Abstract
Many of the major human fungal pathogens are known to undergo morphological changes, which in certain cases are associated with virulence. Although there has been an intense research focus on morphology in fungi, very little is known about how morphology evolved in conjunction with a variety of other virulence properties. However, several recent important discoveries, primarily in Candida species, are beginning to shed light on this important area and answer many longstanding questions. In this minireview, we first provide a description of the major fungal morphologies, as well as the roles of morphology and morphology-associated gene expression in virulence. Next, focusing largely on Candida species, we examine the evolutionary relationships among specific morphological forms. Finally, drawing on recent findings, we begin to address the question of how specific morphological changes came to be associated with virulence of Candida species during evolution.
INTRODUCTION
Many fungal species possess the ability to change their physical shape, or morphology. These species include a wide variety of fungal pathogens, such as Candida albicans, as well as nonpathogenic fungi, such as Saccharomyces cerevisiae. The study of morphology was an area of intense interest for mycologists well before tools were available to identify the molecular pathways and mechanisms that drive morphological change. Some of the earliest sketches of fungi by Helen Beatrix Potter in the late 19th century depict a wide variety of both yeast and mycelial forms (111, 112). In modern times, these studies have taken on additional significance with the discovery that several human fungal pathogens undergo morphological changes that, in certain cases, are associated with virulence (50, 71, 76, 119). Infections by these pathogens are on the rise and have been associated with a significant increase in the number of individuals receiving immunosuppressive therapies, such as cancer patients on chemotherapy and organ transplant recipients (30, 149).
In the past 20 years, a large arsenal of molecular tools has been developed, which has allowed mycologists to gain a better understanding of the relationship between fungal morphology and virulence. While a great deal has been learned, many fundamental questions about the evolution of morphology and virulence in fungi remain unanswered. How and why did the various fungal morphologies evolve? What is the evolutionary relationship among specific morphological forms? What specific contribution does each morphology make to virulence in different species? What mechanisms function to determine fungal morphology, and are these mechanisms evolutionarily conserved? How did morphological changes coevolve with other virulence properties? While answers to many of these questions are not complete, several recent important discoveries, primarily in Candida species, are beginning to provide better clues. In this minireview, we place these discoveries in an evolutionary context with the ultimate goal of providing greater insight into the coevolution of morphology and virulence in human fungal pathogens.
FUNGAL MORPHOLOGIES
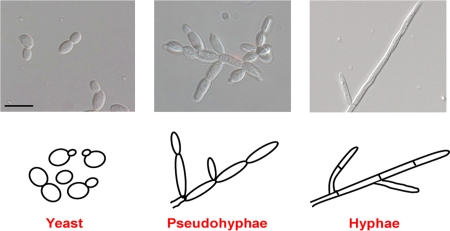
Fungal species can grow in three main cellular morphologies: yeast, pseudohyphae, and hyphae (Fig. 1). Yeasts are single cells that are oval and can exhibit both axial and bipolar budding patterns. Many fungi, such as the model organism S. cerevisiae, are known to grow predominantly in the yeast form. Other fungi, such as Schizosaccharomyces spp., also grow as yeasts but divide by binary fission rather than a budding mechanism (31, 36).
Fig. 1.
Major morphologies of human fungal pathogens. (Top) Images of C. albicans cells as visualized by differential interference contrast (DIC) microscopy (bar = 10 μm). (Bottom) Schematic representation of each morphology.
Pseudohyphae and hyphae are commonly called the “filamentous” morphologies, because cells typically grow in a polarized manner, are elongated in form, and are attached end to end. Pseudohyphal cells are generally ellipsoidal (i.e., their width is larger at the center than at the ends) and have constrictions at the septal junctions (132). In contrast, hyphal cells generally have parallel sides, are uniform in width, and possess true septa lacking constrictions (108) (Fig. 1). Unlike pseudohyphae, hyphal cells also have pores in their septa for cell-cell communication (48, 108). In pseudohyphae and yeast, nuclear division and septum ring formation occur across the mother-bud neck, whereas in hyphae, these events occur entirely within the germ tube (the initial short filament) during the first cell division, based on C. albicans studies (52, 133, 146). In addition, both yeast and pseudohyphal cells grow in synchrony with the cell cycle, whereas in hyphae, initial germ tube formation occurs prior to the G1/S transition (6, 51, 151). Following completion of the first cell cycle, C. albicans hyphal cells remain in G1 phase until they accumulate a cytoplasmic mass sufficient to allow them to enter a second cell cycle; as a consequence, hyphal filaments tend to be less highly branched than those of pseudohyphae (132). Based on studies in Aspergillus nidulans, additional factors, such as sphingolipid biosynthesis and calmodulin, are known to control cell cycle progression in hyphae (21, 115). Hyphal cells are also known to possess several structures that are absent in pseudohyphae. These include true septa, mentioned above, as well as the Spitzenkörper, a specialized organelle that promotes growth at the hyphal tip by coordinating vesicle secretion in a controlled manner (23, 129). (For a more detailed description of the differences among these morphologies, see reference 132.) It is important to bear in mind that while pseudohyphae may appear physically more similar to hyphae, they actually share far more properties with yeasts and might be better described as elongated, attached yeast cells (108, 132). Although not frequently discussed in the literature, intermediate morphologies, such as elongated single yeast cells and pseudohyphal cells with largely parallel sides and minor septal constrictions, are also observed in nature. Yeast, pseudohyphae and hyphae are all vegetative cell types. A nonvegetative morphology, the spherule, is also generated by spores of Coccidioides immitis, the causative agent of valley fever, and is important for invasion of the lungs (55). A final morphological structure observed in fungal pathogens is the chlamydospore. Chlamydospores are large (3 to 4 times the size of yeast), round, thick-walled cells that typically form at the ends of hyphal filaments (128). These cells tend to form in response to nutrient-poor conditions in several fungal pathogens, including C. albicans and Candida dubliniensis, and have also been observed in Cryptococcus neoformans, dermatophytes, Fusarium spp., and classical dimorphic fungi (79, 91, 118, 124, 128, 130). Chlamydospores have only rarely been observed in infected tissue (20, 22), and very little is known about the biological function of these cells. For the purposes of this review, however, we will focus solely on the yeast, pseudohyphal, and hyphal cellular morphologies.
ROLES OF YEAST AND YEAST-ASSOCIATED GENE EXPRESSION IN VIRULENCE
In many fungal pathogens, the yeast form and yeast-associated gene expression are specifically correlated with virulence. This correlation is best observed in a family of six related dimorphic fungal pathogens, which are collectively responsible for over 1 million new infections per year: Histoplasma capsulatum, C. immitis, Paracoccidioides brasiliensis, Blastomyces dermatitidis, Penicillium marneffei, and Sporothrix schenkii (71). These species are typically found in the soil growing in the nonpathogenic hyphal form. Following inhalation of conidia by the host (in the case of S. schenkii, conidia are inoculated into wounds), the organisms convert to the pathogenic yeast form and establish infection. The morphological transition to yeast cells is required for virulence and is known to be triggered by the shift to body temperature, 37°C (71). Causing an irreversible block to this transition by treatment with p-chloromercuriphenylsulfonic acid (PCMS) was shown to render H. capsulatum strains highly attenuated for virulence (89). Previous studies have shown that H. capsulatum yeast cells can evade killing, multiply in macrophages, and use phagocytic cells as a transport vehicle to invade and colonize a variety of organs, including the liver, bone marrow, lymph nodes, and spleen (16, 32, 33). In addition, B. dermatitidis yeast, but not hyphal, cells have been shown to be too large for engulfment by polymorphonuclear neutrophils (PMNs) (28). While not specifically required for virulence in C. albicans, the yeast morphology is known to be important for a variety of virulence-related processes in Candida species, including colonization and rapid dissemination to host tissues, adhesion to host cell surfaces, and biofilm formation (50, 108, 109).
A variety of yeast phase-specific genes that play specific roles in virulence have also been identified. In B. dermatitidis, H. capsulatum, and P. brasiliensis, a direct correlation has been shown between yeast cell wall α-(1,3)-glucan content and virulence (59). A recent study in H. capsulatum suggests that α-(1,3)-glucan in the cell wall functions to mask β-glucan residues from detection by the host macrophage receptor dectin-1, thus evading an immune response (113). Consistent with this hypothesis, disruption of the H. capsulatum yeast-specific gene AGS1, encoding α-(1,3)-glucan synthase, reduces both pathogenesis and the ability of yeast to grow in macrophages in vitro (114). Additional yeast phase-specific virulence factors include B. dermatitidis BAD1, an adhesin that binds complement type 3 receptors, as well as CD14 on lung cells and macrophages, to manipulate the host immune response (12, 98), and H. capsulatum CBP1, a calcium-binding protein required for pathogenicity, in addition to growth in macrophages in vitro (8, 122).
Several key regulators of the hyphal-yeast transition have also been identified in these fungi. In both B. dermatitidis and H. capsulatum, DRK1 encodes a hybrid histidine kinase that senses host signals. DRK1 is required for the hyphal-to-yeast transition, as well as virulence and expression of virulence-specific genes, such as AGS1 (97). Also, in H. capsulatum, RYP1 encodes a transcriptional regulator required for both the transition to yeast phase and expression of the large majority of yeast-specific genes (including several genes important for virulence), as determined by DNA microarray analysis (99).
In addition to the 6 dimorphic ascomycete fungal pathogens discussed above, C. neoformans, a basidiomycete, forms filamentous dikaryons during mating but occurs predominantly in the yeast form during infection (2, 80). C. neoformans basidiospores or desiccated yeast cells are likely to be the initial forms to establish infection in the host, since they are sufficiently small (<3 μm) to accumulate in lung alveoli following inhalation (78); nondesiccated C. neoformans spores are also known to have pathogenic potential (45, 134, 156). During infection, C. neoformans yeast cells are known to replicate in both intracellular and extracellular environments and are likely to survive better in macrophages, dendritic cells, and neutrophils (26, 37, 142). In this fungus, the yeast morphology is also associated with a number of virulence properties, including thermotolerance, capsule formation, and melanin production (57). A specific C. neoformans transcriptional regulator, MGA2, has been linked to thermotolerance (74). In addition, the cyclic AMP (cAMP) pathway is known to be important for both capsule formation and virulence, since a C. neoformans pka1 catalytic mutant is deficient for both of these processes (29). A laccase gene, LAC1, has also been shown to be important for both melanin production and virulence in C. neoformans (116, 117). For a description of additional genes involved in C. neoformans virulence properties, see reference 110.
ROLES OF HYPHAE AND HYPHA-ASSOCIATED GENE EXPRESSION IN VIRULENCE
In several fungal pathogens, the hyphal morphology and expression of hypha-specific genes is critical for virulence. This discussion focuses largely on C. albicans, because significantly more is known about the relationship between hyphae and virulence in that pathogen. In C. albicans, hyphal formation is known to promote virulence by several mechanisms: (i) hyphae can invade epithelial cell layers by exerting mechanical force; invasion can involve growth between epithelial cells, as well as penetration of individual cells (76); (ii) hyphae are capable of breaching and damaging endothelial cells (65, 76, 157); and (iii) following phagocytosis, C. albicans hyphal growth can cause lysis of both macrophages and neutrophils (72, 84). In addition, thigmotropism, or contact sensing, is believed to allow C. albicans hyphae to identify and penetrate small grooves, crevices, and weak points in host tissues during infection (49, 50, 53). In Aspergillus spp., as well as several zygomycetes, hyphae play a critical role in a process known as angioinvasion (42, 43, 123). Once inhaled by the lungs, spores of these species form hyphae that penetrate both pulmonary epithelial and vascular endothelial cell layers; hyphal fragments then disseminate through the bloodstream and eventually establish secondary sites of infection (38).
Multiple lines of evidence suggest that the yeast-hyphal transition is required for virulence in C. albicans. First, a mutant locked in the yeast form is highly attenuated for virulence in a mouse model of systemic candidiasis (84); one caveat, however, is that this strain bears mutations in transcriptional regulators known to control a variety of processes in addition to hyphal formation. Second, a strain that can be genetically engineered to grow exclusively in the yeast form during infection is highly attenuated for virulence; importantly, if this strain is manipulated to undergo the yeast-hyphal transition at various postinfection time points, virulence is restored (121). Finally, it has been shown that virulence is promoted in a strain that is genetically manipulated to increase hyphal formation during infection in a mouse model of systemic candidiasis (19). Interestingly, a recent large-scale analysis of C. albicans homozygous deletion mutants has identified 48 genes that appear to play important roles in infectivity but have no effect on morphology; in addition, out of 24 mutants with significant morphology defects, two-thirds showed normal infectivity (104). Therefore, while the C. albicans yeast-hyphal transition appears to be generally important for virulence, these results challenge the dogma that there is a precise correlation between morphology and virulence and suggest that several factors play independent roles in each process as well.
Based on studies in C. albicans, hyphal formation is well orchestrated, with the expression of genes involved in a variety of virulence-related processes (67, 76, 96). Certain filament-induced genes, such as HGC1, are specifically involved in driving the physical process of hyphal development. HGC1 encodes a cyclin-related protein important for septin phosphorylation, inhibition of cell separation, and activation of the Cdc42 master polarity regulator (47, 125, 144, 145, 154, 155). C. albicans cells deleted for HGC1 are highly attenuated for virulence, indicating a specific requirement for hyphal development in C. albicans pathogenicity. Interestingly, many other hypha-specific genes are also important for pathogenicity but not necessarily involved in hyphal formation per se. For example, ALS3 and HWP1 encode adhesins, which enable C. albicans to move out of the bloodstream, colonize tissues, and form biofilms (34, 101–103, 127, 138, 153). Secretion of degradative enzymes is another virulence property associated with hyphal growth. Several members of the secreted aspartyl proteinase (SAP) gene family (SAP4, SAP5, and SAP6) are induced during the C. albicans yeast-hyphal transition and are known to play a role in host tissue invasion (67, 96, 120). In addition to secreting SAPs, Aspergillus fumigatus, which grows vegetatively in the hyphal form, also produces elastases, enzymes that can break down elastin in lung tissue and that have been suggested to be important for invasive aspergillosis (24). Expression of genes that protect against host defenses is correlated with the hyphal morphology, as well. In C. albicans, SOD5, encoding a superoxide dismutase, is induced during hyphal growth and protects against oxidative stress (87). HYR1, another hypha-specific gene, is important for neutrophil killing and is believed to have a role in antifungal drug resistance (3, 68, 85).
Importantly, during the yeast-hyphal transition, expression of genes involved in a wide variety of virulence processes is coordinately controlled by a common set of signal transduction pathways and downstream transcriptional regulators. In C. albicans host filament-inducing conditions are known to promote hyphal growth by activation of mitogen-activated protein (MAP) kinase, cAMP-protein kinase A (PKA), and GlcNAc, as well as pH- and amino acid-sensing, pathways (10, 15). These pathways target a variety of transcriptional regulators, including Efg1, Ume6, and Nrg1, to control the expression of filament-specific transcripts (11, 35, 152). In A. fumigatus, a MAP kinase pathway has been shown to play an important role in nutrient sensing, as well as conidial germination, which is important for virulence (88, 150). Coordinated expression of genes involved in multiple virulence properties is also likely to be correlated with the hyphal morphology in a variety of additional fungal pathogens (17, 106).
EVOLUTION OF YEAST, PSEUDOHYPHAL, AND HYPHAL MORPHOLOGIES
Early evolution of morphology in fungi.
In order to better understand the evolution of morphology in fungal pathogens, it is important to first examine how morphology evolved in their ancestors. Applying dating methods based on the Sordariomycetes calibration, the first fungi are believed to have appeared approximately 1.5 billion years ago (136). Chytrids are the most ancient fungal species (63). Interestingly, having evolved in an aquatic environment, chytrid zoospores possess flagellar structures and are motile. In addition, chytrids also possess a cell wall containing chitin and can be found in both unicellular and aseptate hyphal forms (Table 1); the hyphae typically form rhizoid-like structures (58, 63, 64, 137). Both zygomycetes and glomeromycetes evolved after chytrids (63). These species are capable of forming mostly aseptate hyphae. The divergence of ascomycetes and basidiomycetes is believed to have occurred at a later point in time, and these species are capable of forming yeast, pseudohyphae, and predominantly septated hyphal morphologies (1, 136). Based on these observations, yeast and aseptate hyphae appear to have been the first fungal morphologies to have evolved. Septated hyphae, as well as pseudohyphae, most likely evolved at a later point in time. These observations also imply that most fungi once possessed basic machinery for growth in both the yeast and hyphal forms. A variety of evolutionary mechanisms, including gene loss, loss-of-function mutations, and alterations in gene expression patterns, may account for the fact that many fungi are known to grow only in one form or the other.
Table 1.
Early evolution of morphology in the fungal kingdom
| Phylum | Morphology |
|---|---|
| Chytridiomycota | Yeast, aseptate hyphae |
| Zygomycota | Yeast, predominantly aseptate hyphae |
| Glomeromycota | Yeast, predominantly aseptate hyphae |
| Basidiomycota | Yeast, pseudohyphae, predominantly septate hyphae |
| Ascomycota | Yeast, pseudohyphae, predominantly septate hyphae |
Evolution of morphology in Candida species.
Given that mammalian hosts appeared long after fungi, it is also useful to examine the more recent evolution of fungal pathogen morphology under host selective pressures. For many years, very little was known about this topic. However, several recent and important discoveries are starting to shed some light on how morphology may have evolved, at least in Candida species. In one of these experiments, expression levels of a filament-specific transcriptional regulator, UME6, were shown to be sufficient to determine C. albicans morphology in a dosage-dependent manner (19). When UME6 is not expressed, cells grow in the yeast form. Low-level UME6 expression generates a largely pseudohyphal population (the remaining cells are mostly yeast). As UME6 levels increase, cells gradually shift from pseudohyphal to hyphal morphology (hybrid pseudohyphal-hyphal filaments are observed). Finally, constitutive high-level UME6 expression is sufficient to generate a nearly complete hyphal population. Importantly, this experiment was carried out in the complete absence of filament-inducing conditions so as to exclude the possibility that morphologies were determined by environmental stimuli. Interestingly, Northern analysis indicated that a subset of filament-specific transcripts were expressed at significantly reduced levels in the largely pseudohyphal population. As UME6 levels rose, there was an increase in both the number of transcripts expressed as well as their levels of induction. These results strongly suggest that a common transcriptional mechanism specifies the shift from yeast to pseudohyphal to hyphal morphology in a dosage-dependent manner and argue against models in which pseudohyphal and hyphal growth is determined by distinct genetic mechanisms. In addition, these results suggest that in the case of Candida species, morphology may have evolved in a stepwise fashion from yeast to pseudohyphae to hyphae (7). Consistent with this hypothesis, while many Candida species are capable of forming yeast and pseudohyphae, only three species, which are phylogenetically closely related (Candida tropicalis, C. dubliniensis, and C. albicans), are known to form hyphae as well (39, 94, 135) (Table 2).
Table 2.
Morphologies of pathogenic Candida species
| Species | Morphology |
|---|---|
| C. glabrata | Yeast, pseudohyphae |
| C. lusitaniae | Yeast, pseudohyphae |
| C. guilliermondii | Yeast, pseudohyphae |
| C. parapsilosis | Yeast, pseudohyphae |
| C. tropicalis | Yeast, pseudohyphae, hyphae |
| C. dubliniensis | Yeast, pseudohyphae, hyphae |
| C. albicans | Yeast, pseudohyphae, hyphae |
The results described above may also imply, at least in the case of Candida species, that the pseudohyphal form is an intermediate morphology between yeast and hyphae. Consistent with this notion, pseudohyphae more closely resemble elongated yeast cells that fail to separate at the end of the cell cycle (108, 132). Specialized structures, such as true septa and the Spitzenkörper, are also absent in pseudohyphae but present in hyphae (23, 133). Of note, specialized structures that are unique to pseudohyphae have not yet been reported. In addition, based on C. albicans studies, weak filament-inducing conditions (e.g., high-phosphate medium) tend to induce more pseudohyphal growth, whereas stronger filament-inducing conditions, such as the combination of serum and 37°C, direct a greater proportion of cells to grow in the hyphal morphology (60, 107). Indeed, a continuum of yeast to pseudohyphal to hyphal morphologies has been observed as inducing signals increase (107, 132). These observations suggest that stepwise evolution of morphology in Candida species may have occurred in response to a variety of environmental stimuli. It is important to note that while the model yeast S. cerevisiae, a distant relative of Candida species, is known to have the ability to transition between yeast and pseudohyphae, this transition does not occur frequently and has been observed only in the presence of a very limited set of environmental cues (46). It is therefore likely that the earliest Candida species also possessed a very weak ability to form pseudohyphae and over time evolved to undergo this morphological transition more frequently in response to a broader array of host environmental conditions and niches.
While the precise mechanisms by which stepwise evolution of morphology in Candida species may have occurred are unknown, recent comparative genomic analyses suggest that gene duplication, rather than horizontal gene transfer, as well as rapidly evolving combinatorial transcription networks appear to have played important roles in the evolution of many properties associated with fungal virulence and/or adaptation to the human host (93, 139, 140). The fact that the most ancient fungi, chytrids, were capable of forming aseptate hyphae (58, 63, 64, 137) strongly suggests that hyphae may have reappeared, rather than emerged de novo, during the evolution of Candida species. Given that pseudohyphae appear to express a subset of hyphal genes at lower levels in C. albicans (19), it is reasonable to hypothesize that mutations which caused an increase in the level and duration of expression of the filamentous growth transcriptional program also played an important role in the evolution of morphology in Candida species.
Is the mechanism of dosage-dependent morphology determination evolutionarily conserved across a wide variety of fungal species? While intriguing, the answer to this question is currently unknown but under active investigation in our laboratory. However, a very recent report indicated that expression of the UME6 homolog in C. dubliniensis, a close relative of C. albicans, is sufficient to promote C. dubliniensis hyphal formation (106). Interestingly, unlike the situation in C. albicans, C.d.UME6 was found to be specifically induced in response to nutrient-poor, rather than nutrient-rich, conditions; a certain core set of filament-induced genes was also found to be conserved in both C. albicans and C. dubliniensis. These findings suggest that while components of the basic machinery important for hyphal growth may be conserved in Candida species, transcriptional regulators and signaling molecules that control the expression of this machinery appear to have been rewired during evolution to respond to specific host environmental cues. Consistent with this notion, in S. cerevisiae, a MAP kinase signal transduction pathway important for mating in haploids was shown to be rewired to mediate diploid pseudohyphal growth in response to nitrogen starvation (83). Components of the same pathway were also shown to play an important role in mediating C. albicans filamentation signals (82). Several components of a Ras/Cdc42/PAK signaling pathway originally implicated in S. cerevisiae mating and pseudohyphal growth are also known to play important roles in morphology and/or pathogenicity of the fungal pathogens C. neoformans, A. fumigatus, and C. albicans; interestingly, this pathway appears to have a conserved role in C. neoformans mating and is also involved in A. fumigatus asexual development (4, 15, 40, 41, 92, 100, 143, 147, 148). In addition to signaling pathways, several key transcriptional regulators are also conserved in various pathogenic fungi. For example, Ryp1, a key regulator of the H. capsulatum hyphal-yeast transition in response to temperature induction, is a member of a family of transcription factors that includes Wor1, an important regulator of C. albicans white-opaque switching and mating (61, 99, 126, 158). Homologs of several S. cerevisiae transcriptional regulators involved in processes not related to filamentation also appear to have been rewired over evolution to play important roles in the regulation of filamentous growth and virulence in C. albicans (5, 9, 13, 66, 70, 95, 131). It is also likely that during the evolution of Candida species certain key filamentous growth signaling and transcriptional response pathways were gradually wired to respond to an increasing repertoire of environmental cues in the host.
It is important to note that, in addition to the stepwise yeast-to-pseudohyphal-to-hyphal evolution model described above, a reductive evolution model, involving the loss of hypha-specific virulence factors, has also been suggested in the case of C. dubliniensis (62). The stepwise yeast-to-pseudohyphal-to-hyphal model also does not explain the existence of dimorphic fungal pathogens, such as H. capsulatum, in which a transition from hyphae to yeast is required for pathogenicity. Furthermore, this model does not explain the existence of fungal pathogens for which a morphological transition is not required for virulence, such as Candida glabrata, which grows primarily in the yeast form, or A. fumigatus, which grows in the hyphal form. In order to answer these questions, we need to examine how the complex relationship between fungal morphology and virulence evolved in response to different environmental selective pressures.
COEVOLUTION OF FUNGAL MORPHOLOGY AND VIRULENCE
It is important to bear in mind that morphological variation is one of many properties that have been shown to contribute to fungal virulence. In certain fungal pathogens, such as C. albicans and H. capsulatum, morphology and morphology-associated gene expression are critical for virulence, whereas in other fungal pathogens, genes important for virulence are expressed without an accompanying transition in morphological form (57). In order to better understand these observations, we need to examine the environmental and selective pressures encountered by different fungal pathogens during evolution.
Coevolution of the hyphal-to-yeast transition with virulence.
The natural reservoir for dimorphic fungal pathogens, such as H. capsulatum, C. immitis, P. brasiliensis, B. dermatitidis, P. marneffei, and S. schenkii, which require the hyphal-to-yeast transition for virulence, is the soil (71). Given that hyphal filaments allow fungi to more rapidly and effectively penetrate the soil in their search for nutrients (hyphae can also produce airborne conidia, which serve as a very effective mechanism for dispersal to new soil habitats [25]), it is logical to assume that evolutionary genetic events, which caused these fungi to grow predominantly in the hyphal form, conferred a clear selective advantage. Molds, which grow exclusively in the hyphal morphology, are the best example of soil and ambient fungi that are very well adapted to compete successfully for nutrients in the environment. It is not surprising, therefore, that dimorphic fungal pathogens, such as H. capsulatum and C. immitis, primarily grow as hyphae in their natural reservoir, the soil (78). Interestingly, however, over evolution, these pathogens have either retained or regained an ability to transition back to the yeast form. This transition occurs primarily in response to environmental conditions (e.g., body temperature, 37°C) once spores have been inhaled by the host and is believed to confer protection from killing by neutrophils, monocytes, and macrophages; the transition to yeast form also allows these pathogens to more rapidly disseminate from the lungs through the bloodstream to establish a systemic infection (71). In turn, the expression of genes encoding proteins important for additional virulence-related properties, such as the AGS1 α-(1,3)-glucan synthase (H. capsulatum) and BAD1 adhesin (B. dermatitidis), is closely orchestrated with the hyphal-to-yeast transition (71). The recent identification of DRK1, a global regulator of both the hypha-yeast transition and virulence gene expression in B. dermatitidis and H. capsulatum (a highly conserved homolog is also present in C. immitis) (71, 97), strongly suggests that these two processes have coevolved to confer a distinct selective advantage for dimorphic fungal pathogens in the host environment.
Coevolution of the yeast-pseudohyphal-hyphal transition with virulence.
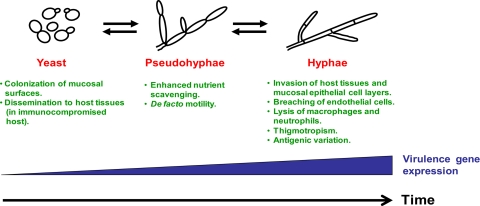
Unlike the dimorphic fungal pathogens discussed above, certain fungal pathogens, primarily Candida species, are not generally found in reservoirs outside the mammalian host. Instead, these opportunistic pathogens reside as commensals in the host oral cavity, gastrointestinal (GI) tract, skin, and/or vagina and are held at bay until the host immune system is compromised (18, 108, 109). Clearly, these species faced very different selective pressures over evolution than their distant soil-dwelling relatives. It is not surprising, therefore, that Candida species are generally found in both yeast and filamentous forms in the host environment (78). In the commensal state, the yeast form allows for rapid transport across mucosal surfaces via bodily fluids (e.g., saliva, vaginal secretions, and water in the GI tract); in contrast, the hyphal form allows for rapid attachment, biofilm formation, and nutrient scavenging in specific regions of the mucosa (108, 109). During a systemic infection, the yeast form is believed to be important for rapid dissemination to target organs via the bloodstream, and the hyphal form has been shown to play an important role in the lysis of macrophages and neutrophils, thigmotropism, biofilm formation, and immune evasion by antigenic variation, as well as the invasion of epithelial cell layers and a variety of tissues (14, 15, 44, 56, 75, 76, 90, 107–109) (Fig. 2).
Fig. 2.
Model for evolution of morphology and virulence in Candida species. In the mammalian host reservoir, the yeast form is believed to have adapted for colonization of mucosal cell surfaces in the oral cavity, gastrointestinal tract, and/or vagina. Stepwise evolution from yeast to pseudohyphae to hyphae is believed to be associated with increased virulence gene expression and the development of a variety of virulence properties. As an evolutionarily intermediate morphology, pseudohyphae were initially most likely important for promoting nutrient scavenging and motility in the host environment, although they may eventually have evolved to possess a weaker version of several virulence properties associated with hyphae. It is important to note that these morphological transitions are typically reversible and that the yeast form has also evolved to play a number of important roles in the virulence process. While the precise time points at which each morphology came to be associated with specific virulence properties are unknown, the entire time since the divergence of C. albicans (which is likely the most highly evolved Candida species) from the nonpathogenic model yeast S. cerevisiae is approximately 841 million years (54). Similar to S. cerevisiae, the most primitive Candida species are likely to have possessed a very weak ability to form pseudohyphae in response to a very limited set of environmental cues. Over evolution, Candida species are believed to have acquired the ability to form pseudohyphae more frequently and in response to a broader range of conditions in the host environment. Finally, it is important to bear in mind that not all fungal pathogens are likely to have evolved virulence properties in association with morphological shifts, as depicted in the model.
In C. albicans, virulence gene expression is closely associated with the yeast-to-hyphal transition. Several transcriptional regulators (e.g., EFG1) simultaneously control the expression of genes important for the physical process of hyphal growth, such as HGC1, as well as genes involved in a wide variety of additional virulence processes, including adhesion and secretion of degradative enzymes (11, 15, 35, 81, 154). Strains deleted for these transcription factors are both defective for filamentous growth and highly attenuated for virulence. A strain overexpressing a key transcriptional repressor of filament-specific genes, NRG1, is locked in the yeast form, shows reduced expression of filament-specific genes, and is highly attenuated for virulence (121, 141). In contrast, a strain expressing high constitutive levels of a transcriptional regulator important for hyphal extension, UME6, causes increased hyphal formation and elevated expression of filament-specific genes and promotes virulence in a mouse model of systemic infection. However, the hyphal morphology is not sufficient for virulence, since the initial inoculum of this strain consisted of yeast cells (19, 152). Combined, these results provide strong evidence for coevolution of the Candida yeast-hyphal transition with virulence.
While the evolutionary advantage of the yeast-hyphal transition for virulence is clear, what is the evolutionary significance and advantage of the pseudohyphal morphology? It is important to note that a variety of significantly less pathogenic Candida species are primarily found in infected tissues as a combination of yeast and pseudohyphae (e.g., C. tropicalis, Candida parapsilosis, Candida guilliermondii, and Candida lusitaniae); indeed, a number of these species either rarely or never appear to form hyphae (77, 135). Recent completion of the genome sequences for several of these species (17) has indicated that many, but not all, of the genes expressed during the yeast-to-filament transition in C. albicans are evolutionarily conserved. We have observed that approximately 85% of the genes originally identified in the C. albicans filamentous-growth transcriptional program (67) have homologs in non-albicans Candida species. Interestingly, a comparative genomic analysis has revealed that Candida species show significant expansion, relative to S. cerevisiae, in a number of gene families associated with virulence, including those encoding cell wall components, secreted proteins, and transporters involved in a variety of processes, such as nutrient acquisition, nutrient uptake, and energy production (17). C. albicans and C. tropicalis, both of which form hyphae, appear to show greater expansion in the number of genes in these families relative to that of other Candida species examined. Consistent with this observation, many less pathogenic non-albicans Candida species, which are only capable of forming yeast and pseudohyphae, have a reduced ability to adhere to buccal epithelial cells and vascular endothelial cells, as well as reduced protease secretion. In contrast, hypha-forming C. albicans, C. dubliniensis, and C. tropicalis appear to have an increased ability to adhere to host cells and secrete proteases relative to other Candida species, which lack the ability to form hyphae (94). As indicated previously, a recent study has also shown that in C. albicans, pseudohyphae appear to express a subset of hyphal genes, including known virulence factors, at significantly reduced levels (19). Combined, these observations are consistent with the stepwise yeast-pseudohyphal-hyphal model for the evolution of morphology in Candida species (Fig. 2) and suggest that the pseudohyphal morphology may be correlated with low-level virulence and reduced expression of genes important for a variety of virulence-related processes. Based on the role of pseudohyphae in S. cerevisiae (46), the initial shift toward increased pseudohyphal growth during the evolution of Candida species is likely to have facilitated nutrient scavenging and promoted de facto motility in the host environment. Over time, it is likely that pseudohyphae may have also acquired a weaker version of several, but most likely not all, of the virulence properties associated with hyphae. Why has the pseudohyphal morphology been conserved throughout evolution in one of the most virulent fungal pathogens, C. albicans? One possibility is that specific microenvironments and niches within the host may require reduced levels of invasion and virulence factor expression. Transitioning to the pseudohyphal morphology may allow C. albicans to modulate its invasion and virulence activity in tissues where nutrient levels and other host resources are not optimal at given time points during infection. In a sense, the pseudohyphal morphology may therefore serve as an evolutionary adaptation for the development of measured niche-specific invasion patterns. Consistent with this hypothesis, certain Candida species that are only capable of forming yeast and pseudohyphae appear to more frequently infect specific sites in the human host (86, 94, 105).
EVOLUTION OF FUNGAL VIRULENCE NOT ASSOCIATED WITH A MORPHOLOGICAL SHIFT
As we discussed above, certain fungal pathogens grow predominantly in the yeast or hyphal form both in their natural reservoirs and during infection in the host. C. glabrata is primarily a yeast form pathogen that resides in host mucosal cavities (27). This pathogen has evolved to express a large family of adhesins important for biofilm formation and virulence in response to niche-specific host environmental cues, such as nicotinic acid limitation in the urogenital tract (69). A. fumigatus, a filamentous fungus that grows vegetatively in the hyphal form, has evolved a variety of virulence traits that are not associated with a morphological shift, including melanin production and thermotolerance, as well as the secretion of proteinases and toxins (24, 38). Interestingly, C. neoformans, a predominantly yeast form pathogen, shares several of these virulence properties (melanin production and the ability to grow well at 37°C) and has also evolved distinct features, such as capsule formation, that promote survival in the host environment (57, 73, 78). Similar to the case of C. albicans, expression of genes important for a number of virulence properties in C. neoformans, including capsule synthesis, is induced in response to body temperature (37°C). However, unlike C. albicans, C. neoformans does not undergo a yeast-to-hyphal transition under these conditions; instead, C. neoformans hyphal growth is initiated during the mating and monokaryotic fruiting processes, which do not occur in vivo (57, 78). These observations suggest that while pathogenic fungi are evolutionarily wired to respond to similar host signals, these signals may trigger different virulence mechanisms. It is important to bear in mind that morphological change is one of many mechanisms that are known to play a role in virulence. Several virulence properties appear to have coevolved with morphological shifts, whereas others have not. Ultimately, selective pressures in both the natural reservoir and the host environment are likely to have played a role in determining whether morphology and virulence coevolved in specific fungal pathogens. In the case of Candida species, these pressures were encountered exclusively in the mammalian host. While our knowledge of the evolutionary relationship between fungal morphology and virulence remains limited, it is hoped that future work, including more extensive cross-species comparative and functional analyses based on newly sequenced fungal pathogen genomes, will shed more light on this important area of study.
ACKNOWLEDGMENTS
We thank Erika Lackey and Brian Wickes for useful comments and suggestions on the manuscript.
This work was supported by a COSTAR predoctoral fellowship (National Institute of Dental and Craniofacial Research [NIDCR] grant T32DE14318-07) to D.S.T. and a Ruth L. Kirschstein National Research Service Award for Individual Predoctoral Fellows (NIDCR grant F31DE020214-02) to P.L.C. and by grants 1R56AI072705 and 1RO1AI083344 from the National Institute of Allergy and Infectious Diseases to D.K.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases, the National Institute of Dental and Craniofacial Research, or the National Institutes of Health.
Footnotes
Published ahead of print on 15 July 2011.
REFERENCES
- 1. Alexopoulos C. J., Mims C. W. 1979. Introductory mycology. Wiley, New York, NY [Google Scholar]
- 2. Alspaugh J. A., Davidson R. C., Heitman J. 2000. Morphogenesis of Cryptococcus neoformans. Contrib. Microbiol. 5:217–238 [DOI] [PubMed] [Google Scholar]
- 3. Bailey D. A., Feldmann P. J., Bovey M., Gow N. A., Brown A. J. 1996. The Candida albicans HYR1 gene, which is activated in response to hyphal development, belongs to a gene family encoding yeast cell wall proteins. J. Bacteriol. 178:5353–5360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ballou E. R., Nichols C. B., Miglia K. J., Kozubowski L., Alspaugh J. A. 2010. Two CDC42 paralogues modulate Cryptococcus neoformans thermotolerance and morphogenesis under host physiological conditions. Mol. Microbiol. 75:763–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Banerjee M., et al. 2008. UME6, a novel filament-specific regulator of Candida albicans hyphal extension and virulence. Mol. Biol. Cell 19:1354–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barelle C. J., et al. 2003. Asynchronous cell cycle and asymmetric vacuolar inheritance in true hyphae of Candida albicans. Eukaryot. Cell 2:398–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bastidas R. J., Heitman J. 2009. Trimorphic stepping stones pave the way to fungal virulence. Proc. Natl. Acad. Sci. U. S. A. 106:351–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Batanghari J. W., Deepe G. S., Jr., Di Cera E., Goldman W. E. 1998. Histoplasma acquisition of calcium and expression of CBP1 during intracellular parasitism. Mol. Microbiol. 27:531–539 [DOI] [PubMed] [Google Scholar]
- 9. Berkey C. D., Vyas V. K., Carlson M. 2004. Nrg1 and Nrg2 transcriptional repressors are differently regulated in response to carbon source. Eukaryot. Cell 3:311–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Biswas K., Morschhauser J. 2005. The Mep2p ammonium permease controls nitrogen starvation-induced filamentous growth in Candida albicans. Mol. Microbiol. 56:649–669 [DOI] [PubMed] [Google Scholar]
- 11. Biswas S., Van Dijck P., Datta A. 2007. Environmental sensing and signal transduction pathways regulating morphopathogenic determinants of Candida albicans. Microbiol. Mol. Biol. Rev. 71:348–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brandhorst T. T., Wuthrich M., Finkel-Jimenez B., Warner T., Klein B. S. 2004. Exploiting type 3 complement receptor for TNF-alpha suppression, immune evasion, and progressive pulmonary fungal infection. J. Immunol. 173:7444–7453 [DOI] [PubMed] [Google Scholar]
- 13. Braun B. R., Kadosh D., Johnson A. D. 2001. NRG1, a repressor of filamentous growth in C. albicans, is down-regulated during filament induction. EMBO J. 20:4753–4761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brown A. J. 2002. Expression of growth form-specific factors during morphogenesis in Candida albicans, p. 87–93 In Calderone R. A. (ed.), Candida and candidiasis. ASM Press, Washington, DC [Google Scholar]
- 15. Brown A. J. 2002. Morphogenetic signaling pathways in Candida albicans, p. 95–106 In Calderone R. A. (ed.), Candida and candidiasis. ASM Press, Washington, DC [Google Scholar]
- 16. Bullock W. E. 1993. Interactions between human phagocytic cells and Histoplasma capsulatum. Arch. Med. Res. 24:219–223 [PubMed] [Google Scholar]
- 17. Butler G., et al. 2009. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459:657–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Calderone R. A. 2002. Introduction and historical perspectives, p. 3–13 In Calderone R. A. (ed.), Candida and candidiasis. ASM Press, Washington, DC [Google Scholar]
- 19. Carlisle P. L., et al. 2009. Expression levels of a filament-specific transcriptional regulator are sufficient to determine Candida albicans morphology and virulence. Proc. Natl. Acad. Sci. U. S. A. 106:599–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chabasse D., Bouchara J. P., de Gentile L., Chennebault J. M. 1988. Candida albicans chlamydospores observed in vivo in a patient with AIDS. Ann. Biol. Clin. (Paris) 46:817–818 [PubMed] [Google Scholar]
- 21. Cheng J., Park T. S., Fischl A. S., Ye X. S. 2001. Cell cycle progression and cell polarity require sphingolipid biosynthesis in Aspergillus nidulans. Mol. Cell. Biol. 21:6198–6209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cole G. T., Seshan K. R., Phaneuf M., Lynn K. T. 1991. Chlamydospore-like cells of Candida albicans in the gastrointestinal tract of infected, immunocompromised mice. Can. J. Microbiol. 37:637–646 [DOI] [PubMed] [Google Scholar]
- 23. Crampin H., et al. 2005. Candida albicans hyphae have a Spitzenkörper that is distinct from the polarisome found in yeast and pseudohyphae. J. Cell Sci. 118:2935–2947 [DOI] [PubMed] [Google Scholar]
- 24. Dagenais T. R., Keller N. P. 2009. Pathogenesis of Aspergillus fumigatus in invasive aspergillosis. Clin. Microbiol. Rev. 22:447–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Deacon J. W. 2006. Fungal biology. Wiley-Blackwell, Oxford, United Kingdom [Google Scholar]
- 26. Del Poeta M. 2004. Role of phagocytosis in the virulence of Cryptococcus neoformans. Eukaryot. Cell 3:1067–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Do Carmo-Sousa L. 1969. Distribution of yeasts in nature, p. 79–105 In Rose A. H., Harrison J. S. (ed.), The yeasts, vol. 1. Academic Press, New York, NY [Google Scholar]
- 28. Drutz D. J., Frey C. L. 1985. Intracellular and extracellular defenses of human phagocytes against Blastomyces dermatitidis conidia and yeasts. J. Lab. Clin. Med. 105:737–750 [PubMed] [Google Scholar]
- 29. D'Souza C. A., et al. 2001. Cyclic AMP-dependent protein kinase controls virulence of the fungal pathogen Cryptococcus neoformans. Mol. Cell. Biol. 21:3179–3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Edmond M. B., et al. 1999. Nosocomial bloodstream infections in United States hospitals: a three-year analysis. Clin. Infect. Dis. 29:239–244 [DOI] [PubMed] [Google Scholar]
- 31. Egel R. 2004. The molecular biology of Schizosaccharomyces pombe. Springer-Verlag, Berlin, Germany [Google Scholar]
- 32. Eissenberg L. G., Goldman W. E. 1991. Histoplasma variation and adaptive strategies for parasitism: new perspectives on histoplasmosis. Clin. Microbiol. Rev. 4:411–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Eissenberg L. G., Goldman W. E. 1994. The interplay between Histoplasma capsulatum and its host cells. Balliere's Clin. Infect. Dis. 1:265–283 [Google Scholar]
- 34. Ene I. V., Bennett R. J. 2009. Hwp1 and related adhesins contribute to both mating and biofilm formation in Candida albicans. Eukaryot. Cell 8:1909–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ernst J. F. 2000. Transcription factors in Candida albicans—environmental control of morphogenesis. Microbiology 146:1763–1774 [DOI] [PubMed] [Google Scholar]
- 36. Feldmann H. 2010. Yeast: molecular and cellular biology. Wiley-VCH Verlag GmbH, Weinheim, Germany [Google Scholar]
- 37. Feldmesser M., Tucker S., Casadevall A. 2001. Intracellular parasitism of macrophages by Cryptococcus neoformans. Trends Microbiol. 9:273–278 [DOI] [PubMed] [Google Scholar]
- 38. Filler S. G., Sheppard D. C. 2006. Fungal invasion of normally non-phagocytic host cells. PLoS Pathog. 2:e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fitzpatrick D. A., Butler G. 2010. Comparative genomic analysis of pathogenic yeasts and the evolution of virulence, p. 1–15 In Ashbee H. R., Bignell E. M. (ed.), Pathogenic yeasts, the yeast handbook. Springer-Verlag, Berlin, Germany [Google Scholar]
- 40. Fortwendel J. R., et al. 2008. Aspergillus fumigatus RasA regulates asexual development and cell wall integrity. Eukaryot. Cell 7:1530–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fortwendel J. R., et al. 2005. A fungus-specific ras homolog contributes to the hyphal growth and virulence of Aspergillus fumigatus. Eukaryot. Cell 4:1982–1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fraser R. S. 1993. Pulmonary aspergillosis: pathologic and pathogenetic features. Pathol. Annu. 28:231–277 [PubMed] [Google Scholar]
- 43. Frater J. L., Hall G. S., Procop G. W. 2001. Histologic features of zygomycosis: emphasis on perineural invasion and fungal morphology. Arch. Pathol. Lab. Med. 125:375–378 [DOI] [PubMed] [Google Scholar]
- 44. Gantner B. N., Simmons R. M., Underhill D. M. 2005. Dectin-1 mediates macrophage recognition of Candida albicans yeast but not filaments. EMBO J. 24:1277–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Giles S. S., Dagenais T. R., Botts M. R., Keller N. P., Hull C. M. 2009. Elucidating the pathogenesis of spores from the human fungal pathogen Cryptococcus neoformans. Infect. Immun. 77:3491–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gimeno C. J., Ljungdahl P. O., Styles C. A., Fink G. R. 1992. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell 68:1077–1090 [DOI] [PubMed] [Google Scholar]
- 47. Gonzalez-Novo A., et al. 2008. Sep7 is essential to modify septin ring dynamics and inhibit cell separation during Candida albicans hyphal growth. Mol. Biol. Cell 19:1509–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gow N. A. 1994. Growth and guidance of the fungal hypha. Microbiology 140:3193–3205 [DOI] [PubMed] [Google Scholar]
- 49. Gow N. A. 1997. Germ tube growth of Candida albicans. Curr. Top. Med. Mycol. 8:43–55 [PubMed] [Google Scholar]
- 50. Gow N. A., Brown A. J., Odds F. C. 2002. Fungal morphogenesis and host invasion. Curr. Opin. Microbiol. 5:366–371 [DOI] [PubMed] [Google Scholar]
- 51. Gow N. A., Gooday G. W. 1984. A model for the germ tube formation and mycelial growth form of Candida albicans. Sabouraudia 22:137–144 [DOI] [PubMed] [Google Scholar]
- 52. Gow N. A., Henderson G., Gooday G. W. 1986. Cytological interrelationships between the cell cycle and duplication cycle of Candida albicans. Microbios 47:97–105 [PubMed] [Google Scholar]
- 53. Gow N. A., et al. 1994. Investigation of touch-sensitive responses by hyphae of the human pathogenic fungus Candida albicans. Scanning Microsc. 8:705–710 [PubMed] [Google Scholar]
- 54. Heckman D. S., et al. 2001. Molecular evidence for the early colonization of land by fungi and plants. Science 293:1129–1133 [DOI] [PubMed] [Google Scholar]
- 55. Hector R. F., Laniado-Laborin R. 2005. Coccidioidomycosis—a fungal disease of the Americas. PLoS Med. 2:e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Heinsbroek S. E., Brown G. D., Gordon S. 2005. Dectin-1 escape by fungal dimorphism. Trends Immunol. 26:352–354 [DOI] [PubMed] [Google Scholar]
- 57. Heitman J., Filler S. G., Edwards J. E., Mitchell A. P. E. 2006. Molecular principles of fungal pathogenesis. ASM Press, Washington, DC [Google Scholar]
- 58. Hibbett D. S., et al. 2007. A higher-level phylogenetic classification of the Fungi. Mycol. Res. 111:509–547 [DOI] [PubMed] [Google Scholar]
- 59. Hogan L. H., Klein B. S. 1994. Altered expression of surface alpha-1,3-glucan in genetically related strains of Blastomyces dermatitidis that differ in virulence. Infect. Immun. 62:3543–3546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hornby J. M., Dumitru R., Nickerson K. W. 2004. High phosphate (up to 600 mM) induces pseudohyphal development in five wild type Candida albicans. J. Microbiol. Methods 56:119–124 [DOI] [PubMed] [Google Scholar]
- 61. Huang G., et al. 2006. Bistable expression of WOR1, a master regulator of white-opaque switching in Candida albicans. Proc. Natl. Acad. Sci. U. S. A. 103:12813–12818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jackson A. P., et al. 2009. Comparative genomics of the fungal pathogens Candida dubliniensis and Candida albicans. Genome Res. 19:2231–2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. James T. Y., et al. 2006. Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature 443:818–822 [DOI] [PubMed] [Google Scholar]
- 64. James T. Y., et al. 2006. A molecular phylogeny of the flagellated fungi (Chytridiomycota) and description of a new phylum (Blastocladiomycota). Mycologia 98:860–871 [DOI] [PubMed] [Google Scholar]
- 65. Jong A. Y., Stins M. F., Huang S. H., Chen S. H., Kim K. S. 2001. Traversal of Candida albicans across human blood-brain barrier in vitro. Infect. Immun. 69:4536–4544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kadosh D., Johnson A. D. 2001. Rfg1, a protein related to the Saccharomyces cerevisiae hypoxic regulator Rox1, controls filamentous growth and virulence in Candida albicans. Mol. Cell. Biol. 21:2496–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kadosh D., Johnson A. D. 2005. Induction of the Candida albicans filamentous growth program by relief of transcriptional repression: a genome-wide analysis. Mol. Biol. Cell 16:2903–2912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Karababa M., Coste A. T., Rognon B., Bille J., Sanglard D. 2004. Comparison of gene expression profiles of Candida albicans azole-resistant clinical isolates and laboratory strains exposed to drugs inducing multidrug transporters. Antimicrob. Agents Chemother. 48:3064–3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kaur R., Domergue R., Zupancic M. L., Cormack B. P. 2005. A yeast by any other name: Candida glabrata and its interaction with the host. Curr. Opin. Microbiol. 8:378–384 [DOI] [PubMed] [Google Scholar]
- 70. Khalaf R. A., Zitomer R. S. 2001. The DNA binding protein Rfg1 is a repressor of filamentation in Candida albicans. Genetics 157:1503–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Klein B. S., Tebbets B. 2007. Dimorphism and virulence in fungi. Curr. Opin. Microbiol. 10:314–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Korting H. C., et al. 2003. Reduced expression of the hyphal-independent Candida albicans proteinase genes SAP1 and SAP3 in the efg1 mutant is associated with attenuated virulence during infection of oral epithelium. J. Med. Microbiol. 52:623–632 [DOI] [PubMed] [Google Scholar]
- 73. Kozubowski L., Lee S. C., Heitman J. 2009. Signalling pathways in the pathogenesis of Cryptococcus. Cell Microbiol. 11:370–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kraus P. R., et al. 2004. Identification of Cryptococcus neoformans temperature-regulated genes with a genomic-DNA microarray. Eukaryot. Cell 3:1249–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kumamoto C. A., Vinces M. D. 2005. Alternative Candida albicans lifestyles: growth on surfaces. Annu. Rev. Microbiol. 59:113–133 [DOI] [PubMed] [Google Scholar]
- 76. Kumamoto C. A., Vinces M. D. 2005. Contributions of hyphae and hypha-co-regulated genes to Candida albicans virulence. Cell Microbiol. 7:1546–1554 [DOI] [PubMed] [Google Scholar]
- 77. Larone D. H. 1995. Medically important fungi—a guide to identification. ASM Press, Washington, DC [Google Scholar]
- 78. Lin X. 2009. Cryptococcus neoformans: morphogenesis, infection, and evolution. Infect. Genet. Evol. 9:401–416 [DOI] [PubMed] [Google Scholar]
- 79. Lin X., Heitman J. 2005. Chlamydospore formation during hyphal growth in Cryptococcus neoformans. Eukaryot. Cell 4:1746–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Lin X., Heitman J. 2006. The biology of the Cryptococcus neoformans species complex. Annu. Rev. Microbiol. 60:69–105 [DOI] [PubMed] [Google Scholar]
- 81. Liu H. 2001. Transcriptional control of dimorphism in Candida albicans. Curr. Opin. Microbiol. 4:728–735 [DOI] [PubMed] [Google Scholar]
- 82. Liu H., Kohler J., Fink G. R. 1994. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 266:1723–1726 [DOI] [PubMed] [Google Scholar]
- 83. Liu H., Styles C. A., Fink G. R. 1993. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science 262:1741–1744 [DOI] [PubMed] [Google Scholar]
- 84. Lo H. J., et al. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90:939–949 [DOI] [PubMed] [Google Scholar]
- 85. Luo G., et al. 2010. Candida albicans Hyr1p confers resistance to neutrophil killing and is a potential vaccine target. J. Infect. Dis. 201:1718–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Mardani M., Hanna H. A., Girgawy E., Raad I. 2000. Nosocomial Candida guilliermondii fungemia in cancer patients. Infect. Control Hosp. Epidemiol. 21:336–337 [DOI] [PubMed] [Google Scholar]
- 87. Martchenko M., Alarco A. M., Harcus D., Whiteway M. 2004. Superoxide dismutases in Candida albicans: transcriptional regulation and functional characterization of the hyphal-induced SOD5 gene. Mol. Biol. Cell 15:456–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. May G. S., Xue T., Kontoyiannis D. P., Gustin M. C. 2005. Mitogen activated protein kinases of Aspergillus fumigatus. Med. Mycol. 43(Suppl. 1):S83–S86 [DOI] [PubMed] [Google Scholar]
- 89. Medoff G., Kobayashi G. S., Painter A., Travis S. 1987. Morphogenesis and pathogenicity of Histoplasma capsulatum. Infect. Immun. 55:1355–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Mitchell A. P. 1998. Dimorphism and virulence in Candida albicans. Curr. Opin. Microbiol. 1:687–692 [DOI] [PubMed] [Google Scholar]
- 91. Miyaji M., Nishimura K. 1977. Investigation on dimorphism of Blastomyces dermatitidis by agar-implantation method. Mycopathologia 60:73–78 [DOI] [PubMed] [Google Scholar]
- 92. Momany M. 2005. Growth control and polarization. Med. Mycol. 43(Suppl. 1):S23–S25 [DOI] [PubMed] [Google Scholar]
- 93. Moran G. P., Coleman D. C., Sullivan D. J. 2011. Comparative genomics and the evolution of pathogenicity in human pathogenic fungi. Eukaryot. Cell 10:34–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Moran G. P., Sullivan D. J., Coleman D. C. 2002. Emergence of non-Candida albicans Candida species as pathogens, p. 37–54 In Calderone R. A. (ed.), Candida and candidiasis. ASM Press, Washinton, DC [Google Scholar]
- 95. Murad A. M. A., et al. 2001. NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J. 20:4742–4752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Nantel A., et al. 2002. Transcription profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Mol. Biol. Cell 13:3452–3465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Nemecek J. C., Wuthrich M., Klein B. S. 2006. Global control of dimorphism and virulence in fungi. Science 312:583–588 [DOI] [PubMed] [Google Scholar]
- 98. Newman S. L., Chaturvedi S., Klein B. S. 1995. The WI-1 antigen of Blastomyces dermatitidis yeasts mediates binding to human macrophage CD11b/CD18 (CR3) and CD14. J. Immunol. 154:753–761 [PubMed] [Google Scholar]
- 99. Nguyen V. Q., Sil A. 2008. Temperature-induced switch to the pathogenic yeast form of Histoplasma capsulatum requires Ryp1, a conserved transcriptional regulator. Proc. Natl. Acad. Sci. U. S. A. 105:4880–4885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Nichols C. B., Perfect Z. H., Alspaugh J. A. 2007. A Ras1-Cdc24 signal transduction pathway mediates thermotolerance in the fungal pathogen Cryptococcus neoformans. Mol. Microbiol. 63:1118–1130 [DOI] [PubMed] [Google Scholar]
- 101. Nobile C. J., Mitchell A. P. 2006. Genetics and genomics of Candida albicans biofilm formation. Cell Microbiol. 8:1382–1391 [DOI] [PubMed] [Google Scholar]
- 102. Nobile C. J., Nett J. E., Andes D. R., Mitchell A. P. 2006. Function of Candida albicans adhesin Hwp1 in biofilm formation. Eukaryot. Cell 5:1604–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Nobile C. J., et al. 2008. Complementary adhesin function in C. albicans biofilm formation. Curr. Biol. 18:1017–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Noble S. M., French S., Kohn L. A., Chen V., Johnson A. D. 2010. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat. Genet. 42:590–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Nosek J., Holesova Z., Kosa P., Gacser A., Tomaska L. 2009. Biology and genetics of the pathogenic yeast Candida parapsilosis. Curr. Genet. 55:497–509 [DOI] [PubMed] [Google Scholar]
- 106. O'Connor L., Caplice N., Coleman D. C., Sullivan D. J., Moran G. P. 2010. Differential filamentation of Candida albicans and Candida dubliniensis is governed by nutrient regulation of UME6 expression. Eukaryot. Cell 9:1383–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Odds F. C. 1985. Morphogenesis in Candida albicans. Crit. Rev. Microbiol. 12:45–93 [DOI] [PubMed] [Google Scholar]
- 108. Odds F. C. 1988. Candida and candidosis. Baillière Tindall, London, United Kingdom [Google Scholar]
- 109. Odds F. C. 1994. Pathogenesis of Candida infections. J. Am. Acad. Dermatol. 31:S2–S5 [DOI] [PubMed] [Google Scholar]
- 110. Perfect J. R. 2006. Cryptococcus neoformans: a sugar-coated killer, p. 281–303 In Heitman J., Filler S. G., Edwards J. E., Mitchell A. P. E. (ed.), Molecular principles of fungal pathogenesis. ASM Press, Washington, DC [Google Scholar]
- 111. Potter B. 1966. The journal of Beatrix Potter, 1881-1897. Frederick Warne, London, United Kingdom [Google Scholar]
- 112. Potter B. 1996. Les champignons. Bibliotheque de L'Image, Paris, France [Google Scholar]
- 113. Rappleye C. A., Eissenberg L. G., Goldman W. E. 2007. Histoplasma capsulatum alpha-(1,3)-glucan blocks innate immune recognition by the beta-glucan receptor. Proc. Natl. Acad. Sci. U. S. A. 104:1366–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Rappleye C. A., Engle J. T., Goldman W. E. 2004. RNA interference in Histoplasma capsulatum demonstrates a role for alpha-(1,3)-glucan in virulence. Mol. Microbiol. 53:153–165 [DOI] [PubMed] [Google Scholar]
- 115. Rasmussen C. D., Lu K. P., Means R. L., Means A. R. 1992. Calmodulin and cell cycle control. J. Physiol. Paris. 86:83–88 [DOI] [PubMed] [Google Scholar]
- 116. Rosas A. L., et al. 2000. Synthesis of polymerized melanin by Cryptococcus neoformans in infected rodents. Infect. Immun. 68:2845–2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Salas S. D., Bennett J. E., Kwon-Chung K. J., Perfect J. R., Williamson P. R. 1996. Effect of the laccase gene CNLAC1, on virulence of Cryptococcus neoformans. J. Exp. Med. 184:377–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Salazar M. E., Restrepo A. 1985. Morphogenesis of the mycelium-to-yeast transformation in Paracoccidioides brasiliensis. Sabouraudia 23:7–11 [DOI] [PubMed] [Google Scholar]
- 119. San-Blas G., et al. 2000. Fungal morphogenesis and virulence. Med. Mycol. 38:79–86 [PubMed] [Google Scholar]
- 120. Sanglard D., Hube B., Monod M., Odds F. C., Gow N. A. 1997. A triple deletion of the secreted aspartyl proteinase genes SAP4, SAP5, and SAP6 of Candida albicans causes attenuated virulence. Infect. Immun. 65:3539–3546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Saville S. P., Lazzell A. L., Monteagudo C., López-Ribot J. L. 2003. Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during infection. Eukaryot. Cell 2:1053–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Sebghati T. S., Engle J. T., Goldman W. E. 2000. Intracellular parasitism by Histoplasma capsulatum: fungal virulence and calcium dependence. Science 290:1368–1372 [DOI] [PubMed] [Google Scholar]
- 123. Shaukat A., et al. 2005. Invasive filamentous fungal infections in allogeneic hematopoietic stem cell transplant recipients after recovery from neutropenia: clinical, radiologic, and pathologic characteristics. Mycopathologia 159:181–188 [DOI] [PubMed] [Google Scholar]
- 124. Simon G., Galgoczy J. 1986. Chlamydospores of dermatophytes. Mykosen 29:469–473 [DOI] [PubMed] [Google Scholar]
- 125. Sinha I., et al. 2007. Cyclin-dependent kinases control septin phosphorylation in Candida albicans hyphal development. Dev. Cell 13:421–432 [DOI] [PubMed] [Google Scholar]
- 126. Srikantha T., et al. 2006. TOS9 regulates white-opaque switching in Candida albicans. Eukaryot. Cell 5:1674–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Staab J. F., Bradway S. D., Fidel P. L., Sundstrom P. 1999. Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwp1. Science 283:1535–1538 [DOI] [PubMed] [Google Scholar]
- 128. Staib P., Morschhauser J. 2007. Chlamydospore formation in Candida albicans and Candida dubliniensis—an enigmatic developmental programme. Mycoses 50:1–12 [DOI] [PubMed] [Google Scholar]
- 129. Steinberg G. 2007. Hyphal growth: a tale of motors, lipids, and the Spitzenkorper. Eukaryot. Cell 6:351–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Stevenson I. L., Becker S. A. 1972. The fine structure and development of chlamydospores of Fusarium oxysporum. Can. J. Microbiol. 18:997–1002 [DOI] [PubMed] [Google Scholar]
- 131. Strich R., et al. 1994. UME6 is a key regulator of nitrogen repression and meiotic development. Genes Dev. 8:796–810 [DOI] [PubMed] [Google Scholar]
- 132. Sudbery P., Gow N., Berman J. 2004. The distinct morphogenic states of Candida albicans. Trends Microbiol. 12:317–324 [DOI] [PubMed] [Google Scholar]
- 133. Sudbery P. E. 2001. The germ tubes of Candida albicans hyphae and pseudohyphae show different patterns of septin ring localization. Mol. Microbiol. 41:19–31 [DOI] [PubMed] [Google Scholar]
- 134. Sukroongreung S., Kitiniyom K., Nilakul C., Tantimavanich S. 1998. Pathogenicity of basidiospores of Filobasidiella neoformans var. neoformans. Med. Mycol. 36:419–424 [PubMed] [Google Scholar]
- 135. Sutton D. A., Fothergill A. W., Rinaldi M. G. 1998. Guide to clinically significant fungi. Williams & Wilkins, Baltimore, MD [Google Scholar]
- 136. Taylor J. W., Berbee M. L. 2006. Dating divergences in the Fungal Tree of Life: review and new analyses. Mycologia 98:838–849 [DOI] [PubMed] [Google Scholar]
- 137. Taylor T. N., Remy W. 1994. Allomyces in the Devonian. Nature 367:601 [Google Scholar]
- 138. Tsuchimori N., et al. 2000. Reduced virulence of HWP1-deficient mutants of Candida albicans and their interactions with host cells. Infect. Immun. 68:1997–2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Tuch B. B., Galgoczy D. J., Hernday A. D., Li H., Johnson A. D. 2008. The evolution of combinatorial gene regulation in fungi. PLoS Biol. 6:e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Tuch B. B., Li H., Johnson A. D. 2008. Evolution of eukaryotic transcription circuits. Science 319:1797–1799 [DOI] [PubMed] [Google Scholar]
- 141. Uppuluri P., et al. 2010. The transcriptional regulator Nrg1p controls Candida albicans biofilm formation and dispersion. Eukaryot. Cell 9:1531–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Urban C. F., Lourido S., Zychlinsky A. 2006. How do microbes evade neutrophil killing? Cell Microbiol. 8:1687–1696 [DOI] [PubMed] [Google Scholar]
- 143. Vallim M. A., Nichols C. B., Fernandes L., Cramer K. L., Alspaugh J. A. 2005. A Rac homolog functions downstream of Ras1 to control hyphal differentiation and high-temperature growth in the pathogenic fungus Cryptococcus neoformans. Eukaryot. Cell 4:1066–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Wang A., Raniga P. P., Lane S., Lu Y., Liu H. 2009. Hyphal chain formation in Candida albicans: Cdc28-Hgc1 phosphorylation of Efg1 represses cell separation genes. Mol. Cell. Biol. 29:4406–4416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Wang Y. 2009. CDKs and the yeast-hyphal decision. Curr. Opin. Microbiol. 12:644–649 [DOI] [PubMed] [Google Scholar]
- 146. Warenda A. J., Konopka J. B. 2002. Septin function in Candida albicans morphogenesis. Mol. Biol. Cell 13:2732–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Waugh M. S., et al. 2002. Ras1 and Ras2 contribute shared and unique roles in physiology and virulence of Cryptococcus neoformans. Microbiology 148:191–201 [DOI] [PubMed] [Google Scholar]
- 148. Waugh M. S., Vallim M. A., Heitman J., Alspaugh J. A. 2003. Ras1 controls pheromone expression and response during mating in Cryptococcus neoformans. Fungal Genet. Biol. 38:110–121 [DOI] [PubMed] [Google Scholar]
- 149. Weig M., Gross U., Muhlschlegel F. 1998. Clinical aspects and pathogenesis of Candida infection. Trends Microbiol. 6:468–470 [DOI] [PubMed] [Google Scholar]
- 150. Xue T., Nguyen C. K., Romans A., May G. S. 2004. A mitogen-activated protein kinase that senses nitrogen regulates conidial germination and growth in Aspergillus fumigatus. Eukaryot. Cell 3:557–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Yokoyama K., Takeo K. 1983. Differences of asymmetrical division between the pseudomycelial and yeast forms of Candida albicans and their effect on multiplication. Arch. Microbiol. 134:251–253 [DOI] [PubMed] [Google Scholar]
- 152. Zeidler U., et al. 2009. UME6 is a crucial downstream target of other transcriptional regulators of true hyphal development in Candida albicans. FEMS Yeast Res. 9:126–142 [DOI] [PubMed] [Google Scholar]
- 153. Zhao X., et al. 2004. ALS3 and ALS8 represent a single locus that encodes a Candida albicans adhesin; functional comparisons between Als3p and Als1p. Microbiology 150:2415–2428 [DOI] [PubMed] [Google Scholar]
- 154. Zheng X., Wang Y., Wang Y. 2004. Hgc1, a novel hypha-specific G1 cyclin-related protein regulates Candida albicans hyphal morphogenesis. EMBO J. 23:1845–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Zheng X. D., Lee R. T., Wang Y. M., Lin Q. S., Wang Y. 2007. Phosphorylation of Rga2, a Cdc42 GAP, by CDK/Hgc1 is crucial for Candida albicans hyphal growth. EMBO J. 26:3760–3769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Zimmer B. L., Hempel H. O., Goodman N. L. 1984. Pathogenicity of the basidiospores of Filobasidiella neoformans. Mycopathologia 85:149–153 [DOI] [PubMed] [Google Scholar]
- 157. Zink S., Nass T., Rosen P., Ernst J. F. 1996. Migration of the fungal pathogen Candida albicans across endothelial monolayers. Infect. Immun. 64:5085–5091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Zordan R. E., Galgoczy D. J., Johnson A. D. 2006. Epigenetic properties of white-opaque switching in Candida albicans are based on a self-sustaining transcriptional feedback loop. Proc. Natl. Acad. Sci. U. S. A. 103:12807–12812 [DOI] [PMC free article] [PubMed] [Google Scholar]