Abstract
TORC1-dependent phosphorylation of Saccharomyces cerevisiae Sch9 was dramatically reduced upon exposure to a protonophore or in respiration-incompetent ρ0 cells but not in respiration-incompetent pet mutants, providing important insight into the molecular mechanisms governing interorganellar signaling in general and retrograde signaling in particular.
TEXT
Saccharomyces cerevisiae is a facultative anaerobic yeast which does not need respiration when a fermentable carbon source is supplied and, hence, is an instrumental model system to understand how cells respond to functional change of mitochondria. In S. cerevisiae, more than 300 nuclear genes are required for respiratory competence (5). Mutant strains lacking any of these nuclear genes are commonly called petite or pet mutants (5, 22). In addition, genes in the mitochondrial genome are also indispensable for respiratory competence. S. cerevisiae strains completely lacking mitochondrial genomes are denoted ρ0, while strains carrying a defective mitochondrial genome are ρ−; strains harboring an intact mitochondrial genome are denoted ρ+ (6). Phenotypically, pet, ρ0, and ρ− mutants all fail to grow on medium containing only nonfermentable carbon sources due to their respiratory incompetence.
The TOR (target of Rapamycin) protein kinase is a Ser/Thr kinase that is highly conserved from yeast to mammals (3, 4, 25). There are 2 distinctive TOR complexes (TORC1 and TORC2 in S. cerevisiae and mTORC1 and mTORC2 in mammals) (16). Like mitochondrial dysfunction, mTORC1 dysfunction has been implicated in a variety of pathologies (e.g., see references 3 and 25). Rapamycin-sensitive TORC1/mTORC1 positively regulates anabolic processes, including ribosome biogenesis and protein translation, while inhibiting stress responses and catabolic processes, such as autophagy. The best-characterized downstream targets of TORC1 and mTORC1 are the protein kinases A, G, and C family (AGC family) kinases Sch9 and S6K1/2, respectively (8, 23). Due to the lack of a specific small-molecule inhibitor, the functions of the rapamycin-insensitive TORC2/mTORC2 are relatively less well understood. Like TORC1, TORC2 and mTORC2 also phosphorylate AGC family kinases—Ypk1/2 in yeast and protein kinase B (PKB)/Akt and serum/glucocorticoid-regulated kinase (SGK) in metazoans (11, 12, 19). Phosphorylation of these AGC kinases serves as a useful proxy for monitoring TOR complex activities. Kim et al. have reported that the treatment of mammalian cells with drugs that compromise mitochondrial function (valinomycin or antimycin A) decreases mTORC1-dependent phosphorylation of S6K (13).
Given the impact of both mTORC1 signaling and mitochondrial function on cellular function and human health, we wished to further probe their molecular connections in the facultative anaerobe S. cerevisiae. To achieve this goal, wild-type (WT, [ρ+]) strains in TB50 and BY backgrounds, RL170-2b and BY4742, respectively, were treated with 25 μg/ml ethidium bromide to create ρ0 mutants (YGSK232 and -233) (see Table S1 in the supplemental material) (6). Both ρ0 mutants exhibited no growth on yeast extract-peptone-2% glycerol (YPG) (18) and slower growth on yeast extract-peptone-2% raffinose (YPR) than the WT (data not shown). The ρ0 phenotypes were confirmed by the growth of diploids produced from crosses with pet mutants (ρ+) (YGSK197 and -248) (see Table S1) on YPG but no growth of diploids produced from crosses with other ρ0 mutants (YGSK238 and -239) (see Table S1) on YPG (data not shown).
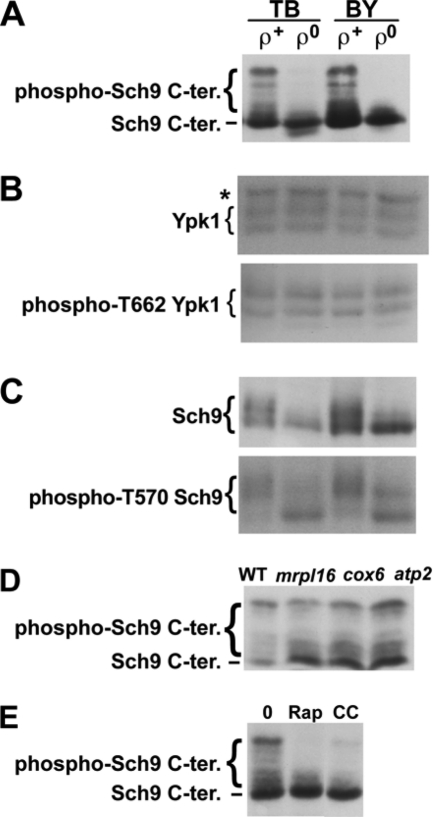
The TORC1-dependent phosphorylation sites (T723, S726, T737, S758, and S765) are located toward the C terminus of Sch9. To monitor the extent of TORC1-dependent Sch9 phosphorylation of these sites in ρ+ and ρ0 cells, cells carrying pJU676 (pRS416; URA3 SCH9-5HA) and pJU450 (pRS415; LEU2 TRP1 HIS3) (23) in TB50 and BY backgrounds (YGSK226, -242, -229, and -243) (see Table S1 in the supplemental material) were diluted to an optical density at 600 nm (OD600) of 0.4 in fresh SD+Asn medium (2% glucose, 10 mM Asn, 0.17% yeast nitrogen base without ammonium sulfate [pH 5.6] plus 50 μg/ml Lys in the case of BY4742 and otherwise with no other amino acids added) and further cultivated for approximately 5 h. Metabolic activity was arrested with the addition of trichloroacetic acid (to 6%), and proteins were extracted and cleaved by treatment with 2-nitro-5-thiocyanatobenzoic acid (NTCB) (23). After SDS-PAGE and electroblotting, the C terminus of Sch9-HA was detected with antihemagglutinin (anti-HA) antibody 12CA5 (23). As shown in Fig. 1A, Sch9 was severely dephosphorylated in ρ0 mutants in both genetic backgrounds. SCH9 2D3E encodes a constitutively active Sch9 in which the TORC1-phosphorylated residues have been replaced with Asp and Glu (23). The slow-growth phenotype of ρ0 cells was not improved by the expression of SCH9 2D3E (data not shown), indicating that the slow-growth phenotype of ρ0 cells is not solely caused by decreased Sch9 phosphorylation. This result is not surprising given that the expression of SCH9 2D3E alone is not sufficient to suppress/bypass the essential functions of TORC1 (9).
Fig. 1.
Phosphorylation of a TORC1 and Pkh1/2-target (Sch9) and a TORC2 target (Ypk1). (A) TORC1-dependent Sch9 phosphorylation in ρ+ and ρ0 cells carrying pJU676 and pJU450 in TB50 (TB [strain RL170-2b]) and BY4742 (BY) backgrounds. Prior to SDS-PAGE/anti-HA blot, protein extracts were treated with NTCB to cleave proteins. (B) Total Ypk1 (upper panel, anti-Ypk1 blot) and phospho-T662 Ypk1 (lower panel, anti-phospho-T659 Ypk2 blot) in uncleaved protein extracts derived from the cells shown in panel A. The band marked with an asterisk (*) represents a non-Ypk1 protein that cross-reacts with the anti-Ypk1 antibody. (C) Total Sch9-HA (upper panel, anti-HA blot) and phospho-T570 Sch9-HA (lower panel, anti-phospho-T570 Sch9 blot) analyzed in uncleaved extracts as described for panel B. Phosphorylation-dependent migration shifts are clearly visible in the upper panel, but these shifts are not due to changes in T570 phosphorylation status (lower panel). (D and E) TORC1-dependent Sch9 phosphorylation, as determined by anti-HA Western blotting of NTCB-treated protein extracts obtained from WT and pet cells (D) and ρ+ cells (YGSK226) (E) after treatment with 200 ng/ml rapamycin (Rap) or 4 μg/ml CCCP (CC) or drug vehicle (0). All treatments were for 70 min. Stocks (1,000×, 200 μg/ml rapamycin and 4 mg/ml CCCP) were prepared in ethanol-Tween 20 (9:1).
To try to see if the Sch9 dephosphorylation observed in ρ0 cells depends on TORC1, we introduced presumably hyperactive TOR1 (TOR1 I1954V) or caffeine-resistant TOR1 (TOR1 I1954V W2176R) alleles (17) on pRS315 into the ρ0 cells in BY4742 backgrounds. Although it has been unclear whether Tor1 I1954V or Tor1 I1954V W2176R gives higher kinase activity toward Sch9 in vivo or in vitro, Tor1 I1954V shows enhanced in vitro kinase activity toward the nonphysiological substrate PHAS-1 (phosphorylated heat- and acid-stable protein 1), and the expression of either allele confers caffeine but not rapamycin resistance (17). However, the introduction of these TOR1 alleles failed to restore Sch9 phosphorylation in ρ0 cells (data not shown). Thus, it is unclear whether the mitochondrial dysfunction signal to Sch9 is transduced via TORC1 or an as-yet-undescribed Sch9 phosphatase.
TORC2 phosphorylates T662 in Ypk1 (12; also Loewith laboratory, unpublished data). Sch9 is also the target of Pkh1/2 (23). Pkh1/2-dependent phosphorylation of T570 in Sch9, like TORC1-dependent phosphorylation, is required for the protein kinase activity of Sch9 (23). To observe these phosphorylation events, proteins were extracted as described above without NTCB treatment. After electroblotting, Ypk1 was detected with anti-Ypk1 antibody (Santa Cruz Biotechnology) or antibody to phospho-T659 Ypk2 (T659 is the TORC2-dependent site), which can cross-react with phospho-T662 Ypk1 (T662 is the TORC2-dependent site) (12; also Loewith laboratory, unpublished). Uncleaved Sch9-HA was also detected with anti-HA antibody 12CA5 or antibody to phospho-T570 Sch9 (T570 is the Pkh1/2-dependent site) (23). As shown in Fig. 1B and C, the phosphorylation of T662 in Ypk1, as well as the magnitude of phosphorylation of T570 in Sch9, were unchanged between ρ+ and ρ0 cells, indicating that mitochondrial genomic dysfunction has no effect on TORC2 signaling or Pkh1/2-dependent Sch9 phosphorylation.
MRPL16, COX6, and ATP2 encode a mitochondrial ribosomal protein, a subunit of cytochrome c oxidase (complex IV), and the β subunit of the F1-ATP synthase complex, respectively (http://www.yeastgenome.org/). The pet mutants (mrpl16Δ, cox6Δ, and atp2Δ in a BY4741 background) showed no growth on YPG (data not shown). Their ρ+ genotypes were confirmed by the growth of diploids produced from crossing with a ρ0 mutant (YGSK233) (see Table S1 in the supplemental material) on YPG (data not shown). To monitor the extent of Sch9 phosphorylation in these pet mutants, cells carrying pJU676 and pJU450 (YGSK247, -248, and -249) were grown as described above (with the addition of 20 μg/ml Met instead of Lys), and proteins were extracted, treated with NTCB, and detected by anti-HA antibody 12CA5 as described above. Despite their respiratory incompetence, the pet mutants exhibited nearly normal Sch9 phosphorylation (Fig. 1D).
In addition to Sch9 phosphorylation, we also monitored glycogen accumulation as an independent readout of TORC1 activity (1, 23). Consistent with our observations with Sch9 phosphorylation, an accumulation of glycogen was observed in ρ0 cells, similar to ρ+ cells treated with rapamycin, but not in ρ+ cells or pet ρ+ cells (see Fig. S1A in the supplemental material). Finally, we examined the effect of the mitochondrial drug (protonophore) carbonyl cyanide 3-chlorophenylhydrazone (CCCP) on Sch9 phosphorylation. As shown in Fig. 1E, similar to rapamycin treatment, Sch9 was dephosphorylated by treatment with CCCP.
In conclusion, we revealed that mitochondrial dysfunction caused by mitochondrial genomic defects or by treatment with a protonophore, but not respiratory incompetence caused in pet mutants, specifically causes dephosphorylation of Sch9, a main target of TORC1; TORC2 and Pkh1/2 activities were not similarly affected. Our results confirm and extend those of a previous report that treatment of mammalian cells with mitochondrial drugs decreases mTORC1 signaling (13). Indeed, our observation that ρ0 mutants and pet mutants differentially affect the phosphorylation state of Sch9 could provide important insight into the molecular mechanisms governing interorganellar signaling in general and mitochondrion-to-nucleus (retrograde) signaling in particular (15). For instance, yeast TORC1 is known to negatively regulate the well-characterized Rtg1/Rtg3 pathway, which is highly induced in ρ0 cells (15). In light of our results, it seems likely that the induction of this pathway in response to mitochondrial stress is due, at least in part, to reduced TORC1 activity.
Microarray analyses performed by Traven et al. demonstrated that, in ρ0 cells, there is an upregulation of Msn2/4-targeted genes, genes encoding mitochondrial biogenesis factors, and genes encoding mitochondrial proteins, such as mitochondrial ribosomal proteins, as well as a downregulation of ribosomal genes (21). Sch9 antagonizes Msn2/4 function and promotes the expression of genes encoding ribosomal proteins (23). Moreover, transcription of the genes encoding proteins important for mitochondrial function and mitochondrial ribosomal proteins is induced in sch9Δ cells (14). Taken together, the behaviors of the genes observed in microarray analysis in ρ0 cells (21) could be attributed to the decreased Sch9 phosphorylation in ρ0 cells (Fig. 1A).
How do mitochondrial defects impinge upon Sch9? Given that the energy-responsive AMP-activated protein kinase (AMPK) inhibits mTORC1 via phosphorylation of S722 and S792 of raptor, a component of mTORC1, in mammalian cells (7), we hypothesized that the yeast AMPK ortholog, Snf1, might be similarly involved in coupling mitochondrial dysfunction (via a potential decrease in energy levels) to TORC1 through phosphorylation of S959 of Kog1. Kog1 is the yeast raptor homolog found in TORC1 and S959 resides in a sequence context similar to that of S792 of raptor; S722 of human raptor is not obviously conserved in yeast Kog1 (7). However, this appears not to be the case, as we observed that Sch9 phosphorylation was normal in yeast kog1Δ strains expressing Kog1 S959A, -D, or -E and in strains where SNF1 is deleted (data not shown). Moreover, the possibility that mitochondrial dysfunction is conveyed to TORC1 by an amino acid availability signal via its known upstream regulator the EGO complex (2) seems unlikely, since Sch9 phosphorylation is still much lower in gtr1Δ ρ0 than in gtr1Δ ρ+ cells that both express hyperactive GTP-locked Gtr1 (GTR1 Q65L) (see Fig. S1B in the supplemental material).
As mitochondrial dysfunction, caused by partial deletion or other mutation in mitochondrial genomic DNA, is related to various human diseases, such as Kearns-Sayre syndrome and chronic progressive external ophthalmoplegia (CPEO) (20, 24), it will be interesting to determine whether this is the consequence of suboptimal mTORC1 function. If this is the case, then small-molecule agonists of mTORC1 function (e.g., LiCl) (10) may be indicated to treat such patients.
Supplementary Material
Acknowledgments
We thank the Kyoto University Foundation for financial support to S.K. and the Swiss National Science Foundation and the Canton of Geneva for financial support to R.L.
We also acknowledge all members of our laboratories for their support.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 12 August 2011.
REFERENCES
- 1. Barbet N. C., et al. 1996. TOR controls translation initiation and early G1 progression in yeast. Mol. Biol. Cell 7:25–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Binda M., et al. 2009. The Vam6 GEF controls TORC1 by activating the EGO complex. Mol. Cell 35:563–573 [DOI] [PubMed] [Google Scholar]
- 3. Bjornsti M. A., Houghton P. J. 2004. The TOR pathway: a target for cancer therapy. Nat. Rev. Cancer 4:335–348 [DOI] [PubMed] [Google Scholar]
- 4. De Virgilio C., Loewith R. 2006. Cell growth control: little eukaryotes make big contributions. Oncogene 25:6392–6415 [DOI] [PubMed] [Google Scholar]
- 5. Dimmer K. S., et al. 2002. Genetic basis of mitochondrial function and morphology in Saccharomyces cerevisiae. Mol. Biol. Cell 13:847–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fox T. D., et al. 1991. Analysis and manipulation of yeast mitochondrial genes. Methods Enzymol. 194:149–165 [DOI] [PubMed] [Google Scholar]
- 7. Gwinn D. M., et al. 2008. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell 30:214–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hay N., Sonenberg N. 2004. Upstream and downstream of mTOR. Genes Dev. 18:1926–1945 [DOI] [PubMed] [Google Scholar]
- 9. Huber A., et al. 2009. Characterization of the rapamycin-sensitive phosphoproteome reveals that Sch9 is a central coordinator of protein synthesis. Genes Dev. 23:1929–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Inoki K., et al. 2006. TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell 126:955–968 [DOI] [PubMed] [Google Scholar]
- 11. Jones K. T., Greer E. R., Pearce D., Ashrafi K. 2009. Rictor/TORC2 regulates Caenorhabditis elegans fat storage, body size, and development through sgk-1. PLoS Biol. 7:e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kamada Y., et al. 2005. Tor2 directly phosphorylates the AGC kinase Ypk2 to regulate actin polarization. Mol. Cell. Biol. 25:7239–7248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kim D. H., et al. 2002. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110:163–175 [DOI] [PubMed] [Google Scholar]
- 14. Lavoie H., Whiteway M. 2008. Increased respiration in the sch9Δ mutant is required for increasing chronological life span but not replicative life span. Eukaryot. Cell 7:1127–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu Z., Butow R. A. 2006. Mitochondrial retrograde signaling. Annu. Rev. Genet. 40:159–185 [DOI] [PubMed] [Google Scholar]
- 16. Loewith R., et al. 2002. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 10:457–468 [DOI] [PubMed] [Google Scholar]
- 17. Reinke A., Chen J. C., Aronova S., Powers T. 2006. Caffeine targets TOR complex I and provides evidence for a regulatory link between the FRB and kinase domains of Tor1p. J. Biol. Chem. 281:31616–31626 [DOI] [PubMed] [Google Scholar]
- 18. Sherman F. 1991. Getting started with yeast. Methods Enzymol. 194:3–21 [DOI] [PubMed] [Google Scholar]
- 19. Soukas A. A., Kane E. A., Carr C. E., Melo J. A., Ruvkun G. 2009. Rictor/TORC2 regulates fat metabolism, feeding, growth, and life span in Caenorhabditis elegans. Genes Dev. 23:496–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Taylor R. W., Turnbull D. M. 2005. Mitochondrial DNA mutations in human disease. Nat. Rev. Genet. 6:389–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Traven A., Wong J. M., Xu D., Sopta M., Ingles C. J. 2001. Interorganellar communication. Altered nuclear gene expression profiles in a yeast mitochondrial DNA mutant. J. Biol. Chem. 276:4020–4027 [DOI] [PubMed] [Google Scholar]
- 22. Tzagoloff A., Dieckmann C. L. 1990. PET genes of Saccharomyces cerevisiae. Microbiol. Rev. 54:211–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Urban J., et al. 2007. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol. Cell 26:663–674 [DOI] [PubMed] [Google Scholar]
- 24. Wallace D. C. 2005. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu. Rev. Genet. 39:359–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wullschleger S., Loewith R., Hall M. N. 2006. TOR signaling in growth and metabolism. Cell 124:471–484 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.