Abstract
The integrase inhibitor (INI) dolutegravir (DTG; S/GSK1349572) has significant activity against HIV-1 isolates with raltegravir (RAL)- and elvitegravir (ELV)-associated resistance mutations. As an initial step in characterizing the different resistance profiles of DTG, RAL, and ELV, we determined the dissociation rates of these INIs with integrase (IN)-DNA complexes containing a broad panel of IN proteins, including IN substitutions corresponding to signature RAL and ELV resistance mutations. DTG dissociates slowly from a wild-type IN-DNA complex at 37°C with an off-rate of 2.7 × 10−6 s−1 and a dissociative half-life (t1/2) of 71 h, significantly longer than the half-lives for RAL (8.8 h) and ELV (2.7 h). Prolonged binding (t1/2, at least 5 h) was observed for DTG with IN-DNA complexes containing E92, Y143, Q148, and N155 substitutions. The addition of a second substitution to either Q148 or N155 typically resulted in an increase in the off-rate compared to that with the single substitution. For all of the IN substitutions tested, the off-rate of DTG from IN-DNA complexes was significantly slower (from 5 to 40 times slower) than the off-rate of RAL or ELV. These data are consistent with the potential for DTG to have a higher genetic barrier to resistance, provide evidence that the INI off-rate may be an important component of the mechanism of INI resistance, and suggest that the slow dissociation of DTG may contribute to its distinctive resistance profile.
INTRODUCTION
Improvements in antiretroviral therapy have resulted in major advances in longevity and quality of life for HIV-infected patients. However, there is still a need to augment the anti-HIV armament: for example, by increasing tolerability and ease of dosing, maximizing potency, providing forgiveness in the face of adherence difficulties, and improving resistance profiles. The latest addition to anti-HIV therapy is inhibition of the essential (31) HIV integrase (IN) enzyme (reviewed in references 1, 4, 7, 17, and 33). IN carries out two key catalytic processes: 3′ end processing, which removes the final 2 nucleotides (e.g., 5′-GT) from each end of the viral cDNA, and insertion of the two viral 3′ DNA ends into opposite strands of the host DNA (strand transfer). Several compounds that specifically block strand transfer of the viral cDNA into the host DNA have been shown to be efficacious in vivo (reviewed in references 34 and 36). Raltegravir (RAL), which was approved by the FDA in 2007, was the first marketed IN inhibitor (INI), and elvitegravir (ELV) and dolutegravir (DTG; S/GSK1349572) are in late stages of development (Fig. 1). All three molecules have a common two-metal-binding motif by which the two essential metals at the IN catalytic site are bound by the inhibitor.
Fig. 1.
HIV-1 integrase inhibitors. (A) Dolutegravir (DTG; S/GSK1349572); (B) raltegravir (RAL); (C) elvitegravir (ELV).
Resistance is typically the primary chink in an antiretroviral agent's armor. Resistance mutations have been identified for both RAL and ELV during in vitro passaging experiments (20, 21, 30, 40, 43, 47) and in clinical studies (5, 35, 36, 45). Primary resistance mutations observed within the IN open reading frame include substitutions at T66 (ELV), E92 (ELV), Y143 (RAL), Q148 (both drugs), and N155 (both drugs) (reviewed in references 3 and 36). Multiple secondary mutations which may increase resistance and/or compensate for fitness defects caused by the primary mutations have also been identified. Overall, both RAL and ELV largely share resistance profiles (3, 30, 36, 37), and it was demonstrated that subjects experiencing virologic failure during therapy with ELV did not respond to treatment with RAL (10). Understanding resistance mechanisms may practically assist in optimizing the design and development of new drugs with improved resistance profiles, help define how to best use anti-HIV agents in the clinic, and inform on the potential that HIV might require multiple mutations to achieve resistance for a drug which may in turn be predictive of a higher barrier to resistance in vivo.
Characterizing the mechanisms of HIV drug resistance can involve many methods, including passage of virus in cell culture in the presence of a drug to select resistance, examination of resistance profiles observed in virus isolates from subjects experiencing virologic failure in the clinic, evaluation of the relative contributions of mutations to resistance and to restoration of viral fitness, and finer-level studies involving biochemical measurements and analysis of molecular structure using crystallography. Of note, biochemical studies have suggested that binding of INIs to HIV-1 IN proceeds by a two-step mechanism with a slow second step, and mutations that increase resistance may alter this mechanism (19, 32). In addition, fast INI dissociation has been proposed to contribute to INI resistance with the N155H, T66I, and Q148R IN substitutions (14, 23).
DTG was designed to have excellent potency, low-mg dosing given once daily without pharmacokinetic boosting, and an improved resistance profile with a potential for a higher barrier to resistance (29). DTG has shown potent antiviral activity in multiple cell types and cell-based assay formats (25, 30) and has demonstrated potent in vivo efficacy in clinical studies to date (39, 42). Extensive in vitro antiviral profiling has shown that DTG has a distinct resistance profile compared to other INIs, with significant antiviral activity against virus with RAL and ELV resistance IN mutations (30). For example, DTG was shown to have wild-type or near wild-type activity in vitro against site-directed mutants with the single primary resistance mutations observed in vivo at Q148 (including Q148H/K/R), at N155, and at Y143 (25, 30) and against clinical isolates harboring genotypes with the Y143 and N155 pathway mutations (46) alone or with additional secondary IN mutations. DTG has also demonstrated in vivo activity in subjects with RAL resistance in the VIKING study (16). As described herein, we investigated the dissociation of DTG, RAL, and ELV from wild-type and mutant IN proteins complexed with DNA to obtain a better understanding of INI dissociation kinetics and the relationship between dissociation rates and INI resistance.
MATERIALS AND METHODS
Reagents.
DTG, RAL, and ELV were synthesized and iodinated at GlaxoSmithKline and then tritium labeled by Quotient Bioresearch (Cardiff, United Kingdom) or Tritec (Teufen, Switzerland), resulting in specific activities of 27 Ci/mmol, 23 Ci/mmol, and 18 Ci/mmol, respectively. Streptavidin-coated scintillation proximity assay (SPA) imaging beads were purchased from PerkinElmer (Boston, MA). Full-length HIV-1 BH10 wild-type and mutant IN enzymes were expressed and purified as described previously (19) with or without 2-mercaptoethanol in the lysis and extraction buffers. Site-directed mutagenesis was used to make mutations in the wild type HIV-1 BH10 IN sequence (41). The oligonucleotides 5′-biotin-ACCCTTTTAGTCAGTGTGGAAAATCTCTAGCA (plus strand, mimics 3′ processed end with GT removed) and 3′-GAAAATCAGTCACACCTTTTAGAGATCGTCA (minus strand) were obtained from Invitrogen (Carlsbad, CA).
Dissociation studies.
Dissociation experiments with INIs and IN-DNA complexes were performed by methods similar to published methods (32). Viral long terminal repeat DNA duplexes were prepared by heating 50 μM 5′-biotinylated, 3′-processed plus-strand DNA and 50 μM minus-strand DNA at 95°C for 5 min in 50 mM 3-(N-morpholino)propanesulfonic acid (MOPS [pH 7.2]), 50 mM NaCl, and 10 mM MgCl2, followed by cooling at room temperature for several hours. The DNA duplexes were attached via the 5′-biotin linker on the plus strand to streptavidin-coated SPA imaging beads by incubating 2 μM biotinylated DNA and 20 mg/ml beads in 25 mM MOPS (pH 7.2) on a Nutator mixer for 80 min at room temperature. Unbound DNA was removed by two rounds of centrifugation and resuspension of the DNA-bead complex in 50 mM MOPS (pH 7.2), 50 mM NaCl, and 10 mM MgCl2. The final bead concentration was 40 mg/ml, and the DNA-bead complex was stored at 4°C until needed.
IN-DNA-bead complexes were formed by incubating 1.0 to 2.8 μM wild-type or mutant IN protein with 5.2 mg/ml DNA-bead complex in 25 mM MOPS (pH 7.2), 23 mM NaCl, 10 mM MgCl2, 10% dimethyl sulfoxide (DMSO), and 10 mM dithiothreitol for 8 min at 37°C. Unbound protein was removed by two rounds of centrifugation and resuspension in 25 mM MOPS (pH 7.2), 23 mM NaCl, and 10 mM MgCl2, with the final bead concentration being 1.5 mg/ml. INI binding was initiated by mixing 140 μl of 1.5 mg/ml IN-DNA-bead complex (assay wells) or 140 μl of 1.5 mg/ml DNA-bead complex (control wells) and 10 μl of 600 nM 3H-labeled DTG, RAL, or ELV in a 96-well microtiter plate. The plate was sealed and incubated overnight at room temperature to obtain maximum binding. The plate was placed in a 37°C incubator for 1 h prior to the addition of unlabeled INI. For the dissociation and dissociation control wells, 10 μl of 640 μM unlabeled INI in 25 mM MOPS (pH 7.2), 23 mM NaCl, 10 mM MgCl2, and 8% DMSO was added to give a final concentration of 40 μM unlabeled DTG, RAL, or ELV. To maintain a consistent DMSO concentration, 10 μl of 25 mM MOPS (pH 7.2), 23 mM NaCl, 10 mM MgCl2, and 8% DMSO was added to the high-signal and high-signal-control wells. Dissociation of 3H-labeled INIs at 37°C was monitored for up to 3 weeks using a ViewLux charge-coupled-device imager (PerkinElmer), with the plate being maintained in a 37°C incubator between time points. The signals for duplicate wells were averaged, and the signal from control wells, which represents nonspecific binding of the 3H-labeled INI to the DNA-bead complex, was subtracted from the high signal (the high-signal control was DNA-bead, 40 nM 3H-labeled INI, and 0.5% DMSO) and the dissociation wells (the dissociation control was DNA-bead, 40 nM 3H-labeled INI, 40 μM INI, and 0.5% DMSO). Relative binding was calculated as the ratio of the background-subtracted dissociation signal to background-subtracted high signal and was fit with the equation RB = EP + ΔRB (e−kofft), using the SigmaPlot (version 8.0) program (Systat Software, Chicago, IL), where RB is the relative binding at time t, EP is the relative binding endpoint, ΔRB is the change in relative binding, and koff is the rate constant for dissociation. The endpoint was allowed to float, except for cases of extremely slow dissociation, when it was fixed at 0.1. Dissociative half-life (t1/2) values were calculated from (ln2)/koff using the mean koff value from multiple experiments. Statistical analysis was performed with the JMP (version 9) program (SAS Institute, Cary, NC). P values were calculated from the log10(koff) using all of the individual off-rate determinations and the least-squares means differences Student's t test (α = 0.05, t = 1.9835).
Strand transfer assay.
To determine the catalytic stability of the IN-DNA complex, IN-DNA-bead complexes were formed as described above for wild-type, E92Q, Y143R, Q148H, N155H, E92Q/N155H, E138K/Q148R, and G140S/Q148H INs. After resuspension of the beads at 1.5 mg/ml, the IN-DNA-bead solution was put into a 37°C incubator and strand transfer activity was determined at various time points. The strand transfer assay was conducted as described previously (2), except that DMSO was omitted and the reaction was quenched after 30 min with 25 mM MOPS (pH 7), 50 mM EDTA, and 500 mM NaCl.
RESULTS
INI dissociation from wild-type IN-DNA complex.
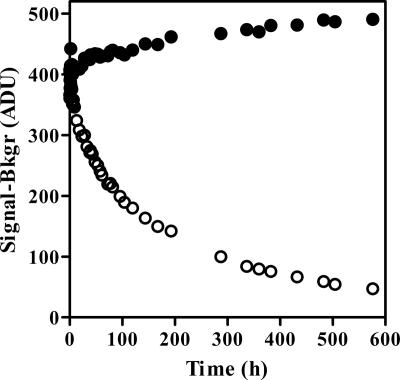
To determine rates of INI dissociation from IN-DNA complexes, [3H]DTG, [3H]RAL, and [3H]ELV were used in an SPA format (22, 23, 32). All experiments were performed at 37°C to approximate physiological conditions. Binding of the 3H-labeled INI to the IN-DNA-bead complex resulted in a high signal, as shown in Fig. 2 for [3H]DTG and a wild-type IN-DNA complex. There was no decrease in the high signal over time, indicating that the [3H]DTG-IN-DNA-bead complex was stable for more than 2 weeks at 37°C with wild-type IN. Dissociation of the 3H-labeled INI was monitored by adding excess unlabeled INI to block rebinding of the released 3H-labeled INI, which resulted in a decrease in signal (Fig. 2 for DTG). Relative binding, the proportion of 3H-labeled INI remaining bound to the IN-DNA-bead complex, was calculated for each time point (see Fig. 3A for wild-type IN). This calculation accounted for changes in the signal due to mixing and settling of the bead complexes and for changes in the signal from nonspecific binding of the 3H-labeled INI to the imaging beads. Separate controls were used for the nonspecific background in the presence and absence of unlabeled INI due to a time-dependent decrease in the signal in the presence of unlabeled INI.
Fig. 2.
DTG dissociation from wild-type IN-DNA-bead complex at 37°C. Signal in the absence (●) or presence (○) of excess unlabeled DTG following subtraction of the appropriate nonspecific binding control. Bkgr, background; ADU, analogue to digital units.
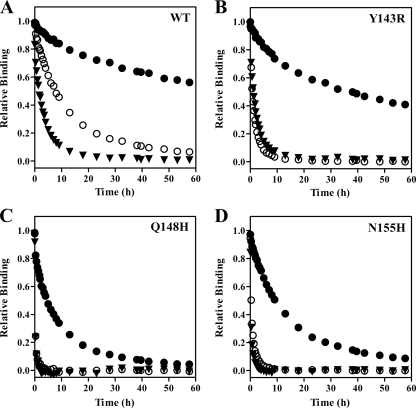
Fig. 3.
INI dissociation from selected IN-DNA complexes. Symbols are the same for all graphs and represent DTG (●), RAL (○), and ELV (▼). IN-DNA complexes contained wild type (WT) IN (A), Y143R IN (B), Q148H IN (C), or N155H IN (D).
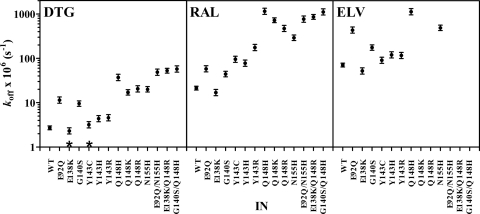
Dissociation of DTG from the wild-type IN-DNA complex was slow, with a koff of (2.7 ± 0.4) × 10−6 s−1 (Table 1). Both RAL and ELV dissociated more quickly (P < 0.0001) than DTG (Fig. 3A), with koff values of (22 ± 2) × 10−6 s−1 for RAL and (71 ± 4) × 10−6 s−1 for ELV (Table 1). The dissociative t1/2 calculated for DTG was 71 h, which was 8 times longer than that of RAL (t1/2, 8.8 h) and 26 times longer than that of ELV (t1/2, 2.7 h) (Table 2). The dissociative half-life value that we measured for RAL with the wild-type IN-DNA complex at 37°C was similar to previously reported values of 7.3 h obtained at 37°C (23) and 11.0 h obtained at 25°C (28). A dissociative half-life value of 11.1 h was previously reported for ELV at 25°C (28).
Table 1.
INI off-rate from IN-DNA complexes at 37°C
| IN |
koffa (s−1) (×10−6) |
Relative koff vs wild type |
||||
|---|---|---|---|---|---|---|
| DTG | RAL | ELV | DTG | RAL | ELV | |
| Wild type | 2.7 ± 0.4 | 22 ± 2 | 71 ± 4 | 1.0 | 1.0 | 1.0 |
| E92Q | 11.4 ± 0.3 | 59 ± 9 | 430 ± 20 | 4.2 | 2.7 | 6.1 |
| E138K | 2.3 ± 0.2 | 17 ± 0.3 | 52 ± 1 | 0.9 | 0.8 | 0.7 |
| G140S | 9.6 ± 0.8 | 44 ± 3 | 180 ± 20 | 3.6 | 2.0 | 2.5 |
| Y143C | 3.2 ± 0.1 | 96 ± 4 | 91 ± 2 | 1.2 | 4.4 | 1.3 |
| Y143H | 4.4 ± 0.2 | 78 ± 2 | 120 ± 6 | 1.6 | 3.5 | 1.7 |
| Y143R | 4.6 ± 0.3 | 176 ± 4 | 116 ± 5 | 1.7 | 8.0 | 1.6 |
| Q148H | 37 ± 3 | 1,160 ± 120 | 1,130 ± 140 | 13.7 | 53 | 16 |
| Q148K | 18 ± 5 | 730 ± 130 | NDb | 6.7 | 33 | |
| Q148R | 21 ± 2 | 480 ± 80 | ND | 7.8 | 22 | |
| N155H | 20 ± 2 | 300 ± 80 | 500 ± 140 | 7.4 | 14 | 7.0 |
| E92Q/N155H | 49 ± 3 | 770 ± 70 | ND | 18 | 35 | |
| E138K/Q148R | 53 ± 10 | 900 ± 340 | ND | 20 | 41 | |
| G140S/Q148H | 58 ± 8 | 1,130 ± 210 | ND | 22 | 51 | |
koff values represent means and standard deviations for 3 to 8 independent experiments.
ND, not determined due to low signal with [3H]ELV.
Table 2.
INI dissociative half-life and antiviral potency
| IN | Dissociative t1/2a (h) |
Fold change in EC50b |
||||
|---|---|---|---|---|---|---|
| DTG | RAL | ELV | DTG | RAL | ELV | |
| Wild type | 71 | 8.8 | 2.7 | 1 | 1 | 1 |
| E92Q | 17 | 3.3 | 0.4 | 1.6 | 3.5 | 19 |
| E138K | 84 | 11 | 3.7 | 0.97 | 1.0 | 0.93 |
| G140S | 20 | 4.4 | 1.1 | 0.86 | 1.1 | 2.7 |
| Y143C | 60 | 2.0 | 2.1 | 0.95 | 3.2 | 1.5 |
| Y143H | 44 | 2.5 | 1.6 | 0.89 | 1.8 | 1.5 |
| Y143R | 42 | 1.1 | 1.7 | 1.4 | 16 | 1.8 |
| Q148H | 5.2 | 0.2 | 0.2 | 0.97 | 13 | 7.3 |
| Q148K | 11 | 0.3 | NDc | 1.1 | 83 | >1,700 |
| Q148R | 9.2 | 0.4 | ND | 1.2 | 47 | 240 |
| N155H | 9.6 | 0.6 | 0.4 | 0.99 | 8.4 | 25 |
| E92Q/N155H | 3.9 | 0.3 | ND | 2.5 | >130 | 320 |
| E138K/Q148R | 3.6 | 0.2 | ND | 4.0 | 110 | 460 |
| G140S/Q148H | 3.3 | 0.2 | ND | 2.6 | >130 | >890 |
Effect of single IN substitutions on INI dissociation from IN-DNA complexes.
Dissociation experiments were also performed with IN-DNA complexes containing different IN substitutions to determine if changes at specific residues impact INI dissociation. Most of the IN substitutions studied here significantly (P < 0.0001) increased the koff of the INIs from the IN-DNA complexes compared to wild type (Table 1; Fig. 4). Despite the effect of single IN substitutions on DTG dissociation from IN-DNA complexes, prolonged binding with t1/2 values of 5.2 to 84 h (Table 2) was observed for DTG with all of the single-residue substitutions that were tested. Of the key INI resistance mutations, the N155H and Q148H/K/R substitutions had the greatest effect on DTG dissociation from IN-DNA complexes, having koff values of 18 × 10−6 to 37 × 10−6 s−1, representing a 6.7- to 13.7-fold increase in koff relative to the wild-type IN-DNA complex (Table 1). The dissociation of RAL and ELV from the N155H and Q148H/K/R IN-DNA complexes was significantly (P < 0.0001) faster than that of DTG (Fig. 3C and D and 4), with koff values of 300 × 10−6 to 1,160 × 10−6 s−1 for RAL and 500 × 10−6 to 1,130 × 10−6 s−1 for ELV (Table 1). The results with RAL and N155H, where the N155H substitution resulted in a 14-fold increase in koff relative to the wild-type IN-DNA complex, are consistent with previously reported observations (18). The Q148H substitution had a greater impact on dissociation of RAL from the IN-DNA complex (53-fold increase in koff) than on dissociation of DTG (13.7-fold increase) or ELV (16-fold increase) (Table 1). In our studies, dissociation of RAL and ELV from the Q148H/K/R substitutions was so fast that more than 50% of the 3H-labeled INI had already dissociated before the first time point at 20 min or an insufficient signal was obtained with [3H]ELV, such that koff could not be measured (Q148K/R). By comparison with wild type, Y143C/H/R substitutions had the least effect on dissociation of DTG (1.2-, 1.6-, and 1.7-fold, respectively) and ELV (1.3-, 1.7-, and 1.6-fold, respectively) from the IN-DNA complexes (Fig. 3B; Table 1). In fact, for DTG the koff for the IN-DNA complex with the Y143C substitution may not be different from the koff for the wild-type IN-DNA complex (P = 0.09) (Fig. 4). The Y143 substitutions had a more substantial impact on dissociation of RAL from IN-DNA complexes, with 4.4-, 3.5-, and 8-fold increases in koff for Y143C/H/R, respectively (Table 1). The E92Q and G140S substitutions did impact the dissociation of all three INIs, but the dissociative half-life for DTG remained 17 to 20 h for these IN-DNA complexes (Table 2). The E138K substitution appeared to decrease the koff with all three INIs, but the difference may not be significant for DTG (P = 0.1).
Fig. 4.
Statistical analysis of INI off-rates from IN-DNA complexes. An analysis of variance model was fit to relate the log-transformed koff values to the inhibitors and proteins. Statistical comparisons were performed, and the resulting P values were calculated using this model. The symbols represent the koff estimate and 95% confidence intervals for each IN-DNA complex containing the indicated IN protein (3 to 8 independent koff values were generated for each INI with each IN-DNA complex). For each INI, the koff values for complexes with the IN substitutions were different (P < 0.05) from the wild type (WT) except where noted (*).
Impact of multiple IN substitutions on INI dissociation from IN-DNA complexes.
While single IN mutations are of interest as the first step in acquiring resistance, most frequently, multiple IN mutations develop during therapy with RAL and ELV (36). For RAL, a broad selection of secondary mutations occurs in the Q148 and N155 pathways (3), such as G140S or E138K with Q148H/K/R and E92Q with N155H. IN proteins with two substitutions, E92Q/N155H, E138K/Q148R, and G140S/Q148H, were prepared, and INI dissociation kinetics were determined. For DTG and RAL, INI dissociation from IN-DNA complexes containing double IN substitutions was faster (P ≤ 0.0003) than dissociation from IN-DNA complexes containing the respective single IN substitution (Table 1; Fig. 4), except for Q148H and G140S/Q148H with RAL (P = 0.76). Despite the increase in koff for DTG, half of the [3H]DTG remained bound to the IN-DNA complexes containing the double IN substitutions for at least 3.3 h, which was much longer than the half-life of RAL (0.2 to 0.3 h). Under the conditions used in these experiments, the signal with [3H]ELV was not sufficient to determine koff for the double IN substitutions.
Stability of IN-DNA complexes at 37°C.
For all IN-DNA-bead complexes where signal was generated from 3H-labeled INI binding, stable high signals were obtained for at least 12 days at 37°C (data not shown). To determine if incubation at 37°C impacted the enzymatic stability of the IN in the IN-DNA complex, strand transfer activity was monitored for IN-DNA complexes containing wild-type IN or the E92Q, Y143R, Q148H, N155H, E92Q/N155H, E138K/Q148R, or G140S/Q148H substitution(s). More than half (52 to 100%) of the initial (time = 1 h) strand transfer activity was observed for all IN-DNA complexes incubated for 24 h at 37°C, for all complexes except the E92Q/N155H IN-DNA complex (44%) incubated for at least 46 h, and for some complexes (N155H and G140S/Q148H) incubated for more than 70 h. For all tested IN-DNA complexes except the wild type, the INI dissociative half-lives were shorter than the time required for the loss of 50% of the strand transfer activity.
Comparison of INI dissociation from IN-DNA complexes and antiviral potency.
In vitro antiviral studies have recently been reported for a broad panel of IN mutants, including mutants with the substitutions used in the dissociation experiments (30). In these studies, INI potency against HIV-1 isolates harboring site-directed NL432-based IN molecular clones was assayed in HeLa-CD4 cells and the fold change (FC) in the INI 50% effective concentration (EC50) versus that for wild-type HIV-1 was determined. For comparison purposes, the dissociative half-life values calculated from koff and the published in vitro antiviral data are listed together in Table 2.
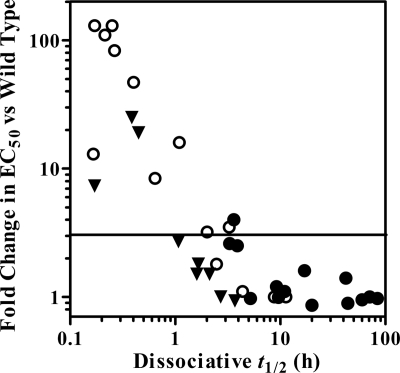
There is no direct correlation between dissociation, antiviral potency, and resistance (Fig. 5). However, IN substitutions that result in short INI dissociative half-lives also tend to have greater loss of antiviral potency. For example, DTG maintained essentially wild-type antiviral potency with all single IN substitutions tested here (FCs, 0.86 to 1.6) and demonstrated prolonged binding (t1/2 ≥ 5.2 h) in the dissociation experiments (Table 2). In contrast, RAL dissociated quickly (t1/2 < 1 h) from the Q148H/K/R and N155H IN-DNA complexes and had decreased antiviral potency against wild-type virus harboring these substitutions (FCs, 8.4 to 83) (30). Consistent with the decreased antiviral potency for ELV with the Q148K/R and E92Q/N155H, E138K/Q148R, and G140S/Q148H substitutions (FCs, 240 to >1,700), increasing the concentration of [3H]ELV 4-fold in the dissociation experiments did not result in higher signals relative to background (data not shown), suggesting that these substitutions may reduce the binding affinity of ELV with these IN-DNA complexes. Overall, there is a qualitatively inverse relationship between the dissociative half-life and in vitro antiviral potency for DTG, RAL, and ELV (Fig. 5). This observation is consistent with results obtained with other INIs (13). Empirical observations based on our data suggest that in vitro resistance (FC in EC50, ≥3 versus wild type [30]) was not observed for IN-DNA complexes/INIs with a dissociative half-life of greater than 4 h and that pronounced in vitro resistance was generally observed for IN-DNA complexes/INIs with a dissociative half-life of less than 1 h.
Fig. 5.
Comparison of dissociative half-life and in vitro antiviral activity. The fold change data were previously reported (30). Symbols represent DTG (●), RAL (○), and ELV (▾). The horizontal line indicates the threshold for resistance that was determined in the cell-based study (FC ≥ 3).
DISCUSSION
Our goal was to compare INI dissociation rates with wild-type and INI-resistant IN-DNA complexes for three INIs that have demonstrated efficacy in clinical studies. Included in this work were DTG (now in phase 3 clinical studies) and the two earlier INIs (RAL, the first FDA-approved INI, and ELV, which is also currently in phase 3 clinical studies). Resistance mutations observed in the clinic during treatment with RAL or ELV were used as a basis to select the IN substitutions for analysis, as no treatment-naïve subject has yet developed resistance to DTG.
For RAL, two main pathways for resistance involve signature mutations which are initially observed at amino acids Q148 and N155 and are almost invariably found with secondary mutations which may increase resistance and impact viral fitness and catalytic efficiency (5, 11, 18, 38, 40). With additional viral replication, mutations may be selected at Y143C and Y143R (12, 44), which then may outcompete the original mutant viruses. Studies have shown that the Y143, Q148, and N155 primary mutations or the addition of Q148 and N155 pathway secondary mutations has less impact on the in vitro enzyme and antiviral potencies of DTG than those of RAL and ELV (25, 30, 48). In our experiments, DTG demonstrated significantly slower dissociation from all IN-DNA complexes than RAL and ELV. The Q148H substitution caused the greatest fold increase in koff for all three INIs. While the addition of the Q148 and N155 secondary mutations did cause an additional increase in koff for DTG compared to the primary substitutions, the effect was less pronounced than it was for RAL. DTG maintained prolonged binding even with the IN-DNA complex containing G140S and Q148H, mutations that are frequently observed in RAL resistance during treatment (36) but have limited impact on the in vitro potency of DTG (25, 30). The in vitro biochemical and antiviral data generated with DTG suggest that an accumulation of IN mutations may be required in these RAL signature resistance pathways to have effects on DTG binding and potency similar to those observed for RAL and ELV.
The crystal structures of prototype foamy virus (PFV) IN in complex with DNA and RAL, ELV, (24), or DTG (25) and HIV-1 IN structural modeling studies (8, 9) provide insight into INI binding modes and how substitutions in the active site could impact INI binding. For RAL, the co-crystal structures show a key π-stacking interaction between RAL's oxadiazole and the side chain of Y212 in PFV IN, which corresponds to Y143 in HIV-1 IN. Since substitution of Y143 with His, Cys, or Arg likely compromises this interaction, RAL's dissociation rates are expected to increase, with the Y143R substitution having the greatest effect, given its flexibility and formal charge. The co-crystal structures with ELV and DTG reveal that these INIs make only limited van der Waals contact with Y143, suggesting that ELV and DTG dissociation rates would be minimally impacted by Y143 substitutions, which is consistent with our dissociation results. From a comparison of the PFV IN-DNA structures, the S217H and N224H (Q148H and N155H, respectively, in HIV-1 IN) substitutions appear to alter the architecture of the structural and catalytic components of the IN active site, which, moreover, appear to perturb the binding of the INIs. We speculate that Q148 may play a role in stabilizing the HIV-1 IN active-site loop into a catalytically active state and that His, Lys, or Arg substitutions at this position may alter the loop, perhaps to a greater extent than is evident in the PFV IN structures, given the added flexibility of the HIV-1 IN loop imparted by G140 (8). Also, the N155H substitution may alter the base of the HIV-1 IN catalytic pocket, the placement of at least the Mg2+ ion coordinated to E152, and the structure of that portion of the α4 helix forming one side of the pocket, causing a minor displacement of the INIs within the pocket. Altogether, these changes would be expected to increase koff for all three INIs, which is in fact observed. Consistent with our findings, an altered loop conformation or a displacement of the coordination complex would be expected to have the greatest negative impact on RAL binding, given the possibility of disrupting its π-stacking interaction with Y143. Additionally, the structural (8) and electronic (9) characteristics of DTG's metal-binding scaffold may contribute to the slower dissociation kinetics of DTG than RAL and ELV. Overall, the dissociation data are consistent with the crystallography and structural modeling results and suggest that key aspects of both the IN active site and the INIs may contribute to the observed differences in INI dissociation rates from the IN-DNA complexes.
The dissociation rate of a drug from its target typically is a major component of residence time, which can be defined as the period during which the ligand (drug) is bound to its receptor (6). A longer residence time on the targeted receptor theoretically provides beneficial characteristics; this is because the ligand has a greater opportunity to have an effect. However, the relationship between in vitro measurements of dissociation and in vivo efficacy is often qualitative rather than quantitative; ligand and receptor effective concentrations may vary substantially during the course of dosing and as a function of ongoing biological processes. As such, evaluation of the in vitro and in vivo relationship should take into account the window or duration of the pharmacological effect. In the case of HIV, there is a window of opportunity for many processes, including those targeted by current anti-HIV agents. For IN strand transfer inhibitors such as the INIs studied here, the window during which binding most likely occurs is between 3′ processing (which generates structural components of the catalytic binding pocket [15, 26] of the HIV cDNA genome ends) and integration into the host genome. Interestingly, in vitro washout experiments found an association between an extended INI dissociative half-life and a more persistent antiviral effect, apparently via the irreversible generation of unintegrated viral cDNA (13). The dissociation rate of INIs from IN-DNA complexes in vitro, which should be reflected in the residence time, might also be an important component of INI efficacy in vivo.
An important consideration is that prolonged binding of DTG to IN-DNA complexes with INI resistance mutations would be suggestive of a higher barrier to resistance. Since INI binding generates nonreplicative forms of unintegrated viral cDNA, such as 2 long terminal repeats (27), there is less opportunity for the virus to undergo further rounds of replication which can generate additional mutations and thereby lead to higher levels of resistance. In the case where these mutations are the first step in a sequence of mutations required to achieve high-level resistance, prolonged INI binding theoretically should provide an improved potential for a higher barrier to resistance.
The comparison of dissociative half-life and antiviral potency suggests that there might be a threshold residence time for INIs which impacts antiviral efficacy in the cell-based system used to determine antiviral potency (30). From our data set, this threshold would be a dissociative half-life of between 1 and 4 h, but additional data would be needed to further refine this relationship and to identify exceptions. It is possible that the threshold is related to the length of the window during HIV replication when IN activity is required. One could speculate that, in particular during periods of nonadherence when drug levels may reach suboptimal levels, an INI with a longer dissociative half-life might remain bound and prevent integration. Additional experiments with systems in which this integration window is significantly longer, such as primary blood-derived monocytes, might be illuminating. In addition, time-of-removal or time-of-addition experiments might reveal whether DTG has a prolonged antiviral effect upon washout similar to what was observed for BMS-878397 compared to RAL (13).
The dissociation data presented here provide additional evidence of differential binding of INIs to wild-type and mutant IN-DNA complexes and suggest that prolonged INI binding may contribute to efficacy. While dissociation kinetics and compound residence time are likely to be important for INIs, other factors such as the rate of compound association (48), drug pharmacokinetics, and viral fitness could also impact efficacy and resistance. Additional work is necessary to elucidate the contribution of each of these components to the emergence of INI resistance. However, DTG's long dissociation half-life with the wild-type and INI-resistant IN-DNA complexes may contribute to its distinct resistance profile and highlight the potential for improved activity against wild-type HIV-1 and clinically relevant INI-resistant viruses. Among the potential important predictions from the preceding discussion is that DTG might have a higher capacity to suppress viral replication should drug levels become suboptimal due to adherence problems (i.e., possess “forgiveness”) and that the slower dissociation may translate to a higher genetic barrier to resistance in vivo. Ultimately, however, such predictions will need to be verified by data from the phase 2b and phase 3 clinical studies which are currently ongoing.
ACKNOWLEDGMENTS
We thank Ken Lawrie and Nick Shipley for synthesizing the iodinated INIs and obtaining the 3H-labeled versions, Lisa Leesnitzer for assistance setting up the SPA method, Steven Novick for performing the statistical analysis, and Angela McKenzie for cataloging the IN proteins.
Footnotes
Published ahead of print on 1 August 2011.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. Asante-Appiah E., Skalka A. M. 1999. HIV-1 integrase: structural organization, conformational changes, and catalysis. Adv. Virus Res. 52:351–369 [DOI] [PubMed] [Google Scholar]
- 2. Boros E. E., Johns B. A., Garvey E. P., Koble C. S., Miller W. H. 2006. Synthesis and HIV-integrase strand transfer inhibition activity of 7-hyroxyl[1,3]thiazolo[5,4-b]pyridin-5(4H)-ones. Bioorg. Med. Chem. Lett. 16:5668–5672 [DOI] [PubMed] [Google Scholar]
- 3. Ceccherini-Silberstein F., et al. 2009. Characterization and structural analysis of HIV-1 integrase conservation. AIDS Rev. 11:17–29 [PubMed] [Google Scholar]
- 4. Chiu T. K., Davies D. R. 2004. Structure and function of HIV-1 integrase. Curr. Top. Med. Chem. 4:965–977 [DOI] [PubMed] [Google Scholar]
- 5. Cooper D. A., et al. 2008. Subgroup and resistance analyses of raltegravir for resistant HIV-1 infection. N. Engl. J. Med. 359:355–365 [DOI] [PubMed] [Google Scholar]
- 6. Copeland R. A., Pompliano D. L., Meek T. D. 2006. Drug-target residence time and its implications for lead optimization. Nat. Rev. Drug Discov. 5:730–739 [DOI] [PubMed] [Google Scholar]
- 7. Craigie R. 2001. HIV integrase, a brief overview from chemistry to therapeutics. J. Biol. Chem. 276:23213–23216 [DOI] [PubMed] [Google Scholar]
- 8. DeAnda F., Hattori K., Yoshinaga T., Kawasuji T., Underwood M. R. 2010. Structural models of HIV-1 integrase and DNA in complex with S/GSK1349572, raltegravir, or elvitegravir: structure-based rationale for INI resistance profiles. Antiviral Ther. 15(Suppl. 2):A73 [Google Scholar]
- 9. DeAnda F., et al. 2010. Structural rationale for slow S/GSK1349572 dissociation from wild type (WT) and raltegravir (RAL) resistant HIV-1 integrase (IN), abstr. H-1166. Abstr. 50th Intersci. Conf. Antimicrob. Agents Chemother American Society for Microbiology, Washington, DC [Google Scholar]
- 10. DeJesus E., et al. 2007. First report of raltegravir (RAL, MK-0518) use after virologic rebound on elvitegravir (EVT, GS 9137), abstr. TUPEB032. Abstr. 4th Int. AIDS Soc. Conf. HIV Pathogenesis, Treatment, Prevention International AIDS Society, Geneva, Switzerland [Google Scholar]
- 11. Delelis O., et al. 2009. The G140S mutation in HIV integrases from raltegravir-resistant patients rescues catalytic defect due to the resistance Q148H mutation. Nucleic Acids Res. 37:1193–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Delelis O., et al. 2010. Impact of Y143 HIV-1 integrase mutations on resistance to raltegravir in vitro and in vivo. Antimicrob. Agents Chemother. 54:491–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dicker I. B., et al. 2011. Longer integrase inhibitor dissociation half-lives induce persistent anti-HIV effects in cell culture, abstr. 521. Abstr. 18th Conf. Retrovir. Opportunistic Infect [Google Scholar]
- 14. Dicker I. B., et al. 2008. Biochemical analysis of HIV-1 integrase variants resistant to strand transfer inhibitors. J. Biol. Chem. 283:23599–23609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dicker I. B., et al. 2007. Changes to the HIV long terminal repeat and to HIV integrase differentially impact HIV integrase assembly, activity, and the binding of strand transfer inhibitors. J. Biol. Chem. 282:31186–31196 [DOI] [PubMed] [Google Scholar]
- 16. Eron J., et al. 2010. Activity of the integrase inhibitor S/GSK1349572 in subjects with HIV exhibiting raltegravir resistance: week 24 results of the VIKING study (ING112961). J. Int. AIDS Soc. 13:O51 [Google Scholar]
- 17. Esposito D., Craigie R. 1999. HIV integrase structure and function. Adv. Virus Res. 52:319–333 [DOI] [PubMed] [Google Scholar]
- 18. Fransen S., et al. 2009. Loss of raltegravir susceptibility by human immunodeficiency virus type 1 is conferred via multiple nonoverlapping genetic pathways. J. Virol. 83:11440–11446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Garvey E. P., et al. 2009. Potent inhibitors of HIV-1 integrase display a two-step, slow-binding inhibition mechanism which is absent in a drug-resistant T66I/M154I mutant. Biochemistry 48:1644–1653 [DOI] [PubMed] [Google Scholar]
- 20. Goethals O., et al. 2010. Primary mutations selected in vitro with raltegravir confer large fold changes in susceptibility to first-generation integrase inhibitors, but minor fold changes to inhibitors with second-generation resistance profiles. Virology 402:338–346 [DOI] [PubMed] [Google Scholar]
- 21. Goethals O., et al. 2008. Resistance mutations in human immunodeficiency virus type 1 integrase selected with elvitegravir confer reduced susceptibility to a wide range of integrase inhibitors. J. Virol. 82:10366–10374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Grobler J. A., Stillmock K. A., Hazuda D. J. 2009. Scintillation proximity assays for mechanistic and pharmacological analyses of HIV-1 integration. Methods 47:249–253 [DOI] [PubMed] [Google Scholar]
- 23. Grobler J. A., et al. 2009. Functionally irreversible inhibition of integration by slowly dissociating strand transfer inhibitors, abstr. O-10. Abstr. 10th Int. Workshop Clin. Pharmacol. HIV Ther. [Google Scholar]
- 24. Hare S., et al. 2010. Molecular mechanisms of retroviral integrase inhibition and the evolution of viral resistance. Proc. Natl. Acad. Sci. U. S. A. 107:20057–20062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hare S., et al. 30 June 2011, posting date Structural and functional analyses of the second-generation integrase strand transfer inhibitor dolutegravir (S/GSK1349572). Mol. Pharmacol. doi:10.1124/mol.111.073189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hare S., Gupta S. S., Valkov E., Engelman A., Cherepanov P. 2010. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature 464:232–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hazuda D. J., et al. 2000. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science 287:646–650 [DOI] [PubMed] [Google Scholar]
- 28. Hluhanich R., et al. 2010. HIV integrase inhibitors (INIs) do not exert a post-antibiotic effect (PAE) despite slow dissociation from IN-DNA complexes in vitro, abstr. H-930. Abstr. 49th Intersci. Conf. Antimicrob. Agents Chemother American Society for Microbiology, Washington, DC [Google Scholar]
- 29. Johns B., et al. 2010. The discovery of S/GSK1349572: a once-daily next generation integrase inhibitor with a superior resistance profile, abstr. 55. Abstr. 17th Conf. Retrovir. Opportunistic Infect. [Google Scholar]
- 30. Kobayashi M., et al. 2011. In vitro virology of S/GSK1349572, a next-generation HIV integrase inhibitor. Antimicrob. Agents Chemother. 55:813–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. LaFemina R. L., et al. 1992. Requirement of active human immunodeficiency virus type 1 integrase enzyme for productive infection of human T-lymphoid cells. J. Virol. 66:7414–7419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Langley D. R., et al. 2008. The terminal (catalytic) adenosine of the HIV LTR controls the kinetics of binding and dissociation of HIV integrase strand transfer inhibitors. Biochemistry 47:13481–13488 [DOI] [PubMed] [Google Scholar]
- 33. Lewinski M. K., Bushman F. D. 2005. Retroviral DNA integration—mechanism and consequences. Adv. Genet. 55:147–181 [DOI] [PubMed] [Google Scholar]
- 34. Liao C., Marchand C., Burke T. R., Jr., Pommier Y., Nicklaus M. C. 2010. Authentic HIV-1 integrase inhibitors. Future Med. Chem. 2:1107–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Markowitz M., et al. 2007. Rapid and durable antiretroviral effect of the HIV-1 integrase inhibitor raltegravir as part of combination therapy in treatment-naive patients with HIV-1 infection: results of a 48-week controlled study. J. Acquir. Immune Defic. Syndr. 46:125–133 [DOI] [PubMed] [Google Scholar]
- 36. McColl D. J., Chen X. 2010. Strand transfer inhibitors of HIV-1 integrase: bringing IN a new era of antiretroviral therapy. Antiviral Res. 85:101–118 [DOI] [PubMed] [Google Scholar]
- 37. Métifiot M., Marchand C., Maddali K., Pommier Y. 2010. Resistance to integrase inhibitors. Viruses 2:1347–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Métifiot M., et al. 2010. Biochemical and pharmacological analyses of HIV-1 integrase flexible loop mutants resistant to raltegravir. Biochemistry 49:3715–3722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Min S., et al. 29 June 2011, posting date Antiviral activity, safety, and pharmacokinetics/pharmacodynamics of dolutegravir as 10-day monotherapy in HIV-1-infected adults. AIDS doi:10.1097/QAD.0b013e32834a1dd9 [DOI] [PubMed] [Google Scholar]
- 40. Nakahara K., et al. 2009. Secondary mutations in viruses resistant to HIV-1 integrase inhibitors that restore viral infectivity and replication kinetics. Antiviral Res. 81:141–146 [DOI] [PubMed] [Google Scholar]
- 41. Ratner L., et al. 1985. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature 313:277–284 [DOI] [PubMed] [Google Scholar]
- 42. Rockstroh J., et al. 2010. Once-daily S/GSK1349572 combination therapy in antiretroviral-naïve adults: rapid and potent 24-week antiviral responses in SPRING-1 (ING112276). J. Int. AIDS Soc. 13:O50 [Google Scholar]
- 43. Shimura K., et al. 2008. Broad antiretroviral activity and resistance profile of the novel human immunodeficiency virus integrase inhibitor elvitegravir (JTK-303/GS-9137). J. Virol. 82:764–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shin C. G., Taddeo B., Haseltine W. A., Farnet C. M. 1994. Genetic analysis of the human immunodeficiency virus type 1 integrase protein. J. Virol. 68:1633–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Steigbigel R. T., et al. 2008. Raltegravir with optimized background therapy for resistant HIV-1 infection. N. Engl. J. Med. 359:339–354 [DOI] [PubMed] [Google Scholar]
- 46. Underwood M. R., Johns B., Sato A., Fujiwara T., Spreen W. 2009. S/GSK1349572: a next generation integrase inhibitor with activity against integrase inhibitor-resistant clinical isolates from patients experiencing virologic failure while on raltegravir therapy, abstr. WEPEA098. Abstr. 5th Int. AIDS Soc. Conf. HIV Pathogenesis, Treatment, Prevention International AIDS Society, Geneva, Switzerland [Google Scholar]
- 47. Witmer M., Danovich R. 2009. Selection and analysis of HIV-1 integrase strand transfer inhibitor resistant mutant viruses. Methods 47:277–282 [DOI] [PubMed] [Google Scholar]
- 48. Yoshinaga T., Kanamori-Koyama M., Seki T., Sato A., Fujiwara T. 2010. Strong inhibition of wild-type and integrase inhibitor (INI)-resistant HIV integrase (IN) strand transfer reaction by the novel INI S/GSK1349572. Antivir. Ther. 15(Suppl. 2):A12 [Google Scholar]