Abstract
Small RNA molecules of about 20 to 30 nucleotides function in gene regulation and genomic defense via conserved eukaryotic RNA interference (RNAi)-related pathways. The RNAi machinery consists of three core components: Dicer, Argonaute, and RNA-dependent RNA polymerase. In fungi, the RNAi-related pathways have three major functions: genomic defense, heterochromatin formation, and gene regulation. Studies of Schizosaccharomyces pombe and Neurospora, and other fungi have uncovered surprisingly diverse small RNA biogenesis pathways, suggesting that fungi utilize RNAi-related pathways in various cellular processes to adapt to different environmental conditions. These studies also provided important insights into how RNAi functions in eukaryotic systems in general. In this review, we will discuss our current understanding of the fungal RNAi-related pathways and their functions, with a focus on filamentous fungi. We will also discuss how RNAi can be used as a tool in fungal research.
INTRODUCTION
RNA interference (RNAi) is a conserved eukaryotic gene-silencing mechanism, and its discovery is one of the most important scientific breakthroughs in the past 20 years. In various RNAi pathways, small noncoding RNAs (sRNAs) of about 20 to 30 nucleotides (nt) act as regulators of cellular processes, such as development, RNA stability and processing, host defense, chromosome segregation, transcription, and translation (11, 12, 27, 34, 57). The silencing effects mediated by sRNAs in different pathways require an Argonaute/Piwi protein as the core component of the RNA-induced silencing complex (RISC) (11, 16). Upon loading of the sRNAs, RISC is guided to the target RNAs, and silencing is triggered.
Eukaryotic sRNAs are generally categorized into three main classes: short interfering RNAs (siRNAs), microRNAs (miRNAs), and Piwi-interacting RNAs (piRNAs). In terms of biogenesis, siRNAs and miRNAs are both derived from double-stranded RNA (dsRNA) precursors that are recognized and processed by Dicer to generate short duplexes (21 to 25 nt). The siRNAs and miRNAs are different in two key aspects. First, siRNAs are derived from exogenous dsRNA (e.g., viral RNA) or endogenous transcripts from repetitive sequences (e.g., transposable elements), or from transcripts that can form long hairpins (12, 34, 41). In many organisms, siRNA-induced silencing requires RNA-dependent RNA polymerases (RdRPs) to generate dsRNA from single-stranded RNA (ssRNA) or to amplify sRNA signals (2, 24, 33). miRNAs, on the other hand, are generated from miRNA-encoding genes that generate ssRNA precursor transcripts that form hairpin structures. Second, siRNAs normally fully match their mRNA targets. The siRNAs trigger RNA cleavage or transcriptional silencing mediated by the Argonaute proteins and generally function in genome defense. In contrast, miRNAs can target mRNAs that are not fully complementary and cause mRNA degradation and translational repression (3, 6).
piRNAs acquired their name due to association of these sRNAs with proteins from the Piwi clade of animal Argonaute proteins; these sRNAs have not been identified in organisms outside the animal kingdom. piRNAs are mostly derived from repetitive elements, transposons, and large piRNA clusters in germ cells (85). They have a distinct biogenesis pathway. piRNAs appear to be processed from single-stranded precursors and can be amplified through a “ping-pong” mechanism mediated by different Piwi family proteins (8, 38, 56, 61). piRNAs play important roles in maintaining germ cell genome stability, although the functions of many piRNAs are still unknown (12, 34, 48, 62, 84, 89)
RNAi components (Dicer, Argonaute, and RdRP) have been identified in all major branches of eukaryotes (16, 79), suggesting that RNAi is an ancient defense and/or regulatory mechanism that existed in the common eukaryotic ancestor. In addition, homologs of the Argonaute proteins, but not other core RNAi components, are found in some eubacteria and archea (86). During evolution, the RNAi pathway has undergone adaptation, leading to novel biogenesis mechanisms and functions in different eukaryotic organisms and lineages. RNAi components are absent from some eukaryotic organisms, most notably Saccharomyces cerevisiae, indicating that the RNAi pathway is dispensable for some single-celled organisms.
The fungi kingdom is a major branch of the eukaryotic organisms and is estimated to include more than 1 million species, with enormous diversity in ecology, morphology, and life cycles. Studies of the filamentous fungus Neurospora crassa and the fission yeast Schizosaccharomyces pombe have uncovered key RNAi components and functions and contributed significantly to our understanding of RNAi in eukaryotes. In this review, we will discuss the RNAi pathways in fungi, mechanistic and functional adaptations in different organisms and environments, and how RNAi can be harnessed for basic research and industrial uses. Since the RNAi studies of S. pombe have been reviewed previously (9, 36, 62, 90), this review will mostly focus on RNAi in filamentous fungi. It is worth noting that in the fission yeast, RNAi mediates gene silencing at the transcriptional level by heterochromatin formation, whereas in filamentous fungi RNAi mostly results in posttranscriptional gene silencing and has no confirmed role in transcriptional gene silencing (19, 30).
RNAi IN FUNGI
The fungi kingdom consists of a large and diverse group of eukaryotes. Current research recognizes seven phyla and one subkingdom that consists of two phyla, Ascomycota and Basidiomycota. Based on current fungal taxonomy, yeasts and filamentous fungi do not form a monophyletic group (44). Most yeasts fall into the phyla Ascomycota and Basidiomycota, whereas most filamentous fungi belong to Ascomycota (37). Since the three core RNAi components (RdRP, Dicer, and Argonaute) have been found in species of different phyla, it is believed that the fungal common ancestor possessed a functional RNAi pathway. In this pathway, an aberrant RNA would be first recognized and converted into dsRNA by RdRPs, subsequently processed by Dicers into siRNA, and loaded onto Argonaute proteins (64). However, this ancestral pathway has experienced various adaptations during fungal evolution. For example, in budding yeasts, the RdRP appears to be universally lost (28, 64): budding yeasts either have noncanonical RNAi or have no RNAi pathway at all. On the other hand, RNAi has largely been retained in all filamentous fungi examined, but it has diverse functions. In the model organism Neurospora crassa, siRNAs and microRNA-like small RNAs (milRNAs) function in genome defense and gene regulation (31, 57). RNAi in Neurospora functions in both vegetative and sexual stages, establishing it as a good paradigm for the diversity of fungal RNAi pathways and functions.
GENOME DEFENSE SYSTEM IN NEUROSPORA CRASSA
RNAi is an ancient host defense mechanism against viruses and transposons (27). The filamentous fungus N. crassa possesses several mechanisms to defend against viral and transposon invasion. Hyphae, the vegetative tissue of filamentous fungi, are typically divided into cells by internal cross-walls called septa. As septa harbor pores large enough for ribosomes, mitochondria, and nuclei to flow through, invading viruses are also able to quickly move from one cell to another (42). To battle against this threat, fungi have evolved an efficient antiviral mechanism. Although no virus has been found in N. crassa laboratory strains so far, dsRNA, an intermediate of viral RNA replication of many viruses, can initiate potent RNAi-mediated gene silencing in these strains (15, 18). In addition, the expression of dsRNA in N. crassa triggers a robust dsRNA-activated transcriptional program that activates the expression of all major RNAi genes, including the Argonaute proteins QDE-2 and Dicer, and many homologs of known antiviral genes (20). The induction of these genes by dsRNA suggests that RNAi is part of the conserved ancient defense response to viral infection.
N. crassa has three mechanisms, two of which are RNAi related, to suppress transposon invasion during the vegetative (asexual) and sexual stages of its life cycle through recognition of repetitive and highly homologous sequences. In the vegetative stage, which accounts for most of the Neurospora life cycle, the introduction of repetitive DNA sequences triggers posttranscriptional gene silencing of all homologous genes, an RNAi silencing phenomenon known as quelling (74). During the sexual cycle, two distinct silencing mechanisms function: repeat-induced point mutation (RIP) and meiotic silencing by unpaired DNA (MSUD; also referred to simply as meiotic silencing) (78, 82). RIP occurs during the early stage of the sexual cycle, when the fused cytoplasm still harbors two haploid nuclei from opposite mating types. RIP causes mutations from C·G to A·T in the repetitive sequences (78). MSUD is an RNAi-related mechanism that occurs later than RIP and is activated when two haploid nuclei are fused into a diploid nucleus. Mechanistic studies of quelling and MSUD have provided important information on how RNAi functions in eukaryotes.
QUELLING, A GENOME DEFENSE MECHANISM OF NEUROSPORA IN THE ASEXUAL CYCLE
Quelling was one of the first RNAi-related phenomena described in eukaryotes, and its study resulted in the identification of some of the earliest known RNAi components (74). In 1992, when Macino and his colleague transformed N. crassa with multiple copies of the structural genes albino-1 (al-1) and albino-3 (al-3) required for caroteniod biosynthesis (responsible for the orange pigment of N. crassa), they obtained some pale yellow/white transformants that had decreased al mRNA levels (74). They named this posttranscriptional gene-silencing phenomenon quelling. In their subsequent studies with forward and reverse genetics approaches, several genes coding for components of the quelling machinery, including quelling deficient element genes qde-1, qde-2, and qde-3 and Dicer protein genes dcl-1 and dcl-2, were identified (13, 15, 23–25). qde-1 encodes an RdRP and was the first RNAi component identified in a eukaryotic organism (24). The identification of QDE-1 soon after the landmark study of Caenorhabditis elegans by Fire, Mello, and colleagues (29) provided strong in vivo evidence that demonstrated the role of dsRNA in RNAi. qde-2, dcl-1, and dcl-2 encode an Argonaute protein and two Dicer-like proteins, respectively (13, 15). These four proteins are the core components in the quelling pathway, and they are highly conserved in most eukaryotes. In short, the aberrant RNAs (aRNAs) produced from a repetitive transgene locus are specifically recognized by QDE-1, which synthesizes dsRNAs. These dsRNAs are then cleaved by DCL proteins into duplex siRNAs of ∼25 nt and subsequently loaded onto QDE-2, the core component of RISC (14, 31). In vegetative cells, DCL-2 is the major Dicer enzyme that processes dsRNA into siRNA, but DCL-1 has a redundant role (15).
The siRNA duplex-bound Argonaute protein is initially inactive. To allow recognition and cleavage of its mRNA targets, the passenger strand of the siRNA duplex must be removed from the complex. In N. crassa, the activation of RISC is a two-step process, requiring the slicer activity of QDE-2 and the QDE-2-interacting protein QIP, an exonuclease (60). First, the QDE-2-bound double-stranded siRNA duplex is nicked by QDE-2 using its slicer activity: QDE-2 treats the passenger strand of the siRNA as a substrate. Second, QIP, which is recruited by QDE-2, recognizes the nicked passenger strand of siRNA and degrades it with its exonuclease activity. QIP was the first identified exonuclease required for RNA-induced silencing in eukaryotes. The importance of cleavage activity of Argonaute proteins in siRNA duplex strand separation was later demonstrated for other organisms (10, 87), and it appears to be an essential step in RNAi pathways. Recently, the Drosophila melanogaster RNase C3PO was shown to remove the nicked siRNA passenger strand to activate the RNAi pathway (59).
Although the mechanism of quelling downstream of dsRNA production is largely clear, how cells generate repetitive transgene-specific dsRNA is still poorly understood. To trigger quelling, the ectopic sequences homologous to the endogenous counterpart must exceed the threshold length of about 130 nt (22). In addition, a transgene per se is not sufficient to trigger quelling unless the transgenic sequences are arranged in large tandem arrays. Based on these observations, it was hypothesized that the large tandem repeats may produce aberrant DNA secondary structures that are recognized by QDE-3, a recQ DNA helicase and an essential component of quelling (25). But how QDE-3 is involved in the production of aRNA and how QDE-1 specifically recognizes the aRNA to produce dsRNA are not known. Studies of the plant RNAi pathway suggest that misspliced or decapped mRNAs may be recognized as aRNAs and trigger RNAi (32, 43). However, the recent studies on DNA damage-induced qiRNA (discussed below) and quelling in N. crassa suggest the existence of a different mechanism. Surprisingly, the production of aRNAs after DNA damage does not require the typical DNA-dependent RNA polymerases (DdRPs), such as polymerase I (Pol I), Pol II, or Pol III (54). Instead, QDE-1, the RdRP, is required for aRNA production (53, 54). Biochemical studies showed that QDE-1 is both an RdRP and a DdRP, and in fact, the DdRP activity of QDE-1 is much stronger than its RdRP activity (53). Therefore, in dsRNA production, QDE-1 has dual roles: it first uses its DdRP activity to produce aRNA from single-stranded DNA (ssDNA) and then uses its RdRP activity to convert aRNA into dsRNA. This aRNA production mechanism explains why QDE-1 specifically recognizes aRNA and not other cellular RNAs.
The function of QDE-1 in aRNA synthesis requires its recruitment to ssDNA. In vitro, QDE-1 produces mostly RNA/DNA hybrids from ssDNA templates (53, 54), indicating the existence of additional factors in aRNA and dsRNA synthesis. Nolan et al. showed that QDE-1 interacts with replication protein A (RPA), an ssDNA binding protein complex important in DNA replication, repair, and recombination pathways (71), but the role of RPA in quelling is not known. Recently, we showed that qiRNA production and aRNA production are completely abolished in an rpa-3 knockout strain (53). Importantly, RPA-3 is also required for quelling, linking the mechanism of qiRNA production to that of quelling. In vitro RNA polymerase assays showed that in the presence of RPA, QDE-1 almost exclusively produces dsRNA from ssDNA, due to the lack of RNA/DNA hybrids. These results suggest that RPA functions in the quelling and qiRNA pathways by recruiting QDE-1 to ssDNA and by preventing the formation of RNA/DNA hybrids.
Taken together, these data support a working model for dsRNA production in the quelling pathway that is summarized in Fig. 1. First, the presence of tandem repetitive DNA sequences results in the formation of aberrant DNA structures that are recognized by QDE-3. QDE-3 unwinds dsDNA to produce ssDNA, which is then stabilized by RPAs. QDE-1, recruited by QDE-3 and RPA to the ssDNA locus, uses its DdRP activity to produce aRNA from ssDNA, and then it converts the aRNA into dsRNA via its RdRP activity.
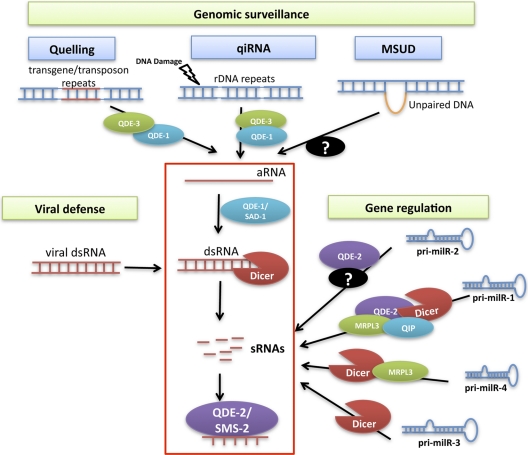
Fig. 1.
sRNA biogenesis pathways and RNAi functions in Neurospora. See text for details.
MEIOTIC SILENCING OF UNPAIRED DNA IN THE SEXUAL CYCLE OF N. CRASSA
Meiotic silencing of unpaired DNA is an RNAi-related phenomenon that can silence unpaired DNA/chromosome during the sexual cycle in N. crassa (4, 46, 63, 81, 82). Meiotic silencing occurs in diploid ascus cells, which are formed by the fusion of two nuclei of opposite mating types (82). The diploid cells eventually divide into eight haploid ascospores after two rounds of meiosis and one round of mitosis. Specifically, meiotic silencing acts in the prophase of first meiosis, when there is a region of unpaired DNA between the two homologous parental chromosomes (i.e., DNA exists in one parental chromosome but not in its pairing partner or heterologous DNA on parental chromosomes) (72, 81, 82). Meiotic silencing triggers the silencing of all homologous sequences in the genome, and this effect can last throughout the rest of the sexual cycle. The unpaired DNA can be caused by the deletion or addition of DNA in one parental strain. Thus, meiotic silencing serves as a complementary mechanism to RIP, which inhibits transposon replication during the sexual cycle. It should be noted that both RIP and meiotic silencing can occur during the sexual stage if repetitive DNA also results in unpaired DNA on the homologous chromosome. In addition, the ability of meiotic silencing to detect and silence transcription from unpaired DNA may also provide a safeguard against genomic rearrangement events, such as deletions, duplications, and translocations, in the sexual cycle.
The fact that meiotic silencing can silence expression of other homologous genes in trans even if those copies are paired (4, 81, 82) suggests that the silencing signal is trans-acting, similar to the case in quelling. Indeed, genetic studies have identified a set of critical genes that are highly homologous to the components of the quelling pathway but are specifically expressed in the meiotic stage. Sad-1 (suppressor of ascus dominance 1) is the first known meiotic silencing component identified by screening for mutants deficient in meiotic silencing. SAD-1 is a putative RdRP homologous to QDE-1 (82). The identification of SAD-1 indicated that, like quelling, the production of dsRNA is an essential step in meiotic silencing. SMS-2 (suppressor of meiotic silencing 2), identified by reverse genetics, is an Argonaute protein homologous to QDE-2 (52). SMS-2 is thought to function by binding to sRNAs originating from the unpaired DNA region to trigger posttranscriptional silencing. Although both DCLs have somewhat redundant functions in the quelling pathway, DCL-2 is the major Dicer enzyme active in the quelling pathway. For meiotic silencing, however, only DCL-1 (also called SMS-3) is required, suggesting that DCL-1/SMS-3 but not DCL-2 is specifically expressed during meiosis (1). SAD-2, a fungus-specific protein with no apparent domain, is required for meiotic silencing and can associate with SAD-1 in vivo. SAD-2 appears to function as a scaffold protein to recruit SAD-1 to the perinuclear region (5, 83). Similarly, DCL-1/SMS-3 and SMS-2 are localized in the perinuclear region, suggesting that this region is the major functional center for MSUD (1, 83).
Although meiotic silencing primarily utilizes a set of meiosis-specific RNAi genes, two recent studies showed that QIP, the exonuclease required in the quelling pathway, is also required for MSUD. These results suggest that quelling and meiotic silencing originated from the same ancestral pathways and are mechanistically linked (51, 92). It is now proposed that MSUD consists of two major steps: meiotic trans-sensing and RNAi-mediated silencing (46). The meiotic trans-sensing mechanism scans for the presence of unpaired DNA regions or significant sequence differences between the two homologous chromosomes during meiosis. Meiotic trans-sensing was initially discovered during studies of the Asm-1 (Ascospore maturation 1) gene (4), which showed that the pairing of the Asm-1 gene on homologous chromosomes is essential for its normal expression, indicating the existence of a trans-sensing mechanism that “senses” the presence/absence of paired DNA on homologous chromosomes. The failure to sense the presence of paired DNA will trigger the production of sRNAs and silencing of the homologous DNA in the genome, a process that is mediated by SAD-1, SAD-2 SMS-2, and DCL-1/SMS-3 (52, 81, 82). Consistent with this model, DNA methylation in N. crassa appears to affect meiotic trans-sensing, but not meiotic silencing (72). The mechanism of meiotic trans-sensing is currently unknown, but genes known to be involved in stable long/short-distance pairing, recombination, and segregation steps of chromosomes during meiosis appear to be dispensable (46). Since the discovery of meiotic silencing in N. crassa, unpaired homologous DNA-triggered silencing in meiosis has also been observed in Caenorhabditis elegans, Drosophila melanogaster, and in mammals, but the silencing mechanisms in Drosophila and mammals appear to be independent from RNAi (46).
NOVEL TYPES OF sRNA AND PRODUCTION PATHWAYS IN N. CRASSA
DNA damage-induced qiRNA.
Treatment of N. crassa cultures with various DNA-damaging agents induces the expression of qde-2 mRNA and QDE-2 protein, suggesting the production of endogenous dsRNA after DNA damage. Analyses of the QDE-2-associated sRNAs revealed a novel class of sRNAs, named qiRNAs, because of their association with QDE-2. qiRNAs most originate from the rDNA locus, and their production requires QDE-1, QDE-3, Dicer, and RPA, but they are different from quelling-induced siRNAs in three aspects: (i) qiRNAs originate from endogenous rDNA loci, (ii) they are about ∼21 nt long, shorter than the typical 25-nt siRNA, and (iii) qiRNAs are induced in response to DNA damage (54). Despite these differences, recent studies suggest that the biogenesis of qiRNAs is similar to that of quelling-induced siRNA (53, 57) (Fig. 1). It is important to note that the rDNA locus is the only highly repetitive DNA region in the genome of wild-type N. crassa strain except when quelling is performed, which introduces another highly repetitive DNA locus. Therefore, the repetitive natures of rDNA and quelled loci are likely to be the common trigger for the production of aRNA and small RNA. The induction of qiRNA by DNA damage, therefore, suggests that replication stress on repetitive transgene loci results in siRNA production and quelling. Mutants deficient in qiRNA production exhibit increased sensitivity to DNA damage and an elevated protein synthesis rate upon DNA damage (54). These results suggest that qiRNA may contribute to the function of DNA damage checkpoints to arrest the cell cycle after DNA damage.
Study of N. crassa miRNAs reveals surprisingly diverse sRNA production pathways.
miRNAs are important regulators of gene expression in animals and plants but were previously thought to be absent from fungi. By analyzing QDE-2-associated sRNAs using deep sequencing, at least 25 milRNAs were identified in N. crassa (55). Like conventional miRNAs, these milRNAs are predominantly derived from one strand of hairpin-like ssRNA precursors with strong preference for U at their 5′ termini (55). In addition, milRNAs can repress the expression of endogenous genes, and the levels of the predicted miRNA target genes are upregulated in RNAi mutants. Furthermore, QDE-2 was found to be specifically associated with mRNAs that are predicted to be the targets of milRNAs. These results indicate that the N. crassa milRNAs are fungal miRNAs. However, the physiological importance of milRNAs remains to be determined.
Animal and plant miRNAs are produced by the well-defined canonical biogenesis pathways (3, 6). Genetic and molecular studies of N. crassa milRNAs, however, revealed surprisingly diverse biogenesis mechanisms (Fig. 1) and shed light on how eukaryotic sRNA can be made (55). These pathways combine different RNAi components, including Dicer, QDE-2, QIP, a putative RNase III domain-containing protein, MRPL3, and other unidentified nucleases, to produce different milRNAs. Among the four most abundant milRNAs, only milR-3 maturation uses the canonical plant miRNA pathway, which only requires Dicers for pre-milRNA processing. Biogenesis of milR-4 requires MRPL3 and is partially dependent on Dicer (55).
The analyses of the maturation of milR-1 and milR-2 milRNAs demonstrated that the Argonaute QDE-2 plays essential but distinct roles in two novel sRNA biogenesis pathways. For milR-1, the hairpin-containing primary milR-1 is first processed by Dicer into double-stranded pre-milR-1. Subsequently, QDE-2 binds to pre-milR-1 and recruits the exonuclease QIP to process the pre-milR-1 into mature milR-1. In this pathway, QDE-2 functions as a scaffold but not a slicer in milRNA maturation. Although the production of most milRNAs is dependent on Dicer, milR-2 is produced independently of Dicer. Instead, the catalytic activity of QDE-2 is necessary for production of mature milR-2, suggesting that QDE-2, after binding to pre-milR-2, cleaves the milRNA* strand to allow the further processing of milRNAs by another unidentified nuclease (55). Interestingly, in both mice and zebrafish, the biogenesis of miR-451 occurs through a similar pathway (17, 21), indicating that the novel sRNA pathways found in fungi also exist in higher eukaryotes.
Dicer-independent small interfering RNAs.
Dicer-independent small interfering RNAs (disiRNAs) are another novel type of sRNA found in Neurospora (55). Like qiRNAs and siRNAs, disiRNAs are about 22 nt long and have a strong 5′ U preference. disiRNAs are mapped to ∼50 nonrepetitive DNA loci which contain genes or intergenic regions with no obvious sequence motifs. Importantly, the biogenesis of disiRNAs is independent of any known RNAi component, including Dicer proteins, indicating the existence of a completely unknown sRNA production pathway. Very little is known about the function of disiRNAs.
RNAi AND SMALL RNAs IN MUCOR CIRCINELLOIDES
M. circinelloides is a basal fungus of the clade zygomycete that has become an important fungal model system for RNAi and sRNA production. Transformation of M. circinelloides with self-replicative plasmids or with vectors for expression of inverted repeat transgenes results in activation of gene silencing pathways and production of sRNAs of two distinct sizes, 21 nt and 25 nt (26, 67–69). Of the two Dicer proteins, DCL-2 plays the major role in siRNA generation (26). Mucor has two RdRPs, and RdRP1 is required for transgene-induced gene silencing (68).
Deep sequencing of M. circinelloides sRNAs recently uncovered several classes of endogenous sRNAs (68). Interestingly, most of these sRNAs have sequences complementary to exons, and thus they are called exonic siRNAs (ex-siRNAs). ex-siRNAs are classified based on differential requirements for RdRPs and DCLs for their production. The biogenesis of most ex-siRNAs depends on RdRP1 and DCL-2, both of which are also required for transgene-induced silencing. These results suggest that RdRP1 may convert the exon-specific transcripts into dsRNAs, which are then processed by DCL-2. The second class of sRNAs requires DCL-2 and RdRP2 for their production, and the third class of ex-siRNAs requires both RdRPs and both Dicer proteins. The fourth class of ex-siRNAs requires both RdRPs but is mostly dependent on DCL-1 for biogenesis. In addition to ex-siRNAs, some DCL-dependent sRNAs originate from putative transposons and repetitive sequences. The differential roles of Dicer proteins and RdRPs in M. circinelloides sRNA biogenesis further indicate the diversity of fungal RNAi processing and sRNA production mechanisms.
SEX-INDUCED SILENCING IN CRYPTOCOCCUS NEOFORMANS
The human fungal pathogen C. neoformans also possesses a fully functional RNAi pathway, and transgene-expressed dsRNA has been successfully used to knock down gene expression in this organism (58). Recently, Heitman and colleagues discovered an RNAi-mediated posttranscriptional gene-silencing mechanism that is specifically activated during sexual reproduction of the fungus, thus, the name, sex-induced silencing (SIS) (91). Like quelling, SIS requires the presence of a tandem array (>2 copies) of the transgene sequence, and the efficiency of SIS correlates with the copy number of the transgene. Although SIS can occur at low frequency during the vegetative growth stage of C. neoformans, its frequency is increased several hundredfold during the sexual phase. Importantly, SIS is abolished in ago1 and Rdp1 single mutants and in dcl1 dcl2 double mutants, indicating SIS is an RNAi-related phenomenon.
Sequencing of the C. neoformans sRNA population revealed that in addition to the siRNAs derived from transgene loci, a significant portion of the siRNAs mapped to repetitive transposable elements and putative centromeric regions, which are rich in repetitive sequences and transposons (91). In the Rdp1, Ago1, Dcr1, and Dcr2 mutants, siRNA production from these loci was abolished. Importantly, the expression of the core RNAi components was highly induced during the sexual cycle, explaining why SIS is activated during the sexual cycle. Supporting the role for SIS in transposon control in C. neoformans, retrotransposons were found to be highly expressed during mating in rdp1 mutants. Although how SIS recognizes repetitive sequences is not clear, its similarity to quelling and meiotic silencing in Neurospora suggests they may be similar pathways.
ROLE OF RNAi IN FUNGAL TRANSPOSON AND VIRAL DEFENSE
Transposon control in N. crassa.
The silencing of repetitive transgenes by quelling suggests that quelling is a host defense against transposon invasion. Most of the examined N. crassa strains, however, lack functional transposons, due to the potent RIP process during the sexual cycle. The only exception is a LINE-1-like transposon, Tad, found in one African Neurospora isolate that was able to transpose in Neurospora (49). By introducing Tad into laboratory N. crassa strains, Nolan et al. showed that the repression of its transposition requires QDE-2 and Dicer but not QDE-1 or QDE-3 (70). These results suggest that Tad transposition may generate inverted repeats, from which dsRNA can be produced without QDE-1 and QDE-3.
Viral defense in Cryphonectria parasitica and Aspergillus nidulans.
The role of fungal RNAi in viral defense was first established in the ascomycete filamentous fungus C. parasitica, the chestnut blight fungus. Nuss and his colleagues found that Cryphonectria hypovirus 1 (CHV1) alleviates the virulence of the fungal host by expression of a papain-like protease, p29. The presence of p29 suppresses hairpin RNA-induced silencing in C. parasitica (76). C. parasitica has two Dicer-like genes, dcl-1 and dcl-2. The dcl-2 mutant shows impaired growth after virus infection, indicating that DCL-2 is involved in virus suppression (77). dcl-2 is essential for production of siRNAs derived from viral transcripts, and its expression is induced by virus infection (94). Although C. parasitica has four Argonaute-like genes, only agl2 is required for the antiviral defense response, and its expression is also induced by viral infection (88). When the fungus was infected by a mutant hypovirus without p29, agl2 and dcl2 mRNAs accumulated to very high levels, suggesting that the virus suppresses RNAi by repressing the expression of the core RNAi genes. In addition to their role in silencing viral replication posttranscriptionally, DCL-2 and AGL2 also promote viral RNA recombination in C. parasitica, resulting in the production of hypovirus defective interfering RNAs. Although the mechanism of the viral recombination is not clear, it is believed to be a viral control mechanism mediated by RNAi components (88, 93, 94).
The model organism Aspergillus nidulans also possesses a fully functional RNAi pathway, and the expression of inverted repeat-containing transgenes in this organism results in robust gene silencing (40). A. nidulans expresses one Dicer and one Argonaute protein, both of which are required for gene silencing. As in N. crassa, the RdRPs are not required for the inverted repeat triggered RNAi (39, 40). By infecting A. nidulans with mycoviruses, it was shown that viral infection suppresses the efficiency of RNAi, suggesting that an RNAi suppressor is encoded by this virus (39). Virus-derived siRNAs were maintained at a high level in the Argonaute mutants, confirming the antiviral role of RNAi.
APPLICATIONS OF RNAi IN FUNGI
RNAi provides an efficient and predictable gene silencing effect: recognition is based on the base pairing between siRNA and target RNA. Since RNAi-mediated silencing normally does not change the genomic structure of target genes, it serves as a good alternative to gene knockout and is extremely useful in studies of gene functions, especially when a gene is essential for cell viability (18). Moreover, RNAi-related methods may be used as tools in biotechnology to facilitate production of certain fungal metabolites by shutting down the expression of enzymes required for synthesis of end products (75). Although the RNAi pathway has been lost sporadically in various fungal species and lineages, most fungi possess this ancient mechanism (64). Three known methods can trigger RNAi-mediated gene silencing in fungi: expression of a hairpin RNA from a transgene, generation of dsRNA by convergent transcription from a transgene, and introduction of siRNA or dsRNA directly into the fungal cell (47, 57). Since the latter approach is not efficient for most fungi, the first two are the most commonly used.
Gene silencing via expression of a long hairpin RNA.
The expression of a hairpin RNA specific for the gene of interest requires the creation of plasmid constructs in which transcription of an inverted repeat (up to 1 kb long) is under the control of a commonly used promoter (18, 35, 58). The expression of the inverted repeats in cells ensures the formation of a long hairpin RNA that is efficiently recognized and processed by Dicer. This approach has been widely used in many fungal species (45, 57, 58, 65). To facilitate the plasmid construction, several vectors have been developed, including a pSilent-1 vector that allows high-throughput RNA silencing (50, 80). The use of an inducible promoter to control the expression of the hairpin RNA offers significant advantages over constitutive promoters, especially when essential genes are the targets. The widely used quinic acid-inducible promoter in N. crassa and the alcohol dehydrogenase promoter in A. nidulans have been very effective in silencing gene expression in an inducer-dependent and dosage-dependent manner (7, 18, 35).
Dual promoters (convergent transcription).
Although the inducible hairpin RNA silencing system is efficient, construction of plasmids containing the inverted repeats can be time-consuming, which limits its application to large-scale gene-silencing analyses. One alternative is to create plasmids with dual promoters. In this system, the gene of interest is flanked by two promoters that drive transcription in opposite directions. Thus, both the sense and antisense RNAs are transcribed simultaneously, and dsRNA should form (95). This method is suitable for high-throughput RNAi studies, in that the construct is relatively easy to make using systems like the Gateway RNAi vector system for fungi (66). The major disadvantage of this approach is that silencing is relatively inefficient compared to that with the inducible hairpin system, perhaps due to the low efficiency of formation of dsRNA (66, 73).
CONCLUSION
Studies of RNAi in fungi have made fundamental contributions to our understanding of RNAi and its functions. The recent discoveries of various small RNA biogenesis mechanisms in different fungi highlight the diversity of small RNAs and adaptation of RNAi pathways to different organisms and environments. Future RNAi studies of various fungal systems will no doubt shed light on the mechanisms, functions, and evolutionary origins of sRNA and RNAi pathways in eukaryotes.
ACKNOWLEDGMENTS
The space limitations of this minireview preclude a comprehensive description of RNAi-related work in fungi, and we apologize to those whose work we were unable to cite.
We thank the members of our laboratory for critical comments on the manuscript.
This research was supported by grants from the National Institutes of Health and the Welch Foundation (I-1560) to Yi Liu.
Footnotes
Published ahead of print on 1 July 2011.
REFERENCES
- 1. Alexander W. G., et al. 2008. DCL-1 colocalizes with other components of the MSUD machinery and is required for silencing. Fungal Genet. Biol. 45:719–727 [DOI] [PubMed] [Google Scholar]
- 2. Allen E., Xie Z., Gustafson A. M., Carrington J. C. 2005. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121:207–221 [DOI] [PubMed] [Google Scholar]
- 3. Ambros V., et al. 2003. A uniform system for microRNA annotation. RNA 9:277–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aramayo R., Metzenberg R. L. 1996. Meiotic transvection in fungi. Cell 86:103–113 [DOI] [PubMed] [Google Scholar]
- 5. Bardiya N., et al. 2008. Characterization of interactions between and among components of the meiotic silencing by unpaired DNA machinery in Neurospora crassa using bimolecular fluorescence complementation. Genetics 178:593–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bartel D. P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297 [DOI] [PubMed] [Google Scholar]
- 7. Barton L. M., Prade R. A. 2008. Inducible RNA interference of brlAβ in Aspergillus nidulans. Eukaryot. Cell 7:2004–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brennecke J., et al. 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128:1089–1103 [DOI] [PubMed] [Google Scholar]
- 9. Buhler M., Moazed D. 2007. Transcription and RNAi in heterochromatic gene silencing. Nat. Struct. Mol. Biol. 14:1041–1048 [DOI] [PubMed] [Google Scholar]
- 10. Buker S. M., et al. 2007. Two different Argonaute complexes are required for siRNA generation and heterochromatin assembly in fission yeast. Nat. Struct. Mol. Biol. 14:200–207 [DOI] [PubMed] [Google Scholar]
- 11. Carmell M. A., Xuan Z., Zhang M. Q., Hannon G. J. 2002. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 16:2733–2742 [DOI] [PubMed] [Google Scholar]
- 12. Carthew R. W., Sontheimer E. J. 2009. Origins and mechanisms of miRNAs and siRNAs. Cell 136:642–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Catalanotto C., Azzalin G., Macino G., Cogoni C. 2000. Gene silencing in worms and fungi. Nature 404:245. [DOI] [PubMed] [Google Scholar]
- 14. Catalanotto C., Azzalin G., Macino G., Cogoni C. 2002. Involvement of small RNAs and role of the qde genes in the gene silencing pathway in Neurospora. Genes Dev. 16:790–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Catalanotto C., et al. 2004. Redundancy of the two dicer genes in transgene-induced posttranscriptional gene silencing in Neurospora crassa. Mol. Cell. Biol. 24:2536–2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cerutti H., Casas-Mollano J. A. 2006. On the origin and functions of RNA-mediated silencing: from protists to man. Curr. Genet. 50:81–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cheloufi S., Dos Santos C. O., Chong M. M., Hannon G. J. 2010. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature 465:584–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cheng P., He Q., Wang L., Liu Y. 2005. Regulation of the Neurospora circadian clock by an RNA helicase. Genes Dev. 19:234–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chicas A., Forrest E. C., Sepich S., Cogoni C., Macino G. 2005. Small interfering RNAs that trigger posttranscriptional gene silencing are not required for the histone H3 Lys9 methylation necessary for transgenic tandem repeat stabilization in Neurospora crassa. Mol. Cell. Biol. 25:3793–3801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Choudhary S., et al. 2007. A double-stranded-RNA response program important for RNA interference efficiency. Mol. Cell. Biol. 27:3995–4005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cifuentes D., et al. 2010. A novel miRNA processing pathway independent of Dicer requires Argonaute 2 catalytic activity. Science 328:1694–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cogoni C., et al. 1996. Transgene silencing of the al-1 gene in vegetative cells of Neurospora is mediated by a cytoplasmic effector and does not depend on DNA-DNA interactions or DNA methylation. EMBO J. 15:3153–3163 [PMC free article] [PubMed] [Google Scholar]
- 23. Cogoni C., Macino G. 1997. Isolation of quelling-defective (qde) mutants impaired in posttranscriptional transgene-induced gene silencing in Neurospora crassa. Proc. Natl. Acad. Sci. U. S. A. 94:10233–10238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cogoni C., Macino G. 1999. Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature 399:166–169 [DOI] [PubMed] [Google Scholar]
- 25. Cogoni C., Macino G. 1999. Posttranscriptional gene silencing in Neurospora by a RecQ DNA helicase. Science 286:2342–2344 [DOI] [PubMed] [Google Scholar]
- 26. de Haro J. P., et al. 2009. A single Dicer gene is required for efficient gene silencing associated with two classes of small antisense RNAs in Mucor circinelloides. Eukaryot. Cell 8:1486–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ding S. W., Voinnet O. 2007. Antiviral immunity directed by small RNAs. Cell 130:413–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Drinnenberg I. A., et al. 2009. RNAi in budding yeast. Science 326:544–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fire A., et al. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806–811 [DOI] [PubMed] [Google Scholar]
- 30. Freitag M., et al. 2004. DNA methylation is independent of RNA interference in Neurospora. Science 304:1939. [DOI] [PubMed] [Google Scholar]
- 31. Fulci V., Macino G. 2007. Quelling: post-transcriptional gene silencing guided by small RNAs in Neurospora crassa. Curr. Opin. Microbiol. 10:199–203 [DOI] [PubMed] [Google Scholar]
- 32. Gazzani S., Lawrenson T., Woodward C., Headon D., Sablowski R. 2004. A link between mRNA turnover and RNA interference in Arabidopsis. Science 306:1046–1048 [DOI] [PubMed] [Google Scholar]
- 33. Gent J. I., et al. 2010. Distinct phases of siRNA synthesis in an endogenous RNAi pathway in C. elegans soma. Mol. Cell 37:679–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ghildiyal M., Zamore P. D. 2009. Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 10:94–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Goldoni M., Azzalin G., Macino G., Cogoni C. 2004. Efficient gene silencing by expression of double stranded RNA in Neurospora crassa. Fungal Genet. Biol. 41:1016–1024 [DOI] [PubMed] [Google Scholar]
- 36. Grewal S. I. 2010. RNAi-dependent formation of heterochromatin and its diverse functions. Curr. Opin. Genet. Dev. 20:134–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Grimm L. H., Kelly S., Krull R., Hempel D. C. 2005. Morphology and productivity of filamentous fungi. Appl. Microbiol. Biotechnol. 69:375–384 [DOI] [PubMed] [Google Scholar]
- 38. Gunawardane L. S., et al. 2007. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315:1587–1590 [DOI] [PubMed] [Google Scholar]
- 39. Hammond T. M., Andrewski M. D., Roossinck M. J., Keller N. P. 2008. Aspergillus mycoviruses are targets and suppressors of RNA silencing. Eukaryot. Cell 7:350–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hammond T. M., Keller N. P. 2005. RNA silencing in Aspergillus nidulans is independent of RNA-dependent RNA polymerases. Genetics 169:607–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hannon G. J. 2002. RNA interference. Nature 418:244–251 [DOI] [PubMed] [Google Scholar]
- 42. Harris S. D. 2001. Septum formation in Aspergillus nidulans. Curr. Opin. Microbiol. 4:736–739 [DOI] [PubMed] [Google Scholar]
- 43. Herr A. J., Molnar A., Jones A., Baulcombe D. C. 2006. Defective RNA processing enhances RNA silencing and influences flowering of Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 103:14994–15001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hibbett D. S., et al. 2007. A higher-level phylogenetic classification of the Fungi. Mycol. Res. 111:509–547 [DOI] [PubMed] [Google Scholar]
- 45. Kadotani N., Nakayashiki H., Tosa Y., Mayama S. 2003. RNA silencing in the phytopathogenic fungus Magnaporthe oryzae. Mol. Plant Microbe Interact. 16:769–776 [DOI] [PubMed] [Google Scholar]
- 46. Kelly W. G., Aramayo R. 2007. Meiotic silencing and the epigenetics of sex. Chromosome Res. 15:633–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Khatri M., Rajam M. V. 2007. Targeting polyamines of Aspergillus nidulans by siRNA specific to fungal ornithine decarboxylase gene. Med. Mycol. 45:211–220 [DOI] [PubMed] [Google Scholar]
- 48. Kim V. N., Han J., Siomi M. C. 2009. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 10:126–139 [DOI] [PubMed] [Google Scholar]
- 49. Kinsey J. A. 1990. Tad, a LINE-like transposable element of Neurospora, can transpose between nuclei in heterokaryons. Genetics 126:317–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Krajaejun T., Gauthier G. M., Rappleye C. A., Sullivan T. D., Klein B. S. 2007. Development and application of a green fluorescent protein sentinel system for identification of RNA interference in Blastomyces dermatitidis illuminates the role of septin in morphogenesis and sporulation. Eukaryot. Cell 6:1299–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lee D. W., Millimaki R., Aramayo R. 2010. QIP, a component of the vegetative RNA silencing pathway, is essential for meiosis and suppresses meiotic silencing in Neurospora crassa. Genetics 186:127–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lee D. W., Pratt R. J., McLaughlin M., Aramayo R. 2003. An Argonaute-like protein is required for meiotic silencing. Genetics 164:821–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lee H. C., et al. 2010. The DNA/RNA-dependent RNA polymerase QDE-1 generates aberrant RNA and dsRNA for RNAi in a process requiring replication protein A and a DNA helicase. PLoS Biol. 8:e1000496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lee H. C., et al. 2009. qiRNA is a new type of small interfering RNA induced by DNA damage. Nature 459:274–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lee H. C., et al. 2010. Diverse pathways generate microRNA-like RNAs and Dicer-independent small interfering RNAs in fungi. Mol. Cell 38:803–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Li C., et al. 2009. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell 137:509–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Li L., Chang S. S., Liu Y. 2010. RNA interference pathways in filamentous fungi. Cell. Mol. Life Sci. 67:3849–3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Liu H., Cottrell T. R., Pierini L. M., Goldman W. E., Doering T. L. 2002. RNA interference in the pathogenic fungus Cryptococcus neoformans. Genetics 160:463–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Liu Y., et al. 2009. C3PO, an endoribonuclease that promotes RNAi by facilitating RISC activation. Science 325:750–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Maiti M., Lee H. C., Liu Y. 2007. QIP, a putative exonuclease, interacts with the Neurospora Argonaute protein and facilitates conversion of duplex siRNA into single strands. Genes Dev. 21:590–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Malone C. D., et al. 2009. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137:522–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Moazed D. 2009. Small RNAs in transcriptional gene silencing and genome defence. Nature 457:413–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Nakayashiki H., et al. 2005. RNA silencing as a tool for exploring gene function in ascomycete fungi. Fungal Genet. Biol. 42:275–283 [DOI] [PubMed] [Google Scholar]
- 64. Nakayashiki H., Kadotani N., Mayama S. 2006. Evolution and diversification of RNA silencing proteins in fungi. J. Mol. Evol. 63:127–135 [DOI] [PubMed] [Google Scholar]
- 65. Namekawa S. H., et al. 2005. Knockdown of LIM15/DMC1 in the mushroom Coprinus cinereus by double-stranded RNA-mediated gene silencing. Microbiology 151:3669–3678 [DOI] [PubMed] [Google Scholar]
- 66. Nguyen Q. B., et al. 2008. Systematic functional analysis of calcium-signalling proteins in the genome of the rice-blast fungus, Magnaporthe oryzae, using a high-throughput RNA-silencing system. Mol. Microbiol. 68:1348–1365 [DOI] [PubMed] [Google Scholar]
- 67. Nicolas F. E., de Haro J. P., Torres-Martinez S., Ruiz-Vazquez R. M. 2007. Mutants defective in a Mucor circinelloides dicer-like gene are not compromised in siRNA silencing but display developmental defects. Fungal Genet. Biol. 44:504–516 [DOI] [PubMed] [Google Scholar]
- 68. Nicolas F. E., et al. 2010. Endogenous short RNAs generated by Dicer 2 and RNA-dependent RNA polymerase 1 regulate mRNAs in the basal fungus Mucor circinelloides. Nucleic Acids Res. 38:5535–5541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Nicolas F. E., Torres-Martinez S., Ruiz-Vazquez R. M. 2003. Two classes of small antisense RNAs in fungal RNA silencing triggered by non-integrative transgenes. EMBO J. 22:3983–3991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Nolan T., et al. 2005. The post-transcriptional gene silencing machinery functions independently of DNA methylation to repress a LINE1-like retrotransposon in Neurospora crassa. Nucleic Acids Res. 33:1564–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Nolan T., et al. 2008. The RNA-dependent RNA polymerase essential for post-transcriptional gene silencing in Neurospora crassa interacts with replication protein A. Nucleic Acids Res. 36:532–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Pratt R. J., Lee D. W., Aramayo R. 2004. DNA methylation affects meiotic trans-sensing, not meiotic silencing, in Neurospora. Genetics 168:1925–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Rappleye C. A., Engle J. T., Goldman W. E. 2004. RNA interference in Histoplasma capsulatum demonstrates a role for alpha-(1,3)-glucan in virulence. Mol. Microbiol. 53:153–165 [DOI] [PubMed] [Google Scholar]
- 74. Romano N., Macino G. 1992. Quelling: transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol. Microbiol. 6:3343–3353 [DOI] [PubMed] [Google Scholar]
- 75. Salame T. M., Ziv C., Hadar Y., Yarden O. 2011. RNAi as a potential tool for biotechnological applications in fungi. Appl. Microbiol. Biotechnol. 89:501–512 [DOI] [PubMed] [Google Scholar]
- 76. Segers G. C., van Wezel R., Zhang X., Hong Y., Nuss D. L. 2006. Hypovirus papain-like protease p29 suppresses RNA silencing in the natural fungal host and in a heterologous plant system. Eukaryot. Cell 5:896–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Segers G. C., Zhang X., Deng F., Sun Q., Nuss D. L. 2007. Evidence that RNA silencing functions as an antiviral defense mechanism in fungi. Proc. Natl. Acad. Sci. U. S. A. 104:12902–12906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Selker E. U., Cambareri E. B., Jensen B. C., Haack K. R. 1987. Rearrangement of duplicated DNA in specialized cells of Neurospora. Cell 51:741–752 [DOI] [PubMed] [Google Scholar]
- 79. Shabalina S. A., Koonin E. V. 2008. Origins and evolution of eukaryotic RNA interference. Trends Ecol. Evol. 23:578–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Shafran H., Miyara I., Eshed R., Prusky D., Sherman A. 2008. Development of new tools for studying gene function in fungi based on the Gateway system. Fungal Genet. Biol. 45:1147–1154 [DOI] [PubMed] [Google Scholar]
- 81. Shiu P. K., Metzenberg R. L. 2002. Meiotic silencing by unpaired DNA: properties, regulation and suppression. Genetics 161:1483–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Shiu P. K., Raju N. B., Zickler D., Metzenberg R. L. 2001. Meiotic silencing by unpaired DNA. Cell 107:905–916 [DOI] [PubMed] [Google Scholar]
- 83. Shiu P. K., Zickler D., Raju N. B., Ruprich-Robert G., Metzenberg R. L. 2006. SAD-2 is required for meiotic silencing by unpaired DNA and perinuclear localization of SAD-1 RNA-directed RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 103:2243–2248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Siomi H., Siomi M. C. 2009. On the road to reading the RNA-interference code. Nature 457:396–404 [DOI] [PubMed] [Google Scholar]
- 85. Siomi M. C., Sato K., Pezic D., Aravin A. A. 2011. PIWI-interacting small RNAs: the vanguard of genome defence. Nat. Rev. Mol. Cell Biol. 12:246–258 [DOI] [PubMed] [Google Scholar]
- 86. Song J. J., Smith S. K., Hannon G. J., Joshua-Tor L. 2004. Crystal structure of Argonaute and its implications for RISC Slicer activity. Science 305:1434–1437 [DOI] [PubMed] [Google Scholar]
- 87. Steiner F. A., Okihara K. L., Hoogstrate S. W., Sijen T., Ketting R. F. 2009. RDE-1 slicer activity is required only for passenger-strand cleavage during RNAi in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 16:207–211 [DOI] [PubMed] [Google Scholar]
- 88. Sun Q., Choi G. H., Nuss D. L. 2009. A single Argonaute gene is required for induction of RNA silencing antiviral defense and promotes viral RNA recombination. Proc. Natl. Acad. Sci. U. S. A. 106:17927–17932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Thomson T., Lin H. 2009. The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annu. Rev. Cell Dev. Biol. 25:355–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Volpe T., Martienssen R. A. 19 January 2011, posting date. RNA interference and heterochromatin assembly. Cold Spring Harb. Perspect. Biol. doi:10.1101/cshperspect.a003731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Wang X., et al. 2010. Sex-induced silencing defends the genome of Cryptococcus neoformans via RNAi. Genes Dev. 24:2566–2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Xiao H., et al. 2010. QIP, a protein that converts duplex siRNA into single strands, is required for meiotic silencing by unpaired DNA. Genetics 186:119–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Zhang X., Nuss D. L. 2008. A host Dicer is required for defective viral RNA production and recombinant virus vector RNA instability for a positive sense RNA virus. Proc. Natl. Acad. Sci. U. S. A. 105:16749–16754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Zhang X., Segers G. C., Sun Q., Deng F., Nuss D. L. 2008. Characterization of hypovirus-derived small RNAs generated in the chestnut blight fungus by an inducible DCL-2-dependent pathway. J. Virol. 82:2613–2619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Ziv C., Yarden O. 2010. Gene silencing for functional analysis: assessing RNAi as a tool for manipulation of gene expression. Methods Mol. Biol. 638:77–100 [DOI] [PubMed] [Google Scholar]