Abstract
Coordinated regulation of gene expression is a hallmark of the Plasmodium falciparum asexual blood-stage development cycle. We report that carbon catabolite repressor protein 4 (CCR4)-associated factor 1 (CAF1) is critical in regulating more than 1,000 genes during malaria parasites' intraerythrocytic stages, especially egress and invasion proteins. CAF1 knockout results in mistimed expression, aberrant accumulation and localization of proteins involved in parasite egress, and invasion of new host cells, leading to premature release of predominantly half-finished merozoites, drastically reducing the intraerythrocytic growth rate of the parasite. This study demonstrates that CAF1 of the CCR4-Not complex is a significant gene regulatory mechanism needed for Plasmodium development within the human host.
INTRODUCTION
Regulation of eukaryotic gene expression is complex, ranging from pretranscriptional to posttranslational regulation. The most stringent control of gene expression is believed to be at the transcriptional level, although posttranscriptional mechanisms have emerged as important modulators (5, 15, 17). Gene regulation in malaria parasite intraerythrocytic development is highly coordinated, with a majority of genes temporally regulated (7, 22). Sequenced Plasmodium genomes reveal a high conservation of basal core eukaryotic transcriptional machinery with few transcription factors (1, 10). Instead, Plasmodium species appear to rely on orthologs of plant Apetala2-like transcription factors (ApiAP2) (2, 14, 26), while evidence for posttranscriptional control is primarily associated with translational repression in sexual stages (25).
The carbon catabolite repressor protein 4 (CCR4)-Not complex is a well-conserved, eukaryotic gene regulatory complex, broadly divided into two modules: CCR4 and CCR4-associated factor 1 (CAF1), regulating mRNA decay, and the Not proteins, regulating transcription and protein degradation (8, 11, 31). Members of the CCR4-Not complex are well conserved in all Plasmodium species, suggestive of their significance in parasite gene regulation as well (10). In this study, we use a Plasmodium falciparum caf1 disruptant (ΔCAF1) mutant, created during the course of a large-scale transposon mutagenesis project of P. falciparum (3), to evaluate the function of the CCR4-Not complex in parasite gene regulation and its significance in intraerythrocytic development.
MATERIALS AND METHODS
Creation and validation of a P. falciparum CAF1 knockout mutant.
Wild-type (WT) NF54 and Δcaf1 clones were cultured in 5% hematocrit (O-positive blood), RPMI 1640 (Invitrogen), 0.5% Albumax, or 10% human AB serum. Δcaf1 clone A was representative of Δcaf1 parasites in all experiments.
For Southern blot analysis, 2 μg genomic DNA was restricted with 10 U EcoRV, separated by gel electrophoresis, depurinated, denatured, and blotted in 1 M ammonium acetate and 0.02 N NaOH to a nylon membrane. The blot was hybridized to 32P-labeled hdhfr, washed, and visualized on film. For Northern blot analysis, 10 μg of total RNA from mixed intraerythrocytic stages was separated by gel electrophoresis, blotted to a nylon membrane, hybridized with 32P-labeled PCR product of caf1 primers (AACTCACCACCTTCTCCATCTTC, CGTTGGCAATATTATTTAAATCG), washed, and visualized on film. For reverse transcription (RT)-PCR analysis (Invitrogen), we analyzed 100 ng DNase I-treated total RNA for caf1 (ATTAGGTTCAACAATTAAAAATTATAACC, TATTATTTGGTGCATTCATGAGC) and maebl (GAACAAGAAGAATATTTTTTAGTACTGC, GGCATGCATGGAGAACTACC) (1 cycle at 42°C followed by 35 cycles of 94°C for 15 s, 50°C for 30 s, and 65°C for 1 min). RT-PCR products were analyzed by gel electrophoresis, cloned, and sequenced.
Attempts to generate a Plasmodium berghei CAF1 gene deletion mutant.
In order to create a P. berghei caf1 (PBANKA_142620) deletion mutant, constructs were generated which would target the gene by double-crossover homologous recombination as previously described (18). These constructs were amplified using an adapted, previously described, anchor-tagging PCR-based method for generation of gene deletion constructs (13). This method employs a two-step PCR, shown in Fig. S1 in the supplemental material. In the first PCR, two-flanking fragments (5′ or 3′ target; sizes are indicated in Fig. S1 in base pairs) were amplified using genomic DNA (cl15cy1) as the template with the primer pairs 4674/4726 (GAACTCGTACTCCTTGGTGACGGGTACCATTGGGGAACTTGTTCAC/CATCTACAAGCATCGTCGACCTCTCACTTTTTGCATACTACAC) or 5′ target 5342/5343 (GAACTCGTACTCCTTGGTGACGGGTACCTCCTTATGTAGCCATTGAC/CATCTACAAGCATCGTCGACCTCCCAAGCCATATATAATACCTG) for pL1518 and pL1585 and 3′ target 4727/4675 (CCTTCAATTTCGGATCCACTAGTGTGGCTTGTGTTAAAGAC/AGGTTGGTCATTGACACTCAGCAGTACTGCCTCTTCCCATTATTCTG). The primers 4726, 5343, and 4727 have 5′-terminal extensions homologous to the hdhfr selectable marker (SM) cassette. This cassette contains hdfr under the control of the ef1a promoter region and the 3′ untranslated region (UTR) pbdhfr/ts and is obtained from plasmid pL0040 by digestion with restriction enzymes XhoI and NotI (pL0040 is available from The Leiden Malaria Research Group). Primers 4674, 5342, and 4675 have a 5′-terminal overhang with an anchor tag suitable for the second PCR. In the second PCR, the fragments were annealed to either side of the hdhfr selectable marker cassette with anchor tag primers 4661/4662 (GAACTCGTACTCCTTGGTGACG/AGGTTGGTCATTGACACTCAGC), resulting in the second PCR fragment with the expected size, i.e., 3.0 kb (1.6 kb of the selectable marker cassette plus two targeting fragments). Sizes and locations of 5′- and 3′-targeting sequences are given in Fig. S1 in the supplemental material. To remove the anchor tag from the final DNA construct, the second PCR fragment was digested with Asp718 and ScaI, as primer 4674/5342 contained an Asp718 restriction enzyme site and 4675 contained an ScaI site. Following ethanol precipitation, the PCR fragment was resuspended in water at 1 μg/μl and used for transfection.
Determination of merozoite number per schizont.
Merozoite numbers were counted in 100 schizonts, three different times, from either WT or ΔCAF1 parasites to determine the average number of merozoites per schizont.
Estimation of P. falciparum asexual cell cycle and proliferation assays.
Parasites were synchronized two times by sorbitol and trophozoites transferred to 96-well plates at 1% parasitemia. For each time point, 200 μl of culture was plated in triplicates, and samples were taken every 2 h for 86 h and processed for flow cytometry. The percentages of schizonts at each time point were fitted into an exponential sine wave function using Kaleidagraph (Synergy Software), and cycle time was estimated as described before (24). Proliferation assays were done as described before at 0.5% starting parasitemia of magnet-purified schizonts, which were allowed to invade fresh red blood cells, and the resulting parasitemia was measured after 24 h (4, 23, 27). Experiments were performed with three biological replicates.
RNA expression analysis by microarray hybridization.
Parasite samples for RNA extractions using a TRI reagent-based RiboPure kit (Applied Biosystems) were collected from three biological replicates of highly synchronized cultures at 6 different time points of intraerythrocytic stages, representing early rings (7 h postinvasion), late rings (14 h), early trophozoites (21 h), late trophozoites (28 h), early schizonts (35 h), and late schizonts (42 h). RNA expression levels were evaluated through hybridization to PFSANGERb520296 Affymetrix arrays as described before (9). Arrays were background adjusted and quantile normalized using the robust multiarray averaging algorithm (16) and analyzed by R/Bioconductor (http://www.bioconductor.org), affy (16), and limma packages (30). Data were P value adjusted to yield a sorted list of differentially expressed genes. Hierarchical clustering was carried out using log-transformed ratios of ΔCAF1 expression values to wild-type expression values in Cluster 3.0 (12) and visualized using Java Treeview (28). A logFC cutoff greater than 1 and less than −1 was imposed to identify genes differentially expressed >2-fold. Raw microarray data will be submitted to the NCBI GEO public database.
Immunoblot analysis.
Twice-synchronized cultures of WT and ΔCAF1 parasites were analyzed starting at mid-trophozoite stages every 2 h. Twenty million magnetically purified parasites were washed in 1× phosphate-buffered saline (PBS) with protease inhibitor (Sigma), resuspended directly in 2× SDS-PAGE sample buffer, boiled for 5 min, flash frozen in liquid nitrogen, stored at −80°C until separation on a 4 to 12% gradient NuPAGE precast gel (Invitrogen), and transferred to a nitrocellulose membrane. Primary antibody dilutions of 1:500, 1:5,000, 1:1,000, and 1:500 for subtilisin 1 (SUB1), serine repeat antigen 5 (SERA5), glideosome-associated protein 45 (GAP45), and erythrocyte binding antigen 175 (EBA175), respectively, were used to probe the membrane.
Immunofluorescence assays.
Purified late-schizont-stage parasites were fixed overnight in 4% paraformaldehyde and 0.0075% glutaraldehyde in RPMI medium, permeabilized in 0.1% Triton X-100 in PBS for 10 min, blocked in 3% bovine serum albumin (BSA) for 10 min, and incubated with a 1:100 dilution of primary antibody for 1 h at 37°C. Following a wash with PBS, parasites were incubated with a 1:70 dilution of secondary antibody for 1 h at room temperature, except in the case of allophycocyanin (APC)-conjugated anti-glycophorin A antibody, which was used at 2 μg/ml and incubated for 15 min. Parasites were washed in 1× PBS, suspended in 1:1 PBS/mounting medium, placed on a glass slide with a poly-l-lysine-coated coverslip, sealed, and visualized by microscopy. Rabbit anti-SUB1 antibody and monoclonal antibody against SERA5 were kind gifts from Michael Blackman and Jean-François Dubremetz, respectively. Affinity-purified anti-GAP45 and anti-EBA175 antibodies were obtained as previously described (6, 19).
Live video microscopy.
Purified late schizonts at 30% parasitemia were viewed with a DeltaVision CORE microscope (Applied Precision). Schizonts were dually labeled with 2 μg/ml anti-glycophorin A antibody–APC (BD Biosciences) and with 10 μg/ml Hoechst 33342 for 10 min and washed in 1× PBS before microscopy.
Transmission electron microscopy.
Purified late schizonts were fixed (2.5% glutaraldehyde, 0.1 M phosphate, pH 7.2), postfixed (1% OsO4, 0.1 M phosphate, pH 7.2), dehydrated in acetone, and embedded in LX112 (EMS). Sections were stained with 8% aqueous uranyl acetate and Reynolds lead citrate before imaging on a JEOL JEM1400 transmission electron microscope.
RESULTS
caf1 knockout parasites display proliferation defects.
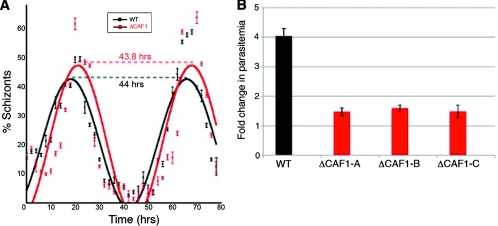
A large-scale transposon mutagenesis study in P. falciparum identified caf1 (PlasmoDB ID MAL8P1.104) as critical for intraerythrocytic growth (3). Disruption of the caf1 (ΔCAF1) coding sequence turned off the transcription of full-length caf1 in intraerythrocytic stages (Fig. 1). Preliminary studies found that the average numbers of merozoites produced per schizont (18.5 and 19, respectively) and cell cycle lengths (Fig. 2A) were similar in ΔCAF1 and wild-type (WT) parasites. Further investigation of this mutant revealed that the growth rate was severely attenuated (Fig. 2B). A significant decrease in proliferation efficiency (two-tailed Student's t test; P = 0.0001) indicated a major defect in parasite egress from the host cell and/or invasion of erythrocytes. ΔCAF1-attenuated growth remained remarkably stable over 50 generations, indicating that survival of a few merozoites each generation was not due to phenotype reversion. Attempts to complement the ΔCAF1 parasites were unsuccessful due to the inability to PCR amplify the approximately 7-kb fragment containing the promoter and coding sequence of caf1.
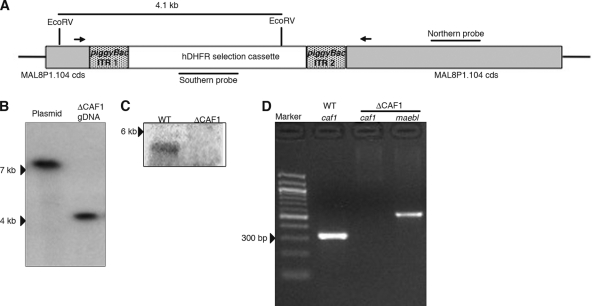
Fig. 1.
Genetic disruption of P. falciparum caf1. (A) A schematic of caf1 disruption showing piggyBac repeats ITR1 and ITR2 flanking hdfr, RT-PCR primers (arrows), probes, and the predicted EcoRV fragment. (B) Southern blot hybridization analysis showed a single integration event in caf1 by the hdhfr probe. (C) Northern hybridization analysis identified a 5-kb transcript for caf1 in the wild-type (WT) parasites but not in the ΔCAF1 clone. (D) RT-PCR analysis detected caf1 in wild-type (WT) parasites but not in ΔCAF1 parasites; maebl was included as the RT-PCR control in ΔCAF1 parasites.
Fig. 2.
Growth phenotype of ΔCAF1 parasites. (A) Intraerythrocytic cycle times of WT and ΔCAF1 parasites were approximately 44 h for both. Error bars indicate standard deviations from three replicates. (B) In vitro assays identified a drastic decrease in proliferation efficiency of three different ΔCAF1 parasite clones. Error bars indicate standard deviations from three replicates.
Transfection experiments to generate a corresponding gene deletion of P. berghei caf1 were attempted with each construct twice, with a total of four independent transfection experiments attempted, and we were not able to obtain mutant parasites (see Fig. S1 in the supplemental material). These multiple unsuccessful attempts to generate a caf1 knockout parasite in the rodent malaria parasite P. berghei demonstrate that the locus is refractory to disruption and suggest essentiality of this gene in vivo.
Disruption of caf1 causes major alterations in parasite gene expression.
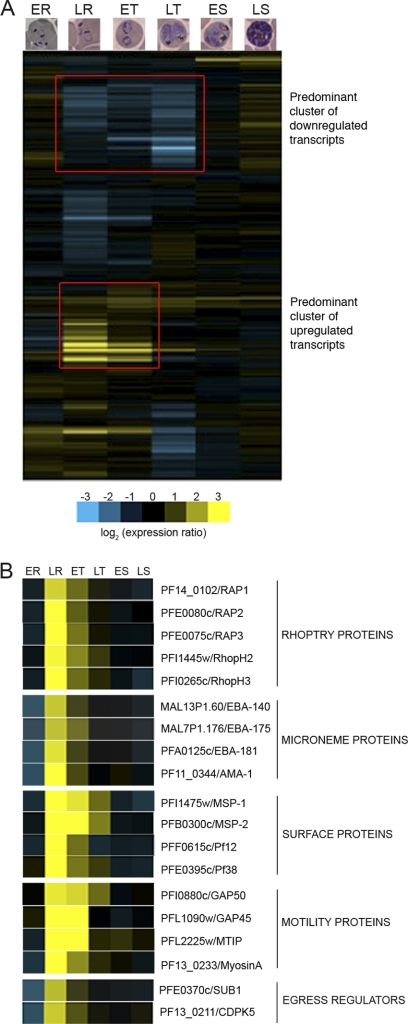
Gene expression analysis of the ΔCAF1-attenuated phenotype compared to that of WT NF54 revealed major transcriptional changes of ΔCAF1 parasites during intraerythrocytic development (Fig. 3A; see also Data Set S1 in the supplemental material). Significant differential expression (P < 0.05) occurred in 1,031 genes in ΔCAF1 parasites, mostly in late-ring through trophozoite stages, with the abundance of transcripts increasing in 366 and decreasing in 665 genes (Table 1; see also Data Set S2 in the supplemental material). The most dramatic effects observed in ΔCAF1 parasites were the increased transcript levels of late schizont genes that are key regulators of merozoite egress and invasion (Fig. 3B).
Fig. 3.
Disruption of P. falciparum caf1 leads to major changes in intraerythrocytic transcription. (A) A hierarchical clustering of log-transformed ratios of ΔCAF1 values to WT values shows a global view of transcriptional changes during intraerythrocytic development in ΔCAF1 (ER, early ring; LR, late ring; ET, early trophozoite; LT, late trophozoite; ES, early schizont; LS, late schizont). Two prominent gene clusters are marked with red rectangles. (B) Disruption of caf1 increased transcript levels of late-schizont-stage genes involved in merozoite egress and invasion. Log fold changes at each stage are color coded (blue, decreased transcript abundance; yellow, increased transcript abundance).
Table 1.
Number of transcripts differentially expressed >2-fold in ΔCAF1 parasites during asexual development (P < 0.05)
| Transcript type | No. of transcripts at parasite stagea: |
Total no. of unique transcripts | |||||
|---|---|---|---|---|---|---|---|
| ER | LR | ET | LT | ES | LS | ||
| Upregulated | 63 | 324 | 183 | 47 | 48 | 65 | 366 |
| Downregulated | 61 | 269 | 98 | 510 | 5 | 4 | 665 |
| Total | 124 | 593 | 281 | 557 | 53 | 69 | 1,031 |
ER, early ring; LR, late ring; ET, early trophozoite; LT, late trophozoite; ES, early schizont; LS, late schizont.
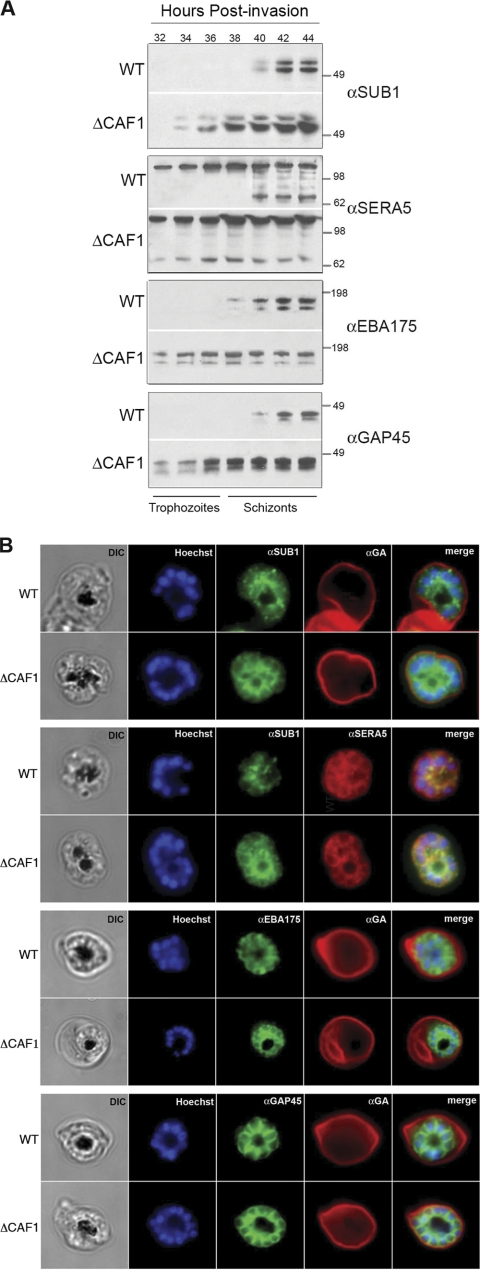
Aberrant expression and localization of late schizont proteins.
A time course analysis of protein expression in ΔCAF1 parasites confirmed premature expression of key merozoite proteins critical for egress (SUB1, SERA5) and invasion (EBA175, GAP45) (Fig. 4A). Importantly, a proteolytic fragment of SERA5 evident much earlier in asexual development in ΔCAF1 parasites indicates that SERA5 is prematurely activated by direct exposure to SUB1. Immunolocalization of SUB1 (Fig. 4B) in the parasitophorous vacuole of ΔCAF1 schizonts, which contrasts with the reported punctate pattern of staining in WT schizonts (32), supports early activation of SERA5 by SUB1. Differential localization was also observed for the transmembrane micronemal ligand, EBA175, which had an apical organelle localization pattern in WT merozoites but was on the surface of ΔCAF1 merozoites (Fig. 4B). The cytoplasmic motor protein GAP45 had a submembrane localization in WT and ΔCAF1 merozoites (Fig. 4B).
Fig. 4.
Mistimed expression and aberrant localization of late-sch-izont-stage proteins in ΔCAF1 parasites. (A) A time course immuno-blot analysis of late schizonts of WT and ΔCAF1 parasites showed premature expression of SUB1, EBA175, and GAP45 and the premature cleavage of SERA5 as early as 32 h postinvasion. Molecular standards (kDa) are shown on the right. (B) IFA of WT and ΔCAF1 schizonts, with glycophorin A (GA)-labeled erythrocytes, localized SUB1 in an apical pattern in WT schizonts but showed parasitophorous vacuole staining in ΔCAF1 schizonts similar to that of SERA5. EBA175 was detected in the apical ends of merozoites in WT parasites but was found distributed on the merozoite surface in ΔCAF1 parasites. GAP45 appeared submembrane in both WT and ΔCAF1 parasites. DIC, differential interference contrast; Hoechst, nucleic acid stain.
ΔCAF1 parasites are defective in egress from host cells.
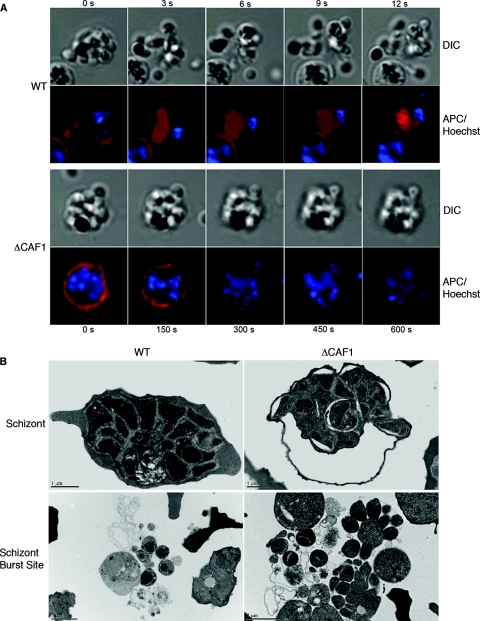
Live video microscopy of WT schizonts displays typical egress as a forceful merozoite release involving an explosion-like lysis of the erythrocyte membrane (Fig. 5A) (see Movies S1 and S3 in the supplemental material). In contrast, ΔCAF1 schizont egress is dysfunctional, occurring as slow degradation of the erythrocyte membrane, blebbing at several locations, with incomplete rupture (Fig. 5A) (see Movies S2 and S4 in the supplemental material). Even without the explosive merozoite release, the erythrocyte plasma membrane around ΔCAF1 schizonts degraded and disappeared (see Fig. S3 in the supplemental material). Physical disruption of ΔCAF1 schizonts failed to increase the invasion rate, suggesting that most merozoites from ΔCAF1 schizonts are not invasive (see Fig. S4 in the supplemental material). Electron microscopy confirmed that most ΔCAF1 schizonts fail to complete development prior to host erythrocyte lysis (Fig. 5B). Merozoites remained attached in clusters incompletely separated from the residual body and with little or no membrane around them (Fig. 5B).
Fig. 5.
ΔCAF1 parasites display defects in egress from host erythrocytes. (A) Visualization of parasite egress from host cells. Live video microscopy of P. falciparum egress stained for nuclei and erythrocyte membrane showed an explosion-like schizont burst and release of merozoites in the wild-type parasite. In the ΔCAF1 parasite, erythrocyte membranes were degraded over a long period, and no forceful release of merozoites was observed. (B) Schizont ultrastructure showed premature erythrocyte membrane lysis and incomplete merozoite maturation in ΔCAF1.
DISCUSSION
Malaria parasites undergo dramatic transformations during intraerythrocytic development, relying on a precise pattern of gene expression to form invasive merozoites at the end of this asexual cycle (Table 2). We find that CAF1 is vital for correct temporal control of gene expression required for appropriate localization and function of critical blood-stage proteins. Unrestricted and premature activation of genes in the CAF1-null mutants resulted in severe disruption of merozoite egress and invasion, resulting in early schizont lysis. Less dramatic are decreases in genes expressed in trophozoite stages involved in basic cellular processes and virulence (var, rifin).
Table 2.
Functional annotation of differentially expressed genes (P < 0.01) in ΔCAF1 parasites during asexual developmenta
| Parasite stageb | GO enrichment of upregulated genes | GO enrichment of downregulated genes |
|---|---|---|
| ER | None | None |
| LR | Chromosome organization and biogenesis; DNA replication; small GTPase-mediated signal transduction; entry into host cell | Pathogenesis (var); RNA splicing; translation |
| ET | DNA replication; small GTPase-mediated signal transduction; entry into host cell | Pathogenesis (var genes) |
| LT | None | Antigenic variation (rifin); mRNA metabolic process; translation; protein transport; small GTPase-mediated signal transduction |
| ES | None | None |
| LS | None | None |
Based on gene ontology, biological process annotation using the MADIBA Web server (21).
ER, early ring; LR, late ring; ET, early trophozoite; LT, late trophozoite; ES, early schizont; LS, late schizont.
Our discovery of the critical regulatory role for CAF1 in the P. falciparum intraerythrocytic cycle provides the first experimental evidence of the importance of the CCR4-Not complex in malaria parasite development. The conservation of CAF1 and other core components of the CCR4-Not complex, and its significance in malaria parasite viability, are consistent with the importance of the CCR4-Not complex found in all eukaryotes where it is studied. This includes the phylogenetically distant parasitic protozoa Trypanosoma brucei, where CAF1 was shown to be the key mediator of mRNA decay and essential for parasite viability (29).
In yeast and other eukaryotes, CAF1 has a major role in determining transcript levels through regulated deadenylation of mRNA. Our data demonstrate that P. falciparum CAF1 has an important role regulating gene-specific transcript abundance in approximately 20% of the parasite genome, especially proteins involved in merozoite egress and invasion. Proteins in these unique apicomplexan processes are targeted to specialized secretory organelles (e.g., micronemes) known as the apical complex and are sequestered there until needed. Since these apical organelles form only in late intraerythrocytic development, premature expression of apical organelle proteins, such as SUB1 and EBA175, results in their aberrant localization. A similar aberrant localization was observed when the temporal expression of apical membrane antigen 1 was altered (20). Therefore, in ΔCAF1 parasites, in the absence of its target organelle, SUB1 is released into the parasitophorous vacuole, resulting in activation of SERA and initiating low-grade host cell lysis that is normally coordinated with the final step of parasite egress. Consequently, CAF1 represents a critical checkpoint, limiting expression until the appropriate stage of maturity, and a key regulator of parasite development.
Until recently, gene expression of malaria parasites was perceived as primarily controlled at the transcription level, and posttranscriptional regulation was relatively unique. We discover that CAF1 of the CCR4-Not complex has a crucial role in P. falciparum gene regulation, influencing the transcript levels of a large number of genes in the intraerythrocytic stages. Such an extensive effect underscores the importance of the CCR4-Not complex in parasite intraerythrocytic development and suggests a much more complicated regulatory network controlling gene expression. Most importantly, this study identifies a novel gene regulatory pathway critical for exponential propagation of the disease-causing, intraerythrocytic stages of the malaria parasite. Further delineation of this gene regulatory pathway should not only augment our comprehension of parasite biology but also provide new avenues for intervention strategies.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by funds from U.S. National Institutes of Health (R01AI033656, R01AI094973), The Wellcome Trust, and The Netherlands Organization for Scientific Research (ZonMw TOP grant number 9120_6135).
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 29 July 2011.
REFERENCES
- 1. Aravind L., Iyer L. M., Wellems T. E., Miller L. H. 2003. Plasmodium biology: genomic gleanings. Cell 115:771–785 [DOI] [PubMed] [Google Scholar]
- 2. Balaji S., Babu M. M., Iyer L. M., Aravind L. 2005. Discovery of the principal specific transcription factors of Apicomplexa and their implication for the evolution of the AP2-integrase DNA binding domains. Nucleic Acids Res. 33:3994–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Balu B., et al. 2009. piggyBac is an effective tool for functional analysis of the Plasmodium falciparum genome. BMC Microbiol. 9:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Balu B., Singh N., Maher S. P., Adams J. H. 2010. A genetic screen for attenuated growth identifies genes crucial for intraerythrocytic development of Plasmodium falciparum. PLoS One 5:e13282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Besse F., Ephrussi A. 2008. Translational control of localized mRNAs: restricting protein synthesis in space and time. Nat. Rev. Mol. Cell Biol. 9:971–980 [DOI] [PubMed] [Google Scholar]
- 6. Blair P. L., Kappe S. H., Maciel J. E., Balu B., Adams J. H. 2002. Plasmodium falciparum MAEBL is a unique member of the ebl family. Mol. Biochem. Parasitol. 122:35–44 [DOI] [PubMed] [Google Scholar]
- 7. Bozdech Z., et al. 2003. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 1:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Collart M. A. 2003. Global control of gene expression in yeast by the Ccr4-Not complex. Gene 313:1–16 [DOI] [PubMed] [Google Scholar]
- 9. Cortes A., et al. 2007. Epigenetic silencing of Plasmodium falciparum genes linked to erythrocyte invasion. PLoS Pathog. 3:e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Coulson R. M., Hall N., Ouzounis C. A. 2004. Comparative genomics of transcriptional control in the human malaria parasite Plasmodium falciparum. Genome Res. 14:1548–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cui Y., et al. 2008. Genome wide expression analysis of the CCR4-NOT complex indicates that it consists of three modules with the NOT module controlling SAGA-responsive genes. Mol. Genet. Genomics 279:323–337 [DOI] [PubMed] [Google Scholar]
- 12. de Hoon M. J., Imoto S., Nolan J., Miyano S. 2004. Open source clustering software. Bioinformatics 20:1453–1454 [DOI] [PubMed] [Google Scholar]
- 13. Ecker A., Moon R., Sinden R. E., Billker O. 2006. Generation of gene targeting constructs for Plasmodium berghei by a PCR-based method amenable to high throughput applications. Mol. Biochem. Parasitol. 145:265–268 [DOI] [PubMed] [Google Scholar]
- 14. Essien K., Stoeckert C. J., Jr 2010. Conservation and divergence of known apicomplexan transcriptional regulons. BMC Genomics 11:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Garneau N. L., Wilusz J., Wilusz C. J. 2007. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 8:113–126 [DOI] [PubMed] [Google Scholar]
- 16. Irizarry R. A., et al. 2003. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jackson R. J., Hellen C. U., Pestova T. V. 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11:113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Janse C. J., Ramesar J., Waters A. P. 2006. High-efficiency transfection and drug selection of genetically transformed blood stages of the rodent malaria parasite Plasmodium berghei. Nat. Protoc. 1:346–356 [DOI] [PubMed] [Google Scholar]
- 19. Jones M. L., Kitson E. L., Rayner J. C. 2006. Plasmodium falciparum erythrocyte invasion: a conserved myosin associated complex. Mol. Biochem. Parasitol. 147:74–84 [DOI] [PubMed] [Google Scholar]
- 20. Kocken C. H. M., et al. 1998. Precise timing of expression of a Plasmodium falciparum-derived transgene in Plasmodium berghei is a critical determinant of subsequent subcellular localization. J. Biol. Chem. 273:15119–15124 [DOI] [PubMed] [Google Scholar]
- 21. Law P. J., Claudel-Renard C., Joubert F., Louw A. I., Berger D. K. 2008. MADIBA: a Web server toolkit for biological interpretation of Plasmodium and plant gene clusters. BMC Genomics 9:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Le Roch K. G., et al. 2003. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science 301:1503–1508 [DOI] [PubMed] [Google Scholar]
- 23. Li Q., et al. 2007. Development and validation of flow cytometric measurement for parasitemia in cultures of P. falciparum vitally stained with YOYO-1. Cytometry A 71:297–307 [DOI] [PubMed] [Google Scholar]
- 24. Liu J., Gluzman I. Y., Drew M. E., Goldberg D. E. 2005. The role of Plasmodium falciparum food vacuole plasmepsins. J. Biol. Chem. 280:1432–1437 [DOI] [PubMed] [Google Scholar]
- 25. Mair G. R., et al. 2010. Universal features of post-transcriptional gene regulation are critical for Plasmodium zygote development. PLoS Pathog. 6:e1000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Painter H. J., Campbell T. L., Llinas M. 2011. The Apicomplexan AP2 family: integral factors regulating Plasmodium development. Mol. Biochem. Parasitol. 176:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Persson K. E., Lee C. T., Marsh K., Beeson J. G. 2006. Development and optimization of high-throughput methods to measure Plasmodium falciparum-specific growth inhibitory antibodies. J. Clin. Microbiol. 44:1665–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Saldanha A. J. 2004. Java Treeview—extensible visualization of microarray data. Bioinformatics 20:3246–3248 [DOI] [PubMed] [Google Scholar]
- 29. Schwede A., et al. 2008. A role for Caf1 in mRNA deadenylation and decay in trypanosomes and human cells. Nucleic Acids Res. 36:3374–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Smyth G. K. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3:Article3 [DOI] [PubMed] [Google Scholar]
- 31. Tucker M., et al. 2001. The transcription factor-associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell 104:377–386 [DOI] [PubMed] [Google Scholar]
- 32. Yeoh S., et al. 2007. Subcellular discharge of a serine protease mediates release of invasive malaria parasites from host erythrocytes. Cell 131:1072–1083 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.