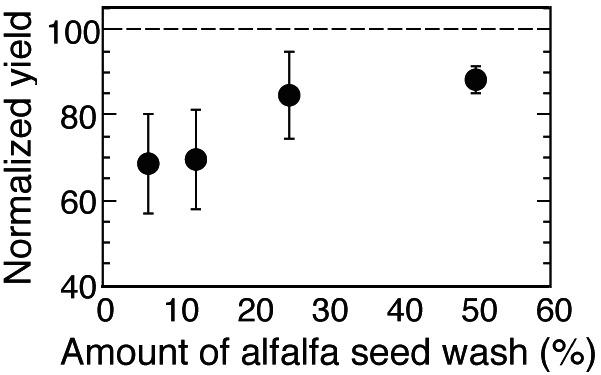
Mean (± SD) growth yield of strain RB6 (agpT-) grown using various concentrations of alfalfa seed wash as a carbon source (n = 4). Yields of strain RB6 (agpT-) are normalized to the yields obtained by the wild-type strain Rm1021. The dashed line shows the value at which the yield of strain RB6 would be equal to the yield of strain Rm1021.
The pmelA-gfp reporter is induced by galactosides in the rhizosphere of alfalfa seedlings grown on artificial medium.
One-day-old alfalfa seedlings grown on BNM and inoculated with strains Rm1021/pRB27 (pmelA-gfp) and Rm1021/pRB29 (control) were monitored by epifluorescence microscopy each day for a week. Control bacteria remained dark (Fig. 4A). Bacteria carrying the pmelA-gfp reporter began to fluoresce brightly between days 3 and 5 after inoculation, indicating that the plant root was releasing galactosides (Fig. 4B). HPLC analysis of sugars exuded from roots of 5-day-old, axenically grown alfalfa seedlings revealed peaks comigrating with galactose, raffinose, and stachyose (data not shown). These compounds are likely responsible for the induction of the pmelA-gfp reporter. Glucose and fructose, as well as several unidentified compounds, were also detected. Why it takes 3 days to see fluorescence is intriguing. Glucose or other carbon compounds such as succinate may have exerted catabolite repression on the pmelA∷gfp reporter immediately after inoculation, which was relieved once these carbon sources were used up by the resident bacteria.
Figure 4.
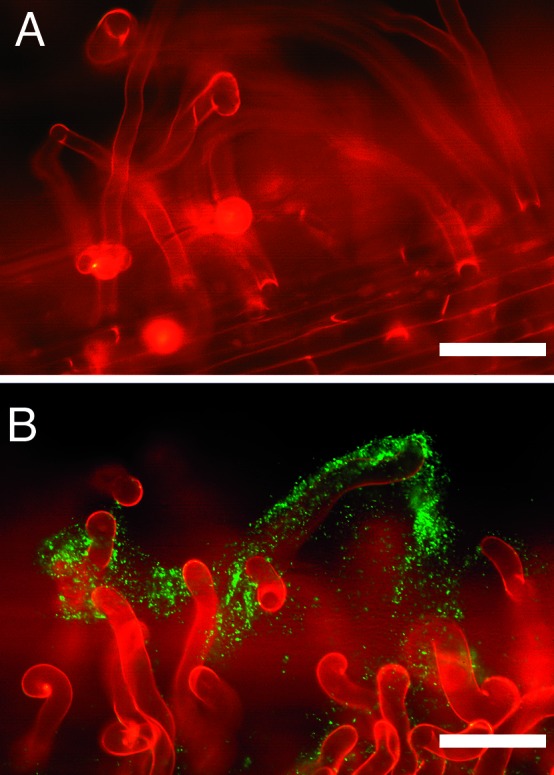
Induction of the pmelA-gfp reporter in the rhizosphere of alfalfa seedlings grown in artificial medium. (A) Fluorescent micrograph of the control strain Rm1021/pRB29 inoculated onto the roots of alfalfa seedlings grown on BNM. (B) Fluorescent micrograph of the pmelA-gfp reporter strain Rm1021/pRB27 inoculated onto the roots of alfalfa seedlings grown on BNM. (Bars = 100 μm.)
The pmelA-gfp reporter is induced in the rhizosphere of legumes and nonlegumes grown in soil.
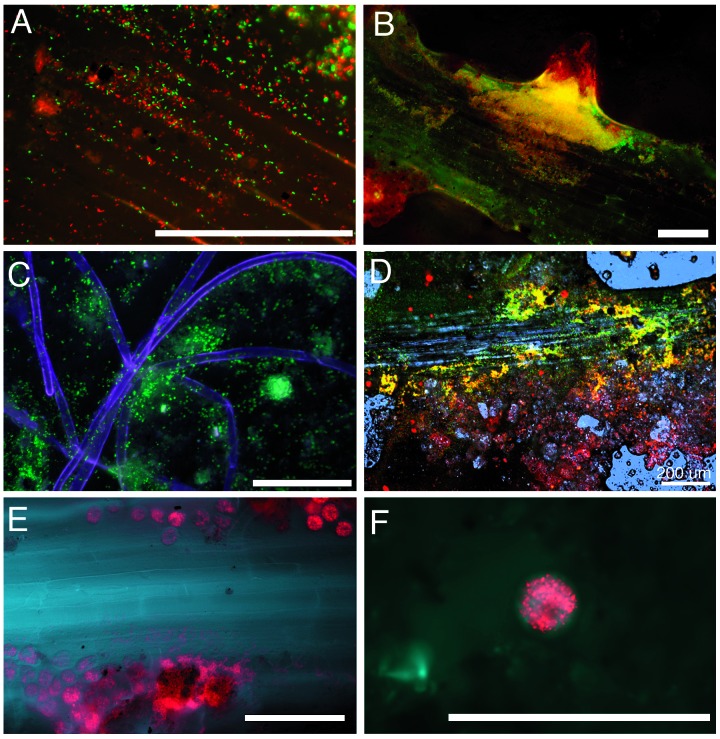
We tested whether galactosides in nonsterile soil around living roots could be detected noninvasively by inoculating microcosms 1–4 days after seed germination with a mixture containing strains Rm1021/pRB27 (pmel∷gfp) and Rm1021/pDG77 (constitutively expressing the red fluorescent protein DsRed). Strain Rm1021/pDG77 was included so that the distribution of inoculated bacteria could be easily seen in soil. Green fluorescent bacteria were seen associated with the root plane (Fig. 5 A, B, and D), on and near root hairs (Fig. 5C), with emerging secondary roots (Fig. 5B), and in the soil near roots (Fig. 5 C and D). In no cases were green fluorescent bacteria seen covering large areas of the root, nor were they associated with root tips (except when lateral root tips were emerging from root axes, e.g., Fig. 5B). In contrast, bacteria constitutively expressing DsRed were distributed continuously in the microcosms and along roots, indicating that inoculation of the two bacterial strains had not been patchy (data not shown). We also observed zones of intense protozoal grazing of introduced bacteria; fluorescent protein from engulfed bacteria could be seen inside active protozoa in the microcosm soil (Fig. 5 E and F).
Figure 5.
Induction of the pmelA-gfp reporter in the rhizosphere of legumes and grasses growing in nonsterilized soil. All panels except C show roots in microcosms inoculated with a mixture of Rm1021/pRB27 (pmelA∷gfp) and Rm1021/pDG77 (constitutive DsRed). C shows root hairs in a microcosm inoculated with Rm1021/pRB27 (pmelA∷gfp) only. In all micrographs, green fluorescence (indicating the presence of galactosides) is colored green, red fluorescence from DsRed is colored red, and in C, E, and F, root autofluorescence (visualized with a brief pulse of UV light) is colored blue. Root autofluorescence was not imaged separately when roots in A and B were examined. Colocalized green and red fluorescence combine to give yellow. [Bars = 100 μm (except in D).] (A) Red and green fluorescent bacteria on the root surface of an Avena sativa seedling. (B) Alfalfa root with red and green fluorescent bacteria. Colocalized red and green bacteria are especially prevalent around an emerging lateral root. (C) Green fluorescent bacteria associated with root hairs of Bromus hordeaceus. (D) Confocal micrograph of an alfalfa root in soil; blue represents reflected confocal laser light revealing root cells, soil structure, and water droplets. (E) Zone of an alfalfa root where protozoa are grazing on red bacteria. (F) Close-up of another alfalfa root showing individual round protozoa with numerous red fluorescent bacteria inside.
Green fluorescent bacteria could no longer be seen 96 h after inoculation. We tested whether the biosensor strain was still present and whether GFP production was still inducible by injecting the α-galactoside melibiose (as a 0.2% solution) into microcosms near root systems. Bacteria at the injection site fluoresced brightly 18 h later, indicating that they were still present and able to respond to the inducer. As a control, sucrose and glucose were also injected into chambers at 96 h; green fluorescence was not induced by these sugars.
Discussion
By using a S. meliloti biosensor strain, we found that galactosides were components of alfalfa, clover, and barrel medic seed wash sugars (Fig. 2 A and B), and that S. meliloti was able to capitalize on α-galactosides to enhance its growth (Fig. 3). Also, galactosides were present in nonsterile soil around the roots of legumes and nonlegumes (Fig. 5). The extent to which the presence of such sugars may promote maintenance or growth of S. meliloti in natural soils will vary, depending on microbial community composition, availability of other growth substrates, etc. However, because S. meliloti can form films along portions of the root (e.g., Fig. 5B), these bacteria are in prime position to capture sugars leaving the root surface.
We observed a patchy distribution of fluorescent galactoside reporter bacteria on and around roots growing in soil microcosms; for example, fluorescence from biosensors was never observed around root tips. This distribution could have stemmed from localized exudation of galactosides or from uneven distribution of the sensor bacteria. The distribution of combined populations of sensor and DsRed-producing bacteria, discerned by visualizing red bacteria, was continuous throughout the chamber, suggesting that the patchy distribution of green fluorescence that developed over several days arose from reporter bacteria encountering the patchy presence of galactosides around the roots. Green fluorescence arising from pmelA∷gfp eventually disappeared. Because GFP is very stable, decay of the GFP signal likely indicates that galactosides were used up and the pmelA promoter shut off. Cells then may have continued to grow and divide using other carbon sources, resulting in a dilution of GFP in the population. Although protozoal grazing of bacteria could also diminish the signal in some zones of the microcosm, it seems unlikely that such grazing would be as extensive as the observed disappearance of the signal throughout the microcosm. Injection of the α-galactoside melibiose into the microcosms revealed that, although gfp expression had ceased several days after inoculation, the biosensors were present and capable of making GFP when the galactoside stimulus reappeared.
The relatively stable green and red fluorescent signals in the inoculated strains persisted after bacteria had been consumed by protozoa (Fig. 5 E and F). Not only will these fluorescent bacteria provide beginning glimpses of available root-derived galactosides in the rhizosphere, but also the persistent fluorescent signal (particularly in the DsRed strain) should allow us to examine whether there are particular zones of intense protistal grazing of these bacteria along roots.
The suite of reporter genes now available to investigate rhizosphere biochemistry and microbial gene activity is very promising. Previously, it had been problematic to identify and describe the compounds released by plant roots to the soil under natural conditions. These compounds can be difficult to extract from soil; they are rapidly used by microbes and other organisms as growth substrates, and it is difficult to sample the root region with high spatial resolution without disturbing the roots. Genetically engineered microbes have been used to monitor bacterial gene activity around roots, but many of these studies have used destructive harvest or have not been in nonsterile soil (16, 38–40). The sensitive bacterial biosensors described recently by Jaeger et al. (16) for use in soil are particularly interesting because they can be used not only to detect but also to quantify tryptophan and sucrose concentrations near root systems. Unfortunately, detection of the ice-nucleation report involves destructive harvest. Our GFP-based biosensor is not easily quantitated in situ, but its expression can be visualized nondestructively with a high degree of spatial resolution. For example, the confocal work (Fig. 5D) revealed that galactosides were in a high enough concentration to trigger the reporter only within 200 μm of the alfalfa root surfaces, indicating that a steep concentration gradient in inducing sugars existed from root surfaces into the rhizosphere soil. Also, in contrast to results for sucrose and fructose from Jaeger et al. (16), we never found galactosides around root tips. Jaeger et al. speculate that apoplastic diffusion of sucrose down the root axis to provision new cell growth in the root tip where phloem is undeveloped (41) provided a ready sucrose source for diffusion into the soil near root tips. Perhaps because galactosides are unlikely to be used to provision growth in the root's meristematic area, we did not see galactosides in this zone.
Whereas the pmelA-gfp reporter was adequate for the studies described in this paper, several modifications would render it more suitable for longer-term studies. Plasmid-based reporters are often lost from their host strains under nonselective conditions. Integrating the reporter into a selectively neutral site in the chromosome would help ensure long-term stability. Also, the gfp gene used in this study encodes a fairly stable protein, and thus the fluorescence of the biosensor strain doesn't decrease rapidly when transcription of the pmelA-gfp reporter decreases. For stronger correlation between fluorescence and transcriptional activity a gfp gene encoding an unstable protein product could be used. Such genes have been engineered, and protein levels from fusions fall quickly in response to decreases in transcription (42).
Using rapidly degraded gfp reporters integrated into the chromosome of a suitable soil bacterium should provide a convenient way to follow noninvasively the spatial and temporal patterns of release of compounds from plant roots. It should be possible to determine zones where the rhizodeposition of particular classes of compounds is high, and to correlate the presence of specific compounds around roots with the development and physiology of the plant. Distributions of particular kinds of sugars, amino acids, and organic acids around roots could, as Jaeger et al. (16) suggest, provide spatially and temporally distinct areas where waves of bacterial, fungal, and protistal populations develop and turn over. Such dynamics may be key for nutrient availability to plants, and, more broadly, carbon and nitrogen cycling in the rhizosphere (43).
Acknowledgments
This work was supported by a University of Connecticut Research Foundation Grant 441996 (to D.J.G.), a Heinz Herrmann Graduate Fellowship in Cell Biology from the Graduate School of the University of Connecticut (to R.B.), and an Andrew W. Mellon Foundation grant (to Z.G.C.).
Abbreviations
- GFP
green fluorescent protein
- BNM
basic nodulation medium
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Mylona P, Pawlowski K, Bisseling T. Plant Cell. 1995;7:869–885. doi: 10.1105/tpc.7.7.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spaink H P. Annu Rev Phytopathol. 1995;33:345–368. doi: 10.1146/annurev.py.33.090195.002021. [DOI] [PubMed] [Google Scholar]
- 3.Hirsch A M. Curr Opin Plant Biol. 1999;2:320–326. doi: 10.1016/S1369-5266(99)80056-9. [DOI] [PubMed] [Google Scholar]
- 4.Gray K M, Pearson J P, Downie J A, Boboye B E A, Greenberg E P. J Bacteriol. 1996;178:372–376. doi: 10.1128/jb.178.2.372-376.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Triplett E, Sadowsky M J. Annu Rev Microbiol. 1992;46:399–428. doi: 10.1146/annurev.mi.46.100192.002151. [DOI] [PubMed] [Google Scholar]
- 6.Segovia L, Piñero D, Palacios R, Martinez-Romero E. Appl Environ Microbiol. 1991;57:426–433. doi: 10.1128/aem.57.2.426-433.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sullivan J T, Ronson C W. Proc Natl Acad Sci USA. 1998;95:5145–5149. doi: 10.1073/pnas.95.9.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broughton W J, Samrey U, Stanley J. FEMS Microbiol Lett. 1987;40:251–255. [Google Scholar]
- 9.Robleto E, Kmiecik K, Oplinger E S, Nienhuis J, Triplett E W. Appl Environ Microbiol. 1998;64:2630–2633. doi: 10.1128/aem.64.7.2630-2633.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Streit W R, Joseph C M, Phillips D A. Mol Plant–Microbe Interact. 1996;9:330–338. doi: 10.1094/mpmi-9-0330. [DOI] [PubMed] [Google Scholar]
- 11.Paulumbo J D, Kado C I, Phillips D A. J Bacteriol. 1998;180:3107–3113. doi: 10.1128/jb.180.12.3107-3113.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whipps J M, Lynch J M. Ann Proc Phytochem Soc Eur. 1985;26:59–71. [Google Scholar]
- 13.Curl E A, Truelove B. The Rhizosphere. Berlin: Springer; 1986. [Google Scholar]
- 14.Bazin M J, Markham P, Scott E M, Lynch J M. In: The Rhizosphere. Lynch J M, editor. West Sussex, U.K.: Wiley; 1990. pp. 99–127. [Google Scholar]
- 15.Cheng W, Zhang Q, Coleman D C, Carroll C R, Hoffman C A. Soil Biol Biochem. 1996;28:1283–1288. [Google Scholar]
- 16.Jaeger C H, Lindow S E, Miller W, Clark E, Firestone M K. Appl Environ Microbiol. 1999;65:2685–2690. doi: 10.1128/aem.65.6.2685-2690.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whipps J M. In: The Rhizosphere. Lynch J M, editor. West Sussex, U.K.: Wiley; 1990. pp. 59–97. [Google Scholar]
- 18.Grayston S J, Vaughan D, Jones D. Appl Soil Ecol. 1996;5:29–56. [Google Scholar]
- 19.Gregory P J, Hinsinger P. Plant Soil. 1999;211:1–9. [Google Scholar]
- 20.Yang C-H, Crowley D E. Appl Environ Microbiol. 2000;66:345–351. doi: 10.1128/aem.66.1.345-351.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griffiths B S, Ritz K, Ebblewhite N, Dobson G. Soil Biol Biochem. 1999;31:145–153. [Google Scholar]
- 22.Johnson N C. Ecol Appl. 1993;3:749–757. doi: 10.2307/1942106. [DOI] [PubMed] [Google Scholar]
- 23.Gagnon H, Ibrahim R K. Mol Plant–Microbe Interact. 1998;11:988–998. [Google Scholar]
- 24.Phillips D A, Wery J, Joseph C M, Jones A D, Teuber L R. Crop Sci. 1995;35:805–808. [Google Scholar]
- 25.Boivin C, Barran L R, Malpica C A, Rosenberg C. J Bacteriol. 1991;173:2809–2817. doi: 10.1128/jb.173.9.2809-2817.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Rudder K E E, Sohlenkamp C, Geiger O. J Biol Chem. 1999;274:20011–20016. doi: 10.1074/jbc.274.28.20011. [DOI] [PubMed] [Google Scholar]
- 27.Phillips D A, Sande E S, Vriezen J A C, De Bruin F J, Le Rudlier D, Joseph C M. Appl Environ Microbiol. 1998;64:3954–3960. doi: 10.1128/aem.64.10.3954-3960.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gordon D M, Ryder M H, Heinrich K, Murphy P J. Appl Environ Microbiol. 1996;62:3991–3996. doi: 10.1128/aem.62.11.3991-3996.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Obendorf R L. Seed Sci Res. 1997;7:63–74. [Google Scholar]
- 30.Gage D J, Long S R. J Bacteriol. 1998;180:5739–5748. doi: 10.1128/jb.180.21.5739-5748.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Charles T C, Finan T M. Genetics. 1991;127:5–20. doi: 10.1093/genetics/127.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Helm R, Cubitt A B, Tsien R Y. Nature (London) 1995;373:663–664. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
- 33.Gage D J, Bobo T, Long S R. J Bacteriol. 1996;178:7159–7164. doi: 10.1128/jb.178.24.7159-7166.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meade H M, Long S R, Ruvkun G B, Brown S E, Ausubel F M. J Bacteriol. 1982;149:114–122. doi: 10.1128/jb.149.1.114-122.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng H-P, Walker G C. J Bacteriol. 1998;180:5183–5191. doi: 10.1128/jb.180.19.5183-5191.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ehrhardt D W, Atkinson E M, Long S R. Science. 1992;256:998–1000. doi: 10.1126/science.10744524. [DOI] [PubMed] [Google Scholar]
- 37.Bringhurst R M, Gage D J. FEMS Microbiol Lett. 2000;188:23–27. doi: 10.1111/j.1574-6968.2000.tb09163.x. [DOI] [PubMed] [Google Scholar]
- 38.Egener T, Hurek T, Reinhold-Hurek B. Mol Plant–Microbe Interact. 1998;11:71–75. doi: 10.1094/MPMI.1998.11.1.71. [DOI] [PubMed] [Google Scholar]
- 39.Ramos C, Molbak L, Molin S. Appl Environ Microbiol. 2000;66:801–809. doi: 10.1128/aem.66.2.801-809.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Overbeek L S, van Elsas J D. Appl Environ Microbiol. 1995;61:890–898. doi: 10.1128/aem.61.3.890-898.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bret-Harte M S, Silk W E. Plant Physiol. 1994;105:19–33. doi: 10.1104/pp.105.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andersen J S, Sternberg C, Poulsen L K, Bjørn S P, Givskov M, Molin S. Appl Environ Microbiol. 1998;64:2240–2246. doi: 10.1128/aem.64.6.2240-2246.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clarholm M. Soil Biol Biochem. 1985;17:181–187. [Google Scholar]