Abstract
A fungus-specific outer kinetochore complex, the Dam1 complex, is essential in Saccharomyces cerevisiae, nonessential in fission yeast, and absent from metazoans. The reason for the reductive evolution of the functionality of this complex remains unknown. Both Candida albicans and Schizosaccharomyces pombe have regional centromeres as opposed to the short-point centromeres of S. cerevisiae. The interaction of one microtubule per kinetochore is established both in S. cerevisiae and C. albicans early during the cell cycle, which is in contrast to the multiple microtubules that bind to a kinetochore only during mitosis in S. pombe. Moreover, the Dam1 complex is associated with the kinetochore throughout the cell cycle in S. cerevisiae and C. albicans but only during mitosis in S. pombe. Here, we show that the Dam1 complex is essential for viability and indispensable for proper mitotic chromosome segregation in C. albicans. The kinetochore localization of the Dam1 complex is independent of the kinetochore-microtubule interaction, but the function of this complex is monitored by a spindle assembly checkpoint. Strikingly, the Dam1 complex is required to prevent precocious spindle elongation in premitotic phases. Thus, constitutive kinetochore localization associated with a one-microtubule-one kinetochore type of interaction, but not the length of a centromere, is correlated with the essentiality of the Dam1 complex.
INTRODUCTION
The separation of sister chromatids during mitosis is driven by a dynamic interaction between two macromolecular structures: (i) the kinetochore (KT), a complex proteinaceous structure built on the centromere DNA (10, 47), and (ii) the mitotic spindle formed by microtubules (MTs). The process of chromosome segregation has been studied extensively in a wide range of organisms, from simple unicellular yeasts to humans. While the major sequence of events required for chromosome segregation remains the same, a few species-specific differences exist. The microtubule organizing centers (MTOCs) in budding yeasts, called spindle pole bodies (SPBs), are embedded in the nuclear envelope and assemble an intranuclear mitotic spindle (5). Thus, the interaction of spindle MTs with KTs does not require the breakdown of the nuclear envelope during closed mitosis in budding yeasts, and the KT-MT interaction is established early in the cell cycle (12, 23). The fission yeast Saccharomyces pombe also undergoes closed mitosis (23), but SPBs remain outside the nuclear envelope during interphase (27) and spindle MTs are nucleated and interact with KTs only when the duplicated SPBs enter the nuclear membrane following mitotic initiation (14). Metazoan cells, in contrast, undergo open mitosis (22, 45) in which the MTs, originating from MTOCs, gain access to KTs only when the nuclear envelope breaks down during mitosis. Finally, the number of MTs that bind to a single chromosome differs in these organisms: in Saccharomyces cerevisiae only one MT binds per KT (52), in S. pombe two or three MTs bind per KT (13), and in metazoans multiple MTs bind per KT (20 to 25 in humans) (34). Thus, considering the timing of the onset of KT-MT interaction, the number of MTs associated with each KT and the pattern of the breakdown of the nuclear envelope, a gradual evolution is evident from budding yeast to fission yeast to metazoans.
The process of chromosome segregation is mediated by nuclear (kinetochore and nonkinetochore/interpolar) and cytoplasmic (astral) MTs that originate from SPBs: kinetochore microtubules (kMTs) that connect SPBs to KTs generate poleward pulling forces for sister chromatid separation during anaphase A. The Dam1 complex (described below) in S. cerevisiae has been shown to be a coupler that transduces MT depolymerization activity into poleward pulling forces. Nonkinetochore/interpolar MTs (IPMTs), which interdigitate at the spindle midzone with the help of plus-end MT binding proteins (4, 32), elongate to generate outward pushing forces to further separate sister chromatids during anaphase B. Cytoplasmic/astral MTs that protrude toward the cytoplasm (41) regulate spindle length and alignment (20). The interaction of astral MTs with the cell cortex generates backward force that acts on spindle poles and keeps them at a specific distance apart from each other (7, 33, 39, 41, 46).
Unlike fission yeast and metazoans, KT-MT interaction starts at a premitotic phase in S. cerevisiae (5). Several lines of evidence suggest that the mitotic spindle in S. cerevisiae is kept short until anaphase onset to facilitate KT-MT attachment and to avoid premature chromosome segregation (2, 30, 31). Thus, a proper KT-MT interaction is important for spindle length maintenance even in the premitotic phases of the budding yeast cell cycle. Improper KT-MT interaction (unattached KTs or improper tension) activates the spindle assembly checkpoint (SAC) system (Mad1p, Mad2p, Mad3p, Bub1p, Bub2p, Bub3p, and Mps1p are part of the SAC in yeast) that prevents metaphase-anaphase transition (36). Eventually the inactivation of SAC by proper KT-MT interaction allows sister chromatid separation.
A fungus-specific 10-protein outer KT complex, namely, the Dam1 complex, has been shown to be involved in KT-MT interaction in S. cerevisiae (1, 18, 26, 38, 49). In this organism, mitotic spindles have been shown to be highly compromised in various mutants of the Dam1 complex (8, 9, 16, 24). All 10 proteins of the Dam1 complex are essential for viability, localized at the KT throughout the cell cycle, and help in chromosome segregation by forming rings around the MTs in S. cerevisiae (48). In contrast, the Dam1 complex is localized at the KT only during mitosis and none of the proteins in this complex is essential for the viability in fission yeast, but mutations in these proteins lead to increased chromosome missegregation. Functional differences of this complex in the conserved process of chromosome segregation raises an interesting question: can redundancy in the essentiality of the Dam1 complex be correlated with the timing of KT-MT interaction (premitotic or mitotic) as well as the localization of this complex (constitutive or mitotic) at the KT, or is it the type of centromere (point or regional) that determines the essentiality of this complex?
The putative homologs of all 10 components of the Dam1 complex are present in Candida albicans (35). C. albicans, the most commonly isolated fungal pathogen, causes candidiasis in immunocompromised patients (15, 21, 50). In C. albicans the nuclear spindle is visible for most of its cell cycle, the nuclear envelope remains intact during mitosis (17), and KT-MT interaction is established early during the cell cycle (42, 44). Similar to S. cerevisiae, only one MT binds to a KT in C. albicans (25). C. albicans has 3- to 5-kb-long CENP-A-rich centromere sequences that are unique and different from each other on various chromosomes (3, 43). The properties of C. albicans centromeres appear to be intermediate between those of S. cerevisiae (125-bp-long centromeres) and S. pombe (40- to 110-kb-long centromeres), thus C. albicans is a good system to study the functional evolution of the Dam1 complex.
In this study, we show that at least four proteins of this evolutionarily conserved outer KT complex, namely, Dam1, Ask1, Dad2, and Spc19, are essential for viability and are localized at the KT throughout the cell cycle in C. albicans. The KT recruitment of the Dam1 complex, however, is independent of MTs. Our results also show that the Dam1 complex is involved in the KT-MT-mediated process of chromosome segregation during mitosis, including spindle length maintenance throughout the cell cycle. Based on our results, we propose that the essentiality of the Dam1 complex can be correlated with the timing of the onset of KT-MT interaction and the number of MTs per KT but not with the type of centromere.
MATERIALS AND METHODS
Strain construction.
Strains used in this study are listed in Table S2 in the supplemental material.
Construction of conditional mutants of DAM1 (Orf19.4837), ASK1 (Orf19.4675), SPC19 (Orf19.4473), and DAD2 (Orf19.3551).
The first copy of DAM1, ASK1, or SPC19 was replaced by HIS1 in the strain SN148. 5′ Sequences of long primers (see Table S3 in the supplemental material) that were homologous to sequences upstream and downstream of each gene and 3′ ends that were homologous to the HIS1 gene were used to construct deletion cassettes by PCR using the plasmid HIS1GFP (19) as the template. Each deletion construct carries 90 to 110 bp of upstream and downstream homology regions flanked by the HIS1 gene. Transformants were selected on complete medium lacking histidine (CM-His medium). In resulting strains J101, J103, and J105, the remaining wild-type allele of DAM1, ASK1, or SPC19, respectively, was placed under the control of the MET3 promoter. We cloned part of the open reading frame (ORF), including the start codon of each gene (that carries a unique restriction site), as BamHI (DAM1 and ASK1) or BamHI/PstI (SPC19) fragments into the corresponding site(s) of pCaDis (6). Each of these resulting plasmids was linearized with ClaI (for DAM1 and ASK1) or SwaI (for SPC19) and transformed into J101, J103, or J105 to give rise to J102, J104, or J106, respectively (see Table S2 in the supplemental material for genotypes of strains used).
To delete the first copy of DAD2, we cloned the HIS1 gene into pBluescript KS II(−) as an EcoRI/BamHI fragment to generate pDad2-1. We then cloned DAD2 upstream (523 bp) and downstream (583 bp) sequences into the sites flanking HIS1 in pDad2Δ1 as BamHI/SacI and HindIII/KpnI fragments, respectively. The resulting plasmid pDad2-3 was digested with SacI and KpnI and transformed into BWP17 (51) to give rise to J107. To place the remaining allele of DAD2 under the control of the PCK1 promoter, URA3 sequence was cloned as a HindIII fragment into the same site of pBluescript to give rise to pDad2-4. Subsequently, pDad2-5 was generated by cloning DAD2 upstream sequence (459 bp) as KpnI and XhoI fragments into pDad2pck1a. The PCK1 promoter was released from pCA01 (29) as a BamHI-BglII fragment and cloned into the BamHI site of pDad2-5 to get pDad2-6. Finally, pDad2-7 was constructed by cloning the DAD2 ORF into pDad2-6 as a BamHI-NotI fragment. A 3.6-kb KpnI-NotI fragment was released from pDad2-7 and used to transform J107 to give rise to J108.
Construction of the mad2 null mutant and conditional double mutants of the Dam1 complex proteins in the mad2 null mutant background.
To delete both copies of MAD2, two deletion cassettes were constructed. MAD2 upstream and downstream sequences were PCR amplified (see Table S1 in the supplemental material) and cloned into BamHI/SmaI and SalI/XhoI sites of pBluescript to generate pMad2-1. The construct for the deletion of the first copy of MAD2 was generated by cloning Candida maltosa LEU2 (37) (PCR amplified from vector pSN42, a gift from Suzanne M. Noble) into EcoRI and SalI sites of pMad-1. The resulting plasmid pMad2-2 was digested with SacII and XhoI and used to transform C. albicans SN148, J102, J104, or J106 to give rise to J109, J111, J113, and J115, respectively. To delete the remaining copy of MAD2, we constructed pMad2-3 by cloning C. albicans ARG4 into SmaI and SalI sites of pMad2-1. The resulting plasmid pMad2-3 was digested with BamHI and XhoI and transformed into J109, J111, J113, or J115 to give rise to J110, J112, J114, and J116, respectively.
Construction of protein A-tagged strains.
A tandem affinity purification (TAP) tag cassette was chromosomally fused to the C terminus of the DAM1 or DAD2 gene. A calmodulin binding domain (CBD) and a protein A domain are the two epitopes present in the TAP tag. The CaURA3-TAP cassette was amplified from the vector pPK335 (11) using long primer pairs (see Table S3 in the supplemental material) homologous to the gene locus (Dam1 or Dad2) at the 5′ end and a CaURA3-TAP cassette at the 3′ end. These TAP-tagging cassettes were transformed into C. albicans J101 or J107, where the first allele of each gene was deleted using CaHIS1 to give rise to J117 or J118. All strains were analyzed for correct integration by confirmatory PCRs using primers listed in Table S3 in the supplemental material.
Media and growth conditions.
Conditional mutant strains J102, J104, and J106, carrying DAM1, ASK1, and SPC19, respectively, under the control of the MET3 promoter were grown in YPDU (1% yeast extract, 2% peptone, 3% glucose, and 0.01% uridine) as a permissive medium and YPDU plus 5 mM cysteine plus 5 mM methionine as a nonpermissive medium. Conditional mutant strain J108, carrying DAD2 under the control of the PCK1 promoter, was grown in YPSU (1% yeast extract, 2% peptone, 2% succinate and 0.01% uridine) as the permissive medium and YPDU as the nonpermissive medium. All C. albicans strains were grown at 30°C.
Viability assays.
Conditional mutants of DAM1, ASK1, SPC19, or DAD2 were grown overnight in inducing medium (YPU-succinate for the PCK1 promoter-driven expression or CM-Met-Cys for the MET3 promoter-driven expression). Cells grown overnight were pelleted down, washed with water, transferred into repressive media (YPU plus 3% glucose PCK1 promoter repression and YPDU plus 5 mM Met plus 5 mM Cys for MET3 promoter repression), and grown at 30°C. Cells were collected at different time points, counted, and then plated on plates containing inducing medium. The number of viable cells that formed colonies was counted after 2 days of incubation at 30°C. Percent viability was plotted at different time points (see Fig. 4; also see Fig. S1 in the supplemental material).
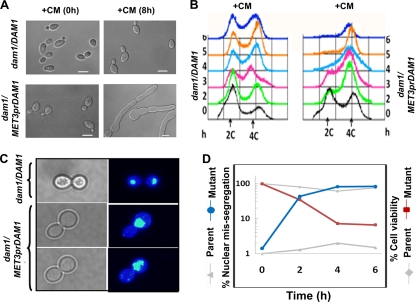
Fig. 4.
Dam1 complex is required for G2/M progression through the cell cycle and completion of mitosis in C. albicans. (A) DIC images showing phenotypes of the parent strain (dam1/DAM1) and the conditional dam1 mutant (dam1/MET3prDAM1) before (0 h) and after (8 h) growth in medium repressive for the MET3 promoter. (B) FACS analysis showing the cellular DNA content of these strains until 6 h of growth in medium repressive for the MET3 promoter. (C) Fixed wild-type cells or cells depleted of Dam1 were stained with DAPI after 4 h of growth in repressive medium. Dam1-depleted cells show single unsegregated nuclei stuck mostly at the mother bud neck, while parent cells undergo proper nuclear segregation. (D) Graphs showing the proportion of cells exhibiting the improper nuclear division of wild-type cells (gray triangles) or conditional mutant cells (blue circles) collected at various time points of growth in nonpermissive medium. Cell viability of wild-type (gray diamonds) and mutant (red squares) strains at corresponding time points also are shown on the same graph. Ask1, Spc19, and Dad2 mutants show similar results (see Fig. S1 in the supplemental material). Bar, 5 μm.
DAPI staining, calcofluor staining, and FACS analysis.
Various C. albicans strains were grown overnight in inducing medium. These cells were pelleted down, washed with water, transferred into repressible media, and grown at 30°C. Cells were harvested at various time points after growth in repressible media and were fixed with 70% alcohol for 10 min. Cells then were pelleted down, suspended in water, and incubated in 4′,6′-diamidino-2-phenylindole (DAPI) solution (50 ng/ml) for 10 min. For calcofluor staining, cells were incubated in calcofluor solution (no. 18909; Sigma) for 5 min and subsequently washed with 10% potassium hydroxide solution. For fluorescent-activated cell sorter (FACS) analysis, cells collected at various time points were fixed, and subsequent steps were followed as described previously (44). Images were captured by using LSM 510 META software using a laser confocal microscope (Carl Zeiss, Germany).
Nocodazole treatment.
For nocodazole treatment, J118 (dad2/DAD2TAP) cells were grown overnight in YPDU and reinoculated in YPDU with an initial optical density at 600 nm (OD600) of 0.2. Nocodazole (no. M1404; Sigma) was added at a concentration of 20 μg/ml when an OD600 of 0.4 (one generation) was achieved. Cells were grown for an additional 4 h before harvesting for immunolocalization and chromatin immunoprecipitation (ChIP) assays.
Subcellular immunolocalization.
Subcellular immunolocalization was performed using protocols described before (44). The dilutions of primary antibodies used were the following: affinity-purified rabbit anti-Dad2 antibody, 1:50; rabbit anti-protein A antibody (no. P3775; Sigma), 1:1,000; affinity-purified rabbit anti-CENP-A antibody (44), 1:500; mouse anti-Myc (no. OP10L; Calbiochem), 1:50; rat anti-tubulin (no. YOL1/34; Invitrogen), 1:100. The fluorescent secondary antibodies were obtained from Invitrogen and used at a dilution of 1:500 for Alexa Fluor goat anti-rabbit IgG 568 (no. A11011), 1:100 for Alexa Fluor goat anti-rat IgG 488 (no. A11006), and 1:500 for Alexa Fluor anti-mouse 488 (no. A11001). Coimmunolocalization experiments were performed using the same protocol.
Microscopy, image capture, and image processing.
All microscopic images were taken by LSM 510 META software using a laser confocal microscope (Carl Zeiss, Germany) with the following lasers for specific flurophores: Ar laser (band pass, 500 to 550 nm) for Alexa Fluor 488, He/Ne laser (band pass, 565 to 615 nm) for Alexa fluor 568, and a two-photon laser near infrared (band pass, ∼780 nm) for DAPI. Palette adjustment was performed to get the optimal intensity for each image. Z stacks were collected at 0.4- to 0.5-μm intervals, and stacked projection images were further processed in Adobe Photoshop.
ChIP assay.
ChIP assays with anti-CENP-A antibodies were performed as described before (40, 43). ChIP assays with anti-protein A antibodies in J117 (Dam1-Prot A) and J118 (Dad2-Prot A) strains were performed using the same protocol except for the following modifications: J117 cells were cross-linked with 1% formaldehyde for 90 min, while J118 cells were cross-linked for 120 min. The DNA recovered after reversing the cross-linking was precipitated using herring sperm DNA and ethanol.
RESULTS
Identification of the Dam1 complex in C. albicans.
Using polypeptide sequences of S. cerevisiae Dam1 complex components as queries in a BLAST analysis, we identified putative homologous open reading frames (ORFs) of each of the 10 proteins that form the Dam1 complex in C. albicans (see Table S1 in the supplemental material). By using a conserved domain search service (NCBI), the conserved motifs in each of these 10 ORFs were identified. These motifs share considerable similarity in amino acid sequence. The overall sequence similarity, however, was found to be low in most cases.
The Dam1 complex is localized to the kinetochore throughout the cell cycle.
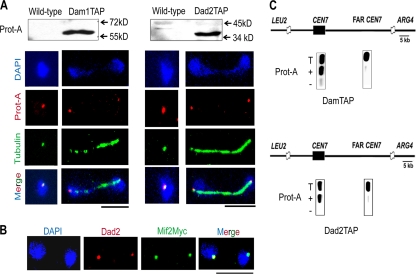
The subcellular localization of Dam1 and Dad2 in C. albicans was performed by indirect immunofluorescence microscopy using anti-protein A antibodies in strains J117 (dam1/DAM1TAP) and J118 (dad2/DAD2TAP), respectively. The only full-length copy of DAM1 or DAD2 present in J117 or J118 was fused with protein A, an epitope of the tandem affinity purification (TAP) tag, at the C terminus of each ORF (strains are listed in Table S2 in the supplemental material). Western blot analysis confirmed the expression of protein A-tagged Dam1 and Dad2 (Fig. 1A, top). We observed bright dot-like signals in all cells that colocalized with DAPI-stained nuclei (Fig. 1A). Each dot represents a cluster of 16 centromeres. Unbudded cells exhibited one dot per cell close to the SPB, while large-budded cells at later stages of the cell cycle exhibited two dots that cosegregated with nuclei connected by mitotic spindles. The localization patterns suggest that Dad2 and Dam1 are present at KTs throughout the cell cycle. The colocalization of Dad2 and a key KT component, Mif2p (Myc tagged), in the strain CAMB1 (43) confirmed that Dad2 is KT localized (Fig. 1B). To determine in vivo associations of these two proteins with the centromeres, we performed standard chromatin immunoprecipitation (ChIP) assays with anti-protein A antibodies in strains J117 and J118. The immunoprecipitated (IP) DNA samples were analyzed by PCR using a specific set of primers designed from CEN7 sequences (primers are listed in Table S3 in the supplemental material). CEN7 regions in J117 and J118 were enriched in the IP DNA, indicating that Dam1 and Dad2 (Fig. 1C) were associated with centromeres. Taken together, these results indicate that Dam1 and Dad2, components of the Dam1 complex, are KT localized throughout the cell cycle in C. albicans.
Fig. 1.
Dam1 complex is localized to the kinetochore throughout the cell cycle in C. albicans. (A) Western blot analysis using total cell lysates from BWP17 (wild type) and J117 (Dam1-Prot A) or J118 (Dad2-Prot A) cells with anti-protein A antibodies (top). DAPI-stained cells of J117 (Dam1TAP) and J118 (Dad2TAP), at different stages of the cell cycle, were immunostained with anti-protein A (red) and anti-tubulin (green) antibodies. (B) Indirect immunostaining with anti-Dad2 (red) and anti-myc (Mif2) (green) antibodies in C. albicans strain CAMB1 expressing a key KT component, Myc-Mif2. (C) Anti-protein A ChIP DNA from J117 or J118 cells was analyzed by PCR with primers specific from CEN7 (assembly 21-CaChr7 coordinates 427369 to 427560) or a region 17 kb away (assembly 21-CaChr7 coordinates 444584 to 444875) from it (FAR CEN7). T, total DNA; +, IP DNA with anti-protein A antibodies; −, bead-only control without antibodies.
Kinetochore recruitment of the Dam1 complex is independent of the presence of microtubules.
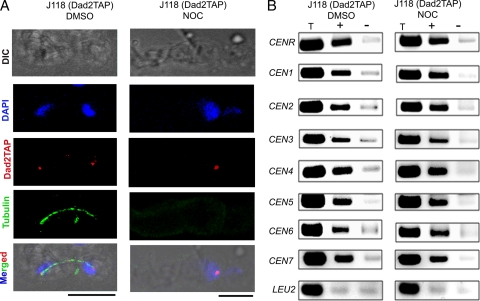
To examine the dependency of the Dam1 complex on spindle MTs for its KT recruitment, we observed the KT localization of Dad2 in the presence of the spindle poison nocodazole (NOC). Indirect immunofluorescence was performed in untreated (DMSO only) and nocodazole-treated J118 (dad2/DAD2TAP) cells using anti-tubulin and anti-protein A antibodies. Nocodazole treatment led to G2/M arrest followed by elongated bud phenotype with no or barely visible tubulin staining (Fig. 2A). The absence of tubulin staining in G2/M-arrested NOC-treated cells confirmed that the mitotic spindle was disrupted. Untreated cells exhibited normal spindles (Fig. 2A). However, the intensities of Dad2 TAP signals in NOC-treated cells were comparable to those in untreated cells, suggesting that the disruption of the spindle did not alter Dad2 localization to the KTs (Fig. 2A). To further confirm this result, we performed ChIP assays in untreated and NOC-treated J118 cells using anti-protein A antibodies. The immunoprecipitated (IP) DNA samples were analyzed by PCR using a specific set of primers designed from CEN regions of all of the chromosomes (primers are listed in Table S3 in the supplemental material). No significant difference in Dad2 TAP binding at any of the CEN regions was observed (Fig. 2B). Taken together, these results confirm that Dad2 localization at the KTs is independent of KT-MT interaction.
Fig. 2.
Kinetochore localization of Dad2 does not depend on integrity of kinetochore-microtubule interaction in C. albicans. (A) Untreated (DMSO) or nocodazole-treated (NOC) J118 (dad2/DAD2TAP) cells were stained by DAPI and anti-protein A and anti-tubulin antibodies. A barely visible signal with anti-tubulin antibodies in NOC-treated cells confirmed the loss of spindle structures. However, Dad2 signals are comparable in DMSO-treated and NOC-treated cells. (B) ChIP assays with anti-protein A antibodies using DMSO-treated and NOC-treated J118 cells confirmed that Dad2 recruitment at the centromeres is independent of kinetochore-microtubule interaction. T, total DNA; +, IP with anti-protein A antibodies; −, bead-only control.
The Dam1 complex is essential for viability in C. albicans.
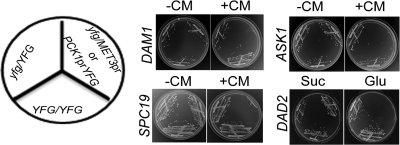
To examine the essentiality of the Dam1 complex in C. albicans, we constructed conditional mutants of four genes, DAM1, ASK1, DAD2, and SPC19. The first copy of each gene was replaced with a HIS1 marker, and the remaining copy was placed under the control of a regulatable promoter (the MET3 promoter for DAM1, ASK1, and SPC19 and the PCK1 promoter for DAD2). The MET3 promoter is repressed in the presence of cysteine (Cys) and methionine (Met) (6), while the PCK1 promoter is repressed when glucose is used as the carbon source (29). Strains J102 (dam1/MET3prDAM1), J104 (ask1/MET3prASK1), and J106 (spc19/MET3prSPC19) failed to grow on plates containing 5 mM Cys and 5 mM Met (Fig. 3). Similarly, the strain J108 (dad2/PCK1prDAD2) failed to grow when glucose was used as the carbon source. The parent strains J101 (dam1/DAM1), J103 (ask1/ASK1), J105 (spc19/SPC19), and J107 (dad2/DAD2) did not show any growth defects. The inability of the conditional mutants to grow under repressible conditions confirmed that each of these four genes is essential for viability.
Fig. 3.
Dam1 complex is essential for viability in C. albicans. C. albicans cells expressing both wild-type copies (YFG/YFG), one deleted copy, and the remaining wild-type copy under the control of either the native promoter (yfg/YFG) or the MET3/PCK1 promoter (yfg/MET3prYFG or yfg/PCK1prYFG) of the indicated gene (DAM1, ASK1, SPC19, or DAD2) were streaked on plates containing inducible (−CM or Suc) or repressible (+CM or Glu) medium (CM, 5 mM cysteine and 5 mM methionine; Suc, succinate; Glu, glucose). The MET3 promoter, which is expressed in the absence of CM (−CM) and repressed in the presence of CM (+CM), was used for the controlled expression of DAM1, ASK1, and SPC19. The PCK1 promoter, which is expressed in the presence of succinate (Suc) and repressed in glucose (Glu), was used to control the expression of DAD2. Plate photographs were taken after 2 to 3 days of incubation at 30°C.
The Dam1 complex is required for G2/M progression through the cell cycle and completion of mitosis in C. albicans.
Conditional mutant strains of DAM1 (J102), ASK1 (J104), SPC19 (J106), and DAD2 (J108) showed the accumulation of large budded cells after 4 h of growth under nonpermissive conditions. A prolonged incubation (>6 h) led to a transition from a large bud to an elongated bud (pseudohyphal) phenotype in each case (Fig. 4A; also see Fig. S1A in the supplemental material). FACS analysis of these mutants confirmed that the majority of the cells had 4N DNA content within 4 h of protein depletion (Fig. 4B; also see Fig. S1B). Almost all arrested cells with large buds (99%) that were depleted of either Dam1 or Ask1 had single undivided DNA masses (Fig. 4C). On the other hand, the depletion of either Spc19 or Dad2 resulted in large-budded cells containing either a single undivided nucleus (75 to 90%) or two unequally divided nuclei (10 to 25%) (Fig. 5). Nuclear missegregation increased drastically between 2 and 6 h of growth under nonpermissive conditions in all cases with a concomitant loss in cell viability, suggesting that chromosome missegregation was the sole cause of loss in cell viability in these mutant cells (Fig. 4D; also see Fig. S1C). The parent strains J101 (Dam1), J103 (Ask1), J105 (Spc19), and J107 (Dad2), grown under similar conditions, exhibited normal cell cycle progression (with no obvious cell cycle arrest), proper nuclear segregation, and unaltered cell viability throughout their growth under similar conditions (Fig. 4D; also see Fig. S1C). All of these results strongly indicate that the Dam1 complex in C. albicans is essential for proper nuclear segregation, G2/M progression through the cell cycle, and the completion of mitosis.
Fig. 5.
Proportion of cells with the indicated phenotypes showing chromosome missegregation in Dam1- or Dad2-depleted cells.
Depletion of an essential kinetochore protein in C. albicans leads to severe spindle defects.
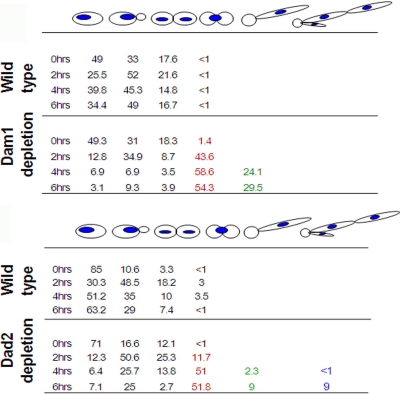
Most of the cells of Dam1 conditional mutants grown under nonpermissive conditions arrested at the large bud stage with unsegregated nucleus in each of the four cases. We examined and compared spindle morphology in known inner KT mutants of CENP-A (Cse4) and CENP-C (Mif2) with conditional mutant strains of the Dam1 complex, namely, J102 (Dam1), J104 (Ask1), J106 (Spc19), and J108 (Dad2), grown under nonpermissive conditions for 4 h (partial depletion) and 8 h (nearly complete depletion). As opposed to the wild type (BWP17) (Fig. 6), conditional mutant strains CAKS3b (CSE4/PCK1prCSE4) and CAMB2 (mif2/PCK1prMIF2), when grown in nonpermissive conditions, showed arrested cells at the G2/M stage with either short (>85%) or medium-long (<15%) spindles (Fig. 6). Two of the four subunits of the Dam1 complex under study, Spc19- and Dad2-depleted cells, showed similar spindle defects (Fig. 6). The spindle defects exhibited due to the depletion of the other two subunits, Dam1 and Ask1, were different from those of the rest of the KT mutants of C. albicans.
Fig. 6.
Depletion of an essential kinetochore protein in C. albicans leads to severe spindle defects. Wild-type cells were grown in YPDU or YPDU plus 10% serum, and conditional mutants of inner kinetochore proteins (CENP-A/Cse4 and CENP-C/Mif2) and two of the four tested outer kinetochore proteins of the Dam1 complex (Dad2 and Spc19) were grown in nonpermissive medium for 4 h (left) or 8 h (right), fixed, and stained with DAPI and anti-tubulin antibodies. The percentage of cells with specific spindle phenotypes associated with each strain was calculated from cells at the elongated bud stage (right). Bar, 5 μm.
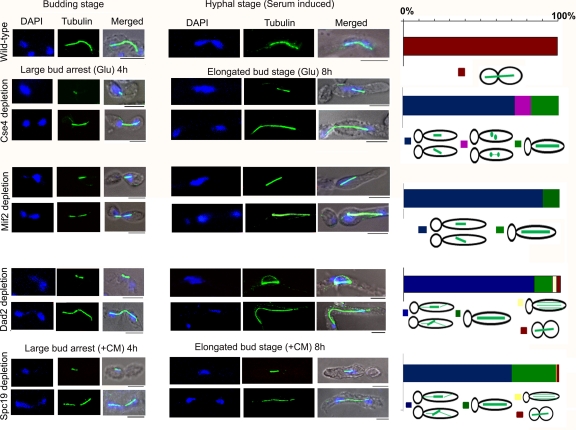
Subunits of the Dam1 complex maintain dynamics of interpolar and astral MTs.
Astral MTs are highly dynamic, and their appearance is mostly undetectable in both wild-type budding and hyphal cells of C. albicans (Fig. 7A and B). Dam1-depleted cells showed a uniform short-spindle phenotype but lacked astral MTs (Fig. 7C and D, upper). More than 45% of dam1 mutant cells exhibited two SPB-like dots situated very close to each other with barely visible MTs between the two dots, indicating an apparent absence of interpolar MTs (Fig. 7C and D, lower). Interestingly, Ask1-depleted cells showed a uniform phenotype of short, often misaligned spindles with a brightly stained extensive network of cytoplasmic (astral) microtubules (Fig. 7E and F).
Fig. 7.
Dam1 and Ask1, two subunits of the Dam1 complex, maintain the polymerization/depolymerization dynamics of interpolar and astral microtubules. Wild-type cells grown in YPDU (A) or YPDU plus 10% serum (B) and conditional mutants of Dam1 (dam1prMET3DAM1/dam1) and Ask1 (ask1prMET3ASK1/ask1) grown in nonpermissive medium for 4 h (C and E) or 8 h (D and F) were fixed and stained with DAPI and anti-tubulin antibodies. More than 50% of short spindles resulting from Dam1 depletion showed no staining of IPMTs at the midzone and thus appeared as two dots (probably representing only SPBs) situated close to each other (C and D, lower). (E and F) Almost all of the short spindles (>99%) caused by Ask1 depletion resulted in the extensive growth of astral microtubules toward the cell cortex. The percentage of cells with specific spindle phenotypes associated with each strain was calculated from cells at the elongated bud stage (extreme right). Bar, 5 μm.
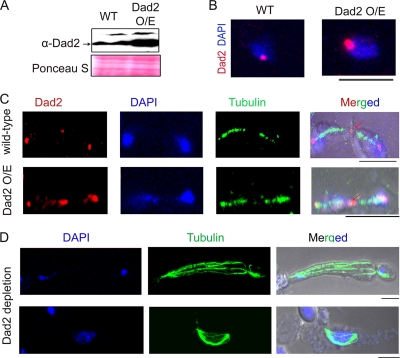
Dad2 also is localized at the spindle midzone.
Western blot analysis and immunostaining with Dad2 antibodies confirmed that strain J108 (dad2/PCK1prDAD2) expressed Dad2 in succinate media at levels higher than those of the wild type (BWP17) (Fig. 8A and B). The overexpression of Dad2 did not alter chromosome segregation fidelity or cell viability compared to those of the wild type (data not shown). Immunostaining with anti-Dad2 antibodies in J108 overexpressing Dad2 also revealed a bright dot-like signal in the midzone in addition to its KT localization (the other two dot-like signals are at the KTs near the SPBs) (Fig. 8C; also see Fig. S2 in the supplemental material). In extremely rare cases (<1% of total population), Dad2 signals were visible throughout the spindle axis (see Fig. S2). However, midzone localization was rarely visible in wild-type BWP17 cells (DAD2/DAD2) (Fig. 8C), probably due to low cellular levels of Dad2 present at the spindle midzone. Thus, in addition to its KT localization, Dad2 probably is localized to the plus end of the nuclear microtubules.
Fig. 8.
Dad2 is localized at the spindle midzone. Dad2 was overexpressed under the control of the PCK1 promoter in succinate-containing medium. (A) Western blot analysis of cell lysates from the wild-type strain (BWP17) and Dad2-overexpressing (O/E) strain J108 (dad2/PCK1prDAD2) grown in succinate using anti-Dad2 or anti-PSTAIRE (loading control) antibody. (B) BWP17 and J108 cells were grown in succinate, fixed, and stained with DAPI and anti-Dad2 antibodies. (C) Succinate-grown wild-type BWP17 and Dad2-overexpressing J108 (dad2/PCK1prDAD2) cells were fixed and stained with DAPI and anti-tubulin and anti-Dad2 antibodies. Apart from the usual KT localization (two spots overlapping with a DAPI-stained nuclear mass), an additional signal at the spindle midzone (shown by an arrow) was visible in anaphase cells. (D) Dad2-depleted cells (J108 grown in glucose) were fixed and stained with DAPI and anti-tubulin antibody. The unbundling of spindle fibers was observed (although rarely) in both long and short spindles. Bar, 5 μm.
In either Dad2- or Spc19-depleted cells we observed, albeit infrequently, unbundled fibers of mitotic spindles connecting two nuclear masses (Fig. 8D).
The spindle assembly checkpoint (SAC) monitors KT-MT interaction mediated by the Dam1 complex.
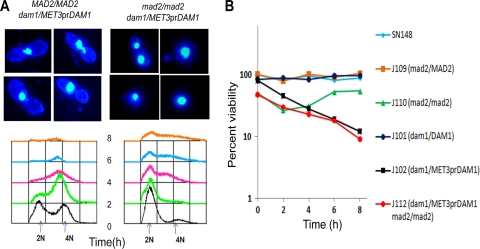
Several lines of evidence described above indicate that the subunits of the Dam1 complex in C. albicans are essential in the KT-MT-mediated process of chromosome segregation. To examine whether the large bud arrest in various Dam1 complex mutants was due to the activation of the SAC, which senses improper KT-MT interaction, we deleted both copies of MAD2, an essential SAC component, in conditional mutant strains J102, J104, and J106 to construct conditional double mutant strains J112 (mad2/mad2 dam1/MET3prDAM1), J114 (mad2/mad2 ask1/MET3prASK1), and J116 (mad2/mad2 spc19/MET3prSPC19). Microscopic observation (differential interference contrast [DIC] imaging) of J112 cells grown in nonpermissive conditions of the MET3 promoter showed no large bud arrest phenotype (Fig. 9A) even after prolonged growth (>8 h). The appearance of unbudded G1 cells with 2N DNA content determined by FACS analysis confirmed that G2/M arrest caused by Dam1 depletion was relieved in the absence of Mad2-mediated checkpoint surveillance (Fig. 9A). In spite of release from G2/M arrest, we observed no improvement in viability as well as growth rate of double mutants compared to those of the conditional single Dam1 complex mutants. The viability of double mutants was marginally less than that of single mutants (Fig. 9B), and no significant change in growth rate was observed (data not shown). These results confirm that the function of the Dam1 complex in mediating KT-MT interactions is under the surveillance of the Mad2-dependent spindle assembly checkpoint in C. albicans. Cells of J114 (mad2/mad2 ask1/MET3prASK1) and J116 (mad2/mad2 spc19/MET3prSPC19) conditional double mutants also showed similar results (data not shown). Dam1-, Ask1-, or Spc19-depleted cells underwent division in the absence of Mad2. However, these cells behaved abnormally in subsequent cell cycle stages. We observed multiple budded cells or chains of cells that were attached to each other with multiple (>2) DAPI-stained nuclear masses (see Fig. S3 in the supplemental material), suggesting defects in budding when any one of the Dam1 complex subunits was depleted in the mad2 mutant strain.
Fig. 9.
Function of the Dam1 complex is under the surveillance of Mad2-mediated spindle assembly checkpoint in C. albicans. (A) J102 (dam1/MET3prDAM1) and J112 (dam1/MET3prDAM1 mad2/mad2) cells were grown in nonpermissive medium for 4 h and stained with DAPI. J102 (dam1 mutant) cells exhibited large bud arrest with unsegregated DNA mass, mostly at the mother-bud neck when the spindle assembly checkpoint was active. J112 (dam1 mad2 double mutant) cells grown under similar conditions, on the other hand, exhibited cells at various stages of the cell cycle, including large budded cells with improperly segregated nuclear masses. FACS analysis in J102 and J112 cells at indicated times of growth in nonpermissive medium are shown as well (lower). (B) J116 (spc19MET3/pr SPC19 mad2/mad2) cells grown in nonpermissive medium for 8 h (depleted Spc19) were stained with either calcofluor white or DAPI. Spc19-depleted cells in the absence of Mad2 led to the formation of cells with multiple buds and fragmented nuclear masses. Ask1 and Dam1 showed similar phenotypes.
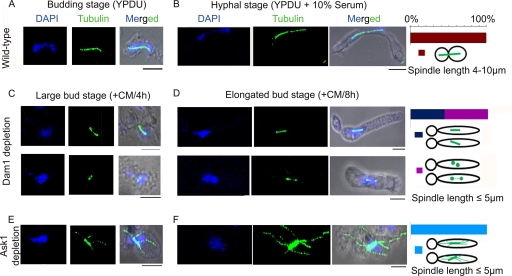
Dam1 complex-dependent KT-MT interaction restricts spindle length in premitotic cells to avoid premature chromosome segregation.
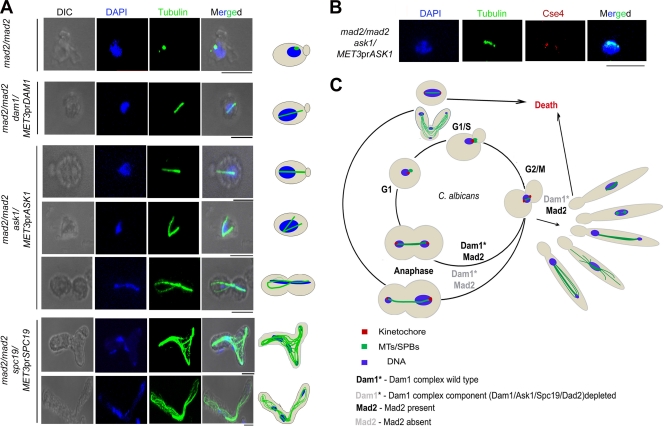
Tubulin staining in J112 (mad2/mad2 dam1/MET3prDAM1), J114 (mad2/mad2 ask1/MET3prASK1), and J116 (mad2/mad2 spc19/MET3prSPC19) strains grown in nonpermissive conditions exhibited long spindles (∼5 μm) in unbudded cells (Fig. 10A). The control parent strain J110 (mad2/mad2) showed a single dot-like signal when stained with anti-tubulin antibodies, most probably representing the SPB, in unbudded cells (Fig. 10A). This result eliminates the possibility of Mad2 in restricting the spindle length in premitotic cells. Immunostaining with anti-Cse4 antibodies in these cells exhibited more than one Cse4 dot-like signal associated with a single nuclear mass, revealing the occurrence of the unclustering of KTs (the precocious separation of the chromosomes) that were either unattached or improperly attached to the MTs (Fig. 10B). These results, along with previous observations (spindle defects in metaphase), together suggest that proper KT-MT interaction mediated by the Dam1 complex is required to restrict spindle elongation in premitotic cells. Figure 10C summarizes various roles of Dam1 complex components at different stages of the cell cycle in C. albicans.
Fig. 10.
Dam1 complex restricts spindle length in premitotic cells to avoid precocious chromosome separation. (A) Cells of the indicated genotypes grown in nonpermissive medium were fixed and stained with DAPI and anti-tubulin antibodies. Unbudded cells exhibited long spindles approaching metaphase length (2 to 5 μm) in Dam1 complex mutants when Mad2 was deleted. The ask1 mad2 double mutant often showed branched spindles in unbudded cells. Unbudded or multiply budded cells formed elongated buds (due to stress response) and often showed aberrantly segregated nuclear masses that were connected by the mitotic spindle. The mad2 mutant cells depleted of Spc19 showed unbundled mitotic spindle fibers connecting aberrantly segregated nuclear masses. (B) Cells of the indicated genotype grown in nonpermissive media were fixed and stained with DAPI and anti-Cse4 and anti-tubulin antibodies. Unbudded cells with long spindles exhibited two Cse4 dots associated with a single nuclear mass, indicating the precocious separation of chromosomes in premitotic cells. (C) A model highlighting the role of the Dam1 complex in the maintenance of spindle length and its morphology throughout the cell cycle. Bar, 5 μm.
DISCUSSION
In this work, we show that four subunits of the Dam1 complex, an evolutionarily conserved fungus-specific protein complex present at the outer KT, are essential for viability in the human pathogenic yeast C. albicans. In spite of a low level of overall homology of the protein sequence with their S. cerevisiae counterparts, the subunits of the Dam1 complex have a conserved function in chromosome segregation. Subcellular localization and ChIP assays confirm that the Dam1 complex is associated with the KT throughout the cell cycle. The depletion of each of the four proteins of the Dam1 complex results in a loss in cell viability with a concomitant increase in chromosome missegregation. As a result, conditional mutant strains in nonpermissive conditions arrest at large bud stage mostly with short spindles. Although the KT localization of the Dam1 complex is independent of the KT-MT interaction, the cell cycle arrest at G2/M is under the surveillance of the Mad2-dependent spindle assembly checkpoint, confirming the evolutionarily conserved role of the Dam1 complex in KT-MT interaction shown previously in S. cerevisiae and S. pombe. The Dam1 complex has been shown to be involved in restricting premature spindle elongation in premitotic cells.
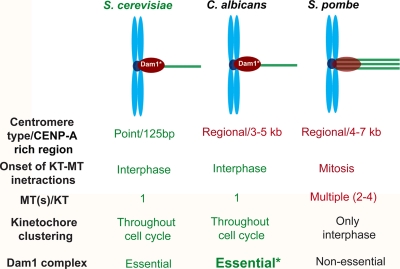
Despite its indispensable role in chromosome segregation in S. cerevisiae, the Dam1 complex is nonessential in fission yeast and even absent from metazoans, indicating that there is reductive evolution of this complex and adaptive evolution of an alternative mechanism that governs KT-MT interaction. Such a dramatic but a gradual change in requirement of the Dam1 complex in chromosome segregation across species can be correlated with the evolution of the complexity of the mechanism of chromosome segregation. In S. cerevisiae and C. albicans, one MT binds per KT, and this KT-MT interaction is established early during the cell cycle, in contrast to the two to three MTs per KT in S. pombe, where KT-MT interaction is established only during mitosis. Interestingly, the Dam1 complex has been shown to be localized at the KTs while they are associated with the spindle (i.e., throughout the cell cycle in S. cerevisiae and C. albicans but only during mitosis in S. pombe). Hence, the essentiality of the Dam1 complex is correlated with maintaining a proper one MT-one KT interaction, which is established during interphase (Fig. 11).
Fig. 11.
Reductive evolution of the Dam1 complex.
The KT localization of S. cerevisiae Dam1 has been shown to be compromised in the absence of spindle MTs (16, 30). However, results from immunofluorescence and ChIP assays in the present study unequivocally prove that the KT recruitment of the Dam1 complex is not dependent upon spindle MTs in C. albicans. This suggests that there are functional differences in the Dam1 complex of S. cerevisiae and C. albicans in terms of their mechanistic roles in KT-MT interaction. Indeed, we found that spindle defects associated with Dam1 and Ask1, two subunits of the Dam1 complex in C. albicans, are unique (discussed below) and have not been observed in S. cerevisiae.
We observed that spindle defects associated with Spc19 and Dad2 are similar to those of inner (Cse4/CENP-A and Mif2/CENP-C; present study) and middle (Mis12/Mtw1 [42]) KT proteins in C. albicans. Mtw1, a protein that is present in the linker layer of the kinetochore, showed similar spindle defects in C. albicans (42). Thus, the majority of the cells with short spindles and a small population with medium-long spindles is found to be a common consequence of kinetochore mutants irrespective of the layer in which they presumably are present. However, two other outer kinetochore proteins in this study, Dam1 and Ask1, exhibited an exclusively short spindle phenotype with a single nuclear mass when depleted from C. albicans cells. This observation suggests a possible functional difference between these two proteins and the rest analyzed from each of the three layers of the kinetochore. Indeed, in S. cerevisiae only Dam1 or Ask1 alone could promote chromosome segregation more efficiently than other KT proteins when tethered onto a noncentromeric sequence either on a plasmid or on a chromosome with a conditionally inactivated centromere (26, 28).
Besides short and long spindles, all four components showed mutant-specific defects in spindle morphology in C. albicans. Most of short spindles in Dam1 mutants were found to have very weak or no staining at the midzone, probably due to a faster depolymerization/destabilization of interpolar MTs. The abnormal growth of astral MTs in the absence of Ask1 might be due to the stabilization of these MTs by inhibiting their rate of depolymerization. We anticipate that these abnormally elongated astral MTs interact with the cell cortex and generate a backward force on SPBs. This may prevent cell cycle stage-specific spindle elongation and alignment, resulting in short and misaligned spindles.
Since the Dam1 complex in C. albicans is localized throughout the cell cycle, we asked whether the Dam1 complex helps in KT-MT interaction during interphase. The Dam1 complex mutants in the absence of Mad2 provided us an opportunity to study the role of this complex in premitotic interphase cells. The presence of long metaphase-like spindles (3 to 5 μm) in these unbudded cells revealed premature spindle elongation in premitotic cells. This led us to propose that the Dam1 complex restricts interphase spindle length elongation. Figure 10 summarizes a comparative analysis of the Dam1 complex from different organisms. Future experiments to test the essentiality of the Dam1 complex in C. albicans by creating conditions that will allow more than one MT binding to each kinetochore may provide a direct link between the function of the Dam1 complex with the number of MTs per KT. The essentiality of the Dam1 complex, along with its exclusive presence across fungal species without a mammalian homolog, makes it an attractive target to develop a safer and more potent antifungal drug to treat candidiasis.
Supplementary Material
ACKNOWLEDGMENTS
We thank B. S. Suma for her help in confocal microscopy.
This work was supported by a grant from the Department of Biotechnology, Government of India, to K.S., and by JNCASR.
J.T. is a senior research fellow supported by the Department of Biotechnology, Government of India.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 13 May 2011.
REFERENCES
- 1. Asbury C. L., Gestaut D. R., Powers A. F., Franck A. D., Davis T. N. 2006. The Dam1 kinetochore complex harnesses microtubule dynamics to produce force and movement. Proc. Natl. Acad. Sci. U. S. A. 103:9873–9878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bachant J., Jessen S. R., Kavanaugh S. E., Fielding C. S. 2005. The yeast S phase checkpoint enables replicating chromosomes to bi-orient and restrain spindle extension during S phase distress. J. Cell Biol. 168:999–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baum M., Sanyal K., Mishra P. K., Thaler N., Carbon J. 2006. Formation of functional centromeric chromatin is specified epigenetically in Candida albicans. Proc. Natl. Acad. Sci. U. S. A. 103:14877–14882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brinkley B. R., Cartwright J. 1971. Ultrastructural analysis of mitotic spindle elongation in mammalian cells in vitro. Direct microtubule counts. J. Cell Biol. 50:416–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Byers B., Goetsch L. 1975. Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J. Bacteriol. 124:511–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Care R. S., Trevethick J., Binley K. M., Sudbery P. E. 1999. The MET3 promoter: a new tool for Candida albicans molecular genetics. Mol. Microbiol. 34:792–798 [DOI] [PubMed] [Google Scholar]
- 7. Carminati J. L., Stearns T. 1997. Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J. Cell Biol. 138:629–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cheeseman I. M., et al. 2001. Implication of a novel multiprotein Dam1p complex in outer kinetochore function. J. Cell Biol. 155:1137–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cheeseman I. M., Enquist-Newman M., Müller-Reichert T., Drubin D. G., Barnes G. 2001. Mitotic spindle integrity and kinetochore function linked by the Duo1p/Dam1p complex. J. Cell Biol. 152:197–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cleveland D. W., Mao Y., Sullivan K. F. 2003. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112:407–421 [DOI] [PubMed] [Google Scholar]
- 11. Corvey C., et al. 2005. Carbon source-dependent assembly of the Snf1p kinase complex in Candida albicans. J. Biol. Chem. 280:25323–25330 [DOI] [PubMed] [Google Scholar]
- 12. De Souza C. P., Osmani S. A. 2007. Mitosis, not just open or closed. Eukaryot. Cell 6:1521–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ding R., McDonald K. L., McIntosh J. R. 1993. Three-dimensional reconstruction and analysis of mitotic spindles from the yeast, Schizosaccharomyces pombe. J. Cell Biol. 120:141–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ding R., West R. R., Morphew D. M., Oakley B. R., McIntosh J. R. 1997. The spindle pole body of Schizosaccharomyces pombe enters and leaves the nuclear envelope as the cell cycle proceeds. Mol. Biol. Cell 8:1461–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Eggimann P., Garbino J., Pittet D. 2003. Epidemiology of Candida species infections in critically ill non-immunosuppressed patients. Lancet Infect. Dis. 3:685–702 [DOI] [PubMed] [Google Scholar]
- 16. Enquist-Newman M., et al. 2001. Dad1p, third component of the Duo1p/Dam1p complex involved in kinetochore function and mitotic spindle integrity. Mol. Biol. Cell 12:2601–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Finley K. R., Berman J. 2005. Microtubules in Candida albicans hyphae drive nuclear dynamics and connect cell cycle progression to morphogenesis. Eukaryot. Cell 4:1697–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Franck A. D., et al. 2007. Tension applied through the Dam1 complex promotes microtubule elongation providing a direct mechanism for length control in mitosis. Nat. Cell Biol. 9:832–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gerami-Nejad M., Berman J., Gale C. A. 2001. Cassettes for PCR-mediated construction of green, yellow, and cyan fluorescent protein fusions in Candida albicans. Yeast 18:859–864 [DOI] [PubMed] [Google Scholar]
- 20. Goshima G., Scholey J. M. 2010. Control of mitotic spindle length. Annu. Rev. Cell Dev. Biol. 26:21–57 [DOI] [PubMed] [Google Scholar]
- 21. Gudlaugsson O., et al. 2003. Attributable mortality of nosocomial candidemia, revisited. Clin. Infect. Dis. 37:1172–1177 [DOI] [PubMed] [Google Scholar]
- 22. Güttinger S., Laurell E., Kutay U. 2009. Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat. Rev. Mol. Cell Biol. 10:178–191 [DOI] [PubMed] [Google Scholar]
- 23. Heath I. B. 1980. Variant mitoses in lower eukaryotes: indicators of the evolution of mitosis. Int. Rev. Cytol. 64:1–80 [DOI] [PubMed] [Google Scholar]
- 24. Hofmann C., et al. 1998. Saccharomyces cerevisiae Duo1p and Dam1p, novel proteins involved in mitotic spindle function. J. Cell Biol. 143:1029–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Joglekar A. P., et al. 2008. Molecular architecture of the kinetochore-microtubule attachment site is conserved between point and regional centromeres. J. Cell Biol. 181:587–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kiermaier E., Woehrer S., Peng Y., Mechtler K., Westermann S. 2009. A Dam1-based artificial kinetochore is sufficient to promote chromosome segregation in budding yeast. Nat. Cell Biol. 11:1109–1115 [DOI] [PubMed] [Google Scholar]
- 27. Kniola B., et al. 2001. The domain structure of centromeres is conserved from fission yeast to humans. Mol. Biol. Cell 12:2767–2775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lacefield S., Lau D. T., Murray A. W. 2009. Recruiting a microtubule-binding complex to DNA directs chromosome segregation in budding yeast. Nat. Cell Biol. 11:1116–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Leuker C. E., Sonneborn A., Delbrück S., Ernst J. F. 1997. Sequence and promoter regulation of the PCK1 gene encoding phosphoenolpyruvate carboxykinase of the fungal pathogen Candida albicans. Gene 192:235–240 [DOI] [PubMed] [Google Scholar]
- 30. Li Y., et al. 2002. The mitotic spindle is required for loading of the DASH complex onto the kinetochore. Genes Dev. 16:183–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu H., Liang F., Jin F., Wang Y. 2008. The coordination of centromere replication, spindle formation, and kinetochore-microtubule interaction in budding yeast. PLoS Genet. 4:e1000262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mastronarde D. N., McDonald K. L., Ding R., McIntosh J. R. 1993. Interpolar spindle microtubules in PTK cells. J. Cell Biol. 123:1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McCollum D. 2002. First things first: spindle orientation and mitotic progression. Nat. Cell Biol. 4:E225–E226 [DOI] [PubMed] [Google Scholar]
- 34. McDonald K. L., O'Toole E. T., Mastronarde D. N., McIntosh J. R. 1992. Kinetochore microtubules in PTK cells. J. Cell Biol. 118:369–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Meraldi P., McAinsh A. D., Rheinbay E., Sorger P. K. 2006. Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol. 7:R23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Musacchio A., Salmon E. D. 2007. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8:379–393 [DOI] [PubMed] [Google Scholar]
- 37. Noble S. M., Johnson A. D. 2005. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot. Cell 4:298–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nogales E., Ramey V. H. 2009. Structure-function insights into the yeast Dam1 kinetochore complex. J. Cell Sci. 122:3831–3836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Oliferenko S., Balasubramanian M. K. 2002. Astral microtubules monitor metaphase spindle alignment in fission yeast. Nat. Cell Biol. 4:816–820 [DOI] [PubMed] [Google Scholar]
- 40. Padmanabhan S., Thakur J., Siddharthan R., Sanyal K. 2008. Rapid evolution of Cse4p-rich centromeric DNA sequences in closely related pathogenic yeasts, Candida albicans and Candida dubliniensis. Proc. Natl. Acad. Sci. U. S. A. 105:19797–19802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Palmer R. E., Sullivan D. S., Huffaker T., Koshland D. 1992. Role of astral microtubules and actin in spindle orientation and migration in the budding yeast, Saccharomyces cerevisiae. J. Cell Biol. 119:583–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Roy B., Burrack L. S., Lone M. A., Berman J., Sanyal K. 2011. CaMtw1, a member of the evolutionarily conserved Mis12 kinetochore protein family, is required for efficient inner kinetochore assembly in the pathogenic yeast Candida albicans. Mol. Microbiol. 80:14–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sanyal K., Baum M., Carbon J. 2004. Centromeric DNA sequences in the pathogenic yeast Candida albicans are all different and unique. Proc. Natl. Acad. Sci. U. S. A. 101:11374–11379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sanyal K., Carbon J. 2002. The CENP-A homolog CaCse4p in the pathogenic yeast Candida albicans is a centromere protein essential for chromosome transmission. Proc. Natl. Acad. Sci. U. S. A. 99:12969–12974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sazer S. 2005. Nuclear envelope: nuclear pore complexity. Curr. Biol. 15:R23–R26 [DOI] [PubMed] [Google Scholar]
- 46. Theesfeld C. L., Irazoqui J. E., Bloom K., Lew D. J. 1999. The role of actin in spindle orientation changes during the Saccharomyces cerevisiae cell cycle. J. Cell Biol. 146:1019–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vos L. J., Famulski J. K., Chan G. K. 2006. How to build a centromere: from centromeric and pericentromeric chromatin to kinetochore assembly. Biochem. Cell Biol. 84:619–639 [DOI] [PubMed] [Google Scholar]
- 48. Westermann S., et al. 2005. Formation of a dynamic kinetochore-microtubule interface through assembly of the Dam1 ring complex. Mol. Cell 17:277–290 [DOI] [PubMed] [Google Scholar]
- 49. Westermann S., et al. 2006. The Dam1 kinetochore ring complex moves processively on depolymerizing microtubule ends. Nature 440:565–569 [DOI] [PubMed] [Google Scholar]
- 50. Wey S. B., Mori M., Pfaller M. A., Woolson R. F., Wenzel R. P. 1988. Hospital-acquired candidemia. The attributable mortality and excess length of stay. Arch. Intern. Med. 148:2642–2645 [DOI] [PubMed] [Google Scholar]
- 51. Wilson R. B., Davis D., Mitchell A. P. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181:1868–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Winey M., et al. 1995. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J. Cell Biol. 129:1601–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.