Abstract
Enterococcus faecium UCN71, isolated from a blood culture, was resistant to low levels of vancomycin (MIC, 16 μg/ml) but susceptible to teicoplanin (MIC, 0.5 μg/ml). No amplification was observed with primers specific for the previously described glycopeptide resistance ligase genes, but a PCR product corresponding to a gene called vanN was obtained using degenerate primers and was sequenced. The deduced VanN protein was related (65% identity) to the d-alanine:d-serine VanL ligase. The organization of the vanN gene cluster, determined using degenerate primers and by thermal asymmetric interlaced (TAIL)-PCR, was similar to that of the vanC operons. A single promoter upstream from the resistance operon was identified by rapid amplification of cDNA ends (RACE)-PCR. The presence of peptidoglycan precursors ending in d-serine and d,d-peptidase activities in the absence of vancomycin indicated constitutive expression of the resistance operon. VanN-type resistance was transferable by conjugation to E. faecium. This is the first report of transferable d-Ala-d-Ser-type resistance in E. faecium.
INTRODUCTION
The glycopeptide antibiotics vancomycin and teicoplanin are widely used for the treatment of severe infections due to Gram-positive bacteria and act by inhibition of peptidoglycan synthesis. Glycopeptides bind to the C-terminal d-alanyl-d-alanine of late peptidoglycan precursors and block the following steps in cell wall assembly (14).
During the past two decades, glycopeptide-resistant enterococci (GRE), in particular Enterococcus faecium, have spread as multiresistant opportunistic pathogens in hospitals worldwide. Since the first descriptions of vancomycin resistance in E. faecium in 1988 (24, 38), eight operons that confer resistance to glycopeptides have been distinguished on the basis of the sequence of the structural gene for the resistance ligase. They are designated according to the gene that encodes either a d-alanyl-d-lactate (vanA, vanB, vanD, and vanM) or a d-alanyl-d-serine (vanC, vanE, vanG, and vanL) ligase (14, 8, 40). The production of modified peptidoglycan precursors ending in d-Ala-d-Lac or d-Ala-d-Ser associated with the elimination of d-Ala-d-Ala-ending precursors is responsible for resistance. This replacement leads to diminished binding affinity of glycopeptides for their targets (1,000-fold lower and 7-fold lower for d-Ala-d-Lac and d-Ala-d-Ser precursors, respectively) (6). The vanA and vanD d-Ala-d-Lac operons are responsible for high and moderate levels of resistance, respectively, to both vancomycin and teicoplanin, whereas the vanB operons are responsible for various resistance levels to vancomycin only. In contrast, the d-Ala-d-Ser operons confer low-level vancomycin resistance, whereas teicoplanin retains activity (11). Three proteins, a ligase, a dehydrogenase (for the d-Ala-d-Lac type) or a serine racemase (for the d-Ala-d-Ser type), and a d,d-dipeptidase are required for resistance (14). The operons encode additional proteins, such as d,d-carboxypeptidases and regulatory proteins, parts of two-component regulatory systems.
Except for VanC resistance, which is intrinsic to Enterococcus gallinarum and Enterococcus casseliflavus, resistance types are acquired. While the d-Ala-d-Lac-type operons may be located either on plasmids or in the chromosome, the d-Ala-d-Ser-type gene clusters, VanG, VanE, and VanL, have so far been detected only in the chromosome of Enterococcus faecalis (8, 11, 14). VanG is the only transferable d-Ala-d-Ser resistance type characterized so far (12).
E. faecium UCN71 was resistant to low levels of vancomycin and susceptible to teicoplanin but did not harbor any known vancomycin resistance ligase gene. We present evidence that resistance to vancomycin in this clinical isolate is due to a new type of determinant, named VanN. Phenotypic and genotypic traits of VanN resistance were identified, and transferability was assessed.
MATERIALS AND METHODS
Strains.
The characteristics of the strains are listed in Table 1. E. faecium UCN71 and E. faecium UCN72 were isolated in 2008 from a blood culture and a rectal swab, respectively, of a patient at the Paoli-Calmettes Institute, Marseille, France. E. faecium BM4107 (24) and E. faecalis JH2-2 (22), which are resistant to fusidic acid and rifampin, were used as recipients in conjugation experiments. VanG-type E. faecalis BM4518 (12) was used as a reference for analysis of peptidoglycan precursors.
Table 1.
Bacterial strains
| Species and strain | Relevant characteristicsa | Reference or source |
|---|---|---|
| E. faecium | ||
| BM4107 | Rifr Fusr | 24 |
| UCN71 | Clinical isolate; Vmr Tes Emr (VanN type) | This study |
| UCN72 | Clinical isolate; Vmr Tes Emr (VanN type) | This study |
| UCN73 | Rifr Fusr Vmr | This study |
| E. faecalis | ||
| JH2-2 | Rifr Fusr | 22 |
| BM4518 | Clinical isolate; Vmr Tes (VanG type) | 12 |
Em, erythromycin; Fus, fusidic acid; r, resistant; Rif, rifampin; s, susceptible; Te, teicoplanin; Vm, vancomycin.
Antibiotic susceptibility testing.
MICs were determined by the broth microdilution technique recommended by the Comité de l'Antibiogramme de la Société Française de Microbiologie (10). Plates were incubated for 24 h at 37°C before being read.
Molecular typing.
Clinical isolates were typed by pulsed-field gel electrophoresis (PFGE) of genomic DNA using a CHEF II Mapper system (Bio-Rad, Marnes-la-Coquette, France) as described previously (36). Total DNA digested with SmaI was subjected to electrophoresis (6 V/cm, 22 h, pulse times of 5 to 30 s at 14°C) in an agarose gel.
Multilocus sequence typing (MLST) of E. faecium was performed as previously reported (20). The alleles and sequence types (STs) were analyzed and determined using the MLST database (http://efaecium.mlst.net/).
PCR.
Amplification was carried out in a final volume of 50 μl containing 40 pmol of each oligonucleotide primer, ca. 100 ng of template DNA, using a GoTaq Flexi DNA polymerase kit (Promega, Madison, WI) as recommended by the manufacturer. The reactions were performed under the following conditions: 30 s at 94°C, 30 s at 50 to 54°C, and 30 s to 2 min at 72°C (30 cycles).
Identification of strains UCN71 and UCN72 at the species level was performed by PCR as previously described (16). A PCR assay with primers specific for resistance genes vanA, vanB, vanC1, and vanC2 (16), vanD (30), vanE (18), vanG (26), vanL (8), and vanM (40) was used to determine the glycopeptide resistance genotype of E. faecium UCN71.
Amplification of fragments internal to genes for related ligases was performed with degenerate V1 and V2 primers as described previously (15). Prior to amplification, total DNA from strain UCN71 was digested with XhoI (Ozyme) in order to cleave the ddl gene and to avoid its amplification. PCR fragments were purified using a NucleoSpin Extract II kit (Macherey-Nagel, Düren, Germany), cloned in pCRTOPO2.1 (Invitrogen, Carlsbad, CA), and sequenced. The entire vanN sequence and adjacent sequences were determined with a series of thermal asymmetric interlaced PCRs (TAIL-PCRs) starting from both sides of the vanN partial gene sequence as described by Liu and Whittier (25) using the primers listed in Table 2. Briefly, the primary PCR mixture consisted of 1× PCR buffer, 2 mM MgCl2, 0.2 mM (each) deoxynucleoside triphosphate (dNTP), 0.2 μM specific primer, an AD primer (5 μM for AD1 and AD2 or 2.5 μM for AD3 and AD4), and 0.8 units of Taq polymerase. For secondary reactions, aliquots of 1/100-diluted primary reaction mixtures were added to secondary PCR mixtures containing 0.2 μM internal specific primer and the same arbitrary primer used in the primary reaction (3 μM for AD1 or AD2 and 1.5 μM for AD3 or AD4). If needed, tertiary PCRs were performed as in the preceding reaction using the inner-most specific primer. Thermal cycling conditions for primary, secondary, and tertiary reactions were used as previously described (25). Finally, a set of degenerate primers complementary to conserved regions of d-Ala-d-Ser operons (8) and vanN operon-specific primers (Table 2) were used to amplify the region downstream from the ligase gene as previously described (8). PCR products were sequenced and concatenated to obtain the entire vanN gene cluster.
Table 2.
Primers used in this study
| Primera | Sequence (5′–3′)b | Position (5′–3′)c | Reference |
|---|---|---|---|
| V1-F | GGIGARGAYGGIWSIHTICARGG | 333–355 | 15 |
| V2-R | TGRAAICCIGGIADIGTRTT | 964–945 | 15 |
| vanN-F | CCTCTGCCATTTGTATGAAT | 403–422 | This study |
| vanN-R | TTGACGCTTGATTTCTCTAC | 851–832 | This study |
| RACE-PCR | |||
| vanNRace1-R | GTCCGTCGATGAGATCAG | 497–480 | This study |
| vanNRace2-R | GCCGCAACCAACATAAGGAAT | 395–375 | This study |
| vanNRace3-R | TCTCCAGTGCCACCGTGTA | 337–319 | This study |
| vanRNRace1-R | CTATGGGAAGGCTGATCG | 4188–4171 | This study |
| vanRNRace2-R | TTCCAGCGGGTTGAAAGGT | 4126–4108 | This study |
| vanRNRace3-R | ATCTGCGCCTAGAGTCAGTC | 4093–4074 | This study |
| Sequencing | |||
| vanTm1-F | CATGGTCGTTGCGATTCACTG | 1679–1699 | This study |
| vanTm2-F | CTGCTTATTACAGCGGTTCACGC | 2042–2064 | This study |
| vanTr-F | CTAGGCTATACACCATCGATACG | 2883–2905 | This study |
| vanR-F | GGAGTGGTAGGGATAATGGATAC | 3790–3812 | This study |
| vanT-U-R | TDATNVDNGCNGGNARRTACCA | 2023–2001 | 8 |
| vanT-U2-R | CCNARNCKRTGCAMNCCNGTRTC | 3034–3012 | 8 |
| RserU1-R | AYRTCNGGNARCATNACRTC | 3978–3959 | 8 |
| SerU1-R | GGNGTNYKNARRTCRTGNGC | 4954–4935 | 8 |
| TAIL-PCR | |||
| AD1 | TGWGNAGWANCASAGA | NA | 25 |
| AD2 | AGWGNAGWANCAWAGG | NA | 25 |
| AD3 | CAWCGICNGAIAWGAA | NA | 25 |
| AD4 | TCSTICGNACITWGGA | NA | 25 |
F, forward; R, reverse.
K for G or T; M for A or C; R for A or G; S for G or C; W for A or T; Y for C or T; D for A, G, or T; V for A, C, or G; I for inosine, and N for any base.
Position from the vanN translation start codon. NA, not applicable.
RNA extraction and mapping of the transcriptional start site.
Total RNA from strain UCN71 grown in Mueller-Hinton broth at 37°C to the exponential growth phase was isolated using an RNeasy minikit (Qiagen, Valencia, CA). The transcriptional start site of vanN was determined using a 5′ rapid amplification of cDNA ends (RACE) system kit (Invitrogen) according to the manufacturer's instructions. The specific RACE-PCR primers designed using the vanN gene sequence and Primer3 software (http://frodo.wi.mit.edu/primer3/) are listed in Table 2.
Conjugation.
Transfer of vancomycin resistance from strain UCN71 to E. faecium BM4107 and E. faecalis JH2-2 was attempted by filter mating (15). Transconjugants were selected on brain heart infusion (BHI) agar plates containing rifampin (60 μg/ml), fusidic acid (50 μg/ml), and vancomycin (10 μg/ml).
Fitness experiments.
The biological cost of vancomycin resistance was evaluated in a microtitration plate growth model. Overnight cultures in BHI broth were inoculated (200 μl) in fresh 10-ml BHI broth, and strains were cultivated at 37°C. Microtitration plates were agitated (10 s at 200 rpm) every 20 min to resuspend the cells, the optical density at 600 nm (OD600) was measured every 10 min, and growth rates were calculated from at least three independent experiments.
Analysis of peptidoglycan precursors.
Analysis of peptidoglycan precursors was performed as described by Reynolds et al. (31). Strain UCN71 was grown in BHI broth with (4 and 8 μg/ml) or without vancomycin to the mid-exponential phase. Ramoplanin (3 μg/ml) was added, and incubation was continued for 20 min. The bacteria were harvested by centrifugation (12,000 × g, 2 min, 4°C), and the accumulated cytoplasmic precursors were extracted with 8% trichloroacetic acid (15 min, 4°C), desalted, and analyzed by high-performance liquid chromatography. Results were expressed as the percentages of total late peptidoglycan precursors represented by UDP-MurNAc-tetrapeptide and UDP-MurNAc-pentapeptide terminating in d-Ala or d-Ser [UDP-MurNAc-pentapeptide(Ala) or UDP-MurNAc-pentapeptide(Ser)]) and were determined from the integrated peak areas.
d,d-peptidase (VanXY) and serine racemase (VanT) activities.
The enzymatic activities in the supernatant and in the resuspended pellet fraction were assayed in UCN71 extracts as described previously (13). Strains were grown until the OD600 reached 0.7 in the absence or presence (4 and 8 μg/ml) of vancomycin. Bacteria were harvested and lysed by treatment with lysozyme (2 mg/ml) at 37°C followed by sonication, and the membrane fraction was pelleted (100,000 × g, 45 min). The supernatant and the resuspended pellet were collected and assayed for d,d-peptidase (VanX or VanY) activity. The amounts of d-Ala released from d-Ala-d-Ala by d,d-dipeptidases and from pentapeptide(Ala) by d,d-carboxypeptidases were determined by using d-amino acid oxidase (5, 33).
The assay for serine racemase activity was carried out in cytoplasmic and membrane fractions as described previously (12). The assay mixture contained bis-Tris propane (50 mM [pH 7.5]), 10 mM l-serine or l-alanine, and the diluted fraction as the enzyme preparation. The d-amino acids produced by racemase activity were assayed using a d-amino acid oxidase assay (27).
The protein concentration was determined according to the method of Bradford (9), with bovine serum albumin as the standard. Specific activity was defined as the number of nanomoles of product formed at 37°C per minute per milligram of protein contained in the extracts.
Nucleotide sequence accession number.
The sequence of vanN was submitted to GenBank and was assigned accession number JF802084.
RESULTS AND DISCUSSION
Characterization of E. faecium UCN71 and UCN72.
E. faecium UCN71 and E. faecium UCN72 were isolated from a blood culture and a rectal swab, respectively. The patient was diagnosed with Burkitt's lymphoma and had received vancomycin for 26 days. Clinical isolates UCN71 and UCN72 were identified as E. faecium by conventional tests and by PCR of the d-Ala:d-Ala ligase ddl gene. Both strains were susceptible to ampicillin and constitutively resistant to erythromycin (MIC, >256 μg/ml) and to low levels of vancomycin (MIC, 16 μg/ml) but remained susceptible to teicoplanin (MIC, 1 μg/ml). No PCR products were obtained using primers specific for the previously described glycopeptide resistance ligases, suggesting that both strains were resistant to vancomycin by acquisition of a new glycopeptide resistance determinant. Since PFGE analysis showed that the restriction profiles of strains UCN71 and UCN72 were indistinguishable (data not shown), further experiments were conducted with only the blood isolate UCN71. MLST indicated that strain UCN71 was of sequence type ST240, which does not belong to the well-characterized clonal complex 17 (CC17) in which hospital-adapted and epidemic E. faecium strains cluster (23, 37). Consistent with this observation, the antibiotic susceptibility profile of UCN71, in particular susceptibility to ampicillin and fluoroquinolones, differed from the usual multidrug resistance profile of vancomycin-resistant E. faecium CC17 responsible for hospital outbreaks.
Identification of the vanN gene.
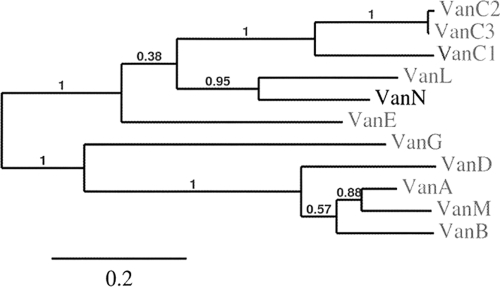
The E. faecium ddl gene has a XhoI restriction site that is absent from all previously described glycopeptide resistance ligase genes. After total UCN71 DNA was digested with XhoI, PCR was performed with the V1 and V2 primers and the ca. 600-bp fragment obtained was cloned into Escherichia coli and sequenced. The deduced sequence was compared with those of the E. faecium Ddl ligase and of the resistance d-Ala:d-Lac and d-Ala:d-Ser ligases. Among the clones analyzed, one had a sequence distantly related to that of the ligase of E. faecium but displayed 65% identity with the corresponding portion of VanL and, to a lesser extent, homology with other d-Ala:d-Ser ligases (Table 3). The upstream and downstream regions were sequenced following TAIL-PCR, and the new gene was named vanN. Identity between VanN and VanC, VanE, VanG, and VanL ligases varied from 41% to 65% (Table 3). All the motifs conserved in the d-Ala:d-Ser ligases were present (17, 39). The EKYQ motif, involved in serine recognition, was present at residues 252 to 255 of VanN. Phylogenetic analysis of the glycopeptide resistance ligase, which allows clustering of the d-Ala-d-Lac or d-Ala-d-Ser resistance types, enabled assignment of VanN to the d-Ala:d-Ser group ligases (Fig. 1).
Table 3.
Percent identity of Van-deduced proteins
| Van type | % identity with indicated proteina |
||||
|---|---|---|---|---|---|
| VanN | VanXYN | VanTN | VanRN | VanSN | |
| VanL | 65 | 61 | 50 and 54b | 74 | 61 |
| VanC | 54 | 50 | 52 | 74 | 52 |
| VanE | 53 | 46 | 45 | 62 | 40 |
| VanG | 41 | 40 | 37 | 49 | 43 |
| VanA | 38 | NA | NA | 51 | 35 |
Numbers in boldface are the highest percentages of identity with VanN-type proteins. NA, not applicable.
The transmembrane and racemase domains of VanTN were compared with those of VanTrL and VanTmL, respectively.
Fig. 1.
Phylogenetic analysis of vancomycin resistance ligases. Branch support values are indicated on the phylogram. The neighbor-joining method (34) was used for sequence comparison. Phylogenetic analysis was conducted using MEGA 4 (35).
Characterization of the vanN operon.
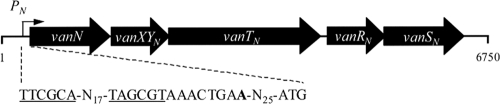
A 6,750-bp sequence was obtained from both sides of the vanN gene using a series of degenerated primers or primers designed for TAIL-PCR. Based on the homology of the deduced sequences to those of proteins encoded by the vanC, vanE, vanG, and vanL gene clusters (Table 3), open reading frames (ORFs) vanN, vanXYN, vanTN, vanRN, and vanSN were identified (Fig. 2). The genetic organization of the vanN operon was similar to that of the vanC, vanE, and vanL gene clusters (14).
Fig. 2.
Schematic representation of the vanN operon. Identified −35 and −10 regions are underlined in the PN promoter region. The transcription start site is indicated in boldface.
VanXYN exhibited 61% identity with VanXYL (Table 3) and contained all the conserved active-site residues of other VanXY proteins that display bifunctional d,d-dipeptidase and d,d-carboxypeptidase activities (32).
The third 2,103-bp vanTN gene putatively encoded a protein similar in length and sequence (52% identity) to VanTc (the racemase of the vanC operon). The N terminus of VanTN contained 10 predicted hydrophobic transmembrane domains homologous with those of VanTC (data not shown) that are likely to be involved in the transport of l-Ser (3). The C terminus of VanTN contained a 13-amino-acid pyridoxal attachment site that binds the phosphate pyridoxal cofactor required for racemase activity (1; also data not shown). In the vanL gene cluster, the transmembrane and racemase domains are located on two adjacent vanTmL and vanTrL genes, respectively (8). The hydrophobic transmembrane and racemase domains of VanTN displayed 50% and 54% identity with those of VanTmL and VanTrL, respectively.
The last two deduced proteins, VanRN and VanSN, were similar to VanR- and VanS-type proteins, of which the two-component regulatory systems found in the van gene clusters are comprised. Deduced protein VanRN exhibited 74% identity with VanRL and VanRC and contained the conserved residues (D10, D53, and K102) found in response regulators from two-component systems in Gram-positive bacteria (28). Deduced protein VanSN exhibited 61% identity with VanSL and contained the five conserved amino acid motifs (H, N, G1, F, and G2 boxes) of the C-terminal transmitter modules of the VanS sensors (21). The H block is responsible for both autophosphorylation and kinase/phosphatase activities, and G1 and G2 correspond to ATP binding blocks (14). In addition, the VanSN N-terminal region contained two transmembrane domains characteristic of the sensor proteins in two-component systems (Fig. 3) (4). Noticeably, VanSN had a serine at position 156, in the H box close to the autophosphorylation site, instead of a proline found at the corresponding position in the other VanS proteins (Fig. 3 and data not shown). Amino acid substitutions in the H domain of VanSB were previously shown to be responsible for constitutive resistance (13, 14). Since dephosphorylation of VanRB is required to prevent transcription of the resistance genes, impaired dephosphorylation of VanRB by VanSB leads to constitutive expression of the vanB cluster (14). Similarly, the substitution in VanSN may affect its phosphatase activity and is likely to be responsible for constitutive expression of resistance. Moreover, VanSN exhibited a glutamine in position 204 instead of an arginine as in VanSL, VanSC and VanSG (Fig. 3). This replacement could play the same role as the R200L substitution associated with the constitutive expression of resistance in VanSC from E. gallinarum (29). These data are consistent with growth curves in the presence of vancomycin that are similar for induced and noninduced cells (data not shown) and analyses of peptidoglycan precursors and d,d-peptidases and racemase activities (see below) that indicate constitutive vancomycin resistance in E. faecium UCN71.
Fig. 3.
Alignment of deduced amino acid sequences of VanSN (E. faecium UCN71), VanSL (E. faecalis N06-0364), VanSC (E. gallinarum BM4174), VanSG (E. faecalis BM4518), and VanSE (E. faecalis N00-410). Identical residues are marked in black, conserved amino acid motifs of histidine kinases (H, N, G1, F, and G2) are indicated by solid lines, and VanSN predicted hydrophobic transmembrane domains are indicated by dotted lines. The P156S and R204Q substitutions in VanSN are indicated by asterisks.
A single promoter, PN, upstream from the ligase gene, was characterized by RACE-PCR (Fig. 2). The presence of the promoter suggests that similarly to the vanC and vanE operons, the genes for the two-component system are cotranscribed with the resistance genes (14, 29).
Characterization of peptidoglycan precursors.
To analyze the cytoplasmic peptidoglycan precursors, cultures of E. faecalis BM4518 and E. faecium UCN71 grown in the presence (4 μg/ml) or in the absence of vancomycin were incubated with ramoplanin to inhibit cell wall synthesis after formation of the precursors. In BM4518 and in the absence of vancomycin, UDP-MurNAc-pentapeptide(Ala) was the main precursor synthesized, whereas incubation with vancomycin led to an increase in the production of UDP-MurNAc-pentapeptide(Ser) but not to elimination of UDP-MurNAc-pentapeptide(Ala) (Table 4). No UDP-MurNAc-tetrapeptide was detected. These data confirm that resistance to vancomycin can be induced in BM4518 by production of precursors terminating in d-Ala-d-Ser (12). In contrast, E. faecium UCN71 synthesized UDP-MurNAc-pentapeptide(Ala) and UDP-MurNAc-pentapeptide(Ser) precursors in equal amounts, even in the absence of induction, indicating constitutive expression of the resistance genes (Table 4). UDP-MurNAc-tetrapeptide (44 to 47%) was also present, due to the d,d-carboxypeptidase activity of the bifunctional VanXYN enzyme that removes the ultimate d-alanine of UDP-MurNAc-pentapeptide(Ala). This d,d-carboxypeptidase acts if the elimination of d-Ala-d-Ala by the d,d-dipeptidase activity of VanXYN is incomplete (7). Similarities could be observed when comparing the proportions of precursors in strain UCN71 with those in VanG- and VanC-type strains (12, 29). Persistent UDP-MurNAc-pentapeptide(Ala) synthesis was previously described in the VanG peptidoglycan analysis, and an important rate of UDP-MurNAc-tetrapeptide synthesis was characterized in a VanC phenotype (12, 29).
Table 4.
Cytoplasmic peptidoglycan precursors synthesized by E. faecium BM4158 (vanG) and E. faecium UCN71 (vanN)
| Strain | Culturea | Amount (%) of indicated peptidoglycan precursorb |
||
|---|---|---|---|---|
| UDP-MurNAc-tetrapeptide | UDP-MurNAc-pentapeptide (d-Ala) | UDP-MurNAc-pentapeptide (d-Ser) | ||
| BM4518 | Uninduced | <1 | 97 ± 1 | 3 ± 1 |
| Vancomycin | 1 ± 3 | 53 ± 3 | 46 ± 4 | |
| UCN71 | Uninduced | 47 ± 3 | 29 ± 1 | 28 ± 1 |
| Vancomycin | 44 ± 4 | 24 ± 1 | 32 ± 2 | |
Four micrograms per milliliter of vancomycin was used.
Peptidoglycan synthesis was inhibited by the addition of ramoplanin to the cultures for 15 min (5).
d,d-dipeptidase and d,d-carboxypeptidase activities.
Expression of the resistance genes in E. faecium UCN71 was studied by analysis of the d,d-dipeptidase and D,D-carboxypeptidase activities of VanXYN, which were assayed by determining the amount of d-Ala released from hydrolysis of the dipeptide d-Ala-d-Ala and of the pentapeptide UDP-Mur-NAc-l-Ala-γ-d-Glu-l-Lys-d-Ala-d-Ala, respectively (Table 5). The d,d-dipeptidase activity VanX was measured after centrifugation at 100,000 × g in the supernatants of lysed bacteria that had been grown in the absence or in the presence of 4 μg/ml of vancomycin as an inducer. In E. faecium UCN71, the d,d-dipeptidase activity in cytoplasmic extracts was at a high level and was constitutive as in BM4174 (VanC) (31) and in contrast to that in BM4405 (VanE) (18) and BM4518 (VanG) (12), which displayed weak and inducible activity. The d,d-carboxypeptidase activity in cytoplasmic fractions of UCN71 was also constitutive and was significantly higher than that in the membrane preparations, consistent with the fact that the VanXYN d,d-carboxypeptidase is a cytoplasmic protein.
Table 5.
d,d-peptidase (VanXYN) and racemase (VanTN) activities in extracts from E. faecium UCN71
| Vancomycin concn (μg/ml) | Activity (nmol min−1 mg protein−1) of indicated protein or enzymea |
|||||
|---|---|---|---|---|---|---|
| Cytoplasmic fraction of d,d-dipeptidaseb |
d,d-carboxypeptidasec |
Membrane fraction of d-serine racemased |
d-alanine racemasec |
|||
| Cytoplasmic fraction | Membrane fraction | Cytoplasmic fraction | Membrane fraction | |||
| 0 | 49 ± 6 | 33 ± 2 | 9 ± 2 | 77 ± 20 | 18 ± 1 | 8 ± 2 |
| 4 | 47 ± 3 | 28 ± 1 | 10 ± 2 | 83 ± 15 | 18 ± 3 | 11 ± 2 |
| 8 | 52 ± 10 | 27 ± 2 | 10 ± 1 | 67 ± 10 | 12 ± 1 | 7 ± 1 |
Results are the means ± standard deviations obtained from a minimum of three independent extracts.
d,d-dipeptidase activity was measured in the supernatants of lysed bacteria after centrifugation at 100,000 × g.
d,d-carboxypeptidase and d-alanine racemase activities were measured in the supernatants and in the resuspended pellet fractions after centrifugation of lysed bacteria at 100,000 × g for 45 min.
d-serine racemase activity was measured in the resuspended pellet fractions after centrifugation of lysed bacteria at 100,000 × g for 45 min.
Racemase activities.
VanC-, VanE-, and VanG-type resistances require three proteins: VanC (VanE, VanG) and VanXYC (VanXYE, VanXYG), which respectively catalyze synthesis of d-Ala-d-Ser and elimination of d-Ala-d-Ala, and VanT (VanTE, VanTG), a membrane-bound serine racemase for production of d-Ser. VanTN serine racemase activity was detected in the membrane fractions from UCN71, and the enzyme was constitutively expressed (Table 5). The activity was similar to that of VanTE in BM4405 (18) except that its expression was inducible. Serine racemase was synthesized constitutively and was present almost exclusively in the cytoplasm (Table 5), a distribution similar to that in BM4174 (VanC) (2), BM4405 (VanE) (18), and BM4518 (VanG) (12).
Transferability of vancomycin resistance.
Resistance to vancomycin could be transferred by conjugation from E. faecium UCN71 to E. faecium BM4107 at a low frequency (on the order of 10−10) but not to E. faecalis JH2-2 (smaller than 10−10). The presence of the vanN gene cluster was confirmed by PCR in one transconjugant, designated UCN73, that was resistant to vancomycin (MIC, 16 μg/ml) and susceptible to teicoplanin, as expected.
The biological cost related to acquisition of VanN resistance was evaluated. In the absence of vancomycin, there was a significant difference (P < 0.05) in the growth of recipient BM4107 (μ = 0.019 ± 0.002 min−1) and transconjugant UCN73 (μ = 0.009 ± 0.003 min−1) (data not shown). A recent study highlighted the major biological cost due to constitutive, in contrast to inducible, expression of vancomycin resistance in enterococci (19). In the transconjugant, the biological cost may be due to acquisition of the mobile genetic element or constitutive expression of resistance to vancomycin by the vanN gene cluster, or both. Although fitness cost is not a barrier to horizontal transfer of VanN-type resistance, it may limit dissemination of enterococci harboring the corresponding genes.
So far, transfer of d-Ala-d-Ser-type resistance has been reported only in E. faecalis. The corresponding determinants are chromosomally located and most are not transferable (11). VanN is the first example of transferable d-Ala-d-Ser-type resistance in E. faecium. A ca. 150-kb plasmid was detected in E. faecium UCN71 and was cotransferred by conjugation together with the vancomycin resistance to the recipient strain of E. faecium BM4147 (data not shown). Plasmid analyses to characterize the vanN mobile genetic element responsible for horizontal transfer are in progress.
In conclusion, transferable VanN-type glycopeptide resistance in E. faecium UCN71 was due to constitutive synthesis of late peptidoglycan precursors ending in d-Ala-d-Ser. The VanC-, VanL-, and VanE-type resistances are biochemically and phenotypically similar. VanN expands the so-called Van alphabet and the capability of enterococci to acquire and to horizontally transfer vancomycin resistance.
ACKNOWLEDGMENTS
We thank M. Auzou for technical assistance.
This work was supported in part by the 7th Framework European Program TROCAR.
Footnotes
Published ahead of print on 1 August 2011.
REFERENCES
- 1. Arias C. A., et al. 1999. Characterization and modelling of VanT: a novel, membrane-bound, serine racemase from vancomycin-resistant Enterococcus gallinarum BM4174. Mol. Microbiol. 31:1653–1664 [DOI] [PubMed] [Google Scholar]
- 2. Arias C. A., Courvalin P., Reynolds P. E. 2000. vanC cluster of vancomycin-resistant Enterococcus gallinarum BM4174. Antimicrob. Agents Chemother. 44:1660-1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arias C. A., Peña J., Panesso D., Reynolds P. 2003. Role of the transmembrane domain of the VanT serine racemase in resistance to vancomycin in Enterococcus gallinarum BM4174. J. Antimicrob. Chemother. 51:557–564 [DOI] [PubMed] [Google Scholar]
- 4. Arthur M., Molinas C., Courvalin P. 1992. The VanS-VanR two-component regulatory system controls synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J. Bacteriol. 174:2582–2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Arthur M., Depardieu F., Reynolds P., Courvalin P. 1996. Quantitative analysis of the metabolism of soluble cytoplasmic peptidoglycan precursors of glycopeptide-resistant enterococci. Mol. Microbiol. 21:33–44 [DOI] [PubMed] [Google Scholar]
- 6. Arthur M., Reynolds P., Courvalin P. 1996. Glycopeptide resistance in enterococci. Trends Microbiol. 4:401–407 [DOI] [PubMed] [Google Scholar]
- 7. Arthur M., Depardieu F., Cabanié L., Reynolds P., Courvalin P. 1998. Requirement of the VanY and VanX d,d-peptidases for glycopeptide resistance in enterococci. Mol. Microbiol. 30:819–830 [DOI] [PubMed] [Google Scholar]
- 8. Boyd D. A., Willey B. M., Fawcett D., Gillani N., Mulvey M. R. 2008. Molecular characterization of Enterococcus faecalis N06-0364 with low-level vancomycin resistance harboring a novel d-Ala-d-Ser gene cluster, vanL. Antimicrob. Agents Chemother. 52:2667–2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bradford M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 10. Comité de l'Antibiogramme de la Société Française de Microbiologie 1996. Technical recommendations for in vitro susceptibility testing. Clin. Microbiol. Infect. 2:(Suppl. 1)S11–S25 [PubMed] [Google Scholar]
- 11. Courvalin P. 2006. Vancomycin resistance in gram-positive cocci. Clin. Infect. Dis. 42(Suppl. 1):S25–S34 [DOI] [PubMed] [Google Scholar]
- 12. Depardieu F., Bonora M. G., Reynolds P. E., Courvalin P. 2003. The vanG glycopeptide resistance operon from Enterococcus faecalis revisited. Mol. Microbiol. 50:931–948 [DOI] [PubMed] [Google Scholar]
- 13. Depardieu F., Courvalin P., Msadek T. 2003. A six amino acid deletion, partially overlapping the VanSB G2 ATP-binding motif, leads to constitutive glycopeptide resistance in VanB-type Enterococcus faecium. Mol. Microbiol. 50:1069–1083 [DOI] [PubMed] [Google Scholar]
- 14. Depardieu F., Podglajen I., Leclercq R., Collatz E., Courvalin P. 2007. Modes and modulations of antibiotic resistance gene expression. Clin. Microbiol. Rev. 20:79–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dutka-Malen S., Molinas C., Arthur M., Courvalin P. 1992. Sequence of the vanC gene of Enterococcus gallinarum BM4174 encoding a d-alanine:d-alanine ligase-related protein necessary for vancomycin resistance. Gene 112:53–58 [DOI] [PubMed] [Google Scholar]
- 16. Dutka-Malen S., Evers S., Courvalin P. 1995. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J. Clin. Microbiol. 33:24–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Evers S., et al. 1996. Evolution of structure and substrate specificity in d-alanine:d-alanine ligases and related enzymes. J. Mol. Evol. 42:706–712 [DOI] [PubMed] [Google Scholar]
- 18. Fines M., Périchon B., Reynolds P., Sahm D. F., Courvalin P. 1999. VanE, a new type of acquired glycopeptide resistance in Enterococcus faecalis BM4405. Antimicrob. Agents Chemother. 43:2161–2164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Foucault M. L., Depardieu F., Courvalin P., Grillot-Courvalin C. 2010. Inducible expression eliminates the fitness cost of vancomycin resistance in enterococci. Proc. Natl. Acad. Sci. U. S. A. 107:16964–16969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Homan W. L., et al. 2002. Multilocus sequence typing scheme for Enterococcus faecium. J. Clin. Microbiol. 40:1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hsing W., Russo F. D., Bernd K. K., Silhavy T. J. 1998. Mutations that alter kinase and phosphatase activities of the two-component sensor EnvZ. J. Bacteriol. 180:4538–4546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jacob A. E., Hobbs S. J. 1974. Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var. zymogenes. J. Bacteriol. 117:360–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Leavis H. L., Bonten M. J., Willems R. J. 2006. Identification of high-risk enterococcal clonal complexes: global dispersion and antibiotic resistance. Curr. Opin. Microbiol. 9:454–460 [DOI] [PubMed] [Google Scholar]
- 24. Leclercq R., Derlot E., Duval J., Courvalin P. 1988. Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. N. Engl. J. Med. 319:157–161 [DOI] [PubMed] [Google Scholar]
- 25. Liu Y. G., Whittier R. F. 1995. Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 25:674–681 [DOI] [PubMed] [Google Scholar]
- 26. McKessar S. J., Berry A. M., Bell J. M., Turnidge J. D., Paton J. C. 2000. Genetic characterization of vanG, a novel vancomycin resistance locus of Enterococcus faecalis. Antimicrob. Agents Chemother. 44:3224–3228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Messer J., Reynolds P. E. 1992. Modified peptidoglycan precursors produced by glycopeptide-resistant enterococci. FEMS Microbiol. Lett. 73:195–200 [DOI] [PubMed] [Google Scholar]
- 28. Msadek T., Knust F., Rapopport G. 1993. Two-component regulatory systems, p. 729–745 In Sonenshein A. L., Hoch J. A., Losick R. (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, DC [Google Scholar]
- 29. Panesso D., et al. 2005. Transcriptional analysis of the vanC cluster from Enterococcus gallinarum strains with constitutive and inducible vancomycin resistance. Antimicrob. Agents Chemother. 49:1060–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Perichon B., Reynolds P., Courvalin P. 1997. VanD-type glycopeptide-resistant Enterococcus faecium BM4339. Antimicrob. Agents Chemother. 41:2016–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Reynolds P. E., Snaith H. A., Maguire A. J., Dutka-Malen S., Courvalin P. 1994. Analysis of peptidoglycan precursors in vancomycin-resistant Enterococcus gallinarum BM4174. Biochem. J. 301:5–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Reynolds P. E., Arias C. A., Courvalin P. 1999. Gene vanXYC encodes d,d-dipeptidase (VanX) and d,d-carboxypeptidase (VanY) activities in vancomycin-resistant Enterococcus gallinarum BM4174. Mol. Microbiol. 34:341–349 [DOI] [PubMed] [Google Scholar]
- 33. Reynolds P. E., Ambur O. H., Casadewall B., Courvalin P. 2001. The VanY(D) DD-carboxypeptidase of Enterococcus faecium BM4339 is a penicillin-binding protein. Microbiology 147:2571–2578 [DOI] [PubMed] [Google Scholar]
- 34. Saitou N., Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 35. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 36. Tenover F. C., et al. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Top J., Willems R., Bonten M. 2008. Emergence of CC17 Enterococcus faecium: from commensal to hospital-adapted pathogen. FEMS Immunol. Med. Microbiol. 52:297–308 [DOI] [PubMed] [Google Scholar]
- 38. Uttley A. H., Collins C. H., Naidoo J., George R. C. 1988. Vancomycin-resistant enterococci. Lancet i:57–58 [DOI] [PubMed] [Google Scholar]
- 39. Weber P., Meziane-Cherif D., Haouz A., Saul F. A., Courvalin P. 2009. Crystallization and preliminary X-ray analysis of a d-Ala:d-Ser ligase associated with VanG-type vancomycin resistance. Acta Crystallogr. Sect. F. Struct. Biol. Cryst. Commun. 65:1024–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xu X., et al. 2010. vanM, a new glycopeptide resistance gene cluster found in Enterococcus faecium. Antimicrob. Agents Chemother. 54:4643–4647 [DOI] [PMC free article] [PubMed] [Google Scholar]