Abstract
Protozoan parasites that profoundly affect mankind represent an exceptionally diverse group of organisms, including Plasmodium, Toxoplasma, Entamoeba, Giardia, trypanosomes, and Leishmania. Despite the overwhelming impact of these parasites, there remain many aspects to be discovered about mechanisms of pathogenesis and how these organisms survive in the host. Combined with the ever-increasing availability of sequenced genomes, RNA interference (RNAi), discovered a mere 13 years ago, has enormously facilitated the analysis of gene function, especially in organisms that are not amenable to classical genetic approaches. Here we review the current status of RNAi in studies of parasitic protozoa, with special emphasis on its use as a postgenomic tool.
INTRODUCTION
Among the many diseases that affect mankind, those caused by protozoan parasites, such as Plasmodium, Leishmania, and the African and South American trypanosomes, are of great public health interest, as they cause a high incidence of morbidity and mortality and predominantly affect low-income populations in developing regions of Africa, Asia, and the Americas, where health resources are very often limited. Moreover, parasitic diseases profoundly affect the quality of human life and have a substantial impact on the economy and development of entire countries. Even though parasitic infections are generally thought to be more common in developing countries, they are also prevalent in developed countries, with trichomoniasis, giardiasis, and toxoplasmosis having a significant impact in the United States. Chemotherapy is the major weapon available against these parasites; however, the success of this approach is undermined by widespread resistance to some of the most effective drugs ever developed, including chloroquine for treatment of malaria. Thus, there is an urgent need to develop new therapeutic regimens based on the identification and validation of potential drug targets.
The extraordinary discovery in 1998 that double-stranded RNA (dsRNA) triggers downregulation of gene expression (24), a process dubbed RNA interference (RNAi), had a profound impact on unraveling novel aspects of eukaryotic biology and, in the postgenomic era, has enormously facilitated the analysis of gene function, especially in organisms that are not amenable to classical genetic approaches. Furthermore, the realization that 20-to-30-nucleotide (nt) small noncoding RNAs guide downregulation of gene expression has opened up a novel and still expanding world of small regulatory RNAs (25, 60) and has generated great expectations that small RNAs may be used as therapeutic agents to ameliorate human diseases.
Here, we provide an up-to-date view of RNAi pathways in protozoan parasites and discuss the current and potential usefulness of RNAi as a tool. We have chosen to focus on organisms that are not only highly relevant with regard to human disease but also have been and continue to be the subject of intense investigations in many laboratories worldwide and, most importantly, have been surveyed for the presence of RNAi. They include (i) the kinetoplastids Trypanosoma brucei, the causative agent of sleeping sickness in humans and nagana in cattle (a wasting disease similar to sleeping sickness) in sub-Saharan Africa, and T. cruzi, which causes Chagas disease in South America, and the Old and New World Leishmania parasites, which cause various forms of leishmaniasis worldwide; (ii) the apicomplexans Plasmodium falciparum, the deadliest of the human malaria parasites, and Toxoplasma gondii, the agent of toxoplasmosis and a major threat in immunocompromised individuals; (iii) the amoebozoa Entamoeba histolytica, the causative agent of amoebic dysentery; and (iv) the diplomonad Giardia intestinalis, which causes giardiasis, also known as beaver fever. These four groups of organisms are evolutionarily very distant from each other (67) and have fundamentally different life styles. Plasmodium and Toxoplasma species are obligate intracellular parasites residing in a parasitophorous vacuole, and T. brucei multiplies in the bloodstream, whereas Leishmania species infect macrophages and other immune cells, where they live in phagolysosomes. T. cruzi infects a variety of cell types transitioning from a parasitophorous vacuole, which is formed at the time of cell invasion, to the cytoplasm, whereas Entamoeba and Giardia grow extracellularly in the intestinal milieu. Importantly, all of the protozoan parasites mentioned above have been adapted to grow under laboratory conditions, techniques have been developed for their genetic manipulation, and their genomes have been or are in the process of being sequenced.
DISCOVERY OF RNA INTERFERENCE AND ITS ABSENCE IN CERTAIN PARASITES
T. brucei was the first protozoan parasite in which RNAi was shown to be functional (44). In 1998, we reported the serendipitous discovery that transfection of long dsRNA homologous to α-tubulin mRNA caused downregulation of the target mRNA and consequent inhibition of the synthesis of the corresponding protein. A similar response was demonstrated for other mRNA targets (44), suggesting that RNAi, which had been reported a few months earlier in a study of Caenorhabditis elegans by the laboratories of Fire and Mello (24), was operational in T. brucei. The existence of the RNAi pathway in this parasite was later confirmed by the identification of endogenous small interfering RNAs, or siRNAs (17), and of two of the classical proteins required for the pathway, namely, TbAGO1, a member of the Argonaute family of proteins (21, 55), and two Dicer-like homologues, TbDCL1 and TbDCL2 (48, 56).
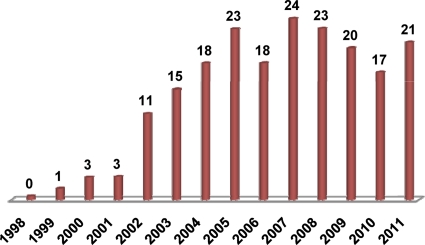
Although the original T. brucei study showed that dsRNA transfection resulted in significant downregulation of target mRNA levels, it was disappointing to discover that, contrary to the results of a C. elegans study (27), the effect was only transient and lasted about one cell cycle (44). Thus, there was no evidence in the T. brucei study for an amplification of the response to dsRNA, a phenomenon that in other organisms has been linked to the activity of RdRp, or RNA-dependent RNA polymerase (60). Indeed, the genomes of T. brucei and of other RNAi-proficient trypanosomatid protozoa do not appear to harbor a potential RdRp gene (Table 1). To overcome the transient nature of RNAi and to provide an experimental methodology to downregulate essential genes, tetracycline-regulated expression of dsRNA was established to exploit the full potential of RNAi as a tool to analyze gene function (see below for more details). Subsequently, conditional expression of dsRNA was transferred from T. brucei to T. congolense (31), which causes nagana in cattle. However, the benefits of the RNAi tool have so far not been exploited in T. congolense. On the other hand, the establishment of conditional dsRNA expression in T. brucei represented a major advance for querying its biology and the RNAi tool has established itself as the method of choice to downregulate gene expression in this organism. Since its initial discovery in 1998, there have been over 500 publications exploiting RNAi in T. brucei for studying basic biological questions and for genome-wide screens (Fig. 1). Importantly, drug target validation at various centers worldwide is currently primarily carried out by the use of RNAi.
Table 1.
RNAi genes and classes of small RNAs identified in protozoan parasites
| Parasite | siRNAs | miRNAs | Argonaute | Dicer-like | RdRp |
|---|---|---|---|---|---|
| T. brucei | + | − | + | + | − |
| L. braziliensis | + | ? | + | + | − |
| E. histolytica | + | ? | + | ? | + |
| G. intestinalis | + | + | + | + | + |
| T. gondii | + | + | + | + | + |
Fig. 1.
The impact of RNAi in T. brucei. PubMed was queried for publications between January 1998 and the end of April 2011 that mention T. brucei. The resulting 3,543 citations were then searched for the terms “RNA interference,” “RNAi,” and “gene silencing”; the searches produced 525 (about 15%) citations. In the graphic shown, the numbers of RNAi citations in T. brucei studies are expressed as percentages of the total number of T. brucei publications in each year. Our analysis does not represent an attempt to capture all relevant publications but simply highlights a noteworthy trend.
The discovery and proven effectiveness of RNAi in T. brucei studies stimulated great interest among laboratories engaged in research investigating other protozoan parasites. Rather disappointingly, direct experimentation showed that neither T. cruzi (16) nor Old World Leishmania major and L. donovani (52) were able to respond to dsRNA, indicating that they lack a functional RNAi pathway. This was confirmed at the genome sequence level, which revealed the absence of RNAi genes (22, 32). On the other hand, a few papers have been published that support the notion that, in Plasmodium falciparum asexual forms, dsRNA could trigger downregulation of gene expression (38, 40, 41). Although these initial findings raised high expectations, they could not be reproduced by several laboratories. Furthermore, the idea of the existence of a functional RNAi pathway in malaria parasites was challenged by investigation of the genome sequence and subsequent rigorous tests of the response to dsRNA: no RNAi genes were identified, and no response to dsRNA could be documented (11). Similarly, reports appeared in the literature documenting the use of dsRNA for gene expression downregulation in T. gondii (2–4, 7, 29, 74), but, once again, these results proved difficult to reproduce in other laboratories. Quite intriguingly, the genome sequence of T. gondii revealed the existence of Dicer, AGO, and RdRp homologues (14) that appeared to have plant/fungal (Dicer/RdRp) and metazoan (AGO) signatures. The apparent discrepancy between the presence of RNAi genes and the inability to exploit RNAi as part of an experimental system has also been reported at meetings on G. intestinalis, although a recent publication challenges this view (51). The available reports on RNAi as a tool in studies of T. gondii and G. intestinalis are at present confusing. Perhaps a careful reevaluation of the approaches, as was previously performed in a Saccharomyces pombe study (59), may help to overcome this hurdle.
For E. histolytica, the first evidence that expression of dsRNA can trigger downregulation of a target mRNA was obtained in 2004 (34, 65), and since then various methods have been developed to implement the RNAi tool in studies of this parasite (reviewed in reference 77), although a direct linkage between dsRNA effects and the RNAi pathway genes has not been forged as yet. Three AGO and one full-length RdRp homologues have been identified in the E. histolytica genome (Table 1), but BLAST searches for potential Dicer homologues revealed only one candidate with a single RNase III domain (1), whereas most Dicers are endowed with two RNase III domains, which form a pseudodimer giving rise to the enzyme's active site (76). Nevertheless, in the budding yeast S. castellii, Dicer also contains a single RNase III domain, which likely undergoes dimerization similar to that seen with its bacterial counterpart (19). Thus, it is possible that the E. histolytica Dicer candidate is indeed an atypical one, but further studies are needed to demonstrate that this protein is required for the RNAi pathway. Then again, the generation of siRNAs in this organism may proceed through a Dicer-independent pathway, in similarity to what has been described for the PIWI-interacting RNAs (61) and certain micro-RNAs (miRNAs) in metazoans (73).
The New World species L. (Viannia) braziliensis, a major cause of cutaneous and mucocutaneous leishmaniasis in South America, represents the latest addition to the list of RNAi-proficient protozoan parasites. When the genome of L. (V.) braziliensis became available in 2007 (49), it was evident that this organism, which is a distant relative of trypanosomes, has a full complement of RNAi pathway genes. Importantly, direct experimentation has proven that the RNAi pathway is functional and that RNAi can be used as a tool to downregulate expression of reporter as well as endogenous genes (37).
With the growing number of sequenced genomes available, it is relevant to ask whether other human protozoan parasites have the hallmarks of the RNAi pathway. So far, we have been able to identify a potential AGO homologue only in Trichomonas vaginalis (NCBI accession number XP_001329688.1), the causative agent of trichomoniasis, a highly prevalent infection of humans in industrialized countries. Searches for possible Dicer and RdRp homologues in T. vaginalis were unsuccessful.
ENDOGENOUS RNAi PATHWAYS AND THEIR BIOLOGICAL ROLE
RNAi pathway in T. brucei.
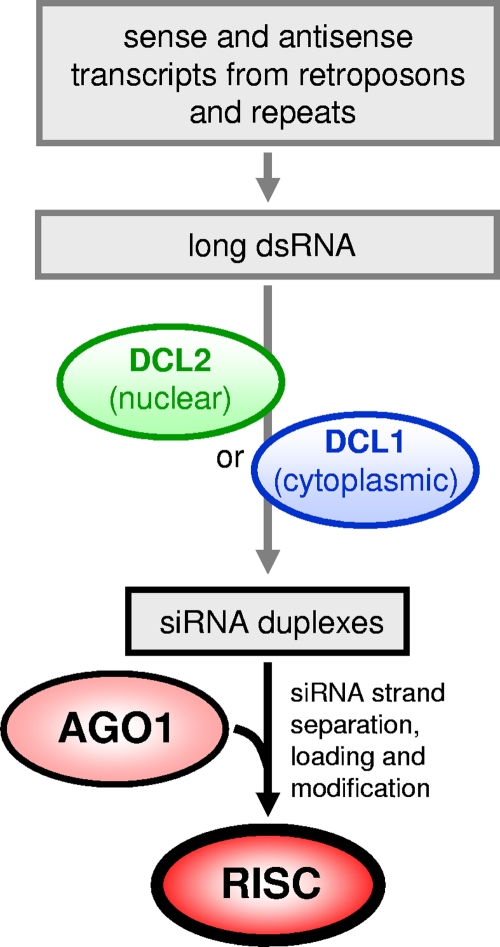
The most detailed analysis of a protozoan parasite RNAi pathway has been done in T. brucei studies (Fig. 2; see reference 8 for details). Briefly, there are two arms of the RNAi pathway, one in the nucleus and the other one in the cytoplasm, which are initiated by nuclear TbDCL2 (48) and cytoplasmic TbDCL1 (56), respectively, and feed into the single AGO1 protein (21, 57). The nuclear pathway is dedicated to downregulation of transcripts derived from retroposons and repeats, seemingly to maintain genome stability by decreasing potential retroposon hopping (47) and possibly by aiding chromosome segregation (21). The role of the cytoplasmic arm of RNAi is less clear, as deletion of TbDCL1 has minor consequences for the endogenous RNAi response. We have put forward the hypothesis that TbDCL1 functions as a sentinel in the cytoplasm to provide a second checkpoint should retroposon and repeat transcripts escape to the cytoplasm (48). We have also reported that TbDCL1 is required for the processing of 40-to-50-nt dsRNA intermediates produced by TbDCL2 (48), an indication that the two enzymes cooperate to bring about a full RNAi response. It is interesting that ablation of TbDCL2 resulted in cells that are hyperresponsive to transfection with long dsRNA as well as with synthetic siRNAs (48), a phenotype consistent with the possibility that the endogenous nuclear pathway is dominant over the cytoplasmic response. In addition, the T. brucei genome codes for a homologue of HEN1, a methyltransferase that in certain organisms modifies the terminal ribose of siRNAs and miRNAs (30), thus protecting the 3′ end from nuclease attack, and we have shown that T. brucei siRNAs have a modified 3′ ribose (48). Intriguingly, HEN1 is not present in L. (V.) braziliensis, suggesting mechanistic diversification of the RNAi pathway among the trypanosomatid protozoa. Whether other protozoan parasites have developed specific adaptations in the repertoire of RNAi genes is at present unknown.
Fig. 2.
RNAi in T. brucei. The Dicer-like enzymes DCL2 and DCL1 process long RNA duplexes (dsRNA) derived from sense and antisense transcripts from retroposons and repeats into siRNA duplexes in the nucleus and the cytoplasm, respectively. Argonaute (AGO1) is programmed with single-stranded “guide” siRNA, following siRNA strand separation, loading, and modification, to form the RNA-induced silencing complex, or RISC.
siRNAs in T. brucei and L. (V.) braziliensis.
The most direct approach to gauge the biological role of the RNAi pathway is to survey the repertoire of small 20-to-30-nt noncoding RNAs (Table 1). In T. brucei, the major classes of siRNAs are derived from retroposons and repeats (17, 48), but recent deep sequencing has expanded the repertoire to include regions of the genome with the potential to produce dsRNA, namely, regions at the ends of convergent transcription units and long inverted repeats (unpublished observations). Furthermore, a recent report has identified siRNA-like molecules derived from variable-surface glycoprotein pseudogenes (69). Thus, in T. brucei, the repertoire of endogenous siRNAs is remarkably similar to that of higher eukaryotes, such as Drosophila and mouse (46, 68). So far, deep sequencing of small RNAs associated with TbAGO1 has not revealed potential miRNAs in insect-form T. brucei (unpublished data), although a few miRNA candidates were identified bioinformatically but were never confirmed to be expressed (39).
In similarity to the results seen with T. brucei, abundant siRNAs derived from the SLACS (Spliced Leader Associated Conserved Sequence) retroposon family accumulate in L. (V.) braziliensis (37). However, the full repertoire of siRNAs has so far not been explored in this organism.
E. histolytica small RNAs.
There is a population of siRNA-like molecules in E. histolytica with an unusual 5′-polyphosphate end (75), a feature that indicates that these molecules are produced via transcription by an RNA polymerase rather than by Dicer cleavage. Consequently, there is the possibility that these small RNAs represent secondary siRNAs, which have so far been described only in C. elegans and plants (10). Most of the amoebic siRNAs are antisense to coding regions, and they have been proposed to downregulate gene expression in a strain-specific fashion. At present, nothing is known about the mechanism generating these putative secondary siRNAs; it remains to be seen whether primary siRNAs exist in E. histolytica.
G. intestinalis small RNAs.
G. intestinalis accumulates siRNA-like molecules derived from one retroposon family enriched at telomeres (64) and has been shown to harbor at least one miRNA (miR2), most likely derived from Dicer processing of a small nucleolar RNA (53), and additional putative miRNAs were identified by deep sequencing (15). The potential in vivo mRNA target(s) of miR2 has been identified only bioinformatically, but reporter gene assays indicate that miR2 inhibits translation, which instead appears to be the primary mode of action of miRNAs in higher eukaryotes, rather than provoking destabilization of the target mRNA (28). Furthermore, the RNAi pathway in G. intestinalis has been linked to antigenic variation and small regulatory RNAs have been implicated in this phenomenon (50). Downregulation of G. intestinalis Dicer or RdRP, via an antisense RNA approach, generated cells expressing two or more variant-specific surface protein genes at one time. Mechanistic details are, however, not yet available.
T. gondii small RNAs.
Deep sequencing of small RNAs in T. gondii has revealed siRNAs derived from repeats and satellite DNA, genomic regions carrying the hallmarks of heterochromatin, and miRNAs with metazoan features (14). Although the association of these small RNAs with TgAGO1 has been confirmed, at present we do not know what the in vivo targets of miRNAs are and what the consequences are of deleting the RNAi pathway in T. gondii.
Conclusions.
Although still sketchy, the emerging picture suggests that, as in other eukaryotes, the RNAi pathway plays diverse roles in the biology of protozoan parasites. The classical and perhaps ancestral role of RNAi in promoting genome stability by silencing mobile elements and repeats is strongly supported in T. brucei and L. (V.) braziliensis and most probably takes place in G. intestinalis. RNAi in these organisms is likely to have a role in heterochromatin formation, as suggested by the observation of chromosome segregation defects in AGO1-null T. brucei (20, 21) and the report from a T. gondii study that the major classes of siRNA are derived from heterochromatic regions enriched in repeats and satellite DNA (14). The presence of 5′ polyphosphate small RNAs in E. histolytica is intriguing (75), but at present hypotheses concerning the biosynthetic pathway and the mode of action in regulating gene expression remain speculative. Finally, miRNAs with regulatory potential have been identified in studies of G. intestinalis (53) and T. gondii (14), suggesting that the single AGO protein in these organisms is likely to perform double duty by controlling genome homeostasis via the siRNA pathway and by regulating gene expression at the posttranscriptional level via the miRNA pathway.
LOSS OF THE RNAi PATHWAY
From the available evidence, it is clear that RNAi deficiency arose independently a number of times through the eukaryotic lineage, for instance, in fungi (19) and in apicomplexan and trypanosomatid protozoa (37). In the latter organisms, data from a protein-based phylogenetic tree support the notion that RNAi pathway genes have been lost at least twice and independently during evolution of the trypanosomatid lineage, once at an unknown point in the branch leading to T. cruzi and a second time after the divergence of the Viannia subgenus from other Leishmania species (37). A similar analysis is not yet available for the apicomplexan parasites, with T. gondii being the only member of this group with a functional RNAi pathway. Inspection of the available trypanosomatid genome sequences reveals that, in T. cruzi, no recognizable remnants of DCL1, DCL2, or AGO1 homologues are present (23), whereas, in Old World Leishmania species, Dicer homologues are absent but the AGO1 locus is occupied by a badly mangled pseudogene (49). While the L. major AGO1 pseudogene has the potential to code for two polypeptides of ∼100 and 200 amino acids, these putative open reading frames (ORFs) are not conserved in the corresponding L. infantum pseudogene (unpublished). Thus, it is unlikely that AGO1 pseudogenes code for functional polypeptides. The mechanisms leading to disappearance of the RNAi genes in organisms where there are no recognizable traces of their past presence are unknown, although chromosomal rearrangements may have contributed to their loss. In the future it will be interesting to survey the genomes of other lesser-known trypanosomatids to determine whether RNAi deficiency arose more than twice in this group of organisms.
It was recognized early on that RNAi has a widespread role in silencing parasitic nucleic acids, such as those of mobile elements and certain RNA viruses, and thus contributes to the maintenance of genome stability and to the prevention of viral spread. It has been argued that, in organisms where mobile elements have decayed over long evolutionary times and have become nonfunctional, the RNAi pathway no longer offers a selective advantage and can be lost without major consequences (19). Indeed, among the protozoan parasites there appears to be a correlation between the absence of mobile elements in the genome and RNAi deficiency. For instance, RNAi-negative Old World Leishmania and P. falciparum are devoid of such elements. In contrast, the genomes of African trypanosomes and L. (V.) braziliensis are rich in retroposon elements, some of which appear to be intact and have the potential for retroposition. Interestingly, genetic ablation of the RNAi pathway in T. brucei resulted in genomic rearrangements of the SLACS retroposon elements (47). The exception to this rule is RNAi-deficient T. cruzi, whose genome is rich in sequences resembling mobile elements. T. cruzi may be in a “transition” state where transposition/retroposition is minimal or not taking place, or perhaps other mechanisms are in place to inhibit movement of mobile elements.
Viral invasion has been proposed as a selective force to drive loss of the RNAi pathway both in yeast species (19) and in Old World Leishmania species (12, 37). Additionally, in Leishmania, loss of the pathway may have contributed to increase genome plasticity and to alter virulence (see reference 37 for an in-depth discussion of this topic).
RNAi AS A TOOL
RNAi proved quickly to be an immensely useful tool for studying gene function in almost all organisms that possess the required set of proteins of the interference pathway. Following the initial discovery of RNAi in T. brucei (44), researchers took immediate advantage by developing RNAi technology as the method of choice for achieving downregulation of gene expression, either for individual genes or for the entire genome. The fast-paced evolution of RNAi technology for T. brucei was driven in part by the development of vectors for inducible and heritable expression of toxic gene products (initially designed for proteins and later for long double-stranded RNA) in the laboratories of Christine Clayton (70) and George A. M. Cross (71) and by studying the endogenous components required to achieve efficient RNAi (21, 48, 55, 56).
The use of RNAi in T. brucei for reverse genetics approaches has exploded (Fig. 1), and there were two initial studies that are deserving of special attention for the efforts of the researchers to expand the method to a small-scale screen. A multilaboratory collaboration (62) heroically performed a systematic RNAi gene function screen for the entirety of chromosome 1 of T. brucei bloodstream parasites. They analyzed 210 genes, and RNAi knockdown of 33% of the genes produced a significant phenotype. Growth and cell cycle defects were observed for 23% and 16% of the genes, respectively, and 6% of the knockdowns displayed alterations in both growth and cell cycle progression. RNAi targeting of 12% of the studied ORFs was lethal. The second screen, which was performed in the laboratory of Christine Clayton (72), targeted 37 genes containing an RNA-recognition motif (RRM) in both procyclic and bloodstream-form trypanosomes. Knockdowns of 8 of these RRM proteins resulted in a strong growth defect in bloodstream T. brucei cells, and downregulation of 9 others produced mild growth inhibition. RNAi knockdown for 7 of the studied genes modestly affected growth in procyclic cells.
On the other hand, the potential of RNAi for forward genetics screens in studies of T. brucei and other RNAi-positive protozoan parasites has not been fully unleashed. Even though the genome-wide approach was pioneered in T. brucei studies (43), to date the use of such screens for gene function discovery and drug target validation has been limited and trails behind the ever-faster and more widely ranging RNAi genome-wide screen methodologies for mammalian cells, Drosophila, and nematodes (13).
The very first genome-wide RNAi screen was performed in the laboratory of Paul Englund in a search for genes that modulate expression and posttranslational modification of the surface coat proteins EP procyclins in T. brucei (43). It relied on the generation of an RNAi plasmid library for producing double-stranded RNA from genomic DNA fragments. The DNA was fragmented by sonication to an average length of ∼1 kbp and was placed between opposing T7 RNA polymerase promoters. In the resulting library, the average size of plasmid inserts was ∼0.66 kbp. Astonishingly, the hexokinase 1 gene was identified as a regulator for the switch in expression from the glycosylated EP procyclins to the unglycosylated GPEET procyclins and revealed an unanticipated link between glycolysis and surface coat protein expression. This RNAi library was subsequently used in several screens addressing different aspects of T. brucei biology. First, knocking down hexose transporters rendered trypanosomes resistant to the drug tubercidin, and this drug acted by inhibiting the glycolytic pathway (18). Second, by isolation of 1,400 trypanosome clones and screening for loss of kinetoplast DNA (kDNA), a protein component (named p166) of the kDNA segregation molecular machinery was identified and characterized (78). Third, 204 clonal cell lines were analyzed to identify novel regulators of the T. brucei cell cycle (42), which resulted in the identification of TOR1- and TOR2-like kinases. Finally, a screen to identify genes responsible for maintaining mitochondrial membrane potential selected 20 clones, with the surprising result that only a telomerase-associated protein emerged as a possible candidate for the observed phenotype (66).
The only other genome-wide RNAi library for T. brucei was generated recently in the laboratory of Isabel Roditi (54) by digestion of genomic DNA with restriction enzymes and insertion of 0.5-to-2-kbp fragments into a vector with opposing T7 promoters. It was used to identify the TbAT1 adenosine transporter and the AAT6 amino acid transporter as responsible for the uptake of the drugs melarsoprol and eflornithine, respectively. The involvement of AAT6 in uptake of eflornithine was also verified in a separate screen using the Englund RNAi library (9), a study that also validated previous observations that the gene coding for nitroreductase is involved in rendering the drug nifurtimox toxic to trypanosomes.
RIT-Seq.
A giant leap forward in the evolution of RNAi screening technology for trypanosomes was the development in the laboratory of David Horn of the RIT-Seq method (RNA Interference Target Sequencing) (6). The strategy artfully combines the powers of genome-wide RNAi screens (in this particular case, employing the Englund RNAi library) with the strength of Illumina genome sequencing for identifying, at the same time and in the same pool of cells, all genes potentially associated with loss of fitness. It relies on massively parallel sequencing of tags from the RNAi library present in the selected cell population to reveal gaps (or “cold spots”) of genomic coverage that correspond to genes whose knockdown is detrimental to the parasite under the conditions tested. Using this powerful new tool, Alsford and colleagues identified 1,972 genes with a loss-of-fitness knockdown phenotype in procyclic trypanosomes and 1,908 and 2,724 genes with a loss-of-fitness phenotype in bloodstream trypanosomes cultured for 3 and 6 days after RNAi induction, respectively, as well as 2,677 genes whose downregulation led to loss of fitness during the differentiation from bloodstream to procyclic forms (6). Valuable insights concerning the function of specific gene groups were obtained in the course of the analysis. This revolutionary method with exceptional potential could possibly be used to link a specific phenotype (if an appropriate selection procedure is devised to enrich for cells exhibiting the feature) with downregulation of specific genes, resulting in what would be considered a “hot spot” in the genomic sequencing coverage of RNAi library tags.
Perspectives on RNAi screens.
What improvements in efficiency of RNAi screens are needed in the near future? The current genomic DNA insert libraries (43, 54) need to provide severalfold coverage of the genome (5-to 10-fold for the plasmid library and ∼5-fold for the transfected trypanosome cells), resulting in a requirement for a large number of trypanosome clones on the order of 3 × 105. There have already been improvements in the efficiency of transfection protocols and the use of strains with better plasmid integration efficacy, such as the I-SceI meganuclease-expressing cell line generated in David Horn's laboratory (6, 26). Another avenue for exploration is the generation of genome-wide systematic RNAi libraries, with the help of the recently determined boundaries of genes obtained through the use of RNA-Seq (35, 45, 58). The number of individual cell lines needed to provide systematic genome-wide coverage (∼104), as well as the number of transformants in pooled experiments (∼5 × 104 for 5-fold coverage), thus will be greatly reduced. The systematic libraries would also cover the 1,114 transcripts newly identified by Kolev et al. (35) that are currently not distinguished from neighboring genes when scoring for genes with a knockdown phenotype. The integration site should be a single locus characterized in the T. brucei genome that negates all of the different epigenetic effects of the surrounding chromatin on expression of the double-stranded RNA construct (5). The trigger for RNAi should ideally be generated as a hairpin instead of as sense and antisense transcripts produced from opposing T7 RNA polymerase promoters. Tet-regulatable T7 promoters may be “leaky,” a problem that is not as pronounced with the RNA polymerase I promoters used to inducibly express long RNA hairpins. A new method for single-step hairpin cloning is required to facilitate the generation of a long-hairpin RNAi library, which would allow these superior RNAi triggers to be expressed from any integratable (or episomal) plasmid construct that provides tightly controlled expression. Recently, a protocol that combines a single cloning step with an in vitro recombination reaction in a specially constructed plasmid vector (pTrypRNAiGate) was described and tested using bloodstream-form T. brucei and is also suitable for knockdowns in procyclic cells (33). Last, the specific goals (or screening parameters) for genome-wide screens should be better and more narrowly defined, which would make the results easier to interpret and lead to faster assignment of function to specific gene products (proteins or noncoding RNAs).
It was recently demonstrated that RNAi is functional in some Leishmania species (37). Development of RNAi tools in these parasites has been lagging, primarily for the single reason that there is currently no Leishmania system available for inducible expression of toxic gene products (proteins or dsRNA). The development of such an enormously valuable resource would undoubtedly allow research on Leishmania gene function to expeditiously adopt and benefit from the RNAi methods developed for T. brucei functional genomics.
Research into the mechanism of RNAi and attempts to use it as a tool for functional genetics in G. intestinalis and T. gondii are also in its infancy. The only other protozoan parasite with a well-studied but still not completely understood RNAi system is E. histolytica (75). The emerging theme is that long dsRNA (or the expression of long antisense transcripts that base pair with the target to form long dsRNAs) is an excellent trigger for RNAi not only in trypanosomes but also in E. histolytica (77), G. intestinalis (51), and T. gondii (4).
RECONSTRUCTION OF THE RNAi PATHWAY IN RNAi-NEGATIVE ORGANISMS
Perhaps the most exciting (and, at the same time, the most challenging) opportunity for new applications of RNAi technology is the transformation of RNAi-negative protozoan parasites into RNAi-positive organisms by the introduction of a required minimal set of RNAi machinery transgenes. This would inevitably energize any functional genetics/genomics studies of such species. Although for protozoan parasites this is still in the realm of hypothesis, there is a precedent for a successful conversion to RNAi proficiency in Saccharomyces cerevisiae studies (19, 63). This was achieved through the use of two different sources of RNAi machinery genes. In the first approach, introduction of only two genes (Dicer and Argonaute from a related budding yeast species, S. castellii) was sufficient for the successful downregulation of both a reporter transcript targeted by siRNAs (even in yeast species, long hairpin dsRNA produces siRNAs more efficiently than sense and antisense RNA pairs synthesized from opposing promoters) and endogenous retrotransposons (19). In the second approach, the genes encoding human Dicer, AGO2, and TRBP (TAR RNA-Binding Protein) were capable of reconstituting RNAi for use against reporter green fluorescent protein (GFP) mRNA (63).
Successful engineering of RNAi in deficient protozoan parasites may require many different attempts. One approach could be the introduction of budding-yeast Dicer and Argonaute (the apparently most minimalist RNAi machinery known to date), although the possibility exists that additional factors (e.g., dsRNA-binding proteins) present in the RNAi-negative recipient may be required to cooperate with transplanted Dicer and AGO. The introduction of RNAi machinery genes from closely related parasites may be a better option; however, the precise number of essential RNAi genes for any protozoan parasite is at present unknown, and the number of characterized genes recognized to be involved in the mechanism of RNAi is still growing. Expression levels of the RNAi transgenes should be coordinated in a way that reflects relative protein abundances in the donor parasite. Alternative approaches, for instance, expression of double or even triple fusions (36) of RNAi factors, e.g., Dicer-AGO or Dicer-AGO-dsRNA binding protein, can be used to achieve this goal.
Obviously, reconstruction of the RNAi pathway in organisms such as Plasmodium species and L. major presents a challenge but would bring about enormous benefits, as clearly manifested by the major impact RNAi has had in the past decade in T. brucei studies (Fig. 1). At the same time, protozoan parasites with RNAi machinery must be explored to the fullest to achieve a comprehensive understanding of gene function.
ACKNOWLEDGMENTS
Work in the authors' laboratory is funded by grants AI28798 and AI56333 from the National Institutes of Health.
Footnotes
Published ahead of print on 15 July 2011.
REFERENCES
- 1. Abed M., Ankri S. 2005. Molecular characterization of Entamoeba histolytica RNase III and AGO2, two RNA interference hallmark proteins. Exp. Parasitol. 110:265–269 [DOI] [PubMed] [Google Scholar]
- 2. Al-Anouti F., Ananvoranich S. 2002. Comparative analysis of antisense RNA, double-stranded RNA, and delta ribozyme-mediated gene regulation in Toxoplasma gondii. Antisense Nucleic Acid Drug Dev. 12:275–281 [DOI] [PubMed] [Google Scholar]
- 3. Al-Anouti F., Quach T., Ananvoranich S. 2003. Double-stranded RNA can mediate the suppression of uracil phosphoribosyltransferase expression in Toxoplasma gondii. Biochem. Biophys. Res. Commun. 302:316–323 [DOI] [PubMed] [Google Scholar]
- 4. Al Riyahi A., Al-Anouti F., Al-Rayes M., Ananvoranich S. 2006. Single argonaute protein from Toxoplasma gondii is involved in the double-stranded RNA induced gene silencing. Int. J. Parasitol. 36:1003–1014 [DOI] [PubMed] [Google Scholar]
- 5. Alsford S., Horn D. 2008. Single-locus targeting constructs for reliable regulated RNAi and transgene expression in Trypanosoma brucei. Mol. Biochem. Parasitol. 161:76–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Alsford S., et al. 2011. High-throughput phenotyping using parallel sequencing of RNA interference targets in the African trypanosome. Genome Res. 21:915–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ananvoranich S., Al Rayes M., Al Riyahi A., Wang X. 2006. RNA silencing of glycolysis pathway in Toxoplasma gondii. J. Eukaryot. Microbiol. 53(Suppl. 1):S162–S163 [DOI] [PubMed] [Google Scholar]
- 8. Atayde V. D., Tschudi C., Ullu E. 2011. The emerging world of small silencing RNAs in protozoan parasites. Trends Parasitol. 27:321–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Baker N., Alsford S., Horn D. 2011. Genome-wide RNAi screens in African trypanosomes identify the nifurtimox activator NTR and the eflornithine transporter AAT6. Mol. Biochem. Parasitol. 176:55–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Baulcombe D. C. 2007. Molecular biology. Amplified silencing. Science 315:199–200 [DOI] [PubMed] [Google Scholar]
- 11. Baum J., et al. 2009. Molecular genetics and comparative genomics reveal RNAi is not functional in malaria parasites. Nucleic Acids Res. 37:3788–3798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Beverley S. M. 2003. Protozomics: trypanosomatid parasite genetics comes of age. Nat. Rev. Genet. 4:11–19 [DOI] [PubMed] [Google Scholar]
- 13. Boutros M., Ahringer J. 2008. The art and design of genetic screens: RNA interference. Nat. Rev. Genet. 9:554–566 [DOI] [PubMed] [Google Scholar]
- 14. Braun L., et al. 2010. A complex small RNA repertoire is generated by a plant/fungal-like machinery and effected by a metazoan-like Argonaute in the single-cell human parasite Toxoplasma gondii. PLoS Pathog. 6:e1000920 doi:10.1371/journal.ppat.1000920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen X. S., Collins L. J., Biggs P. J., Penny D. 2009. High throughput genome-wide survey of small RNAs from the parasitic protists Giardia intestinalis and Trichomonas vaginalis. Genome Biol. Evol. 1:165–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. DaRocha W. D., Otsu K., Teixeira S. M., Donelson J. E. 2004. Tests of cytoplasmic RNA interference (RNAi) and construction of a tetracycline-inducible T7 promoter system in Trypanosoma cruzi. Mol. Biochem. Parasitol. 133:175–186 [DOI] [PubMed] [Google Scholar]
- 17. Djikeng A., Shi H., Tschudi C., Ullu E. 2001. RNA interference in Trypanosoma brucei: cloning of small interfering RNAs provides evidence for retroposon-derived 24-26-nucleotide RNAs. RNA 7:1522–1530 [PMC free article] [PubMed] [Google Scholar]
- 18. Drew M. E., et al. 2003. The adenosine analog tubercidin inhibits glycolysis in Trypanosoma brucei as revealed by an RNA interference library. J. Biol. Chem. 278:46596–46600 [DOI] [PubMed] [Google Scholar]
- 19. Drinnenberg I. A., et al. 2009. RNAi in budding yeast. Science 326:544–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Durand-Dubief M., et al. 2007. The Argonaute protein TbAGO1 contributes to large and mini-chromosome segregation and is required for control of RIME retroposons and RHS pseudogene-associated transcripts. Mol. Biochem. Parasitol. 156:144–153 [DOI] [PubMed] [Google Scholar]
- 21. Durand-Dubief M., Bastin P. 2003. TbAGO1, an Argonaute protein required for RNA interference, is involved in mitosis and chromosome segregation in Trypanosoma brucei. BMC Biol. 1:2 doi:10.1186/1741-7007-1181-1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. El-Sayed N. M., et al. 2005. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science 309:409–415 [DOI] [PubMed] [Google Scholar]
- 23. El-Sayed N. M., et al. 2005. Comparative genomics of trypanosomatid parasitic protozoa. Science 309:404–409 [DOI] [PubMed] [Google Scholar]
- 24. Fire A., et al. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806–811 [DOI] [PubMed] [Google Scholar]
- 25. Ghildiyal M., Zamore P. D. 2009. Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 10:94–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Glover L., Horn D. 2009. Site-specific DNA double-strand breaks greatly increase stable transformation efficiency in Trypanosoma brucei. Mol. Biochem. Parasitol. 166:194–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Grishok A. 2005. RNAi mechanisms in Caenorhabditis elegans. FEBS Lett. 579:5932–5939 [DOI] [PubMed] [Google Scholar]
- 28. Guo H., Ingolia N. T., Weissman J. S., Bartel D. P. 2010. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466:835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Holmes M., et al. 2010. Silencing of tachyzoite enolase 2 alters nuclear targeting of bradyzoite enolase 1 in Toxoplasma gondii. Microbes Infect. 12:19–27 [DOI] [PubMed] [Google Scholar]
- 30. Horwich M. D., et al. 2007. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr. Biol. 17:1265–1272 [DOI] [PubMed] [Google Scholar]
- 31. Inoue N., Otsu K., Ferraro D. M., Donelson J. E. 2002. Tetracycline-regulated RNA interference in Trypanosoma congolense. Mol. Biochem. Parasitol. 120:309–313 [DOI] [PubMed] [Google Scholar]
- 32. Ivens A. C., et al. 2005. The genome of the kinetoplastid parasite, Leishmania major. Science 309:436–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kalidas S., Li Q., Phillips M. A. 2011. A Gateway((R)) compatible vector for gene silencing in bloodstream form Trypanosoma brucei. Mol. Biochem. Parasitol. 178:51–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kaur G., Lohia A. 2004. Inhibition of gene expression with double strand RNA interference in Entamoeba histolytica. Biochem. Biophys. Res. Commun. 320:1118–1122 [DOI] [PubMed] [Google Scholar]
- 35. Kolev N. G., et al. 2010. The transcriptome of the human pathogen Trypanosoma brucei at single-nucleotide resolution. PLoS Pathog. 6:e1001090 doi:10.1371/journal.ppat.1001090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kolev N. G., Yario T. A., Benson E., Steitz J. A. 2008. Conserved motifs in both CPSF73 and CPSF100 are required to assemble the active endonuclease for histone mRNA 3′-end maturation. EMBO Rep. 9:1013–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lye L. F., et al. 2010. Retention and loss of RNA interference pathways in trypanosomatid protozoans. PLoS Pathog. 6:e1001161 doi:10.1371/journal.ppat.1001161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Malhotra P., et al. 2002. Double-stranded RNA-mediated gene silencing of cysteine proteases (falcipain-1 and -2) of Plasmodium falciparum. Mol. Microbiol. 45:1245–1254 [DOI] [PubMed] [Google Scholar]
- 39. Mallick B., Ghosh Z., Chakrabarti J. 2008. MicroRNA switches in Trypanosoma brucei. Biochem. Biophys. Res. Commun. 372:459–463 [DOI] [PubMed] [Google Scholar]
- 40. McRobert L., McConkey G. A. 2002. RNA interference (RNAi) inhibits growth of Plasmodium falciparum. Mol. Biochem. Parasitol. 119:273–278 [DOI] [PubMed] [Google Scholar]
- 41. Mohmmed A., Dasaradhi P. V., Bhatnagar R. K., Chauhan V. S., Malhotra P. 2003. In vivo gene silencing in Plasmodium berghei—a mouse malaria model. Biochem. Biophys. Res. Commun. 309:506–511 [DOI] [PubMed] [Google Scholar]
- 42. Monnerat S., Clucas C., Brown E., Mottram J. C., Hammarton T. C. 2009. Searching for novel cell cycle regulators in Trypanosoma brucei with an RNA interference screen. BMC Res. Notes 2:46 doi:10.1186/1756-0500-1182-1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Morris J. C., Wang Z., Drew M. E., Englund P. T. 2002. Glycolysis modulates trypanosome glycoprotein expression as revealed by an RNAi library. EMBO J. 21:4429–4438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ngô H., Tschudi C., Gull K., Ullu E. 1998. Double-stranded RNA induces mRNA degradation in Trypanosoma brucei. Proc. Natl. Acad. Sci. U. S. A. 95:14687–14692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nilsson D., et al. 2010. Spliced leader trapping reveals widespread alternative splicing patterns in the highly dynamic transcriptome of Trypanosoma brucei. PLoS Pathog. 6:e1001037 doi:10.1371/journal.ppat.1001037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Okamura K., et al. 2008. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature 453:803–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Patrick K. L., et al. 2008. Genomic rearrangements and transcriptional analysis of the spliced leader-associated retrotransposon in RNA interference-deficient Trypanosoma brucei. Mol. Microbiol. 67:435–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Patrick K. L., et al. 2009. Distinct and overlapping roles for two Dicer-like proteins in the RNA interference pathways of the ancient eukaryote Trypanosoma brucei. Proc. Natl. Acad. Sci. U. S. A. 106:17933–17938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Peacock C. S., et al. 2007. Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nat. Genet. 39:839–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Prucca C. G., et al. 2008. Antigenic variation in Giardia lamblia is regulated by RNA interference. Nature 456:750–754 [DOI] [PubMed] [Google Scholar]
- 51. Rivero M. R., Kulakova L., Touz M. C. 2010. Long double-stranded RNA produces specific gene downregulation in Giardia lamblia. J. Parasitol. 96:815–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Robinson K. A., Beverley S. M. 2003. Improvements in transfection efficiency and tests of RNA interference (RNAi) approaches in the protozoan parasite Leishmania. Mol. Biochem. Parasitol. 128:217–228 [DOI] [PubMed] [Google Scholar]
- 53. Saraiya A. A., Wang C. C. 2008. snoRNA, a novel precursor of microRNA in Giardia lamblia. PLoS Pathog. 4:e1000224 doi:10.1371/journal.ppat.1000224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schumann Burkard G., Jutzi P., Roditi I. 2011. Genome-wide RNAi screens in bloodstream form trypanosomes identify drug transporters. Mol. Biochem. Parasitol. 175:91–94 [DOI] [PubMed] [Google Scholar]
- 55. Shi H., Djikeng A., Tschudi C., Ullu E. 2004. Argonaute protein in the early divergent eukaryote Trypanosoma brucei: control of small interfering RNA accumulation and retroposon transcript abundance. Mol. Cell. Biol. 24:420–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shi H., Tschudi C., Ullu E. 2006. An unusual Dicer-like1 protein fuels the RNA interference pathway in Trypanosoma brucei. RNA 12:2063–2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shi H., Ullu E., Tschudi C. 2004. Function of the trypanosome Argonaute 1 protein in RNA interference requires the N-terminal RGG domain and arginine 735 in the Piwi domain. J. Biol. Chem. 279:49889–49893 [DOI] [PubMed] [Google Scholar]
- 58. Siegel T. N., Hekstra D. R., Wang X., Dewell S., Cross G. A. 2010. Genome-wide analysis of mRNA abundance in two life-cycle stages of Trypanosoma brucei and identification of splicing and polyadenylation sites. Nucleic Acids Res. 38:4946–4957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sigova A., Rhind N., Zamore P. D. 2004. A single Argonaute protein mediates both transcriptional and posttranscriptional silencing in Schizosaccharomyces pombe. Genes Dev. 18:2359–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Siomi H., Siomi M. C. 2009. On the road to reading the RNA-interference code. Nature 457:396–404 [DOI] [PubMed] [Google Scholar]
- 61. Siomi M. C., Sato K., Pezic D., Aravin A. A. 2011. PIWI-interacting small RNAs: the vanguard of genome defence. Nat. Rev. Mol. Cell Biol. 12:246–258 [DOI] [PubMed] [Google Scholar]
- 62. Subramaniam C., et al. 2006. Chromosome-wide analysis of gene function by RNA interference in the African trypanosome. Eukaryot. Cell 5:1539–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Suk K., et al. 2011. Reconstitution of human RNA interference in budding yeast. Nucleic Acids Res. 39:e43 doi:10.1093/nar/gkq1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ullu E., Lujan H. D., Tschudi C. 2005. Small sense and antisense RNAs derived from a telomeric retroposon family in Giardia intestinalis. Eukaryot. Cell 4:1155–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Vayssié L., Vargas M., Weber C., Guillen N. 2004. Double-stranded RNA mediates homology-dependent gene silencing of gamma-tubulin in the human parasite Entamoeba histolytica. Mol. Biochem. Parasitol. 138:21–28 [DOI] [PubMed] [Google Scholar]
- 66. Verner Z., Paris Z., Lukes J. 2010. Mitochondrial membrane potential-based genome-wide RNAi screen of Trypanosoma brucei. Parasitol. Res. 106:1241–1244 [DOI] [PubMed] [Google Scholar]
- 67. Walker G., Dorrell R. G., Schlacht A., Dacks J. B. 2011. Eukaryotic systematics: a user's guide for cell biologists and parasitologists. Parasitology 15:1–26 [DOI] [PubMed] [Google Scholar]
- 68. Watanabe T., et al. 2008. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 453:539–543 [DOI] [PubMed] [Google Scholar]
- 69. Wen Y. Z., et al. 2011. Pseudogene-derived small interference RNAs regulate gene expression in African Trypanosoma brucei. Proc. Natl. Acad. Sci. U. S. A. 108:8345–8350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wirtz E., Clayton C. 1995. Inducible gene expression in trypanosomes mediated by a prokaryotic repressor. Science 268:1179–1183 [DOI] [PubMed] [Google Scholar]
- 71. Wirtz E., Leal S., Ochatt C., Cross G. A. 1999. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99:89–101 [DOI] [PubMed] [Google Scholar]
- 72. Wurst M., et al. 2009. An RNAi screen of the RRM-domain proteins of Trypanosoma brucei. Mol. Biochem. Parasitol. 163:61–65 [DOI] [PubMed] [Google Scholar]
- 73. Yang J. S., Lai E. C. 2010. Dicer-independent, Ago2-mediated microRNA biogenesis in vertebrates. Cell Cycle 9:4455–4460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yu L., Gao Y. F., Li X., Qiao Z. P., Shen J. L. 2009. Double-stranded RNA specific to adenosine kinase and hypoxanthine-xanthine-guanine-phosphoribosyltransferase retards growth of Toxoplasma gondii. Parasitol. Res. 104:377–383 [DOI] [PubMed] [Google Scholar]
- 75. Zhang H., Ehrenkaufer G. M., Pompey J. M., Hackney J. A., Singh U. 2008. Small RNAs with 5′-polyphosphate termini associate with a Piwi-related protein and regulate gene expression in the single-celled eukaryote Entamoeba histolytica. PLoS Pathog. 4:e1000219 doi:10.1371/journal.ppat.1000219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zhang H., Kolb F. A., Jaskiewicz L., Westhof E., Filipowicz W. 2004. Single processing center models for human Dicer and bacterial RNase III. Cell 118:57–68 [DOI] [PubMed] [Google Scholar]
- 77. Zhang H., Pompey J. M., Singh U. 2011. RNA interference in Entamoeba histolytica: implications for parasite biology and gene silencing. Future Microbiol. 6:103–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zhao Z., Lindsay M. E., Roy Chowdhury A., Robinson D. R., Englund P. T. 2008. p166, a link between the trypanosome mitochondrial DNA and flagellum, mediates genome segregation. EMBO J. 27:143–154 [DOI] [PMC free article] [PubMed] [Google Scholar]