Abstract
The role of fomites in infectious disease transmission relative to other exposure routes is difficult to discern due, in part, to the lack of information on the level and distribution of virus contamination on surfaces. Comparisons of studies intending to fill this gap are difficult because multiple different sampling methods are employed and authors rarely report their method's lower limit of detection. In the present study, we compare a subset of sampling methods identified from a literature review to demonstrate that sampling method significantly influences study outcomes. We then compare a subset of methods identified from the review to determine the most efficient methods for recovering virus from surfaces in a laboratory trial using MS2 bacteriophage as a model virus. Recoveries of infective MS2 and MS2 RNA are determined using both a plaque assay and quantitative reverse transcription-PCR, respectively. We conclude that the method that most effectively recovers virus from nonporous fomites uses polyester-tipped swabs prewetted in either one-quarter-strength Ringer's solution or saline solution. This method recovers a median fraction for infective MS2 of 0.40 and for MS2 RNA of 0.07. Use of the proposed method for virus recovery in future fomite sampling studies would provide opportunities to compare findings across multiple studies.
INTRODUCTION
Preclusion of infection is the most effective method to combat the respiratory and gastrointestinal diseases that cause over 6 million annual deaths worldwide (12, 59). Successful interventions to reduce disease burden include hand and environmental hygiene (9, 76), but the impact of these interventions is difficult to quantify, as the importance of contact with contaminated surfaces, or fomites, relative to other transmission routes is uncertain (17, 62).
Evidence of the importance of fomites comes from both laboratory and field studies. Laboratory studies have demonstrated that the handling of either artificially or naturally contaminated fomites by susceptible hosts indoors results in subsequent infection (41, 43). Additionally, virus can be transferred between hands and fomites on contact and survive on fomites for hours or days (1, 8, 72). As such, it is not surprising that fomites, such as carpets (29, 65), towels, and medication cart items (60) have been implicated as the primary cause of multiple outbreaks. Environmental hygiene intended to mitigate fomite-mediated transmission has been shown to significantly reduce illness-related absences in classrooms (18). Despite this evidence, questions remain regarding both the efficacy of fomite-mediated transmission relative to other exposure routes (5, 46) and the likelihood of virus transfer from fomites to hosts (66).
To better understand the role of fomites in disease transmission, characterization of the level and distribution of virus on surfaces is required. Surface contamination is most often described by the positivity rate, defined as the fraction of total samples collected on which the organism is detectable (19). However, the positivity rate does not provide an indication of infection risk, which depends on exposure magnitude (42). To estimate the risk of infection, information about the quantity of virus on the surface is required. Virus density, expressed as the number of virions or number of virus equivalents per unit surface area, has been measured only in a few studies (10, 68, 73). Both positivity rate and virus density are influenced by the sampling method and detection assay: more sensitive sampling methods and detection assays will yield increases in positivity rates and higher measured concentrations even though the actual level of virus contamination may be unchanged. Use of a sensitive, standard method would limit bias introduced by the use of multiple different sampling methods.
Three previous studies have compared or evaluated virus surface sampling methods and suggested that implement type (the tool used to collect the sample, such as a swab) and eluent type (the liquid used to aid in removal, such as saline solution) significantly influence virus recovery. Carducci et al. (20) recovered a greater fraction of hepatitis C virus from a seeded surface using beef extract than using bovine serum albumin when swabbing with a cotton-tipped applicator. The study demonstrated that eluent type can significantly impact virus recovery from surfaces. Similarly, Taku et al. (79) demonstrated the impact of implement type by comparing calicivirus recovery from food surfaces for four sampling methods. Rinsing a surface in 0.05 M glycine buffer, rubbing with a cell scraper, and then aspirating the buffer was recommended over (i) rinsing the surface in buffer and then aspirating, (ii) swabbing the surface with a cotton-tipped applicator, or (iii) swabbing the surface with a nylon filter. However, Taku et al.'s (79) recommended method is not easily adapted to the geometry of most fomites. Finally, Scherer et al. (75) evaluated one implement-eluent combination (cotton-tipped swabs wetted in phosphate-buffered saline [PBS]) for efficacy of rotavirus and norovirus RNA recovery and reported a mean recovery of 7 to 53%, dependent on surface type and initial seeded titer. However, they did not compare the method to any other methods. Further research is needed to refine implement and eluent choice for sampling fomites to maximize virus recovery.
In the present study, we reviewed the literature on virus sampling of fomites and used a laboratory-based trial to compare methods of virus detection on surfaces. We identified, summarized, and analyzed 59 articles that include unique data sets on virus detection on surfaces. A subset of the sampling methods identified from the meta-analysis were compared in a laboratory-based study of the removal of bacteriophage MS2, as measured using both culture-dependent and culture-independent (quantitative reverse-transcription PCR [qRT-PCR]) methods, from plastic and stainless steel surfaces. Based on both the literature review and experimental results, we identified the implement and eluent combinations that most effectively remove infective virus and virus RNA from nonporous fomites.
MATERIALS AND METHODS
Review of virus surface sampling literature.
Literature on virus surface sampling was identified by searching PubMed and Embase databases for records added on or before 28 June 2011 and reviewing the references of the identified studies. For further details, see the supplemental material.
For analysis, data from the articles were separated into data sets according to the virus, the presence/absence of a clinically infected individual, and the location of sampling. If the authors included clinical or food samples, those samples were removed from data analysis. Further details on the criteria of the separation of articles into data sets are presented in the supplemental material. In summary, 98 data sets from 59 articles were obtained.
The positivity rate was determined, as the outcome variable, for each data set. The positivity rate was the only feasible outcome variable, as most of the studies identified report only the presence/absence of virus on surfaces. Only a fraction (9 of 98, or 9%) reported quantitative data. To allow for logit transformation, the positivity rate for studies that detected the virus on none or all of the samples was adjusted to a detection limit of 1/n or (n − 1)/n, respectively, where n is the study's total number of samples collected. The positivity rate is an inherently biased outcome variable because the lower limit of detection (LLOD) varies across studies for reasons described previously. As few studies (30%) reported either the quantitative concentration of the virus or the LLOD of the sampling method, the positivity rate could not be adjusted to account for the bias.
We assessed the influence of the implement and eluent used to collect and analyze the samples on the positivity rate. Similar implements and eluents were grouped for data analysis. Polyester and Dacron swabs were both categorized as polyester. The eluent used was categorized into one of four groups: media (defined here as any eluent with a carbon source and includes Amies medium, beef extract, brain heart infusion broth, Letheen broth, minimum essential medium, RPMI 1640, and tryptose phosphate broth with 0.5% gelatin), saline (defined as any isotonic eluent without a carbon source and includes phosphate-buffered saline, 0.8% saline, and Ringer's solution), water, or unreported. Additives and constituents of eluents, such as antibiotics, were ignored for data analysis to avoid overparameterization.
Statistics.
All statistical analyses were performed using R (version 2.9.0; R Foundation for Statistical Computing, Vienna, Austria). The normality of transformed data was assessed using the Kolmogorov-Smirnoff test. Two bivariate linear models were used to determine the statistical significance of implement and eluent, separately, on the transformed positivity rate; the positivity rate was weighted by the total number of samples in each study.
Laboratory-based surface sampling method comparison.
In a laboratory-based trial, we compared fractions of virus recovered from surfaces for a subset of the implement and eluent choices identified in the literature.
Virus and preparation of inoculum.
MS2 bacteriophage was obtained from the American Type Culture Collection (ATCC). MS2 (ATCC 15597-B1) is a positive-sense single-stranded RNA (ssRNA) virus with an icosahedral, tailless capsid approximately 27 nm in diameter. The isoelectric point (pI) of MS2 is 3.9. MS2 was chosen because of its prior use as a surrogate for human viruses, such as norovirus (26), and the availability of plaque assay and qRT-PCR methods to enumerate both infective phage and copies of nucleic acids (63, 80). Escherichia coli HS(pFamp)R (ATCC 700891) was used to propagate and enumerate viable MS2, measured as the number of PFU.
The inoculum was prepared using the method described by Pecson et al. (67). In brief, the log-phase E. coli host was inoculated with MS2 at a multiplicity of infection of 0.1 and incubated at 37°C for 4 to 6 h. Chloroform was added to complete cell lysis, and the sample was clarified by centrifugation for 15 min at 4,000 × g. The supernatant was stored at 4°C overnight in 10% polyethylene glycol (PEG 6000; Sigma-Aldrich, St. Louis, MO) and 0.5 M NaCl. The solution was centrifuged at 7,000 × g for 45 min, and the pellet was resuspended in dilution buffer (5 mM NaH2PO4, 10 mM NaCl, pH 7.4). A chloroform-only extraction was used to remove the remaining polyethylene glycol (PEG), and the sample was concentrated in an Amicon Ultra centrifugal filter device (100,000 nominal molecular-weight limit; Millipore, Billerica, MA), triple washed with dilution buffer, and then filtered through a 0.1-μm-pore-size polyvinylidene difluoride filter (Millipore, Billerica, MA). The propagated virus was then enumerated using the double-agar-layer method and diluted in the dilution buffer to a virus stock of 1 × 106 PFU/ml. Immediately before being seeded on the surface, the virus stock was mixed with tryptic soy broth (TSB) to form a 50% solution.
Implements and eluents tested.
The implements tested were cotton-tipped (Thermo Fisher Scientific, Waltham, MA) and polyester-tipped (Thermo Fisher Scientific, Waltham, MA) swabs as well as antistatic cloth (Bel-Art Products, Pequannock, NJ). The antistatic cloth used in the study is a microfiber cloth wet by the manufacturer with 1,1-difluoroethane. Prior to sampling, the cloth was cut into single-ply square swatches of approximately 9 cm2 and stored in sterile, sealed 5-ml containers for 5 to 10 days.
The eluents tested include 0.85% saline, one-quarter-strength Ringer's solution (EMD Chemicals, Inc., Gibbstown, NJ), and viral transport media (Copan Diagnostics, Murietta, CA). A fourth eluent, termed acid/base, was added to assess an eluent adapted from a method to concentrate virus from environmental water samples (48). The acid/base eluent relies on knowledge of the virus surface charge to improve recovery from fomites. Briefly, a weakly acidic (0.5 mM dihydrogen sulfate, H2SO4) eluent is used to wet the implement prior to sampling. Viruses with low isoelectric points adsorb to negatively charged surfaces (like cotton) under acidic conditions (48). After sampling, the implement is placed into a weakly basic (1 mM sodium hydroxide, pH 10.5 to 10.8) eluent which reverses the surface charge of the virus to elute the virus from the implement. The implements/eluents were chosen based on a subjective analysis of the literature. Specifically, we included implements/eluents that were either commonly used (e.g., cotton and saline) or had the potential to improve virus recovery (e.g., antistatic cloth and acid/base).
Surfaces tested.
To determine the method most effective in removing virus from surfaces, we compared recovery from both high-temperature polyvinyl chloride (PVC) plastic (part no. 8748K21) and type 304 stainless steel with a mirror-like finish (part no. 9785K11), both obtained from McMaster-Carr (Santa Fe Springs, CA). Many of the surfaces identified in the literature review that were frequently contaminated (e.g., door knobs, faucet handles, drains, medical instruments, toys, playmats, computer parts, and telephones) were composed of plastic or metal. PVC plastic and stainless steel were chosen as representative samples, as it was infeasible to test every potential type of surface material. Between 8 and 10 replicates for each eluent and implement combination were tested on both surfaces. In total, 230 samples were collected (3 implements, 4 eluents, 2 surfaces, and 8 to 10 replicates). All 230 samples were tested using the double-agar-layer plaque assay method and a subset (213 samples) using qRT-PCR.
Study design.
For both plastic and stainless steel surfaces, a 5-μl inoculum of MS2 stock was seeded in the center of 120 5-cm by 5-cm surface swatches, resulting in a seeded surface density of 3.7 log10 PFU per swatch with a standard deviation of 0.13 log10 PFU per swatch, equivalent to 5.4 log10 RNA target copies per swatch with a standard deviation of 0.16 log10 RNA target copies per swatch. An additional 8 surface swatches were seeded with bacteriophage-free TSB to act as negative controls to confirm that there was no cross-contamination of samples (e.g., due to aerosolization and deposition of MS2). The seeded aliquot was dried for 45 ± 1 min under ambient conditions (temperature of 20 to 22°C and relative humidity of 45 to 60%, determined by a thermometer and hygrometer; Springfield Precision Instruments, Wood Ridge, NJ), outside a laminar flow hood. The order of implement and eluent combinations used to recover MS2 from the surfaces was randomized prior to the start of the study.
Centrifuge tubes (15 ml; BD Biosciences, San Jose, CA) were filled with 1.5 ml of 0.85% saline, viral transport media, Ringer's solution, or 1 mM sodium hydroxide. To sample, the polyester- or cotton-tipped swabs were wetted in the eluent (or in 0.5 mM dihydrogen sulfate, for acid/base) and then rubbed with moderate and consistent pressure across the surface first horizontally, then vertically, and then diagonally for 10 s. The swab was then placed into the centrifuge tube, and the tube was capped and stored on ice for 4 h to mimic typical transportation time. Antistatic cloth, otherwise following the same procedure, was not wetted prior to sampling.
After storage, the samples were vortexed for 60 s. An aliquot of 100 μl was used to assay the samples for infective MS2 using the double-agar-layer method (80). In brief, each sample was hand mixed with 4 ml of 0.7% tryptic soy agar (BD Biosciences, San Jose, CA) with 0.0015% ampicillin sodium salt (Sigma-Aldrich, St. Louis, MO) and 0.0015% streptomycin sulfate (Sigma-Aldrich, St. Louis, MO) at 42°C for 5 to 10 s. The sample was then poured into a 100- by 15-mm petri dish (Thermo Fisher Scientific, Waltham, MA) with a solidified base of 1.5% tryptic soy agar with the same antibiotic concentrations. After the sample solidified, it was incubated inverted at 37°C for 16 to 24 h; the plaques were then counted. One aliquot was assayed using the double-agar-layer method per sample. The remaining sample was stored at −80°C.
qRT-PCR.
Viral recovery from the samples was also determined using qRT-PCR. Two hundred thirteen samples, 6 to 10 samples for each combination of implement and eluent, were assayed using qRT-PCR. RNA was extracted and quantified from 200 μl of sample volume, after storage at −80°C for 15 to 180 days following sample collection.
To extract viral RNA, we used the Invitrogen PureLink viral RNA/DNA extraction kit (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions using 200-μl samples eluted in 20 μl of DNase/RNase-free water. Genomic RNA was enumerated using qRT-PCR. The reagents, primers, and cycling conditions of O'Connell et al. (63) for a 25-μl reaction with a 5-μl template were used. The location in the MS2 genome of the primers, probe, and target (the sequence of the qRT-PCR amplicon) is the RNA replicase β chain. The forward primer (5′-GCTCTGAGAGCGGCTCTATTG-3′), reverse primer (5′-CGTTATAGCGGACCGCGT-3′), and probe (5′-[FAM]-CCGAGACCAATGTGCGCCGTG-[TAMRA]-3′) were obtained from Eurofins MWG operon (Huntsville, AL) (63). Extracts were stored at −80°C for less than 48 h prior to qRT-PCR, which was performed using a StepOnePlus real-time PCR system (Applied Biosystems, Carlsbad, CA). All samples and standards were run in triplicate.
RNA standards were created from total genomic RNA extracted without the aid of carrier RNA from a high titer of purified MS2. The extracted RNA was enumerated as 20 ng/μl using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA) and diluted to three standards, at 10-fold dilution, between 1 fg/μl (500 genome equivalents/μl), and 100 fg/μl (50,000 genome equivalents/μl). A genome of 3,569 nucleotides with an average molecular mass of 330 Da per nucleotide was assumed to convert the RNA concentration to genome equivalents (63). The threshold cycles (CT) for the standards from 12 sets were combined, and linear regression was used to create a pooled master curve relating the CT to genome equivalents. The master curve had an R2 of 0.987 and an efficiency of 100.8%.
Statistics.
Descriptive statistics (mean, median, and standard deviation) are provided for the recovery of infective MS2 from each surface using each method. The fraction of MS2 recovered was defined as the number of MS2 virions recovered (estimated using the number of either infective MS2 or MS2 RNA target copies) divided by the number of MS2 virions initially seeded. Significance was determined if the P value was <0.05. To assess the efficacy of implement and eluent choice, n-way analysis of variance (ANOVA) was used, where the fraction of recovered MS2 was the dependent variable and surface, implement, and eluent were the independent variables. The Kolmogorov-Smirnoff test was used to assess the normality of variables. As described in Results, variables were transformed as needed. Linear regression was used to model the relationship of the number of target copies estimated from qRT-PCR as a function of the number of infective MS2 virions. All statistics were performed using R (version 2.9.0; R Foundation for Statistical Computing, Vienna, Austria).
RESULTS
Literature review.
Fifty-nine relevant articles on sampling of surfaces for virus were identified and separated into 98 data sets. For further discussion of the identified articles, including the separation into data sets, implement, eluent, and assay used, positivity rate, and locale, see the supplemental methods and Tables S1 and S2 in the supplemental material. In summary, environmental contamination was assessed for 24 different etiologic agents, including causative agents of gastrointestinal, respiratory, blood-borne, and/or sexually transmitted diseases. The positivity rates of the studies, when logit transformed, were normally distributed (P = 0.87, Kolmogorov-Smirnov). In total, 9,686 samples were collected, with detectable virus on 1,855, for an overall positivity rate of 0.192.
Thirteen different eluents (excluding additives) were used in the 98 identified studies. In 7 (7%) of the studies, the authors did not identify the eluent. The authors of 45 of the studies (46%) used media, while 41 (42%) used a saline solution. Studies in which eluent was unreported were grouped into a “not reported” category and included in the linear model. The linear model demonstrated no significant influence of eluent category on the positivity rate, when the positivity rate was weighted by total samples collected (R2 = 0.04, P = 0.28). Table 1 provides a summary of the studies, aggregated by eluent type, and includes the number of samples collected, number with detectable virus, and fraction with detectable virus for each eluent.
Table 1.
Summary of the eluent types used in the reviewed articles, including categorization for analysis, associated number of samples collected, number of samples with detectable virus, and fraction of samples with detectable virus
| Eluent | No. of studies | No. of samples | No. of positive samples | Fraction positive | References |
|---|---|---|---|---|---|
| Media | 45 | 5,292 | 888 | 0.168 | 3, 4, 11, 13, 14, 18, 20, 21, 23, 24, 27, 31, 38, 39, 40, 41, 47, 50, 51, 66, 70, 73, 74, 77, 83, 84, 87 |
| Saline solutions | 41 | 3,757 | 783 | 0.208 | 6, 7, 10, 11, 15, 16, 19, 25, 30, 32, 33, 34, 35, 36, 49, 52, 53, 54, 55, 56, 57, 68, 78, 81, 82, 85, 86 |
| Water | 5 | 359 | 86 | 0.240 | 61, 71 |
| Not reported | 7 | 278 | 98 | 0.353 | 22, 37, 44, 58 |
| Total | 98 | 9,686 | 1,855 | 0.192 |
Four implement types were used in the studies. Studies in which the implement was unreported or reported as an unspecified swab type (18% of identified studies) were grouped into a “not reported” category and included in the linear model. A division of studies by implement type, including the number of studies for each, the total samples collected, the number with detectable virus, and the fraction with detectable virus for each implement type are provided in Table 2. Implement type explained 18% of the variation in the positivity rate (R2 = 0.18, P < 0.001) according to the linear model using the logit-transformed positivity rate weighted by total sample number as the dependent variable. Compared to that for cotton-tipped swabs, the positivity rate was significantly higher for polyester swabs (P = 0.006) and significantly lower for rayon-tipped swabs (P = 0.03). There was no significant difference in positivity between cotton-tipped swabs and antistatic cloths (P = 0.21), although antistatic cloths had the highest positivity rate (0.408), likely due to the small sample size (n = 2).
Table 2.
Summary of the implement types used in the reviewed articles, including the number of studies, total number of samples, the samples with detectable virus, and the fraction of samples with detectable virus
| Implement | No. of studies | No. of samples | No. of positive samples | Fraction positive | References |
|---|---|---|---|---|---|
| Cotton | 58 | 5,660 | 1,025 | 0.181 | 4, 6, 19, 20, 21, 23, 24, 25, 31, 32, 33, 34, 35, 36, 38, 39, 40, 41, 49, 50, 51, 52, 53, 61, 66, 68, 70, 77, 78, 82, 83, 84, 85, 86, 87 |
| Polyester | 16 | 2,110 | 609 | 0.289 | 7, 11, 13, 27, 30, 54, 56, 57, 73, 74 |
| Rayon | 4 | 571 | 36 | 0.063 | 10, 18, 47 |
| Antistatic | 2 | 125 | 51 | 0.408 | 15, 16 |
| Not reported | 18 | 1,220 | 134 | 0.110 | 3, 37, 44, 55, 58, 71, 81 |
| Total | 98 | 9,686 | 1,855 | 0.192 |
Laboratory-based recovery of infective MS2.
Results of the recovery of infective MS2 from stainless steel and plastic for each implement-eluent combination are provided in Table 3. The fraction of infective MS2 recovered for all experimental conditions ranged from <0.003 to 0.97. The mean, median, and interquartile range of fraction recovered for all experimental conditions were 0.29, 0.31, and 0.11 to 0.44. The recovery of infective MS2 was at or below the detection limit (0.003) in 40 of the 230 samples (17%). Most (n = 39) of the nondetects were obtained when using antistatic cloth. Due to the high percentage of nondetects, the distribution of the fraction recovered differed significantly from normal (Kolmogorov-Smirnoff test, P < 0.001).
Table 3.
Summary of the fraction of infective MS2 and MS2 RNA target copies recovered using each implement-eluent combination from stainless steel and plastic surfacesa
| Implement eluentb | Fraction of infective MS2 recovered from: |
Fraction of MS2 RNA target copies recovered from: |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Stainless steel |
Plastic |
Stainless steel |
Plastic |
|||||||||||||
| No. | μ | Median | σ | No. | μ | Median | σ | No. | μ | Median | σ | No. | μ | Median | σ | |
| Antistatic | ||||||||||||||||
| Saline | 10 | 0.15 | 0.13 | 0.15 | 9 | 0.01 | 0.003 | 0.01 | 9 | 0.70 | 0.40 | 0.81 | 8 | 0.22 | 0.12 | 0.28 |
| Ringer's | 10 | 0.16 | 0.10 | 0.18 | 9 | 0.03 | 0.003 | 0.08 | 8 | 0.23 | 0.20 | 0.10 | 8 | 0.07 | 0.04 | 0.07 |
| VTM | 10 | 0.10 | 0.07 | 0.12 | 10 | 0.01 | 0.003 | 0.01 | 9 | 0.34 | 0.28 | 0.27 | 8 | 0.15 | 0.03 | 0.21 |
| Acid/base | 10 | 0.23 | 0.24 | 0.14 | 8 | 0.003 | 0.003 | 0.001 | 10 | 0.99 | 0.42 | 1.47 | 6 | 0.08 | 0.07 | 0.07 |
| Cotton | ||||||||||||||||
| Saline | 10 | 0.38 | 0.38 | 0.15 | 9 | 0.39 | 0.45 | 0.16 | 10 | 0.27 | 0.14 | 0.34 | 7 | 0.13 | 0.08 | 0.19 |
| Ringer's | 10 | 0.33 | 0.34 | 0.11 | 10 | 0.54 | 0.56 | 0.16 | 10 | 0.05 | 0.05 | 0.03 | 10 | 0.07 | 0.07 | 0.04 |
| VTM | 10 | 0.35 | 0.38 | 0.13 | 10 | 0.36 | 0.34 | 0.07 | 10 | 0.18 | 0.20 | 0.11 | 9 | 0.17 | 0.18 | 0.07 |
| Acid/base | 10 | 0.32 | 0.32 | 0.12 | 9 | 0.37 | 0.33 | 0.15 | 10 | 0.27 | 0.07 | 0.05 | 9 | 0.07 | 0.08 | 0.04 |
| Polyester | ||||||||||||||||
| Saline | 10 | 0.39 | 0.38 | 0.17 | 9 | 0.39 | 0.41 | 0.12 | 10 | 0.09 | 0.05 | 0.09 | 8 | 0.07 | 0.06 | 0.06 |
| Ringer's | 10 | 0.39 | 0.38 | 0.13 | 9 | 0.59 | 0.56 | 0.21 | 10 | 0.09 | 0.08 | 0.04 | 9 | 1.41 | 0.10 | 2.96 |
| VTM | 10 | 0.29 | 0.30 | 0.13 | 9 | 0.39 | 0.37 | 0.13 | 8 | 0.12 | 0.11 | 0.07 | 9 | 0.32 | 0.09 | 0.52 |
| Acid/base | 10 | 0.39 | 0.38 | 0.17 | 9 | 0.48 | 0.48 | 0.11 | 10 | 0.07 | 0.07 | 0.03 | 8 | 0.06 | 0.05 | 0.04 |
The number of samples, mean (μ), median, and standard deviation (σ) are reported.
VTM, viral transport media.
The influence of independent variables (surface sampled, implement, and eluent) on the recovery of infective MS2 was determined using n-way ANOVA on ranked values. Based on the analysis, only the implement significantly influenced recovery (P < 0.001). Post hoc Tukey's pairwise comparison demonstrated that antistatic cloth, with an overall recovery fraction of 0.09, recovered significantly less infective MS2 than both polyester swabs (mean recovery fraction = 0.40, P < 0.001) and cotton swabs (mean recovery fraction = 0.38, P < 0.001). Polyester swab recovery and cotton swab recovery were not significantly different (P = 0.37).
Neither the surface sampled (P = 0.96) nor eluent type (P = 0.05) significantly influenced the fraction of infective MS2 recovered. The eluent type that recovered the largest fraction was one-quarter-strength Ringer's solution, with a mean fraction of 0.24, followed by saline (mean recovery fraction = 0.20) and acid/base (mean recovery fraction = 0.19). Viral transport media recovered the lowest fraction of virus (mean recovery fraction = 0.17).
The interaction effect of the implement and eluent combination was not significant (P = 0.48). The combination of polyester swab and Ringer's resulted in the largest mean fraction recovered (mean recovery fraction = 0.48), though it was not significantly different (P > 0.05) from polyester and any other eluent or Ringer's and any other implement besides antistatic cloth, based on Tukey's post hoc tests.
LLOD of infective MS2.
The median and interquartile range of the fractional recovery of infective MS2 using polyester/Ringer's were 0.45 and 0.33 to 0.58 respectively. Using the mean and interquartile range, along with the assumption that the bacteriophage double-agar-layer method enumerates ≥1 PFU, the theoretical lower limit of detection is 2.2 PFU per area sampled, with an interquartile range of 1.7 to 3.0 PFU per area sampled.
Laboratory-based recovery of MS2 RNA.
Results of the recovery of MS2 RNA target copies from stainless steel and plastic for each implement/eluent combination are provided in Table 3. The fraction of MS2 RNA target copies recovered ranged from 0.01 to 8.87. Recovery exceeded 1.0 (or 100%) for 9 of the 213 samples. The distribution of the fraction of target copies recovered, after the fraction was log10 transformed, was approximately normal (Kolmogorov-Smirnoff test, P = 0.079). The mean, median, and interquartile range of the log10-transformed fraction of target RNA recovered were −1.00 (equivalent to 10.0% recovery), −1.07 (8.7%), and −1.33 (4.7%) to −0.74 (18.2%), respectively. No product was detected in the extraction blanks or template controls.
To assess the influence of surface sampled, implement, and eluent on the log10-transformed fraction of MS2 RNA target copies recovered (an approximately normally distributed dependent variable), n-way ANOVA was used. The fraction of MS2 RNA target copies recovered was significantly influenced by surface (P = 0.005) and implement (P = 0.001), but not eluent type (P = 0.21). Specifically, recovery was greater from stainless steel than from plastic surfaces (P = 0.005), and, as identified using Tukey's post hoc test, antistatic cloth removed a significantly greater fraction than both cotton-tipped (P = 0.002) and polyester-tipped (P = 0.007) swabs. Although polyester-tipped swabs recovered a greater fraction of MS2 RNA target copies than cotton-tipped swabs, the difference was not significant (P = 0.98).
LLOD of MS2 RNA.
The median and interquartile range of fractional recovery of MS2 RNA using polyester/Ringer's were 0.06 and 0.03 to 0.13, respectively. Assuming that the qRT-PCR method has a lower limit of quantification of ≥250 genome equivalents (63), then the theoretical lower limit of quantification for the polyester-tipped swab/Ringer's combination is 4,200 genome equivalents per area sampled, with an interquartile range of 1,900 to 8,300 genome equivalents per area sampled.
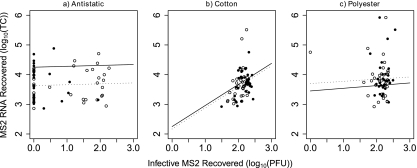
Infective MS2 (log10 PFU) recovered was not significantly associated with target copies of MS2 RNA recovered (log10 genome equivalents), as shown using both nonparametric correlation (Spearman's ρ = 0.044, P = 0.51) and linear regression. The infective MS2 recovered was plotted against target copies of MS2 RNA recovered, in Fig. 1 for each implement separately, with best-fit lines for recovery from plastic and stainless steel surfaces as determined using linear regression (see Table S3 in the supplemental material). Surface type and recovery of infective MS2 explained approximately 30%, 2%, and 8% of the total variability in the recovery of target copies of MS2 RNA for antistatic cloth (R2 = 0.296), cotton (R2 = 0.022), and polyester (R2 = 0.076), respectively.
Fig. 1.
The relationship between MS2 RNA target copies (TC) recovered and infective MS2 (PFU) recovered using antistatic cloth (a), cotton-tipped swabs (b), and polyester-tipped swabs (c) from plastic (•) and stainless steel (○) surfaces. Best-fit lines for recovery from plastic (dotted lines) and stainless steel (solid lines) for each implement are based on linear models described in the supplemental material.
DISCUSSION
Measuring viruses on surfaces is important for understanding the distribution of infectious agents in the environment and the role of fomites in disease transmission. Presently, researchers are using diverse techniques to recover and quantify viruses on surfaces. We demonstrate through a combined literature review and laboratory trial that the sampling method significantly affects virus recovery from surfaces. Variation in sampling method may contribute to the wide range in positivity rates reported across studies.
Recoveries reported here are similar to those previously reported by others. The median fractional recovery of MS2 RNA of 7% is low in the range of recovery of other viruses reported by Taku et al. (79) of 6 to 90%, Carducci et al. (20) of 13% to 76%, and Scherer et al. (75) of 7 to 53%. The authors attributed the range in fractional recovery within each study to different sampling methods, surface types, and/or dimensions of the area sampled. Contributions to differences in recovery across the studies might also include the use of different viruses or suspension media, variations in drying times, and/or differences in the ratio of exogenous RNA to intact virions in seeded inoculum.
Polyester-tipped swabs appear to be the most effective sampling implement for recovering virus from fomites. As shown by the meta-analysis, studies using polyester-tipped swabs reported a significantly higher overall positivity rate relative to studies using either cotton- or rayon-tipped swabs. In the laboratory comparison, polyester-tipped swabs recovered a greater fraction of infective MS2 and MS2 RNA than cotton-tipped swabs. Similarly, polyester-tipped swabs recovered a greater fraction of infective MS2 than antistatic cloth. Standardization of a sampling method to the polyester-tipped swabs may facilitate cross-study comparisons and allow for quantification of viruses using both culture- and molecularly based methods. The recommendation to use polyester swabs is consistent with the recommendation of the Centers for Disease Control and Prevention to use synthetic fibers for clinical sample collection (http://www.cdc.gov/h1n1flu/specimencollection.htm), as cotton-tipped swabs may contain trace contaminants (28, 69) and irregular fiber arrangements that reduce recovery of bacteria (64). Although antistatic cloth recovered the greatest fraction of MS2 RNA from surfaces, it recovered the lowest fraction of infective MS2. Because of the apparent inhibitory effect of antistatic cloth on infective MS2 recovery, we do not recommend its use.
Eluent type does not significantly influence the recovery of virus from surfaces. As shown in the meta-analysis and laboratory comparisons, eluent type did not significantly influence the positivity rate, fraction of infective MS2 recovered, or fraction of MS2 RNA target copies recovered. In the future, eluent should be chosen based on the study design. For example, investigations sampling for infective virus should consider eluents compatible with tissue culture, and investigations sampling for nucleic acid should consider eluents compatible with nucleic acid extraction and subsequent PCR. In general, we suggest one-quarter-strength Ringer's because the combination of polyester-tipped swabs and Ringer's recovered the greatest fraction of infective MS2 (0.48) and the second greatest fraction (besides antistatic cloth combinations) of MS2 RNA target copies (0.10).
Conclusions drawn from the meta-analysis are consistent with conclusions drawn from the laboratory comparison. Both the meta-analysis and the laboratory comparison suggest that eluent does not influence virus recovery, as eluent did not significantly influence the positivity rate or fraction of virus recovered. Similarly, both the meta-analysis and laboratory comparison demonstrated that the implement influences recovery. Specifically, polyester-tipped swabs are associated with greater recovery than cotton-tipped swabs. Finally, the meta-analysis suggested that antistatic cloths recover a greater fraction than swabs, based on two studies sampling for norovirus (15, 16). The laboratory comparison demonstrated that when molecular methods are used to assay for virus, as they were in the aforementioned studies, antistatic cloth removes a significantly greater fraction than swabs.
Comparisons of culture-based (i.e., infective MS2) and molecularly based (i.e., MS2 RNA target copies) recovery suggest that (i) the fraction of infective MS2 recovered is greater than the fraction of MS2 RNA and (ii) there is no significant association between the two metrics. The lower fractional recovery of MS2 RNA relative to infective MS2 may be because the estimate of seeded RNA target copies included exogenous RNA, and the exogenous RNA may degrade on fomites more rapidly than intact virions. A second possibility is that the seeded titer contained viral aggregates. Because the infectivity assay (i.e., double-agar-layer method) cannot distinguish between individual virions and virus aggregates, the degradation of a subset of the virus in aggregates could subsequently reduce recovery of RNA target copies without impacting apparent infective MS2 recovery. The lack of an association between the two metrics is a result of variability in recovery relative to variability in the initial seeded MS2 titer. Specifically, surfaces were seeded with the same initial MS2 titer, so random variability, the sampling method, and/or the enumeration method likely contributed to variability in recovery that masked variability in seeded titer. Understanding the relationship between the density of infective virus on surfaces and the density of nucleic acid target copies requires future research either employing multiple detection methods (e.g., cell culture and qRT-PCR) in field studies or examining a range of seeded titers in laboratory studies.
Although a standardized sampling method is recommended to allow cross-comparison of studies reporting positivity rates, there may be limitations. The specific recommendation to use polyester swabs is based on results of both a laboratory-scale study and a review of literature. The laboratory-scale study focused on recovery of one virus (MS2) from two surfaces (high-temperature PVC plastic and type 304 stainless steel with a mirror-like finish). Pathogenic viruses, however, have wide variation in physicochemical properties (such as size, shape, and isoelectric point) that may influence recovery by a standardized method. Similarly, the morphology and composition of the fomites' surfaces may also influence recovery. PVC plastic and stainless steel are appropriate representative samples, as both are widely used in consumer products (2, 45). Finally, testing additional eluents or eluent additives (such as antibiotics, surfactants, or disinfectant inactivators) may also influence recovery. Nevertheless, a standardized method is required for cross-comparison of studies, and our findings suggest that polyester-tipped swabs perform best.
Use of a standard method with a known recovery fraction will facilitate the extrapolation of measured virus densities to exposure and risk estimates. Priorities in future research include quantifying virus concentrations on surfaces and linking surface contamination to adverse health outcomes. Knowledge of virus quantity is an important step toward linking fomites to health risk, as exposures to greater concentrations result in greater risk of infection (42). To link fomites to health risk, longitudinal studies could track health and surface contamination, similar to the work of Bright et al. (18), Boxman et al. (15), and Gallimore et al. (34). Polyester-tipped swabs, as evidenced by this study, are compatible with quantification of virus using both the plaque assay to measure infectivity and qRT-PCR to measure nucleic acids.
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of the Boehm lab, who assisted with the work and/or provided suggestions for improving the manuscript. Three anonymous reviewers provided constructive comments that improved the manuscript.
The research has been funded, in part, by the UPS Foundation and the U.S. Environmental Protection Agency (EPA) under the Science to Achieve Results (STAR) Graduate Fellowship Program, Assistance Agreement no. F07D30757. F.J.T. was supported by NSF awards BES-0641406 and SES-0827384.
The EPA has not officially endorsed this publication, and the views expressed herein may not reflect the views of the EPA.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 5 August 2011.
REFERENCES
- 1. Abad F. X., Pinto R. M., Bosch A. 1994. Survival of enteric viruses on environmental fomites. Appl. Environ. Microbiol. 60:3704–3710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Adams R. O. 2009. A review of the stainless steel surface. J. Vac. Sci. Technol. A 1:12–18 [Google Scholar]
- 3. Akhter J., al Hajjar S., Myint S., Qadri S. M. 1995. Viral contamination of environmental surfaces on a general paediatric ward and playroom in a major referral centre in Riyadh. Eur. J. Epidemiol. 11:587–590 [DOI] [PubMed] [Google Scholar]
- 4. Asano Y., et al. 1999. Rapid contamination with varicella-zoster virus DNA to the throat of a daycare attendee and environmental surfaces from a child with varicella. Pediatr. Int. 41:233–236 [DOI] [PubMed] [Google Scholar]
- 5. Atkinson M. P., Wein L. M. 2008. Quantifying the routes of transmission for pandemic influenza. Bull. Math. Biol. 70:820–867 [DOI] [PubMed] [Google Scholar]
- 6. Bagutti C., et al. 2011. Detection of adeno and lentiviral (HIV1) contaminations on laboratory surfaces as a tool for the surveillance of biosafety standards. J. Appl. Microbiol. 111:70–82 [DOI] [PubMed] [Google Scholar]
- 7. Bausch D. G., et al. 2007. Assessment of the risk of Ebola virus transmission from bodily fluids and fomites. J. Infect. Dis. 196:142–147 [DOI] [PubMed] [Google Scholar]
- 8. Bean B., et al. 1982. Survival of influenza viruses on environmental surfaces. J. Infect. Dis. 146:47–51 [DOI] [PubMed] [Google Scholar]
- 9. Bell D. M. 2006. Non-pharmaceutical interventions for pandemic influenza, international measures. Emerg. Infect. Dis. 12:81–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bellamy K., Laban K. L., Barrett K. E., Talbot D. C. 1998. Detection of viruses and body fluids which may contain viruses in the domestic environment. Epidemiol. Infect. 121:673–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Boone S. A., Gerba C. P. 2005. The occurrence of influenza A virus on household and day care center fomites. J. Infect. 51:103–109 [DOI] [PubMed] [Google Scholar]
- 12. Boone S. A., Gerba C. P. 2007. Significance of fomites in the spread of respiratory and enteric viral disease. Appl. Environ. Microbiol. 73:1687–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Boone S. A., Gerba C. P. 2010. The prevalence of human parainfluenza virus 1 on indoor office fomites. Food Environ. Virol. 2:41–46 [Google Scholar]
- 14. Booth T. F., et al. 2005. Detection of airborne severe acute respiratory syndrome (SARS) coronavirus and environmental contamination in SARS outbreak units. J. Infect. Dis. 191:1472–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Boxman I., et al. 2009. Norovirus on swabs taken from hands illustrate route of transmission: a case study. J. Food Prot. 72:1753–1755 [DOI] [PubMed] [Google Scholar]
- 16. Boxman I. L., et al. 2009. Environmental swabs as a tool in norovirus outbreak investigation, including outbreaks on cruise ships. J. Food Prot. 72:111–119 [DOI] [PubMed] [Google Scholar]
- 17. Brankston G., Gitterman L., Hirji Z., Lemieux C., Gardam M. 2007. Transmission of influenza A in human beings. Lancet Infect. Dis. 7:257–265 [DOI] [PubMed] [Google Scholar]
- 18. Bright K. R., Boone S. A., Gerba C. P. 2010. Occurrence of bacteria and viruses on elementary classroom surfaces and the potential role of classroom hygiene in the spread of infectious diseases. J. Sch. Nurs. 26:33–41 [DOI] [PubMed] [Google Scholar]
- 19. Butz A. M., Fosarelli P., Dick J., Cusack T., Yolken R. 1993. Prevalence of rotavirus on high-risk fomites in day-care facilities. Pediatrics 92:202–205 [PubMed] [Google Scholar]
- 20. Carducci A., et al. 2002. Detection and potential indicators of the presence of hepatitis C virus on surfaces in hospital settings. Lett. Appl. Microbiol. 34:189–193 [DOI] [PubMed] [Google Scholar]
- 21. Carducci A., Verani M., Lombardi R., Casini B., Privitera G. 2011. Environmental survey to assess viral contamination of air and surfaces in hospital settings. J. Hosp. Infect. 77:242–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. CDC 2008. Norovirus outbreak in an elementary school—District of Columbia, February 2007. MMWR Morb. Mortal. Wkly. Rep. 56:1341–1343 [PubMed] [Google Scholar]
- 23. Cheesbrough J. S., Green J., Gallimore C. I., Wright P. A., Brown D. W. 2000. Widespread environmental contamination with Norwalk-like viruses (NLV) detected in a prolonged hotel outbreak of gastroenteritis. Epidemiol. Infect. 125:93–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen Y. C., et al. 2004. SARS in hospital emergency room. Emerg. Infect. Dis. 10:782–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dankert J., Uitentuis J., Houwen B., Tegzess A. M., Van Der Hem G. K. 1976. Hepatitis B surface antigen in environmental samples from hemodialysis units. J. Infect. Dis. 134:123. [DOI] [PubMed] [Google Scholar]
- 26. Dawson D. J., Paish A., Staffell L. M., Seymour I. J., Appleton H. 2005. Survival of viruses on fresh produce, using MS2 as a surrogate for norovirus. J. Appl. Microbiol. 98:203–209 [DOI] [PubMed] [Google Scholar]
- 27. Dowell S. F., et al. 2004. Severe acute respiratory syndrome coronavirus on hospital surfaces. Clin. Infect. Dis. 39:652–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ellner P. D., Ellner C. J. 1966. Survival of bacteria on swabs. J. Bacteriol. 91:905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Evans M. R., et al. 2002. An outbreak of viral gastroenteritis following environmental contamination at a concert hall. Epidemiol. Infect. 129:355–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ferenczy A., Bergeron C., Richart R. M. 1989. Human papillomavirus DNA in fomites on objects used for the management of patients with genital human papillomavirus infections. Obstet. Gynecol. 74:950–954 [PubMed] [Google Scholar]
- 31. Fischer B., Powis J., Firestone Cruz M., Rudzinski K., Rehm J. 2008. Hepatitis C virus transmission among oral crack users: viral detection on crack paraphernalia. Eur. J. Gastroenterol. Hepatol. 20:29–32 [DOI] [PubMed] [Google Scholar]
- 32. Foulongne V., Courgnaud V., Champeau W., Segondy M. 2011. Detection of Merkel cell polyomavirus on environmental surfaces. J. Med. Virol. 83:1435–1439 [DOI] [PubMed] [Google Scholar]
- 33. Froio N., et al. 2003. Contamination by hepatitis B and C viruses in the dialysis setting. Am. J. Kidney Dis. 42:546–550 [DOI] [PubMed] [Google Scholar]
- 34. Gallimore C. I., et al. 2006. Environmental monitoring for gastroenteric viruses in a pediatric primary immunodeficiency unit. J. Clin. Microbiol. 44:395–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gallimore C. I., et al. 2005. Use of a heminested reverse transcriptase PCR assay for detection of astrovirus in environmental swabs from an outbreak of gastroenteritis in a pediatric primary immunodeficiency unit. J. Clin. Microbiol. 43:3890–3894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gallimore C. I., et al. 2008. Contamination of the hospital environment with gastroenteric viruses: comparison of two pediatric wards over a winter season. J. Clin. Microbiol. 46:3112–3115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Girou E., et al. 2008. Determinant roles of environmental contamination and noncompliance with standard precautions in the risk of hepatitis C virus transmission in a hemodialysis unit. Clin. Infect. Dis. 47:627–633 [DOI] [PubMed] [Google Scholar]
- 38. Goldhammer K. A., Dooley D. P., Ayala E., Zera W., Hill B. L. 2006. Prospective study of bacterial and viral contamination of exercise equipment. Clin. J. Sport Med. 16:34–38 [DOI] [PubMed] [Google Scholar]
- 39. Green J., et al. 1998. The role of environmental contamination with small round structured viruses in a hospital outbreak investigated by reverse-transcriptase polymerase chain reaction assay. J. Hosp. Infect. 39:39–45 [DOI] [PubMed] [Google Scholar]
- 40. Gurley E. S., et al. 2007. Person-to-person transmission of Nipah virus in a Bangladeshi community. Emerg. Infect. Dis. 13:1031–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gwaltney J. M. 1982. Transmission of experimental rhinovirus infection by contaminated surfaces. Am. J. Epidemiol. 116:828–833 [DOI] [PubMed] [Google Scholar]
- 42. Haas C., Rose J., Gerba C. 1999. Quantitative microbial risk assessment. John Wiley and Sons, Inc., New York, NY [Google Scholar]
- 43. Hall C. B., Douglas R. G., Geiman J. M. 1980. Possible transmission by fomites of respiratory syncytial virus. J. Infect. Dis. 141:98–102 [DOI] [PubMed] [Google Scholar]
- 44. Hamada N., et al. 2008. Nosocomial outbreak of epidemic keratoconjunctivitis accompanying environmental contamination with adenoviruses. J. Hosp. Infect. 68:262–268 [DOI] [PubMed] [Google Scholar]
- 45. Heudorf U., Mersch-Sundermann V., Angerer J. 2007. Phthalates: toxicology and exposure. Int. J. Hyg. Environ. Health 210:623–634 [DOI] [PubMed] [Google Scholar]
- 46. Jennings L. C., Dick E. C. 1987. Transmission and control of rhinovirus colds. Eur. J. Epidemiol. 3:327–335 [DOI] [PubMed] [Google Scholar]
- 47. Jones E. L., Kramer A., Gaither M., Gerba C. P. 2007. Role of fomite contamination during an outbreak of norovirus on houseboats. Int. J. Environ. Health Res. 17:123–131 [DOI] [PubMed] [Google Scholar]
- 48. Katayama H., Shimasaki A., Ohgaki S. 2002. Development of a virus concentration method and its application to detection of enterovirus and Norwalk virus from coastal seawater. Appl. Environ. Microbiol. 68:1033–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kawahara A., Yoshida M. 2009. Detection of viral DNA in nonlesional skin of patients with molluscum contagiosum and on environmental fomites. Br. J. Dermatol. 160:1357–1359 [DOI] [PubMed] [Google Scholar]
- 50. Keswick B. H., Pickering L. K., DuPont H., Woodward W. E. 1983. Survival and detection of rotaviruses on environmental surfaces in day care centers. Appl. Environ. Microbiol. 46:813–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Killingley B., et al. 2010. Virus shedding and environmental deposition of novel A (H1N1) pandemic influenza virus: interim findings. Health Technol. Assess. 14:237–354 [DOI] [PubMed] [Google Scholar]
- 52. Kuusi M., et al. 2002. A prolonged outbreak of Norwalk-like calicivirus (NLV) gastroenteritis in a rehabilitation centre due to environmental contamination. Epidemiol. Infect. 129:133–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lauer J. L., VanDrunen N. A., Washburn J. W., Balfour H. H. 1979. Transmission of hepatitis B virus in clinical laboratory areas. J. Infect. Dis. 140:513–516 [DOI] [PubMed] [Google Scholar]
- 54. Lederman E., et al. 2009. Eczema vaccinatum resulting from the transmission of vaccinia virus from a smallpox vaccinee: an investigation of potential fomites in the home environment. Vaccine 27:375–377 [DOI] [PubMed] [Google Scholar]
- 55. Lessa F. C., et al. 2009. Health care transmission of a newly emergent adenovirus serotype in health care personnel at a military hospital in Texas, 2007. J. Infect. Dis. 200:1759–1765 [DOI] [PubMed] [Google Scholar]
- 56. Leung J., et al. 2010. A 2009 varicella outbreak in a Connecticut residential facility for adults with intellectual disability. J. Infect. Dis. 202:1486–1491 [DOI] [PubMed] [Google Scholar]
- 57. Lopez A. S., et al. 2008. Transmission of a newly characterized strain of varicella zoster virus from a patient with herpes zoster in a long term care facility, West Virginia, 2004. J. Infect. Dis. 197:646–653 [DOI] [PubMed] [Google Scholar]
- 58. Lyman W. H., et al. 2009. Prospective study of etiologic agents of acute gastroenteritis outbreaks in child care centers. J. Pediatr. 154:253–257 [DOI] [PubMed] [Google Scholar]
- 59. Mathers C. D., Boerma T., Ma Fat D. 2008. The global burden of disease: 2004 update. World Health Organization, Geneva, Switzerland [Google Scholar]
- 60. Morens D. M., Rash V. M. 1995. Lessons from a nursing home outbreak of influenza A. Infect. Control Hosp. Epidemiol. 16:275–280 [DOI] [PubMed] [Google Scholar]
- 61. Morter S., et al. 2011. Norovirus in the hospital setting: virus introduction and spread within the hospital environment. J. Hosp. Infect. 77:106–122 [DOI] [PubMed] [Google Scholar]
- 62. Mubareka S., et al. 2009. Transmission of influenza virus via aerosols and fomites in the guinea pig model. J. Infect. Dis. 199:858–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. O'Connell K. P., et al. 2006. Real-time fluorogenic reverse transcription-PCR assays for detection of bacteriophage MS2. Appl. Environ. Microbiol. 72:478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Osterblad M., et al. 2003. Evaluation of a new cellulose sponge-tipped swab for microbiological sampling: a laboratory and clinical investigation. J. Clin. Microbiol. 41:1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Osterholm M. T., Max B. J., Hanson M., Polesky H. F. 1979. Potential risk of salivary-mediated viral hepatitis type B transmission from oral exposure to fomites. J. Hyg. (Lond.) 83:487–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Pappas D. E., Hendley J. O., Schwartz R. H. 2010. Respiratory viral RNA on toys in pediatric office waiting rooms. Pediatr. Infect. Dis. J. 29:102–104 [DOI] [PubMed] [Google Scholar]
- 67. Pecson B. M., Martin L. V., Kohn T. 2009. Quantitative PCR for determining the infectivity of bacteriophage MS2 upon inactivation by heat, UVB and singlet oxygen: advantages and limitations of an enzymatic treatment to reduce false-positive results. Appl. Environ. Microbiol. 75:5544–5554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Piazza M., et al. 1987. HBsAg (hepatitis B virus surface antigen) contamination in the dental environment. Minerva Stomatol. 36:903–907 [PubMed] [Google Scholar]
- 69. Pollock M. R. 1947. The growth of H. pertussis on media without blood. Br. J. Exp. Pathol. 28:295–307 [PMC free article] [PubMed] [Google Scholar]
- 70. Ramani S., et al. 2008. Investigation of the environment and of mothers in transmission of rotavirus infections in the neonatal nursery. J. Med. Virol. 80:1099–1105 [DOI] [PubMed] [Google Scholar]
- 71. Runner J. C. 2007. Bacterial and viral contamination of reusable sharps containers in a community hospital setting. Am. J. Infect. Control 35:527–530 [DOI] [PubMed] [Google Scholar]
- 72. Rusin P., Maxwell S., Gerba C. 2002. Comparative surface-to-hand and fingertip-to-mouth transfer efficiency of Gram-positive bacteria, Gram-negative bacteria, and phage. J. Appl. Microbiol. 93:585–592 [DOI] [PubMed] [Google Scholar]
- 73. Russell K. L., et al. 2006. Transmission dynamics and prospective environmental sampling of adenovirus in a military recruit setting. J. Infect. Dis. 194:877–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sandora T. J., Shih M. C., Goldmann D. A. 2008. Reducing absenteeism from gastrointestinal and respiratory illness in elementary school students: a randomized, controlled trial of an infection-control intervention. Pediatrics 121:e1555. [DOI] [PubMed] [Google Scholar]
- 75. Scherer K., et al. 2009. Application of a swab sampling method for the detection of norovirus and rotavirus on artificially contaminated food and environmental surfaces. Food Environ. Virol. 1:42–49 [Google Scholar]
- 76. Siegel J. D., Rhinehart E., Jackson M. 2007. 2007 guideline for isolation precautions: preventing transmission of infectious agents in healthcare settings. Centers for Disease Control and Prevention, Atlanta, GA: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Soule H., et al. 1999. Monitoring rotavirus environmental contamination in a pediatric unit using polymerase chain reaction. Infect. Control Hosp. Epidemiol. 20:432–434 [DOI] [PubMed] [Google Scholar]
- 78. Strauss S., Sastry P., Sonnex C., Edwards S., Gray J. 2002. Contamination of environmental surfaces by genital human papillomaviruses. Sex. Transm. Infect. 78:135–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Taku A., et al. 2002. Concentration and detection of caliciviruses from food contact surfaces. J. Food Prot. 65:999–1004 [DOI] [PubMed] [Google Scholar]
- 80. U.S. EPA 2001. EPA Method 1602: male-specific (F+) and somatic coliphage in water by single agar layer (SAL) procedure. EPA 821-R-01-029. U.S. Environmental Protection Agency, Washington, DC [Google Scholar]
- 81. Wadl M., et al. 2010. Food-borne norovirus-outbreak at a military base, Germany, 2009. BMC Infect. Dis. 10:30–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Widdowson M. A., et al. 2002. An outbreak of diarrhea in a neonatal medium care unit caused by a novel strain of rotavirus: investigation using both epidemiologic and microbiological methods. Infect. Control Hosp. Epidemiol. 23:665–670 [DOI] [PubMed] [Google Scholar]
- 83. Wilde J., Van R., Pickering L., Eiden J., Yolken R. 1992. Detection of rotaviruses in the day care environment by reverse-transcriptase polymerase chain reaction. J. Infect. Dis. 166:507–511 [DOI] [PubMed] [Google Scholar]
- 84. Winther B., Alper C. M., Mandel E. M., Doyle W. J., Hendley J. O. 2007. Temporal relationships between colds, upper respiratory viruses detected by polymerase chain reaction, and otitis media in young children followed through a typical cold season. Pediatrics 119:1069. [DOI] [PubMed] [Google Scholar]
- 85. Wu H. M., et al. 2005. A norovirus outbreak at a long-term-care facility: the role of environmental surface contamination. Infect. Control Hosp. Epidemiol. 26:802–810 [DOI] [PubMed] [Google Scholar]
- 86. Xerry J., Gallimore C. I., Cubitt D., Gray J. J. 2010. Tracking environmental norovirus contamination in a paediatric primary immunodeficiency unit. J. Clin. Microbiol. 48:2552–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Yoshikawa T., et al. 2001. Rapid contamination of the environments with varicella-zoster virus DNA from a patient with herpes zoster. J. Med. Virol. 63:64–66 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.