Abstract
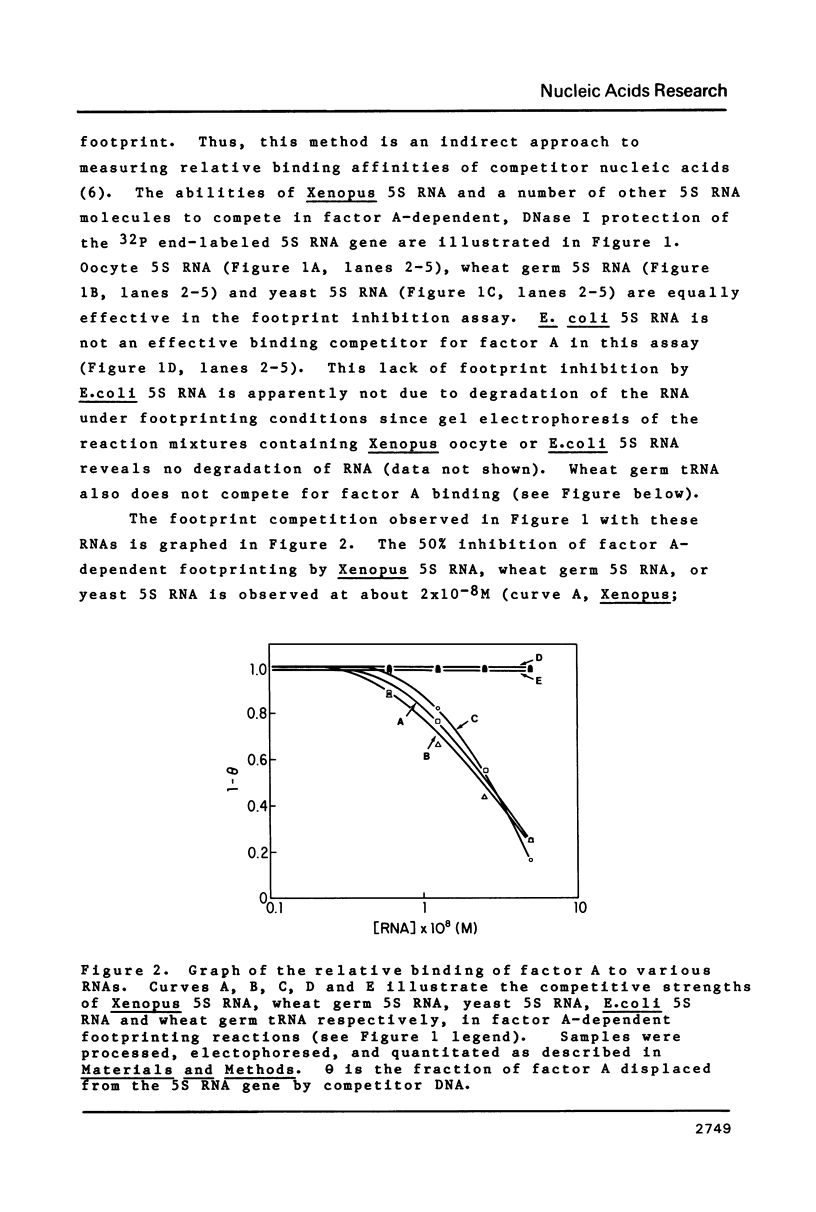
Footprint competition assays are utilized to study the binding of Xenopus transcription factor A to a variety of single-stranded nucleic acids. The addition of Xenopus oocyte, yeast, or wheat germ 5S RNA as footprint competitors reveals that factor A binds these 5S RNAs with similar affinity. In contrast, factor A does not bind to E.coli 5S RNA or wheat germ tRNA in this assay. Factor A binding to single stranded DNA is also examined using footprint competition. Factor A binds preferentially to non-specific single stranded (M13) DNA versus double stranded (pBR322) DNA. Factor A binds equally well to single stranded DNA fragments containing either the coding or non-coding strands of the 5S RNA gene. Using single stranded M13 DNA as a competitor, the factor A-5S RNA gene complex is found to dissociate with a half-life of 5-6 min.
Full text
PDF




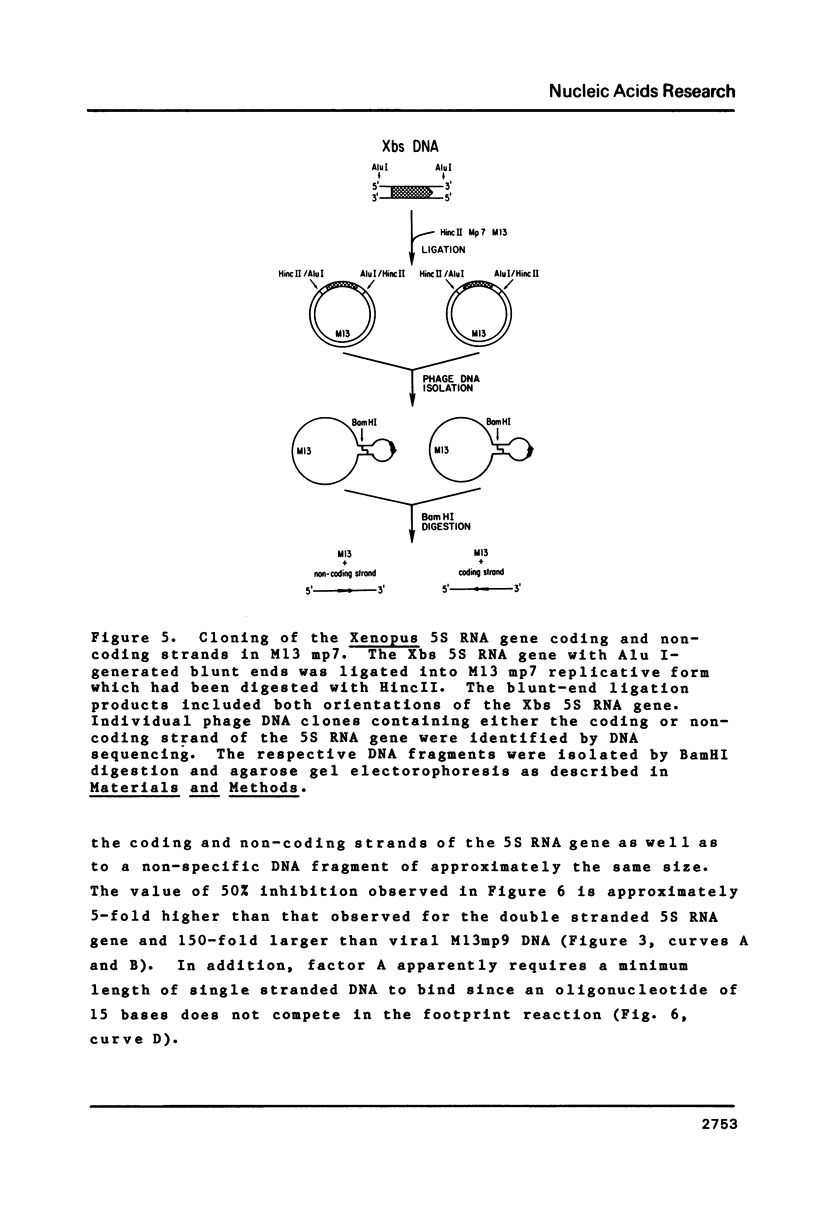
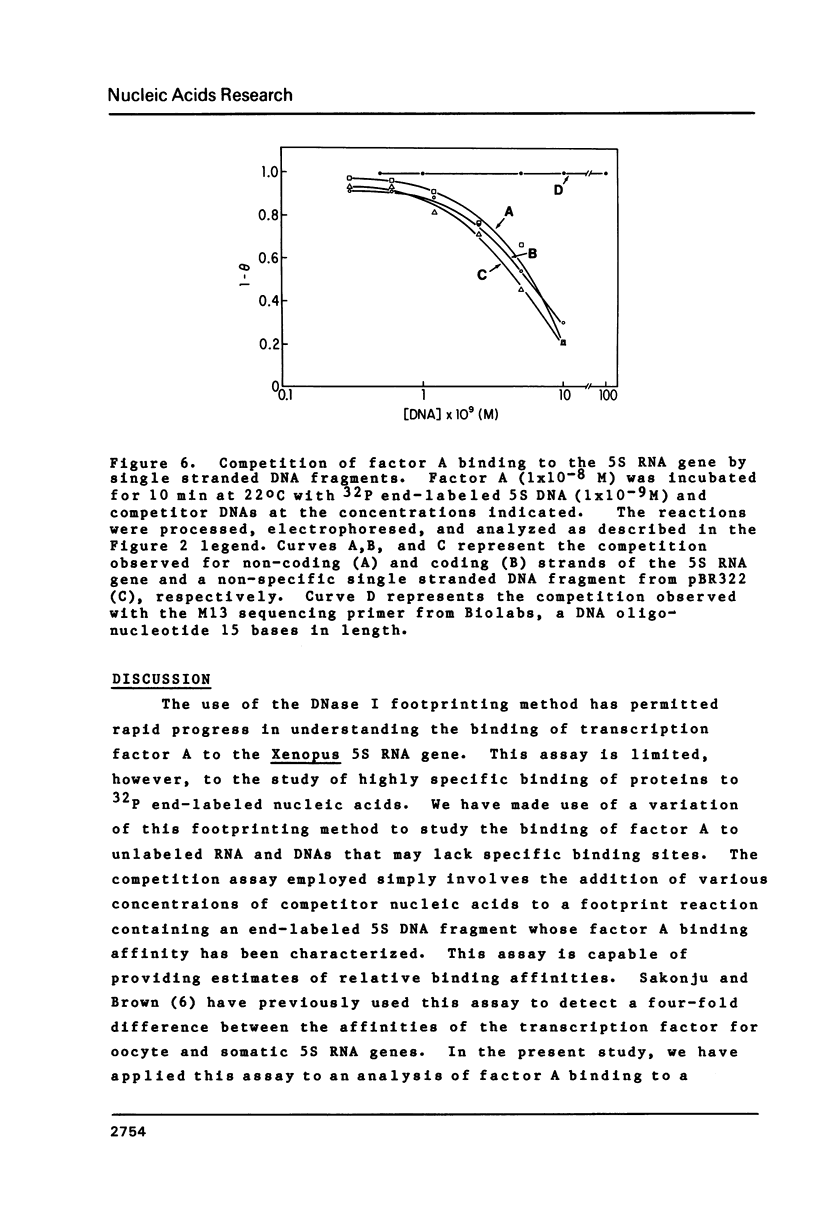




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birkenmeier E. H., Brown D. D., Jordan E. A nuclear extract of Xenopus laevis oocytes that accurately transcribes 5S RNA genes. Cell. 1978 Nov;15(3):1077–1086. doi: 10.1016/0092-8674(78)90291-x. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D. F., Sakonju S., Brown D. D. A control region in the center of the 5S RNA gene directs specific initiation of transcription: II. The 3' border of the region. Cell. 1980 Jan;19(1):27–35. doi: 10.1016/0092-8674(80)90385-2. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D. F., Wormington W. M., Brown D. D. Stable transcription complexes of Xenopus 5S RNA genes: a means to maintain the differentiated state. Cell. 1982 Feb;28(2):413–421. doi: 10.1016/0092-8674(82)90359-2. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brownlee G. G., Sanger F., Barrell B. G. The sequence of 5 s ribosomal ribonucleic acid. J Mol Biol. 1968 Jun 28;34(3):379–412. doi: 10.1016/0022-2836(68)90168-x. [DOI] [PubMed] [Google Scholar]
- Engelke D. R., Ng S. Y., Shastry B. S., Roeder R. G. Specific interaction of a purified transcription factor with an internal control region of 5S RNA genes. Cell. 1980 Mar;19(3):717–728. doi: 10.1016/s0092-8674(80)80048-1. [DOI] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanas J. S., Bogenhagen D. F., Wu C. W. Cooperative model for the binding of Xenopus transcription factor A to the 5S RNA gene. Proc Natl Acad Sci U S A. 1983 Apr;80(8):2142–2145. doi: 10.1073/pnas.80.8.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn L. J., Gurdon J. B. The reactivation of developmentally inert 5S genes in somatic nuclei injected into Xenopus oocytes. Nature. 1981 Feb 5;289(5797):461–465. doi: 10.1038/289461a0. [DOI] [PubMed] [Google Scholar]
- Mackay R. M., Spencer D. F., Doolittle W. F., Gray M. W. Nucleotide sequences of wheat-embryo cytosol 5-S and 5.8-S ribosomal ribonucleic acids. Eur J Biochem. 1980 Dec;112(3):561–576. doi: 10.1111/j.1432-1033.1980.tb06122.x. [DOI] [PubMed] [Google Scholar]
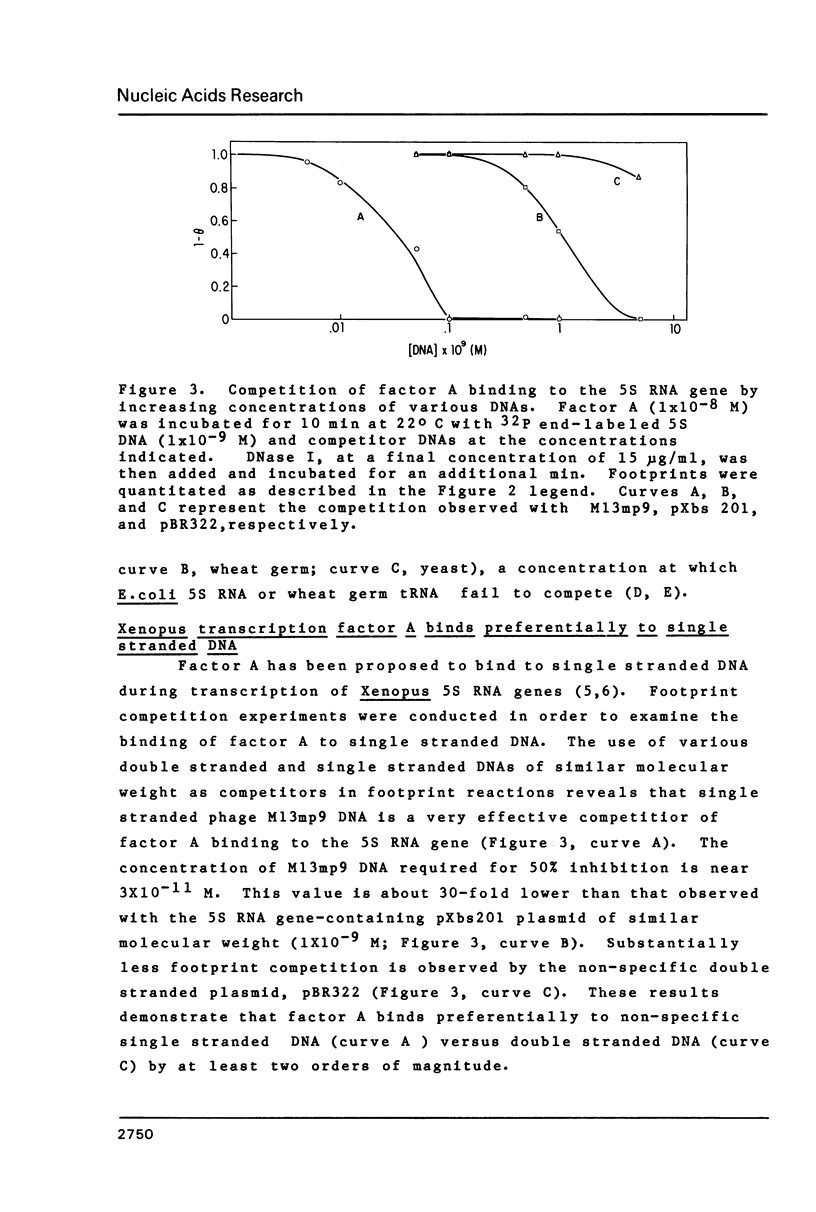
- Patton J. R., Chae C. B. A method for isolation of a large amount of a single-stranded DNA fragment. Anal Biochem. 1982 Oct;126(1):231–234. doi: 10.1016/0003-2697(82)90134-8. [DOI] [PubMed] [Google Scholar]
- Pelham H. R., Brown D. D. A specific transcription factor that can bind either the 5S RNA gene or 5S RNA. Proc Natl Acad Sci U S A. 1980 Jul;77(7):4170–4174. doi: 10.1073/pnas.77.7.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds W. F., Gottesfeld J. M. 5S rRNA gene transcription factor IIIA alters the helical configuration of DNA. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1862–1866. doi: 10.1073/pnas.80.7.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakonju S., Bogenhagen D. F., Brown D. D. A control region in the center of the 5S RNA gene directs specific initiation of transcription: I. The 5' border of the region. Cell. 1980 Jan;19(1):13–25. doi: 10.1016/0092-8674(80)90384-0. [DOI] [PubMed] [Google Scholar]
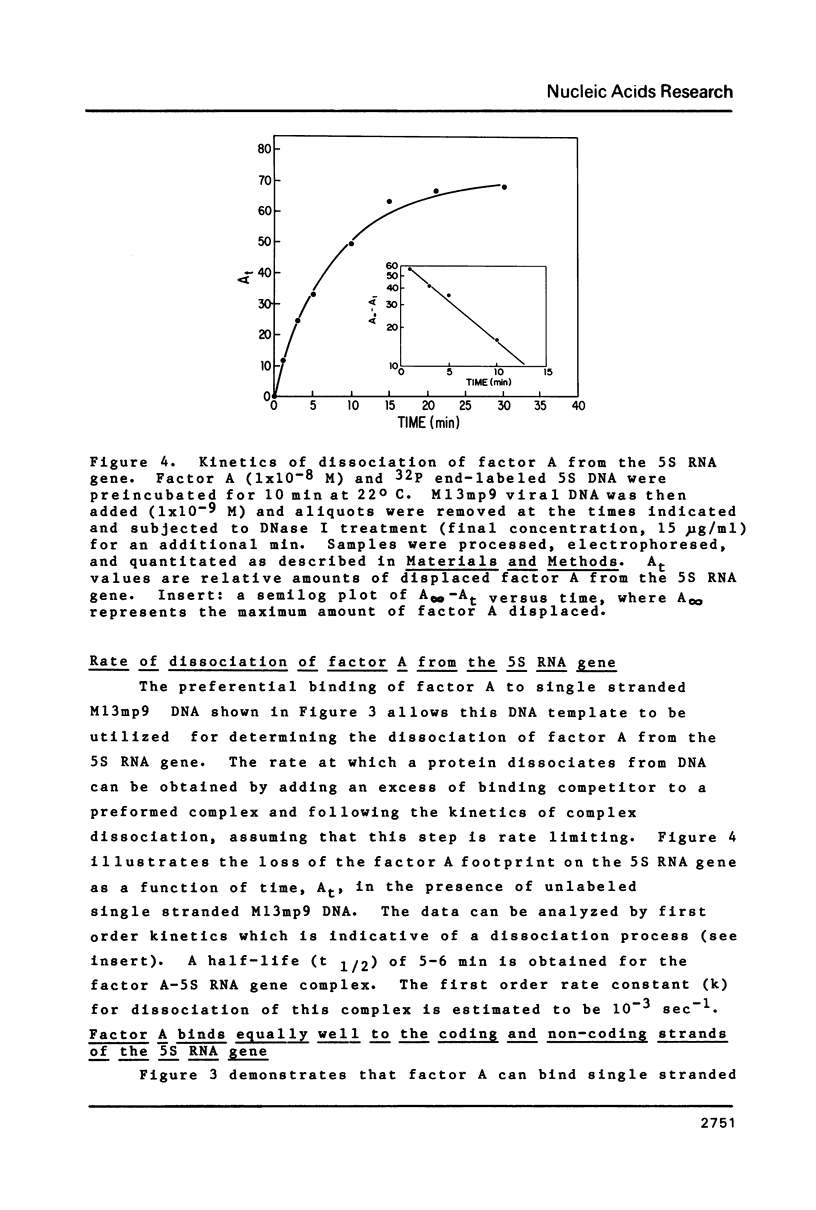
- Sakonju S., Brown D. D. Contact points between a positive transcription factor and the Xenopus 5S RNA gene. Cell. 1982 Dec;31(2 Pt 1):395–405. doi: 10.1016/0092-8674(82)90133-7. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Scheele G., Blackburn P. Role of mammalian RNase inhibitor in cell-free protein synthesis. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4898–4902. doi: 10.1073/pnas.76.10.4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela P., Bell G. I., Masiarz F. R., DeGennaro L. J., Rutter W. J. Nucleotide sequence of the yeast 5S ribosomal RNA gene and adjacent putative control regions. Nature. 1977 Jun 16;267(5612):641–643. doi: 10.1038/267641a0. [DOI] [PubMed] [Google Scholar]
- Wegnez M., Monier R., Denis H. Sequence heterogeneity of 5 S RNA in Xenopus laevis. FEBS Lett. 1972 Sep 1;25(1):13–20. doi: 10.1016/0014-5793(72)80443-5. [DOI] [PubMed] [Google Scholar]