Abstract
The objective of the present study was to compare the effects of the modified live porcine reproductive and respiratory syndrome virus (PRRSV) vaccine (Ingelvac PRRS MLV; Boehringer Ingelheim Animal Health, St. Joseph, MO) on European and North American PRRSV shedding in the semen of experimentally infected boars. The boars were randomly divided into six groups. Vaccinated boars shed the North American PRRSV at the rate of 100.1 to 101.0 viral genome copies per ml and 3.63 to 101.1 50% tissue culture infective doses (TCID50)/ml, respectively, in semen, whereas nonvaccinated boars shed the North American PRRSV at the rate of 100.2 to 104.7 viral genome copies per ml and 1.14 to 103.07 TCID50/ml, respectively, in semen. Vaccinated boars shed the European PRRSV at the rate of 100.1 to 104.57 viral genome copies per ml and 1.66 to 103.10 TCID50/ml, respectively, in semen, whereas nonvaccinated boars shed the European PRRSV at the rate of 100.3 to 105.14 viral genome copies per ml and 1.69 to 103.17 TCID50/ml, respectively, in semen. The number of genomic copies of the European PRRSV in semen samples was not significantly different between vaccinated and nonvaccinated challenged European PRRSV boars. The present study demonstrated that boar vaccination using commercial modified live PRRSV vaccine was able to decrease subsequent shedding of North American PRRSV in semen after challenge but was unable to decrease shedding of European PRRSV in semen after challenge.
INTRODUCTION
Porcine reproductive and respiratory syndrome virus (PRRSV) is a member of the genus Arterivirus, the family Arteriviridae, and the order Nidovirales. The virus contains a positive single-stranded polyadenylated RNA approximately 15 kb in length that contains nine open reading frames (ORFs) (2, 20). Two distinct PRRSV genotypes have been identified: European (type I) and North American (type II) (11, 19). Infection by PRRSV causes respiratory disease in nursery and growth-completing pigs and reproductive failure in sows and boars (3, 4, 6, 7, 9, 24, 27).
Although PRRSV was first isolated in Korea in 1994 (14) and all PRRSV isolates corresponded to the North American genotype until 2000 (5), European PRRSV has recently emerged in Korea (13, 17, 21). The company claims that modified live PRRSV vaccine can cross-protect against challenge of both European and North American genotypes (Ingelvac PRRS MLV package insert; Boehringer Ingelheim Animal Health, St. Joseph, MO [http://bi-vetmedica.com//sites/default/files/ingelvac_PRRS_MLV_rp.pdf]). However, a large number of abortions caused by the European PRRSV genotype occurred in a swine herd in which sows and gilts had been vaccinated for 3 years under a whole-herd vaccination program. All pregnant and nonpregnant sows and all gilts were vaccinated every 4 months according to the manufacturer's recommendation (personal observation). Although this outbreak was not related to European genotype PRRSV-contaminated semen, this incident raised the possibility that the modified live PRRSV vaccine could not protect against the European PRRSV genotype isolated in Korea. Furthermore, studies have shown mixed results regarding the efficacy of this commercial vaccine against the genetically diverse field strains of PRRSV (18). Hence, the impact of European PRRSV genotype-contaminated semen is enhanced due to its widespread distribution throughout the Korean swine industry, even though boars in commercial artificial insemination centers have already been vaccinated with modified live PRRSV. Although it has been reported that modified live PRRSV vaccination reduced the subsequent shedding of European and North American genotypes of PRRSV in boars (8, 22), the objective of the present study was to compare the effects of boar vaccination on North American and European genotype challenges, including the reduction of subsequent virus shedding in semen.
MATERIALS AND METHODS
Commercial vaccine and PRRSV inocula.
The commercial modified live PRRSV vaccine (Ingelvac PRRS MLV; Boehringer Ingelheim Animal Health) was used in this study. Boars were vaccinated with a 2.0-ml dose intramuscularly as previously described (8, 22).
European (SNU090485) and North American (SNU090851) PRRSV strains were used as inocula. The European PRRSV strain was isolated in lung samples from an aborted fetus and neonatal pigs in 2009 in Kyounggi Province. The North American PRRSV strain was isolated in lung samples from a postweaning pig in 2009 in Chungcheung Province. The nucleotide sequence homology in ORF5 between the European PRRSV strain (GenBank accession number JN315686) and the vaccine strain (GenBank accession number AF535152) is 68%, and between the North American PRRSV strain (GenBank accession number JN315685) and the vaccine strain it is 86%, using BioEdit, version 7.0.0 (Ibis Biosciences, Carlsbad, CA [http://www.mbio.ncsu.edu/BioEdit/bioedit.html]). Each virus (passage 6) was propagated in MARC-145 cells to a titer of 1 × 106 50% tissue culture infective doses (TCID50)/ml.
Experimental design.
At 8 months of age, 30 purebred male Landrace pigs were purchased from a PRRSV-free commercial farm. All boars were negative for porcine circovirus type 2 (PCV2) and PRRSV according to routine serological testing prior to delivery and on arrival. All boars were individually housed throughout the experiment in an environmentally controlled building with pens over completely slatted floors.
The boars were randomly divided into six groups. The boars in group 1 ([T01] n = 5) were immunized with modified live PRRSV vaccine with single 2.0-ml doses. Six weeks after vaccination (42 days postinoculation [dpi]), these boars were inoculated with European PRRSV intranasally (1 ml) with an infectious titer of 106 TCID50 per ml. The boars in group 2 ([T02] n = 5) were immunized with modified live PRRSV vaccine with single 2.0-ml doses. Six weeks after vaccination (−42 dpi), these boars were inoculated with North American PRRSV intranasally (1 ml) with an infectious titer of 106 TCID50 per ml (0 dpi). The boars in group 3 ([T03] n = 5) were inoculated with European PRRSV intranasally (1 ml) with an infectious titer of 106 TCID50 per ml (0 dpi). The boars in group 4 ([T04] n = 5) were inoculated with North American PRRSV intranasally (1 ml) with an infectious titer of 106 TCID50 per ml (0 dpi). The boars in group 5 ([T05] n = 5) were immunized with modified live PRRSV vaccine with single 2.0-ml doses. The boars in group 6 ([T06] n = 5) served as negative controls and were exposed to neither vaccine nor virus. Following PRRSV inoculation, the physical conditions of the boars were monitored daily, and their rectal temperatures were taken. All of the methods were previously approved by the Seoul National University Institutional Animal Care and Use Committee.
Serology.
Blood samples from each pig were collected by jugular venipuncture at −42, −21, 0, 14, 21, 28, 35, 42, 49, and 60 dpi, and the sera were stored at −20°C. The serum samples were tested using a commercially available PRRSV enzyme-linked immunosorbent assay ([ELISA] HerdCheck PRRS 2XR; Idexx Laboratories Inc., Westbrook, ME).
Virus isolation.
Blood and semen (raw) samples were collected for virus isolation at −42, −21, −7, 0, 4, 7, 10, 14, 18, 21, 25, 28, 32, 35, 39, 42, 46, 49, 53, 56, and 60 dpi from all boars used in this study. PRRSV was isolated from serum and semen as previously described (9, 12). Virus titrations were also performed in confluent monolayers of MARC-145 cells in 96-well plates as previously described (12).
Sequence analysis.
The PRRSV isolates from semen were further analyzed for the ORF5 sequence. RNA was extracted from PRRSV-infected MARC-145 cell lines (4) and amplified from the ORF5 region by reverse transcription-PCR (RT-PCR) (23). Sequencing was performed on the purified RT-PCR products of amplified ORF5.
Real-time PCR.
Real-time PCR for the European and North American PRRSVs was performed as previously described (26). Real-time PCR for the vaccine strain was designed in this study based on ORF5 because the nucleotide sequence homology in ORF5 between the challenging North American PRRSV strain (SNU090851) and the vaccine strain is 86%, using BioEdit, version 7.0.0 (Ibis Biosciences). The primers and probes were used for PRRSV quantitation in real-time PCR with hybridization probes that were based on the sequence of the vaccine strain (GenBank accession number AF535152): forward, 5′-CTAACAAATTTGATTGGGCAG-3′; reverse, 5′-AGGACATGCAATTCTTTGCAA-3′; probe. The probe for TaqMan PCR (5′-AGTCGCTTTAGTCACTGTCT-3′) was labeled with FAM (6-carboxyfluorescein) as a fluorescent reporter dye at the 5′ end and with TAMRA (6-carboxytetramethylrhodamine) as a quencher-fluorescent dye at the 3′ end. A region of 231 bp (13958 to 14188) was amplified from ORF5 of the vaccine strain. The vaccine strain real-time PCR assay consisted of 94°C for 30 s, 58.5°C for 60 s, and 72°C for 60 s.
Quantification of PRRSV RNA.
RNA extractions from the semen (raw) and serum samples were collected −42, −21, −7, 0, 4, 7, 10, 14, 18, 21, 25, 28, 32, 35, 39, 42, 46, 49, 53, 56, and 60 dpi from all boars used in this study and performed as previously described (26). Real-time PCR for the European and North American PRRSVs and the vaccine strain were used to quantify PRRSV genomic cDNA copy numbers using RNA extraction from semen and serum samples performed as previously described (26). The real-time PCR was considered to be positive if the cycle threshold (CT) level was obtained at ≤45 cycles, as previously described (26).
Standard curve.
To construct a standard curve, real-time PCR was performed in quadruplicate in two different assays: (i) using 10-fold serial dilutions of the PRRSV plasmid as the standard, with concentrations ranging from 1010 to 103 copies/ml, and (ii) using 10-fold serial dilutions of the European and North American PRRSVs cultured in MARC-145 cells from 106 TCID50/ml to 10−1 TCID50/ml or 10-fold serial dilutions of the vaccine strain cultured in MARC-145 cells from 104.5 TCID50/ml to 10−3.5 TCID50/ml. The PRRSV plasmid was prepared as described previously (10). Briefly, the transcript cDNA product was cloned into the pCR2.1 plasmid (Invitrogen, Carlsbad, CA). The recombinant plasmid was purified using a plasmid Miniprep Kit (Qiagen, Valencia, CA) according to the manufacturer's instructions, and the concentration of the purified plasmid was determined using a spectrophotometer.
Interference.
Potential interference from detecting the two virus types simultaneously was examined by mixing their RNAs together in one reaction tube. The sensitivity for the detection of one or both virus types was evaluated.
Statistical analysis.
Summary statistics were calculated for the six groups to assess the overall quality of the data, including normality. For a single comparison of PRRSV RNA quantification, Student's t test for paired samples (European and North American PRRSV RNA quantification) was used to estimate the difference at each time point. Continuous data for PRRSV serology over time between the groups were analyzed at each time point using a Mann-Whitney U test. The Pearson correlation coefficient was used to assess the relationship of PRRSV RNA load between blood and semen. A value of 0.05 was considered significant.
RESULTS
Clinical signs.
Vaccinated boars (groups T01, T02, and T05) and negative-control boars (T06) were clinically normal in health and rectal temperatures (38 to 39.5°C) throughout the experiment, whereas in nonvaccinated boars (T03 and T04) daily rectal temperatures increased (39.1 to 39.3°C) after PRRSV challenge. Some nonvaccinated boars (T03 and T04) were depressed and anorectic for approximately 3 days after PRRSV challenge.
Serology of PRRSV.
Anti-PRRSV antibodies were detected in serum samples at 3 weeks postvaccination (−21 dpi) in vaccinated boars (groups T01, T02, and T05) only. In nonvaccinated boars (T03 and T04), no anti-PRRSV antibodies were detected in serum samples until challenge (0 dpi). As expected, no anti-PRRSV antibodies were detected in serum from the negative-control boars (T06) throughout the experiment (data not shown).
Specificity of real-time PCR.
Primers from the vaccine strain did not react with challenging European (SNU090485) and North American (SNU090851) PRRSV strains. Primers from the vaccine strain did not react with the North American PRRSV strain, but primers from the North American PRRSV strain did react with the vaccine strain (data not shown).
Standard curve.
Standard curves were constructed by plotting the logarithm of the copy number of the 10-fold serially diluted plasmid against the measured CT values. The linear correlation (R2) between the CT and the logarithm of the plasmid copy number were repeatedly greater than 0.998 for the European PRRSV, 0.997 for the North American PRRSV, and 0.995 for the vaccine strain (data not shown).
Interference.
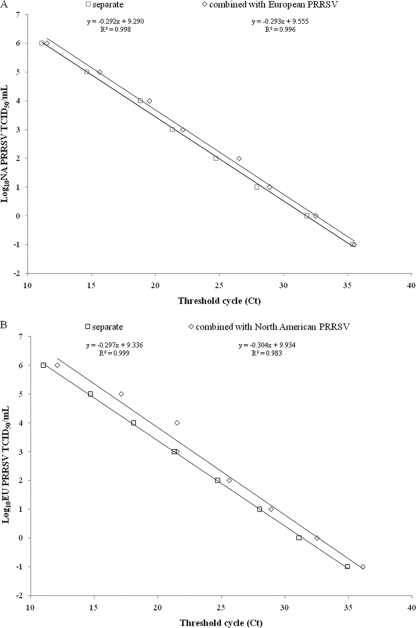
To determine whether having the two types of viruses present in the same sample interferes with the standard curve, detection of the viral copies of one or the other type was performed. The detection of the North American PRRSV was found to be efficient and sensitive, independent of a combination of the North American PRRSV with the same TCID50/ml of the European PRRSV per tube (Fig. 1 A), whereas the detection of the European PRRSV was somehow less sensitive when the same TCID50/ml of the North American PRRSV per tube was present, which demonstrated a reduction in the correlation coefficient of the slope of the RNA standard to 98.4% for European PRRSV detection (Fig. 1B). The detection of the vaccine strain was found to be efficient and sensitive, independent of a combination of the vaccine strain with the same TCID50/ml of European PRRSV per tube, whereas the detection of the European PRRSV was somehow less sensitive when the same TCID50/ml of vaccine strain per tube was present, which demonstrated a reduction in the correlation coefficient of the slope of the RNA standard to 98.8% for European PRRSV detection. The detection of the vaccine strain was somehow less sensitive when the same TCID50/ml of North American PRRSV per tube was present, which demonstrated a reduction in the correlation coefficient of the slope of the RNA standard to 98.2% for vaccine strain detection.
Fig. 1.
Quantification curves to study the occurrence of interference in the detection of the European PRRSV and North American PRRSV when the two virus types are combined. (A) RNA corresponding to 106 TCID50/ml of North American PRRSV per tube was added in addition to European PRRSV RNA. (B) RNA corresponding to 106 TCID50/ml of North American PRRSV per tube was added in addition to European PRRSV RNA.
Virus isolation and sequence analysis in blood and semen.
Attempts were made to isolate and identify the European and North American PRRSVs and the vaccine strain from the serum and semen of the five groups (Table 1). No PRRSV was isolated from the serum and semen of the negative-control boars (group T06). The vaccine strain was isolated only from the serum and semen of the vaccinated boars (groups T01, T02, and T05) before challenge (0 dpi), and thereafter vaccine strains were not isolated from any serum and semen samples from the vaccinated boars (groups T01, T02, and T05). All isolated PRRSV was confirmed to be the same propagating virus in the challenge stock by sequence analysis.
Table 1.
Isolation of PRRSV in serum and semen samples from PRRSV-inoculated and negative-control boars
| Group (n)a | PRRSV type | Sample type | No. of positive boars at the indicated day postinfection |
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| −42 | −21 | −7 | 0 | 4 | 7 | 10 | 14 | 18 | 21 | 25 | 28 | 32 | 35 | 39 | 42 | 46 | 49 | 53 | 56 | 60 | |||
| T01 (5) | European | Serum | 0 | 0 | 0 | 0 | 5 | 5 | 5 | 5 | 4 | 3 | 4 | 4 | 2 | 2 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| European | Semen | 0 | 0 | 0 | 0 | 5 | 5 | 5 | 4 | 3 | 3 | 2 | 3 | 2 | 0 | 3 | 3 | 2 | 0 | 2 | 0 | 0 | |
| Vaccine | Serum | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Vaccine | Semen | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| T02 (5) | North American | Serum | 0 | 0 | 0 | 0 | 5 | 5 | 5 | 4 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| North American | Semen | 0 | 0 | 0 | 0 | 5 | 5 | 5 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Vaccine | Serum | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Vaccine | Semen | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| T03 (5) | European | Serum | 0 | 0 | 0 | 0 | 5 | 5 | 5 | 5 | 4 | 4 | 3 | 2 | 3 | 2 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| European | Semen | 0 | 0 | 0 | 0 | 5 | 5 | 5 | 4 | 3 | 2 | 4 | 0 | 3 | 0 | 2 | 0 | 3 | 0 | 3 | 0 | 0 | |
| T04 (5) | North American | Serum | 0 | 0 | 0 | 0 | 5 | 5 | 5 | 5 | 5 | 4 | 3 | 2 | 4 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| North American | Semen | 0 | 0 | 0 | 0 | 5 | 5 | 5 | 3 | 3 | 3 | 3 | 0 | 4 | 2 | 4 | 3 | 0 | 2 | 3 | 0 | 0 | |
| T05 (5) | Vaccine | Serum | 0 | 3 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Vaccine | Semen | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
n, number of boars in group.
Log10 TCID50/ml quantification and real-time PCR of PRRSV RNA in blood.
Genomic copies of the European, North American, and vaccine PRRSV strains were not detected in the serum samples at −42 dpi for all groups. No genomic copies of the European PRRSV were observed in the serum samples from vaccinated, challenged North American PRRSV boars (T02), from nonvaccinated, challenged North American PRRSV boars (T04), or from vaccinated, nonchallenged boars (T05). We measured no genomic copies of the North American PRRSV in the serum samples from the vaccinated, challenged European PRRSV boars (T01), from the nonvaccinated, challenged European PRRSV boars (T03), or from the vaccinated, nonchallenged boars (T05). The vaccine strain was detected until −14 dpi only in the serum samples from vaccinated, challenged European PRRSV boars (T01), from vaccinated, challenged North American PRRSV boars (T02), and from vaccinated, nonchallenged boars (T05) (Fig. 2 A and B). No PRRSV was observed in the serum samples from the negative control (T06) boars throughout the experiment.
Fig. 2.
Detection of PRRSV in blood sample. Mean group log10 European or North American PRRSV RNA load (A) and mean group log10 TCID50/ml European or North American PRRSV (B) from the different treatment groups were determined. T01, boars immunized with modified live PRRSV vaccine and challenged with the European PRRSV; T02, boars immunized with modified live PRRSV vaccine and challenged with the North American PRRSV; T03, boars challenged with the European PRRSV only; T04, boars challenged with the North American PRRSV only. *, significant difference (P ≤ 0.001) between T02 and T04 at the same time point.
For the intergroup comparison, the number of genomic copies and the log10 TCID50/ml of the European PRRSV were not significantly different in the serum samples from vaccinated, challenged European PRRSV boars (T01) from values in samples from nonvaccinated, challenged European PRRSV boars (T03) (Fig. 2). The mean viral titers in serum expressed as log10 TCID50/ml and the number of genomic copies of the North American PRRSV were lower in the serum samples from the vaccinated, challenged North American PRRSV boars (T02) than in the serum samples from the nonvaccinated, challenged North American PRRSV boars (T04) at 7 (P = 0.005), 10 (P = 0.002), 14 (P = 0.005), 18 (P < 0.000), and 21 (P = 0.001) dpi for TCID50/ml and at 4 (P = 0.002), 7 (P = 0.001), 10 (P = 0.001), 14 (P = 0.001), 18 (P = 0.001), 21 (P = 0.001), 25 (P < 0.000), 28 (P = 0.008), and 32 (P = 0.004) dpi for the number of genomic viral copies.
Log10 TCID50/ml quantification and real-time PCR of PRRSV RNA in semen.
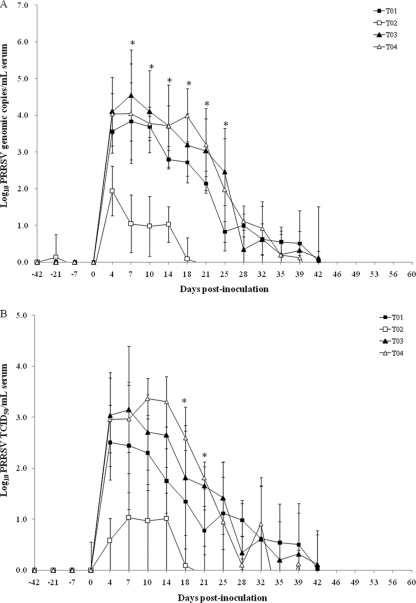
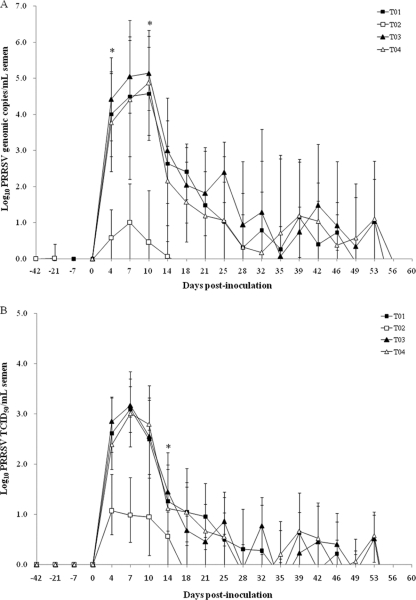
We failed to detect genomic copies of European, North American, and vaccine PRRSV strains in the semen samples at −42 dpi in all groups. No genomic copies of the European PRRSV were detected in the semen samples from vaccinated, challenged North American PRRSV boars (T02), from nonvaccinated, challenged North American PRRSV boars (T04), or from vaccinated, nonchallenged boars (T05). Likewise, no genomic copies of the North American PRRSV were found in the semen samples from vaccinated, challenged European PRRSV boars (T01), from nonvaccinated, challenged European PRRSV boars (T03), or from vaccinated, nonchallenged boars (T05). We measured no genomic copies of the vaccine strain in the semen samples from nonvaccinated, challenged boars (T03 and T04). The vaccine strain was detected until −21 dpi only in semen from the vaccinated, challenged European PRRSV boars (T01), from the vaccinated, challenged North American PRRSV boars (T02), and from the vaccinated, nonchallenged boars (T05) (Fig. 3). No PRRSV was detected in the semen samples from the negative-control (T06) boars throughout the course of the experiment.
Fig. 3.
Detection of PRRSV in the semen sample. Mean group log10 European or North American PRRSV RNA load (A) and mean group log10 TCID50/ml European or North American PRRSV (B) from the different treatment groups were determined. T01, boars immunized with modified live PRRSV vaccine and challenged with the European PRRSV; T02, boars immunized with modified live PRRSV vaccine and challenged with the North American PRRSV; T03, boars challenged with the European PRRSV only; T04, boars challenged with the North American PRRSV only. *, significant difference (P ≤ 0.001) between T02 and T04 at the same time point.
For the intergroup comparison, the numbers of genomic copies and the log10 TCID50/ml of the European PRRSV in the semen samples did not significantly differ between the vaccinated, challenged European PRRSV boars (T01) and the nonvaccinated, challenged European PRRSV boars (T03) (Fig. 3). The mean viral titers in the semen expressed as log10 TCID50/ml and the number of genomic copies of the North American PRRSV were significantly lower in the semen samples from vaccinated, challenged (North American PRRSV) boars (T02) than in the semen samples from nonvaccinated, challenged (North American PRRSV) boars (T04) at 14 (P = 0.001), 18 (P = 0.003), 21 (P = 0.002), 25 (P = 0.013), and 28 (P = 0.017) dpi for TCID50/ml and at 4 (P = 0.001), 7 (P = 0.003), 10 (P = 0.001), 14 (P = 0.024), 18 (P = 0.015), 39 (P = 0.028),42 (P = 0.031), and 56 (P = 0.017) dpi for the number of genomic viral copies.
The number of genomic copies of PRRSV RNA in the semen samples did not correlate with the blood genomic copy number in vaccinated, challenged European PRRSV boars (T01; Spearman correlation coefficient [rs] = 0.542), in vaccinated, challenged North American PRRSV boars (T02; rs = 0.499), in nonvaccinated, challenged European PRRSV boars (T03; rs = 0.694), or in nonvaccinated, challenged North American PRRSV boars (T04; rs = 0.688).
DISCUSSION
The present study has demonstrated that the vaccination of boars by commercial modified live vaccine decreased subsequent shedding of the North American PRRSV after challenge but was unable to decrease the shedding of the European PRRSV in the semen after challenge. These results indicated that the North American PRRSV-based modified live vaccine is more effective against homologous challenges than against heterologous challenges. These results agree with previous findings in which less heterologous protection was observed in a trial that included the challenge of pregnant sows than in homologous challenge with the same vaccine (16). Moreover, vaccination with this vaccine barely reduced the level of viremia after challenge with the European PRRSV in preweaning pigs (25). In the present study, vaccinated boars (groups T01, T02, and T05) shed the vaccine PRRSV strain only during the first 21 days postvaccination. These results are similar to those of a previous study (8).
Vaccinated boars shed the North American PRRSV at the rate of 100.1 to 101.0 viral genome copies per ml and 3.63 to 101.1 50% tissue culture infective doses (TCID50)/ml, respectively, in semen, whereas nonvaccinated boars shed the North American PRRSV at the rate of 100.2 to 104.7 viral genome copies per ml and 1.14 to 103.07 TCID50/ml, respectively, in semen. Thus, vaccination of boars reduced the shedding of North American PRRSV by approximately 99.7% in the semen compared to nonvaccinated boars. These results agree with a previous study that showed that boars are protected from the North American PRRSV shedding in the semen after vaccination (8). The reduction in PRRSV cDNA shedding is meaningful because the dose of viruses in semen plays a major role in the transmissibility of the PRRSV. For example, the North American PRRSV can be transmitted through extended semen depending on the dose of the virus. One out of five gilts (20%) inseminated only at doses of 40 and 400 TCID50/ml of seroconverted extended semen compared to 100% seroconversion in animals given extended semen doses >4,000 TCID50/ml (1). Hence, the amount of North American PRRSV present in semen from vaccinated boars may minimize the transmission of North American PRRSV to sows via artificial insemination.
Vaccinated boars shed the European PRRSV at the rate of 100.1 to 104.57 viral genome copies per ml and 1.66 to 103.10 TCID50/ml, respectively, in semen, whereas nonvaccinated boars shed the European PRRSV at the rate of 100.3 to 105.14 viral genome copies per ml and 1.69 to 103.17 TCID50/ml, respectively, in semen. Thus, vaccination of boars reduced the shedding of European PRRSV by only 11.1% in the semen compared to nonvaccinated boars. These observations are in contrast with a Danish study (22) in which European PRRSV (Danish isolate) shedding was significantly reduced after heterologous challenge in boars that were immunized with the same vaccine used in this study (22). We have no clear explanation for this discrepancy, but it may be due to antigenic variation between two European genotypes. Because the ORF5 and ORF7 nucleotide sequences in the Korean isolate (European genotype) are 88% and 91% identical, respectively, to the Danish European PRRSV isolate (PRRSV 18794/93; GenBank accession numbers AY035906.1 for ORF5 and AY035951.1 for ORF7) used in a previous study (22), this genetic difference may indicate that the two European PRRSVs are antigenically different. In addition, the genetic diversity within the European PRRSV may affect the efficacy of the European-type vaccine (15). However, in the present study, the antigenic difference was not proven. Further study is needed to determine the antigenic difference between Korean and Danish European PRRSV strains to explain the various degrees of shedding reduction in the semen that result from the application of the same North American-based commercial vaccine against the different European genotype PRRSV strains.
The low efficiency of the North American PRRSV-based vaccine used in this study for reducing European PRRSV shedding is clinically significant information although this study used extra label for the vaccine. Practitioners and producers should note that under field conditions, a modified live vaccine may not efficiently reduce the shedding of some European PRRSV strains in boars even if the boars are vaccinated in a whole-herd vaccination program in which all boars are vaccinated every 3 to 4 months, similar to a sow vaccination program. Therefore, it is strongly recommended that regular surveillance of the European PRRSV genotype in the semen is undertaken in countries where both genotypes of PRRSV coexist.
ACKNOWLEDGMENTS
This research was supported by contract research funds of the Research Institute for Veterinary Science from the College of Veterinary Medicine and by Brain Korea 21 Program for Veterinary Science in the Republic of Korea.
Footnotes
Published ahead of print on 10 August 2011.
REFERENCES
- 1. Benfield D., Nelson C. M., Steffen, Rowland R. R. R. 2000. Transmission of PRRSV by artificial insemination using extended semen seeded with different concentrations of PRRSV, p. 405–408 In Proceedings of the 31st Annual Meeting of the American Association of Swine Practitioners, Indianapolis, IN [Google Scholar]
- 2. Cavanagh D. 1997. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 142:629–633 [PubMed] [Google Scholar]
- 3. Cheon D. S., Chae C. 1999. Distribution of a Korean strain of porcine reproductive and respiratory syndrome virus in experimentally infected pigs, as demonstrated immunohistochemically and by in-situ hybridization. J. Comp. Pathol. 120:79–88 [DOI] [PubMed] [Google Scholar]
- 4. Cheon D. S., Chae C. 2000. Comparison of virus isolation, reverse transcription-polymerase chain reaction, immunohistochemistry, and in situ hybridization for the detection of porcine reproductive and respiratory syndrome virus from naturally aborted fetuses and stillborn piglets. J. Vet. Diagn. Invest. 12:582–587 [DOI] [PubMed] [Google Scholar]
- 5. Cheon D. S., Chae C. 2000. Antigenic variation and genotype of isolates of porcine reproductive and respiratory syndrome virus in Korea. Vet. Rec. 147:215–218 [DOI] [PubMed] [Google Scholar]
- 6. Cheon D. S., Chae C. 2001. Distribution of porcine reproductive and respiratory syndrome virus in stillborn and liveborn piglets from experimentally infected sows. J. Comp. Pathol. 124:231–237 [DOI] [PubMed] [Google Scholar]
- 7. Cheon D. S., Chae C., Lee Y. S. 1997. Detection of nucleic acids of porcine reproductive and respiratory syndrome virus in the lungs of naturally infected piglets as determined by in situ hybridization. J. Comp. Pathol. 117:157–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Christopher-Hennings J., Nelson E., Nelson J., Benfield D. 1997. Effects of a modified-live vaccine against porcine reproductive and respiratory syndrome virus in boars. Am. J. Vet. Res. 58:40–45 [PubMed] [Google Scholar]
- 9. Christopher-Hennings J., et al. 1995. Detection of porcine reproductive and respiratory syndrome virus in boar semen by PCR. J. Clin. Microbiol. 33:1730–1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gagnon C. A., et al. 2008. Development and use of a multiplex real-time quantitative polymerase chain reaction assay for detection and differentiation of porcine Circovirus-2 genotypes 2a and 2b in an epidemiological survey. J. Vet. Diagn. Invest. 20:545–558 [DOI] [PubMed] [Google Scholar]
- 11. Gilbert S. A., Larochelle R., Magar R., Cho H. J., Deregt D. 1997. Typing of porcine reproductive and respiratory syndrome viruses by a multiplex PCR assay. J. Clin. Microbiol. 35:264–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hermann J. R., Munoz-Zanzi C. A., Roof M. B., Burkhart K., Zimmerman J. J. 2005. Probability of porcine reproductive and respiratory syndrome (PRRS) virus infection as a function of exposure route and dose. Vet. Microbiol. 110:7–16 [DOI] [PubMed] [Google Scholar]
- 13. Kim S. H., et al. 2010. A molecular analysis of European porcine reproductive and respiratory syndrome virus isolated in South Korea. Vet. Microbiol. 143:394–400 [DOI] [PubMed] [Google Scholar]
- 14. Kweon C. H., et al. 1994. Isolation of porcine reproductive and respiratory syndrome virus (PRRSV) in Korea. Korean J. Vet. Res. 34:77–83 [Google Scholar]
- 15. Labarque G., et al. 2004. Impact of genetic diversity of European-type porcine reproductive and respiratory syndrome virus strains on vaccine efficacy. Vaccine 22:4183–4190 [DOI] [PubMed] [Google Scholar]
- 16. Lager K. M., Mengeling W. L., Brockrneier S. L. 1995. Limited cross-protection between two strains of porcine reproductive and respiratory syndrome virus in pregnant swine, p. 10 Abstr. Second Int. Symp. Porcine Reprod. Respir. Syndr., Copenhagen, Denmark, 5 to 8 August 1995 [Google Scholar]
- 17. Lee C., et al. 2010. Prevalence and phylogenetic analysis of the isolated type I porcine reproductive and respiratory syndrome virus from 2007 to 2008 in Korea. Virus Genes 40:225–230 [DOI] [PubMed] [Google Scholar]
- 18. Meng X. J. 2000. Heterogeneity of porcine reproductive and respiratory syndrome virus: implication for current vaccine efficacy and future vaccine development. Vet. Microbiol. 74:309–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Meng X. J., Paul P. S., Halbur P. G., Lum M. A. 1995. Phylogenetic analyses of the putative M (ORF 6) and N (ORF 7) genes of porcine reproductive and respiratory syndrome virus (PRRSV): implication for the existence of two genotypes of PRRSV in the U.S.A. and Europe. Arch. Virol. 140:745–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Meulenberg J. J. M., et al. 1993. Lelystad virus, the causative agent of porcine epidemic abortion and respiratory syndrome (PEARS), is related to LDV and EAV. Virology 192:62–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nam E., et al. 2009. Complete genomic characterization of a European type 1 porcine reproductive and respiratory syndrome virus isolate in Korea. Arch. Virol. 154:629–638 [DOI] [PubMed] [Google Scholar]
- 22. Nielsen T. L., et al. 1997. Examination of virus shedding in semen from vaccinated and from previously infected boars after experimental challenge with porcine reproductive and respiratory syndrome virus. Vet. Microbiol. 54:101–112 [DOI] [PubMed] [Google Scholar]
- 23. Oleksiewicz M. B., Botner A., Madsen K. G., Storgaard T. 1998. Sensitive detection and typing of porcine reproductive and respiratory syndrome virus by RT-PCR amplification of whole viral genes. Vet. Microbiol. 64:7–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sur J. H., et al. 1997. Porcine reproductive and respiratory syndrome virus replicates in testicular germ cells, alters spermatogenesis, and induces germ cell death by apoptosis. J. Virol. 71:9170–9179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. van Woensel P. A. M., Liefkens K., Denmmaret S. 1998. Effect on viraemia of an American and a European serotype PRRSV vaccine after challenge with European wild-type strains of the virus. Vet. Rec. 142:510–512 [DOI] [PubMed] [Google Scholar]
- 26. Wasilk A., et al. 2004. Detection of U.S., Lelystad, and European-like porcine reproductive and respiratory syndrome viruses and relative quantitation in boar semen and serum samples by real-time PCR. J. Clin. Microbiol. 42:4453–4461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zimmerman J., et al. 2006. Porcine reproductive and respiratory syndrome virus (porcine arterivirus), p. 387–417 In Straw B. E., Zimmerman J. J., D'allaire S., Taylor D. J. (ed.), Diseases of swine, 9th ed. Blackwell Publishing, Ames, IA [Google Scholar]