Abstract
The titan cell is a recently described morphological form of the pathogenic fungus Cryptococcus neoformans. Occurring during the earliest stages of lung infection, titan cells are 5 to 10 times larger than the normal yeast-like cells, thereby resisting engulfment by lung phagocytes and favoring the persistence of infection. These enlarged cells exhibit an altered capsule structure, a thickened cell wall, increased ploidy, and resistance to nitrosative and oxidative stresses. We demonstrate that two G-protein-coupled receptors are important for induction of the titan cell phenotype: the Ste3a pheromone receptor (in mating type a cells) and the Gpr5 protein. Both receptors control titan cell formation through elements of the cyclic AMP (cAMP)/protein kinase A (PKA) pathway. This conserved signaling pathway, in turn, mediates its effect on titan cells through the PKA-regulated Rim101 transcription factor. Additional downstream effectors required for titan cell formation include the G1 cyclin Pcl103, the Rho104 GTPase, and two GTPase-activating proteins, Gap1 and Cnc1560. These observations support developing models in which the PKA signaling pathway coordinately regulates many virulence-associated phenotypes in diverse human pathogens.
INTRODUCTION
Cryptococcus neoformans is an opportunistic fungal pathogen most commonly observed in immunocompromised patient populations. C. neoformans spores or desiccated yeast cells are inhaled into the lungs, where they are small enough to lodge in the alveoli. In healthy patients, the cryptococcal pulmonary infection is cleared or contained in granulomas; however, in immunocompromised patients, C. neoformans is able to disseminate from the lungs to cause a systemic infection (19). At late stages of infection, C. neoformans penetrates the blood-brain barrier (BBB) and causes infections of the central nervous system. Due to an increase in the number of HIV+ individuals, C. neoformans has become the third leading cause of death in adults in sub-Saharan Africa (40). Each year, C. neoformans is responsible for approximately 1 million infections worldwide, resulting in more than 600,000 deaths (40).
C. neoformans has several well-studied virulence factors, including polysaccharide capsule, melanin production, and mating type (7, 19). The cryptococcal polysaccharide capsule has been shown to modulate the host immune response (7, 19). Capsule material can bind both antibody and complement, preventing phagocytosis by host macrophages. The production of melanin protects cryptococcal cells from oxidative stress produced by the host immune system in response to infection and phagocytosis (7, 19). The C. neoformans mating type has also been shown to be a factor in pathogenesis. Cryptococcus has two mating types: a and α. Of these, mating type α cells account for the majority of clinical isolates (7, 19). In experiments where both mating types were used to coinfect mice, mating type a cells were unable to penetrate the blood-brain barrier efficiently (35, 36). Previously, we demonstrated that dramatic differences in the rate of titan cell formation between mating types accounted for the difference in BBB penetration during coinfections (37).
Many Cryptococcus virulence factors are regulated by the cyclic AMP (cAMP)/protein kinase A (PKA) pathway (1, 2, 16, 41). The Gα protein Gpa1 activates the cAMP pathway, and strains with mutations in the GPA1 gene display alterations in the surface capsule, melanin production, and virulence (1, 16). Disruption of other positive regulators of the cAMP/PKA pathway, including Pka1 and Cac1, also results in an avirulent phenotype (1, 2, 9, 16). The Rim101 transcription factor is downstream of Pka1 in C. neoformans (38). Signaling through both the cAMP/PKA pathway and the pH/Rim pathway activates Rim101 under host conditions (38). In the pathogenic fungus Candida albicans, Rim101 regulates the yeast-to-hypha switch in response to iron starvation and increased pH, which are relevant conditions for infection (14, 15). The role of Rim101 in morphological changes in C. neoformans under host conditions was previously uncharacterized.
C. neoformans can produce enlarged “titan” cells in response to the host lung environment. Titan cells can reach diameters of 50 to 100 μm, representing a 5- to 10-fold increase over the size of cells grown in vitro (37, 51). During pulmonary infection, titan cells account for approximately 20% of the cryptococcal cell population. Characterization of titan cells has revealed that they are inefficiently engulfed by host immune cells (37). In addition, titan cells are more resistant to the oxidative and nitrosative antimicrobial mechanisms produced by the host immune response (37). Titan cells also exhibit alterations in the cell wall and capsule. The cell wall is thickened, and the capsule is highly cross-linked and cannot be sheared from the cell by chemical or physical means (51). Analysis of nuclear content has shown that titan cells have increased ploidy and can be tetraploid or octoploid (37).
Titan cell production is regulated by elements of the pheromone signaling pathway in mating type a cells. In coinfections with both mating types, mating type a cells double their production of titan cells to approximately 40%, while mating type α cells remain at the basal titan cell production level of 20% (37). Additionally, increased titan cell formation has been correlated with decreased penetration of the BBB by mating type a cells during coinfection, suggesting that pheromone signaling leading to titan cell formation affects virulence (37). C. neoformans pheromone peptides are sensed by the G-protein-coupled receptors (GPCRs) Ste3a and Ste3α (12, 26, 43). Ste3α interacts with the Gα proteins Gpa2 and Gpa3 to signal via a mitogen-activated protein kinase (MAPK) signal transduction pathway to regulate mating (12, 13, 22). Disruption of pheromone sensing in mating type a cells during coinfection by deletion of the pheromone receptor Ste3a reduced titan cell formation in mating type a cells to the basal level of 20% (37). Thus, increased production of titan cells during mixed infections is dependent on intact Ste3a signaling.
Previous work identified and characterized titan cell formation during early pulmonary infection (37, 51). However, the mechanism by which pheromone signaling affected titan cell production in mating type a cells but not mating type α cells remained unclear. Additionally, the signaling pathway necessary for production of the basal level of titan cells during a typical infection had not been identified. Here we sought to identify receptors that sense the host environment so as to regulate titan cell production. We also identified Gα proteins downstream of these receptors that are important for titan cell formation. Additionally, we sought to identify downstream effectors of the signaling pathway and potential transcription factors responsible for titan cell induction. We determined that the conserved cAMP/PKA signaling pathway controls the dramatic titan cell morphological change through activation of the Rim101 transcription factor.
MATERIALS AND METHODS
Ethics statement.
All animals were handled in strict accordance with good animal practice as defined by the relevant national and/or local animal welfare bodies, and all animal work was approved by the Institutional Animal Care and Use Committee (IACUC) under protocols 0712A22250 and 1010A91133.
Strains and media.
The Cryptococcus neoformans var. grubii strains used in this study are described in Table 1. Strains were stored as glycerol stocks at −80°C and were grown at 30°C in yeast extract-peptone-dextrose (YPD) agar or broth medium (BD Biosciences, Sparks, MD).
Table 1.
Strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| KN99a | MATa | 36 |
| KN99α | MATα | 36 |
| YSB83 | MATα gpa1Δ::NAT | 49 |
| CDX153 | MATα gpa1Δ::NATura5 + GDP1-GPA1(Q284L)URA5 | 49 |
| YSB85 | MATagpa1Δ::NEO | 49 |
| CDX40 | MATagpa1Δ::NATura5 + GDP1-GPA1(Q284L)URA5 | 49 |
| CDX42 | MATagpa1Δ::NEOura5 + GDP1-GPA1wtURA5 | 49 |
| YSB25 | MATα gpa2Δ::NEO | 22 |
| LHO6 | MATα gpa3Δ::NEO | This study |
| LHO8 | MATα gpa2Δ::NAT gpa3Δ::NEO | This study |
| YSB110 | MATagpa2Δ::NAT | 22 |
| LHO5 | MATagpa3Δ::NEO | This study |
| LHO7 | MATagpa2Δ::NAT gpa3Δ::NEO | This study |
| YPH308 | MATα gpa2Δ::NAT gpa3Δ::NEO | 22 |
| YPH106 | MATagpa2Δ::NAT gpa3Δ::NEO | 22 |
| CDX1 | MATα gpr1Δ::NAT | 49 |
| CDX3 | MATα gpr2Δ::NEO | 49 |
| CDX5 | MATα gpr3Δ::NEO | 49 |
| CDX6 | MATα gpr4Δ::NEO | 49 |
| CDX11 | MATα gpr5Δ::NAT | 49 |
| CDX18 | MATα gpr4Δ::NEO gpr5Δ::NAT | 49 |
| LHO17 | MATα gpr4Δ::NEO + GPR5 | This study |
| CDX9 | MATagpr4Δ::NAT | 49 |
| CDX10 | MATagpr5Δ::NAT | 49 |
| CDX19 | MATagpr4Δ::NEO gpr5Δ::NAT | 49 |
| LHO18 | MATagpr4Δ::NEO + GPR5 | This study |
| CDX14 | MATα gpr8Δ::NAT | This study |
| CDX16 | MATα gpr9Δ::NAT | This study |
| CDC7 | MATα pkr1Δ::URA5 | 16 |
| JKH7 | MATα pka1Δ::URA5 | 20 |
| RPC3 | MATα cac1Δ::URA5 | 2 |
| TOC2 | MATα rim101Δ::NAT | 38 |
| TOC4 | MATα rim101Δ::NAT + RIM101::NEO | 38 |
| CBN32 | MATα cdc24Δ::NAT | 34 |
| ERB010 | MATα cdc42Δ::NAT | 5 |
| ERB007 | MATα cdc420Δ::NEO | 5 |
| ERB012 | MATα cdc420Δ::NAT + CDC420::NEO | 5 |
| KLC109 | MATanrg1Δ::NEO | 9 |
| KLC113 | MATα nrg1Δ::NEO | 9 |
| NB007 | MATaste3Δ::NAT | 37 |
| XL1598 | MATα mat2Δ::NEO | 31 |
| XL1601 | MATα znf2Δ::NEO | 31 |
| YSB345 | MATα ste7Δ::NEO | 31 |
| CHY776 | MATα sxi1αΔ::NAT | 24 |
| YSB317 | MATα ste50Δ::NAT-STM#296 | 25 |
| D782 | zap102Δ (CNAG_03366) | 32 |
| D1480 | sks1Δ (CNAG_06568) | 32 |
| D1477 | cnf0270Δ (CNAG_06554) | 32 |
| D989 | cnh0970Δ (CNAG_04455) | 32 |
| D439 | Expressed sequence AA960436Δ (CNAG_01624) | 32 |
| D1059 | ypl260WΔ (CNAG_05236) | 32 |
| D1131 | yor378W07-AΔ (CNAG_03713) | 32 |
| D1139 | tpo105Δ (CNAG_04546) | 32 |
| D1258 | rim10102Δ (CNAG_05431) | 32 |
| D1023 | fms102Δ (CNAG_05027) | 32 |
| D949 | cnh3700Δ (CNAG_04248) | 32 |
| D986 | yak103Δ (CNAG_04433) | 32 |
| D1462 | cnb1770-AΔ (CNAG_06457) | 32 |
| D521 | mlh1Δ (CNAG_02073) | 32 |
| D88 | zap103Δ (CNAG_04352) | 32 |
| D488 | ksp1Δ (CNAG_01905) | 32 |
| D545 | hap3Δ (CNAG_02215) | 32 |
| D425 | bem2Δ (CNAG_01533) | 32 |
| D689 | cnc1560Δ (CNAG_02953) | 32 |
| D1381 | ykl071WΔ (CNAG_06074) | 32 |
| D1068 | crn102Δ (CNAG_05301) | 32 |
| D359 | lsb1Δ (CNAG_01183) | 32 |
| D156 | rck2-AΔ (CNAG_00130) | 32 |
| D1179 | liv16Δ (CNAG_07193) | 32 |
| D722 | upb6Δ (CNAG_03109) | 32 |
| D671 | rsp3102Δ (CNAG_02827) | 32 |
| D1490 | rho104-AΔ (CNAG_06606) | 32 |
| D785 | pcl103Δ (CNAG_03385) | 32 |
| D61 | bni4Δ (CNAG_02071) | 32 |
| D173 | ubp1Δ (CNAG_00187) | 32 |
| D1340 | gap1Δ/rdg1Δ (CNAG_05838) | 32 |
| D1255 | ypl230W03-CΔ (CNAG_05420) | 32 |
Double mutant strains were isolated from the progeny of matings between YSB25 and YSB105 for gpa2Δ gpa3Δ double mutant strains LHO7 and LHO8. Single gpa3Δ strains LHO5 and LHO6 were isolated from the progeny of matings between YPH308 (22) and KN99a. Briefly, matings were performed on V8 agar, and basidiospores were micromanipulated as described previously (21). Strains were isolated using YPD agar containing 200 μg/ml nourseothricin (NAT) and/or 200 μg/ml neomycin (NEO). Mutant strains were then screened by PCR using the primers listed in Table S1 in the supplemental material to verify gene deletions and marker insertion into the appropriate loci. gpr8Δ and gpr9Δ mutants were generated in the H99 strain background by overlap PCR as described previously (10). The 5′ and 3′ regions of the GPR8 gene were amplified from H99 genomic DNA with primers JH13288/JH13289 and JH13290/JH13291, respectively. The flanking regions of the GPR9 gene were amplified with primers JH13295/JH13296 and JH13297/JH13298, respectively. The dominant selectable marker (NEOr) was amplified with the M13 primers (M13F and M13R) from plasmid pJAF1 (17). The target gene replacement cassettes of GPR8 and GPR9 were generated by overlap PCR with primers JH13288/JH13291 and JH13295/JH13298, respectively. Purified overlap PCR products were precipitated onto 10-μl gold microcarrier beads (diameter, 0.6 μm; Bio-Rad), and strain H99 was biolistically transformed as described previously (11). Stable transformants were selected on YPD medium containing G418 (200 mg/liter). To screen for mutants of GPR8 and GPR9, diagnostic PCRs were performed by analyzing the 5′ junctions of the disrupted mutant alleles with primers JH13287/JH8994 (GPR8) and JH13294/JH8994 (GPR9) (see Table S1 in the supplemental material). Positive transformants identified by PCR screening were further confirmed by Southern blot analysis.
Gene complementation in the double mutant strains was performed by isolating progeny from matings between CDX18 and KN99α or CDX19 and KN99a. Briefly, matings were performed on V8 agar, and basidiospores were micromanipulated as described previously (21). Strains were isolated using YPD agar containing 200 μg/ml NAT or NEO. Single mutant strains were then screened by PCR using the primers listed in Table S1 in the supplemental material to verify gene deletions and marker insertions into the appropriate loci.
In vivo titan cell assay.
C. neoformans cells were cultured overnight in YPD broth. The resulting yeast cells were pelleted and resuspended in sterile phosphate-buffered saline (PBS) at a concentration of 1 × 108 cells/ml based on the hemocytometer count. Groups of 6- to 8-week-old female A/J mice (Jackson Laboratory, Bar Harbor, ME) were anesthetized by intraperitoneal pentobarbital injection. One mouse (preliminary experiments) or three to five mice per treatment were infected intranasally with 2 × 107 cells in 50 μl PBS (Lonza, Rockland, ME). At 3 days postinfection, mice were sacrificed by CO2 inhalation. Lungs were lavaged with 1.5 ml sterile PBS three times using a 20-gauge needle placed in the trachea. Cells in the lavage fluid were pelleted at 16,000 × g, resuspended in 3.7% formaldehyde, and incubated at room temperature for 30 min. Cells were then washed once with PBS, and >300 cells per animal were analyzed for size by microscopy (AxioImager; Carl Zeiss, Inc.). Cell body sizes were measured, and cells were classified as small cells (diameter, <10 μm) or titan cells (diameter, >10 μm).
Flow cytometry.
KN99α was grown overnight in YPD medium to log phase. Cells were centrifuged at 16,000 × g for 1 min, and the supernatant was discarded. Cells were washed three times in sterile PBS, resuspended in 3.7% formaldehyde in PBS, and incubated for 30 min at room temperature. Bronchoalveolar lavage (BAL) samples were washed once in 0.05% sodium dodecyl sulfate (SDS) to lyse mammalian cells and were then washed twice in sterile water. Cells in the lavage fluid were pelleted at 16,000 × g, resuspended in 3.7% formaldehyde, and incubated at room temperature for 30 min. All samples were then washed once with PBS and were resuspended in PBS containing 300 ng/ml 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen). Samples were incubated at room temperature for 15 min, washed with PBS, and resuspended in PBS. Cells were examined for cell size by forward scatter (FSC) and for nuclear content by DAPI using an LSRII flow cytometer with FACSDiva software (BD Biosciences). Cell populations were analyzed using FlowJo software (Tree Star, Inc., Ashland, OR) (see Fig. S2 in the supplemental material).
Yeast two-hybrid assay.
The split-ubiquitin system has been designed to specifically detect the potential interactions between membrane proteins (24a). This system was utilized to investigate the interaction between several G-protein-coupled receptor proteins (Ste3a, Ste3α, Gpr5, and Gpr4) and Gα proteins (Gpa1, Gpa2, and Gpa3). Vectors and yeast strains were included in DUALmembrane kit 2 (Dualsystems Biotech, Zürich, Switzerland). Full-length cDNAs of STE3a, STE3α, GPR5, and GPR4 were cloned separately into pCCW (the C-terminal half of ubiquitin [Cub] was fused to the C terminus of each protein). Full-length GPA1, GPA2, and GPA3 cDNAs were cloned into the pDL2XN vector (the mutated N-terminal half of ubiquitin [NubG protein] was fused to the C terminus of each G protein). A GPA1 dominant active allele (GPA1Q284L) and a dominant negative allele (GPA1G283A) were also cloned into pDL2XL. All cDNA sequences were confirmed by DNA sequencing. Cub and NubG fusion constructs were cotransformed into the Saccharomyces cerevisiae host strain NMY32. Interaction was determined by the growth of yeast transformants on a medium lacking histidine or adenine and also by measurement of β-galactosidase activity.
Coinfections.
Four mice per treatment were infected as described above with 2 × 107 Alexa Fluor 488-labeled cells of strain KN99a, strain KN99α, or the gpr4Δ gpr5Δ mating type a or α mutant, or with an approximately 1:1 ratio of one stained and one unstained strain. Infected mice were sacrificed at 3 days postinfection by CO2 inhalation. Lungs were lavaged with 1.5 ml sterile PBS three times using a 20-gauge needle placed in the trachea. Cells in the lavage fluid were pelleted at 16,000 × g, resuspended in 3.7% formaldehyde, and incubated at room temperature for 30 min. Cells were then washed once with PBS and were resuspended in PBS. More than 300 cells per animal were analyzed for size and fluorescence by microscopy (AxioImager; Carl Zeiss, Inc.).
RNA preparation.
RNA was prepared as described previously (38). Briefly, strains were incubated to mid-log phase, washed twice, and then incubated in Dulbecco's modified Eagle's medium (DMEM) at 37°C under 5% CO2 for 3 h. The cells were then washed twice before centrifugation, freezing on dry ice, and lyophilization. Total RNA was extracted using the Qiagen (Valencia, CA) RNeasy Plant Mini kit.
RNA sequencing and transcript analysis.
Clonal library preparation, mRNA enrichment through poly(A) capture, and Illumina sequencing on a GAII Illumina genome analyzer were performed from total RNA at the Duke Sequencing Core Facility (Duke University). Briefly, total RNA was purified using Sera-Mag oligo(dT) magnetic beads according to the manufacturer's protocol (Illumina, San Diego, CA). The mRNA was fragmented and used as a template for first-strand cDNA synthesis. Second-strand synthesis was then performed, and the resulting double-stranded cDNA was end repaired, adenylated, and ligated into the paired-end (PE) adaptors, according to the manufacturer's protocols (Illumina). Afterwards, the cDNA templates were purified, enriched, and used for sequencing on the Illumina GAII genome analyzer. The wild-type (strain H99) sample was sequenced with 72-bp paired-end reads, and the rim101Δ sample was sequenced with 36-bp single-end reads.
All reads were then mapped to the C. neoformans reference genome provided by the Broad Institute (Cryptococcus neoformans var. grubii H99 Sequencing Project, Broad Institute of Harvard and MIT; http://www.broadinstitute.org/) using TopHat, version 1.3.0 (28, 44). The aligned reads were then analyzed for FPKM (fragments per kilobase of transcript per million mapped reads) using Cufflinks, version 1.0.3. Statistically significant differences between strains were determined by CuffDiff (45).
Promoter analysis.
The DNA sequence 1,000 bp upstream of the start codon, as defined by the Broad Institute Cryptococcus neoformans var. grubii H99 Sequencing Project, was examined for the presence of the predicted Rim101 binding site (GCCAAG) on either strand (38).
Statistical analysis.
All analyses were performed using Analyze-It (Analyze-it Ltd., Leeds, England). One-way analysis of variance (ANOVA) was used to analyze differences in titan cell production, and P values of <0.02 were considered significant.
RESULTS
G-protein-coupled receptor signaling regulates titan cell formation.
Previously, we showed that signaling through Ste3a in mating type a cells stimulates titan cell production during coinfection with cells of the opposite mating type (37). While Ste3a signaling increases titan cell production over the basal level, disruption of Ste3a does not completely abolish titan cell production. Thus, another receptor likely senses environmental cues that stimulate titan cell production. Because G-protein-coupled receptors play an integral role in environmental sensing, and Ste3a, a G-protein-coupled receptor, is known to regulate titan cell production, we hypothesized that there is at least one additional G-protein-coupled receptor responsible for titan cell regulation.
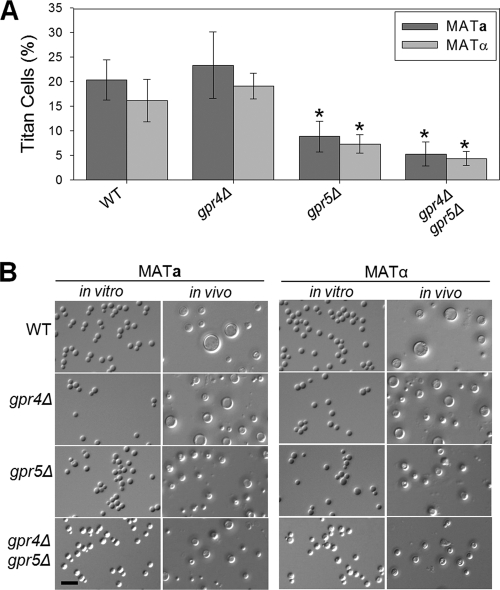
Therefore, we screened a library of G-protein-coupled receptor mutants for alterations in titan cell production by using an intranasal model of cryptococcal infection (see Fig. S1 in the supplemental material). Fungal cells were collected by bronchoalveolar lavage (BAL) at 3 days postinfection, fixed immediately, and examined by microscopy for cell size and by flow cytometry for cell ploidy (see Fig. S2 in the supplemental material). Of the G-protein-coupled receptor mutant strains, only gpr5Δ mutants showed a decrease in titan cell formation. Pulmonary infections with the wild-type strains resulted in 20.4% titan cells in the mating type a strain and 16.1% titan cells in the mating type α strain. In contrast, infections with gpr5Δ mutants resulted in 2-fold reductions in titan cell formation in both mating types, to 8.8% titan cells in mating type a and 7.3% in mating type α (P, 0.004 and 0.013, respectively) (Fig. 1A). Interestingly, titan cells produced by the gpr5Δ mutant strains rarely exceeded 12 μm in diameter (Fig. 1B). The gpr5Δ mutants had no defects in capsule, melanin, urease, or growth at high temperatures in vitro (data not shown).
Fig. 1.
Titan cell signaling in vivo is regulated by the Gpr5 receptor. Mice were infected intranasally with 2 × 107 cells of wild-type KN99a or KN99α or with 2 × 107 cells of the gpr4Δ, gpr5Δ, or gpr4Δ gpr5Δ mating type a or α mutant. At 3 days postinfection, BAL was performed. (A) Small cells (cell body diameter, <10 μm) and titan cells (cell body diameter, >10 μm) were quantified by microscopy. More than 300 cells per treatment per mouse were counted. Error bars indicate standard deviations for 3 to 5 mice per treatment. Statistical analysis of the difference between each mutant strain and the wild-type strain of the same mating type was performed. *, P < 0.02. (B) In vitro cells were grown overnight in rich medium (YPD), fixed in 3.7% formaldehyde, and examined by microscopy. In vivo BAL samples isolated at 3 days postinfection were fixed in 3.7% formaldehyde and were examined by microscopy. Bar, 20 μm.
Previous studies showed that Gpr4 and Gpr5 share a high degree of sequence homology (49). To further assess the relative contributions of these two GPCRs to titan cell production, we directly compared titan cell production in gpr5Δ, gpr4Δ, and gpr4Δ gpr5Δ mutants of both mating types. Deletion of GPR4 alone had no effect on titan cell formation. The gpr4Δ gpr5Δ double mutants showed significant decreases in titan cell production, with averages of 5.3% titan cells in mating type a and 4.4% in mating type α (P, 0.001 and 0.011, respectively, for the comparison to the corresponding wild-type mating type) (Fig. 1A). However, there was no statistically significant difference between the double mutant and the gpr5Δ single mutant (P, 0.435). Complementation of either the gpr5Δ or the gpr4Δ gpr5Δ mutant with Gpr5 restored titan cell production to wild-type levels (data not shown). In addition, neither the presence nor the absence of various amino acids stimulated titan cell formation in vitro (data not shown). Similarly, no difference in cell enlargement between the wild type and the gpr5Δ mutant in response to phosphatidylcholine treatment was observed (data not shown). These data suggest that signaling through the GPCR Gpr5, and not the related protein Gpr4, controls titan cell production in both mating type a and mating type α cells. None of the titan cell defects in these GPCR mutants were affected by the mating type, demonstrating that the regulation of basal titan cell production by Gpr5 is separate from the induction of titan cells by the mating pathway.
Gpa1 signaling results in titan cell production.
C. neoformans has three known Gα proteins that can interact with G-protein-coupled receptors. Gpa2 and Gpa3 regulate signaling through Ste3α in mating type α cells by acting in opposition to each other (22). Mutation of either Gpa2 or Gpa3 alone did not cause a defect in virulence in a mating type α background, but the double mutant was slightly attenuated (29). Based on the Ste3α data and the role of Ste3a in titan cell induction, we hypothesized that Gpa2 and Gpa3 would interact with Ste3a to regulate titan cell production. We anticipated that deletion of Gpa2 and/or Gpa3 in mating type a cells would alter the rate of titan cell formation.
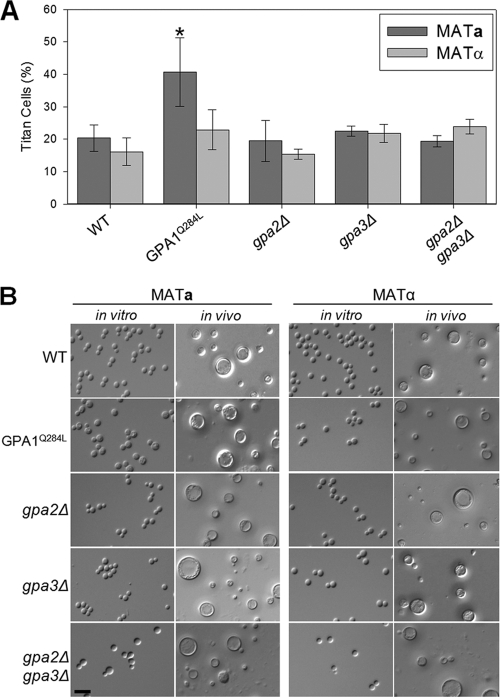
Therefore, we determined the degree of titan cell production in gpa2Δ and gpa3Δ mutants in each mating type. In both mating types, the gpa2Δ mutant strains exhibited no significant alteration in titan cell production (Fig. 2) from that for the corresponding wild-type strains (P, 0.812 and 0.752). Similarly, the gpa3Δ mutant strains in both mating types showed normal titan cell production (P, 0.379 and 0.087). The gpa2Δ gpa3Δ double mutant strains also showed no significant alteration in titan cell production (P, 0.708 and 0.075), demonstrating that neither Gpa2 nor Gpa3 plays a role in the regulation of titan cell production.
Fig. 2.
Signaling via the Gα protein Gpa1 regulates titan cell production. Mice were infected intranasally with 2 × 107 cells of wild-type KN99a or KN99α or with 2 × 107 cells of the GPA1Q284L, gpa2Δ, gpa3Δ, or gpa2Δ gpa3Δ mating type a or α mutant. At 3 days postinfection, BAL samples were collected. (A) Small cells (cell body diameter, <10 μm) and titan cells (cell body diameter, >10 μm) were quantified by microscopy. More than 300 cells were counted per treatment per mouse. Error bars indicate standard deviations for 3 to 5 mice per treatment. Statistical analysis of the difference between each mutant strain and the wild-type strain of the same mating type was performed. *, P < 0.02. (B) In vitro cells were grown overnight in rich medium (YPD), fixed in 3.7% formaldehyde, and examined by microscopy. In vivo BAL samples isolated at 3 days postinfection were fixed in 3.7% formaldehyde and were examined by microscopy. Bar, 20 μm.
Since the two Gα proteins predicted to interact with Ste3a did not affect titan cell production, we then hypothesized that the Gα protein Gpa1 might regulate titan cell formation. Gpa1 plays an integral role in the regulation of many virulence factors, including capsule and melanin (1, 2). Because Gpa1 controls the production of these virulence factors, gpa1Δ strains are avirulent (1, 2). As early as 24 h postinfection, the vast majority of gpa1Δ cells are phagocytosed and killed (data not shown), preventing us from quantifying in vivo titan cell production by the deletion mutants. Instead, we used a constitutively active GPA1 mutant allele (GPA1Q284L) to elucidate the role of Gpa1 in titan cell formation (49). Infections with mating type a cells expressing the dominant active GPA1Q284L allele resulted in a significant increase in titan cell production to approximately 40%, representing a 2-fold increase in the proportion of titan cells over that in wild-type mating type a strains (P = 0.012) (Fig. 2). Interestingly, the mating type α strain expressing the GPA1Q284L mutant allele showed no difference in titan cell production from the wild-type mating type α strain (P = 0.172). These results are consistent with a model in which the Gpa1 protein interacts with the Gpr5 and Ste3a receptors to induce titan cell formation.
Ste3a interacts with Gpa1 but not with Gpa2 or Gpa3.
Our data suggest that in mating type a cells, Ste3a may interact with Gpa1, rather than with Gpa2 or Gpa3, to regulate titan cell production. Therefore, we assessed the interactions between Ste3a and the Gα proteins using a split-ubiquitin yeast two-hybrid system. In this system, membrane protein interactions release an artificial transcription factor made up of three proteins: protein A, LexA, and VP16 (24a). Upon release, the transcription factor translocates to the nucleus and activates reporter genes.
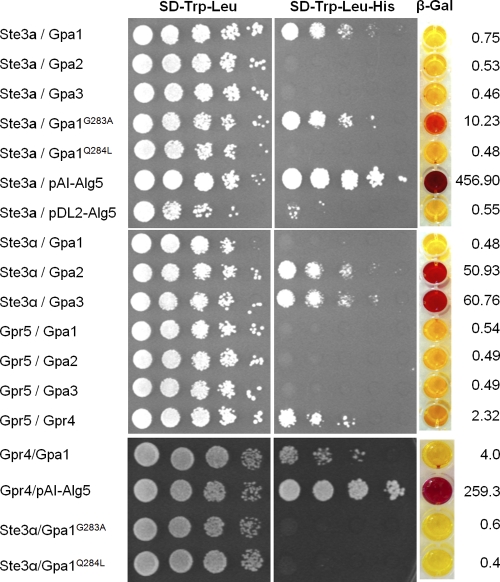
Three versions of Gpa1 were screened for possible interactions with Ste3a and Ste3α: (i) wild-type Gpa1, (ii) Gpa1G283A, which contains a dominant negative mutation, and (iii) Gpa1Q284L, which renders the protein constitutively active. In addition, we examined interactions with Gpa2 and Gpa3 (49). Our results show that Ste3a interacts with both the wild-type Gpa1 allele and the Gpa1 dominant negative allele, but no interaction was detected with the constitutively active Gpa1 allele, Gpa2, or Gpa3 (Fig. 3). G-protein-coupled receptors normally serve as guanine nucleotide exchange factors (GEF) and bind to the inactive form of Gα proteins to activate the G protein. Therefore, the stronger interaction between Ste3a and Gpa1G283A than between Ste3a and the wild-type Gpa1 construct indicated that the weak interaction in this assay with the wild-type Gpa1 protein was likely valid.
Fig. 3.
Ste3a interacts with Gpa1 in the split-ubiquitin yeast two-hybrid system. The C-terminal half of ubiquitin (Cub) was fused to the C termini of the full-length cDNAs of Ste3a (Ste3a::Cub), Ste3α (Ste3α::Cub), and Gpr5 (Gpr5::Cub). The N-terminal half of ubiquitin (NubG) was fused to the C termini of full-length cDNAs of Gpa1 (Gpa1::NubG), Gpa2 (Gpa2::NubG), Gpa3 (Gpa3::NubG), Gpa1G283A (Gpa1G283A::NubG), and Gpa1Q284L (Gpa1Q284L::NubG). The interaction of Ste3a::Cub with the control vector pAI-Alg5 served as a positive control to ensure the correct topology of the Ste3a::Cub fusion protein; the interaction of Ste3a::Cub with the empty vector pDL2-Alg5 served as a negative control. Yeast transformants contained both a Cub fusion and a NubG fusion construct and were grown on selective medium lacking histidine or adenine after serial dilution. β-Galactosidase activity assays were performed to further verify interactions. Interactions between Ste3a (top) or Ste3α (center) and the Gα proteins are shown on selective media, as are control interactions (bottom).
As a control, we also measured the interactions between Ste3α and the Gα proteins. We confirmed robust interactions between Ste3α and Gpa2 or Gpa3, but not Gpa1. These data are consistent with previously published results demonstrating that Ste3α interacts with Gpa2 and Gpa3 to control the mating pathway (22, 23). In contrast to the findings for Ste3α, we were unable to detect any interactions between Ste3a and either Gpa2 or Gpa3.
Previous studies found that the G-protein-coupled receptor Gpr4 is important for activating Gpa1 in response to methionine (49). The same studies showed that Gpr5 shares sequence similarity with Gpr4 but does not respond to methionine. Thus, the function of Gpr5 remained unknown (49). In the split-ubiquitin assays, we detected no interactions between Gpr5 and any of the Gα proteins (Fig. 3). In contrast, Gpr5 and Gpr4 demonstrated a robust interaction in this system, indicating that these predicted G-protein-coupled receptors can form heterodimer or oligomer structures.
Signaling through Ste3a is sufficient for titan cell formation during coinfection.
Previously, we showed that coinfection with both mating types results in increased titan cell formation in mating type a cells and that this increase is dependent on Ste3a (37). We also demonstrate here that titan cell production in both mating types is regulated by signaling through the Gpr5 receptor. Therefore, we wanted to determine whether titan cell signaling through Ste3a is dependent on simultaneous signaling via Gpr5 or whether Ste3a signaling is sufficient to produce titan cells in the presence of mating type α cells.
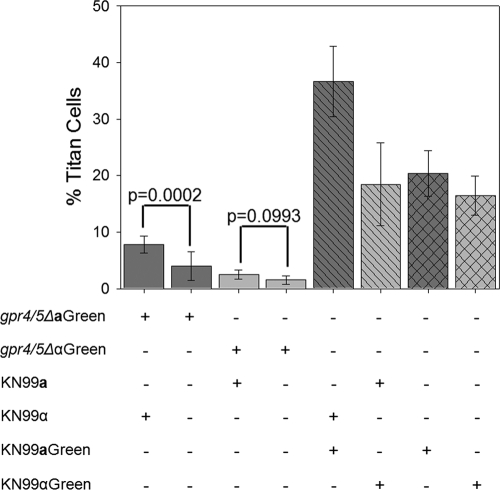
To assess whether Ste3a titan cell signaling is dependent on Gpr5 signaling, BAL samples were obtained 3 days postinfection from mice either infected with the fluorescently labeled gpr4Δ gpr5Δ MATa or gpr4Δ gpr5Δ MATα mutant only, coinfected with unlabeled KN99a and the fluorescently labeled gpr4Δ gpr5Δ MATα mutant or with the fluorescently labeled gpr4Δ gpr5Δ MATa mutant and unlabeled KN99α, infected with fluorescently labeled KN99α or KN99a only, or coinfected with fluorescently labeled KN99a and unlabeled KN99α or with unlabeled KN99a and fluorescently labeled KN99α (Fig. 4). BAL samples were immediately fixed, and the proportion of titan cells with green fluorescence was compared to the total population of fluorescently marked cells by microscopy. Consistent with previous findings, infections with either individual wild-type strain, KN99a or KN99α, produced approximately 20% titan cells. Coinfection with both mating types yielded an increase in titan cell production in mating type a cells to approximately 40%, while mating type α cells were unaffected, remaining at the basal level of 20%. Infection with a gpr4Δ gpr5Δ double mutant strain of either mating type alone resulted in a dramatic reduction in titan cell formation (Fig. 1 and 4). The mating type α gpr4Δ gpr5Δ mutant was unaffected by coinfection; titan cell production remained low, at 1.54%. Surprisingly, coinfection with the fluorescently labeled mating type a gpr4Δ gpr5Δ double mutant and the unlabeled wild-type mating type α strain resulted in a significant increase in titan cell production by the gpr4Δ gpr5Δ MATa mutant, to an average of 7.79% (P < 0.001), demonstrating that the mating type a strain was still able to undergo induction of titan cell production. Thus, the Ste3a signal is sufficient for titan cell induction, independent of the signals for basal titan cell production. However, signaling though both Ste3a and Gpr5 is required for optimal titan cell production during coinfection.
Fig. 4.
Effects of coinfection on titan cell formation. Mice were intranasally infected with 2 × 107 wild-type (KN99a or KN99α) or gpr4Δ gpr5Δ mutant cells labeled with Alexa Fluor 488 (Green) or were coinfected with one labeled and one unlabeled strain. Cells obtained by BAL were fixed and examined by microscopy for green fluorescence (cell type) and cell size. More than 300 cells were examined per animal, and the small cells (cell body diameter, <10 μm) and titan cells (cell body diameter, >10 μm) were quantified. Error bars indicate standard deviations for 3 to 4 mice per treatment.
Rim101 and Cdc420 regulate titan cell formation.
We have shown that pheromone sensing via Ste3a is able to induce titan cell production in mating type a cells. Our data also show that Ste3a does not interact with the Gα proteins, Gpa2 and Gpa3, that are known to regulate the MAPK mating pathway in C. neoformans. To determine what role, if any, the MAPK mating pathway plays in titan cell production, we characterized titan cell formation in strains with gene deletion mutations in the MAPK mating pathway members Ste20, Ste50, Ste7, and Ste12. Additionally, we examined the downstream transcription factors Sxi1α, Znf2, and Mat2 (24–26, 31). All mutant strains showed wild-type levels of titan cell production (data not shown). These data show that the MAPK mating pathway does not regulate titan cell production.
Instead, Ste3a was shown to interact with Gpa1, a member of the cAMP/PKA pathway, to regulate titan cells. Thus, we examined transcription factors downstream of PKA to determine their role in titan cell production. Rim101 is a transcription factor regulated by the PKA pathway in C. neoformans, and deletion of Rim101 affects capsule attachment (38). Additionally, this transcription factor is responsible for the major morphological switch from yeast to hyphae during systemic infection by C. albicans (14). Based on these observations, we tested a rim101Δ mutant strain for its ability to produce titan cells in order to determine whether Rim101 is involved in titan cell formation.
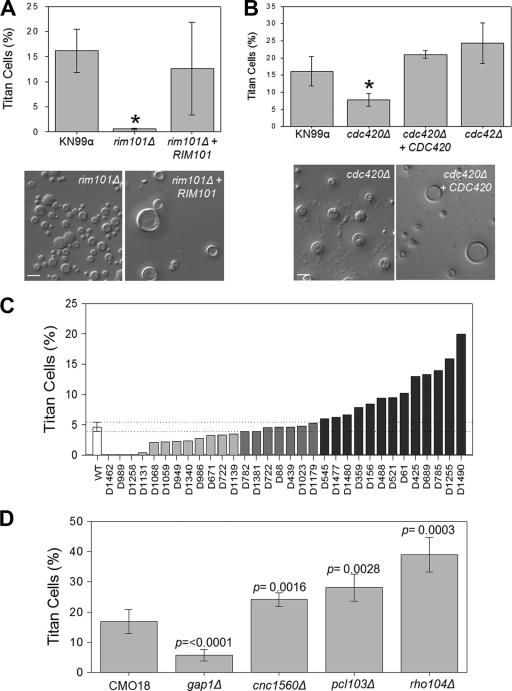
Unlike the gpa1Δ mutant and other activating mutants in the PKA pathway (such as pka1Δ or cac1Δ mutants), the rim101Δ strain does not have a virulence defect. Therefore, rim101Δ cells can be recovered from infected lungs and analyzed for titan cell production (Fig. 5A). The rim101Δ mutant strain exhibited a dramatic reduction in titan cell formation, with a rate of 0.46%, compared with approximately 17% in the wild type (Fig. 5A) (P < 0.001). Complementation of Rim101 restored titan cell production to wild-type levels (P = 0.579). These results suggest that Rim101 is a major regulator of titan cell production.
Fig. 5.
Regulators of titan cell signaling. Mice were infected intranasally with 2 × 107 cryptococcal cells. At 3 days postinfection, BAL samples were collected and fixed. Small cells (cell body diameter, <10 μm) and titan cells (cell body diameter, >10 μm) were quantified by microscopy. More than 300 cells were counted per treatment per mouse. (A) The rim101Δ mutant and a complemented (rim101Δ + RIM101) strain were assessed for titan cell formation. Error bars indicate standard deviations for 3 to 5 mice per treatment. The asterisk indicates a significant difference (P < 0.02) from the wild type. (B) cdc420Δ, cdc420Δ + CDC420, and cdc42Δ strains were assessed for titan cell formation. Error bars indicate standard deviations for 3 to 5 mice per treatment. The asterisk indicates a significant difference (P < 0.02) from the wild type. (C) Thirty-two mutant strains were assayed for titan cell formation. The standard deviation (dotted lines) for the wild type (open bar) was used to identify mutant strains that exhibit altered titan cell formation. (D) Four of the mutant strains for which results are shown in panel C were further analyzed for titan cell formation. For all these strains, the levels of titan cell formation are statistically significantly different from that of the wild type (P < 0.02). Error bars indicate standard deviations for 3 to 5 mice per treatment.
Morphological changes have also been shown to be controlled by the GTPase Cdc42 in other fungi (42). Unlike other pathogenic fungi, Cryptococcus has two paralogues for Cdc42: Cdc42 and Cdc420. In C. neoformans, Cdc42 confers thermotolerance and regulates morphogenesis during mating. Both paralogues can also regulate morphological changes in response to increased temperatures. Based on their roles in regulating morphology in response to host body temperature, we tested the effects of Cdc42 and Cdc420 deletions on titan cell production (Fig. 5B). The cdc42Δ mutant exhibited an average titan cell production level of 24.3%, similar to that of the wild type (P = 0.101). In contrast, the cdc420Δ mutant showed a reduction in titan cell production to 7.81%, significantly lower than wild-type levels (P = 0.017). Complementation of Cdc420 restored titan cell production to wild-type levels (P = 0.131). Thus, Cdc420 but not Cdc42 stimulates titan cell formation.
Additional effectors of titan cell induction.
To identify additional members of the titan cell signaling pathway, we analyzed strains from a large collection of C. neoformans mutant strains that exhibited alterations in pathogenesis (32). BAL samples were collected at 3 days postinfection, fixed, and examined by microscopy. This preliminary screen identified several strains in which titan cell formation was altered from that in the wild-type strain background (CMO18) used to create this mutant collection (Fig. 5C). From the strains with altered titan cell formation, we chose 4 strains with mutations in genes with known functions for further analysis (Fig. 5D). One strain exhibited a lower level of titan cell production than that of the wild type and contained a mutation in the coding sequences for the GTPase-activating protein (GAP) Gap1. Infection with the gap1Δ mutant resulted in only 5.79% titan cell formation, significantly lower than the wild-type level of 16.9% (P < 0.0001). Three mutants demonstrated an increase in titan cell formation: those with mutations in a GAP gene, Cnc1560 (P = 0.002); a rho family GTPase, Rho104 (P < 0.001); and a cell cycle regulation gene, Pcl103 (P = 0.003). Each protein encoded by one of these genes is predicted to control aspects of morphogenesis in other organisms.
To determine if the expression levels of genes involved in titan cell formation are regulated by Rim101, we analyzed the promoter regions for predicted Rim101 binding sites (38) and examined their expression through RNA-Seq experiments (Table 2). CNC1560 and RHO104 both contain predicted Rim101 binding sites in their promoters, and both exhibited Rim101-dependent alterations in expression. Additionally, the CDC420 gene contains a Rim101 binding site in its promoter, while CDC42 does not. No alteration in the expression of CDC420 was observed in the rim101Δ mutant, although titan cell production in the wild-type strain was also not observed under the RNA-Seq conditions, suggesting that Rim101 binding may not be the only signal leading to alterations in CDC420 expression. Taken together, these data suggest that titan cell formation is regulated by Rim101 and acts through multiple effectors controlling aspects of cell morphogenesis.
Table 2.
Changes in gene expression in the rim101Δ mutant
| Gene | Function | Rim101 binding site | Fold change in expression |
|---|---|---|---|
| CNC1560 | GTPase-activating protein | Yes | +4.7 |
| PCL103 | G1 cyclin E | No | −17.7 |
| GAP1 | GTPase-activating protein | Yes | 0.0 |
| RHO104 | Rho-GTPase | Yes | −3.5 |
| CDC420 | GTPase | Yes | +1.3 |
| CDC42 | GTPase | No | +1.4 |
| GPA1 | Gα protein | Yes | +2.4 |
| CAC1 | Adenylyl cyclase | Yes | +1.5 |
| PKA1 | Protein kinase A | Yes | 0.0 |
| PKR1 | PKA regulatory subunit | No | −1.6 |
DISCUSSION
Titan cells play an important role in the initial pulmonary phase of C. neoformans infections. The morphological transition from yeast cell to titan cell can be induced by several conditions. We showed previously that titan cell formation in both mating types results from signals in the host lung environment (37). Titan cell production can also be stimulated in vitro by spent media from macrophage cell lines (37). Recent studies have shown that treatment of cryptococcal cells grown in vitro with phosphatidylcholine can stimulate cell enlargement, suggesting that phospholipids may be at least one component of the host-derived signal (8). We also demonstrated that cryptococcal titan cell formation is enhanced in mating type a cells by activation of the Ste3a pheromone receptor in response to the opposite mating type. Here we further explore the mating type-dependent and mating type-independent signals inducing titan cells.
We have shown that the Gpr5 receptor regulates titan cell production in response to the host lung environment. Deletion of the Gpr5 receptor significantly decreased titan cell formation during infection. The gpr5Δ mutant exhibits no defects in other known virulence factors, suggesting that signaling through this receptor specifically targets titan cell production. Interestingly, Gpr5 has high sequence homology to the methionine-sensing Gpr4 receptor (49). Deletion of GPR4 had no effect on titan cell formation, and alterations in amino acid prevalence did not stimulate titan cell formation in vitro. Similarly, we observed no difference in cell enlargement between the wild type and the gpr5Δ mutant in response to phosphatidylcholine treatment in vitro. Thus, the ligand for Gpr5 has not yet been defined. The observation that pheromone signaling, phospholipids, and another signal recognized by Gpr5 can independently stimulate titan cell formation suggests that additional, as yet uncharacterized receptors and signals are involved in titan cell formation under various conditions.
Our prior observations revealed that infection with both C. neoformans mating types resulted in enhanced titan cell formation, suggesting that the pheromone response pathway might play a role in this process (37). However, our recent data do not support a simple model of pheromone pathway activation leading to increased titan cells. For example, only mating type a cells undergo enhanced titan cell formation in response to the mating partner. Also, mutations in downstream components of the pheromone response pathway do not affect titan cell induction in the setting of a lung infection. Moreover, our data suggest that the pheromone receptors may interact with distinct downstream effectors in the different mating types. The Ste3a pheromone receptor did not interact in the yeast two-hybrid system with Gpa2 or Gpa3, the Gα proteins that mediate pheromone response in mating type α cells. Instead, Ste3a demonstrates in vitro interactions with the Gα protein Gpa1. The interaction between Ste3a and Gpa1 was transient and less stable than interactions between Ste3α and Gpa2 or Gpa3. These data suggest that Ste3a interacts only weakly with Gα proteins in our yeast two-hybrid assays. Thus, we cannot eliminate the possibility that Ste3a is able to interact with Gpa2 and/or Gpa3 under native conditions or that additional cofactors not present in the yeast two-hybrid assay are required to stabilize interactions between Ste3a and Gpa2 or Gpa3. Yet the interactions between Ste3a and Gpa1 suggest very different signaling paradigms for the two mating types. Also, dominant, activating mutations in Gpa1 result in striking increases in levels of titan cell formation in mating type a cells only. Together, these data likely explain the mating type a-specific enhancement of titan cell formation in mixed infections.
It is also clear that mating type α and mating type a cells have very different morphological responses in other settings. For example, in confrontation assays between serotype D strains, mating type α cells primarily undergo filamentous changes in response to a mating type a partner; in contrast, mating type a cells primarily undergo cell enlargement when confronted with the α mating partner (43). Therefore, the morphological response in mating is quite distinct for the two C. neoformans mating types. Here we observed a similar divergence in the morphological response to the mating partner in titan cell formation. Mating type a cells undergo accelerated titan cell formation in vivo in the presence of mating type α cells; mating type α cells do not. Therefore, it is plausible that mating type-specific signaling rewiring has occurred to allow such a different cellular response between mating type α and mating type a cells in a number of settings.
These data raise many questions regarding current signaling models for C. neoformans. For instance, if the primary role of Gpa1 is simply to activate cAMP/PKA signaling, why does this protein not activate titan cell formation in both mating types? How have conserved pheromone receptors adapted to interact with distinct Gα proteins to mediate very different downstream processes—pheromone recognition for both Ste3a and Ste3α, and titan cell induction only for Ste3a? It is clear that these signaling proteins often act in multiprotein complexes. Therefore, it is possible that downstream effectors of titan cell formation are activated only by Ste3a and Gpa1 in a complex, and not merely by accentuation of local cAMP/PKA activity. Additionally, mating type a and α cells may have different levels of basal Gpa1 activity. If mating type α cells are already utilizing maximal Gpa1 activity in vivo, the addition of a constitutively active form of Gpa1 will not affect the level of downstream signaling.
Our yeast two-hybrid data also showed no interactions between Gpr5 and any of the three cryptococcal Gα proteins. As with Ste3a, the interactions between Gpr5 and the Gα proteins may be transient and not detectable by use of our yeast two-hybrid system. Alternatively, interactions between Gpr5 and the Gα protein may require a multiprotein complex to stabilize the interaction. We were able to detect interactions between Gpr4 and Gpr5 in our system, showing that Gpr5 is capable of forming multiprotein complexes.
Gpa1 is a major regulator of the PKA pathway (1, 2, 16, 41). The PKA pathway regulates many virulence factors in C. neoformans, including capsule and melanin (2, 16, 41). The role of cAMP signaling in titan cell formation is strongly supported by the enhanced morphological changes induced by dominant active Gpa1. The involvement of this pathway is further supported by the striking role of the PKA-responsive transcription factor Rim101. Analysis of other cAMP pathway components is currently limited by the poor viability of these strains in vivo, the site of optimal titan cell induction. Emerging in vitro systems for titan cell formation will hopefully allow us to address the role of the cAMP pathway in this process more extensively.
Downstream of the PKA pathway is the transcription factor Rim101, which is well known to regulate morphology changes and virulence in other fungal species (4, 14, 15, 50). For example, in C. albicans, Rim101 regulates the yeast-to-hypha switch in response to pH changes and is critical to pathogenesis (14, 15). Our data show that Rim101 is the major regulator of titan cell production downstream of the PKA pathway. Deletion of the RIM101 gene abolishes titan cell production. The rim101Δ mutant had a more severe defect in titan cell production than the gpr5Δ mutant, suggesting that Gpr5 is likely only one of many signals acting through Rim101. Rim101 activity in several fungi, including Ustilago maydis, C. albicans, and C. neoformans, is regulated primarily by changes in pH (3, 6, 14, 15, 38). At alkaline pHs, Rim101 directs the expression of genes encoding cell wall proteins, proteases, and iron acquisition proteins, in addition to mediating morphological changes in response to pH (3, 4, 14, 15, 38). No effect of pH alone on titan cell formation in vitro has been observed (L. H. Okagaki and K. Nielsen, unpublished data), but it remains unclear whether pH regulation of Rim101 in vivo plays a role in titan cell production. Several pathways are known to regulate Rim101 activity. In C. albicans, Rim101 can be activated by a number of upstream pathways, including pH sensing by membrane proteins such as Dgf16p (6), as well as the ESCRT pathway (4, 47, 48). In C. neoformans, Rim101 has been shown to be regulated in a PKA-dependent manner (38). In addition to changes in RIM101 gene expression, Rim101 activity is regulated posttranslationally (14, 15, 18, 30, 38). Thus, it seems likely that multiple, as yet uncharacterized signals modulate the activity of Rim101 in vivo to promote titan cell production.
Rim101 has numerous direct and indirect downstream targets, and it will be important to determine which targets are the most important effectors for the formation of titan cells. We have begun an initial evaluation of potential effectors of titan cell formation by using existing mutant collections. Our first selection of mutants was biased toward those with predicted roles in morphogenesis and genome duplication, given the known phenotypic changes of titan cells from the wild type. This allowed us to test a focused group of strains in the complicated in vivo induction system. Using this method, we have identified genes whose products either enhance or suppress titan cell formation in vivo, including the cell cycle regulators Cdc420 and Pcl103 and the GTPase-activating proteins Rho104, Cnc1560, and Gap1, suggesting possible mechanisms for the cell enlargement and ploidy increases associated with titan cells. Many of these genes, as well as additional cyclins, DNA synthases, and other cell cycle-regulatory genes that have not been examined yet for their effects on titan cell production, are either directly or indirectly regulated by Rim101 (38). Although some of the gene promoters have putative Rim101 binding sites, this level of analysis is not sufficient to truly define direct targets of this transcription factor. Moreover, it is clear from the rim101 comparative transcriptional profiling experiments, and from the effects of these putative effector proteins on titan cell formation in vivo, that merely implicating Rim101 in the control of these proteins is overly simplistic. For example, the cell cycle regulator Pcl103 exhibited a 17.7-fold decrease in gene expression in the rim101Δ mutant from that in the wild type, suggesting that it might coregulate titan cell transitions with Rim101. However, in contrast to that of Rim101, deletion of Pcl103 dramatically increased titan cell formation.
In Saccharomyces cerevisiae, Pcl1 is a critical G1 cyclin that is known to interact with cell polarization proteins and cell cycle regulators, including Cdc42 (33, 52). Additionally, protein localization studies have shown that Pcl1 is important for the proper localization of the bud neck protein Bni4 (33, 52), a critical step in the cell cycle. In C. neoformans, the Pcl1 homologue Pcl103 may play a similar role, directing cell polarization during budding. Downregulation of Pcl103 may limit budding. Reduced budding may lengthen the G1 phase or alter the cell cycle, resulting in endoreplication and increased ploidy. Endoreplication, an altered cell cycle that lacks the M phase, in S. cerevisiae results in genome duplication without cell division and generates enlarged cells with increased ploidy (27) similar to that observed in C. neoformans titan cells.
C. neoformans contains multiple paralogues of several proteins that are critical for cell cycle regulation and the stress response (5, 46). Cdc420 is a paralogue of Cdc42, which regulates the cell cycle by directing cell polarity (5, 42). The duplicated CDC42 genes are unique to Cryptococcus. However, before we defined its role in titan cell formation, Cdc420 did not have a clear, unique role in C. neoformans physiology (5). In addition to Cdc42/Cdc420, C. neoformans also has paralogues of Ras and Rac, which are members of stress response pathways involved in morphological transitions. The duplicated RAC genes are also unique to C. neoformans, suggesting that they might be needed for an altered developmental stage of the cell cycle in response to stress, such as titan cell formation.
Taken together, our studies develop the foundation for a signal transduction pathway leading to titan cell production. Titan cell production can be regulated by the Ste3a and Gpr5 receptors, yet other receptors that might sense pH, iron limitation, and phospholipids also likely play a role in transducing signals that either stimulate or repress titan cell formation. The differences in titan cell production between mating type a and α cells during coinfection is likely due to rewiring of the mating pathway between the two cell types and/or utilization of the Ste3a receptor for purposes other than mating. Titan cell signaling is coregulated with other cryptococcal virulence factors via the PKA pathway, suggesting that titan cell formation is a novel virulence factor. Understanding of the basic signal transduction pathway leading to titan cell formation will facilitate further exploration of the downstream changes in gene expression that modulate titan cell formation and will allow further exploration of the role this virulence factor plays in the pathogenesis of C. neoformans.
Supplementary Material
ACKNOWLEDGMENTS
We thank Joe Heitman, Hiten Madhani, Christina Hull, and Xiaorong Lin for providing cryptococcal strains for our studies.
This work was supported by NIH grants AI08027, AI070152, and AI089244 to K.N., AI050158 and AI074677 to J.A.A., and UMDNJ foundation grant PC38-11 to C.X. L.H.O. was also supported by the Dennis W. Watson Fellowship, Department of Microbiology, University of Minnesota, and T.R.O. by an American Heart Association MAA Spring 09 Predoctoral Fellowship (09PRE2010039).
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 5 August 2011.
REFERENCES
- 1. Alspaugh J. A., Perfect J. R., Heitman J. 1997. Cryptococcus neoformans mating and virulence are regulated by the G-protein α subunit GPA1 and cAMP. Genes Dev. 11:3206–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alspaugh J. A., et al. 2002. Adenylyl cyclase functions downstream of the Gα protein Gpa1 and controls mating and pathogenicity of Cryptococcus neoformans. Eukaryot. Cell 1:75–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arechiga-Carvajal E. T., Ruiz-Herrera J. 2005. The RIM101/pacC homologue from the basidiomycete Ustilago maydis is functional in multiple pH-sensitive phenomena. Eukaryot. Cell 4:999–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baek Y. U., Martin S. J., Davis D. A. 2006. Evidence for novel pH-dependent regulation of Candida albicans Rim101, a direct transcriptional repressor of the cell wall β-glycosidase Phr2. Eukaryot. Cell 5:1550–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ballou E. R., Nichols C. B., Miglia K. J., Kozubowski L., Alspaugh J. A. 2010. Two CDC42 paralogues modulate Cryptococcus neoformans thermotolerance and morphogenesis under host physiological conditions. Mol. Microbiol. 75:763–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barwell K. J., Boysen J. H., Xu W., Mitchell A. P. 2005. Relationship of DFG16 to the Rim101p pH response pathway in Saccharomyces cerevisiae and Candida albicans. Eukaryot. Cell 4:890–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Casadevall A., Perfect J. R. 1998. Cryptococcus neoformans. ASM Press, Washington, DC [Google Scholar]
- 8. Chrisman C. J., Albuquerque P., Guimaraes A. J., Nieves E., Casadevall A. 2011. Phospholipids trigger Cryptococcus neoformans capsular enlargement during interactions with amoebae and macrophages. PLoS Pathog. 7:e1002047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cramer K. L., Gerrald Q. D., Nichols C. B., Price M. S., Alspaugh J. A. 2006. Transcription factor Nrg1 mediates capsule formation, stress response, and pathogenesis in Cryptococcus neoformans. Eukaryot. Cell 5:1147–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Davidson R. C., et al. 2002. A PCR-based strategy to generate integrative targeting alleles with large regions of homology. Microbiology 148:2607–2615 [DOI] [PubMed] [Google Scholar]
- 11. Davidson R. C., et al. 2000. Gene disruption by biolistic transformation in serotype D strains of Cryptococcus neoformans. Fungal Genet. Biol. 29:38–48 [DOI] [PubMed] [Google Scholar]
- 12. Davidson R. C., Moore T. D., Odom A. R., Heitman J. 2000. Characterization of the MFα pheromone of the human fungal pathogen Cryptococcus neoformans. Mol. Microbiol. 38:1017–1026 [DOI] [PubMed] [Google Scholar]
- 13. Davidson R. C., Nichols C. B., Cox G. M., Perfect J. R., Heitman J. 2003. A MAP kinase cascade composed of cell type specific and non-specific elements controls mating and differentiation of the fungal pathogen Cryptococcus neoformans. Mol. Microbiol. 49:469–485 [DOI] [PubMed] [Google Scholar]
- 14. Davis D., Edwards J. E., Jr., Mitchell A. P., Ibrahim A. S. 2000. Candida albicans RIM101 pH response pathway is required for host-pathogen interactions. Infect. Immun. 68:5953–5959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Davis D., Wilson R. B., Mitchell A. P. 2000. RIM101-dependent and -independent pathways govern pH responses in Candida albicans. Mol. Cell. Biol. 20:971–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. D'Souza C. A., et al. 2001. Cyclic AMP-dependent protein kinase controls virulence of the fungal pathogen Cryptococcus neoformans. Mol. Cell. Biol. 21:3179–3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fraser J. A., Subaran R. L., Nichols C. B., Heitman J. 2003. Recapitulation of the sexual cycle of the primary fungal pathogen Cryptococcus neoformans var. gattii: implications for an outbreak on Vancouver Island, Canada. Eukaryot. Cell 2:1036–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Futai E., et al. 1999. The protease activity of a calpain-like cysteine protease in Saccharomyces cerevisiae is required for alkaline adaptation and sporulation. Mol. Gen. Genet. 260:559–568 [DOI] [PubMed] [Google Scholar]
- 19. Heitman J., Kozel T. R., Kwon-Chung K. J., Perfect J. R., Casadevall A. (ed.). 2010. Cryptococcus: from human pathogen to model yeast. ASM Press, Washington, DC [Google Scholar]
- 20. Hicks J. K., Bahn Y. S., Heitman J. 2005. Pde1 phosphodiesterase modulates cyclic AMP levels through a protein kinase A-mediated negative feedback loop in Cryptococcus neoformans. Eukaryot. Cell 4:1971–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hsueh Y. P., Idnurm A., Heitman J. 2006. Recombination hotspots flank the Cryptococcus mating-type locus: implications for the evolution of a fungal sex chromosome. PLoS Genet. 2:e184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hsueh Y. P., Xue C., Heitman J. 2007. G protein signaling governing cell fate decisions involves opposing Gα subunits in Cryptococcus neoformans. Mol. Biol. Cell 18:3237–3249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hsueh Y. P., Xue C., Heitman J. 2009. A constitutively active GPCR governs morphogenic transitions in Cryptococcus neoformans. EMBO J. 28:1220–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hull C. M., Davidson R. C., Heitman J. 2002. Cell identity and sexual development in Cryptococcus neoformans are controlled by the mating-type-specific homeodomain protein Sxi1α. Genes Dev. 16:3046–3060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24a. Iyer K., et al. 2005. Utilizing the split-ubiquitin membrane yeast two-hybrid system to identify protein-protein interactions of integral membrane proteins. Sci. STKE 275:3. [DOI] [PubMed] [Google Scholar]
- 25. Jung K. W., Kim S. Y., Okagaki L. H., Nielsen K., Bahn Y. S. 2011. Ste50 adaptor protein governs sexual differentiation of Cryptococcus neoformans via the pheromone-response MAPK signaling pathway. Fungal Genet. Biol. 48:154–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Karos M., et al. 2000. Mapping of the Cryptococcus neoformans MATα locus: presence of mating type-specific mitogen-activated protein kinase cascade homologs. J. Bacteriol. 182:6222–6227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kondorosi E., Roudier F., Gendreau E. 2000. Plant cell-size control: growing by ploidy? Curr. Opin. Plant Biol. 3:488–492 [DOI] [PubMed] [Google Scholar]
- 28. Langmead B., Trapnell C., Pop M., Salzberg S. L. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li L., et al. 2007. Canonical heterotrimeric G proteins regulating mating and virulence of Cryptococcus neoformans. Mol. Biol. Cell 18:4201–4209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li M., Martin S. J., Bruno V. M., Mitchell A. P., Davis D. A. 2004. Candida albicans Rim13p, a protease required for Rim101p processing at acidic and alkaline pHs. Eukaryot. Cell 3:741–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lin X., Jackson J. C., Feretzaki M., Xue C., Heitman J. 2010. Transcription factors Mat2 and Znf2 operate cellular circuits orchestrating opposite- and same-sex mating in Cryptococcus neoformans. PLoS Genet. 6:e1000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu O. W., et al. 2008. Systematic genetic analysis of virulence in the human fungal pathogen Cryptococcus neoformans. Cell 135:174–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Moffat J., Andrews B. 2004. Late-G1 cyclin-CDK activity is essential for control of cell morphogenesis in budding yeast. Nat. Cell Biol. 6:59–66 [DOI] [PubMed] [Google Scholar]
- 34. Nichols C. B., Perfect Z. H., Alspaugh J. A. 2007. A Ras1-Cdc24 signal transduction pathway mediates thermotolerance in the fungal pathogen Cryptococcus neoformans. Mol. Microbiol. 63:1118–1130 [DOI] [PubMed] [Google Scholar]
- 35. Nielsen K., et al. 2005. Cryptococcus neoformans α strains preferentially disseminate to the central nervous system during coinfection. Infect. Immun. 73:4922–4933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nielsen K., et al. 2003. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and α isolates. Infect. Immun. 71:4831–4841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Okagaki L. H., et al. 2010. Cryptococcal cell morphology affects host cell interactions and pathogenicity. PLoS Pathog. 6:e1000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. O'Meara T. R., et al. 2010. Interaction of Cryptococcus neoformans Rim101 and protein kinase A regulates capsule. PLoS Pathog. 6:e1000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Reference deleted.
- 40. Park B. J., et al. 2009. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS 23:525–530 [DOI] [PubMed] [Google Scholar]
- 41. Pukkila-Worley R., et al. 2005. Transcriptional network of multiple capsule and melanin genes governed by the Cryptococcus neoformans cyclic AMP cascade. Eukaryot. Cell 4:190–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rincon S., Coll P. M., Perez P. 2007. Spatial regulation of Cdc42 during cytokinesis. Cell Cycle 6:1687–1691 [DOI] [PubMed] [Google Scholar]
- 43. Shen W. C., Davidson R. C., Cox G. M., Heitman J. 2002. Pheromones stimulate mating and differentiation via paracrine and autocrine signaling in Cryptococcus neoformans. Eukaryot. Cell 1:366–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Trapnell C., Pachter L., Salzberg S. L. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25:1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Trapnell C., et al. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28:511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Waugh M. S., et al. 2002. Ras1 and Ras2 contribute shared and unique roles in physiology and virulence of Cryptococcus neoformans. Microbiology 148:191–201 [DOI] [PubMed] [Google Scholar]
- 47. Wolf J. M., Davis D. A. 2010. Mutational analysis of Candida albicans SNF7 reveals genetically separable Rim101 and ESCRT functions and demonstrates divergence in bro1-domain protein interactions. Genetics 184:673–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wolf J. M., Johnson D. J., Chmielewski D., Davis D. A. 2010. The Candida albicans ESCRT pathway makes Rim101-dependent and -independent contributions to pathogenesis. Eukaryot. Cell 9:1203–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xue C., Bahn Y. S., Cox G. M., Heitman J. 2006. G protein-coupled receptor Gpr4 senses amino acids and activates the cAMP-PKA pathway in Cryptococcus neoformans. Mol. Biol. Cell 17:667–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yuan X., Mitchell B. M., Hua X., Davis D. A., Wilhelmus K. R. 2010. The RIM101 signal transduction pathway regulates Candida albicans virulence during experimental keratomycosis. Invest. Ophthalmol. Vis. Sci. 51:4668–4676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zaragoza O., et al. 2010. Fungal cell gigantism during mammalian infection. PLoS Pathog. 6:e1000945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zou J., et al. 2009. Regulation of cell polarity through phosphorylation of Bni4 by Pho85 G1 cyclin-dependent kinases in Saccharomyces cerevisiae. Mol. Biol. Cell 20:3239–3250 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.