Abstract
The Saccharomyces cerevisiae RNA-binding protein Khd1/Hek2 associates with hundreds of potential mRNA targets preferentially, including the mRNAs encoding proteins localized to the cell wall and plasma membrane. We have previously revealed that Khd1 positively regulates expression of MTL1 mRNA encoding a membrane sensor in the cell wall integrity (CWI) pathway. However, a khd1Δ mutation has no detectable phenotype on cell wall synthesis. Here we show that the khd1Δ mutation causes a severe cell lysis when combined with the deletion of the CCR4 gene encoding a cytoplasmic deadenylase. We identified the ROM2 mRNA, encoding a guanine nucleotide exchange factor (GEF) for Rho1, as a target for Khd1 and Ccr4. The ROM2 mRNA level was decreased in the khd1Δ ccr4Δ mutant, and ROM2 overexpression suppressed the cell lysis of the khd1Δ ccr4Δ mutant. We also found that Ccr4 negatively regulates expression of the LRG1 mRNA encoding a GTPase-activating protein (GAP) for Rho1. The LRG1 mRNA level was increased in the ccr4Δ and khd1Δ ccr4Δ mutants, and deletion of LRG1 suppressed the cell lysis of the khd1Δ ccr4Δ mutant. Our results presented here suggest that Khd1 and Ccr4 modulate a signal from Rho1 in the CWI pathway by regulating the expression of RhoGEF and RhoGAP.
INTRODUCTION
Regulation of gene expression is achieved not only at the transcriptional level but also at the posttranscriptional level. In the nucleus, the primary transcript is processed by the addition of 5′-cap structure, the removal of introns, and the addition of a polyadenosine [poly(A)] tail. Upon export to the cytoplasm, the mature mRNA is subject to quality control and occasionally localized to particular subcellular compartments for translation. Finally, mRNAs are degraded in processing bodies (P-bodies), a process that must also be tightly controlled. The integration of all these regulatory steps ensures that a given mRNA produces the proper proteins at the right time and place. RNA-binding proteins play important roles in posttranscriptional regulation of gene expression (14, 16, 19, 23, 41). Eukaryotic cells encode a large number of RNA-binding proteins, each of which has unique RNA-binding activity and protein-protein interaction characteristics.
A Saccharomyces cerevisiae RNA-binding protein, K homology (KH) domain protein 1 (Khd1), also known as a heterogeneous nuclear RNP K-like gene (Hek2), contains three K homology (KH) RNA-binding domains (here after we use Khd1) and is most related to mammalian heterogeneous nuclear ribonucleoprotein K (hnRNP K) and poly(C)-binding proteins (PCBP1 to -4) (5, 10, 21, 28). Khd1 is required for efficient localization of ASH1 mRNA to the bud tip in dividing yeast by possibly acting as a repressor of translation during mRNA transport (21, 35). ASH1 encodes a transcriptional repressor for HO endonuclease and thus prevents mating type switching in daughter cells (4, 40). Khd1 is also involved in the regulation of the telomeric position effect and telomere length (10, 11). More recently, Khd1 appears to regulate asymmetric expression of FLO11 to determine daughter cell fate during filamentous growth (46). We have previously identified target mRNAs for Khd1 on a genome-wide level and found that Khd1 associates with hundreds of mRNAs, comprising almost 20% of the yeast's transcriptome (17). A significant fraction of the potential Khd1 mRNA targets encode proteins localized to the cell periphery, such as the cell wall and plasma membrane, and also nuclear proteins involved in transcriptional regulation. Despite these many mRNA targets, the khd1Δ mutant has no drastic phenotype in rich media except for the partial delocalization of ASH1 mRNA and protein, resulting in slightly reduced expression of HO and the reduced expression of Mtl1, a membrane sensor in the cell wall integrity (CWI) signaling pathway (17, 21, 35). Thus, physiological functions of Khd1 in the CWI pathway and other processes still remain unclear.
The cell wall of the budding yeast Saccharomyces cerevisiae is required to maintain the cell shape and integrity (24). The cell must remodel this rigid structure during vegetative growth and during pheromone-induced morphogenesis. Cell wall remodeling is monitored and regulated by the CWI signaling pathway, which activates the protein kinase C (Pkc1)-activated Mpk1/Slt2 mitogen-activated protein (MAP) kinase (MAPK) cascade and a glucan synthesis (26). In the CWI signaling pathway, signals are initiated at the plasma membrane through the cell surface sensors Wsc1, Wsc2, Wsc3, Mid2, and Mtl1. Together with phosphatidylinositol-4,5-bisphosphate (PI4,5P2), which recruits the Rom1/2 guanine nucleotide exchange factors (GEFs) to the plasma membrane, the sensors stimulate nucleotide exchange on a small G-protein Rho1. Rho1 activates five effectors, including the Pkc1-MAPK cascade, the β1,3-glucan synthase, the Bni1 formin protein, the exocyst component Sec3, and the Skn7 transcription factor. The MAP kinase cascade, which is comprised of Bck1, Mkk1/2, and Mpk1, is activated by Pkc1. Two transcription factors, Rlm1 and the SBF complex (Swi4/Swi6), are targets of the MAP kinase. Loss of function of any protein kinase downstream of Pkc1 (or both Mkk1 and Mkk2) results in cell lysis at elevated growth temperature. The growth defects of these mutants are osmoremedial (e.g., with 1 M sorbitol), consistent with a primary defect in cell wall biogenesis. Loss of PKC1 results in osmoremedial cell lysis at all growth temperatures. Unlike loss of Pkc1, loss of Rho1 or loss of both of the redundant catalytic subunits of glucan synthase, Fks1 and Fks2, does not result in osmoremedial cell lysis but is lethal, since cell wall synthesis is completely impaired in the rho1Δ and fks1Δ fks2Δ mutants.
In this study, we show that the khd1Δ mutation causes cell lysis when combined with deletion of the CCR4 gene encoding a cytoplasmic deadenylase (6). While the ccr4Δ single mutant showed a weak cell lysis as previously reported (18), the khd1Δ ccr4Δ double mutant showed more severe cell lysis. We then searched the target mRNAs for Khd1 and Ccr4 in the CWI pathway and found the ROM2 mRNA encoding the RhoGEF as one of the target mRNA. We also found that Ccr4 negatively regulates expression of the LRG1 mRNA encoding a GTPase-activating protein (GAP) for Rho1. Our results indicate that Khd1 and Ccr4 modulate a signal from Rho1 in the CWI pathway by regulating the expression of RhoGEF and RhoGAP.
MATERIALS AND METHODS
Strains and general methods.
Escherichia coli DH5a was used for DNA manipulations. Strains used in this study are described in Table 1. Standard procedures were followed for yeast manipulations (22). The media used in this study included rich medium, synthetic complete (SC) medium, and synthetic minimal (SD) medium (22). SC media lacking amino acids or other nutrients (e.g., SC-Ura corresponds to SC medium lacking uracil) were used to select transformants. Recombinant DNA procedures were carried out as described previously (37).
Table 1.
Strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| 10B | MATα ade2 trp1 can1 leu2 his3 ura3 GAL psi+HOp-ADE2-HO 3′UTR | 21 |
| 10BD | MATa/MATα ade2/ade2 trp1/trp1 can1/can1 leu2/leu2 his3/his3 ura3/ura3 | 21 |
| 10BD-c163 | MATa/MATα ade2/ade2 trp1/trp1 can1/can1 leu2/leu2 his3/his3 ura3/ura3 KHD1/khd1Δ::CgHIS3 CCR4/ccr4Δ::CgLEU2 | This study |
| 10BD-p163 | MATa/MATα ade2/ade2 trp1/trp1 can1/can1 leu2/leu2 his3/his3 ura3/ura3 KHD1/khd1Δ::CgHIS3 CAF1/caf1Δ::CgLEU2 | This study |
| 10BD-cp163 | MATa/MATα ade2/ade2 trp1/trp1 can1/can1 leu2/leu2 his3/his3 ura3/ura3 KHD1/khd1Δ::CgTRP1 CCR4/ccr4Δ::CgLEU2 CAF1/caf1Δ::CgHIS3 | This study |
| 10BD-163-n3 | MATa/MATα ade2/ade2 trp1/trp1 can1/can1 leu2/leu2 his3/his3 ura3/ura3 KHD1/khd1Δ::CgHIS3 NOT3/not3Δ::CgLEU2 | This study |
| 10BD-163-n4 | MATa/MATα ade2/ade2 trp1/trp1 can1/can1 leu2/leu2 his3/his3 ura3/ura3 KHD1/khd1Δ::CgHIS3 NOT4/not4Δ::CgLEU2 | This study |
| 10BD-163-c40 | MATa/MATα ade2/ade2 trp1/trp1 can1/can1 leu2/leu2 his3/his3 ura3/ura3 KHD1/khd1Δ::CgHIS3 CAF40/caf40Δ::CgLEU2 | This study |
| 10BD-163-c130 | MATa/MATα ade2/ade2 trp1/trp1 can1/can1 leu2/leu2 his3/his3 ura3/ura3 KHD1/khd1Δ::CgHIS3 CAF130/caf130Δ::CgLEU2 | This study |
| 10BD-163-p2 | MATa/MATα ade2/ade2 trp1/trp1 can1/can1 leu2/leu2 his3/his3 ura3/ura3 KHD1/khd1Δ::CgHIS3 PAN2/pan2Δ::CgLEU2 | This study |
| 10BD-163-p3 | MATa/MATα ade2/ade2 trp1/trp1 can1/can1 leu2/leu2 his3/his3 ura3/ura3 KHD1/khd1Δ::CgHIS3 PAN3/pan3Δ::CgLEU2 | This study |
| 10BD-c163-r1 | MATa/MATα ade2/ade2 trp1/trp1 can1/can1 leu2/leu2 his3/his3 ura3/ura3 KHD1/khd1Δ::CgTRP1 CCR4/ccr4Δ::CgLEU2 ROM1/rom1Δ::CgHIS3 | This study |
| 10BD-c163-r2 | MATa/MATα ade2/ade2 trp1/trp1 can1/can1 leu2/leu2 his3/his3 ura3/ura3 KHD1/khd1Δ::CgTRP1 CCR4/ccr4Δ::CgLEU2 ROM2/rom2Δ::CgHIS3 | This study |
| 10BD-c163-l1 | MATa/MATα ade2/ade2 trp1/trp1 can1/can1 leu2/leu2 his3/his3 ura3/ura3 KHD1/khd1Δ::CgTRP1 CCR4/ccr4Δ::CgLEU2 LRG1/lrg1Δ::CgHIS3 | This study |
| c1H-1A | MATα ade2 trp1 can1 leu2 his3 ura3 | This study |
| c1H-1B | MATα ade2 trp1 can1 leu2 his3 ura3 khd1Δ::CgHIS3 ccr4Δ::CgLEU2 | This study |
| c1H-1C | MATa ade2 trp1 can1 leu2 his3 ura3 khd1Δ::CgHIS3 | This study |
| c1H-1D | MATa ade2 trp1 can1 leu2 his3 ura3 ccr4Δ::CgLEU2 | This study |
| p1H-2A | MATα ade2 trp1 can1 leu2 his3 ura3 | This study |
| p1H-2B | MATα ade2 trp1 can1 leu2 his3 ura3 caf1Δ::CgLEU2 | This study |
| p1H-2C | MATa ade2 trp1 can1 leu2 his3 ura3 khd1Δ::CgHIS3 caf1Δ::CgLEU2 | This study |
| p1H-2D | MATa ade2 trp1 can1 leu2 his3 ura3 khd1Δ::CgHIS3 | This study |
| cp3-2C | MATa ade2 trp1 can1 leu2 his3 ura3 ccr4Δ::CgLEU2 caf1Δ::CgHIS3 | This study |
| cp3-5A | MATa ade2 trp1 can1 leu2 his3 ura3 khd1Δ::CgTRP1 ccr4Δ::CgLEU2 caf1Δ::CgHIS3 | This study |
| cl4-1B | MATα ade2 trp1 can1 leu2 his3 ura3 khd1Δ::CgTRP1 ccr4Δ::CgLEU2 lrg1Δ::CgHIS3 | This study |
| KY803 | MATa gcn4-Δ1 leu2 ura3 trp1 | 7 |
| KY803-36 | MATa gcn4-Δ1 leu2 ura3 trp1 not2-1 | 7 |
| KY803-39 | MATa gcn4-Δ1 leu2 ura3 trp1 not1-2 | 7 |
| KY803k | MATa gcn4-Δ1 leu2 ura3 trp1 khd1Δ::CgLEU2 | This study |
| KY803-36k | MATa gcn4-Δ1 leu2 ura3 trp1 not2-1 khd1Δ::CgLEU2 | This study |
| KY803-39k | MATa gcn4-Δ1 leu2 ura3 trp1 not1-2 khd1Δ::CgLEU2 | This study |
Plasmids.
Plasmids used in this study are described in Table 2. Plasmids YCplac33-KHD1FLAG and YCplac-KHD1FLAG-L284N were used for the immunoprecipitation. Plasmid YCplac-RHO1-Q68L expressing the RHO1(Q68L) allele was kindly provided by Y. Ohya and S. Nogami (39). YCplac33-PKC1-R398P expresses the PKC1(R398P) allele (32). Plasmids pCgLEU2, pCgHIS3, and pCgTRP1 are pUC19 carrying the Candida glabrata LEU2, HIS3, and TRP1 genes, respectively (36).
Table 2.
Plasmids used in this study
| Plasmid | Relevant markers | Source or reference |
|---|---|---|
| YCplac33 | URA3 CEN-ARS | 13 |
| YCplac33-KHD1FLAG | URA3 CEN-ARS KHD1-FLAG | This study |
| YCplac33-KHD1FLAG-L284N | URA3 CEN-ARS KHD(L284N)-FLAG | This study |
| YEplac195 | URA 2μm | 13 |
| YEplac195-ROM2 | URA3 2μm ROM2 | This study |
| YEplac195-WSC2 | URA3 2μm WSC2 | This study |
| YCplac33-RHO1-Q68L | URA3 CEN-ARS RHO1(Q68L) | 39 |
| pRS316-PKC1-R398P | URA3 CEN-ARS PKC1(R398P) | 32 |
| pCgLEU2 | C. glabrata LEU2 carried by pUC19 | 36 |
| pCgHIS3 | C. glabrata HIS3 carried by pUC19 | 36 |
| pCgTRP1 | C. glabrata TRP1 carried by pUC19 | 36 |
| pFA6a-13myc-kanMX6 | 13myc-ADH 3′UTR-kanMX6 | 27 |
Gene deletion and protein tagging.
Deletions of KHD1, CCR4, POP2, NOT3, NOT4, NOT5, CAF40, CAF130, PAN2, PAN3, ROM1, ROM2, and LRG1 were constructed by a PCR-based gene deletion method (2, 36, 38, 43). Primer sets were designed such that 46 bases at the 5′ ends of the primers were complementary to those at the corresponding region of the target gene and 20 bases at their 3′ ends were complementary to the pUC19 sequence outside the polylinker region in the plasmid pCgLEU2, pCgHIS3, or pCgTRP1. Primer sets for PCR were designed to delete the open reading frame (ORF) completely. The PCR products were transformed into the wild-type strain and selected for Leu+, His+, or Trp+.
Determination of cell lysis.
Aniline blue staining was done as previously described (39). Cell lysis was determined for aliquots of cell cultures as previously described (25) using propidium iodide staining. A minimum of 200 cells were counted for each sample.
Northern blot analysis.
Total RNA was prepared from cells using ISOGEN reagent (Nippon Gene) and the RNeasy minikit (Qiagen). RNA samples were separated by 1.5% denatured agarose gel electrophoresis and transferred to a nylon membrane. Then, RNA was hybridized using a digoxigenin (DIG)-labeled antisense probe. The primer set j298 (TGACGATATGATGAGCTCCTCCTTACGTCA) and j297 (TTAACCCCAGAAATCTAACGACG) and the primer set j259 (ATGATTCAAAATTCTGCTGGTTA) and j260 (GCCAATATTTATGAATTCCATAAC) were used to detect transcript containing ROM2 and LRG1, respectively. After washing and blocking, the membrane was incubated with alkaline phosphatase-conjugated anti-DIG antibody, and the signal was detected by enhanced chemiluminescence.
Screening for multicopy suppressors of the growth defect of the khd1Δ ccr4Δ mutant.
The khd1Δ ccr4Δ strain (c1H-1B) was transformed with a genomic library carried in YEp24, a multicopy vector marked with URA3. Transformants were plated on SC-Ura plates containing 10% sorbitol and incubated at room temperature for 4 days. The plates were replica plated to yeast extract-peptone-dextrose (YPD) plates and continuously incubated at 37°C for 3 days. Eight transformants that formed colonies at 37°C were identified. The corresponding plasmids were isolated from the transformants, and those that conferred the ability to proliferate at 37°C to the khd1Δ ccr4Δ strain were identified by retransformation. Sequencing of the insert DNAs of the eight recovered plasmids revealed that four contained the CCR4 gene, two contained the SSD1 gene, one contained the ROM2 gene, and one contained the WSC2 gene. Regional analysis of the suppression ability confirmed that the CCR4, SSD1, ROM2, and WSC2 genes were responsible to ensure the growth of the khd1Δ ccr4Δ strain.
Immunoprecipitation of Khd1-FLAG and RT-PCR analysis.
Cells were grown in SC-Ura at 30°C to mid-log phase and harvested by centrifugation. The cells were washed twice in XT buffer (50 mM HEPES-KOH at pH 7.3, 20 mM potassium acetate, 2 mM EDTA, 0.1% Triton X-100, 5% glycerol) and resuspended in XT buffer containing protease inhibitors, phenylmethylsulfonyl fluoride (PMSF), aprotinin, and leupeptin. Glass beads were added, and the cells were broken by rigid vortexing at 4°C (4 times at 3,500 rpm for 30 s each). The supernatant was removed and centrifuged for 10 min at 5,000 × g. To immunoprecipitate FLAG-tagged Khd1 (Khd1-FLAG), 200 μl of extract was incubated with anti-FLAG antibody (M2) coupled to protein G Sepharose beads (20 μl; GE Healthcare) for 2 h at 4°C. Beads were washed three times with 400 μl XT buffer, and bound material was eluted with 50 μl elution buffer (0.1 μg/μl 3× FLAG peptide in XT buffer) for 10 min at 4°C. Western blots were performed using anti-FLAG antibody (M2). Reverse transcription (RT)-PCR analyses with RNAs isolated from total extracts and from Khd1-FLAG-immunoprecipitates were performed as previously described (21). The number of amplification cycles was adjusted to avoid reaching a plateau phase during PCR.
RESULTS
The khd1Δ mutation displays a synthetic growth defect with the ccr4Δ mutation.
Despite the many mRNA targets, the khd1Δ mutant has no drastic phenotype in rich media except for the partial delocalization of ASH1 mRNA and protein, resulting in slightly reduced expression of HO and reduced expression of Mtl1 (17, 21, 35). One possibility is that Khd1 shares its redundant function(s) with some other gene products. During the analysis of MTL1 mRNA stability control by Khd1, we found that the khd1Δ ccr4Δ mutant cells grow slower than each single mutant cell, and this phenotype is clear in the W303 background (data not shown). Ccr4 is a catalytic subunits of the major cytoplasmic deadenylase involved in mRNA poly(A) tail shortening, and is also a component of the Ccr4-Not complex (6). To clarify this result, we performed a standard genetic analysis of diploid strain 10BD-c163 that was heterozygous for khd1Δ and ccr4Δ alleles in the W303 background. Tetrad analysis revealed that the khd1Δ mutant cells grew similarly to wild-type cells, the ccr4Δ mutant cells grew slower than wild-type cells, and the khd1Δ ccr4Δ double mutant cells grew much more slowly than the ccr4Δ mutant cells (Fig. 1A). The khd1Δ ccr4Δ double mutant cells grew very slowly at 25°C but failed to grow at elevated temperatures of 37°C (data not shown). Thus, the khd1Δ mutation showed a synthetic growth defect with the ccr4Δ mutation. We then confirmed the synthetic growth defect of the khd1Δ ccr4Δ double mutant using the KHD1 and CCR4 genes expressed on the plasmid. The KHD1 and CCR4 genes expressed on the plasmid complemented the slow growth of the khd1Δ ccr4Δ double mutant (data not shown).
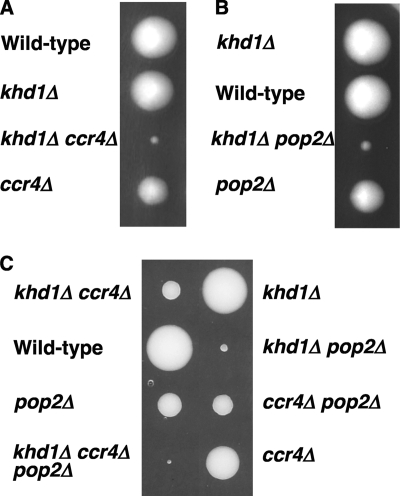
Fig. 1.
Growth of the khd1Δ ccr4Δ and khd1Δ pop2Δ mutant strains. (A) Strain 10BD-c163 that was heterozygous for the khd1Δ and ccr4Δ alleles was sporulated, and the tetrad was dissected onto a YPD plate. Growth after 4 days at 25°C is shown. Genotypes are indicated on the left side. More than 50 tetrads were dissected, and representative data are shown. (B) Strain 10BD-p163 that was heterozygous for the khd1Δ and pop2Δ alleles was sporulated, and the tetrad was dissected onto a YPD plate. Growth after 4 days at 25°C is shown. Genotypes are indicated on the left side. More than 50 tetrads were dissected, and representative data are shown. (C) Strain 10BD-cp163 that was heterozygous for the khd1Δ, ccr4Δ, and pop2Δ alleles was sporulated, and the tetrad was dissected onto YPD containing 10% sorbitol. Growth after 6 days at 25°C is shown. Genotypes are indicated on the both sides. More than 30 tetrads were dissected, and representative data are shown.
The khd1Δ ccr4Δ mutant shows severe cell lysis.
Previous studies demonstrated that the ccr4Δ mutant showed a temperature-sensitive cell lysis (18). Then, we next examined whether the khd1Δ ccr4Δ double mutants showed more severe cell lysis than the ccr4Δ single mutant. Microscopy analysis revealed that the khd1Δ ccr4Δ double mutant cells showed larger cell sizes and aberrant morphology, and some of the cells were lysed ghost cells (data not shown). Such aberrant morphology and lysed ghost cells were not observed in wild-type and khd1Δ single mutant cells. The ccr4Δ single mutant cells were a little larger than wild-type cells as previously reported (30), and few ccr4Δ cells showed lysed ghost, but the population was less than that of the khd1Δ ccr4Δ double mutant. The quantitative analysis of cell lysis using propidium iodide (see Materials and Methods) revealed that the khd1Δ ccr4Δ double mutants indeed showed severe cell lysis (Fig. 2). Aniline blue staining with 1,3-β-glucan also revealed that the khd1Δ ccr4Δ double mutant cells had abnormal deposition of the staining (data not shown). These results suggest that Khd1 together with Ccr4 has a role in the CWI signaling pathway.
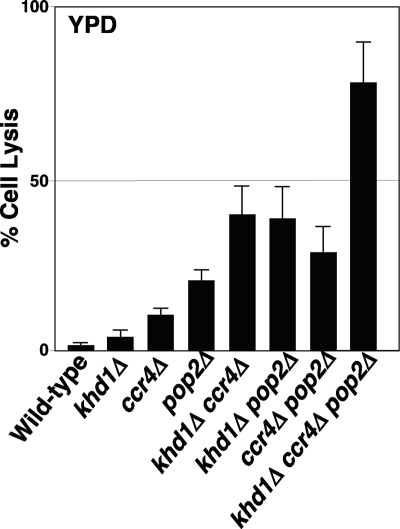
Fig. 2.
The khd1Δ ccr4Δ double mutant cells show severe cell lysis. Cell lysis assay in YPD medium. Wild-type (c1H-1A), khd1Δ (c1H-1C), ccr4Δ (c1H-1D), khd1Δ ccr4Δ (c1H-1B), pop2Δ (p1H-2B), khd1Δ ccr4Δ (p1H-2C), ccr4Δ pop2Δ (cp3-2C), and khd1Δ ccr4Δ pop2Δ (cp3-5A) cells were grown in YPD medium to mid-log phase at 25°C and shifted to 37°C for 4 h. Cell lysis was monitored by propidium iodide staining. The graph represents averages from three independent cultures for each strain. Error bars depict the standard deviations. At least 200 cells were counted per each strain.
The khd1Δ mutant displays a synthetic growth defect, with mutation in POP2 as well as CCR4, but not other components of the Ccr4-Not complex.
Ccr4 is one of the components in the Ccr4-Not complex involved in various cellular processes such as transcriptional control and mRNA degradation (6). Therefore, we then examined whether the khd1Δ mutation also showed a synthetic growth defect with mutations in other components of the Ccr4-Not complex, POP2, NOT1/CDC39, NOT2/CDC36, NOT3, NOT4, CAF40, and CAF130. Tetrad analysis revealed that the khd1Δ mutation also showed a synthetic growth defect with a deletion mutation in POP2 that encodes another catalytic subunit of cytoplasmic deadenylase (Fig. 1B). Similar to the khd1Δ ccr4Δ double mutant, the khd1Δ pop2Δ double mutants also showed a more marked cell lysis than the pop2Δ single mutant (Fig. 2). It should be noted that the khd1Δ pop2Δ double mutant grew slower than the khd1Δ ccr4Δ double mutant. The khd1Δ ccr4Δ pop2Δ triple mutants indeed showed slower growth and more severe cell lysis (Fig. 1C and 2), suggesting that Ccr4 and Pop2 may act independently. The khd1Δ mutation did not show a synthetic growth defect with a deletion mutation in the other components of the Ccr4-Not complex, NOT3, NOT4, CAF40, and CAF130 (data not shown). Since deletions of NOT1/CDC39, NOT2/CDC36, and NOT5 were lethal in our strain background, we examined the synthetic growth defect of the khd1Δ mutation with conditional not1-2 and not2-1 alleles (7). A conditional allele of NOT5 was not available in our hand. The cell growth of the khd1Δ not1-2 and khd1Δ not2-1 double mutant cells was similar to that of the not1-2 and not2-1 single mutants (data not shown), respectively, indicating that the khd1Δ mutation did not show a synthetic growth defect with these alleles. These results suggest that Khd1 shares its function on cell wall synthesis with Ccr4 and Pop2 but not other components of the Ccr4-Not complex.
Ccr4 and Pop2 are catalytic subunits of the major cytoplasmic deadenylase involved in mRNA degradation (8). Therefore, we next examined whether the khd1Δ mutation showed a synthetic growth defect with deletion mutations in PAN2 and PAN3, encoding other cytoplasmic deadenylases. The pan2Δ and pan3Δ single mutant cells showed normal growth, and the khd1Δ mutation did not affect their growth (data not shown). These results suggest that Khd1 shares its function in the CWI pathway with Ccr4 and Pop2 but not with Pan2 and Pan3.
Isolation of multicopy suppressors of the khd1Δ ccr4Δ double mutant.
Khd1 regulates the expression of Mtl1, one of the membrane sensors in the CWI pathway (17), implicating the possibility that the khd1Δ mutation enhances the cell lysis of the ccr4Δ mutant by reducing the upstream signal from Mtl1p. Then, we examined the growth of the mtl1Δ ccr4Δ double mutant and found that the mtl1Δ ccr4Δ double mutant grew similarly to the ccr4Δ single mutant (data not shown). Thus, only the reduced expression of Mtl1 by the khd1Δ mutation is not responsible for the severe cell lysis phenotype of the khd1Δ ccr4Δ mutant.
Khd1 and Ccr4 carry an RNA-binding protein and a cytoplasmic deadenylase, respectively, suggesting that Khd1 and Ccr4 function in the CWI pathway through regulating mRNA levels of some components that are required for the maintenance of the CWI pathway. To identify a possible target for Khd1 and Ccr4, we screened a genomic library for genes that, when overexpressed, suppressed the growth defect of the khd1Δ ccr4Δ double mutant. We identified ROM2 and WSC2 as multicopy suppressors (for details of the screening, see Materials and Methods). We also identified SSD1 in this screening, but we did not study this gene further here since the W303 strains carry a defective SSD1 allele. ROM2 encodes a GEF for Rho1 in the CWI pathway (33). WSC2 encodes a membrane sensor (26). As shown in Fig. 3A, overexpression of ROM2 and WSC2 suppressed the growth defect of the khd1Δ ccr4Δ double mutants. Since Rom2 and Wsc2 are upstream activators for Rho1 and Pkc1 in the CWI signaling pathway, we next examined whether the activated forms of RHO1 and PKC1 suppressed the growth defect of the khd1Δ ccr4Δ double mutant. Indeed, a constitutively active RHO1 allele, RHO1(Q68L), and a constitutively active PKC1 allele, PKC1(R398P), were able to suppress the growth defect of the khd1Δ ccr4Δ double mutant (Fig. 3B). These results, together with the observation that ROM2 and WSC2 act as the multicopy suppressors, suggest that the signaling upstream of Rho1 is impaired in the khd1Δ ccr4Δ double mutants.
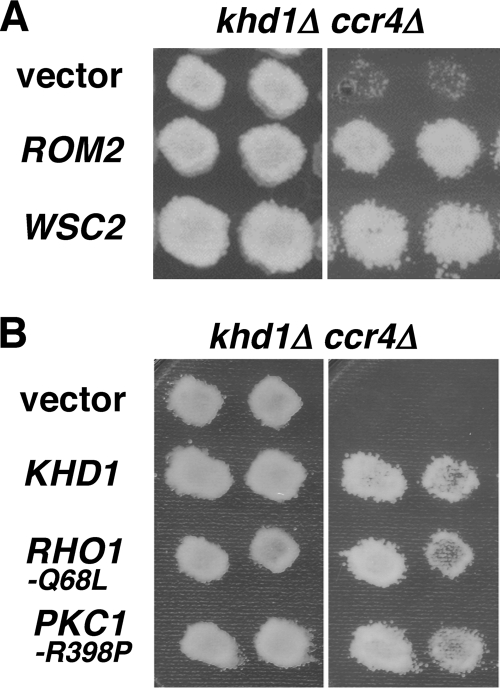
Fig. 3.
Overexpression of ROM2 and WSC2 suppresses the growth defect of khd1Δ ccr4Δ double mutants. (A) Multicopy suppressors of khd1Δ ccr4Δ. Transformants of the khd1Δ ccr4Δ strain (c1H-1B) carrying the indicated plasmids were streaked onto YPD medium and incubated at 25°C (left) or 37°C (right). Each patch represents an independent transformant. Plasmids used were YEplac195 (vector), YEplac195-ROM2 (ROM2), and YEplac195-WSC2 (WSC2). (B) Activated Rho1 and Pkc1 alleles in khd1Δ ccr4Δ. Transformants of the khd1Δ ccr4Δ strain (c1H-1B) carrying the indicated plasmids were streaked onto YPD medium and incubated at 25°C (left) or 37°C (right). Each patch represents an independent transformant. Plasmids used were YCplac33 (vector), YCplac-RHO1-Q68L, and YCplac33-PKC1-R398P.
ROM2 is a candidate target transcript linking Khd1 and Ccr4 in the CWI signaling pathway.
Suppression of the khd1Δ ccr4Δ mutants by ROM2 or WSC2 overexpression suggests the following two possibilities. One possibility is that these genes are the direct targets for Khd1 and Ccr4, and their expression is regulated by Khd1 and Ccr4. The other possibility is that these genes are not the direct targets for Khd1 and Ccr4, but ROM2 or WSC2 overexpression suppressed the cell lysis by activating Rho1 and Pkc1. Our previous genome-wide analysis of Khd1-associated mRNAs revealed that both ROM2 and WSC2 are potential target mRNAs for Khd1 (17), and we have confirmed that the WSC2 mRNA is associated with Khd1 in vivo (17). To ensure that Khd1 associates with the ROM2 mRNA, we then performed immunoprecipitation and RT-PCR analyses with the immunoprecipitates (Fig. 4). For this purpose, we utilized plasmids expressing FLAG-tagged Khd1 (Khd1-FLAG) and Khd1(L284N) mutant protein [Khd1(L284N)-FLAG] under the control of the endogenous promoter. A replacement of Leu-284 to Asn in the third KH domain of Khd1 reduced its RNA-binding activity (17). Khd1-FLAG and Khd1(L284N)-FLAG were immunoprecipitated with anti-FLAG antibodies, and the coprecipitated mRNAs were analyzed with RT-PCR. The ROM2 mRNA was clearly detected in RNA samples from the immunoprecipitated Khd1-FLAG, less detected in those from Khd1(L284N)-FLAG, but not detectable in untagged isolates (Fig. 4). This result suggests that Khd1 associates with the ROM2 mRNA and that the third KH domain is required for the efficient binding of Khd1 to the RNA.
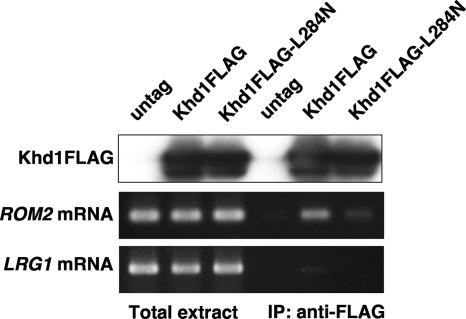
Fig. 4.
Khd1 associates with the ROM2 mRNA. Total extracts were prepared from the cells harboring the empty vector, the FLAG-tagged Khd1 plasmid, and the FLAG-tagged Khd1(L284N) plasmid. Khd1-FLAG and Khd1(L284N)-FLAG were immunoprecipitated (IP) with anti-FLAG antibodies. Samples were analyzed by immunoblot analysis with anti-FLAG to test for the presence of Khd1-FLAG proteins. The RNAs were isolated from the total extracts and from the immunoprecipitates. ROM2 and LRG1 mRNAs were detected by RT-PCR.
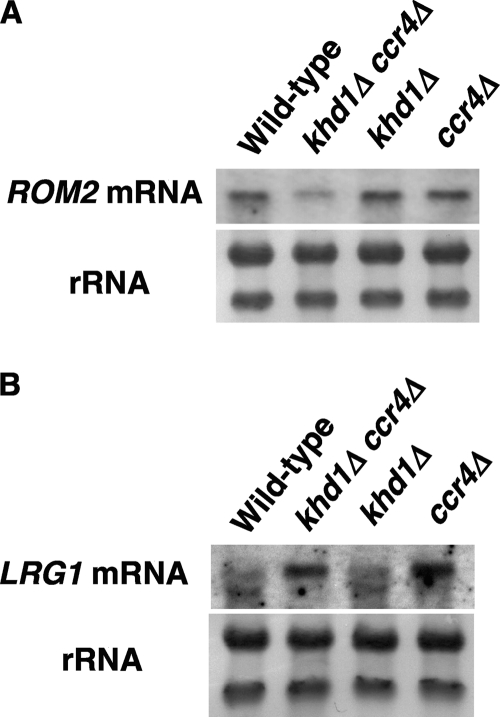
To determine whether Khd1 and Ccr4 affect the expression of ROM2 and WSC2, we next examined the expression levels of ROM2 and WSC2 mRNAs in wild-type, khd1Δ, ccr4Δ, and khd1Δ ccr4Δ mutant cells (Fig. 5A). The ROM2 mRNA level was not altered in the khd1Δ mutant compared to that in wild-type cells. In contrast, the ROM2 mRNA level was decreased in the ccr4Δ mutant and more decreased in khd1Δ ccr4Δ double mutant cells. In contrast, the WSC2 mRNA levels were not affected by the khd1Δ, ccr4Δ, or khd1Δ ccr4Δ mutations (data not shown). Together with the observations that ROM2 overexpression suppressed the growth defect of the khd1Δ ccr4Δ mutant and that the ROM2 mRNA is associated with Khd1, these results suggest that ROM2 mRNA is a target mRNA for Khd1 and Ccr4.
Fig. 5.
Khd1 and Ccr4 regulate the ROM2 mRNA level. (A) The ROM2 mRNA levels in wild-type, khd1Δ, ccr4Δ, and khd1Δ ccr4Δ cells. Wild-type (c1H-1A), khd1Δ (c1H-1C), ccr4Δ (c1H-1D), and khd1Δ ccr4Δ (c1H-1B) cells were cultured to mid-logarithmic phase in YPD medium at room temperature and collected, and total RNA was prepared. The ROM2 transcripts were quantified by Northern blotting, as described in Materials and Methods. rRNA was included as a quantity control. (B) The LRG1 mRNA levels in wild-type, khd1Δ, ccr4Δ, and khd1Δ ccr4Δ cells. Wild-type (c1H-1A), khd1Δ (c1H-1C), ccr4Δ (c1H-1D), and khd1Δ ccr4Δ (c1H-1B) cells were cultured to mid-logarithmic phase in YPD medium at room temperature and collected, and total RNA was prepared. The LRG1 transcripts were quantified by Northern blotting, as described in Materials and Methods. rRNA was included as a quantity control.
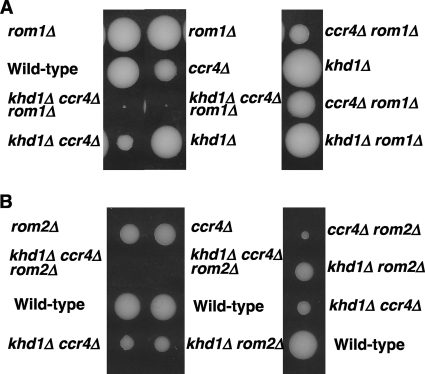
Rom2 and Rom1 comprise a redundant pair of GEFs for Rho1 (33). Loss of ROM2 function results in temperature-sensitive growth, whereas loss of both ROM2 and ROM1 is lethal. We found that the ROM1 mRNA level was not affected by the khd1Δ, ccr4Δ, or khd1Δ ccr4Δ mutations (data not shown). Therefore, if the ROM2 function were impaired in the khd1Δ ccr4Δ mutant, the khd1Δ ccr4Δ rom1Δ triple mutant would show a more severe growth defect than the khd1Δ ccr4Δ, khd1Δ rom1Δ, and ccr4Δ rom1Δ double mutants. Indeed, the khd1Δ ccr4Δ rom1Δ triple mutant cells showed much slower growth than the double mutant cells (Fig. 6A).
Fig. 6.
Growth of the khd1Δ ccr4Δ, khd1Δ ccr4Δ rom1Δ, and khd1Δ ccr4Δ rom2Δ mutant strains. (A) Strain 10BD-c163r1 that was heterozygous for the khd1Δ, ccr4Δ, and rom1Δ alleles was sporulated, and the tetrad was dissected onto YPD containing 10% sorbitol. Growth after 6 days at 25°C is shown. Genotypes are indicated on both sides. More than 20 tetrads were dissected, and representative data are shown. (B) Strain 10BD-c163r2 that was heterozygous for the khd1Δ, ccr4Δ, and rom2Δ alleles was sporulated, and the tetrad was dissected onto YPD containing 10% sorbitol. Growth after 4 days at 25°C is shown. Genotypes are indicated on both sides. More than 20 tetrads were dissected, and representative data are shown.
We then examined the cell growth of the khd1Δ ccr4Δ rom2Δ triple mutant cells. If ROM2 were a sole target mRNA for Khd1 and Ccr4, the khd1Δ ccr4Δ rom2Δ triple mutant cells would grow similarly to that of the rom2Δ single or khd1Δ ccr4Δ double mutant. Surprising, the khd1Δ ccr4Δ rom2Δ triple mutant cells were inviable (Fig. 6B). Unlike the rom1Δ mutant cells that grew similarly to wild-type cells, the rom2Δ mutant cells grew slower than the wild-type cells, and the ccr4Δ rom2Δ double mutant cells grew more slowly than each single mutant cell (Fig. 6B). The khd1Δ rom2Δ double mutant cells grew similarly to the rom2Δ single mutant cells. These additive effects of the khd1Δ, ccr4Δ, and rom2Δ mutations suggest that Khd1 and Ccr4 have additional target mRNAs that regulate the CWI signaling pathway.
LRG1 is another target transcript linking Ccr4 to the CWI pathway.
The activation of Rho1 is regulated by both GEFs and GAPs acting in opposition. Therefore, we next examined the possibility that Khd1 and Ccr4 regulated the levels of the mRNAs encoding RhoGAPs. Among the 11 RhoGAPs identified in S. cerevisiae, four GAPs, Bem2, Sac7, Bag7, and Lrg1, have been shown to act on Rho1 both in vitro and in vivo (26). We then examined the expression of RhoGAPs in wild-type, khd1Δ, ccr4Δ, and khd1Δ ccr4Δ mutant cells. We found that the LRG1 mRNA level in the ccr4Δ and khd1Δ ccr4Δ mutants was increased compared to that in the wild-type cells (Fig. 5B). On the other hand, the expression level of BEM2, SAC7, or BAG7 mRNAs was not altered significantly in the ccr4Δ and khd1Δ ccr4Δ mutants (data not shown). Consistent with the fact that LRG1 was not a potential target mRNA for Khd1 in our previous analysis (17), the LRG1 mRNA was not enriched in the RNA samples from the Khd1-FLAG immunoprecipitates (Fig. 4). Thus, Khd1 is unlikely to be involved in LRG1 expression.
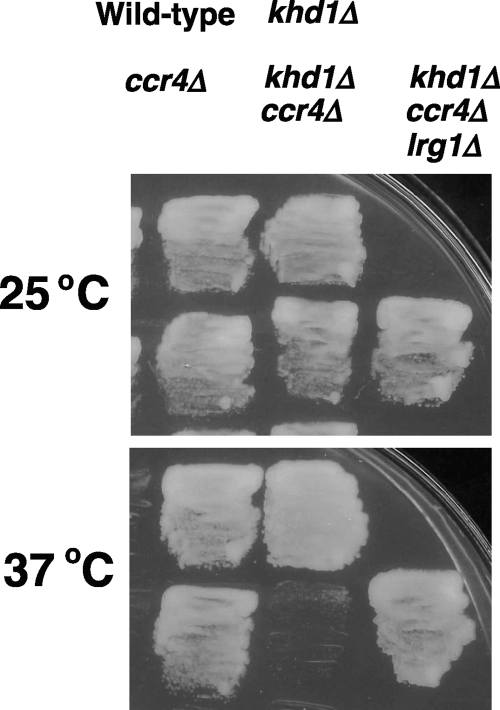
If the increased Lrg1 protein level had contributed to the cell lysis in the khd1Δ ccr4Δ mutant, a deletion of LRG1 should suppress the growth defect of the khd1Δ ccr4Δ mutants. To test this possibility, we constructed the khd1Δ ccr4Δ lrg1Δ triple mutant by a standard genetic manipulation. As shown in Fig. 7, a deletion of LRG1 efficiently suppressed the growth defect of the khd1Δ ccr4Δ mutant. These results suggest that the LRG1 mRNA is a target mRNA for Ccr4 and that the elevated level of LRG1 contributes to the cell lysis of the khd1Δ ccr4Δ mutant. Alternatively, the elevated level of LRG1 in the khd1Δ ccr4Δ mutant may be a consequence of the decreased ROM2 expression caused by the khd1Δ ccr4Δ double mutation. The lrg1Δ mutation that conferred to an increase of Rho1-GTP could compensate for the decreased CWI signaling caused by the khd1Δ ccr4Δ double mutation.
Fig. 7.
Loss of LRG1 suppresses the cell lysis of the khd1Δ ccr4Δ mutant. Wild-type (c1H-1A), khd1Δ (c1H-1C), ccr4Δ (c1H-1D), khd1Δ ccr4Δ (c1H-1B), and khd1Δ ccr4Δ lrg1Δ (cl4-1B) cells were plated on YPD medium plates and grown at either 25°C or 37°C for 3 days.
DISCUSSION
In this study, we first showed the role of Khd1 in the CWI signaling pathway. Whereas the khd1Δ single mutant cells showed no detectable phenotype on cell wall synthesis, the khd1Δ mutation showed a severe cell wall defect when combined with the ccr4Δ or pop2Δ mutation that is known to confer a weak cell lysis (18). Although it has been reported that mutations in several genes, such as RAD53 and SPT15, show a synthetic lethal or synthetic growth defect with the ccr4Δ and pop2Δ mutation (3, 34, 44), this is a first report showing that CCR4 and POP2 genetically interact with KHD1. The khd1Δ mutation did not show a synthetic growth defect with mutation in the other components of the Ccr4-Not complex. Genome-wide analyses of the roles of each subunit of the Ccr4-Not complex have revealed that the ccr4Δ and pop2Δ mutants show similar gene expression patterns that are different from those of other not deletions (1, 9). Thus, Khd1 shares a role in the expression of particular genes with Ccr4 and Pop2.
The CWI pathway consists of several membrane sensors, Wsc1, Wsc2, Wsc3, Mid2, and Mtl1 (26). Among these sensors, Wsc1 and Mid2 are reported to be the most important in the CWI signaling, and the wsc1Δ mid2Δ double mutant shows the cell lysis phenotype. Mtl1 has a minor role, and the mtl1Δ mutant has a normal response for the CWI signaling. Only the reduced expression of Mtl1 by the khd1Δ mutation is not responsible for the severe cell lysis phenotype of the khd1Δ ccr4Δ mutant, since the mtl1Δ ccr4Δ double mutant grew similarly to the ccr4Δ single mutant. On the other hand, overexpression of MTL1 as well as WSC2 suppressed the cell lysis of the khd1Δ ccr4Δ double mutant (data not shown), suggesting that overexpression of the sensors can activate the CWI signaling and overcome the cell lysis of the khd1Δ ccr4Δ double mutant presumably by Rho1 activation. The WSC1, WSC2, WSC3, MID2, and MTL1 mRNAs are all bound to Khd1 (17). However, only the levels of MTL1 mRNA and Mtl1 were regulated by Khd1. MTL1 mRNA has a cis-acting region containing CNN repeats that direct Khd1p binding, and the cis-acting region is responsible for Khd1-mediated control (31). The cis-acting region of MTL1 mRNA translates into an amino acid sequence containing stretches of serine residues. Although MID2 and WSC mRNAs also have CNN repeats and Khd1 indeed bound to the repeats, the khd1Δ mutation does not affect their expression (17). The synthetic growth defect between the khd1Δ and ccr4Δ mutations implicates the possibility that the levels of Mid2 and Wsc1 to -3 besides Mtl1 are regulated by Ccr4 or by both Ccr4 and Khd1. However, it is not the case, the Mid2 or Wsc2 protein level was not affected by the khd1Δ ccr4Δ double mutation (data not shown). Physiological relevance of the binding of Khd1 to MID2 or WSC mRNAs remains to be elucidated.
We show the ROM2 mRNA encoding RhoGEF, as one of the targets for Khd1 and Ccr4 in the CWI signaling pathway. ROM2 has been shown to be a potential target mRNA for Khd1 and contains three CNN repeats in the coding sequence (17). We then confirmed that Khd1 associates with the ROM2 mRNA in a manner dependent on the RNA-binding activity of Khd1. We also show that ROM2 overexpression suppresses the cell lysis of the khd1Δ ccr4Δ mutant. The ROM2 mRNA level was slightly decreased in the ccr4Δ mutant and further decreased in the khd1Δ ccr4Δ double mutant. Consistent with the decrease in ROM2 expression, the khd1Δ ccr4Δ double mutant cells showed a severe cell lysis defect probably due to the decreased Rho1 activity. Since Ccr4 is a deadenylase, previously known target mRNAs for Ccr4p such as WHI5 and CRT1 are negatively regulated by Ccr4 through the poly(A) shorting (30, 47). How Khd1 and Ccr4 positively regulate the expression of ROM2 should be elucidated. In case of the regulation of MTL1 mRNA stability by Khd1, MTL1 mRNA itself bears the multiple CNN repeats involved in destabilization by the decapping enzyme Dcp1/2 and the 5′-3′ exonuclease Xrn1, and Khd1 stabilizes MTL1 mRNA through binding to this element (31). Similarly, Khd1 may stabilize ROM2 mRNA through binding to the CNN repeats within the coding sequence of the ROM2 mRNA.
The ccr4Δ mutant shows pleiotropic phenotypes, including a weak cell lysis, a defect in checkpoint control, a defect in cell cycle progression, and abnormal morphology (18, 30, 44, 47). Previous studies have revealed the target mRNAs for Ccr4 that account for these phenotypes of the ccr4Δ mutant. For checkpoint control, the CRT1 mRNA was shown to be negatively regulated by Ccr4 (47). Deletion of CRT1 suppresses the checkpoint defect of the ccr4Δ mutant. For cell cycle progression, the WHI5 mRNA was shown to be negatively regulated by Ccr4. Loss of WHI5 suppresses the cell cycle defect of the ccr4Δ mutant (30). However, it has remained unknown how cell lysis occurs in the ccr4Δ cells and what the target for Ccr4 is. In the present study, we identified the target for Ccr4, the LRG1 mRNA. Similarly to CRT1 and WHI5 mRNAs, the LRG1 mRNA level is negatively regulated by Ccr4. Loss of LRG1 suppressed the cell lysis of the khd1Δ ccr4Δ double mutant. It should be noted that loss of CRT1 did not suppress the cell lysis of the khd1Δ ccr4Δ double mutant (data not shown), suggesting that each mRNA target is responsible for each of the several phenotypes of the ccr4Δ mutant. Consistent with the fact that the LRG1 mRNA was not a target mRNA for Khd1, the LRG1 mRNA levels in the ccr4Δ single mutant and the khd1Δ ccr4Δ double mutant are similar. Thus, Khd1 does not seem to be involved in the LRG1 expression. Stewart et al. (42) have indicated that another RNA-binding protein Mpt5/Puf5 negatively regulates the LRG1 mRNA level and that the lrg1Δ mutation suppresses the growth defect of the mpt5Δ mutation. Since Mpt5 directly binds to the 3′ untranslated region (3′UTR) of the LRG1 mRNA (12) and physically interacts with Ccr4 and Pop2 (15, 20), Ccr4 may regulate LRG1 mRNA in a 3′UTR-dependent manner together with Mpt5. On the other hand, since the ccr4Δ mpt5Δ double mutant grew slower than each single mutant and the ccr4Δ mpt5Δ khd1Δ triple mutant was lethal (our unpublished data), Mpt5 and Ccr4 may act independently in the CWI pathway.
In this paper, we found that Khd1 and Ccr4 function in the CWI signaling pathway through regulation of the ROM2 and LRG1 mRNAs. What is the relevance of the regulation? Khd1 has been shown to bind to bud-localized mRNAs such as ASH1 and MTL1 (17, 21, 35). Rho1 is known to be spatially and temporally regulated, and the regulators for the Rho1, Rom2, and Lrg1 proteins are also localized at the bud tip and bud neck (29, 45). The regulation of ROM2 and LRG1 mRNAs by Khd1 and Ccr4 may be involved in such spatial and temporal expressions of the encoded proteins, presumably affecting the spatial and temporal activation of Rho1.
ACKNOWLEDGMENTS
We thank Yoshikazu Ohya and Satoru Nogami (University of Tokyo) for providing the plasmid YCplac-RHO1-Q68L.
T.M. and K.I. are supported by grants-in-aid for Scientific Research from the Ministry of Education, Science, Sports, Culture, and Technology, Japan (2009 to 2011). T.M. is supported by the Nakajima Science Foundation. X.L. is supported by the JSPS Postdoctoral Fellowship for Foreign Researchers.
Footnotes
Published ahead of print on 26 August 2011.
REFERENCES
- 1. Azzouz N., et al. 2009. Specific roles for the Ccr4-Not complex subunits in expression of the genome. RNA 15:377–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baudin A., Ozier K. O., Denouel A., Lacroute F., Cullin C. 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21:3329–3330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Biswas D., Yu Y., Mitra D., Stillman D. J. 2006. Genetic interactions between Nhp6 and Gcn5 with Mot1 and the Ccr4-Not complex that regulate binding of TATA-binding protein in Saccharomyces cerevisiae. Genetics 172:837–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bobola N., Jansen R. P., Shin T. H., Nasmyth K. 1996. Asymmetric accumulation of Ash1p in postanaphase nuclei depends on a myosin and restricts yeast mating-type switching to mother cells. Cell 84:699–709 [DOI] [PubMed] [Google Scholar]
- 5. Bomsztyk K., Denisenko O., Ostrowski J. 2004. hnRNP K: one protein multiple processes. Bioessays 26:629–638 [DOI] [PubMed] [Google Scholar]
- 6. Collart M. A. 2003. Global control of gene expression in yeast by the Ccr4-Not complex. Gene 313:1–16 [DOI] [PubMed] [Google Scholar]
- 7. Collart M. A., Struhl K. 1994. NOT1(CDC39), NOT2(CDC36), NOT3, and NOT4 encode a global-negative regulator of transcription that differentially affects TATA-element utilization. Genes Dev. 8:525–537 [DOI] [PubMed] [Google Scholar]
- 8. Coller J., Parker R. 2004. Eukaryotic mRNA decapping. Annu. Rev. Biochem. 73:861–890 [DOI] [PubMed] [Google Scholar]
- 9. Cui Y., et al. 2008. Genome wide expression analysis of the CCR4-NOT complex indicates that it consists of three modules with the NOT module controlling SAGA-responsive genes. Mol. Genet. Genomics 279:323–337 [DOI] [PubMed] [Google Scholar]
- 10. Denisenko O., Bomsztyk K. 2002. Yeast hnRNP K-like genes are involved in regulation of the teromeric position effect and telomere length. Mol. Cell. Biol. 22:286–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Denisenko O., Bomsztyk K. 2008. Epistatic interaction between the K-homology domain protein HEK2 and SIR1 at HMR and telomeres in yeast. J. Mol. Biol. 375:1178–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gerber A. P., Herschlag D., Brown P. O. 2004. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2:342–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gietz R. D., Sugino A. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527–534 [DOI] [PubMed] [Google Scholar]
- 14. Glisovic T., Bachorik J. L., Yong J., Dreyfuss G. 2008. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 14:1977–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Goldstrohm A. C., Hook B. A., Seay D. J., Wickens M. 2006. PUF proteins bind Pop2p to regulate messenger RNAs. Nat. Struct. Mol. Biol. 13:533–539 [DOI] [PubMed] [Google Scholar]
- 16. Halbeisen R. E., Galgano A., Scherrer T., Gerber A. P. 2008. Post-transcriptional gene regulation: From genome-wide studies to principles. Cell. Mol. Life Sci. 65:798–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hasegawa Y., Irie K., Gerber A. P. 2008. Distinct roles for Khd1p in the localization and expression of bud-localized mRNAs in yeast. RNA 14:2333–2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hata H., et al. 1998. Dhh1p, a putative RNA helicase, associates with the general transcription factors Pop2p and Ccr4p from Saccharomyces cerevisiae. Genetics 148:571–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hogan D. J., Riordan D. P., Gerber A. P., Herschlag D., Brown P. O. 2008. Diverse RNA-Binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol. 6:2297–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hook B. A., Goldstrohm A. C., Seay D. J., Wickens M. 2007. Two yeast PUF proteins negatively regulate a single mRNA. J. Biol. Chem. 282:15430–15438 [DOI] [PubMed] [Google Scholar]
- 21. Irie K., et al. 2002. The Khd1 protein, which has three KH RNA-binding motifs, is required for proper localization of ASH1 mRNA in yeast. EMBO J. 21:1158–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kaiser C. A., Adams A., Gottschling D. E. 1994. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 23. Keene J. D. 2007. RNA regulons: coordination of post-transcriptional events. Nat. Rev. Genet. 8:533–543 [DOI] [PubMed] [Google Scholar]
- 24. Klis F. M. 1994. Review: cell wall assembly in yeast. Yeast 10:851–869 [DOI] [PubMed] [Google Scholar]
- 25. Krause S. A., Gray J. V. 2002. The protein kinase C pathway is required for viability in quiescence in Saccharomyces cerevisiae. Curr. Biol. 12:588–593 [DOI] [PubMed] [Google Scholar]
- 26. Levin D. E. 2005. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69:262–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Longtine M. S., et al. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953–961 [DOI] [PubMed] [Google Scholar]
- 28. Makeyev A. V., Liebhaber S. A. 2002. The Poly(C)-binding proteins: a multiplicity of functions and a search for mechanisms. RNA 8:265–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Manning B. D., Padmanabha R., Snyder M. 1997. The Rho-GEF Rom2p localizes to sites of polarized cell growth and participates in cytoskeletal functions in Saccharomyces cerevisiae. Mol. Biol. Cell 8:1829–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Manukyan A., et al. 2008. Ccr4 alters cell size in yeast by modulating the timing of CLN1 and CLN2 expression. Genetics 179:345–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mauchi N., Ohtake Y., Irie K. 2010. Stability control of MTL1 mRNA by the RNA-binding protein Khd1p in yeast. Cell Struct. Funct. 35:95–105 [DOI] [PubMed] [Google Scholar]
- 32. Nonaka H., et al. 1995. A downstream target of RHO1 small GTP-binding protein is PKC1, a homolog of protein kinase C, which leads to activation of the MAP kinase cascade in Saccharomyces cerevisiae. EMBO J. 14:5931–5938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ozaki K., et al. 1996. Rom1p and Rom2p are GDP/GTP exchange proteins (GEPs) for the Rho1p small GTP binding protein in Saccharomyces cerevisiae. EMBO J. 15:2196–2207 [PMC free article] [PubMed] [Google Scholar]
- 34. Pan X., et al. 2006. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell 124:1069–1081 [DOI] [PubMed] [Google Scholar]
- 35. Paquin N., et al. 2007. Local activation of yeast ASH1 mRNA translation through phosphorylation of Khd1p by the casein kinase Yck1p. Mol. Cell 26:795–809 [DOI] [PubMed] [Google Scholar]
- 36. Sakumoto N., et al. 1999. A series of protein phosphatase gene disruptants in Saccharomyces cerevisiae. Yeast 15:1669–1679 [DOI] [PubMed] [Google Scholar]
- 37. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 38. Schneider B. L., Steiner B., Seufert W., Futcher A. B. 1996. pMPY-ZAP: a reusable polymerase chain reaction-directed gene disruption cassette for Saccharomyces cerevisiae. Yeast 12:129–134 [DOI] [PubMed] [Google Scholar]
- 39. Sekiya-Kawasaki M., et al. 2002. Dissection of upstream regulatory components of the Rho1p effector, 1,3-beta-glucan synthase, in Saccharomyces cerevisiae. Genetics 162:663–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sil A., Herskowitz I. 1996. Identification of asymmetrically localized determinant, Ash1p, required for lineage-specific transcription of the yeast HO gene. Cell 84:711–722 [DOI] [PubMed] [Google Scholar]
- 41. Siomi H., Dreyfuss G. 1997. RNA-binding proteins as regulators of gene expression. Curr. Opin. Genet. Dev. 3:345–353 [DOI] [PubMed] [Google Scholar]
- 42. Stewart M. S., Krause S. A., McGhie J., Gray J. V. 2007. Mpt5p, a stress tolerance- and lifespan-promoting PUF protein in Saccharomyces cerevisiae, acts upstream of the cell wall integrity pathway. Eukaryot. Cell 6:262–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tadauchi T., Matsumoto K., Herskowitz I., Irie K. 2001. Post-transcriptional regulation through the HO 3′-UTR by Mpt5, a yeast homolog of Pumilio and FBF. EMBO J. 20:552–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Traven A., et al. 2009. The Ccr4-Pop2-NOT mRNA deadenylase contributes to septin organization in Saccharomyces cerevisiae. Genetics 182:955–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Watanabe D., Abe M., Ohya Y. 2001. Yeast Lrg1p acts as a specialized RhoGAP regulating 1,3-beta-glucan synthesis. Yeast 18:943–951 [DOI] [PubMed] [Google Scholar]
- 46. Wolf J. J., et al. 2010. Feed-forward regulation of a cell fate determinant by an RNA-binding protein generates asymmetry in yeast. Genetics 185:513–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Woolstencroft R. N., et al. 2006. Ccr4 contributes to tolerance of replication stress through control of CRT1 mRNA poly(A) tail length. J. Cell Sci. 119:5178–5192 [DOI] [PubMed] [Google Scholar]