Abstract
Deadenylation is often the rate-limiting event in regulating the turnover of cellular mRNAs in eukaryotes. Removal of the poly(A) tail initiates mRNA degradation by one of several decay pathways, including deadenylation-dependent decapping, followed by 5′ to 3′ exonuclease decay or 3′ to 5′ exosome-mediated decay. In trypanosomatids, mRNA degradation is important in controlling the expression of differentially expressed genes. Genomic annotation studies have revealed several potential deadenylases. Poly(A)-specific RNase (PARN) is a key deadenylase involved in regulating gene expression in mammals, Xenopus oocytes, and higher plants. Trypanosomatids possess three different PARN genes, PARN-1, -2, and -3, each of which is expressed at the mRNA level in two life-cycle stages of the human parasite Trypanosoma brucei. Here we show that T. brucei PARN-1 is an active deadenylase. To determine the role of PARN-1 on mRNA stability in vivo, we overexpressed this protein and analyzed perturbations in mRNA steady-state levels as well as mRNA half-life. Interestingly, a subset of mRNAs was affected, including a family of mRNAs that encode stage-specific coat proteins. These data suggest that PARN-1 functions in stage-specific protein production.
INTRODUCTION
Regulation of gene expression in the protozoan parasite Trypanosoma brucei allows the organism to adapt and survive during its life cycle in two very different environments, the mammalian bloodstream and the tsetse fly. Expression of numerous protein-coding genes is regulated posttranscriptionally, particularly at the level of mRNA stability (4, 11, 25). For example, differential mRNA stability accounts for the stage-specific expression of procyclins, hexose transporters, and phosphoglycerate kinases (6, 20, 27, 28, 53).
In the well-studied Saccharomyces and mammalian systems, mRNA decay is a tightly controlled, multistep, and multipathway process. Various cis-acting elements, embedded in specific mRNAs, are recognized by RNA-binding proteins (7, 52, 55), which stabilize mRNAs or recruit RNases to carry out mRNA degradation and inhibit translation (8, 34, 39, 47). The removal of the mRNA 3′ poly(A) tail by 3′ to 5′ exoribonucleases (deadenylases) is often the rate-limiting step in mRNA degradation in vertebrates and thus a key point in regulation of mRNA turnover (19, 40).
A number of different deadenylases exist in eukaryotes, including poly(A)-specific RNase (PARN), the CCR4/CAF1/NOT complex, and the PAN2/PAN3 complex (reviewed in reference 23). The specific role of each of these proteins remains largely unknown, although evidence suggests that each enzyme may recognize a particular set of mRNA substrates (46). PARN functions in the targeted degradation of specific mRNAs in humans, Xenopus, and higher plants (1, 3, 22, 31, 32, 36, 58). To date, no PARN-encoding genes have been characterized in any single-cell eukaryote (56). In humans, PARN initiates decay of mRNAs containing AU-rich elements or nonsense codons. In Xenopus, PARN regulates oocyte maturation, whereas in Arabidopsis, PARN regulates embryogenesis.
The trypanosome genome encodes homologs to the deadenylation enzymes PARN, CAF1/NOT, and PAN2/PAN3, although a CCR4 homolog is absent (50, 51). Investigation of T. brucei CAF1/NOT1 showed that these two proteins were essential, whereas PAN2 depletion studies were less conclusive (50).
We set out to study PARN in T. brucei on the basis of our discovery of deadenylation activity in cytoplasmic extracts from this organism (42). T. brucei possesses three PARN homologs (PARN-1, -2, and -3), each of which is transcribed in both the procyclic (Pro) and bloodstream form (BF) stages of the parasite life cycle (30; this work). We chose PARN-1 to initiate our studies of PARN-dependent mRNA decay in trypanosomes. We verified that PARN-1 is a functional deadenylase, and we overexpressed PARN-1 in situ to identify transcripts that are targeted for degradation by this enzyme. A subset of mRNAs targeted for PARN-1-dependent degradation in T. brucei was identified using microarray studies and quantitative real-time PCR (qRT-PCR). Analysis of the genes coding for several of these mRNAs suggests that PARN-1 contributes to regulating differential gene expression.
MATERIALS AND METHODS
Culturing and transfection of parasites.
T. brucei Lister 427 procyclic cells were cultured in SDM-79 medium containing 10% fetal calf serum (FCS) at 26°C in 5% CO2. Procyclic 29-13 cells were cultured in the presence of 15 μg/ml G418 and 50 μg/ml hygromycin to maintain expression of T7 RNA polymerase and the tetracycline (Tet) repressor (57). Transfected parasites containing the integrated plasmid were selected for by the addition of 2.5 μg/ml phleomycin (2). Lister 427 BF cells were grown in HMI-9 medium containing 10% FCS and 10% Serum Plus (SAFC Biosciences) at 37°C in 5% CO2 (26). Single-marker BF cells (a gift from G. Cross) were cultured in the presence of 2.5 μg/ml G418 to maintain the T7 RNA polymerase and Tet repressor (57), and transfected parasites were obtained using an Amaxa system and selected for using 2.5 μg/ml phleomycin.
Plasmid constructs.
The pSAP1 vector containing the streptavidin-binding protein-protein A tandem affinity purification (TAP) tag was a generous gift from Larry Simpson. The 630-bp open reading frame (ORF) was PCR amplified to add HindIII and KpnI sites upstream and a BamHI site downstream of the tag. The product was ligated into the pLEW111 expression vector using the HindIII and BamHI sites to produce pLEW111-TAP. The PARN-1 ORF was PCR amplified, and KpnI sites were added to each end. PARN-1 was ligated into pLEW111-TAP using KpnI to produce a Tet-inducible, TAP-tagged PARN-1 protein expression vector.
RNA analysis.
Total RNA was extracted from procyclic cells grown to 8 × 106 cells/ml and BF cells grown to 1 × 106 cells/ml using Qiagen's RNeasy minikit. Poly(A)+ RNA was isolated from the total RNA using Qiagen's Oligotex mRNA midikit.
For Northern blot analysis, 10 μg of total RNA was separated on a formaldehyde-1.2% agarose gel in morpholinepropanesulfonic acid (MOPS) buffer (49). RNA was hydrolyzed and transferred to a nylon membrane overnight by capillary diffusion in 3 M NaCl–0.01 N NaOH. Radiolabeled probes were generated from 25 ng of PCR product using a Megaprime DNA labeling system (Amersham). Each probe was specific for a particular PARN gene; gene-specific primers for amplification were, for PARN-1, 5′-AACTGCCTAGCGGACGTTCT-3′ and 5′-TCACCTTCTTAGTGCTTTGC-3′, amplifying the region between nucleotides (nt) 960 and 1761 of the ORF; for PARN-2, 5′-GGTTTCAACATTTTTGGAAG-3′ and 5′-CTACCCACTAAGACGGTAAA-3′, amplifying the region between nt 710 and 1530 of the ORF; and for PARN-3, 5′-GTTACCACCGGAGGCTACGACTC-3′ and 5′-CTACTTCTCAGTCAAATGTT-3′, amplifying the region between nt 1032 and 1872 of the ORF. Probes were hybridized to the membrane overnight in hybridization buffer (5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% sodium pyruvate, 5× Denhardt's solution, 0.5% SDS, 100 μg/ml heparin, 0.1% diethyl pyrocarbonate) and washed three times (0.5× SSC, 15 min). Radiolabeled bands were analyzed by a PhosphorImager apparatus (Molecular Dynamics).
S100 protein extracts.
Extracts were prepared as described previously (42). Briefly, 2 liters of T. brucei procyclic cells were grown to 2 × 107 cells/ml. Cell cultures were harvested by centrifugation (1,100 × g, 20 min, 4°C). Cell pellets were washed three times in cold 1× phosphate-buffered saline (PBS) and resuspended in three pellet volumes of hypotonic lysis buffer (10 mM HEPES-KOH, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 1 mM dithiothreitol [DTT], 0.05% NP-40, 1 μM protease inhibitors), incubated on ice for 10 min, and then sheared using a tight-fitting 7-ml Dounce homogenizer. The cellular suspension was centrifuged (12,000 × g, 10 min, 4°C), and the supernatant was transferred to a new tube. The supernatant was adjusted to 275 mM KCl and centrifuged (100,000 × g, 1 h, 4°C). The S100 sample was collected and dialyzed in buffer D (20 mM HEPES-KOH, pH 7.9, 50 mM KCl, 0.2 mM EDTA, 20% glycerol, 1 mM DTT, 1 μM protease inhibitors) for 4 h at 4°C. The sample was aliquoted into 200-μl samples, quick-frozen, and stored at −80°C. Protein concentrations were determined by Bradford assay. Extracts typically yield 3 ml at 10 μg/μl.
TAP purification.
Procyclic parasites overexpressing PARN-1 (PARN-1 OvEx cells) were grown to 1 × 107 cells/ml in 1 liter culture medium. Expression of TAP-tagged PARN protein was induced with 500 μg/ml tetracycline for 18 h. Cells were harvested by centrifugation, and S100 protein extracts were prepared as described above. TAP-tagged protein was purified as described previously (12, 29). All steps were performed at 4°C. Briefly, 2 ml of S100 extract was adjusted to 10 ml with IgG-binding buffer (10 mM Tris HCl, pH 8.0, 150 mM NaCl, 2 mM EDTA, 0.1% NP-40, 1 mM DTT, 10% glycerol, Roche complete protease inhibitor cocktail tablet). One milliliter rabbit IgG agarose beads (Sigma) was washed with 10 ml IgG-binding buffer and incubated (1 h) in a 0.8- by 4-cm Poly-prep chromatography column (Bio-Rad). After incubation, the column was placed in a stand and opened to allow unbound protein to flow through. The protein-bead mixture was washed twice with 10 ml IgG-binding buffer and once with 10 ml tobacco etch virus (TEV)-cleavage buffer (10 mM Tris HCl, pH 8.0, 150 mM NaCl, 0.5 mM EDTA, 0.1% NP-40, 1 mM DTT, 5% glycerol). Beads were resuspended with 3 ml TEV-cleavage buffer containing 200 units of AcTEV protease (Invitrogen) and incubated (3 h). Protein was eluted from the column by gravity flow, and 1 ml TEV-cleavage buffer was added to the column to elute any residual protein. Four milliliters of eluted protein was adjusted to 10 ml with IgG-binding buffer. One milliliter streptavidin Sepharose high-performance beads (GE Healthcare) was washed with 10 ml IgG-binding buffer and incubated with the extracts (1 h) in the Poly-prep chromatography column. The column was then drained and washed twice with 10 ml IgG-binding buffer. Beads were resuspended in 1.5 ml elution buffer (40 mM Tris HCl, pH 8.0, 150 mM NaCl, 0.5 mM EDTA, 0.1% NP-40, 1 mM DTT, 10% glycerol, 2 mM biotin [Sigma]). Eluted protein was collected, and elution was repeated with another 1.5 ml of elution buffer. Protein samples were dialyzed in 1 liter buffer D (4 h). The samples were aliquoted, quick-frozen, and stored at −80°C.
Nuclear fractionation.
Log-phase procyclic cells were harvested by centrifugation (900 × g, 15 min, 4°C), and the pellet was washed twice with wash buffer (20 mM Tris-HCl, pH 7.4, 100 mM NaCl, 3 mM MgCl2, 2.5 mM DTT, 1 μM protease inhibitors). Pellet was resuspended in hypotonic buffer (10 mM HEPES, pH 7.9, 10 mM KCl, 2.5 mM MgCl2, 1 mM EDTA, 2.5 mM DTT, 1 μM protease inhibitors) and incubated on ice for 10 min. Cells were lysed in 0.2% NP-40 followed Dounce homogenization in a tight-fitting 7-ml Dounce homogenizer. An aliquot was saved as total lysate for immunoblots. Total lysate was layered on hypotonic buffer plus 0.8 M sucrose and centrifuged in a swing-out rotor (8,000 × g, 10 min, 4°C). The top layer was collected and saved as the cytoplasmic fraction for immunoblots, and the pellet was collected and saved as the nuclear fraction for immunoblots. Figure 2A contains protein fractions from equal number of cells (∼7 × 106) in each of the nine lanes.
Fig. 2.
PARN-1 is primarily cytoplasmic. (A) PARN-1 is absent from the nucleus. Fractionation of procyclic cells by sucrose cushion separates the nuclear fraction from the cytoplasmic fraction. Antibody against the RNA polymerase II largest subunit (RPB1) was used as a nuclear control, and antibody against translation elongation factor 2 (EF-2) was used as a nuclear control. (B) PARN-1 is absent from the mitochondria in wild-type cells. Fractionation of procyclic cells by a Percoll gradient separates the mitochondrial fraction from the cytoplasmic fraction. Antibody against elongation factor 2 was used as a cytoplasmic control, and antibody against guide RNA-binding complex proteins 1 and 2 (GRBC1 and -2) was used as a mitochondrial control. (C) TAP-tagged PARN-1 is primarily cytoplasmic in cells in which PARN-1 is overexpressed. Antibody against GRBC1 and -2 was used as a mitochondrial protein control.
Mitochondrial fractionation.
Mitochondria were isolated from T. brucei cells as described previously, with some exceptions (45). Briefly, 1 liter of procyclic cells was harvested by centrifugation (6,000 × g, 10 min, 4°C) and washed twice in PBS. Cells were resuspended in hypotonic lysis buffer (1 mM Tris-Cl, pH 8.0, 1 mM EDTA), incubated on ice for 5 min, and then sheared using a tight-fitting 7-ml Dounce homogenizer. An aliquot was saved as total lysate for immunoblots. Sucrose was added to the lysate at a final concentration of 0.25 M, and the lysate was centrifuged at 15,000 × g for 10 min at 4°C. Supernatant was removed and saved as the cytoplasmic fraction for immunoblots. The pellet was resuspended in STM buffer (20 mM Tris-Cl, pH 8.0, 250 mM sucrose, 2 mM MgCl2), and the suspension was incubated with 1/200 volume DNase I for 60 min. Two milliliters of STE buffer (20 mM Tris-Cl, pH 8.0, 250 mM sucrose, 2 mM EDTA) was added to the lysate to stop the DNase I reaction, and lysate was centrifuged at 15,000 × g for 10 min at 4°C. The supernatant was removed, and the pellet was resuspended in 0.4 ml of 70% Percoll and homogenized in a 2-ml Dounce homogenizer. The resuspended pellet was layered under a 15 to 40% linear Percoll gradient and centrifuged at 103,000 × g for 60 min at 4°C. The middle layer was collected and was washed twice with STE buffer by centrifugation (32,530 × g for 15 min at 4°C). The washed pellet was saved as the mitochondrial fraction for immunoblots. In Fig. 2B and C, each of the lanes containing total lysate represents 2 × 105 cell equivalents, each of the lanes containing cytoplasmic extract represents 2 × 105 cell equivalents, and each of the lanes containing mitochondrial proteins represents a 50-fold higher cell equivalent to compensate for the decreased protein yield during mitochondrial fractionation.
Antibodies, Western analysis, and immunodepletion.
The C-terminal 133 amino acids of the PARN-1 open reading frame were cloned into the pGEX-6P-1 bacterial expression vector and transformed into Escherichia coli BL21. Protein expression was induced overnight and purified by a glutathione S-transferase column. The purified protein was used to generate polyclonal antibody in rabbits (Lampire). Antibody was purified from sera by affinity chromatography with purified bacterium-expressed protein. Western analysis was performed by separating proteins on a 10% SDS-polyacrylamide gel and transferring the proteins to a polyvinylidene difluoride membrane. PARN-1 protein was identified using purified polyclonal antibody, and the TAP tag alone was identified using peroxidase antiperoxidase antibody (Sigma). Bound antibody was detected with anti-rabbit IgG (from donkey) linked to horseradish peroxidase, followed by chemiluminescence (Amersham). For PARN-1 immunodepletion experiments, 100 μl of resuspended protein A-Sepharose beads was incubated with 150 μl of polyclonal antibody. Beads were washed and split into three tubes. Approximately 1 μg of TAP tag-purified PARN-1 protein was added to the first set of beads and the mixture was incubated (1 h, 4°C). The antibody-bead complex was then removed from the sample by centrifugation, and the extract was depleted for two additional rounds. The depleted extracts were aliquoted into 50-μl samples, quick-frozen, and stored at −80°C.
Preparation of RNA substrates.
RNA substrates were prepared as previously described (18). DNA templates were prepared from the pGEM4 vector (Promega). Vector was digested with HindIII, and an A60-T60 homopolymer containing a 22-nt adapter sequence was ligated downstream. A 65-bp sequence from the pGEM vector containing the SP6 promoter was PCR amplified along with the 3′ A60 sequence. The 22-nt adapter sequence was removed by SspI digestion for the pGEMA60 substrate but was not removed for the pGEMA60–22-nt adapter sequence substrate. For the A0 substrate lacking a poly(A) tail, an HindIII-digested vector was used.
One microgram of DNA template was used to make internally labeled RNA with a Riboprobe in vitro transcription system (Promega). RNA was capped by the addition of 5 mM m7GTP cap analog and labeled with 50 μCi [alpha-32P]UTP. The reaction mixtures were incubated (1 h, 37°C), and the RNA products were purified by phenol-chloroform extraction, followed by ethanol precipitation. Samples were resuspended in 10 μl TBE (Tris-borate-acetate)-urea loading buffer and separated on a 5% acrylamide–7 M urea gel. Radiolabeled RNA bands were excised from the gel and eluted in 400 μl HSCB buffer (50 mM Tris-HCl, pH 7.6, 400 mM NaCl, 0.1% SDS) with 50 μg proteinase K overnight at room temperature. Samples were phenol-chloroform extracted, ethanol precipitated, and resuspended in RNase-free water.
In vitro deadenylase assays.
Assays were adapted from the previously described protocol (17). For assays using S100 protein extracts, 14-μl reaction mixtures containing 80 μg dialyzed S100 extract, 100,000 cpm labeled RNA (∼50 fmol), 2.3% polyvinyl alcohol, 3.2 mM MgCl2, and 500 ng poly(A) were prepared. For assays using TAP tag-purified PARN-1 protein, 15-μl reaction mixtures contained ∼50 ng purified PARN-1 protein, 100,000 cpm labeled RNA (∼50 fmol), 2.2% polyvinyl alcohol, 3 mM MgCl2, 2 μg bovine serum albumin (BSA), and 16 units RNase inhibitor (Amersham). All reaction mixtures were incubated at 26°C, and the reactions were stopped by the addition of 300 μl HSCB2 (20 mM Tris, pH 8.0, 400 mM NaCl, 0.1% SDS, 20 mM EDTA, 10 μg glycogen). RNA products were purified by phenol-chloroform extraction, followed by ethanol precipitation. RNA was resuspended in 10 μl TBE-urea loading buffer and separated on a 5% acrylamide–7 M urea gel. Gels were analyzed by a PhosphorImager apparatus (Molecular Dynamics) and quantitated by ImageQuant software.
Microarray analysis.
Procyclic cells were grown to a density of 8 × 106 cells/ml. PARN-1 and TAP tag overexpression was induced for 48 h with 500 ng/ml tetracycline. RNA was extracted as previously described. Four micrograms of poly(A)+ RNA was reverse transcribed for 16 h at 42°C in a reaction mixture containing 12 μg of random hexamers (Invitrogen), 50 mM Tris-HCl, pH 8.3, 75 mM KCl, 3 mM MgCl2, 10 mM DTT, 0.5 mM dATP, 0.5 mM dCTP, 0.5 mM dGTP, 0.17 mM dTTP, 1 mM 5-(3-aminoallyl)-dUTP, 0.33 mM KPO4, 80 U RNaseOUT (Invitrogen), and 800 U SuperScript III reverse transcriptase (Invitrogen). The steady-state lengths of poly(A) tails on T. brucei mRNAs are 150 to 200 nt. The RNA in the cDNA-RNA mixture was hydrolyzed in 100 mM EDTA and 200 mM NaOH. The reaction mixture was incubated (15 min, 65°C), and then the pH was adjusted to ∼7.0 with 333 mM Tris (pH 7.0). cDNA was purified using a QIAquick PCR purification kit (Qiagen) and dried to ∼1 μl by a SpeedVac apparatus. cDNA was resuspended in 100 mM sodium carbonate buffer (pH 9.3) and labeled with Cy3 or Cy5 monoreactive dye (GE Healthcare). Two reaction mixtures were incubated for 2 h at room temperature and stopped with 80 mM sodium acetate (pH 5.2). Labeled cDNAs were purified using the QIAquick PCR purification kit, combined, and dried to ∼1 μl by the SpeedVac apparatus. Samples were resuspended in hybridization buffer containing 40% formamide, 5× SSC, 0.1% SDS, and 0.6 μg/μl salmon sperm DNA and heated (twice, 5 min, 95°C). T. brucei DNA microarrays, version 3, obtained from the Pathogen Functional Genomic Research Center, were prehybridized at (2 h, 42°C) in 5× SSC, 0.1% SDS, and 1% BSA. Slides were washed in water and then dried using centrifugation. Samples were applied to the microarray slides, covered with a 24- by 60-cm glass coverslip (Fisher), and placed in a chamber (HybChamber; GeneMachines). The chamber was placed in a 42°C water bath, covered in aluminum foil, and incubated for 16 h. Slides were washed (2× SSC and 0.1% SDS, 0.1× SSC and 0.1% SDS, 0.1× SSC) and dried by centrifugation. Slides were scanned using a GenePix 4000A microarray scanner (Axon Instruments), and data were analyzed using GenePixPro (version 6.0; Axon Instruments) and MultiExperiment Viewer (version 4.1) software (the Institute for Genomic Research [TIGR]). Using this analysis, we determined that PARN-1 mRNA levels were increased ∼27-fold following induction with tetracycline.
Quantitative real-time PCR.
qRT-PCR experiments were carried out using an iScript one-step RT-PCR kit with SYBR green (Bio-Rad). The Primer sets are shown in Fig. S3 in the supplemental material. Briefly, 100 ng total RNA was added to a 25-μl reaction mixture containing 1× SYBR green reaction mixture, 300 nM forward and reverse primers, and 0.5 μl iScript reverse transcriptase. Reactions were run in a RotorGene 3000 real-time cycler (Corbett Research). Cycling conditions were as follows: 50°C for 10 min for cDNA synthesis, 5 min at 95°C for reverse transcriptase inactivation, and 40 cycles of 10 s at 95°C and 30 s at 55°C, followed by melt curve analysis from 55°C to 99°C. To assay for mRNA stability, cells were treated with 10 μg/ml actinomycin D, total RNA was extracted at 0, 30, 60, 120, and 180 min, and mRNA levels were analyzed by qRT-PCR. All mRNA levels were normalized to that of 7SL RNA, and the percentage of mRNA remaining from the starting amount (time zero) was calculated. For the mRNA decay kinetics, the percentage of mRNA remaining was plotted versus time. An exponential trendline was fit to each set of data, and half-lives were calculated using the trendline equation (y = ex).
Generation of double-knockout cell lines.
The 500-bp region upstream of the PARN-1 ORF was PCR amplified from genomic DNA with the addition of a BglII site downstream with primers JM300 (5′-CACCGAATTCGTACTCTTCTCTAAATTCGTTTC-3′) and JM301 (5′-CACCAGATCTAGTTACACTTGGGCTAATGC-3′). The 500-bp region downstream of the PARN-1 ORF was PCR amplified with the addition of a MluI site upstream with primers JM302 (5′-CACCACGCGTAGAAGGGTGTTTAAAAAAAAAAAAATTCGC-3′) and JM303 (5′-CACCGAATTCGGACGTTGGTCTTATGAAC-3′). The phleomycin resistance ORF was PCR amplified with a BglII site upstream and MluI site downstream of the ORF using primers JM304 (5′-CACCAGATCTATGGCCAAGTTGACCAGTGCC-3′) and JM305 (5′-CACCACGCGTTCAGTCCTGCTCCTCGGCCAC-3′). The neomycin resistance open reading frame was PCR amplified with a BglII site upstream and MluI site downstream of the ORF using primers JM204 (5′-CACCAGATCTCGGAAAGGGAGAGAAAC-3′) and JM205 (5′-CACCACGCGCCCAGTGGATTGAATTG-3′). Upstream and downstream sequences were ligated to either the phleomycin or neomycin resistance open reading frames using the BglII and MluI sites. The ligated constructs were PCR amplified using primers JM300 and JM305, and the PCR product was inserted in TOPO vector pCR 2.1.
The phleomycin resistance plasmid was digested with EcoRI and transfected into 427 procyclic T. brucei cells. After 24 h, 1 μg/ml phleomycin was added to cultures to select transfected cells. After these single-knockout cell lines were verified by restriction digestion, cells were transfected with a NotI-digested neomycin resistance plasmid. After 24 h, 15 μg/ml G418 was added to the cultures to screen for cells containing deletions of both PARN-1 alleles. Double-knockout strains were confirmed by PCR of genomic DNA and Northern blotting.
Microarray data accession number.
Microarray data were deposited into the NCBI GEO database (accession no. GSE20593).
RESULTS
T. brucei encodes three PARN homologs.
The amino acid sequence of Homo sapiens PARN (HsPARN) was used to identify possible PARN homologs in the T. brucei genome (Fig. 1A) (5). Three genes were designated T. brucei PARN-1 (TbPARN-1), TbPARN-2, and TbPARN-3 on the basis of their relative homology to HsPARN. PARN proteins are members of the DEDD exoribonuclease superfamily and the DEDDh family, characterized by (i) three exonuclease (Exo) motifs (Exo I, II, and III; Fig. 1A, gray boxes; see Fig. S1A in the supplemental material), (ii) four invariant acid amino acids necessary for catalytic activity (Fig. 1A, asterisks; see Fig. S1A in the supplemental material), (iii) a fifth invariant acidic residue located between Exo II and III (DTK, where the boldface D indicates the fifth invariant acidic residue; Fig. 1A, triangle; see Fig. S1A in the supplemental material), and (iv) a conserved histidine within the Exo III motif (Fig. 1A, filled circle; see Fig. S1A in the supplemental material) (48, 59). Because the TbPARNs and HsPARN are highly divergent within their C termini, we restricted the comparison to the N termini. The N terminus of TbPARN-1 is 30% identical and 17% similar in amino acid sequence to the HsPARN sequence, TbPARN-2 is 27% identical and 15% similar, and TbPARN-3 is 18% identical and 11% similar.
Fig. 1.
T. brucei PARN-1 is a member of a small family of related PARN proteins. (A) Representation of the amino acid sequence of the open reading frames shows that each of the three proteins contains the conserved Exo I, Exo II, and Exo III domains (gray boxes). The four residues of the DEDD motif required for catalytic activity are noted with asterisks, and a fifth conserved aspartate residue is shown by open triangles. The filled circles indicate the histidine in Exo III indicative of DEDDh subfamily members (59). The divergent C termini are shown as a crosshatched box (PARN-1), a hatched box (PARN-2), and a stippled box (PARN-3). The drawing is to scale. Chr, chromosome. (B) Northern blot of total RNA shows transcript levels of the three PARN genes in Pro and BF cells. Arrows indicate that PARN-1 migrates as a 2.1-kb mRNA, PARN-2 as a 2.05-kb mRNA, and PARN-3 as a 3.0-kb mRNA. Ethidium bromide staining of the formaldehyde gel shows 18S and the two main fragments of 28S rRNAs used as loading controls. The TriTryp gene numbers are as follows: PARN-1, Tb927.8.2850; PARN-2, Tb927.10.8360; PARN-3, Tb09.211.4350.
We determined whether each of the three T. brucei PARN-coding genes produces mRNAs in parasites using Northern blot analysis (Fig. 1B). DNA probes were targeted to the divergent, terminal 800 bp of each ORF to distinguish among the three PARN genes. Northern blot analysis showed that PARN-1, 2-, and -3 are transcribed in procyclic parasites, which is the life-cycle stage in the tsetse fly midgut, and in the bloodstream-form parasites, which is the replicating life-cycle stage in the mammalian host. In addition, the steady-state levels of each PARN mRNA do not differ in a stage-specific manner. PARN mRNA is present at low steady-state levels compared to beta-tubulin mRNA quantities, as determined in the Northern blot and qRT-PCR analyses (Fig. 1B and data not shown). The absolute levels of PARN-1, -2, and -3 proteins were not compared. We conclude that T. brucei constitutively expresses multiple PARN genes.
We aligned TbPARN-1 with the HsPARN and Arabidopsis thaliana PARN (AtPARN) to uncover similarities that might suggest functional conservation, unrelated to the three exonuclease motifs, among PARNs from different organisms (see Fig. S1 in the supplemental material). TbPARN-1, as well as TbPARN-2 and -3, lacks the RNA recognition motif (RRM) and the nuclear localization signal (NLS) found in HsPARN. The RRM in HsPARN enhances deadenylase activity by interacting with the 5′ cap of mRNA (14, 43). The absence of an RRM in TbPARN proteins is consistent with our observation that deadenylation in the cytoplasm does not require a 5′ cap on the RNA substrate (42). Moreover, the absence of an NLS in all three TbPARN proteins is consistent with our observations that TbPARN-1 resides primarily in the cytoplasm in procyclic parasites (Fig. 2). The lack of an NLS in AtPARN correlates with its predominantly cytoplasmic localization (9, 48). Thus, TbPARN-1 likely functions without a 5′-cap dependency and in the parasite's cytoplasm.
PARN-1 is nonessential for parasite viability.
To identify the role of PARN-1 in T. brucei, we deleted the two allelic copies of this gene using homologous recombination (see Fig. S2A in the supplemental material). Each allele was replaced with a drug resistance cassette. Northern blot analysis demonstrated the absence of PARN-1 mRNA in the double knockout (see Fig. S2B in the supplemental material). Western analysis demonstrated the absence of PARN-1 protein in the double knockout (see Fig. S2C in the supplemental material). Cell viability, growth rate, and microscopic analysis of two clonal cell lines showed no growth alterations compared to wild-type parasites (see Fig. S2D in the supplemental material). These results prove that PARN-1 is a nonessential gene in cultured procyclic T. brucei.
To explore the combined necessity of PARN-1, -2, and -3 in parasites, we simultaneously depleted all three PARNs using RNA interference in procyclic and bloodstream-form parasites (data not shown). PARN-1 and PARN-2 were shown to be decreased at the mRNA level by Northern analysis, and PARN-3 was shown to be decreased at the protein level by Western analysis using anti-PARN-3 antibody (data not shown). Neither growth rates nor gross morphology was affected in PARN-depleted procyclic forms, and growth rates were only slightly affected (∼10% slower than control rates) in PARN-depleted BF parasites. Because decreased amounts of all three PARNs did not affect cell growth, we conclude that PARN proteins either are not essential for T. brucei viability or are sufficient to sustain cell viability at low levels.
PARN-1 is a deadenylase in vitro.
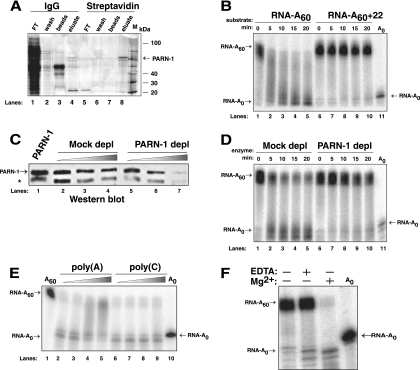
Purified PARN-1 was used in RNase assays to assess its deadenylase activity. To obtain purified protein, the PARN-1 ORF was tagged with a TAP tag and transfected on a Tet-inducible expression vector to produce the PARN-1 OvEx cell line. TAP-tagged PARN-1 was expressed and purified from S100 extracts using a two-step affinity method that employed IgG and streptavidin chromatography (Fig. 3A). A synthetic, radiolabeled 60-nt poly(A) tail (RNA-A60) was used as substrate. As expected, RNA-A60 was trimmed, in an apparently distributive manner, in the presence of purified PARN-1 (fraction shown in Fig. 3A, lane 8) to produce an RNA lacking a poly(A) tail, RNA-A0, after 10 min (Fig. 3B, lanes 1 to 5). To ensure that we were specifically assaying PARN-1 activity, we tested eluate (Fig. 3A, lane 8, and C, lane 1) that was PARN-1 depleted using PARN-1-specific polyclonal antibody or mock depleted using nonspecific antibody (Fig. 3C, lanes 2 to 7). Following depletion of PARN-1 protein using PARN-1-specific antibodies, RNA-A60 degradation was abolished (Fig. 3D, lanes 6 to 10). This was not the case in the mock depletion of PARN-1 (Fig. 3D, lanes 1 to 5). Thus, PARN-1 has deadenylase activity.
Fig. 3.
Purified PARN-1 has deadenylase activity in vitro. (A) A silver-stained 10% SDS-acrylamide gel shows the purification of TAP-tagged PARN-1 from procyclic PARN-1 OvEx cell S100 protein extract. Fractions applied to the IgG resin (lanes 1 to 4) flowed through the column (FT) and eluted in the wash or after TEV protease cleavage (eluate). Eluted protein was applied to streptavidin resin (lanes 5 to 8), and the flowthrough, wash, and protein eluted with biotin are shown. In each case, the material retained on the resin following elution is shown (beads). Highly enriched PARN-1 is indicated by an arrow. A molecular size marker (lane M) is shown. (B) Time course analysis of purified PARN-1 incubated with radiolabeled RNAs. Lanes 1 to 5, RNA-A60 substrate incubated with 50 ng enzyme; lanes 6 to 10, RNA-A60+22 substrate incubated with 50 ng enzyme; lane 11, deadenylated RNA (RNA-A0). About 1% of input material contained an unblocked 3′ end, due to substrate purification procedures, and was deadenylated as expected. (C) A Western blot, using anti-PARN-1 antibody, identifies purified PARN-1 samples that underwent three rounds of depletion (depl) with either PARN-1-specific antibody (lanes 5 to 7) or nonspecific antibody (lanes 2 to 4). Lane 1, starting sample; asterisk, clipped PARN-1 protein that has lost its streptavidin-binding protein tag. (D) Time course analysis of depleted PARN-1 samples incubated with radiolabeled RNA. Lanes 1 to 5, RNA-A60 substrate incubated with mock-depleted sample; lanes 6 to 10, RNA-A60 substrate incubated with PARN-1-depleted sample. (E) PARN-1 activity in the presence of poly(A) and poly(C) competitor. Lanes 2 to 5, deadenylation of RNA-A60 substrate in the presence of increased concentrations of poly(A) after 30 min; lanes 6 to 9, deadenylation of RNA-A60 substrate in the presence of increased concentrations of poly(C) after 30 min. Amounts of competitor added are 0 ng, 50 ng, 100 ng, and 500 ng in lanes 2 to 5, respectively, and lanes 6 to 9, respectively. (F) PARN-1 activity in the presence (+) and absence (−) of Mg2+ and EDTA after 30 min. Reaction products from deadenylase assays (B, D, E, and F) were separated using a 7 M urea–5% polyacrylamide denaturing gel. There are no radioactive signals running faster than the RNA-A0 species in the experiments. In panels B, D, E, and F the RNA-A0 migration was slower near the outside edges of the gels. In each case, the RNA-A0 is clearly marked.
To test whether PARN-1 was an adenosine-specific exonuclease, PARN-1 activity on RNA-A60 was evaluated in the presence of poly(A) or poly(C) competitor (Fig. 3E). Poly(A) competitor inhibited RNA-A60 deadenylation (Fig. 3E, lanes 2 to 5), whereas poly(C) competitor had no effect on RNA-A60 deadenylation (Fig. 3E, lanes 6 to 9). In addition, we tested PARN-1 activity using a 22-heteronucleotide sequence added 3′ to the poly(A) tail of RNA-A60 (RNA-A60+22) as substrate (Fig. 3B, lanes 6 to 10). PARN-1 did not degrade this RNA. Thus, PARN-1 appears to be exclusively a deadenylase.
The catalytic activity of most deadenylases requires divalent cations (13, 15, 59). To determine whether PARN-1 activity has this requirement, enzyme activity was tested in the presence and absence of Mg2+ (Fig. 3F). When Mg2+ was absent, RNA-A60 deadenylation was inhibited, demonstrating that TbPARN-1 activity is dependent upon a divalent cation, such as Mg2+, for its deadenylation activity.
Cells overexpressing PARN-1 have increased deadenylase activity.
To examine the role of PARN-1 in procyclic T. brucei, we determined whether overexpression of PARN-1 resulted in increased deadenylation. PARN-1 OvEx cells and a control culture expressing the TAP tag alone were induced, and S100 protein extracts were prepared (Fig. 4B). Both cultures grew at the same rate, indicating that there was no gross effect of PARN-1 overexpression on cell growth (data not shown). Deadenylation rates were measured using the in vitro deadenylation assay (Fig. 4A). The PARN-1 OvEx cell extract rapidly deadenylated RNA-A60, and nearly all RNA-A60 substrate was converted to RNA-A0 after 15 min (Fig. 4A, lanes 5 to 8). In contrast, control extracts deadenylated RNA-A60 at normal rates, and 45 min was required to convert nearly all RNA-A60 substrate to RNA-A0 (Fig. 4A, lanes 1 to 4). A graphic representation of the data is shown in Fig. 4C. Other deadenylases were unaffected by PARN-1 overexpression; thus, their activities were the same in PARN-1 OvEx cell and control extracts. Therefore, these data indicate that induced overexpression of PARN-1 protein in parasites enhances deadenylase activity.
Fig. 4.
Overexpression of PARN-1 enhances deadenylation in cytoplasmic extracts. (A) Time course of PARN-1 OvEx cell and control S100 extracts incubated with radiolabeled RNA. Enzyme activity was assayed from TAP tag alone and TAP-tagged PARN-1 protein in S100 extracts from Tet-induced (OvEx) and uninduced (basal) cultures. Lanes 1 to 4, RNA-A60 substrate incubated with S100 extracts from control cells induced with Tet (control cells produce a Tet-regulated streptavidin-binding protein and protein A TAP tag alone); lanes 5 to 8, RNA-A60 substrate incubated with S100 extracts from PARN-1 OvEx cells after Tet induction. Reaction products were separated on a 7 M urea–5% polyacrylamide denaturing gel. (B) Western blot showing protein amounts in uninduced and induced (basal and OvEx) S100 extracts. Control and PARN-1 OvEx cell extracts are included. Control cells contain a conditionally expressed TAP tag, which is detected with secondary antibody. Loading controls show that equal amounts of protein were applied to each gel lane. (C) The distance (in millimeters) that the majority of the RNA migrated, relative to the input RNA-A0 marker migration, is graphed as a function of deadenylation assay reaction time. The experiments were done three or more times, and the results of a representative experiment are shown.
A subset of procyclic mRNAs is reduced in PARN-1 OvEx parasites.
To determine the effect of PARN-1 overexpression on global mRNA steady-state levels, the mRNA expression profile of PARN-1 OvEx cells was examined by microarray analysis. Three clones each from the PARN-1 OvEx and control cell lines were used for analysis, and each experiment was run in duplicate, with dye labeling reversed between duplicates. Heat maps of the six arrays are shown (Fig. 5A). Twenty-nine protein-coding genes had their mRNAs decreased (Fig. 5B). Within this gene set, 4 genes encode T. brucei alanine-rich proteins (BARPs), 2 genes encode acidic phosphatases, 2 genes encode bona fide ribosomal subunits, and 12 genes encode hypothetical proteins. Eight protein-coding genes had their mRNAs increased. This set includes PARN-1, as expected. All of the misregulated mRNAs appear to be RNA polymerase II-dependent genes, except for Tb927.4.1200, a putative expression site-associated gene (ESAG), which is usually transcribed by RNA polymerase I. Thus, a limited number of mRNAs expressed in procyclic parasites are regulated, at least in part, by PARN-1.
Fig. 5.
Overexpression of PARN-1 decreases the steady-state mRNA levels of a subset of genes in T. brucei. (A) Heat map showing the change in steady-state mRNA levels after induction of PARN-1 deadenylase. Data were determined by microarray analysis. The data are arranged by the systematic gene name, which is based on chromosomal gene location. Columns 1 to 6 present the results for the 6 independent experiments. RNAs are represented as lines colored relative to their expression levels, as indicated in the heat map key on the left of the map. Green indicates an increase in mRNA levels in PARN-1 OvEx cells, red indicates a decrease in mRNA levels in PARN-1 OvEx cells, and black indicates no change. Each gene on the microarray was represented in duplicate; thus, for each experiment, mRNA levels were obtained from both points and averaged. In the case of dye flips, values were multiplied by −1 to allow numerical comparisons of all six arrays. Heat maps were generated in the MultiExperiment Viewer (MeV) program (version 4.1; TIGR), with genes arranged by identifier. The locus on chromosome (Chr) 9 containing the BARP genes is indicated with an arrow. (B) Summary of the gene loci encoding the mRNAs that were decreased or increased in PARN-1 OvEx cells. Column 1, systematic gene name, as designated in GeneDB; column 2, description of the gene product; column 3, number of independent experiments (n/6) in which the gene-encoding mRNA was decreased at least 1.5-fold; column 4, average fold decrease in PARN-1 OvEx cells among the six experiments; rows 1 to 24, genes located on the locus on chromosome 9 between Tb09.244.2400 and Tb09.244.2860; asterisks, genes further analyzed by qRT-PCR experiments; shaded gray, genes with reduced mRNA levels in procyclic parasites relative to other life-cycle stages (30, 44, 54). Microarray data were deposited into the NCBI GEO database (accession no. GSE20593). Eleven of the 14 BARP ORFs were present in triplicate on the microarray slide.
PARN-1 affects the steady-state level and decay rate of at least four different procyclic mRNAs.
To confirm the microarray data indicating that the levels of specific mRNAs were reduced in PARN-1 OvEx parasites, qRT-PCR was performed on a subset of genes (Fig. 5B, asterisks, and 6A). Primer sets and amplified RNA regions are shown in Fig. S4 in the supplemental material. BARP mRNA levels were reduced ∼3.2-fold in cells overexpressing PARN-1, determined using a primer set that recognized sequences common to all BARP isoforms (Fig. 6A). mRNA levels of the BARP isoform Tb09.244.2520 were reduced ∼5.9-fold in cells overexpressing PARN-1. The mRNA levels of two conserved hypothetical proteins (designated p28 and p16 to reflect their molecular masses) were reduced ∼2-fold in PARN-1 OvEx cells. The acidic phosphatase (Acid phos) mRNA transcribed from Tb927.5.630 was reduced ∼2-fold in PARN-1 OvEx cells. Dihydroxyacetone phosphate acyltransferase (DHAPAT) mRNA steady-state levels, which were unchanged in the microarray data set, were the same in PARN-1 OvEx and control cell lines. Thus, the qRT-PCR data and the microarray data are consistent with each other and confirm that a subset of mRNAs is regulated by PARN-1.
Fig. 6.
PARN-1 overexpression affects the steady-state levels and mRNA decay of specific genes. (A) Steady-state mRNA levels of genes in PARN-1 OvEx cells (light bars) and control cells (dark bars) determined by qRT-PCR. mRNA amounts were normalized to that of 7SL RNA, and levels from control cells were set equal to 100%. BARP (all), primers used to recognize all BARP isoforms; BARP (2520), BARP isoform Tb09.244.2520. (B) Percent mRNA remaining before actinomycin D treatment (dark bars) and 2 h after actinomycin D treatment (light bars), as determined by qRT-PCR. (C) A log scale shows the degradation kinetics of BARP, p28, and DHAPAT (control) mRNAs in PARN-1 OvEx cells. Actinomycin D (10 μg/ml) was added at time zero, and mRNA was isolated at the indicated time points and quantitated using qRT-PCR. Control cells in each graph show cells induced to overexpress the TAP tag alone. The quantitated results are expressed as the mean and 1 standard deviation for at least three independent experiments performed using two different RNA preparations. Although the degradation kinetics appear to be biphasic, overall half-lives (t1/2s) are calculated on the basis of changes in total mRNA levels and are therefore plotted on a semilog scale.
To determine whether the reduced levels of BARP, p28, p16, and Acid phos mRNAs in PARN-1 OvEx cells were caused by increased rates of mRNA decay, we measured mRNA amounts at 0 and 2 h following RNA synthesis inhibition by actinomycin D (Fig. 6B). DHAPAT mRNA was used to represent the mRNAs that were unaffected by PARN-1 overexpression in the microarray study. As expected, the amount of DHAPAT mRNA remaining was similar in the PARN-1 OvEx and control cell lines (30% versus 31%). The amount of mRNA remaining from all BARP isoforms was lower in PARN-1 OvEx cells (8%) than control cells (18%) after 2 h. Similarly, the amount of mRNA remaining from the BARP isoform Tb09.244.2520 was also decreased in PARN-1 OvEx cells (2%) relative to control cells (6%). In addition, the amount of p28 mRNA remaining was lower in PARN-1 OvEx cells (8%) than control cells (21%). Unexpectedly, the p16 mRNA amounts remaining in PARN-1 OvEx cells (23%) were close to those in control cells (19%), and the Acid phos mRNA amounts remaining in PARN-1 OvEx cells (37%) was greater than those remaining in control cells (27%). A direct measure of mRNA half-life was done for BARP (using the primers specific for the isoform Tb09.244.2520), p28, and control message DHAPAT (Fig. 6C). The degradation kinetics were exponential, as expected, and are shown for BARP mRNA (see Fig. S4 in the supplemental material). The half-life of BARP mRNA was decreased from 22 min to 15 min and the half-life of p28 mRNA was decreased from 60 min to 33 min after PARN-1 overexpression. DHAPAT, the control mRNA, was unaffected by PARN-1 overexpression, maintaining a half-life of 100 min under the two different conditions. In summary, the overexpression of PARN-1, as analyzed by qRT-PCR and mRNA decay profiles, resulted in an increase in the mRNA decay rates of BARP and p28 mRNAs.
DISCUSSION
Herein we show that trypanosomatids possess three different PARN genes, PARN-1, -2, and -3. Each PARN gene is expressed at the mRNA level in two life-cycle stages of the human parasite Trypanosoma brucei. PARN-1 is an active deadenylase and appears to regulate a subset of mRNAs, including a family of stage-specific coat proteins, the BARPs.
PARN proteins are members of the DEDD RNase superfamily, characterized by three exonuclease motifs that contain four invariant acidic amino acids. Most eukaryotes possess a single PARN gene. The three T. brucei PARNs, PARN-1, -2, and -3, each contain all three exonuclease motifs. Moreover, each possesses the conserved acidic amino acids, suggesting that all three PARNs are active in deadenylation. Other members of the trypanosomatid family also possess three PARN homologs, indicating multiple roles for PARNs in mRNA regulation during the complex life cycle of these organisms.
The sequences at the C terminus of all three T. brucei PARNs and that of the active AtPARN are highly divergent from the sequence at the C terminus of human PARN. In human PARN, the C terminus binds cap 0 (m7GpppG) of mRNA during deadenylation (14, 21, 38). Trypanosome mRNAs contain a cap 4 (a hypermethylated form of m7GpppAACU). Thus, the divergence from the human PARN in the C termini of the three T. brucei PARNs may reflect an interaction between at least one of them and the unique trypanosome cap structure.
Deletion of PARN-1 was not lethal to cultured procyclic parasites. This result was not surprising, as functional PARN is also not essential for viability in cultured HeLa cells, Xenopus oocytes, or Saccharomyces pombe (10, 31). However, PARN may be required for cellular processes involved in development. For example, embryonic development is stymied in Arabidopsis lacking PARN (48).
T. brucei PARN-1 participates in regulating specific mRNAs, as determined by our microarray and qRT-PCR data. Over-expression of PARN-1 affected BARP and p28 mRNA abundance and decay. In humans, PARN plays a role in regulating specific mRNAs via targeting to AU-rich element-containing mRNAs (35). In Xenopus and Arabidopsis, PARN plays a role in embryogenesis, targeting different subsets of mRNAs at specific stages of development (31, 48). Similarly, PARN-1 may regulate BARPs in a life-stage-specific manner in T. brucei. BARP mRNA is present only at low levels in the procyclic life stage, where we have confirmed that the PARN-1 protein is present (data not shown). These low BARP levels are possibly due to PARN-1-mediated decay. In contrast, BARP protein is highly expressed in epimastigotes (54), and we predict that PARN-1-mediated decay of BARP mRNA is decreased in this life-cycle stage.
Other mRNA decay studies in trypanosomes have begun to characterize CAF1, PAN2, DHH1, XRNA, DCP1/2, and DCPS-like activities and the exosome in T. brucei (16, 24, 33, 37, 41, 50, 51). CAF1 and PAN2 appear to affect the decay of stable mRNAs since depletion of either protein resulted in an increased poly(A) tail length in bloodstream-form parasites (50, 51). In addition, depletion of the CAF1, PAN2, or XRNA protein decreased the decay rate of the unstable EP1 procyclin mRNA in bloodstream-form cells. Also, recent studies of DHH1 show that this RNA helicase has a selective role in modulating levels of developmentally regulated mRNAs (33).
On the basis of our findings that PARN-1 is a nonessential protein for cultured procyclic parasites and appears to regulate the steady-state levels of a subpopulation of cellular mRNAs, we propose that PARN-1 operates as a regulatory enzyme to control the levels of a subset of mRNAs during the parasite life cycle. The paradigm for differential deadenylase targeting of specific mRNAs includes RNA-binding proteins that recruit deadenylases via cis-acting elements in specific mRNAs. Interestingly, Roditi and colleagues have shown that the BARP 3′ untranslated region (UTR) contains cis-acting elements that contribute to mRNA instability in procyclic parasites (54). PARN-1 may participate in the regulation of BARP mRNA turnover by interacting with 3′ UTR-protein complexes. Thus, it will be interesting to see if the set of three PARN deadenylases, recruited by different RNA-binding proteins, serves to modulate stage-specific mRNA decay during the parasite life cycle.
In summary, we present herein the first characterization of PARN in a single-cell eukaryote. PARN-mediated degradation of stage-specific BARP messages suggests a role for PARN in T. brucei development much like the developmental role of PARN in multicell eukaryotes. Interestingly, T. cruzi and Leishmania spp. also encode PARN homologues. We speculate that these human-infective trypanosomes, and possibly other pathogenic single-cell eukaryotes, utilize PARN to regulate gene expression during development.
Supplementary Material
ACKNOWLEDGMENTS
We thank David Wah and Mahi Banday for critical reading of the manuscript. We thank Ruslan Aphasizhev for anti-guide RNA-binding complex protein antibodies. We gratefully appreciate the figure illustrations by Timothy Linteau and Han Wu. We appreciate the work of Harleen Jammu for anti-PARN-1 production.
This work was supported by National Institutes of Health, NIAID (grant AI535835 to V.B.), and a Foundation of UMDNJ grant to V.B.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 8 July 2011.
REFERENCES
- 1. Balatsos N. A., Nilsson P., Mazza C., Cusack S., Virtanen A. 2006. Inhibition of mRNA deadenylation by the nuclear cap binding complex (CBC). J. Biol. Chem. 281:4517–4522 [DOI] [PubMed] [Google Scholar]
- 2. Banerjee H., et al. 2009. Identification of the HIT-45 protein from Trypanosoma brucei as an FHIT protein/dinucleoside triphosphatase: substrate specificity studies on the recombinant and endogenous proteins. RNA 15:1554–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Belostotsky D. A., Sieburth L. E. 2009. Kill the messenger: mRNA decay and plant development. Curr. Opin. Plant Biol. 12:96–102 [DOI] [PubMed] [Google Scholar]
- 4. Berberof M., et al. 1995. The 3′-terminal region of the mRNAs for VSG and procyclin can confer stage specificity to gene expression in Trypanosoma brucei. EMBO J. 14:2925–2934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Berriman M., et al. 2005. The genome of the African trypanosome Trypanosoma brucei. Science 309:416–422 [DOI] [PubMed] [Google Scholar]
- 6. Blattner J., Clayton C. E. 1995. The 3′-untranslated regions from the Trypanosoma brucei phosphoglycerate kinase-encoding genes mediate developmental regulation. Gene 162:153–156 [DOI] [PubMed] [Google Scholar]
- 7. Brewer G. 1991. An A + U-rich element RNA-binding factor regulates c-myc mRNA stability in vitro. Mol. Cell. Biol. 11:2460–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen C. Y., Shyu A. B. 1995. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 20:465–470 [DOI] [PubMed] [Google Scholar]
- 9. Chiba Y., et al. 2004. AtPARN is an essential poly(A) RNase in Arabidopsis. Gene 328:95–102 [DOI] [PubMed] [Google Scholar]
- 10. Chou C. F., et al. 2006. Tethering KSRP, a decay-promoting AU-rich element-binding protein, to mRNAs elicits mRNA decay. Mol. Cell. Biol. 26:3695–3706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Clayton C. E. 2002. Life without transcriptional control? From fly to man and back again. EMBO J. 21:1881–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Das A., Bellofatto V. 2009. The non-canonical CTD of RNAP-II is essential for productive RNA synthesis in Trypanosoma brucei. PLoS One 4:e6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Daugeron M. C., Mauxion F., Seraphin B. 2001. The yeast POP2 gene encodes a nuclease involved in mRNA deadenylation. Nucleic Acids Res. 29:2448–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dehlin E., Wormington M., Korner C. G., Wahle E. 2000. Cap-dependent deadenylation of mRNA. EMBO J. 19:1079–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dlakic M. 2000. Functionally unrelated signalling proteins contain a fold similar to Mg2+-dependent endonucleases. Trends Biochem. Sci. 25:272–273 [DOI] [PubMed] [Google Scholar]
- 16. Estevez A. M., Kempf T., Clayton C. 2001. The exosome of Trypanosoma brucei. EMBO J. 20:3831–3839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ford L. P., Watson J., Keene J. D., Wilusz J. 1999. ELAV proteins stabilize deadenylated intermediates in a novel in vitro mRNA deadenylation/degradation system. Genes Dev. 13:188–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ford L. P., Wilusz J. 1999. An in vitro system using HeLa cytoplasmic extracts that reproduces regulated mRNA stability. Methods 17:21–27 [DOI] [PubMed] [Google Scholar]
- 19. Fritz D. T., Bergman N., Kilpatrick W. J., Wilusz C. J., Wilusz J. 2004. mRNA decay in mammalian cells: the exonuclease perspective. Cell Biochem. Biophys. 41:265–278 [DOI] [PubMed] [Google Scholar]
- 20. Furger A., Schurch N., Kurath U., Roditi I. 1997. Elements in the 3′ untranslated region of procyclin mRNA regulate expression in insect forms of Trypanosoma brucei by modulating RNA stability and translation. Mol. Cell. Biol. 17:4372–4380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gao M., Fritz D. T., Ford L. P., Wilusz J. 2000. Interaction between a poly(A)-specific RNase and the 5′ cap influences mRNA deadenylation rates in vitro. Mol. Cell 5:479–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Garneau N. L., Wilusz J., Wilusz C. J. 2007. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 8:113–126 [DOI] [PubMed] [Google Scholar]
- 23. Goldstrohm A. C., Hook B. A., Wickens M. 2008. Regulated deadenylation in vitro. Methods Enzymol. 448:77–106 [DOI] [PubMed] [Google Scholar]
- 24. Haile S., Estevez A. M., Clayton C. 2003. A role for the exosome in the in vivo degradation of unstable mRNAs. RNA 9:1491–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hendricks E. F., Matthews K. 2007. Post-transcriptional control of gene expression in African trypanosomes, p. 209–237. In Barry J. D., McCulloch R., Mottram J. C., Acosta-Serrano A. (ed.), Trypanosomes; after the genome. Horizon Bioscience, Norwich, United Kingdom [Google Scholar]
- 26. Hirumi H., Hirumi K. 1989. Continuous cultivation of Trypanosoma brucei blood stream forms in a medium containing a low concentration of serum protein without feeder cell layers. J. Parasitol. 75:985–989 [PubMed] [Google Scholar]
- 27. Hotz H. R., Hartmann C., Huober K., Hug M., Clayton C. 1997. Mechanisms of developmental regulation in Trypanosoma brucei: a polypyrimidine tract in the 3′-untranslated region of a surface protein mRNA affects RNA abundance and translation. Nucleic Acids Res. 25:3017–3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hotz H. R., Lorenz P., Fischer R., Krieger S., Clayton C. 1995. Role of 3′-untranslated regions in the regulation of hexose transporter mRNAs in Trypanosoma brucei. Mol. Biochem. Parasitol. 75:1–14 [DOI] [PubMed] [Google Scholar]
- 29. Ibrahim B. S., et al. 2009. Structure of the C-terminal domain of transcription factor IIB from Trypanosoma brucei. Proc. Natl. Acad. Sci. U. S. A. 106:13242–13247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jensen B. C., Sivam D., Kifer C. T., Myler P. J., Parsons M. 2009. Widespread variation in transcript abundance within and across developmental stages of Trypanosoma brucei. BMC Genomics 10:482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim J. H., Richter J. D. 2006. Opposing polymerase-deadenylase activities regulate cytoplasmic polyadenylation. Mol. Cell 24:173–183 [DOI] [PubMed] [Google Scholar]
- 32. Korner C. G., et al. 1998. The deadenylating nuclease (DAN) is involved in poly(A) tail removal during the meiotic maturation of Xenopus oocytes. EMBO J. 17:5427–5437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kramer S., et al. 2010. The RNA helicase DHH1 is central to the correct expression of many developmentally regulated mRNAs in trypanosomes. J. Cell Sci. 123:699–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lai W. S., et al. 1999. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol. Cell. Biol. 19:4311–4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lai W. S., Kennington E. A., Blackshear P. J. 2003. Tristetraprolin and its family members can promote the cell-free deadenylation of AU-rich element-containing mRNAs by poly(A) RNase. Mol. Cell. Biol. 23:3798–3812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lejeune F., Li X., Maquat L. E. 2003. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol. Cell 12:675–687 [DOI] [PubMed] [Google Scholar]
- 37. Li C. H., et al. 2006. Roles of a Trypanosoma brucei 5′ → 3′ exoribonuclease homolog in mRNA degradation. RNA 12:2171–2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Martinez J., Ren Y. G., Nilsson P., Ehrenberg M., Virtanen A. 2001. The mRNA cap structure stimulates rate of poly(A) removal and amplifies processivity of degradation. J. Biol. Chem. 276:27923–27929 [DOI] [PubMed] [Google Scholar]
- 39. Mauxion F., Faux C., Seraphin B. 2008. The BTG2 protein is a general activator of mRNA deadenylation. EMBO J. 27:1039–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Meyer S., Temme C., Wahle E. 2004. mRNA turnover in eukaryotes: pathways and enzymes. Crit. Rev. Biochem. Mol. Biol. 39:197–216 [DOI] [PubMed] [Google Scholar]
- 41. Milone J., Wilusz J., Bellofatto V. 2002. Identification of mRNA decapping activities and an ARE-regulated 3′ to 5′ exonuclease activity in trypanosome extracts. Nucleic Acids Res. 30:4040–4050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Milone J., Wilusz J., Bellofatto V. 2004. Characterization of deadenylation in trypanosome extracts and its inhibition by poly(A)-binding protein Pab1p. RNA 10:448–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nilsson P., et al. 2007. A multifunctional RNA recognition motif in poly(A)-specific RNase with cap and poly(A) binding properties. J. Biol. Chem. 282:32902–32911 [DOI] [PubMed] [Google Scholar]
- 44. Nolan D. P., et al. 2000. Characterization of a novel alanine-rich protein located in surface microdomains in Trypanosoma brucei. J. Biol. Chem. 275:4072–4080 [DOI] [PubMed] [Google Scholar]
- 45. Panigrahi A. K., Schnaufer A., Stuart K. D. 2007. Isolation and compositional analysis of trypanosomatid editosomes. Methods Enzymol. 424:1–24 [DOI] [PubMed] [Google Scholar]
- 46. Parker R., Song H. 2004. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 11:121–127 [DOI] [PubMed] [Google Scholar]
- 47. Peng S. S., Chen C. Y., Xu N., Shyu A. B. 1998. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 17:3461–3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Reverdatto S. V., Dutko J. A., Chekanova J. A., Hamilton D. A., Belostotsky D. A. 2004. mRNA deadenylation by PARN is essential for embryogenesis in higher plants. RNA 10:1200–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sambrook J., Russell D. W. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 50. Schwede A., et al. 2008. A role for Caf1 in mRNA deadenylation and decay in trypanosomes and human cells. Nucleic Acids Res. 36:3374–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schwede A., et al. 2009. The role of deadenylation in the degradation of unstable mRNAs in trypanosomes. Nucleic Acids Res. 37:5511–5528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tadauchi T., Matsumoto K., Herskowitz I., Irie K. 2001. Post-transcriptional regulation through the HO 3′-UTR by Mpt5, a yeast homolog of Pumilio and FBF. EMBO J. 20:552–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tetaud E., Barrett M. P., Bringaud F., Baltz T. 1997. Kinetoplastid glucose transporters. Biochem. J. 325(Pt 3):569–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Urwyler S., Studer E., Renggli C. K., Roditi I. 2007. A family of stage-specific alanine-rich proteins on the surface of epimastigote forms of Trypanosoma brucei. Mol. Microbiol. 63:218–228 [DOI] [PubMed] [Google Scholar]
- 55. Vlasova I. A., Bohjanen P. R. 2008. Posttranscriptional regulation of gene networks by GU-rich elements and CELF proteins. RNA Biol. 5:201–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wiederhold K., Passmore L. A. 2010. Cytoplasmic deadenylation: regulation of mRNA fate. Biochem. Soc. Trans. 38:1531–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wirtz E., Leal S., Ochatt C., Cross G. A. 1999. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99:89–101 [DOI] [PubMed] [Google Scholar]
- 58. Yamashita A., et al. 2005. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat. Struct. Mol. Biol. 12:1054–1063 [DOI] [PubMed] [Google Scholar]
- 59. Zuo Y., Deutscher M. P. 2001. Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acids Res. 29:1017–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.