Abstract
Type II Toxoplasma gondii KU80 knockouts (Δku80) deficient in nonhomologous end joining were developed to delete the dominant pathway mediating random integration of targeting episomes. Gene targeting frequency in the type II Δku80 Δhxgprt strain measured at the orotate (OPRT) and the uracil (UPRT) phosphoribosyltransferase loci was highly efficient. To assess the potential of the type II Δku80 Δhxgprt strain to examine gene function affecting cyst biology and latent stages of infection, we targeted the deletion of four parasite antigen genes (GRA4, GRA6, ROP7, and tgd057) that encode characterized CD8+ T cell epitopes that elicit corresponding antigen-specific CD8+ T cell populations associated with control of infection. Cyst development in these type II mutant strains was not found to be strictly dependent on antigen-specific CD8+ T cell host responses. In contrast, a significant biological role was revealed for the dense granule proteins GRA4 and GRA6 in cyst development since brain tissue cyst burdens were drastically reduced specifically in mutant strains with GRA4 and/or GRA6 deleted. Complementation of the Δgra4 and Δgra6 mutant strains using a functional allele of the deleted GRA coding region placed under the control of the endogenous UPRT locus was found to significantly restore brain cyst burdens. These results reveal that GRA proteins play a functional role in establishing cyst burdens and latent infection. Collectively, our results suggest that a type II Δku80 Δhxgprt genetic background enables a higher-throughput functional analysis of the parasite genome to reveal fundamental aspects of parasite biology controlling virulence, pathogenesis, and transmission.
INTRODUCTION
Toxoplasma gondii is an extremely widespread obligate intracellular protozoan pathogen of virtually all warm-blooded animals (19). Infection is initiated following oral ingestion of transmissible parasite stages included within cysts contained in infected meat or within oocysts released in the environment by cat feces. Primary T. gondii infection rarely causes significant disease in the nonimmunocompromised individual, unless infection is transmitted in utero (50). Latent T. gondii infection is characterized by the presence of tissue cysts containing nonreplicating bradyzoites. The cysts persist primarily in muscles, the eyes, and the brain (19). For unknown reasons, latent tissue cysts occasionally rupture, and this cyst reactivation can lead to recrudescent disease. By example, infection in utero is associated with the highest risk of later development of ocular toxoplasmosis (66), a significant recurrent retinal infection (36). Immunosuppression markedly increases the likelihood of cyst rupture, conversion of latent bradyzoites to rapidly dividing tachyzoites, and the development of reactivated disease (51). Immune control of latent infection fails in AIDS and other severe immunodeficiencies, resulting in a recrudescent acute infection and causing a potentially lethal toxoplasmic encephalitis (12).
T. gondii has rapidly developed as an outstanding model organism for obligate intracellular eukaryotic pathogens (40, 53). Within the single genus and species T. gondii, three major strain types were defined by their virulence phenotype in mice (58). Currently, type I strains of T. gondii are excellent in vitro models, but this acutely virulent strain type does not readily develop tissue cysts or latent infection in laboratory mice. In contrast, type II strains of T. gondii easily establish latent infections in mice that are characterized by the presence of tissue cysts, the key obligate developmental and latent stage required for the remarkably high transmission potential of this parasite (52). Oral transmission appears to have driven the recent clonal expansion of strains that predominate in North America and Europe (62). Type II strains represent the most prevalent strain type chronically infecting North American and European populations (33). After oral ingestion of a tissue cyst in a naive host, the encysted bradyzoite reactivates and converts back to the rapidly dividing tachyzoite form that causes disseminated acute infection, followed later by cyst development and the establishment of a latent infection in the host (19). While induction of tachyzoite to bradyzoite conversion in vitro can be triggered by various stress responses (temperature, high pH, chemical stress, nutrient stress, and cytokines) (67), little is currently known regarding fundamental parasite biology underlying acute and latent phases of infection in vivo.
Host-parasite interaction in acute and latent phases of type II T. gondii infection is dynamic. During acute infection T. gondii selectively invades host cell types primarily of the dendritic, macrophage, and neutrophil lineages (4, 11, 31). By invading host cell types that are critical to mounting effective innate responses to infection, the parasite actively seeks to manipulate the host through several mechanisms (14). Despite or because of this strategy, the host rapidly mounts highly effective CD8+ T cell responses associated with strong interferon gamma (IFN-γ) responses (26, 58). These innate and adaptive immune responses are required to achieve control of acute infection and may also trigger tachyzoite to bradyzoite conversion and development of the latent transmissible tissue cyst (29, 63, 67).
Several landmark studies have recently identified four parasite antigen genes (GRA4, GRA6, ROP7, and tgd057) that possess CD8+ T cell epitopes that elicit corresponding antigen-specific CD8+ T cells associated with the immune control of T. gondii infection (5, 27, 42, 68). The ROP7 protein of unknown function belongs to a family of rhoptry proteins localized to the parasite rhoptry organelles. These Apicomplexa-specific secretory organelles discharge their contents into the host cytosol during invasion of the host cell (7). The parasite dense granule (GRA) proteins GRA4 and GRA6 are members of the Toxoplasma prominent dense granule protein family (46, 49). Dense granule proteins are highly expressed proteins. The dense granule secretory organelles discharge their contents into the parasitophorous vacuole (PV) space following host cell invasion and the initial formation of the parasitophorous vacuole membrane (PVM) (9). A newly proposed member of the dense granule protein family called GRA15 localized to the dense granules, the PV space, the space outside of the PVM, and was also associated with evacuoles, suggesting that GRA15 is also a rhoptry-like protein in potentially being secreted during invasion of the host cell (54). GRA15 plays a significant role in host cell NF-κB nuclear translocation and NF-κB-mediated transcription and regulates the induction of IL-12 secretion in infected mouse macrophages (54). Various potential biological roles for GRA proteins have been previously proposed and significant studies have been performed on GRA protein secretion, traffic, and interaction biology (10, 49). At the tachyzoite stage, most of the GRA proteins associate with the PV membranes, meaning either the PVM and its digitations or the intravacuolar network of membranous tubules (49). At the bradyzoite stage, most of the GRA proteins were localized at the cyst wall or in the cyst matrix (64).
Type I mutants deleted in GRA2 or GRA6 showed a phenotype of reduced acute virulence during infection in mice (48; C. Mercier, unpublished data). However, to date, the only GRA proteins successfully disrupted in the type II background are GRA3 (13) and GRA15 (54). Disruption of type II GRA3 resulted in a phenotype of reduced acute virulence (13). Disruption of type II GRA15 increased parasite burdens in infected mice without influencing virulence (54). These previous studies did not address a functional role for GRA proteins in cyst development during infection. While GRA proteins appear to be central to the host-parasite relationship, it is currently unknown whether GRA proteins are required to establish latent infection.
Tachyzoite-to-bradyzoite conversion, cyst development, and latent infection are critical to parasite survival and transmission. Nine distinct targeted gene deletions (1, 6, 13, 32, 38, 54, 55, 57, 65, 69) have been successfully developed in type II strains since genetic transformation of T. gondii was reported nearly 20 years ago (18, 39, 61). In contrast, recently reported type I KU80 knockout strains that exhibit highly efficient gene replacement frequencies has significantly accelerated the development of targeted gene deletions (2, 3, 15, 20, 23, 26, 34, 37, 44). Increased efficiency of double-crossover homologous recombination at targeted loci in KU80 knockouts is due to the functional loss of the nonhomologous end-joining DNA repair pathway that mediates a major mechanism underlying frequent random insertion of linear episomes in T. gondii (26). We report here the development of KU80 knockouts (Δku80) in type II T. gondii. Eight genetic loci were targeted for the development of mutant strains with targeted single-gene deletions or multiple-gene deletions to functionally examine candidate genes required for cyst development and latent infection. In particular, we examined the hypothesis that a host response to parasite antigen genes that elicit antigen-specific CD8+ T cell responses during host infection may be essential to cyst development. Our results support a critical role for dense granule proteins GRA4 and GRA6 in fundamental biology at the host-parasite interface that is essential for cyst development and transmission of T. gondii.
MATERIALS AND METHODS
Primers.
All oligonucleotide primers used in the development of plasmids for targeting gene deletions are listed in Table S1 in the supplemental material. All oligonucleotide primers used in validation of mutants with gene deletions are listed in Table S2 in the supplemental material.
Plasmid constructs.
All plasmids were developed using the yeast shuttle vector pRS416 and yeast recombination cloning which fused three distinct genetic elements (an ∼1-kb 5′ target flank, an ∼2-kb hypoxanthine-xanthine-guanine phosphoribosyltransferase [HXGPRT] selectable marker cassette, and an ∼1-kb 3′ target flank, all 5′ to 3′ and in this order) with pRS416 using 31- to 33-bp homologous crossovers at recombination junctions (23). DNA elements for yeast recombination were amplified from type II Prugniaud (Pru) genomic DNA or type I RH genomic DNA as indicated. Targeting plasmids constructed in pRS416 were verified by using restriction enzyme digests, and target DNA flanks were sequenced to verify DNA sequence homology.
Plasmid pΔTPR1 was constructed to delete nucleotides 2741952 to 2737843 in the Pru KU80 locus, defined as TGME49_112510 on chrXI of the ToxoDB database version 6.0 (www.toxodb.org). This deletion strategy deletes 1,228 bp of the KU80 5′ upstream putative gene regulatory region and the first three exons of the predicted four protein coding exons of the KU80 gene. The HXGPRT minigene cassette was fused between a 2,997-bp 5′ genomic targeting flank and a 3,304-bp 3′ genomic targeting flank amplified from Pru genomic DNA.
Plasmid pΔTPFC1 was constructed by NotI and PmeI digestion of pΔTPR1 at the 3′ end of the 3′ targeting flank, followed by ligation with the cytosine deaminase (CD) selectable marker contained on a NotI/PmeI restriction fragment isolated from plasmid pMCD4.4 (26).
Plasmid pΔTPRC2 was constructed by digesting pΔTPR1 with KspI, followed by self-ligation to delete the HXGPRT minicassette fragment from pΔTPR1.
Plasmid pΔUPRP was constructed to delete nucleotides 2709350 to 2713372 of the uracil phosphoribosyltransferase (UPRT) chromosomal locus on chrXI annotated as TGME49_112480. The HXGPRT minigene cassette was fused between a 1,131-bp 5′ UPRT genomic targeting flank and a 1,119-bp 3′ UPRT genomic targeting flank amplified from Pru genomic DNA.
Plasmid pΔUPP was constructed to delete nucleotides 1491873 to 1488688 of the uridine phosphorylase (UP) chromosomal locus (23) on chrXI annotated as TGME49_110640. The HXGPRT minigene cassette was fused between a 1,141-bp 5′ UP genomic targeting flank and a 955-bp 3′ UP genomic targeting flank amplified from Pru genomic DNA.
Plasmid pΔOPT was constructed to delete nucleotides 2733578 to 2735556 of the orotate phosphoribosyltransferase (OPRT) locus defined as TGGT1_010360 on chrVIIb. The HXGPRT minigene cassette was fused between a 1,050-bp 5′ OPRT genomic targeting flank and a 1,118-bp 3′ OPRT genomic targeting flank amplified from RH genomic DNA.
Plasmid pΔOPP was constructed to delete nucleotides 2719166 to 2721148 of the OPRT locus defined as TGME49_059660 on chrVIIb. The HXGPRT minigene cassette was fused between a 1,050-bp 5′ OPRT genomic targeting flank and a 1,118-bp 3′ OPRT genomic targeting flank amplified from Pru genomic DNA.
Plasmid pΔOPPC was constructed by digesting pΔOPP with SacII, followed by self-ligation to delete the HXGPRT minicassette.
Plasmid pΔGRA4P was constructed to delete nucleotides 1581347 to 1570147 of the GRA4 locus on chrXI annotated as TGME49_110780. The HXGPRT minigene cassette was fused between a 1,124-bp 5′ GRA4 genomic targeting flank and a 989-bp 3′ GRA4 genomic targeting flank amplified from Pru genomic DNA.
Plasmid pΔGRA4PC was constructed by digesting pΔGRA4 with SpeI, followed by self-ligation to delete the HXGPRT minicassette.
Plasmid pΔGRA6P was constructed to delete nucleotides 7216123 to 7215219 of the GRA6 locus on chrX annotated as TGME49_075440. The HXGPRT minigene cassette was fused between a 1,048-bp 5′ GRA6 genomic targeting flank and a 955-bp 3′ GRA6 genomic targeting flank amplified from Pru genomic DNA.
Plasmid pΔGRA6PC was constructed by digesting pΔGRA6 with SpeI, followed by self-ligation to delete the HXGPRT minicassette.
Plasmid pΔROP4/7P was constructed to delete nucleotides 1414364 to 1404628 of the ROP4/7 locus on chr1a annotated as TGME49_095110. This gene locus is incomplete in the genome database. Analysis of the Toxodb database identified common flanking and unique DNA surrounding the ROP4/7 locus and suggested the gene structure for the ROP4/7 locus consists of a 5′ ROP4 gene, followed by a complete ROP7A allele and ROP7B allele (with identical coding DNA but minor polymorphisms in their UTRs) and a 3′ truncated and incomplete ROP7C allele (L. M. Rommereim and D. J. Bzik, unpublished data). Consequently, we targeted the deletion of the entire locus (∼14,297 bp) and all ROP4 and ROP7 alleles by using target DNA flanks derived from unique DNA sequences that reside just 5′ or just 3′ of the ROP4/7 locus. The HXGPRT minigene cassette was fused between a 1,179-bp 5′ ROP4/7 genomic targeting flank and a 989-bp 3′ ROP4/7 genomic targeting flank amplified from Pru genomic DNA.
Plasmid pΔTGD057P was constructed to delete nucleotides 6486828 to 6487525 of the tgd057 locus on chrXI annotated as TGME49_015980. The HXGPRT minigene cassette was fused between an 800-bp 5′ tgd057 genomic targeting flank and a 1,259-bp 3′ tgd057 genomic targeting flank amplified from Pru genomic DNA.
Plasmid pGRA4X was constructed to insert the coding region of GRA4 into the UPRT locus under the control of the UPRT 5′ transcribed, untranslated region (5′UTR). Targeted insertion of the pGRA4X cassette deleted nucleotides 2709350 to 2713372 of the coding region of the UPRT chromosomal locus on chrXI annotated as TGME49_112480. The GRA4 coding region (plus 12 bp 5′ of the ATG) and 635 bp of the GRA4 3′UTR (nucleotides 1579415 to 1581100 on ChrXI) was amplified from Pru genomic DNA and inserted between the 5′ UPRT target flank and the HXGPRT marker of plasmid pΔUPRP.
Plasmid pGRA6X was constructed to insert the coding region of GRA6 into the UPRT locus under the control of the UPRT 5′UTR. Targeted insertion of the pGRA6X cassette deleted nucleotides 2709350 to 2713372 of the coding region of the UPRT chromosomal locus on chrXI annotated as TGME49_112480. The GRA6 coding region (plus 15 bp 5′ of the ATG) and 542 bp of the GRA6 3′UTR (nucleotides 7214586 to 7215818 on ChrX) was amplified from Pru genomic DNA and inserted between the 5′ UPRT target flank and the HXGPRT marker of plasmid pΔUPRP.
Culture conditions and strains.
All parasite strains were continuously maintained in vitro by serial passage in Eagle modified essential medium supplemented with 1% fetal bovine serum in diploid human foreskin fibroblasts (HFF) at 36°C (26). Pyrimidine auxotrophs were supplemented with uracil (250 μM). The parental Pru strain (Δhxgprt) was previously made transgenic for green fluorescent protein (GFP) under the control of the LDH2 bradyzoite stage-specific promoter and was designated BSG-4 (60). The T. gondii strains used and developed in the present study are shown in Table 1. We previously reported the RHΔku80Δhxgprt strain (26).
Table 1.
T. gondii strains used and developed in this studya
| Strain | Parent strain (source or reference) | Genotype |
|---|---|---|
| PruΔku80::HXGPRT | PruΔhxgprt (BSG-4) (60) | Δku80 |
| PruΔku80Δhxgprt | PruΔku80::HXGPRT | Δku80 Δhxgprt |
| PruΔku80Δuprt::HXGPRT | PruΔku80Δhxgprt | Δku80 Δuprt::HXGPRT |
| RHΔku80Δoprt::HXGPRT | RHΔku80Δhxgprt (26) | Δku80 Δoprt::HXGPRT |
| PruΔku80Δoprt::HXGPRT | PruΔku80Δhxgprt | Δku80 Δoprt::HXGPRT |
| PruΔku80ΔoprtΔhxgprt | PruΔku80Δoprt::HXGPRT | Δku80 Δoprt Δhxgprt |
| PruΔku80Δup::HXGPRT | PruΔku80Δhxgprt | Δku80 Δup::HXGPRT |
| PruΔku80Δgra4::HXGPRT | PruΔku80Δhxgprt | Δku80 Δgra4::HXGPRT |
| PruΔku80Δgra4Δhxgprt | PruΔku80Δgra4::HXGPRT | Δku80 Δgra4 Δhxgprt |
| PruΔku80Δgra6::HXGPRT | PruΔku80Δhxgprt | Δku80 Δgra6::HXGPRT |
| PruΔku80Δgra6Δhxgprt | PruΔku80Δgra6::HXGPRT | Δku80 Δgra6 Δhxgprt |
| PruΔku80Δtgd057::HXGPRT | PruΔku80Δhxgprt | Δku80 Δtgd057::HXGPRT |
| PruΔku80Δrop4/7::HXGPRT | PruΔku80Δhxgprt | Δku80 Δrop4/7::HXGPRT |
| PruΔku80Δgra4Δtgd057::HXGPRT | PruΔku80Δgra4Δhxgprt | Δku80 Δgra4 Δtgd057::HXGPRT |
| PruΔku80Δgra6Δtgd057::HXGPRT | PruΔku80Δgra6Δhxgprt | Δku80 Δgra6 Δtgd057::HXGPRT |
| PruΔku80Δgra6Δgra4::HXGPRT | PruΔku80Δgra6Δhxgprt | Δku80 Δgra6 Δgra4::HXGPRT |
| PruΔku80Δgra6Δgra4Δhxgprt | PruΔku80Δgra6Δgra4::HXGPRT | Δku80 Δgra6 Δgra4 Δhxgprt |
| PruΔku80Δgra4Δuprt::gra4coding region + 3′UTRHXGPRT | PruΔku80Δgra4Δhxgprt | Δku80 Δgra4 Δuprt::gra4coding region + 3′UTRHXGPRT |
| PruΔku80Δgra6Δuprt::gra6coding region + 3′UTRHXGPRT | PruΔku80Δgra6Δhxgprt | Δku80 Δgra6 Δuprt::gra6coding region + 3′UTRHXGPRT |
All strains used and developed in this study are transgenic for LDH2-GFP and the chloramphenicol drug resistance marker (CAT) randomly integrated into the parental Pru strain at undefined genetic loci (60).
Transformation, selection, and gene deletion verification strategy.
Electroporations were performed using a model BTX600 electroporator and previously described methods (26). All transfected plasmids were linearized 5′ of the 5′ target DNA flank prior to transfection using unique restriction enzyme sites designed into the targeting plasmids. Forward selections to integrate the pminiHXGPRT selectable marker were performed in mycophenolic acid (MPA; 25 μg/ml) and xanthine (50 μg/ml) (16, 26). Negative selections to excise HXGPRT were performed in 6-thioxanthine (6TX) (200 μg/ml) (23, 26). Negative selections using the cytosine deaminase (CD) selectable marker were performed in 5-fluorocytosine (5FC; 50 μM) (22, 26). Negative selections to delete UPRT were performed in 5 μM 5-fluorodeoxyuridine (FUDR) (17). After transfection, parasites were allowed to replicate for 24 h without selection to allow replication and ramp up homologous recombination, and then selections were launched and continuously maintained through verification steps of cloned isolates. The strategy for verification of mutant strains was previously described (23, 26).
Genomic DNA isolation and PCR.
Genomic DNA purifications used a DNA blood minikit (Qiagen) and were performed with a Qiacube automated robotic work station (Qiagen). PCR products were amplified using a 1:1 mixture of Taq DNA polymerase and an Expand long-template PCR (Roche).
Determination of gene replacement frequencies and statistical analysis.
PFU assays were used to determine gene replacement frequencies. Three replicate PFU flasks were prepared for each titration point and selection condition. A Student t test analysis was used to calculate the standard error of the mean (SEM). PFU assays were performed at various times after transfection to determine the gene replacement frequency at the UPRT locus based on the fraction of parasites that had dual resistance to MPA and FUDR compared to the fraction of parasites that were resistant only to MPA (26). PFU assays were performed at various times after transfection to determine the gene replacement frequency at the OPRT locus based on the fraction of parasites that exhibited resistance to MPA and grew with uracil supplementation compared to the fraction of parasites that were resistant to MPA without uracil supplementation (25).
Determination of the replication rate of mutant strains in vitro.
The intracellular growth rate of various type II strains was measured in HFFs in a 45-h growth assay using previously described methods (24). Briefly, freshly lysed tachyzoites were Nuclepore filtered to isolate individual tachyzoites that were used to infect HFF monolayers at a multiplicity of infection of ∼0.1. After 1 h of invasion, the culture was vigorously washed four times in phosphate-buffered saline (PBS) to remove extracellular tachyzoites (verified by microscopy). Cultures were then returned to growth medium, and the number of parasites per vacuole was scored from at least 50 vacuoles per sample at 45 h postinfection. Cultures were prepared in duplicate, and the growth assay was repeated twice. The data samples were subjected to a Student t test and are represented as the means (parasites per vacuole) ± the SEM. Differences in parasites per vacuole were compared for parental and mutant strains matched for the presence or absence of HXGPRT. A P value of <0.05 was considered significant.
Mice.
Female 7- to 9-week-old C57BL/6 (H-2Kb), CBA (H-2Kk), and BALB/c (H-2Ld) mice were purchased from Jackson laboratories (Bar Harbor, ME) and maintained at the Dartmouth-Hitchcock Medical Center mouse facility. All mice were cared for and handled according to Animal Care and Use Program of Dartmouth College using National Institutes of Health (NIH)-approved institutional animal care and use committee guidelines. Groups of mice (as indicated) were infected by oral gavage with 10 cysts or were infected by intraperitoneal injection with 0.2 ml (200 tachyzoites) of various Pru genotypes in PBS. Each tachyzoite preparation was subjected to a PFU assay to determine viability. All mice infected with tachyzoites received 130 to 252 PFU. Infected mice were then monitored daily for wellness.
Cyst isolation, visualization, enumeration, and size determination.
Brains from mice inoculated with various genotypes of the Pru background were harvested and homogenized by using a Dounce homogenizer in 2 ml of sterile PBS (30). All cyst counts were performed immediately after the isolation of brain homogenates. In most experiments, 20 slides (10%) of each brain were scored for cysts, and any deviation from this counting scheme was performed only in cases when cyst numbers were low and additional slides were scored. Cysts were scored under an inverted fluorescence phase-contrast microscope (Olympus CKX41) to count GFP-positive (GFP+) cysts at a magnification of ×150, which provided the highest sensitivity for the detection of GFP+ cysts. Average cyst size was measured as a relative unit of diameter. Average cyst size was determined at a magnification of ×150 by measuring bradyzoite diameter units (BDU). BDU were determined for each randomly selected cyst by counting the number of GFP+ bradyzoites residing in a direct line from one edge of the cyst wall to the opposite edge of the cyst wall at the maximum point of the cyst diameter. BDU were determined for 25 randomly selected cysts from each mouse brain tissue sample. At least five mice were used to determine BDU for each parasite strain tested. Cyst data samples were subjected to a Student t test and are represented as means ± the SEM. Differences in cyst burdens between groups of infected mice were significant if the P value was <0.05. Cysts were also evaluated using confocal microscopy. Images were acquired using a Zeiss LSM 510 Meta laser scanning confocal microscope equipped with a 20× Plan Apo NA 0.75 objective lens (Carl Zeiss Microimaging, Thornwood, NY). Confocal settings were set to 1 Airy unit (1.8-μm optical section). A zoom setting of six generated a pixel size of 0.15 by 0.15 μm. LSM 510 software version 3.2 was used.
RESULTS
Generation of T. gondii strains PruΔku80::HXGPRT and PruΔku80Δhxgprt.
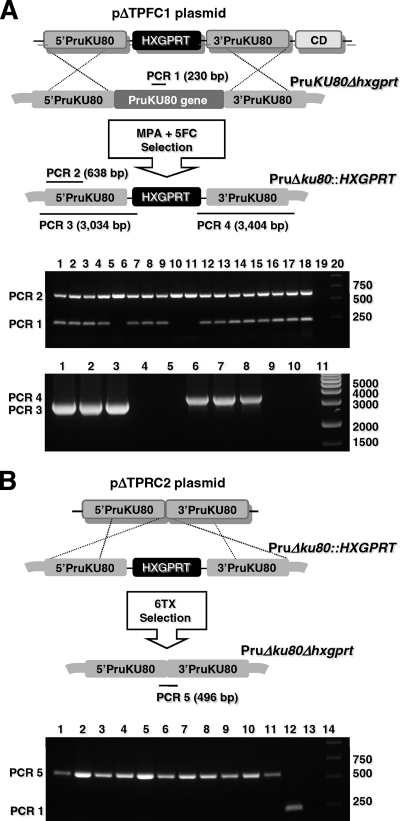
Alignments of the predicted type I (26) and type II (TGME49_112510, 583.m05492 [www.toxodb.org] version 4.0) KU80 loci revealed frequent strain specific nucleotide polymorphisms. Plasmid pΔTPR1 was developed to functionally delete the type II KU80 gene. Multiple independent transfections of various type II strains, selections, and screens of more than ∼800 MPA resistant clones failed to identify a targeted KU80 knockout in any type II strain.
We constructed plasmid pΔTPFC1 with the cytosine deaminase (CD) gene inserted 3′ of the 3′ KU80 target DNA flank (Fig. 1A). Inclusion of the CD gene provided a more robust selection strategy for double crossovers at the KU80 locus by using negative selection to eliminate transformants that still retained expression of CD by growth of the transfected parasite population in 5-fluorocytosine (Fig. 1A). After continued attempts using the CD negative selection strategy, we detected a low frequency of KU80 knockout clones (PruΔku80::HXGPRT) that were positive for PCR 2 and were negative for PCR 1 that probes for the presence of the targeted deletion in the KU80 gene (Fig. 1A, lanes 5, 9, and 10, top gel panel). Three PCR 1-negative clones were then validated as targeted KU80 knockouts using PCR 3 and PCR 4 to demonstrate precise 5′ and 3′ integration of HXGPRT at the deleted KU80 locus (Fig. 1A, bottom gel panel). To delete the HXGPRT marker from the PruΔku80::HXGPRT strain, two clones were transfected with plasmid pΔTPRC2, which is identical to plasmid pΔTPR1 but lacks the HXGPRT gene (Fig. 1B). 6TX-resistant parasite clones were obtained, and these clones uniformly exhibited the genotype Δku80 Δhxgprt based on PCR analysis using PCR 1 and PCR 5 that was designed to span the deleted region of KU80 and to demonstrate the targeted deletion of the 2-kb HXGPRT marker (Fig. 1B).
Fig. 1.
Construction of the type II Δku80 Δhxgprt genetic background. (A) Strategy for disrupting the KU80 gene in the parental Pru strain via integration of the HXGPRT marker. Approximate locations of PCR products using primer pairs to verify the genotype at the KU80 locus (not to scale) and the expected PCR product sizes are shown. (Top agarose gel panel) PCR 1 and PCR 2 products: 17 randomly selected clones isolated after selection (lanes 1 to 17), parental Pru (lane 18), no template (lane 19), DNA size ladder (lane 20). Clones in lanes 5, 9, and 10 appear negative for PCR 1 showing deletion of the KU80 gene. (Bottom agarose gel panel) PCR 3: clones from lanes 5, 9, and 10 (lanes 1 to 3), parental Pru (lane 4), no template (lane 5); PCR 4: clones from lanes 5, 9, and 10 (lanes 6 to 8), parental Pru (lane 9), no template (lane 10). DNA size ladder (lane 11). A targeted KU80 knockout is positive for the PCR 2, PCR 3, and PCR 4 products and negative for the PCR 1 product. Genotypes with an intact KU80 locus are positive for PCR 1 and PCR 2 and negative for PCR 3 and PCR 4. (B) Excision of the HXGPRT marker from the KU80 locus in the PruΔku80::HXGPRT strain. The strategy for excision of HXGPRT is depicted with negative selection in 6TX after transfection with plasmid pΔTPRC2. Approximate locations of PCR products using primer pairs to verify genotype are depicted (not to scale). The expected PCR product sizes are shown for a positive result. (Agarose gel panel) PCR 1 and PCR 5 products: 11 clones isolated after 6TX selection (lanes 1 to 11), parental Pru (lane 12), parental PruΔku80::HXGPRT (lane 13), DNA size ladder (lane 14). The PruΔku80Δhxgprt strain is positive for PCR 5 and negative for PCR 1 (lanes 1 to 11), the parental Pru strain is positive for PCR 1 and negative for PCR 5 (lane 12), and the parental PruΔku80::HXGPRT strain is negative for PCR 1 (496-bp product) and PCR 5 (lane 13).
Influence of DNA sequence homology on gene replacement frequency at the OPRT locus.
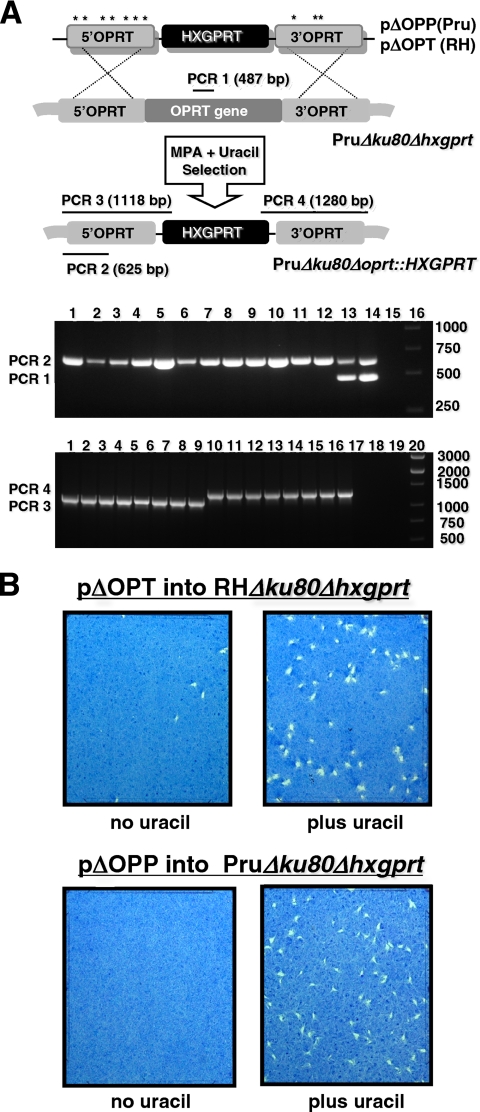
Disruption of de novo pyrimidine synthesis induces uracil auxotrophy (23, 24). Here, the OPRT locus was targeted for deletion to induce uracil auxotrophy as a phenotype. Type II OPRT deletion was targeted using plasmid pΔOPP and type I OPRT deletion was targeted using plasmid pΔOPT (Fig. 2A). OPRT knockouts were obtained in type I and type II backgrounds based on the identification of clones that were positive for PCR 2 and negative for PCR 1 that probes for the presence of the targeted deletion in the OPRT gene (Fig. 2A, top gel panel). Using PCR 3 and PCR 4, four of these clones from each strain type were then shown to be targeted OPRT deletions based on precise 5′ and 3′ integration of the HXGPRT marker at the OPRT locus (Fig. 2A, bottom gel panel).
Fig. 2.
Targeted gene replacement at the orotate phosphoribosyltransferase (OPRT) locus. (A) Strategy for disruption of OPRT by double-crossover homologous recombination in type II strain PruΔku80Δhxgprt using plasmid pΔOPP and in type I strain RHΔku80Δhxgprt using plasmid pΔOPT. Compared to plasmid pΔOPP, plasmid pΔOPT contains seven single-nucleotide polymorphisms (indicated by “*”) in the 5′ targeting DNA flank and three nucleotide polymorphisms in the 3′ targeting DNA flank. The PCR strategy for genotype verification of deletion of OPRT is depicted (not to scale). The expected PCR product sizes are shown for a positive result. (Top agarose gel panel) PCR 1 and PCR 2 products: 6 clones isolated after targeting type I OPRT deletion (lanes 1 to 6), 6 clones isolated after targeting type II OPRT deletion (lanes 7 to 12), parental type I RHΔku80Δhxgprt (lane 13), parental type II PruΔku80Δhxgprt (lane 14), no template (lane 15) DNA size ladder (lane 16). All six type I clones and all six type II clones appear to be OPRT knockouts (PCR 1 negative and PCR 2 positive). Type I and type II clones were evaluated in PCR 3 and PCR 4 to verify integration of HXGPRT at the deleted OPRT locus. (Bottom agarose gel panel) PCR 3 products: type I clones (lanes 1 to 4), type II clones (lanes 5 to 8); PCR 4 products: type I clones (lanes 9 to 12), type II clones (lanes 13 to 16); PCR 3 and PCR 4 on parental type I RHΔku80Δhxgprt (lane 17), PCR 3 and PCR 4 on parental type II PruΔku80Δhxgprt (lane 18), PCR 3 and PCR 4 using no template (lane 19), DNA size ladder (lane 20). (B) PFU assays were performed at various times after transfection of pΔOPT in the type I RHΔku80Δhxgprt (top) or after transfection of plasmid pΔOPP into the type II PruΔku80Δhxgprt (bottom) to determine the gene replacement frequency at the OPRT locus based on the fraction of parasites that exhibited resistance to MPA and grew with uracil supplementation compared to the fraction of parasites that were resistant to MPA without uracil supplementation (Table 2). The PFU assays shown were sampled 25 days posttransfection with or without uracil. All PFU assays contained MPA selection medium.
The efficiency of gene targeting at the OPRT locus in the type I versus the type II Δku80 Δhxgprt strain was measured using target DNA flanks with a strain-specific DNA homology of 100% (Fig. 2A) (Table 2). PFU assays performed on the MPA-selected population of parasites at 18 and again at 25 days posttransfection of the targeting plasmids revealed that targeted disruption of OPRT created the uracil auxotroph phenotype at a high frequency in both the type I and the type II Δku80 Δhxgprt strains (Fig. 2B). The gene targeting frequency at the OPRT locus was found to be slightly higher in the type II strain PruΔku80Δhxgprt (99.8% ± 0.1%) compared to the type I strain RHΔku80Δhxgprt (92.9% ± 1.5%) (Table 2).
Table 2.
Gene replacement frequency at the OPRT locus in type I and type II KU80 knockouts
| Transfected strain | Plasmida | Day assayed | Gene replacement (%) at the OPRT locus |
|
|---|---|---|---|---|
| Expt 1 | Expt 2 | |||
| RHΔku80Δhxgprt | pΔOPT | 18 | 92.1 | 93.0 |
| RHΔku80Δhxgprt | pΔOPT | 25 | 91.4 | 94.3 |
| PruΔku80Δhxgprt | pΔOPP | 18 | 98.8 | 99.3 |
| PruΔku80Δhxgprt | pΔOPP | 25 | 99.7 | 99.8 |
| PruΔku80Δhxgprt | pΔOPT | 18 | 13.2 | 12.4 |
| PruΔku80Δhxgprt | pΔOPT | 25 | 10.6 | 14.9 |
pΔOPT is derived from type I RH, and pΔOPP is derived from type II Pru.
Naturally occurring nucleotide polymorphisms present within the 5′ and 3′ noncoding DNA between type I and type II strains enabled a test of homology requirements necessary for efficient gene targeting at the OPRT locus in T. gondii (Fig. 2A). These nucleotide polymorphisms divided the 5′ target flank into segments of 94, 162, 53, 127, 39, 240, 88, and 247 bp of homology and the 3′ target flank into segments of 530, 22, 359, and 208 bp of homology (sequence alignments between type I and type II OPRT are not shown). Type I targeting plasmid pΔOPT with 7 nucleotide mismatches in the 5′ target DNA flank and 3 mismatches in the 3′ target DNA flank relative to type II was inefficient in targeting gene deletion at the type II OPRT locus (Table 2).
Targeted disruption of the key enzymes of the pyrimidine salvage pathway.
Disruption of UPRT was used to measure gene replacement efficiency in strain PruΔku80Δhxgprt (26). The frequency of gene replacement at the UPRT locus was determined at different time points after transfection of plasmid pΔUPRP by plating equal numbers of parasites either in MPA or in MPA plus FUDR selection. The nonreverting PruΔku80Δuprt::HXGPRT genotype was also confirmed in several MPA resistant clones (data not shown). The frequency of gene replacement at the UPRT locus in strain PruΔku80Δhxgprt was found to be only 3.3% ± 0.4% at 10 days posttransfection, but this frequency steadily increased to 98.2% ± 1.1% by day 32 posttransfection (Table 3). In contrast, the gene replacement efficiency in the parental Pru strain was <0.10% at 32 days postinfection (Table 3).
Table 3.
Gene replacement frequency at the UPRT locus
| Transfected strain | Plasmid | Day assayed | Gene replacement (%) at the UPRT locus |
|
|---|---|---|---|---|
| Expt 1 | Expt 2 | |||
| PruΔhxgprt | pΔUPRP | 25 | 0.03 | 0.00 |
| PruΔhxgprt | pΔUPRP | 32 | 0.08 | 0.00 |
| PruΔku80Δhxgprt | pΔUPRP | 10 | 3.74 | 2.90 |
| PruΔku80Δhxgprt | pΔUPRP | 18 | 46.7 | 55.4 |
| PruΔku80Δhxgprt | pΔUPRP | 25 | 75.9 | 82.1 |
| PruΔku80Δhxgprt | pΔUPRP | 32 | 97.1 | 99.2 |
It is currently unknown whether the two critical enzymes of the pyrimidine salvage pathway (23) (UPRT and uridine phosphorylase [UP]) are required for cyst development and latent T. gondii infection. To complete a genetic dissection of the major pyrimidine salvage activities in the type II T. gondii and to assess the potential role of the salvage pathway in cyst development, we deleted the UP gene using plasmid pΔUPP (data not shown) (Table 1). We found that 23 of 24 randomly selected MPA-resistant clones (96%) had the UP gene deleted.
Efficient targeted excision of the HXGPRT selectable marker from PruΔku80 mutant strains.
HXGPRT was targeted for deletion from the OPRT locus in strain PruΔku80Δoprt::HXGPRT using plasmid pΔOPPC and negative selection in 6TX. Strain PruΔku80Δoprt was easily isolated and validated by PCR analysis (data not shown) (Table 1).
PruΔku80Δhxgprt stably maintains the ability to develop cysts and chronic infection in mice.
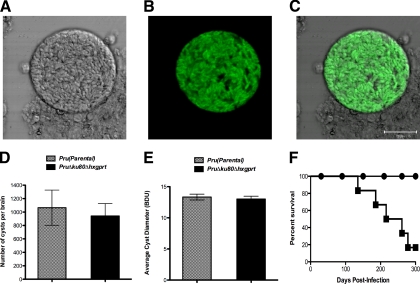
We observed no significant difference (P = 0.20) between the in vitro intracellular replication rates of strain PruΔku80Δhxgprt (14.2 ± 0.50 parasites/vacuole) compared to the parental Pru strain (15.0 ± 0.15 parasites/vacuole) (Table 4). The type II strain PruΔku80Δhxgprt stably maintained the ability to develop brain tissue cyst burdens in mice after more than 16 months of continuous in vitro culture. Brain tissue cysts obtained from mice infected with the strain PruΔku80Δhxgprt revealed a cyst wall structure (Fig. 3 A), as well as the expected bradyzoite stage-specific expression of GFP (Fig. 3B and C) (60). In contrast, GFP expression was not detected in strain PruΔku80Δhxgprt during in vitro culture of the tachyzoite stages (data not shown). In addition, brain cyst burdens in C57BL/6 mice at 3 weeks postinfection with strain PruΔku80Δhxgprt (942 ± 184) were essentially identical (P = 0.70) to the brain cyst burdens observed after infection with the parental Pru strain (1,065 ± 262) (Fig. 3D). Furthermore, cyst sizes in strain PruΔku80Δhxgprt (13.0 ± 0.46 BDU) were essentially identical (P = 0.63) to the cyst sizes measured in the parental Pru strain (13.3 ± 0.47 BDU) (Fig. 3E). Finally, peroral infection of the genetically susceptible C57BL/6 background with 10 brain cysts of strain PruΔku80Δhxgprt caused an acute infection with morbidity, and infected mice recovered and then developed the expected chronic infection that eventually led to mortality (Fig. 3F). Deletion of the KU80 gene in strain PruΔku80Δhxgprt did not lead to any significant defects in regard to the intracellular growth rate, the development of brain cyst burdens, the cyst size, or the establishment of a chronic or latent infection.
Table 4.
Intracellular replication rate of selected strains used in this study
| Strain (reference) | Mean no. of parasites per vacuolea ± SEM |
P (compared to the control strain)b |
||
|---|---|---|---|---|
| P | Significance | Control strain genotype | ||
| PruΔku80Δgra4::HXGPRT | 14.1 ± 0.48 | 0.74 | NS | Δku80::HXGPRT |
| PruΔku80Δgra6::HXGPRT | 13.9 ± 0.29 | 1 | NS | Δku80::HXGPRT |
| PruΔku80Δgra6Δgra4::HXGPRT | 13.2 ± 0.14 | 0.09 | NS | Δku80::HXGPRT |
| PruΔku80Δuprt::HXGPRT | 14.2 ± 0.13 | 0.33 | NS | Δku80::HXGPRT |
| PruΔku80Δtgd057::HXGPRT | 15.2 ± 0.31 | 0.02 | S | Δku80::HXGPRT |
| PruΔku80Δrop4/7::HXGPRT | 14.9 ± 0.20 | 0.03 | S | Δku80::HXGPRT |
| PruΔku80::HXGPRT | 13.9 ± 0.31 | ND | ||
| PruΔhxgprt (parental) (60) | 15.0 ± 0.17 | ND | ||
| PruΔku80Δhxgprt | 14.2 ± 0.57 | 0.20 | NS | Δhxgprt (parental) |
| PruΔku80Δgra4Δhxgprt | 14.5 ± 0.36 | 0.70 | NS | Δku80 Δhxgprt |
| PruΔku80Δgra6Δhxgprt | 13.8 ± 0.23 | 0.51 | NS | Δku80 Δhxgprt |
| PruΔku80Δgra6Δgra4Δhxgprt | 13.5 ± 0.36 | 0.36 | NS | Δku80 Δhxgprt |
Parasites per vacuole were scored after 45 h of intracellular replication.
To determine the P value, strains were matched for the presence or absence of HXGPRT. S, significant; NS, not significant; ND, not determined.
Fig. 3.
The PruΔku80Δhxgprt strain elicits GFP+ cysts and chronic and/or latent infection in genetically susceptible C57BL/6 mice. (A to E) C57BL/6 mice were infected by intraperitoneal inoculation of 200 of the tachyzoites of PruΔku80Δhxgprt strain or the parental Pru strain. (A, B, and C) An example of a cyst observed at 5 weeks postinfection of C57BL/6 mice with strain PruΔku80Δhxgprt. (A) Bright-field laser confocal microscopy with light collected from a 1.8-μm-thick section. (B) GFP+ fluorescence from the same sample shown in panel A. (C) Merge of panels A and B. (D) Brain cyst burdens were measured in the parental Pru strain and the PruΔku80Δhxgprt strain at 3 weeks postinfection of C57BL/6 mice. (E) The average size of brain cysts was determined in the parental Pru strain and the PruΔku80Δhxgprt strain at 3 weeks postinfection of C57BL/6 mice as a relative measure of cyst diameter (BDU [see Materials and Methods]). (F) C57BL/6 mice were perorally infected with 10 cysts of the PruΔku80Δhxgprt strain. A survival curve of C57BL/6 mice (n = 6) infected perorally with 10 brain cysts of strain PruΔku80Δhxgprt (solid squares), or uninfected mice (n = 4) inoculated with PBS (solid circles).
Targeted deletions at the GRA4, GRA6, ROP4/7, and tgd057 loci.
To assess the interplay of host response and cyst development, targeted deletions were developed at the four gene loci known to encode CD8+ T cell epitopes that elicit corresponding antigen-specific CD8+ T cell populations during T. gondii infection. Targeting plasmids pΔGRA4P, pΔGRA6P, pΔROP4/7P, and pΔTGD057P were used to develop mutant strains with deletions of the GRA4, GRA6, ROP4/7, or tgd057 loci. Plasmids pΔGRA4PC and pΔGRA6PC were then used to target the removal of HXGPRT from the disrupted GRA4 and GRA6 loci in 6TX-negative selections. Finally, the GRA4 and GRA6 strains with HXGPRT deleted were used to develop several mutant strains with targeted deletion of two genetic loci (Δku80 Δgra6 Δgra4::HXGPRT, Δku80 Δgra4 Δtgd057::HXGPRT, and Δku80 Δgra6 Δtgd057::HXGPRT) (Table 1).
Determination of the replication rate of mutant strains in vitro.
None of the mutant strains examined exhibited any significant decrease in their intracellular growth rate in a 45 h growth assay (parasites per vacuole) compared to the control strain matched for the presence or absence of HXGPRT (Table 4). Two of the mutant strains, Δku80 Δtgd057::HXGPRT (P = 0.020) and Δku80 Δrop4/7::HXGPRT (P = 0.032), were found to exhibit a significant increase in the number of parasites per vacuole in the 45-h in vitro growth assay.
Measurement of brain tissue cyst burdens in mice infected with mutant strains.
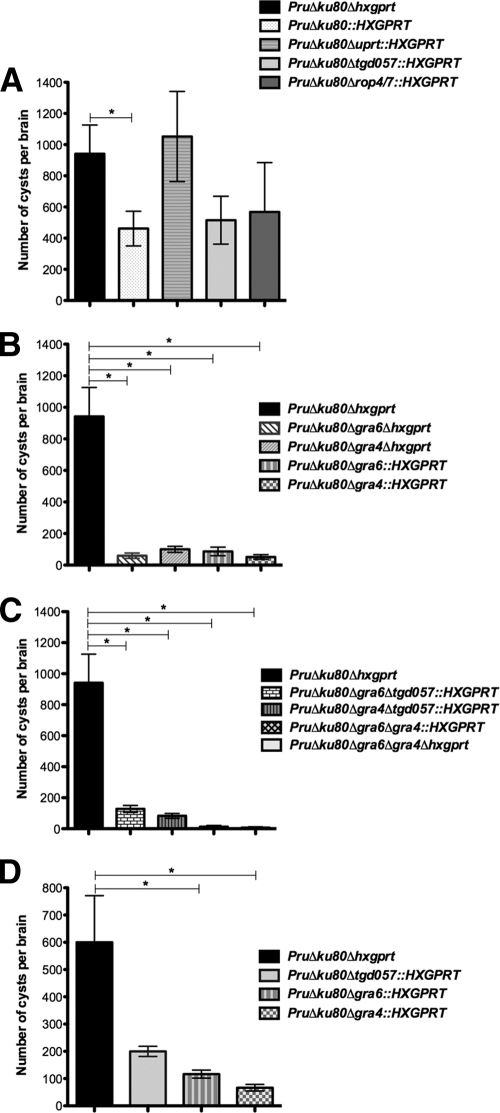
Cyst development was examined in the genetically susceptible C57BL/6 murine background that exhibits higher cyst burdens, reactivation, and chronic infection compared to the genetically resistant BALB/c murine background (30, 45, 56). Cyst burdens measured at 3 weeks postinfection of C57BL/6 mice were 462 ± 111 for the PruΔku80::HXGPRT strain and 942 ± 184 for the PruΔku80Δhxgprt strain (Fig. 4A; P = 0.0499). Nonetheless, all of the knockouts reported in the present study were developed in the PruΔku80Δhxgprt parent strain that is normal in regard to intracellular replication rate, cyst development, and cyst burdens (Fig. 3). The mutant strains developed in the PruΔku80Δhxgprt parent share the identical disrupted KU80 locus and differ only in regard to the presence of HXGPRT that is inserted at the gene locus targeted for deletion. Consequently, several gene knockouts developed in the PruΔku80Δhxgprt strain using HXGPRT selection were subsequently deleted of HXGPRT to allow more optimal assessment of phenotypes in comparison to the parental PruΔku80Δhxgprt strain.
Fig. 4.
Brain cyst burdens measured at 3 weeks postinfection. C57BL/6 or CBA mice were infected by intraperitoneal inoculation of 200 tachyzoites of mutant or parental strains. (A, B, and C) Brain cyst burdens in C57BL/6 mice. (A) Cyst burdens were measured at 3 weeks postinfection of C57BL/6 mice infected with the strain genotypes Δku80::HXGPRT (6 mice), Δku80 Δhxgprt (6 mice), Δku80 Δuprt::HXGPRT (5 mice), Δku80 Δtgd057::HXGPRT (6 mice), and Δku80 Δrop4/7::HXGPRT (6 mice). (B) Cyst burdens were measured at 3 weeks postinfection of C57BL/6 mice infected with the strain genotypes Δku80 Δhxgprt (6 mice), Δku80 Δgra4::HXGPRT (6 mice), Δku80 Δgra6::HXGPRT (6 mice), Δku80 Δgra4 Δhxgprt (4 mice), and Δku80 Δgra6 Δhxgprt (4 mice). (C) Cyst burdens were measured at 3 weeks postinfection of C57BL/6 mice infected with the strain genotypes Δku80 Δhxgprt (6 mice), Δku80 Δgra4 Δtgd057::HXGPRT (6 mice), Δku80 Δgra6 Δtgd057::HXGPRT (6 mice), Δku80 Δgra6 Δgra4::HXGPRT (6 mice), and Δku80 Δgra6 Δgra4 Δhxgprt (4 mice). (D) Brain cyst burdens in CBA mice. Cyst burdens were measured at 3 weeks postinfection of CBA mice infected with strain genotypes Δku80 Δhxgprt (6 mice), Δku80 Δgra4::HXGPRT (6 mice), Δku80 Δgra6::HXGPRT (6 mice), and Δku80 Δtgd057::HXGPRT (4 mice). Cyst data samples were subjected to a Student t test and are represented as the means ± the SEM. Cyst burdens determined in mutant strains were compared to cyst burdens in the parental Δku80 Δhxgprt strain to establish significance (P < 0.05).
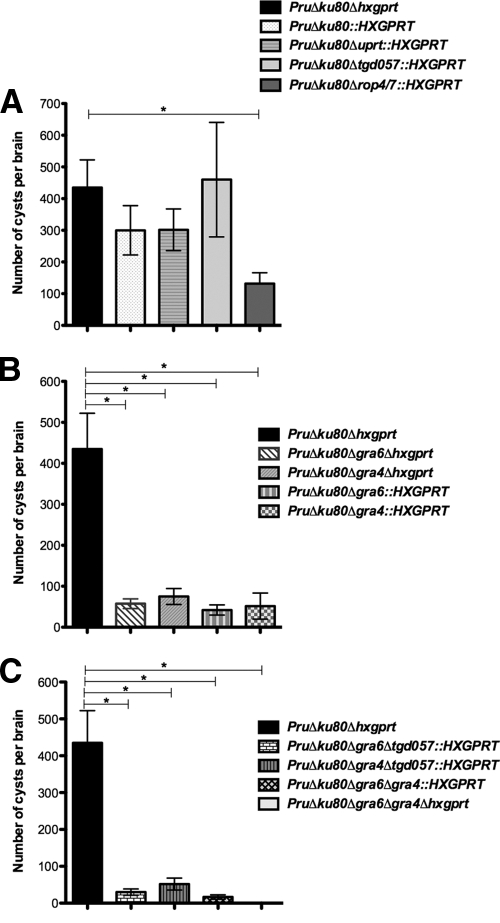
Several mutant strains, including PruΔku80Δup::hxgprt (data not shown), PruΔku80Δuprt::hxgprt (P = 0.75), PruΔku80Δrop4/7::hxgprt (P = 0.33), and PruΔku80Δtgd057::hxgprt (P = 0.11), exhibited no significant difference in cyst burdens compared to the parental PruΔku80Δhxgprt strain at 3 weeks postinfection (Fig. 4A). In contrast, at 3 weeks postinfection of C57BL/6 mice, a 91% reduction (P = 0.0010) in cyst burden was observed in the Δgra6 mutant with HXGPRT (Δku80 Δgra6::HXGPRT) and a 91% reduction (P = 0.0010) in cyst burden was observed in the Δgra4 mutant with HXGPRT (Δku80 Δgra4::HXGPRT) (Fig. 4B). Similarly, a 93% reduction (P = 0.0051) in cyst burden was observed in the Δgra6 mutant with HXGPRT deleted (Δku80 Δgra6 Δhxgprt) and an 89% reduction (P = 0.0066) in cyst burden was observed in the Δgra4 mutant with HXGPRT deleted (Δku80 Δgra4 Δhxgprt) (Fig. 4B). Double mutants involving the GRA4 and/or the GRA6 loci were also examined for cyst development in C57BL/6 mice. Although the PruΔku80Δgra4Δtgd057::HXGPRT (91% reduction; P = 0.0009) and PruΔku80Δgra6Δtgd057::HXGPRT (86%; P = 0.0032) mutant strains revealed similar reductions in cyst burden (Fig. 4C) compared to the Δgra4 and Δgra6 single mutants with or without the HXGPRT marker (Fig. 4B), the double-mutant strain PruΔku80Δgra6Δgra4::HXGPRT revealed a markedly more severe defect in cyst burdens at 3 weeks postinfection (99% reduction; P = 0.0005) (Fig. 4C). This more significant defect in cyst burden in the Δku80 Δgra6 Δgra4::hxgprt double mutant was then verified by construction of the Δku80 Δgra6 Δgra4 Δhxgprt strain that exhibited a 99.8% reduction (P = 0.0037) in cyst burden (Fig. 4C). Collectively, these results demonstrate a significant defect in the development of brain cyst burdens in mutant strains with GRA4 and/or GRA6 deleted in C57BL/6 mice.
We also measured cyst burdens in the CBA murine (H-2Kk) background because the four currently characterized CD8+ T cell epitopes encoded by the GRA4, GRA6, ROP7, and tgd057 genes are not recognized by the H-2Kk-restricted CBA background. In the CBA murine background a 77% reduction (P = 0.018) in cyst burden was observed in the Δgra6 mutant and an 87% reduction (P = 0.011) in cyst burden was observed in the Δgra4 mutant at 3 weeks postinfection (Fig. 4D). In contrast, no significant difference in cyst burden was observed in the Δtgd057 mutant (P = 0.099).
In addition, the cyst burdens for many of the mutant strains were also measured at 5 weeks postinfection of C57BL/6 mice. The PruΔku80::HXGPRT (P = 0.28), PruΔku80Δuprt::HXGPRT (P = 0.25), and PruΔku80Δtgd057::HXGPRT (P = 0.90) strains exhibited no significant difference in cyst burdens compared to the PruΔku80Δhxgprt strain at 5 weeks postinfection (Fig. 5A). In contrast, the PruΔku80Δrop4/7::HXGPRT mutant exhibited a 70% reduction in cyst burden at 5 weeks postinfection (P = 0.0089) (Fig. 5A). Significant reductions in cyst burdens compared to the parental Δku80 Δhxgprt strain were also observed in each of the Δgra4 and Δgra6 mutant strains examined at 5 weeks postinfection (PruΔku80Δgra4::HXGPRT [P = 0.0020], PruΔku80Δgra6::HXGPRT [P = 0.0029], PruΔku80Δgra4Δhxgprt [P = 0.011], PruΔku80Δgra6Δhxgprt [P = 0.0087], PruΔku80Δgra4Δtgd057::HXGPRT [P = 0.0015], PruΔku80Δgra6Δtgd057::HXGPRT [P = 0.0010], PruΔku80Δgra6Δgra4::HXGPRT [P = 0.0007], PruΔku80Δgra6Δgra4Δhxgprt [P = 0.0040]) (Fig. 5B and C).
Fig. 5.
Brain cyst burdens measured at 5 weeks postinfection. C57BL/6 mice were infected by intraperitoneal inoculation of 200 tachyzoites of mutant or parental strains. (A, B, and C) Brain cyst burdens in C57BL/6 mice. (A) Cyst burdens were measured at 5 weeks postinfection of C57BL/6 mice infected with the strain genotypes Δku80::HXGPRT (6 mice), Δku80 Δhxgprt (6 mice), Δku80 Δuprt::HXGPRT (6 mice), Δku80 Δtgd057::HXGPRT (6 mice), and Δku80 Δrop4/7::HXGPRT (6 mice). (B) Cyst burdens were measured at 5 weeks postinfection of C57BL/6 mice infected with the strain genotypes Δku80 Δhxgprt (6 mice), Δku80 Δgra4::HXGPRT (6 mice), Δku80 Δgra6::HXGPRT (6 mice), Δku80 Δgra4 Δhxgprt (4 mice), and Δku80 Δgra6 Δhxgprt (4 mice). (C) Cyst burdens were measured at 5 weeks postinfection of C57BL/6 mice infected with the strain genotypes Δku80 Δhxgprt (6 mice), Δku80 Δgra4 Δtgd057::HXGPRT (6 mice), Δku80 Δgra6 Δtgd057::HXGPRT (6 mice), Δku80 Δgra6 Δgra4::HXGPRT (6 mice), and Δku80 Δgra6 Δgra4 Δhxgprt (4 mice). Cyst data samples were subjected to a Student t test and are represented as means ± the SEM. Cyst burdens determined in mutant strains were compared to cyst burdens in the parental Δku80 Δhxgprt strain to establish significance (P < 0.05).
Complementation of the Δgra4 and Δgra6 mutants.
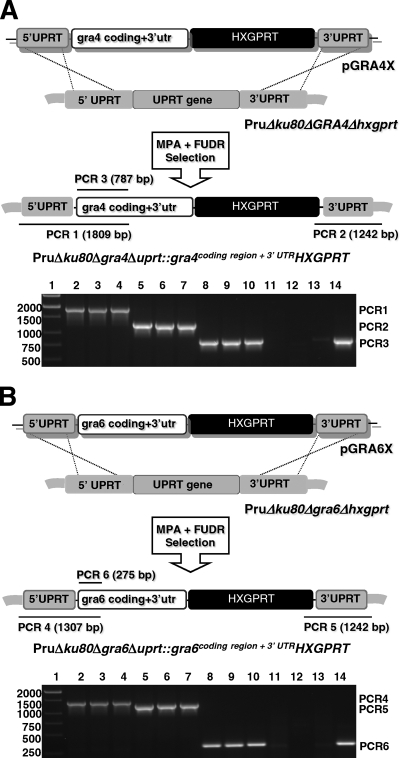
To more clearly establish that the defect in cyst burdens observed in the Δgra4 and Δgra6 mutant strains was due to the deletion of GRA protein function, we complemented the Δgra4 and Δgra6 mutants by reinserting a functional allele of the deleted GRA gene. Rather than simply recreating the parental strain with an intact GRA locus, we complemented the Δku80 Δgra4 Δhxgprt and the Δku80 Δgra6 Δhxgprt mutants by targeting the insertion of the GRA coding region and the HXGPRT marker into the UPRT locus, deleting the coding region of the UPRT gene (Fig. 6A and B). This strategy was based on our observation of normal cyst burdens in the Δku80 Δuprt::hxgprt mutant (Fig. 4A). In addition, by design we deleted the extremely strong GRA4 and GRA6 transcriptional promoters and their 5′ transcribed, untranslated region (5′ GRA UTRs), and via homologous recombination placed the GRA protein coding region(s) under the control of the endogenous UPRT locus 5′UTR which ranks at the ∼ 78% for expression level percentile (microarray data [www.toxodb.org]) (Fig. 6). In contrast, the relative expression level percentile ranking is ∼ 94.2% for the endogenous GRA4 locus and ∼99.2% for the endogenous GRA6 locus. Parasites transfected with the pGRA4X and pGRA6X complementation cassettes were selected in MPA for 10 days then the population was selected in FUDR. Complemented clones were examined using a PCR strategy (Fig. 6) and correctly targeted clones with the genotypes Δku80 Δgra4 Δuprt::gra4coding region + 3′UTR HXGPRT and Δku80 Δgra6 Δuprt::gra6coding region + 3′UTR HXGPRT were validated (Fig. 6).
Fig. 6.
Genetic strategy for complementation of the Δgra4 and Δgra6 deletion mutants. The coding region of the deleted GRA gene and the associated GRA 3′UTR was placed under regulatory control of the endogenous UPRT locus 5′UTR. (A) Plasmid pGRA4X was transfected into the PruΔku80Δgra4Δhxgprt strain, and parasites were selected in MPA and then selected in FUDR to select for the Δuprt phenotype. Clones isolated from this selection were examined using PCR 1, PCR 2, and PCR 3 to verify precise integration of the GRA4 coding region at the deleted uprt locus. PCR 1 measures correct 5′ integration, PCR 2 measures correct 3′ integration, and PCR 3 measures the presence of the GRA4 coding region. (Agarose gel panel) PCR 1, PCR 2, and PCR 3 products: DNA size ladder (lane 1), PCR 1 from 3 complemented clones (lanes 2 to 4), PCR 2 from 3 complemented clones (lanes 5 to 7), PCR 3 from 3 complemented clones (lanes 8 to 10), PCR 1 from parental PruΔku80Δgra4Δhxgprt (lane 11), PCR 2 from parental PruΔku80Δgra4Δhxgprt (lane 12), PCR 3 from parental PruΔku80Δgra4Δhxgprt (lane 13), PCR 3 from PruΔku80Δhxgprt (lane 14). (B) Plasmid pGRA6X was transfected into the PruΔku80Δgra6Δhxgprt strain, and parasites were selected in MPA and then selected in FUDR to select for the Δuprt phenotype. Clones isolated from this selection were examined using PCR 4, PCR 5, and PCR 6 to verify precise integration of the GRA6 coding region at the deleted uprt locus. PCR 4 measures correct 5′ integration, PCR 5 measures correct 3′ integration, and PCR 6 measures the presence of the GRA4 coding region. (Agarose gel panel) PCR 4, PCR 5, and PCR 6 products: DNA size ladder (lane 1), PCR 4 from three complemented clones (lanes 2 to 4), PCR 5 from three complemented clones (lanes 5 to 7), PCR 6 from three complemented clones (lanes 8 to 10), PCR 4 from parental PruΔku80Δgra6Δhxgprt (lane 11), PCR 5 from parental PruΔku80Δgra6Δhxgprt (lane 12), PCR 6 from parental PruΔku80Δgra6Δhxgprt (lane 13), PCR 6 from PruΔku80Δhxgprt (lane 14).
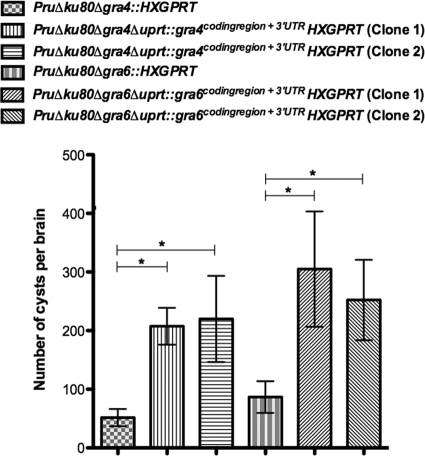
Two independent cloned isolates of each complemented strain were examined for cyst burden in C57BL/6 mice at 3 weeks postinfection. GRA4 complemented clones exhibited a 402 to 426% increase in cyst burdens compared to the Δgra4 mutant strain also lacking HXGPRT (GRA4, P = 0.0010 and P = 0.024) (Fig. 7). GRA6 complemented clones exhibited a 291 to 352% increase in cyst burdens compared to the Δgra6 mutant strain also lacking HXGPRT (GRA6, P = 0.032 and P = 0.031) (Fig. 7).
Fig. 7.
Cyst burdens are significantly increased in the GRA-complemented strains. C57BL/6 mice were infected by intraperitoneal inoculation of 200 tachyzoites of mutant parental or complemented strains. Cyst burdens were measured at 3 weeks postinfection of C57BL/6 mice infected with the strains PruΔku80Δgra4::HXGPRT (6 mice), PruΔku80Δgra4Δuprt::gra4coding region + 3′UTRHXGPRT (clone 1, 4 mice, and clone 2, 4 mice), PruΔku80Δgra6::HXGPRT (6 mice), PruΔku80Δgra6Δuprt::gra6coding region + 3′UTRHXGPRT (clone 1, 4 mice, and clone 2, 4 mice). Cyst data samples were subjected to a Student t test and are represented as means ± the SEM. Cyst burdens determined in complemented strains were compared to cyst burdens in the parental Δgra strain to establish significance (P < 0.05).
DISCUSSION
Type II isolates of T. gondii represent a prevalent strain type infecting humans and causing life-long infections characterized by the presence of the bradyzoite stage encysted tissue cyst. Previously, targeted genetic dissection of type II T. gondii strains was limited by inefficient homologous recombination pathways relative to highly efficient pathways for random integration of targeting episomes. By eliminating the dominant pathway of nonhomologous recombination via knockout and functional disruption of the KU80 gene (26), we now report a new genetic model that enables efficient gene replacements and the reliable development of targeted gene deletions in type II T. gondii.
The PruΔku80Δhxgprt strain exhibited efficient gene targeting at the UPRT locus, at the OPRT locus, and at all other targeted loci. Efficient gene targeting was demonstrated to be dependent on perfect DNA sequence homology. We successfully targeted seven genetic loci for deletion (UPRT, UP, OPRT, GRA4, GRA6, ROP4/7, and tgd057), we retargeted the deletion of HXGPRT inserted at three disrupted loci (OPRT, GRA4, GRA6), we constructed three mutant strains with two targeted gene deletions (Δgra4 Δgra6, Δgra4 Δtgd057, and Δgra6 Δtgd057), and we functionally complemented the GRA4 and GRA6 knockouts. These results show that the type II Δku80 Δhxgprt genetic background is amenable for rapid and reliable development of targeted gene deletions, and other more complex genetic manipulations involving sequential targeting, removal of HXGPRT at the deleted locus, and retargeting using HXGPRT. The ability of the PruΔku80Δhxgprt strain to develop cyst burdens and latent infection was maintained after long-term in vitro culture or multiple genetic manipulations. In addition, the PruΔku80Δhxgprt strain retained a normal intracellular replication rate compared to the parental Pru strain, and this strain also retains normal acute virulence in mice (data not shown). The type I KU80 knockout strain RHΔku80Δhxgprt retains a normal intracellular replication rate, acute virulence in mice (26), and this strain also exhibits an identical profile of gene expression during the cell cycle as parental RH (3). Collectively, these results support the PruΔku80Δhxgprt genetic background as an improved genetic model for dissecting type II T. gondii biology.
Type II mutants deleted in pyrimidine salvage activities for UPRT and UP were not defective in cyst development, demonstrating that pyrimidine salvage through the UPRT or UP enzyme activities is not required for cyst development or the establishment of latent infection. These results suggest that de novo pyrimidine synthesis is functional and essential during bradyzoite stage conversion, cyst development, and latent infection.
We examined the hypothesis that host CD8+ T cell responses directed against specific parasite antigens containing CD8+ T cell epitopes (GRA4, GRA6, ROP7, and tgd057) (5, 27, 42, 68) may influence cyst development. Our analysis was hindered by the low cyst burdens (<60 cysts per brain) that we observed in the H-2Ld-restricted BALB/c background. The H-2Ld-restricted BALB/c background is genetically resistant to infection and establishes a latent infection with reduced cyst burdens compared to genetically susceptible murine backgrounds such as C57BL/6, which exhibit higher cyst burdens, reactivation, and chronic infection (30, 45, 56). Recent observations suggest that infection of genetically susceptible C57BL/6 mice with the Pru strain produces the highest brain tissue cyst burden at 3 weeks postinfection (56), then cysts rupture (reactivate) and are lost at a higher rate than any potential rate of cyst reformation during chronic infection. Immune control of type II T. gondii infection has been proposed to be dependent on type II strain-specific H-2Ld-restricted cytotoxic T cells (35). This hypothesis is supported by the identification of the immunodominant and protective decapeptide HF10 epitope within the type II GRA6 protein that is presented in the context of the H-2Ld major histocompatibility complex class I molecule (5).
The previously identified GRA4, GRA6, and ROP7 CD8+ T cell epitopes are H-2Ld-restricted (5, 27), whereas the tdg057 CD8+ T cell epitope is H-2Kb restricted. Consequently, in C57BL/6 mice a CD8+ T cell response is mounted against the known tdg057 epitope (68), but not against the known GRA4, GRA6, or ROP7 epitopes. At 3 weeks postinfection of C57BL/6 mice, we observed no significant differences in cyst burdens in either the Δrop4/7 or the Δtgd057 mutants compared to the Δku80 Δhxgprt parent. The absence of CD8+ T cell response to the tgd057 epitope, as well as the loss of tgd057 protein function in the Δtgd057 mutant, did not significantly affect cyst burdens. Similarly, the absence of CD8+ T cell response to ROP7 in both the parental strain and the Δrop4/7 mutant, along with the loss of ROP4/7 protein functions, did not significantly affect cyst burdens at 3 weeks postinfection. In contrast, while the CD8+ T cell response to the GRA4 and GRA6 epitopes is absent in the parental strain in the C57BL/6 murine background, the deletion of the GRA4 (Δgra4) and GRA6 (Δgra6) genes markedly reduced cyst burdens independently of any potential CD8+ T cell response to the known epitopes within these GRA proteins. However, we cannot conclusively rule out the possibility that the GRA4 and/or GRA6 genes encode as-yet-uncharacterized H-2Kb-restricted epitopes. We also observed a significant defect in cyst burdens in the Δgra4 and Δgra6 mutants in the CBA murine background (H-2Kk-restricted). These results suggest that the GRA4 and GRA6 proteins directly mediate functions necessary for the development of normal cyst burdens, rather than cyst development being strictly dependent on the host response to these proteins.
Previous studies have also reported defects in cyst development following gene disruption in type II strains. Disruption of the SRS9 gene or the SAG2CDXY gene locus revealed a phenotype of normal cyst burdens at 4 weeks postinfection, and then reduced cyst burdens were observed later during latent infection (41, 55). Disruption of a pseudouridine synthase homologue (PUS1) revealed a phenotype of slightly smaller cyst size and slightly increased cyst burdens (1). Targeted disruption of the heat shock protein BAG1 gene (6, 69) resulted in a minor defect in cyst development. A significant defect in cyst burden was observed at 5 weeks postinfection in the Δrop4/7 mutant. Interestingly, the Δrop4/7 mutant and the Δtgd057 mutant exhibited an increase in their in vitro growth rate. The recently reported type II GRA15 knockout was also reported to exhibit an increased growth rate in vitro (54).
Cross-linking studies suggest that a multimeric protein complex of GRA2, GRA4, and GRA6 exists within the nanotubular network of membranes within the parasitophorous vacuole space (43). However, a recent study of protein interactions using a more comprehensive panel of HA-FLAG-tagged GRA proteins did not reveal a direct interaction of GRA2, GRA4, and GRA6 in the membrane fraction (8). Disruption of GRA6 in type I tachyzoites revealed an altered intravacuolar network of membranes characterized by small vesicles instead of elongated nanotubules (47). The N terminus of GRA6 was also recently shown to be a critical domain in interacting with negatively charged lipids and is necessary for association of the protein with the vacuolar membranous network of nanotubules (28).
Type II mutants with GRA4 or GRA6 deleted exhibited drastically reduced cyst burdens during T. gondii infection. The defect in cyst development in these strains was independent of the presence or absence of HXGPRT. Thus, these GRA proteins appear to be involved in some aspect of cyst development that significantly reduces the likelihood of cyst formation rather than completely abrogating the parasites biological ability to convert to the bradyzoite stage and create a tissue cyst. Complementation of the Δgra4 and Δgra6 mutants using a functional allele of the corresponding GRA protein significantly restored cyst burdens. However, cyst burdens were not completely restored to the expected levels when GRA4 and GRA6 expression was placed under regulatory control of the UPRT locus. The relative strengths of the GRA4 and GRA6 promoters (GRA4, ∼94.2%; GRA6, ∼99.2% [www.toxodb.org]) are significantly higher than that of the UPRT locus (∼78%) used in the complementation study. These observations suggest that an extremely high level of expression of GRA4 and GRA6 may be necessary for the development of normal cyst burdens.
A more severe defect in cyst burden in the Δku80Δgra6Δgra4::HXGPRT and the Δku80 Δgra6 Δgra4 Δhxgprt mutants compared to mutants with single deletions of GRA4 or GRA6 suggests that GRA4 and GRA6 most likely play independent functional roles required for cyst development during infection. Ultrastructural localization is also consistent with the likelihood of independent roles for GRA4 and GRA6 in cyst development since, while GRA4 is abundant in the tachyzoite stage, PV it is not detectable in the bradyzoite stage cyst wall, whereas GRA6 is easily detected in both stages (21). Since the cyst wall structure is thought to arise directly from modifications to the PVM during tachyzoite to bradyzoite conversion (59, 67), the function of GRA4 in cyst development is likely to be exerted prior to or during formation of the cyst wall.
Our study did not specifically address the mechanisms by which GRA4 or GRA6 influence cyst development. With the exception of uracil auxotrophy of the Δoprt mutants, no significant decreases were observed in the in vitro replication rate of any of the mutants isolated in the present study. The defect in cyst burden in the Δgra4 and Δgra6 mutants may be due to a more successful innate or adaptive host response that suppresses parasite burden or dissemination necessary for development of brain tissue cysts. Alternatively, GRA4 and GRA6 may play a direct role in parasite biology associated with the probability of successful tachyzoite to bradyzoite conversion or cyst development. Additional studies are therefore necessary to assess acute virulence, parasite tissue burdens, dissemination patterns, and host response during infection to further gauge potential mechanisms underlying the defect in cyst development in the Δgra4 and Δgra6 mutants.
The GRA protein family is extremely interesting because they are a remarkable family of highly expressed and compartmentalized proteins restricted thus far to the cyst-forming parasites Toxoplasma and Neospora (49). No identifiable ortholog can be discerned even in closely related Conoidasida such as Cryptosporidium or Eimeria or Aconoidasida such as Plasmodium, which do not form tissue cysts (10). Our results suggest that in addition to GRA4 and GRA6 other members of the prominent GRA protein family may also play critical roles in biology underlying successful cyst development and latent infection.
The type II Δku80 Δhxgprt strain developed in the present study provides a reliable genetic model for targeted genetic dissection of parasite biology occurring during T. gondii infection. The ability to now control both parasite genes and host cell genes can reveal the complex interplay of parasite biology and host response during T. gondii infection. These genetic approaches will accelerate the development of improved strategies for vaccines, immunological interventions, and other therapeutics. Targeted genetic approaches using a type II Δku80 Δhxgprt genetic background has the immediate potential to functionally reveal parasite genes playing critical biological roles that, when intercepted during in vivo infection, block parasite development, pathogenesis, or transmission.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to numerous Toxoplasma research laboratories that provided helpful advice and suggestions during the course of this study. This project was made possible by the work of the developers of the Toxoplasma gondii Genome Resource (at www.ToxoDB.org), and their work is gratefully acknowledged. ToxoDB, PlasmoDB, and EuPathDB are part of the National Institutes of Health/National Institute of Allergy and Infectious Disease (NIH/NIAID)-funded Bioinformatics Resource Center. Type II strains were kindly provided by John Boothroyd (Stanford University), Jeroen Saeij (Massachusetts Institute of Technology), and Elmer Pfefferkorn (Dartmouth College). This study was supported by the use of facilities that were provided by the Imaging Shared Resource of the Dartmouth-Hitchcock Norris Cotton Cancer Center.
L.M.R. was a predoctoral trainee on NIH training grant T32AI007519 on Host-Microbe Interactions. This study was supported by the NIH (grant AI039454 to L.M.W. and grants AI41930, AI073142, AI075931, and AI091461 to D.J.B.).
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 29 April 2011.
REFERENCES
- 1. Anderson M. Z., Brewer J., Singh U., Boothroyd J. C. 2009. A pseudouridine synthase homologue is critical to cellular differentiation in Toxoplasma gondii. Eukaryot. Cell 8:398–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beck J. R., et al. 2010. A novel family of Toxoplasma IMC proteins displays a hierarchical organization and functions in coordinating parasite division. PLoS Pathog. 6:e1001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Behnke M. S., et al. 2010. Coordinated progression through two subtranscriptomes underlies the tachyzoite cycle of Toxoplasma gondii. PLoS One 5:e12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bierly A. L., Shufesky W. J., Sukhumavasi W., Morelli A. E., Denkers E. Y. 2008. Dendritic cells expressing plasmacytoid marker PDCA-1 are Trojan horses during Toxoplasma gondii infection. J. Immunol. 181:8485–8491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blanchard N., et al. 2008. Immunodominant, protective response to the parasite Toxoplasma gondii requires antigen processing in the endoplasmic reticulum. Nat. Immunol. 9:937–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bohne W., et al. 1998. Targeted disruption of the bradyzoite-specific gene BAG1 does not prevent tissue cyst formation in Toxoplasma gondii. Mol. Biochem. Parasitol. 92:291–301 [DOI] [PubMed] [Google Scholar]
- 7. Boothroyd J. C., Dubremetz J. F. 2008. Kiss and spit: the dual roles of Toxoplasma rhoptries. Nat. Rev. Microbiol. 6:79–88 [DOI] [PubMed] [Google Scholar]
- 8. Braun L., et al. 2008. Purification of Toxoplasma dense granule proteins reveals that they are in complexes throughout the secretory pathway. Mol. Biochem. Parasitol. 157:13–21 [DOI] [PubMed] [Google Scholar]
- 9. Carruthers V. B., Sibley L. D. 1997. Sequential protein secretion from three distinct organelles of Toxoplasma gondii accompanies invasion of human fibroblasts. Eur. J. Cell Biol. 73:114–123 [PubMed] [Google Scholar]
- 10. Cesbron-Delauw M. F., Gendrin C., Travier L., Ruffiot P., Mercier C. 2008. Apicomplexa in mammalian cells: trafficking to the parasitophorous vacuole. Traffic 9:657–664 [DOI] [PubMed] [Google Scholar]
- 11. Chtanova T., et al. 2008. Dynamics of neutrophil migration in lymph nodes during infection. Immunity 29:487–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Contini C. 2008. Clinical and diagnostic management of toxoplasmosis in the immunocompromised patient. Parasitologia 50:45–50 [PubMed] [Google Scholar]
- 13. Craver M. P., Knoll L. J. 2007. Increased efficiency of homologous recombination in Toxoplasma gondii dense granule protein 3 demonstrates that GRA3 is not necessary in cell culture but does contribute to virulence. Mol. Biochem. Parasitol. 153:149–157 [DOI] [PubMed] [Google Scholar]
- 14. Denkers E. Y., Butcher B. A. 2005. Sabotage and exploitation in macrophages parasitized by intracellular protozoans. Trends Parasitol. 21:35–41 [DOI] [PubMed] [Google Scholar]
- 15. Dixon S. E., Bhatti M. M., Uversky V. N., Dunker A. K., Sullivan W. J., Jr 2011. Regions of intrinsic disorder help identify a novel nuclear localization signal in Toxoplasma gondii histone acetyltransferase TgGCN5-B. Mol. Biochem. Parasitol. 175:192–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Donald R. G., Carter D., Ullman B., Roos D. S. 1996. Insertional tagging, cloning, and expression of the Toxoplasma gondii hypoxanthine-xanthine-guanine phosphoribosyltransferase gene. Use as a selectable marker for stable transformation. J. Biol. Chem. 271:14010–14019 [DOI] [PubMed] [Google Scholar]
- 17. Donald R. G., Roos D. S. 1995. Insertional mutagenesis and marker rescue in a protozoan parasite: cloning of the uracil phosphoribosyltransferase locus from Toxoplasma gondii. Proc. Natl. Acad. Sci. U. S. A. 92:5749–5753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Donald R. G., Roos D. S. 1993. Stable molecular transformation of Toxoplasma gondii: a selectable dihydrofolate reductase-thymidylate synthase marker based on drug-resistance mutations in malaria. Proc. Natl. Acad. Sci. U. S. A. 90:11703–11707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dubey J. P. 2007. The history and life cycle of Toxoplasma gondii, p. 1–18 In Weiss L. M., et al. (ed.), Toxoplasma gondii: the model apicomplexan parasite: perspectives and methods. Elsevier, London, United Kingdom [Google Scholar]
- 20. Fentress S. J., et al. 2010. Phosphorylation of immunity-related GTPases by a Toxoplasma gondii-secreted kinase promotes macrophage survival and virulence. Cell Host Microbe 8:484–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ferguson D. J. 2004. Use of molecular and ultrastructural markers to evaluate stage conversion of Toxoplasma gondii in both the intermediate and definitive host. Int. J. Parasitol. 34:347–360 [DOI] [PubMed] [Google Scholar]
- 22. Fox B. A., Belperron A. A., Bzik D. J. 1999. Stable transformation of Toxoplasma gondii based on a pyrimethamine-resistant trifunctional dihydrofolate reductase-cytosine deaminase-thymidylate synthase gene that confers sensitivity to 5-fluorocytosine. Mol. Biochem. Parasitol. 98:93–103 [DOI] [PubMed] [Google Scholar]
- 23. Fox B. A., Bzik D. J. 2010. Avirulent uracil auxotrophs based on disruption of orotidine-5′-monophosphate decarboxylase elicit protective immunity to Toxoplasma gondii. Infect. Immun. 78:3744–3752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fox B. A., Bzik D. J. 2002. De novo pyrimidine biosynthesis is required for virulence of Toxoplasma gondii. Nature 415:926–929 [DOI] [PubMed] [Google Scholar]
- 25. Fox B. A., Ristuccia J. G., Bzik D. J. 2009. Genetic identification of essential indels and domains in carbamoyl phosphate synthetase II of Toxoplasma gondii. Int. J. Parasitol. 39:533–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fox B. A., Ristuccia J. G., Gigley J. P., Bzik D. J. 2009. Efficient gene replacements in Toxoplasma gondii strains deficient for nonhomologous end joining. Eukaryot. Cell 8:520–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Frickel E. M., et al. 2008. Parasite stage-specific recognition of endogenous Toxoplasma gondii-derived CD8+ T cell epitopes. J. Infect. Dis. 198:1625–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gendrin C., Bittame A., Mercier C., Cesbron-Delauw M. F. 2010. Post-translational membrane sorting of the Toxoplasma gondii GRA6 protein into the parasite-containing vacuole is driven by its N-terminal domain. Int. J. Parasitol. 40:1325–1334 [DOI] [PubMed] [Google Scholar]
- 29. Gigley J. P., Fox B. A., Bzik D. J. 2009. Cell-mediated immunity to Toxoplasma gondii develops primarily by local Th1 host immune responses in the absence of parasite replication. J. Immunol. 182:1069–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gigley J. P., Fox B. A., Bzik D. J. 2009. Long-term immunity to lethal acute or chronic type II Toxoplasma gondii infection is effectively induced in genetically susceptible C57BL/6 mice by immunization with an attenuated type I vaccine strain. Infect. Immun. 77:5380–5388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gubbels M. J., Striepen B., Shastri N., Turkoz M., Robey E. A. 2005. Class I major histocompatibility complex presentation of antigens that escape from the parasitophorous vacuole of Toxoplasma gondii. Infect. Immun. 73:703–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Holpert M., Gross U., Bohne W. 2006. Disruption of the bradyzoite-specific P-type (H+)-ATPase PMA1 in Toxoplasma gondii leads to decreased bradyzoite differentiation after stress stimuli but does not interfere with mature tissue cyst formation. Mol. Biochem. Parasitol. 146:129–133 [DOI] [PubMed] [Google Scholar]
- 33. Howe D. K., Honore S., Derouin F., Sibley L. D. 1997. Determination of genotypes of Toxoplasma gondii strains isolated from patients with toxoplasmosis. J. Clin. Microbiol. 35:1411–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huynh M. H., Carruthers V. B. 2009. Tagging of endogenous genes in a Toxoplasma gondii strain lacking Ku80. Eukaryot. Cell 8:530–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Johnson J. J., et al. 2002. In vitro correlates of Ld-restricted resistance to toxoplasmic encephalitis and their critical dependence on parasite strain. J. Immunol. 169:966–973 [DOI] [PubMed] [Google Scholar]
- 36. Jones J. L., Holland G. N. 2010. Annu. burden of ocular toxoplasmosis in the US. Am. J. Trop. Med. Hyg. 82:464–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Joyce B. R., Queener S. F., Wek R. C., Sullivan W. J., Jr 2010. Phosphorylation of eukaryotic initiation factor-2α promotes the extracellular survival of obligate intracellular parasite Toxoplasma gondii. Proc. Natl. Acad. Sci. U. S. A. 107:17200–17205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Karasov A. O., Boothroyd J. C., Arrizabalaga G. 2005. Identification and disruption of a rhoptry-localized homologue of sodium hydrogen exchangers in Toxoplasma gondii. Int. J. Parasitol. 35:285–291 [DOI] [PubMed] [Google Scholar]
- 39. Kim K., Soldati D., Boothroyd J. C. 1993. Gene replacement in Toxoplasma gondii with chloramphenicol acetyltransferase as selectable marker. Science 262:911–914 [DOI] [PubMed] [Google Scholar]
- 40. Kim K., Weiss L. M. 2008. Toxoplasma: the next 100 years. Microbes Infect. 10:978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kim S. K., Karasov A., Boothroyd J. C. 2007. Bradyzoite-specific surface antigen SRS9 plays a role in maintaining Toxoplasma gondii persistence in the brain and in host control of parasite replication in the intestine. Infect. Immun. 75:1626–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kirak O., et al. 2010. Transnuclear mice with predefined T cell receptor specificities against Toxoplasma gondii obtained via SCNT. Science 328:243–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Labruyere E., Lingnau M., Mercier C., Sibley L. D. 1999. Differential membrane targeting of the secretory proteins GRA4 and GRA6 within the parasitophorous vacuole formed by Toxoplasma gondii. Mol. Biochem. Parasitol. 102:311–324 [DOI] [PubMed] [Google Scholar]
- 44. Larson E. T., et al. 2009. Toxoplasma gondii cathepsin L is the primary target of the invasion-inhibitory compound morpholinurea-leucyl-homophenyl-vinyl sulfone phenyl. J. Biol. Chem. 284:26839–26850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. McLeod R., Skamene E., Brown C. R., Eisenhauer P. B., Mack D. G. 1989. Genetic regulation of early survival and cyst number after peroral Toxoplasma gondii infection of A×B/B×A recombinant inbred and B10 congenic mice. J. Immunol. 143:3031–3034 [PubMed] [Google Scholar]
- 46. Mercier C., Cesbron-Delauw M. F., Ferguson D. J. P. 2007. Dense granules of the infectious stages of Toxoplasma gondii: their central role in the host/parasite relationship, p. 475–492 In Ajioka D. S., et al. (ed.), Toxoplasma: molecular and cellular biology. Horizon Bioscience, Norfolk, United Kingdom [Google Scholar]
- 47. Mercier C., et al. 2002. Biogenesis of nanotubular network in Toxoplasma parasitophorous vacuole induced by parasite proteins. Mol. Biol. Cell 13:2397–2409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mercier C., Howe D. K., Mordue D., Lingnau M., Sibley L. D. 1998. Targeted disruption of the GRA2 locus in Toxoplasma gondii decreases acute virulence in mice. Infect. Immun. 66:4176–4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mercier C., Travier L., Bittame A., Gendrin C., Cesbraun-Delauw M. F. 2010. The dense granule proteins of Toxoplasma gondii, p. 1–31 In De Bruyn O., Peeters S. (ed.), Parasitology research trends. Nova Science Publishers, Hauppage, NY [Google Scholar]
- 50. Montoya J. G., Remington J. S. 2008. Management of Toxoplasma gondii infection during pregnancy. Clin. Infect. Dis. 47:554–566 [DOI] [PubMed] [Google Scholar]
- 51. Odaert H., Soete M., Fortier B., Camus D., Dubremetz J. F. 1996. Stage conversion of Toxoplasma gondii in mouse brain during infection and immunodepression. Parasitol. Res. 82:28–31 [DOI] [PubMed] [Google Scholar]
- 52. Robben P. M., Sibley L. D. 2004. Food- and waterborne pathogens: you are (infected by) what you eat! Microbes Infect. 6:406–413 [DOI] [PubMed] [Google Scholar]
- 53. Roos D. S. 2005. Genetics: themes and variations in apicomplexan parasite biology. Science 309:72–73 [DOI] [PubMed] [Google Scholar]
- 54. Rosowski E. E., et al. 2011. Strain-specific activation of the NF-κB pathway by GRA15, a novel Toxoplasma gondii dense granule protein. J. Exp. Med. 208:195–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Saeij J. P., Arrizabalaga G., Boothroyd J. C. 2008. A cluster of four surface antigen genes specifically expressed in bradyzoites, SAG2CDXY, plays an important role in Toxoplasma gondii persistence. Infect. Immun. 76:2402–2410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Schaeffer M., et al. 2009. Dynamic imaging of T cell-parasite interactions in the brains of mice chronically infected with Toxoplasma gondii. J. Immunol. 182:6379–6393 [DOI] [PubMed] [Google Scholar]
- 57. Schwarz J. A., Fouts A. E., Cummings C. A., Ferguson D. J., Boothroyd J. C. 2005. A novel rhoptry protein in Toxoplasma gondii bradyzoites and merozoites. Mol. Biochem. Parasitol. 144:159–166 [DOI] [PubMed] [Google Scholar]
- 58. Sibley L. D., Ajioka J. W. 2008. Population structure of Toxoplasma gondii: clonal expansion driven by infrequent recombination and selective sweeps. Annu. Rev. Microbiol. 62:329–351 [DOI] [PubMed] [Google Scholar]
- 59. Sinai A. P. 2008. Biogenesis of and activities at the Toxoplasma gondii parasitophorous vacuole membrane. Subcell. Biochem. 47:155–164 [DOI] [PubMed] [Google Scholar]
- 60. Singh U., Brewer J. L., Boothroyd J. C. 2002. Genetic analysis of tachyzoite to bradyzoite differentiation mutants in Toxoplasma gondii reveals a hierarchy of gene induction. Mol. Microbiol. 44:721–733 [DOI] [PubMed] [Google Scholar]
- 61. Soldati D., Boothroyd J. C. 1993. Transient transfection and expression in the obligate intracellular parasite Toxoplasma gondii. Science 260:349–352 [DOI] [PubMed] [Google Scholar]
- 62. Su C., et al. 2003. Recent expansion of Toxoplasma through enhanced oral transmission. Science 299:414–416 [DOI] [PubMed] [Google Scholar]
- 63. Suzuki Y., Remington J. S. 1988. Dual regulation of resistance against Toxoplasma gondii infection by Lyt-2+ and Lyt-1+, L3T4+ T cells in mice. J. Immunol. 140:3943–3946 [PubMed] [Google Scholar]
- 64. Torpier G., et al. 1993. Toxoplasma gondii: differential location of antigens secreted from encysted bradyzoites. Exp. Parasitol. 77:13–22 [DOI] [PubMed] [Google Scholar]
- 65. Vanchinathan P., Brewer J. L., Harb O. S., Boothroyd J. C., Singh U. 2005. Disruption of a locus encoding a nucleolar zinc finger protein decreases tachyzoite-to-bradyzoite differentiation in Toxoplasma gondii. Infect. Immun. 73:6680–6688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wallace G. R., Stanford M. R. 2008. Immunity and Toxoplasma retinochoroiditis. Clin. Exp. Immunol. 153:309–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Weiss L. M., Kim K. 2000. The development and biology of bradyzoites of Toxoplasma gondii. Front. Biosci. 5:D391–D405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wilson D. C., et al. 2010. Differential regulation of effector- and central-memory responses to Toxoplasma gondii infection by IL-12 revealed by tracking of Tgd057-specific CD8+ T cells. PLoS Pathog. 6:e1000815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhang Y. W., et al. 1999. Disruption of the Toxoplasma gondii bradyzoite-specific gene BAG1 decreases in vivo cyst formation. Mol. Microbiol. 31:691–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.