Abstract
The transcriptional silencing of the cryptic mating-type loci in Saccharomyces cerevisiae is one of the best-studied models of repressive heterochromatin. However, this type of heterochromatin, which is mediated by the Sir proteins, has a distinct molecular composition compared to the more ubiquitous type of heterochromatin found in Schizosaccharomyces pombe, other fungi, animals, and plants and characterized by the presence of HP1 (heterochromatin protein 1). This review discusses how the loss of important heterochromatin proteins, including HP1, in the budding yeast lineage presented an evolutionary opportunity for the development and diversification of alternative varieties of heterochromatin, in which the conserved deacetylase Sir2 and the replication protein Orc1 play key roles. In addition, we highlight how this diversification has been facilitated by gene duplications and has contributed to adaptations in lifestyle.
INTRODUCTION
In Saccharomyces cerevisiae, repressive chromatin forms at the cryptic mating-type loci, HMRa and HMLα, and prevents the expression of extra copies of the genes that determine mating-type (reviewed in reference 99). These two loci enable mating-type switching but must remain silenced to maintain cell type identity and the capacity to mate. A related type of repressive chromatin forms at the telomeres, where it serves a structural role and represses subtelomeric genes. The tandem rDNA array is also embedded in a distinct type of chromatin that serves to suppress unequal sister chromatid exchange. However, the molecular composition of this chromatin is distinct from the cryptic mating-type loci and telomeres.
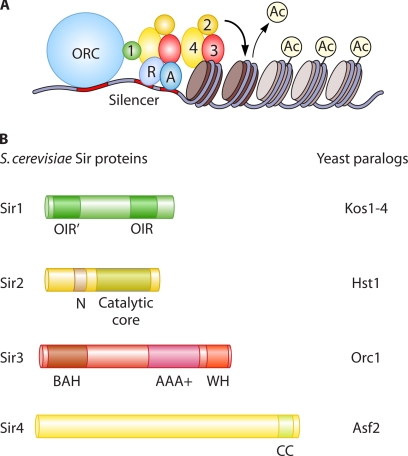
Silencing is initiated at specific DNA sequences termed silencers. HMRa and HMLα are each flanked by silencers, termed E and I, which have binding sites for the origin recognition complex (ORC), as well as Rap1, Abf1, or both (Fig. 1). Together, these silencer binding proteins recruit the main structural components of silenced chromatin, the Sir (silent information regulator) proteins. At telomeres, Rap1 binding sites embedded in the degenerate telomeric repeat sequence recruit the Sir proteins. The Ku complex also stabilizes the association of Sir proteins with telo- meres.
Fig. 1.
Functions of Sir proteins. (A) The silencer binding proteins ORC, Rap1 (R), and Abf1 (A) recruit the Sir proteins (1 to 4) to the silencer. Sir2 deacetylates neighboring nucleosomes, generating binding sites for Sir3 and Sir4. Reiterations of this cycle enable spreading. (B) Conserved domains of the four Sir proteins in S. cerevisiae are indicated. For each protein, the names of paralogs are listed.
The assembly of Sir proteins into silenced chromatin involves two phases, nucleation and spreading. First, the Sir proteins assemble at the mating-type silencers or chromosome ends through interactions with silencer binding proteins. Subsequently, Sir2, Sir3, and Sir4 spread along the chromosome via interactions with histones (Fig. 1). Sir1 is a silencer-associated protein that stabilizes the other Sir proteins at the mating-type silencers. Sir2 is a NAD+-dependent deacetylase, and its enzymatic activity is required for the spreading of the Sir proteins (60, 98). Sir3 and Sir4 bind preferentially to deacetylated histones H3 and H4 (20, 52, 79, 89). Sir4 also serves as a scaffold, interacting with Sir2, Sir3, and silencer-associated proteins. These observations inspire a sequential deacetylation model in which Sir2 deacetylates nearby nucleosomes, creating new high-affinity binding sites for Sir3 and Sir4, which in turn recruit additional Sir2 to the newly deacetylated nucleosome (Fig. 1). However, recent studies suggest that spreading may not always occur in a linear fashion. Instead, assembly may be focused in regions of the chromatin fiber brought together by silencers (75, 76, 118).
THE SUBPHYLUM SACCHAROMYCOTINA
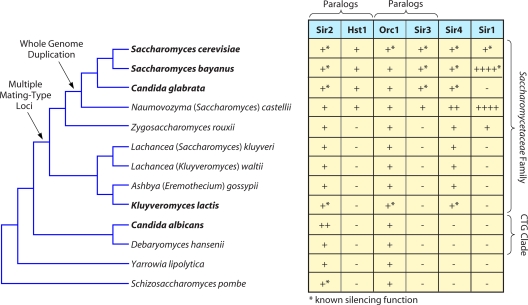
Given that the molecular composition of heterochromatin in S. cerevisiae is distinct from that of other well-studied organisms, it is important to understand when and how this unique silencing mechanism evolved. Thus, we focus on the fungal subphylum Saccharomycotina, which consists primarily of budding yeasts. These species are also referred to as hemiascomycetes. Comparisons of average protein sequence identity suggest that the diversity within this subphylum is slightly greater than that among the chordates (33). The phylogenetic relationships of the species discussed in this review are illustrated in Fig. 2. The family Saccharomycetaceae includes S. cerevisiae and is punctuated by a whole-genome duplication that occurred approximately 100 million years ago. The CTG clade, which includes the opportunistic human pathogen Candida albicans, is characterized by a change in the genetic code, such that CTG encodes serine rather than leucine. The fission yeast Schizosaccharomyces pombe belongs to a different subphylum, Taprhinomycotina, which is thought to have diverged from Saccharomycotina around a billion years ago (53, 54).
Fig. 2.
Distribution of Sir proteins in species discussed. For each species, the presence (+) or absence (−) of silencing proteins is indicated. The paralogs Sir2/Hst1 and Orc1/Sir3 are separated for clarity. In other cases, multiple paralogs are indicated by the number of + symbols. Species in boldface have been subject to experimental investigations of silencing. Asterisks identify proteins known to function in silencing. The tree represents the relative relationships of species and is based on the consensus in the field (18, 67, 68, 106).
SACCHAROMYCOTINA SPECIES LACK KEY COMPONENTS OF HP1-MEDIATED HETEROCHROMATIN
S. pombe has been another important model organism for studying heterochromatin formation (reviewed in reference 48), particularly because many key proteins are conserved between S. pombe and metazoans. In S. pombe, heterochromatin forms at pericentromeric regions, telomeres, and the cryptic mating-type loci. Two important heterochromatin proteins are a methyltransferase, Clr4, which specifically methylates lysine 9 of histone H3, and a chromodomain-containing protein, Swi6, which binds preferentially to H3-K9me. These proteins are well-conserved among eukaryotes and are generally known as SuVar3-9 (the methyltransferase) and HP1 (the chromodomain-containing protein). However, these proteins are missing from the genomes of S. cerevisiae and other Saccharomycotina species.
The formation of heterochromatin in S. pombe is often initiated via a mechanism that involves small, noncoding RNAs. These RNAs are part of a protein-RNA complex known as RITS (RNA-induced transcriptional silencing) that is related to the RISC posttranscriptional silencing complex (88, 111). Indeed, the RNA interference (RNAi) proteins argonaute and dicer are required for heterochromatin formation in S. pombe. However, argonaute and dicer are missing in S. cerevisiae and many Saccharomycotina species (86), although argonaute and noncanonical dicer proteins have recently been identified in a few Saccharomycotina species, including Naumovozyma castellii and C. albicans (31). These RNAi proteins have been suggested to silence retrotransposons, but a potential role in nucleating heterochromatin-like structures, as occurs in S. pombe and metazoans, has not been explored.
In summary, many of the key components of heterochromatin in S. pombe and other eukaryotes are missing in the Saccharomycotina species. It is unclear what led to the loss of these proteins, but their absence presented an evolutionary opportunity for the development and diversification of alternative silencing mechanisms.
DISTRIBUTION AND FUNCTION OF Sir PROTEINS AMONG BUDDING YEAST SPECIES
SIR2.
The deacetylase Sir2 is the most widespread and well-conserved of the Sir proteins (Fig. 2). In fact, unlike the other Sir proteins, which are restricted to budding yeasts, Sir2 has homologs among all domains of life, including eubacteria and archaea (reviewed in references 47, 102, and 110). Furthermore, many species have multiple Sir2 family members. For example, in S. cerevisiae there are five Sir2 deacetylases (SIR2 and HST1 to HST4 [HST1-4]) (15), but only Sir2 functions in silencing.
The Sir2 family is defined by a conserved catalytic domain (Fig. 1), which employs a mechanism distinct from that of other deacetylases. In particular, deacetylation is coupled to the lysis of NAD+, potentially linking the activity of these enzymes to the metabolic state of the cell. S. cerevisiae Sir2 (ScSir2) and its orthologs in the Saccharomycotina have a second conserved domain, which likely enables these proteins to interact with specific partners, such as Sir4.
Orthologs of ScSir2 have been identified in all examined Saccharomycotina species (36, 97), and the silencing function of these orthologs is conserved in the few species that have been investigated, namely, Kluyveromyces lactis (5, 23, 57), Candida glabrata (30, 94, 96), and Saccharomyces bayanus (118). In fact, Sir2 has a role in silencing beyond the Saccharomycotina. In S. pombe, SpSir2 contributes to silencing at centromeres, telomeres, and mating-type loci, where it deacetylates H3-K9, thereby promoting methylation of this lysine and the association of Swi6 (39, 103). In Drosophila melanogaster, mutations in DmSir2 affect position effect variegation mediated by HP1 (4, 87) and repression mediated by polycomb group proteins (43). Moreover, in distant protozoan species, such as Trypanosoma brucei and Plasmodium falciparum, Sir2 homologs are also associated with subtelomeric chromatin (2, 34, 40). Therefore, the Sir2 deacetylase most likely had an ancient role in silencing that has gained prominence in budding yeasts. In addition, Sir2 associates with the rDNA independently of the other Sir proteins, and this function has been conserved in other eukaryotes, including mammals (84).
SIR3.
The histone-binding protein Sir3 arose in the whole-genome duplication from ORC1, a subunit of the origin recognition complex found in all eukaryotes (12, 19, 28, 65). Consequently, distinct Sir3 proteins are found only in species that descended from the whole-genome duplication (Fig. 2).
Both Sir3 and Orc1 have three conserved domains (Fig. 1). The N-terminal BAH (bromo-adjacent homology) domain binds nucleosomes and in ScSir3 is required for silencing in vivo and the formation of SIR-nucleosome filaments in vitro (12, 16, 89, 100). In addition, the BAH domain of ScOrc1 binds ScSir1 (108, 116). The AAA+ domain (ATPases associated with diverse activities) belongs to a functionally diverse superfamily that hydrolyzes ATP and harnesses the released energy for assembly and disassembly of macromolecular complexes. This domain is poorly conserved in Sir3 and lacks residues critical for binding ATP but nevertheless can be aligned with Orc1 along its entire length, indicating that the integrity of the domain is important. Finally, the winged helix domain is predicted to bind DNA. The AAA+ and winged helix domains of ScSir3 overlap with regions of the protein shown to make critical contacts with other silencing proteins, including histone tails (20, 52), Sir4 (35, 82, 83, 91), and Rap1 (83).
Orthologs of Sir3 contribute to silencing of subtelomeric domains in C. glabrata (21, 26) and associate with cryptic mating-type loci in S. bayanus (44, 118), indicating that the silencing function of Sir3 is conserved. Moreover, the nonduplicated Orc1 protein fulfills the role of Sir3 in K. lactis and perhaps in Lachancea kluyveri (58, 109). However, it is unknown whether Orc1 acts with the deacetylase Sir2 to generate heterochromatin outside the Saccharomycetaceae family. Therefore, Orc1 either acquired a Sir3-like role in silencing within the Saccharomycotina subphylum or already had such a role in heterochromatin formation and became more critical in the absence of HP1 and other heterochromatin proteins. As discussed below, the second model is consistent with connections between ORC and heterochromatin in a wide range of species.
SIR4.
The scaffold protein Sir4 displays extremely low sequence conservation (36, 118), and in some genomes the identification of SIR4 is based on synteny rather than homology. Consequently, SIR4 has been identified only in the Saccharomycetaceae family and is either absent or highly diverged and nonsyntenic in the CTG clade (36, 97) (our unpublished analysis). An unresolved yet important issue is how Sir2 is targeted to silenced domains in species that apparently lack Sir4, as ScSir4 is required for the recruitment of ScSir2 and ScSir3 to silencers and telomeres (72, 98).
A consistent structural feature of Sir4 is a coiled-coil domain (6, 36) (Fig. 1), which interacts with Sir3 and is essential for silencing in S. cerevisiae (22, 85). A functionally defined PAD (partitioning and anchoring) domain enables ScSir4 to associate with the nuclear periphery (3), and other less well-defined regions of Sir4 interact with Sir1, Sir2, and Rap1. Orthologs of Sir4 contribute to silencing in S. bayanus (44), C. glabrata (62) and K. lactis (6, 57), indicating a conserved function. However, ScSir4 appears to have lost an ancestral function, as it cannot complement a sir4Δ mutation in the closely related species S. bayanus (118), although both S. bayanus Sir4 (SbSir4) and K. lactis Sir4 (KlSir4) complement a sir4Δ mutation in S. cerevisiae (6, 118).
SIR1.
The silencer-associated protein Sir1 has a restricted distribution, with Zygosaccharomyces rouxii being the species most distant from S. cerevisiae in which a Sir1-like protein has been identified (Fig. 2). The SIR1 gene family has undergone dramatic expansions and contractions. Consequently, some species, such as C. glabrata, have lost Sir1, whereas others, such as S. bayanus, encode multiple Sir1-like proteins, termed Kos (kin of Sir1) proteins (44). SIR1 and many of the KOS genes are located in subtelomeric regions, and this placement likely contributed to the rapid gains and losses of the SIR1 family.
ScSir1 contains a functionally defined OIR (ORC-interacting region) domain that associates with the ScOrc1 BAH domain and with ScSir4 to stabilize the SIR complex at silencers (45). This domain is conserved across species, and two-hybrid analyses confirm that it consistently interacts with Orc1 (14). Interestingly, a second OIR-like domain also occurs in Sir1 (Fig. 1), indicating that there was an internal duplication within the SIR1 gene.
In addition to S. cerevisiae, SIR1-like genes have been examined experimentally in S. bayanus (44), where there are four family members, SIR1, KOS1, KOS2, and KOS3. All four paralogs contribute to silencing at the cryptic mating-type loci. However, the exact contributions of the different paralogs remain to be determined.
IMPACT OF GENE DUPLICATIONS ON SILENCING PROTEINS
All four of the SIR genes, as defined in S. cerevisiae, have undergone duplications within the Saccharomycetaceae family, and it is important to understand how these duplications have led to partitioning and specialization of the functions of the Sir proteins. The duplications of SIR2 and SIR3 occurred in conjunction with the whole-genome duplication (Fig. 2) (28, 65, 112, 113). Subsequent to this event, most genes returned to single-copy status. However, about 10% of S. cerevisiae genes, including SIR2 and SIR3, are retained paralogs. Consequently, the nonduplicated orthologs of Sir2 and Sir3 have additional functions, as outlined below.
Divergence of function after duplication can occur through neofunctionalization or subfunctionalization (24, 50), and both ScSIR2 and ScSIR3 are products of subfunctionalization. One mechanism of subfunctionalization is duplication, degeneration, and complementation, in which duplicated genes each lose one of the original functions and together retain the entire set of ancestral functions (38). Subfunctionalization can also occur through specialization, in which the divergence of functions among paralogs also involves the accumulation of advantageous mutations in at least one of the duplicated genes, enabling it to outperform the ancestral gene (24, 50, 51, 74).
Duplication, degeneration, and complementation of Sir2.
The paralog of the deacetylase Sir2 is Hst1 (homolog of Sir2), a component of the SUM1 transcriptional repressor complex that represses middle sporulation, NAD+-biosynthetic, and α-specific genes in S. cerevisiae (11, 115, 117). Similarly, in C. glabrata, Hst1 regulates midsporulation genes as well as genes necessary for high-affinity uptake of NAD+ precursors (77). Interestingly, Sir2 and Hst1 generate distinct types of chromatin. Unlike the SIR complex, the SUM1 complex does not form extended domains of silenced chromatin but instead functions in a promoter-specific manner to repress its target genes.
Characterization of the nonduplicated Sir2 ortholog from K. lactis reveals that KlSir2 has both Hst1-like and Sir2-like properties, indicating that subfunctionalization occurred after duplication (57). Consistent with this idea, KlSir2 complements an hst1Δ deletion in S. cerevisiae (56) and partially suppresses a sir2Δ mating defect (23). Studies of chimeric ScSir2-Hst1 molecules indicate that distinct regions of these deacetylases enable them to associate with the SIR or SUM1 complexes (41, 56, 80), and these interaction domains are conserved in KlSir2 (41). The most parsimonious model is that the ancestral Sir2 also utilized these interaction domains and that after duplication the paralogs acquired complementary inactivating mutations that reduced their affinities for one of the two complexes. Thus, Sir2 and Hst1 represent an example of the duplication, degeneration, and complementation mechanism of subfunctionalization.
Although the initial subfunctionalization of Sir2 simply retained its ancestral functions, the division may ultimately have been beneficial. For example, ScHst1 has a lower affinity than ScSir2 for the cofactor NAD+ (11), and at slightly reduced concentrations of NAD+, Hst1-repressed genes are induced but Sir2-repressed genes are not (11, 77). Consequently, as NAD+ levels start to fall, ScHst1-repressed NAD+ biosynthetic genes are upregulated to restore NAD+ pools, without compromising ScSir2 function.
Potential specialization of Orc1.
The paralog of Sir3 is Orc1, the largest subunit of the origin recognition complex (ORC). ORC binds to origins of DNA replication, is found throughout eukaryotes, and has orthologs in prokaryotes (DnaA) and archaea (Orc1/Cdc6). Orc1 likely had a silencing function long before it gave rise to Sir3, as connections between ORC and heterochromatin have been observed in a wide variety of species (7, 27, 70, 78, 90, 93). It has generally been assumed that ORC acts as a landing pad to recruit silencing factors to heterochromatic domains, based on the paradigm from S. cerevisiae, in which ScOrc1 stabilizes the SIR complex at silencers by interacting with ScSir1. However, Orc1 could also act like ScSir3 to facilitate the spreading of silencing proteins by binding nucleosomes.
Nonduplicated orthologs of Orc1/Sir3 show more sequence similarity to the duplicated Orc1 than to Sir3, initially leading researchers to propose that the silencing functions of Sir3 arose after duplication (65). However, the nonduplicated Orc1 from L. kluyveri weakly complements a sir3 mutation in S. cerevisiae (109), and the nonduplicated Orc1 from K. lactis has the ability to spread across and silence a cryptic mating-type locus in K. lactis in a Sir3-like manner (58). The capacity of KlOrc1 to spread and promote the spreading of other silencing proteins implies that the common ancestor of KlOrc1 and ScSir3 had a similar ability and that subfunctionalization of the replication and spreading functions of Orc1 occurred after duplication. This conclusion is consistent with the existence of an ancient partnership of Orc1 and Sir2 to generate extended heterochromatic domains.
Curiously, KlOrc1 does not appear to act like ScOrc1 to nucleate silencing. KlOrc1 is not a silencer binding protein (58, 105) and SIR1 is not detected in the K. lactis genome (36, 44). Therefore, Orc1 either lost its silencer-binding function in the K. lactis lineage or gained this property in the S. cerevisiae lineage. SIR1 is first identifiable in Z. rouxii, a species with a nonduplicated Orc1 (Fig. 2), so Orc1 likely acquired the ability to function as a silencer binding protein prior to the whole-genome duplication.
An important unanswered question is whether SIR3 continued to evolve after duplication, such that it acquired new properties that improved its silencing ability. The accelerated sequence divergence of SIR3 compared to ORC1 may indicate that SIR3 acquired new properties or may reflect relaxed selection. It is also unclear whether there is an adaptive advantage in utilizing both Sir3 and Orc1 in different capacities to achieve silencing.
An ancient tandem duplication of SIR4.
SIR4 is an ancient paralog of the gene ASF2 (anti-silencing factor), which occurs in tandem with SIR4 in species of the Saccharomycetaceae family that did not undergo the whole-genome duplication (19). Little is known about the function of Asf2, except that it antagonizes silencing in both S. cerevisiae and K. lactis (57, 69) and copurifies with ScSir2 (16). Thus, Asf2 may compete with Sir4 for binding to Sir2. Studies on the evolutionary histories of these two rapidly changing proteins would be interesting, especially in light of the absence of Sir4-like proteins outside the Saccharomycetaceae family.
Internal duplication of SIR1.
SIR1 displays two types of duplication, expansions and contractions of subtelomeric SIR1-like (KOS) genes and an internal duplication resulting in two tandem OIR-like domains. Phylogenetic analysis of OIR-like domains reveals a clear separation of the N- and C-terminal domains, indicating that the internal duplication occurred once during evolution (44). Kos3, which has a single OIR domain, is thought to be the ancestral form of the protein (44). After the internal duplication occurred, the resultant SIR1-like gene was subsequently duplicated in its entirety and diversified, yielding SIR1, KOS1, KOS2, and KOS4.
An important unanswered question is how the tandem duplication of the OIR domain contributes to the function of Sir1. In S. cerevisiae, the C-terminal domain interacts with ScOrc1 and is important for the recruitment of the SIR complex to silencers. In contrast, the function of the N-terminal domain (OIR′) is unclear, although it is hypothesized to interact with Sir3 or another BAH domain-containing protein (25, 61). An intriguing possibility is that the OIR domain duplication was coupled to the duplication and divergence of its interaction partner Orc1/Sir3 (44). A second unanswered question is whether the multiple Sir1-like proteins found in some species have distinct or overlapping functions.
GENOMIC LOCATIONS ASSOCIATED WITH Sir PROTEINS
The Sir proteins were originally identified as transcriptional repressors of the cryptic mating-type loci. In budding yeasts, mating-type is determined by the MAT or MTL locus, which has two idiomorphs, a and α, encoding transcription factors that regulate the expression of cell-type-specific genes. Additional copies of mating-type cassettes enable switching of mating-type and emerged in the Saccharomycetaceae family (Fig. 2) (17). In S. cerevisiae, all three mating-type loci are on the same chromosome—two SIR-silenced loci located near the telomeres and an active locus situated more internally on the chromosome. However, there is plasticity in the number and placement of mating-type loci. For example, the mating-type loci are not all on the same chromosome in C. glabrata and K. lactis. Lachancea waltii has two adjacent mating-type loci near a single telomere (29), Ashbya gossypii has three mating-type loci near telomeres and a fourth more internally located (Fred Dietrich, personal communication), and L. kluyveri has only a single mating-type locus and can no longer undergo mating-type switching. In addition to this variability in organization of the mating-type loci, the extra mating-type loci are not always silenced as expected. In C. glabrata, MTL2 and MTL3 are both located near telomeres, but only MTL3 is silenced (94), and in K. lactis HMRa is repressed by the SUM1 complex instead of the SIR complex (57, 58).
Although the Sir proteins were first identified as repressors of mating-type loci, this was probably not their original function. Species outside the Saccharomycetaceae family contain a single, active mating-type locus, for which there is no evidence of regulation by Sir2 (18, 95). Nevertheless, Sir2 is present in these species and must have another function. Two candidate regions at which Sir2 may act are the telomeres and centromeres, which are silenced by HP1-containing heterochromatin in other eukaryotes.
An ancient role for Sir2 in generating subtelomeric heterochromatin seems highly likely given its presence at telomeres in S. pombe (39, 103) as well as S. cerevisiae, S. bayanus (118), C. glabrata (21, 62), and K. lactis (49, 58). However, a considerable reorganization of telomere structure did occur early in the Saccharomycotina subphylum (reviewed in reference 71). Compared to most eukaryotes, these yeasts display longer and more varied telomere repeat units, within which are embedded binding sites for Rap1, the protein responsible for recruiting Sir proteins to telomeres. Thus, the way in which Sir2 is recruited to telomeres is distinct in Saccharomycotina species.
Centromeres are often associated with heterochromatin. In most eukaryotes, including S. pombe, centromeres are flanked by repetitive sequences that are incorporated into heterochromatin, which is required for faithful chromosome segregation. In contrast, Saccharomycotina species lack HP1 and must either employ an alternative type of pericentromeric chromatin or have evolved other mechanisms to preclude the requirement for pericentromeric heterochromatin. There are two types of centromeres observed in budding yeasts. Species in the Saccharomycetaceae family generally have “point” centromeres, in which a relatively short DNA sequence (<500 bp) specifies the centromere (32). A specialized pericentromeric heterochromatin structure has not been observed at the centromeres in S. cerevisiae, and Sir2, 3, and 4 do not associate with centromeres (104). Curiously, Sir1 is found at centromeres (104), although its function is unknown. Thus, the development of point centromeres may have circumvented the requirement for a specialized chromatin structure. In contrast, species in the CTG clade have more complex centromeres that span 3 to 5 kb and are epigenetically inherited (10, 73, 101). Although the flanking chromatin structure of C. albicans centromeres has not been characterized, these centromeres are highly efficient origins of replication and bind ORC (66). It will be interesting to investigate whether the association of ORC facilitates the formation of a specialized chromatin structure containing Sir2.
RAPID SEQUENCE EVOLUTION OF SILENCERS
The silencers that recruit the SIR complex to the cryptic mating-type loci have evolved much more rapidly than the Sir proteins themselves. For example, in K. lactis, the identified silencers do not contain binding sites for ORC or Rap1 but instead bind Reb1 and Ume6 (9, 105). Interestingly, Reb1 and Rap1 are related myb domain-containing proteins, suggesting that this family of proteins may be well-suited to function as silencer binding proteins. In C. glabrata, silencing of the MTL3 locus is apparently not nucleated at a silencer sequence at all but is instead subject to subtelomeric silencing (94). This loss of silencers is consistent with the absence of the silencer-associated Sir1 protein in this species (44). Thus, substitutions of one silencer binding protein for another, and even the complete loss of silencers, have occurred over the course of evolution, but nevertheless the primary role of the Sir proteins in forming silenced chromatin has been preserved in species of the Saccharomycetaceae family.
Comparisons of silencers in S. cerevisiae and closely related (sensu stricto) species provide insights into how silencers diverge. Although the same proteins bind to the silencers in these species, DNA sequences between the protein binding sites display elevated sequence divergence compared to other noncoding regions of the genome (107). This observation suggests that silencing impairs the fidelity of DNA replication or repair, thereby increasing the likelihood of acquiring or losing protein binding sites in these regions.
SPECIES-SPECIFIC ADAPTATIONS OF SILENCING
Saccharomyces bayanus.
S. bayanus is the species most closely related to S. cerevisiae in which silencing has been examined experimentally. This yeast has fermentative capabilities similar to those of S. cerevisiae and is often identified in spontaneously fermented wines and ciders. In addition, S. bayanus and S. cerevisiae can mate and subsequently undergo meiosis, although the resulting spores are inviable (46). The maintenance of transcriptional silencing, mediated by Sir2, Sir3, and Sir4, occurs similarly in the two species. However, there are intriguing differences in the nucleation of silencing. S. bayanus has four paralogs of Sir1, whereas S. cerevisiae has one. Curiously, all four paralogs of Sir1 in S. bayanus contribute to transcriptional silencing, with Kos3 playing a distinct role compared to the other paralogs (44). Furthermore, the Sir1-interacting protein Sir4 has properties in S. bayanus not found in ScSir4. ScSir4 cannot efficiently associate with SbHMR, whereas SbSir4 does stably associate with ScHMR (118). Finally, the overall sequences of the silencers have diverged significantly between these two species, although these silencers appear to bind the same proteins (ORC, Rap1, and Abf1) in both species (107, 118). Thus, there has been a coordinated evolution of Sir1, Sir4, and silencer sequences, resulting in species-specific requirements for the assembly of silenced chromatin. These observations raise the possibility that the coexistence of incompatible proteins in hybrid cells interferes with silencing and thus contributes to reproductive barriers between species.
Candida glabrata.
C. glabrata is more distantly related to S. cerevisiae yet is descended from the whole-genome duplication. This yeast is a commensal, opportunistic pathogen and is a common cause of yeast infection in humans, along with C. albicans (37, 64). Although both of these species bear the name Candida, they are phylogenetically distant (Fig. 2).
A striking difference between C. glabrata and S. cerevisiae is that silencing is regulated by environmental conditions in C. glabrata, whereas it is constitutive in S. cerevisiae. Nevertheless, the general mechanism of silencing appears to be conserved (26, 30, 62). The environmental regulation of silencing in C. glabrata is possible because C. glabrata is auxotrophic for niacin, a precursor of NAD+ (30), which is required for Sir2 deacetylation. In niacin-poor environments, the cellular level of NAD+ drops, thereby reducing the function of C. glabrata Sir2 (CgSir2) and hence silencing (30, 77). Loss of silencing leads to the induction of several subtelomeric EPA adhesin genes, enabling the cells to adhere to epithelial cells (21, 30). Regulating silencing in this way may facilitate colonization of the urinary tract, an environment poor in niacin. Subtelomeric EPA adhesin genes are also induced under conditions that favor biofilm formation (62).
C. glabrata reproduces mitotically and is not known to have a sexual stage in its life cycle, despite maintaining three mating-type loci and genes important for meiosis (114). The adaptation to a predominantly asexual mode of reproduction may have enabled Sir-mediated silencing to become regulatable. Perturbations in CgSir2 function would have deleterious effects if cells needed to maintain cell type identity in readiness for mating.
Kluyveromyces lactis.
Among species whose genomes were not duplicated, silencing has been most extensively studied in K. lactis. This species was originally isolated from milk-derived products, although it grows on a wide range of carbon sources. Interest in cultivating K. lactis for biotechnology led to the development of its genetics, and its divergence prior to the whole-genome duplication makes K. lactis a convenient proxy for the ancestral nonduplicated state.
K. lactis has orthologs of Sir2 and Sir4, which function similarly to their S. cerevisiae homologs (5, 6, 57). In contrast, there is no distinct Sir3 in K. lactis but rather a single nonduplicated Orc1/Sir3 protein. KlOrc1 acts in conjunction with Sir2 and Sir4 to generate specialized chromatin structures at telomeres and HMLα. Furthermore KlOrc1 depends on its nucleosome-binding BAH domain to facilitate spreading of these proteins, much as ScSir3 does (58). Thus, a SIR-like complex does exist in this nonduplicated species.
Silencing in K. lactis involves additional factors beyond those characterized in S. cerevisiae. In particular, the KlSUM1 complex acts in concert with the SIR complex to silence the cryptic mating-type locus HMLα (57). Since KlSir2 associates with both the SUM1 and SIR complexes, its role in silencing is probably mediated through both complexes. Interestingly, the KlSUM1 complex also represses HMRa in the absence of Sir4 or Orc1. Thus, although the role of Sir2 in silencing mating-type loci is conserved in K. lactis, Sir2 does not always act as part of the SIR complex. In contrast to the mating-type loci, telomeres in K. lactis associate with components of the SIR-like complex but not KlSum1 (58). The different protein compositions of the chromatin at these three loci probably confer distinct properties. We speculate that the Sum1-Sir2 complex has a greater role in repressing transcription, because deletion of KlSUM1 results in a greater induction of HMLα genes than does deletion of KlSIR4 (57). In contrast, the Sir4-Sir2-Orc1 complex, which still assembles in the absence of KlSum1, may fulfill a different function, such as preventing inappropriate switching events at HMLα or recombination at telomeres.
In addition to repressing the mating-type loci, the KlSUM1 complex acts in a promoter-specific fashion to repress many of the same sporulation genes regulated by the ScSUM1 complex, as well as other cell-type-specific genes required for mating, such as the pheromone MFα1 and the G protein γ subunit STE18 (57). Consequently, Sir2 may be a critical factor preventing K. lactis from mating in certain conditions. K. lactis haploid cells delay mating until nutrients become scarce (8, 13, 55) unlike S. cerevisiae cells, which mate in nutrient-rich conditions. This difference is explained in part by a requirement for the transcription factor Rme1 (also known as KlMts1), which is induced in low-nutrient conditions and activates expression of some genes necessary for mating (13). However, Rme1/Mts1 induction alone may be insufficient to complete mating, as Sir2 represses some cell-type-specific and pheromone-induced genes not induced by Rme1/Mts1. Although Sir2-mediated repression may need to be relieved for progression of mating, Sir2-mediated silencing of HMLα and HMRa favors mating by maintaining cell identity. However, unlike S. cerevisiae, K. lactis cells lacking Sir proteins are not completely sterile. Thus, the time at which Sir2-mediated repression is relieved may govern proper progression of the K. lactis sexual cycle and link it to nutrient availability. An interesting question for future studies is whether variations in Sir2-mediated repression alter the ways species coordinate life cycle transitions with environmental changes.
Candida albicans.
Silencing and Sir2 function in the CTG clade are poorly understood. Species from this clade are responsible for the majority of human yeast infections, with C. albicans being the most common pathogen. These species are phenotypically diverse and vary in their abilities to mate, sporulate, and colonize mammalian hosts. The only conserved Sir proteins in these species are Sir2 and Orc1. C. albicans has five homologs of Sir2. One homolog, orf19.1992, was identified prior to genome sequencing and was annotated as SIR2 (92). However, another Sir2 homolog, orf19.4761, has better sequence conservation with Sir2 orthologs. The functions of these two homologs have not been clarified such that one can be definitively annotated as Sir2. Therefore, both genes will be referred to by their systematic names from the SC5314 strain.
Gene dosage of orf19.4761 correlates with the replicative life span of C. albicans, such that cells with more SIR2 genes have longer life spans and display asymmetric distribution of oxidized and damaged proteins during cell division (42). These observations are consistent with the role of SIR2 in aging in S. cerevisiae (1, 63). Deletion of the other Sir2 homolog, orf19.1992, lowers the frequency of phenotypic switching from the opaque to the white state (59). As only C. albicans opaque cells are competent to mate (81), orf19.1992 may reduce mating by favoring the white state. It is not clear whether either of these phenotypes is related to the presumed transcriptional repression activities of Sir2 proteins.
Further investigations into silencing in the CTG clade will answer important questions regarding the evolution of Sir2-mediated silencing. Does silenced chromatin form at the telomeres or near the centromeres? If so, do Sir2 and Orc1 act together to generate this silenced chromatin and are other proteins, perhaps related to Sir4 or Sum1, involved? How have the mechanism of silencing and the loss of mating and meiosis influenced one another? Might the lack of a highly specialized silenced chromatin correlate with the increased genome plasticity observed in these species?
MODEL FOR EVOLUTION OF Sir-MEDIATED SILENCING
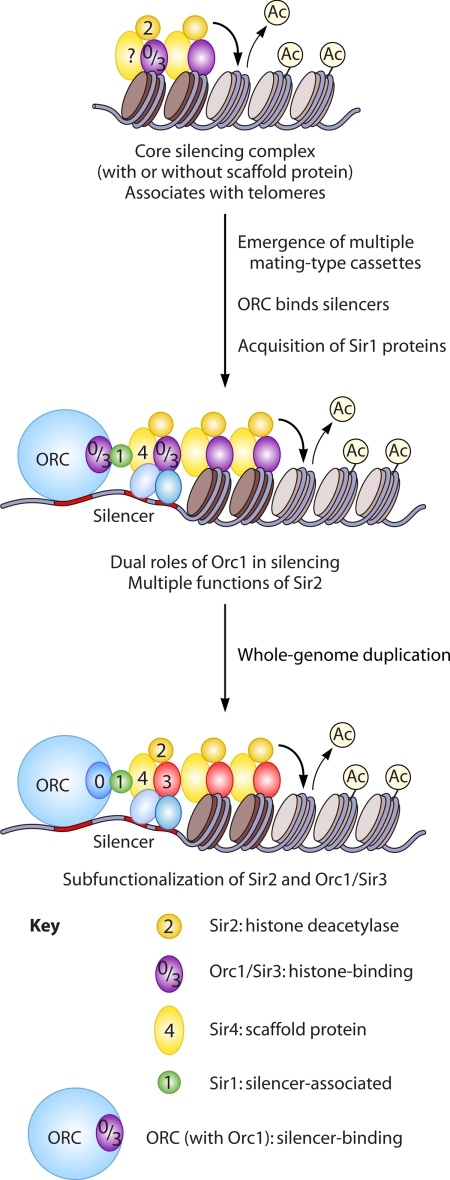
Based on the studies outlined above, we propose the following model for the evolution of Sir-mediated silencing (Fig. 3). First, HP1-containing heterochromatin was lost early in the Saccharomycotina subphylum. This loss necessitated adaptations, such as the development of alternative chromatin structures at telomeres and centromeres to maintain genome stability. At the core of such alternative silencing mechanisms was the conserved deacetylase Sir2, which participates in the formation of repressive chromatin in a variety of fungal and nonfungal species. We speculate that Sir2 partnered with the conserved replication protein Orc1 to generate repressive chromatin, with Sir2 deacetylating nucleosomes that could be bound by the BAH domain of Orc1. It is not clear whether the joint action of Sir2 and Orc1 is evolutionarily ancient and increased in importance after the loss of HP1 or whether it originated after the loss of HP1. However, the participation of Orc1 and Sir2 in subtelomeric heterochromatin in the protozoan parasite Plasmodium falciparum (78) is consistent with an ancient relationship. It is also unclear whether a Sir4-like protein was part of this proposed ancestral silencing complex, as SIR4 is rapidly evolving and could still be found outside the Saccharomycetaceae family. Mechanistic studies of Sir2 function in Candida species will help resolve these issues by clarifying the role of Orc1 and identifying Sir2-interacting proteins.
Fig. 3.
Model for the evolution of Sir-mediated silencing. (Top) In the ancestral state, the deacetylase Sir2 partnered with the histone binding protein Orc1. (Middle) An intermediate stage involved dual roles for Orc1, binding silencers and nucleosomes. (Bottom) After the whole-genome duplication, subfunctionalization of Sir2/Hst1 and Orc1/Sir3 occurred.
In the Saccharomycetaceae family, changes in mating-type architecture and protein function led to the development of Sir-mediated silencing, as characterized in S. cerevisiae (Fig. 2). One important change was the emergence of silent mating-type cassettes in telomere-proximal locations that could exploit the preexisting silenced domains at the ends of chromosomes. The Sir1 family of proteins emerged relatively recently, and the ability of these proteins to interact with Orc1 may have expanded the role of Orc1 in the establishment of silencing. Finally, the whole-genome duplication enabled the partitioning and specialization of Sir2/Hst1 and Orc1/Sir3 functions. In addition, the Sir proteins likely evolved different adaptive functions in yeast species not yet examined. For example, in N. castellii, paralogs of SUM1 and SIR4 have been retained (19), suggesting the existence of multiple varieties of silencing complexes in this species.
ACKNOWLEDGMENTS
We thank Brendan Cormack, Joseph Heitman, and Oliver Zill for comments on this manuscript and Cletus Kurtzman and Kenneth Wolfe for guidance on yeast taxonomy.
Research in the Rusche lab is supported by a grant from the NIH (GM073991).
Footnotes
Published ahead of print on 15 July 2011.
REFERENCES
- 1. Aguilaniu H., Gustafsson L., Rigoulet M., Nystrom T. 2003. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science 299:1751–1753 [DOI] [PubMed] [Google Scholar]
- 2. Alsford S., Kawahara T., Isamah C., Horn D. 2007. A sirtuin in the African trypanosome is involved in both DNA repair and telomeric gene silencing but is not required for antigenic variation. Mol. Microbiol. 63:724–736 [DOI] [PubMed] [Google Scholar]
- 3. Ansari A., Gartenberg M. R. 1997. The yeast silent information regulator Sir4p anchors and partitions plasmids. Mol. Cell. Biol. 17:7061–7068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Astrom S. U., Cline T. W., Rine J. 2003. The Drosophila melanogaster sir2+ gene is nonessential and has only minor effects on position-effect variegation. Genetics 163:931–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Astrom S. U., Kegel A., Sjostrand J. O., Rine J. 2000. Kluyveromyces lactis Sir2p regulates cation sensitivity and maintains a specialized chromatin structure at the cryptic alpha-locus. Genetics 156:81–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Astrom S. U., Rine J. 1998. Theme and variation among silencing proteins in Saccharomyces cerevisiae and Kluyveromyces lactis. Genetics 148:1021–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Auth T., Kunkel E., Grummt F. 2006. Interaction between HP1alpha and replication proteins in mammalian cells. Exp. Cell Res. 312:3349–3359 [DOI] [PubMed] [Google Scholar]
- 8. Barsoum E., Martinez P., Astrom S. U. 2010. Alpha3, a transposable element that promotes host sexual reproduction. Genes Dev. 24:33–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Barsoum E., Sjostrand J. O., Astrom S. U. 2010. Ume6 is required for the MATa/MATalpha-cellular identity and transcriptional silencing in Kluyveromyces lactis. Genetics 184:999–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Baum M., Sanyal K., Mishra P. K., Thaler N., Carbon J. 2006. Formation of functional centromeric chromatin is specified epigenetically in Candida albicans. Proc. Natl. Acad. Sci. U. S. A. 103:14877–14882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bedalov A., Hirao M., Posakony J., Nelson M., Simon J. A. 2003. NAD+-dependent deacetylase Hst1p controls biosynthesis and cellular NAD+ levels in Saccharomyces cerevisiae. Mol. Cell. Biol. 23:7044–7054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bell S. P., Mitchell J., Leber J., Kobayashi R., Stillman B. 1995. The multidomain structure of Orc1p reveals similarity to regulators of DNA replication and transcriptional silencing. Cell 83:563–568 [DOI] [PubMed] [Google Scholar]
- 13. Booth L. N., Tuch B. B., Johnson A. D. 2010. Intercalation of a new tier of transcription regulation into an ancient circuit. Nature 468:959–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bose M. E., et al. 2004. The Origin Recognition Complex and Sir4 protein recruit Sir1p to yeast silent chromatin through independent interactions requiring a common Sir1p domain. Mol. Cell. Biol. 24:774–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brachmann C. B., et al. 1995. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev. 9:2888–2902 [DOI] [PubMed] [Google Scholar]
- 16. Buchberger J. R., et al. 2008. Sir3-nucleosome interactions in spreading of silent chromatin in Saccharomyces cerevisiae. Mol. Cell. Biol. 28:6903–6918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Butler G., et al. 2004. Evolution of the MAT locus and its Ho endonuclease in yeast species. Proc. Natl. Acad. Sci. U. S. A. 101:1632–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Butler G., et al. 2009. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459:657–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Byrne K. P., Wolfe K. H. 2005. The Yeast Gene Order Browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res. 15:1456–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Carmen A. A., Milne L., Grunstein M. 2002. Acetylation of the yeast histone H4 N terminus regulates its binding to heterochromatin protein SIR3. J. Biol. Chem. 277:4778–4781 [DOI] [PubMed] [Google Scholar]
- 21. Castano I., et al. 2005. Telomere length control and transcriptional regulation of subtelomeric adhesins in Candida glabrata. Mol. Microbiol. 55:1246–1258 [DOI] [PubMed] [Google Scholar]
- 22. Chang J. F., et al. 2003. Structure of the coiled-coil dimerization motif of Sir4 and its interaction with Sir3. Structure 11:637–649 [DOI] [PubMed] [Google Scholar]
- 23. Chen X. J., Clark-Walker G. D. 1994. sir2 mutants of Kluyveromyces lactis are hypersensitive to DNA-targeting drugs. Mol. Cell. Biol. 14:4501–4508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Conant G. C., Wolfe K. H. 2008. Turning a hobby into a job: how duplicated genes find new functions. Nat. Rev. Genet. 9:938–950 [DOI] [PubMed] [Google Scholar]
- 25. Connelly J. J., et al. 2006. Structure and function of the Saccharomyces cerevisiae Sir3 BAH domain. Mol. Cell. Biol. 26:3256–3265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. De Las Penas A., et al. 2003. Virulence-related surface glycoproteins in the yeast pathogen Candida glabrata are encoded in subtelomeric clusters and subject to RAP1- and SIR-dependent transcriptional silencing. Genes Dev. 17:2245–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Deng Z., Dheekollu J., Broccoli D., Dutta A., Lieberman P. M. 2007. The origin recognition complex localizes to telomere repeats and prevents telomere-circle formation. Curr. Biol. 17:1989–1995 [DOI] [PubMed] [Google Scholar]
- 28. Dietrich F. S., et al. 2004. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science 304:304–307 [DOI] [PubMed] [Google Scholar]
- 29. Di Rienzi S. C., et al. 2011. Genetic, genomic, and molecular tools for studying the protoploid yeast, L. waltii. Yeast 28:137–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Domergue R., et al. 2005. Nicotinic acid limitation regulates silencing of Candida adhesins during UTI. Science 308:866–870 [DOI] [PubMed] [Google Scholar]
- 31. Drinnenberg I. A., et al. 2009. RNAi in budding yeast. Science 326:544–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dujon B. 2005. Hemiascomycetous yeasts at the forefront of comparative genomics. Curr. Opin. Genet. Dev. 15:614–620 [DOI] [PubMed] [Google Scholar]
- 33. Dujon B. 2006. Yeasts illustrate the molecular mechanisms of eukaryotic genome evolution. Trends Genet. 22:375–387 [DOI] [PubMed] [Google Scholar]
- 34. Duraisingh M. T., et al. 2005. Heterochromatin silencing and locus repositioning linked to regulation of virulence genes in Plasmodium falciparum. Cell 121:13–24 [DOI] [PubMed] [Google Scholar]
- 35. Enomoto S., Johnston S. D., Berman J. 2000. Identification of a novel allele of SIR3 defective in the maintenance, but not the establishment, of silencing in Saccharomyces cerevisiae. Genetics 155:523–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fabre E., et al. 2005. Comparative genomics in hemiascomycete yeasts: evolution of sex, silencing, and subtelomeres. Mol. Biol. Evol. 22:856–873 [DOI] [PubMed] [Google Scholar]
- 37. Fidel P. L., Jr., Vazquez J. A., Sobel J. D. 1999. Candida glabrata: review of epidemiology, pathogenesis, and clinical disease with comparison to C. albicans. Clin. Microbiol. Rev. 12:80–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Force A., et al. 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151:1531–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Freeman-Cook L. L., et al. 2005. Conserved locus-specific silencing functions of Schizosaccharomyces pombe sir2+. Genetics 169:1243–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Freitas-Junior L. H., et al. 2005. Telomeric heterochromatin propagation and histone acetylation control mutually exclusive expression of antigenic variation genes in malaria parasites. Cell 121:25–36 [DOI] [PubMed] [Google Scholar]
- 41. Froyd C. A., Rusche L. N. 2011. The duplicated deacetylases Sir2 and Hst1 subfunctionalized by acquiring complementary inactivating mutations. Mol. Cell. Biol. 31:3351–3365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fu X. H., Meng F. L., Hu Y., Zhou J. Q. 2008. Candida albicans, a distinctive fungal model for cellular aging study. Aging Cell 7:746–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Furuyama T., Banerjee R., Breen T. R., Harte P. J. 2004. SIR2 is required for polycomb silencing and is associated with an E(Z) histone methyltransferase complex. Curr. Biol. 14:1812–1821 [DOI] [PubMed] [Google Scholar]
- 44. Gallagher J. E., Babiarz J. E., Teytelman L., Wolfe K. H., Rine J. 2009. Elaboration, diversification and regulation of the Sir1 family of silencing proteins in Saccharomyces. Genetics 181:1477–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gardner K. A., Rine J., Fox C. A. 1999. A region of the Sir1 protein dedicated to recognition of a silencer and required for interaction with the Orc1 protein in Saccharomyces cerevisiae. Genetics 151:31–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Greig D. 2009. Reproductive isolation in Saccharomyces. Heredity 102:39–44 [DOI] [PubMed] [Google Scholar]
- 47. Greiss S., Gartner A. 2009. Sirtuin/Sir2 phylogeny, evolutionary considerations and structural conservation. Mol. Cells 28:407–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Grewal S. I., Rice J. C. 2004. Regulation of heterochromatin by histone methylation and small RNAs. Curr. Opin. Cell Biol. 16:230–238 [DOI] [PubMed] [Google Scholar]
- 49. Gurevich R., Smolikov S., Maddar H., Krauskopf A. 2003. Mutant telomeres inhibit transcriptional silencing at native telomeres of the yeast Kluyveromyces lactis. Mol. Genet. Genomics 268:729–738 [DOI] [PubMed] [Google Scholar]
- 50. Hahn M. W. 2009. Distinguishing among evolutionary models for the maintenance of gene duplicates. J. Hered. 100:605–617 [DOI] [PubMed] [Google Scholar]
- 51. He X., Zhang J. 2005. Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics 169:1157–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hecht A., Laroche T., Strahl-Bolsinger S., Gasser S. M., Grunstein M. 1995. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell 80:583–592 [DOI] [PubMed] [Google Scholar]
- 53. Heckman D. S., et al. 2001. Molecular evidence for the early colonization of land by fungi and plants. Science 293:1129–1133 [DOI] [PubMed] [Google Scholar]
- 54. Hedges S. B. 2002. The origin and evolution of model organisms. Nat. Rev. Genet. 3:838–849 [DOI] [PubMed] [Google Scholar]
- 55. Herman A., Roman H. 1966. Allele specific determinants of homothallism in Saccharomyces lactis. Genetics 53:727–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hickman M. A., Rusche L. N. 2007. Substitution as a mechanism for genetic robustness: the duplicated deacetylases Hst1p and Sir2p in Saccharomyces cerevisiae. PLoS Genet. 3:e126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hickman M. A., Rusche L. N. 2009. The Sir2-Sum1 complex represses transcription using both promoter-specific and long-range mechanisms to regulate cell identity and sexual cycle in the yeast Kluyveromyces lactis. PLoS Genet. 5:e1000710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hickman M. A., Rusche L. N. 2010. Transcriptional silencing functions of the yeast protein Orc1/Sir3 subfunctionalized after gene duplication. Proc. Natl. Acad. Sci. U. S. A. 107:19384–19389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hnisz D., Schwarzmuller T., Kuchler K. 2009. Transcriptional loops meet chromatin: a dual-layer network controls white-opaque switching in Candida albicans. Mol. Microbiol. 74:1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hoppe G. J., et al. 2002. Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation. Mol. Cell. Biol. 22:4167–4180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hou Z., et al. 2009. Phylogenetic conservation and homology modeling help reveal a novel domain within the budding yeast heterochromatin protein Sir1. Mol. Cell. Biol. 29:687–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Iraqui I., et al. 2005. The Yak1p kinase controls expression of adhesins and biofilm formation in Candida glabrata in a Sir4p-dependent pathway. Mol. Microbiol. 55:1259–1271 [DOI] [PubMed] [Google Scholar]
- 63. Kaeberlein M., McVey M., Guarente L. 1999. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 13:2570–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kaur R., Domergue R., Zupancic M. L., Cormack B. P. 2005. A yeast by any other name: Candida glabrata and its interaction with the host. Curr. Opin. Microbiol. 8:378–384 [DOI] [PubMed] [Google Scholar]
- 65. Kellis M., Birren B. W., Lander E. S. 2004. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428:617–624 [DOI] [PubMed] [Google Scholar]
- 66. Koren A., et al. 2010. Epigenetically-inherited centromere and neocentromere DNA replicates earliest in S-phase. PLoS Genet. 6:e1001068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kurtzman C. P. 2003. Phylogenetic circumscription of Saccharomyces, Kluyveromyces and other members of the Saccharomycetaceae, and the proposal of the new genera Lachancea, Nakaseomyces, Naumovia, Vanderwaltozyma and Zygotorulaspora. FEMS Yeast Res. 4:233–245 [DOI] [PubMed] [Google Scholar]
- 68. Kurtzman C. P. 2011. Discussion of teleomorphic and anamorphic ascomycetous yeasts and yeast-like taxa, p. 293–307 In Kurtzman C. P., Fell J. W., Boekhout T. (ed.), The yeasts, a taxonomic study, 5th ed. Elsevier, Amsterdam, The Netherlands [Google Scholar]
- 69. Le S., Davis C., Konopka J. B., Sternglanz R. 1997. Two new S-phase-specific genes from Saccharomyces cerevisiae. Yeast 13:1029–1042 [DOI] [PubMed] [Google Scholar]
- 70. Leatherwood J., Vas A. 2003. Connecting ORC and heterochromatin: why? Cell Cycle 2:573–575 [PubMed] [Google Scholar]
- 71. Lue N. F. 2010. Plasticity of telomere maintenance mechanisms in yeast. Trends Biochem. Sci. 35:8–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Luo K., Vega-Palas M. A., Grunstein M. 2002. Rap1-Sir4 binding independent of other Sir, yKu, or histone interactions initiates the assembly of telomeric heterochromatin in yeast. Genes Dev. 16:1528–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lynch D. B., Logue M. E., Butler G., Wolfe K. H. 2010. Chromosomal G + C content evolution in yeasts: systematic interspecies differences, and GC-poor troughs at centromeres. Genome Biol. Evol. 2:572–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lynch M., Katju V. 2004. The altered evolutionary trajectories of gene duplicates. Trends Genet. 20:544–549 [DOI] [PubMed] [Google Scholar]
- 75. Lynch P. J., Rusche L. N. 2009. A silencer promotes the assembly of silenced chromatin independently of recruitment. Mol. Cell. Biol. 29:43–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lynch P. J., Rusche L. N. 2010. An auxiliary silencer and a boundary element maintain high levels of silencing proteins at HMR in Saccharomyces cerevisiae. Genetics 185:113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ma B., et al. 2009. High-affinity transporters for NAD+ precursors in Candida glabrata are regulated by Hst1 and induced in response to niacin limitation. Mol. Cell. Biol. 29:4067–4079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Mancio-Silva L., Rojas-Meza A. P., Vargas M., Scherf A., Hernandez-Rivas R. 2008. Differential association of Orc1 and Sir2 proteins to telomeric domains in Plasmodium falciparum. J. Cell Sci. 121:2046–2053 [DOI] [PubMed] [Google Scholar]
- 79. Martino F., et al. 2009. Reconstitution of yeast silent chromatin: multiple contact sites and O-AADPR binding load SIR complexes onto nucleosomes in vitro. Mol. Cell 33:323–334 [DOI] [PubMed] [Google Scholar]
- 80. Mead J., et al. 2007. Swapping the gene-specific and regional silencing specificities of the Hst1 and Sir2 histone deacetylases. Mol. Cell. Biol. 27:2466–2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Miller M. G., Johnson A. D. 2002. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell 110:293–302 [DOI] [PubMed] [Google Scholar]
- 82. Moazed D., Kistler A., Axelrod A., Rine J., Johnson A. D. 1997. Silent information regulator protein complexes in Saccharomyces cerevisiae: a SIR2/SIR4 complex and evidence for a regulatory domain in SIR4 that inhibits its interaction with SIR3. Proc. Natl. Acad. Sci. U. S. A. 94:2186–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Moretti P., Freeman K., Coodly L., Shore D. 1994. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev. 8:2257–2269 [DOI] [PubMed] [Google Scholar]
- 84. Murayama A., et al. 2008. Epigenetic control of rDNA loci in response to intracellular energy status. Cell 133:627–639 [DOI] [PubMed] [Google Scholar]
- 85. Murphy G. A., et al. 2003. The Sir4 C-terminal coiled coil is required for telomeric and mating-type silencing in Saccharomyces cerevisiae. J. Mol. Biol. 334:769–780 [DOI] [PubMed] [Google Scholar]
- 86. Nakayashiki H. 2005. RNA silencing in fungi: mechanisms and applications. FEBS Lett. 579:5950–5957 [DOI] [PubMed] [Google Scholar]
- 87. Newman B. L., Lundblad J. R., Chen Y., Smolik S. M. 2002. A Drosophila homologue of Sir2 modifies position-effect variegation but does not affect life span. Genetics 162:1675–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Noma K., et al. 2004. RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat. Genet. 36:1174–1180 [DOI] [PubMed] [Google Scholar]
- 89. Onishi M., Liou G.-G., Buchberger J. R., Walz T., Moazed D. 2007. Role of the conserved Sir3-BAH domain in nucleosome binding and silent chromatin assembly. Mol. Cell 28:1015–1028 [DOI] [PubMed] [Google Scholar]
- 90. Pak D. T., et al. 1997. Association of the origin recognition complex with heterochromatin and HP1 in higher eukaryotes. Cell 91:311–323 [DOI] [PubMed] [Google Scholar]
- 91. Park Y., Hanish J., Lustig A. J. 1998. Sir3p domains involved in the initiation of telomeric silencing in Saccharomyces cerevisiae. Genetics 150:977–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Perez-Martin J., Uria J. A., Johnson A. D. 1999. Phenotypic switching in Candida albicans is controlled by a SIR2 gene. EMBO J. 18:2580–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Prasanth S. G., Prasanth K. V., Siddiqui K., Spector D. L., Stillman B. 2004. Human Orc2 localizes to centrosomes, centromeres and heterochromatin during chromosome inheritance. EMBO J. 23:2651–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Ramirez-Zavaleta C. Y., Salas-Delgado G. E., De Las Penas A., Castano I. 2010. Subtelomeric silencing of the MTL3 locus of Candida glabrata requires yKu70, yKu80, and Rif1 proteins. Eukaryot. Cell 9:1602–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Reedy J. L., Floyd A. M., Heitman J. 2009. Mechanistic plasticity of sexual reproduction and meiosis in the Candida pathogenic species complex. Curr. Biol. 19:891–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Rosas-Hernandez L. L., et al. 2008. yKu70/yKu80 and Rif1 regulate silencing differentially at telomeres in Candida glabrata. Eukaryot. Cell 7:2168–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Rusche L. N., Hickman M. A. 2007. Evolution of silencing at the mating-type loci in hemiascomycetes, p. 189–200 In Heitman J., Casselton L., Kronstand J. (ed.), Sex in fungi: molecular determination and evolutionary implications. American Society for Microbiology, Washington, DC [Google Scholar]
- 98. Rusche L. N., Kirchmaier A. L., Rine J. 2002. Ordered nucleation and spreading of silenced chromatin in Saccharomyces cerevisiae. Mol. Biol. Cell 13:2207–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Rusche L. N., Kirchmaier A. L., Rine J. 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72:481–516 [DOI] [PubMed] [Google Scholar]
- 100. Sampath V., et al. 2009. Mutational analysis of the Sir3 BAH domain reveals multiple points of interaction with nucleosomes. Mol. Cell. Biol. 29:2532–2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Sanyal K., Baum M., Carbon J. 2004. Centromeric DNA sequences in the pathogenic yeast Candida albicans are all different and unique. Proc. Natl. Acad. Sci. U. S. A. 101:11374–11379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Sauve A. A., Wolberger C., Schramm V. L., Boeke J. D. 2006. The biochemistry of sirtuins. Annu. Rev. Biochem. 75:435–465 [DOI] [PubMed] [Google Scholar]
- 103. Shankaranarayana G. D., Motamedi M. R., Moazed D., Grewal S. I. 2003. Sir2 regulates histone H3 lysine 9 methylation and heterochromatin assembly in fission yeast. Curr. Biol. 13:1240–1246 [DOI] [PubMed] [Google Scholar]
- 104. Sharp J. A., Krawitz D. C., Gardner K. A., Fox C. A., Kaufman P. D. 2003. The budding yeast silencing protein Sir1 is a functional component of centromeric chromatin. Genes Dev. 17:2356–2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Sjostrand J. O., Kegel A., Astrom S. U. 2002. Functional diversity of silencers in budding yeasts. Eukaryot. Cell 1:548–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Souciet J. L., et al. 2009. Comparative genomics of protoploid Saccharomycetaceae. Genome Res. 19:1696–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Teytelman L., Eisen M. B., Rine J. 2008. Silent but not static: accelerated base-pair substitution in silenced chromatin of budding yeasts. PLoS Genet. 4:e1000247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Triolo T., Sternglanz R. 1996. Role of interactions between the origin recognition complex and SIR1 in transcriptional silencing. Nature 381:251–253 [DOI] [PubMed] [Google Scholar]
- 109. van Hoof A. 2005. Conserved functions of yeast genes support the duplication, degeneration and complementation model for gene duplication. Genetics 171:1455–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Vaquero A. 2009. The conserved role of sirtuins in chromatin regulation. Int. J. Dev. Biol. 53:303–322 [DOI] [PubMed] [Google Scholar]
- 111. Verdel A., et al. 2004. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303:672–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Wolfe K. H., Shields D. C. 1997. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387:708–713 [DOI] [PubMed] [Google Scholar]
- 113. Wong S., Butler G., Wolfe K. H. 2002. Gene order evolution and paleopolyploidy in hemiascomycete yeasts. Proc. Natl. Acad. Sci. U. S. A. 99:9272–9277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Wong S., Fares M. A., Zimmermann W., Butler G., Wolfe K. H. 2003. Evidence from comparative genomics for a complete sexual cycle in the ‘asexual’ pathogenic yeast Candida glabrata. Genome Biol. 4:R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Xie J., et al. 1999. Sum1 and Hst1 repress middle sporulation-specific gene expression during mitosis in Saccharomyces cerevisiae. EMBO J. 18:6448–6454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Zhang Z., Hayashi M. K., Merkel O., Stillman B., Xu R. M. 2002. Structure and function of the BAH-containing domain of Orc1p in epigenetic silencing. EMBO J. 21:4600–4611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Zill O. A., Rine J. 2008. Interspecies variation reveals a conserved repressor of alpha-specific genes in Saccharomyces yeasts. Genes Dev. 22:1704–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Zill O. A., Scannell D., Teytelman L., Rine J. 2010. Co-evolution of transcriptional silencing proteins and the DNA elements specifying their assembly. PLoS Biol. 8:e1000550. [DOI] [PMC free article] [PubMed] [Google Scholar]