Abstract
Miniaturization and high-throughput screening are currently the focus of emerging research areas such as systems biology and systems biotechnology. A fluorescence-based screening assay for the online monitoring of oxygen and pH and a numerical method to mine the resulting online process data are described. The assay employs commercial phosphorescent oxygen- and pH-sensitive probes in standard 48- or 96-well plates on a plate reader equipped with a shaker. In addition to dual parametric analysis of both pH and oxygen in a single well, the assay allows monitoring of growth, as measured by absorbance. Validation of the assay is presented and compared with commercially available plates equipped with optical sensors for oxygen and pH. By using model-free fitting to the readily available online measurements, the length and rate of each phase such as the duration of lag and transition phase or acidification, growth, and oxygen consumption rates are automatically detected. In total, nine physiological descriptors, which can be used for further statistical and comparison analysis, are extracted from the pH, oxygen partial pressure (pO2), and optical density (OD) profiles. The combination of a simple mix-and-measure procedure with an automatic data mining method allows high sample throughput and good reproducibility while providing a physiological state identification and characterization of test cells. As a proof of concept, the utility of the workflow in assessing the physiological response of Escherichia coli to environmental and genetic perturbations is demonstrated.
INTRODUCTION
The availability of high-throughput (HT) and high-information-content screening procedures is one of the technical prerequisites for enhanced development of therapeutic drugs, new biological products, and growth media or strain improvement through metabolic engineering or directed evolution. HT operation can be easily accomplished on a small scale in disposable multiwell plates. Unfortunately, plates are often not equipped to accurately and continuously provide data about the status of the cultivation. Such data are nevertheless critical in assessing the physiological behavior of the cells for better understanding cellular processes and adaptation to environmental or genetic perturbations. Biomolecular components such as enzymes, DNA, RNA, and intra- and extracellular metabolites are undoubtedly the most informative variables to provide a mechanistic understanding of the physiological status of cells. Regrettably, such variables are typically available only via offline measurements, and samplings in plates interfere strongly with the cultivation by further reducing the volume. In contrast to the offline parameters that are measured periodically, many online parameters such as oxygen partial pressure (pO2), pH, and biomass are measured noninvasively and continuously with respect to the time scale of the production cycle. In addition, these major physiological parameters provide information on cellular metabolism. For instances, the trend of pH values may be a qualitative indicator of phenomena such as organic acid production or consumption (50) and the course of pO2 may point out substrate or oxygen limitations, product inhibitions, or diauxic growth (2). For these reasons, online measurements are favorable for small-scale process monitoring.
The development of fluorescence-based sensing technologies provides an attractive solution to enable small-scale process monitoring. The approach relies on the use of fluorescent probes sensitive to oxygen or pH. Optical oxygen sensing is based on fluorescence quenching by molecular oxygen, while pH sensing is based on the interdependence of optical properties with respect to the equilibrium between the acidic and basic forms of a reagent resulting from pH changes in the medium. These, in turn, may be monitored by changes in fluorescence intensity. Using solid-state sensors, 96-well plates that are capable of detecting biological oxygen consumption or extracellular acidification or basification have been successfully applied to the analysis of a variety of bacteria, mammalian cells, and fungi (3, 13, 30). Each sensor allows the measurement of one parameter—either pH or O2—and is provided as a patch occupying the entire bottom of wells in 96-well plates. Such a design prevents the measurement of cell growth via optical density (OD), as well as the parallel measurement of pH and O2. Dual devices containing two sensors, one for pH and one for O2, are currently available in 24- and 96-well plates, allowing simultaneous optical monitoring of oxygen and pH during bacterial or mammalian cell cultivation (17, 35, 38) (SXF-96 analyzer; Seahorse Bioscience). Fully integrated systems using multiwell plates and measuring simultaneously a third parameter such as growth (34), CO2 (SXF-24_3 analyzer; Seahorse Bioscience), or temperature (28) have also been reported. These two latter systems can suffer from certain limitations, including cost and the requirement for dedicated instrumentation (i.e., reader and plate). More recently, the development of water-soluble probes that can be dispensed directly into samples has overcome many of these limitations and allows the dual measurement of oxygen and pH and the use of plates with several formats (i.e., from 6- to 384-well plates). Examples using dispensable water-soluble probes in physiological samples include extracellular assessment of oxygen and/or pH (24–26, 36, 43) and measurement of intracellular pH (9, 49) in eukaryotic and bacterial cells. The most frequently used indicators are 8-hydroxypyrene-1,3,6-trisulfonic acid sodium salt (HPTS), carbxyfluorescein derivatives (e.g., mono- and dichlorocarboxyfluorescein), and seminaphthorhodafluor (SNARF) for pH measurement and ruthenium complexes and platinum (or palladium)-porphyrins for oxygen assessment (3).
While fluorescence-based technologies can provide real online data about cultivation, these measurements do not give direct information on the physiological status of cells (52). Indeed, online parameters are often recorded every few minutes for cultivation periods that can last from a few days to a couple of weeks, resulting in large data sets. Further, online data are quite heterogeneous in time scales and data types. Finally, noise due to the instrument limitation may be present in the raw online measurement (10). In order to capture the key physiological descriptors (e.g., growth rate, lag phase, and acidification rate) from such complex data sets, raw data have to be compiled, noise in the measurements must be eliminated, and the results must be introduced into appropriate models. There are a large number of techniques available to formulate such models, each varying in complexity and ease of development from the others. The choice of a particular type of model generally depends on the existing amount of a priori knowledge about the system to be modeled. This includes linear models (e.g., multiple linear regression) (21, 53), nonlinear models (e.g., artificial neural networks) (31, 32, 53), statistical techniques (e.g., principal component analysis) (14, 31, 32), and artificial intelligence-based approaches (e.g., knowledge-based systems) (31). However, the problem of finding the most appropriate model that best fits the data, which requires a priori information on the bioprocess, thereby limits its applicability for routine and automatic detection of physiological descriptors.
In this study, we describe a simple, high-throughput, robust workflow for multiplex monitoring of both pH and oxygen using fluorescence and biomass via absorbance and for mining resulting online parameters in small-scale processes. The approach is flexible with regard to the user choice of probes, plate, detection mode, and data analysis. It consists of 3 steps. First, water-soluble pH and oxygen dyes are used as the basis of a simple mix-and-measure assay to record in parallel both oxygen consumption and extracellular acidification or basification in addition to growth. Second, a numerical method is applied to smooth the online data and to extract the physiological descriptors. Third, statistical analysis of the resulting physiological descriptors is used to discriminate distinct physiological patterns and cluster the data into meaningful groups. General performance evaluation of this approach is presented and followed by an illustration of how this workflow may be used in the examination of the physiological response of Escherichia coli to environmental and genetic perturbations.
MATERIALS AND METHODS
Bacterial strains and cultivation media.
The K-12 Escherichia coli strains MG1655 and BW25113 and 94 single-gene knockout (KO) mutants from the KEIO collection (4) were used in this study. All E. coli strains were grown on minimal synthetic medium containing 48 mM Na2HPO4, 22 mM KH2PO4, 9 mM NaCl, 19 mM NH4Cl, 2 mM MgSO4, 0.1 mM CaCl2, 0.1 g/liter of thiamine, and 2.5 g/liter of carbon source. In this study, d-cellobiose, d-glucose, d-xylose, d-mannose, l- and d-arabinose, l-rhamnose, l-fucose, d-galacturonic acid, d-glucuronic acid, d-sorbitol, dulcitol, arbutin, d-salicin, d-psicose, and d-allose were used as carbon sources and were obtained from Sigma-Aldrich, France. Magnesium sulfate and calcium chloride were autoclaved separately. Carbon sources and thiamine were sterilized by filtration.
Measurement device.
Cells were cultivated in a FLUOstar Optima plate reader (BMG Labtech, Offenburg, Germany) at 37°C and 600 rpm (orbital) with a shaking diameter of 1 mm. The shaking and measurement procedures were as follows: shake duration, 300 s; fluorescent measurement duration, 123 s per 48 wells, 20 flashes; shaking duration, 60 s; absorbance measurement duration, 50 s per 48 wells, 20 flashes; 180 cycles, 24 h, flash. Biomass was determined by measurement of optical density (OD) at 600 nm. The experiments were carried out with standard round 96- or 48-well plates with flat bottoms from Starlab, France (catalog number 1830048), covered with lids. If not otherwise specified, the experiments were conducted with a 500-μl working volume of medium.
Measurement of pH.
The pH was measured by adding a sterile solution of BCECF [2′,7′-bis-(2-carboxyethyl)-5-(and -6)-carboxyfluorescein; catalog number C3411; Sigma-Aldrich, France] to synthetic medium before inoculation with cells. The soluble fluorescent pH indicator was applied in a final concentration of 2 μM in the fermentation medium. This indicator was excited with a wavelength of 450 nm and 485 nm, and the emission was detected for both excitation wavelengths at 540 nm. The pH values were derived from a calibration curve generated with buffers containing BCECF at the same concentration (i.e., 2 μM) as that in the culture medium. Buffers ranging from pH 4.0 to 9.0 were applied to calibrate the measurement device. For each buffer condition, the intensity ratio IR was calculated as follows: IR=(Iem540 nm·ex485 nm)/(Iem540 nm·ex450 nm). After determining IR for the different buffers, IR is plotted versus pH. The resulting sigmoidal curve is fitted by using the Boltzmann equation (3): pH=pHo + dpH × ln [(IRmin − IR)/(IR − IRmax)], where pHo is the point of inflection and therefore matches the pKa value; IR is the measured intensity; IRmin and IRmax are the minimal and maximal values of the sigmoidal curve, respectively; and dpH is the slope at the point of inflection. The calibration parameters pHo, dpH, IRmin, and IRmax were calculated using Sigmaplot software (SPSS Inc.).
Measurement of the oxygen partial pressure (pO2).
Oxygen partial pressure (pO2) in percent air saturation was measured by adding a sterile solution of RuII(bpy)3 [Tris(2,2′-bipyridyl)dichlororuthenium(II) hexahydrate; catalog number 224758; Sigma-Aldrich, France] to synthetic medium before inoculation with cells. The soluble fluorescent oxygen indicator was applied at a final concentration of 20 μM in the cultivation medium and measured with an excitation filter of 450 nm and an emission filter of 600 nm. The oxygen measurements were calibrated with solutions containing RuII(bpy)3 at the same concentration (i.e., 20 μM) as that in the culture medium. Doubly distilled water was used for the calibration value of 100% air saturation, and a solution of 1% sodium sulfite was used for 0% air saturation.
The oxygen values were correlated with a rearranged Stern-Volmer equation (40) as follows:
where Ii is the emission intensity at the initial oxygen concentration [O2]i, prior to the reaction initiation (t=0), in air- or oxygen-saturated solution; It is the emission intensity at some intermediate time t, for which the oxygen concentration has decreased to [O2]t; and Io is the final emission intensity in the absence of oxygen.
Assay validation.
Selection of required dye concentrations was performed as follows. Stock solutions of 100× RuII(bpy)3 and 200× BCECF were prepared in doubly distilled water. Various volumes of these stocks were added to wells containing minimal synthetic medium (pH 7.0) to obtain the desired final dye concentrations. Cells were added to the medium to obtain a final OD at 600 nm of 1. The total volume of each well was 200 μl. Two replicates per dye concentration were measured. The fluorescence emission at a particular dye concentration was measured after 5 min of orbital shaking. For the pH assay, the signal-to-noise ratio (S/N) was calculated by dividing the intensity ratio (IR) of the pH-sensitive wavelength (490 nm) and the pH-insensitive wavelength (440 nm) by the IR of the medium with or without cells. For the oxygen assay, S/N was calculated by dividing the fluorescence intensity at 450 nm of the dye by the fluorescence intensity at 450 nm of the medium with or without cells.
For BCECF and Hydroplate (PreSens), the accuracy was determined by calculating the difference between the measured mean of 8 wells per pH unit and the corresponding pH measured for each condition with a Eutcher pH meter. The repeatability was defined as relative standard deviation (RSD) of 8 wells per pH unit measured during two consecutive days. The pH drift was estimated by monitoring wells containing pH buffer solutions and 2 μM BCECF over 1 h. For RuII(bpy)3 and Oxoplate (PreSens), the accuracy was determined by calculating the difference between the average of 8 wells per condition (i.e., air-saturated medium or air-saturated water or deoxygenated water) and the corresponding pO2 of the solutions measured with a polarographic oxygen sensor (Metler Toledo). Each solution was measured twice during two consecutive days. The repeatability was defined as RSD of 8 wells per condition, measured during two consecutive days. The oxygen drift was estimated by monitoring wells containing either air-saturated medium or air-saturated water or deoxygenated water over 1 h. The fluorescence emissions were measured for 1 h with 1 min of orbital shaking between each measurement. All data points were considered for calculations.
Analysis of glucose and acetate levels.
Glucose and acetate concentrations were determined by high-performance liquid chromatography (HPLC) using an Agilent 1100 Series chromatograph (Santa Clara, CA). An Aminex HPX-87H ion-exchange column (Bio-Rad, Hercules, CA) and a refractive index detector (RI-101; Shodex, Munich, Germany) were used for separation and detection, respectively. The column temperature was 48°C, and 5 mM H2SO4 was used as a mobile phase at a flow rate of 0.5 ml/min.
Calculations of physiological descriptors.
Physiological descriptors, i.e., the maximum specific growth rate on the first substrate (μ1) and on the second substrate (μ2), the oxygen consumption rate on the first substrate (RO2), the acidification (RAc) and the basification (RBa) rates, the length of lag phase (LLP), the length of the first growth phase (LEx1), the length of transition phase (LTP), and the length of the second growth phase (LEx2), were calculated from the time profiles of biomass, pH, and oxygen using a home-made program developed in R (48). The first substrate corresponds to the carbon source provided at the beginning of the cultivation, and the second substrate is the product (often acetate but not always) produced from the first substrate and consumed only after the first carbon source has been exhausted. The data were smoothed using simple moving average methods as described in the Pastecs package, an R package dedicated to time series analysis (27). Growth data were log transformed. To estimate descriptors from each time profile, model-free spline fits were applied. The model-free fit applies a smoothed cubic spline as implemented in the R function smooth.spline. A spline is a piecewise-polynomial real function that does not assume a simple functional relationship between the x and y variables for the considered curve. Instead, the curve is represented as a superposition of elementary functions, e.g., cubic spline here. For each type of profile (growth, pH, and oxygen), the maximum slopes that provided a measure of μ1 and RBa and the minimum slopes that provide a measure of RO2 and RAc were estimated from the spline fit by taking the maximum and the minimum, respectively, of the numerical derivative with respect to time. Using the equation of the tangent of the fitted curve at the maximum or minimum slope and the maximal or minimal value of variable considered, starting and ending time points of each linear part of the curves were extracted. Using these data, it was possible to calculate the length of each phase. LLP and LEx1 were derived from the growth curve, while LTP and LEx2 were derived from the pH curve. Within the interval corresponding to the second growth, the slope was used to derive μ2.
Z score calculation and hierarchical cluster analysis.
The data set obtained from the environmental perturbation experiment can be viewed as a matrix Xij where indices i=1, …M and j=1, …Nrun across experimental conditions (i.e., environmental conditions) and the physiological descriptor dimensions, respectively. In our experiment M=19, that is 9 different carbon sources × 2 interplate replicates + 1 additional interplate replicate for the glucose condition and n=9 (i.e., 9 different carbon sources).
The Z score for each experiment was computed as follows: ZXij=(Xij − μXj)/σXj. μXj and σXj are the mean and the standard deviation of the physiological descriptor j across the M experiments, respectively.
The hierarchical cluster analysis on Z score values was performed using the hclust function as described in the R Stats package (48). The Manhattan function was used as distance metric, and Ward's linkage was used as the clustering algorithm.
RESULTS
pH assay design.
The soluble fluorescent dye BCECF, one of the most widely used fluorescent indicators for intracellular pH, was chosen to monitor the extracellular pH. This dye exhibits a pKa value of approximately 7 and is hydrophilic. The dye is membrane impermeant unless an acid shock treatment is applied to load BCECF within the cells (49). Although the concentration of dye to use as an intracellular pH indicator is known, no data were available for using BCECF as an extracellular pH indicator. Hence, we determined the optimal amount of dye to add in the medium to obtain adequate signal intensity, for BCECF concentrations ranging from 0.5 to 10 μM (concentrations generally used in the literature) (Table 1). For both conditions (i.e., with and without cells), use of 2 to 4 μM BCEF produced maximal signal-to-noise ratio (S/N), and dye concentrations below or above these values resulted in decreased S/N. Because BCECF has a pH-insensitive isosbestic point at about 440 nm that allows correction of pH changes for differences in medium composition, no drastic changes in S/N were observed between medium with cells and that without cells. A BCECF concentration of 2 μM was thus chosen for subsequent experiments. Next, the accuracy and repeatability of the pH measurement were evaluated and compared with data collected in a 96-well plate equipped with optodes sensitive to pH (Hydroplate; PreSens) (Table 2). For pHs between 5 and 8, BCECF showed accuracy comparable to, and higher repeatability than, that of Hydroplate. In general, the pH drifts measured for BCECF were lower than the ones measured for the Hydroplate and fell within the accuracy of the BCECF sensor. Finally, the possible cytotoxic effect caused by BCECF was examined by comparing growth of an E. coli strain with 2 μM dye and its growth without the dye (data not shown). It was found that such an amount of dye did not significantly affect bacterial growth.
Table 1.
Changes of S/N as a function of BCECF concentrations (varying from 0.5 to 10 μM) and Ru(II)(bpy)3 concentrations (varying from 2.5 to 50 μM) with (final OD at 600 nm of 1) or without cells in M9 medium at pH 7
| Assay, dye, and concn (μM) | S/Na |
|
|---|---|---|
| Without cells | With cells | |
| pH assay, BCECF | ||
| 0.5 | 2.63 ± 0.05 | 2.77 ± 0.14 |
| 1 | 2.63 ± 0.03 | 2.79 ± 0.13 |
| 2 | 2.65 ± 0.05 | 2.81 ± 0.10 |
| 4 | 2.64 ± 0.04 | 2.82 ± 0.06 |
| 8 | 2.59 ± 0.04 | 2.77 ± 0.07 |
| 10 | 2.55 ± 0.03 | 2.71 ± 0.05 |
| Oxygen assay, Ru(II)(bpy)3 | ||
| 2.5 | 21.7 ± 7.2 | 6.3 ± 1.0 |
| 5 | 31.1 ± 10.7 | 8.0 ± 1.4 |
| 10 | 54.5 ± 19.2 | 14.2 ± 2.4 |
| 20 | 94.7 ± 32.5 | 25.2 ± 3.9 |
| 40 | 148.3 ± 52.2 | 43.6 ± 6.9 |
| 50 | 169.0 ± 60.3 | 51.3 ± 8.1 |
S/N was calculated by dividing the fluorescence signal of the dye by the background fluorescence of the medium with or without cells. Values are averages ± standard deviations of duplicates per condition.
Table 2.
Accuracy, repeatability, and drift per hour of pH assays performed with BCECF and Hydroplate
| pH of buffer solution | Accuracy (pH) |
Repeatability (RSDa) |
Drift (pH/h) |
|||
|---|---|---|---|---|---|---|
| BCECF | Hydroplate | BCECF | Hydroplate | BCECF | Hydroplate | |
| 3 | 0.11 | 0.08 | 1.86 | 15.01 | 0.16 | 0.23 |
| 4 | 0.51 | 0.08 | 0.46 | 1.75 | 0.02 | 0.03 |
| 5 | 0.06 | 0.06 | 0.24 | 0.50 | 0.02 | 0.05 |
| 6 | 0.05 | 0.05 | 0.24 | 0.33 | 0.00 | 0.03 |
| 7 | 0.00 | 0.04 | 0.29 | 0.38 | 0.04 | 0.03 |
| 8 | 0.10 | 0.08 | 0.65 | 1.07 | 0.00 | 0.16 |
| 9 | 0.34 | 0.29 | 1.48 | 2.80 | 0.04 | 0.11 |
RSD, relative standard deviation (percent).
Oxygen assay design.
The soluble fluorescent probe RuII(bpy)3 was chosen for its potential for multiplexing with the BCECF probe, thereby allowing dual parameter metabolic analysis of extracellular pH and oxygen consumption. The overlay spectra (data not shown) indicate that both probes may be excited using a single excitation band (440 to 500), whereas emission can be discriminated using appropriate filters. The concentration of dye to add in the medium was selected for RuII(bpy)3 concentrations varying from 2.5 to 50 μM (concentrations generally used in the literature) (Table 1). In contrast to BCECF, Ru(II)(bpy)3 does not possess an isosbestic point, and S/N was strongly affected by the medium composition (i.e., cells and dye concentrations). While the fluorescence level of Ru(II)(bpy)3 was constant for the same concentration, the background fluorescence of the medium increased when cells were added. This effect is likely due to biogenic fluorophores such as proteins, coenzymes, and vitamins. However, the fluorescence intensity that was specifically due to the dye was 6 to 51 times above the background fluorescence signal (Table 1), depending on the dye concentration. From these, a RuII(bpy)3 concentration of 20 μM, which gives a good S/N, was selected. In Table 3, the accuracy and repeatability of the oxygen measurements were evaluated and compared with a 96-well plate equipped with oxygen-sensitive optodes (Oxoplate; PreSens). Compared with Oxoplate, RuII(bpy)3 showed comparable accuracy and lower repeatability. However, these data are acceptable for most screening applications, especially when working with synthetic minimal medium, for which good accuracy and repeatability were observed. The oxygen drifts measured in water solutions were higher than the ones obtained with the Oxoplate, but they were largely lower for synthetic minimal medium. The cytotoxicity and cell permeability of RuII(bpy)3 were also tested for the selected concentration. Growth of E. coli cells was not significantly affected upon cultivation with 20 μM RuII(bpy)3, demonstrating the absence of toxicity of the fluorescent dye (data not shown). Penetration of RuII complex through cell membrane was investigated by incubating E. coli cells with RuII(bpy)3 under standard growth conditions. As shown in Table 4, most of the RuII(bpy)3 was detected in the supernatant (extracellular) after centrifugation of the cell suspension and only a negligible percentage (below 1%) was found in the cells. This was likely due to nonspecific fluorescence since similar results were obtained when no RuII(bpy)3 was added in the medium (Table 4). These data indicated that RuII(bpy)3 is apparently not transported into the cells and is not cytotoxic.
Table 3.
Accuracy, repeatability, and drift per hour of oxygen assay performed with Ru(II)(bpy)3 and Oxoplate
| Solution | Accuracy (% air saturation) |
Repeatability (% air saturation) |
Drift (% air saturation/h) |
|||
|---|---|---|---|---|---|---|
| Ru(II)(bpy)3 | Oxoplate | Ru(II)(bpy)3 | Oxoplate | Ru(II)(bpy)3 | Oxoplate | |
| Air-saturated water | 0.9 | 5.5 | 5.1 | 0.69 | 13.2 | 0.2 |
| Deoxygenated water | 3.5 | 0.0 | 2.1 | 0.08 | 5.6 | 0.3 |
| Air-saturated medium | 2.3 | 3.4 | 1.3 | 0.90 | 0.1 | 1.8 |
Table 4.
Retention of RuII(bpy)3 in E. coli cells
| RuII(bpy)3 concn (μM) | Relative fluorescence at 450 nm of cells grown with RuII(bpy)3a |
|
|---|---|---|
| Extracellular | Intracellular | |
| 20 | 99.3 | 0.7 |
| 0 | 99.0 | 1.0 |
Data were corrected for the background fluorescence (signal) of the medium. Before measurement, the cells were washed twice and resuspended in M9 medium, pH 7.
Multiplexing pH and oxygen assays to infer cellular metabolism.
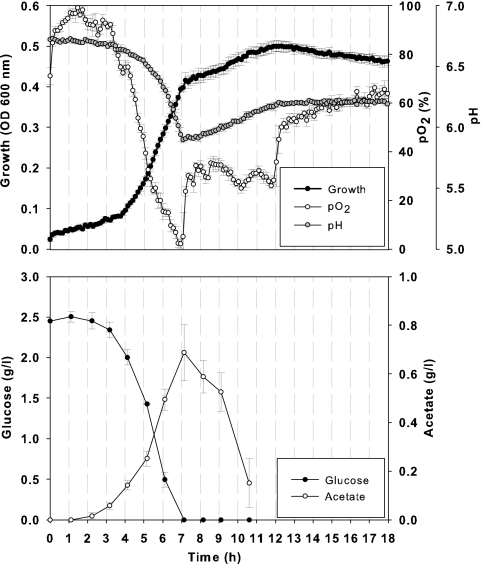
The spectral compatibility of both BCECF and RuII(bpy)3 probes opens up the possibility of dual parametric analysis of both pH and oxygen in a single well. In addition, growth, as measured by absorbance, might not interfere with the dual fluorescence signals of BCECF and RuII(bpy)3. Thus, it should be possible to monitor pH, oxygen, and growth in parallel. As a validation study, we tested the fluorescence-based assay to examine E. coli MG1665 cells growing on glucose. The behavior of E. coli under this condition is well known, and typically, six phases can be observed (21). There is a lag phase, followed by a first growth phase where cells use the glucose and produce acetate. After glucose is depleted, cells then switch to previously produced acetate (i.e., transition phase) and start a second growth phase. To achieve this, E. coli cells were grown in a 48-well plate and growth, pH, pO2, glucose, and acetate levels were followed (Fig. 1). The results showed that profiles of growth, substrate uptake, and product formation were consistent with the expected behavior of E. coli under the tested condition. Most importantly, each specific phase of culture growth was nicely captured by the multiplex measurement of pH, pO2, and growth. The high reproducibility of the technique was notable, with the relative standard deviation for all data points being below 1% for pH assay and below 5% for oxygen assay.
Fig. 1.
Online measurements of pO2, pH, and growth (OD at 600 nm) and offline measurements of glucose and acetate levels in E. coli MG1655 cultivations. Cells were grown in a minimal synthetic medium with 2.5 g/liter of glucose, 20 μM RuII(bpy)3, and 2 μM BCECF in a 48-well plate with 500 μl filling volume, at 37°C, with a 700-rpm shaking frequency and a 1-mm shaking diameter. Averages and standard deviations of 4 replicates per condition are shown.
Extracting physiological descriptors from online measurements.
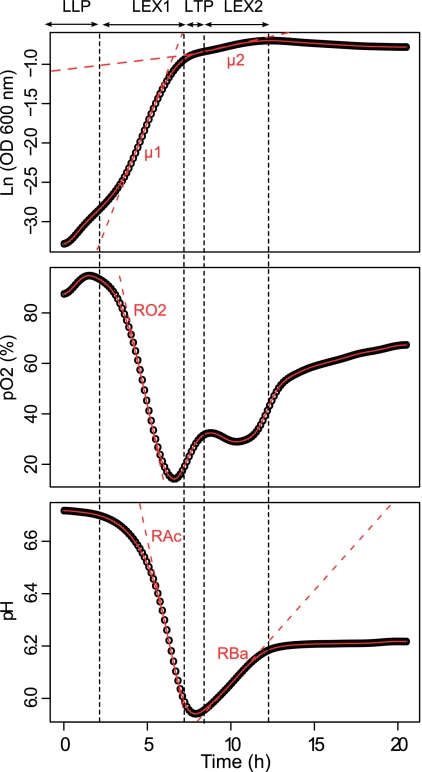
The above approach enables the user to monitor cultivations without taking samples. Depending on the format of the plate used, a large number of data can be generated and have to be processed and interpreted. To this end, we developed a numerical approach based on a cubic spline interpolation of the 3 online parameter profiles, i.e., growth, pH, and pO2. By using this approach, it was possible to automatically extract the values of the nine following physiological descriptors: μ1=0.54 h−1, μ2=0.051 h−1, RO2=−30.7% air saturation·h−1, RAc=−0.30 pH·h−1, RBa=0.071 pH·h−1, LLP=2.14 h, LEx1=5.07 h, LTP=1.17 h, and LEx2=3.87 h (Fig. 2). Briefly, parameters μ1, μ2, RO2, RAc, and RBa were numerically derived from the spline fitted curves (red solid line) and used to calculate the equations of the tangents (red dashed lines) of fitted curves. Based on these equations, online profiles were segmented into different time intervals (vertical black dashed line), which were used to extract LLP, LEx1, LTP, and LEx2. The intervals were not fixed but were strictly data dependent. Direct comparison of the numerical values obtained for RAc, RBa, and RO2 with glucose consumption rate and acetate production and consumption rates, respectively, was not possible since the units were not comparable. However, the Pearson correlations between the relevant tangents were equal to 1, indicating a strong linear relationship between online and offline variables in terms of rates. Lengths of each phase calculated using our approach are indicated in Fig. 2 and correlated nicely with the cultivation phases discussed in Fig. 1.
Fig. 2.
Examples of fitting growth, oxygen, and pH curves. The circles represent the smoothed data of optical densities, oxygen levels, and pHs measured at successive time points. The spline fits are indicated by the red solid line and were used to derive μ1 and μ2 (first and second growth rates), RAc and RBa (acidification and basification rates), and RO2 (oxygen consumption rate). The red dashed lines are tangents to the fitted curves with a slope equal to alternatively μ1, μ2, RAc, RBa, and RO2. The vertical dashed black lines indicate the starting and ending points of the four time-related parameters LLP (length of lag phase), LEx1 (length of the first growth phase), LTP (length of transition phase), and LEx2 (length of the second growth phase).
Use of physiological descriptors to monitor the response of E. coli to environmental perturbations.
To prove the applicability of the proposed workflow for investigating physiological changes of biological systems to environmental challenges, the response of E. coli grown on various carbon sources was assessed. In total, we recorded the online profiles for more than 130 samples encompassing 16 carbon sources, 8 biological replicates (i.e., 4 intraplate and 2 interplate), and relevant controls. Before processing the data, intraplate replicates were averaged and a set of 7 carbon sources was removed. This set includes d-arabinose, d-cellobiose, arbutin, d-salicin, dulcitol, d-psicose, and d-allose, i.e., all carbon sources for which no significant growth was detected after 20 h of cultivation. These results were in agreement with previously published data (16) where no growth was reported for d-arabinose, d-cellobiose, arbutin, d-salicin, and dulcitol and a weak growth was observed for d-psicose. In these experiments, growth was observed for d-allose. In our case, the first phase of growth on d-allose mostly escaped the screening window since it has been shown that growth of E. coli on d-allose induced a long lag phase of about 19 h (5). Physiological descriptors were extracted as described above. In order to standardize physiological descriptors on a unique scale across the wide range of experimental conditions, the variables were transformed to zero mean and unit variance using Z score calculation. To identify groups of carbon sources that result in similarities or differences in physiological states, the resulting collection of Z scores was used for unsupervised hierarchical cluster analysis. Standardization based on Z score calculation was applied before clustering to equalize the data sets on the same scale and to ensure that the clustering result was not dominated by one or a small number of variables.
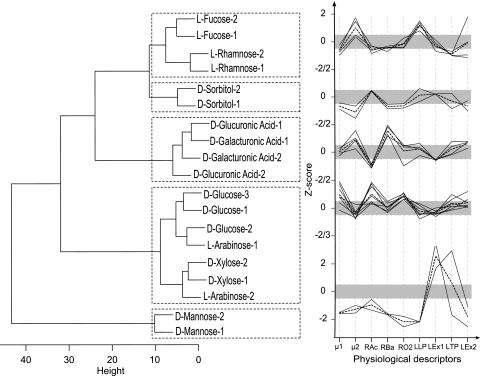
The results are presented in Fig. 3. As expected from the high reproducibility of the method, interplate replicates clustered together. In addition, carbon sources with similar physiological effects showed similar Z score patterns and thereby clustered together. The cluster analysis grouped together carbon sources that are metabolized through parallel pathways, converge to the same products, and/or present regulatory cross talk as illustrated for l-fucose with l-rhamnose (11) and for d-glucuronic acid with d-galacturonic acid (44). Surprisingly, the two PTS (phosphoenolpyruvate:carbohydrate phosphotransferase system) substrates, d-sorbitol and d-mannose, which are both metabolized into fructose-6-phosphate, exhibited very different Z score profiles. While most of the Z scores for physiological descriptors of cells growing on d-sorbitol fell within the noise envelope, growth on d-mannose produced strong changes. This is consistent with the finding of Bettenbrock and colleagues, who demonstrated that under extreme environmental conditions such as growth on d-sorbitol, the PTS regulation is most likely overridden by other global antistress regulatory networks (6). The sugars showing behaviors closest to that of d-glucose were the two non-PTS carbon sources d-xylose and l-arabinose. Both sugars are metabolized via xylulose 5-P and have a joint regulation (12). Although their metabolic fate is different from that of d-glucose, the physiological descriptors revealed glucose-like behavior. Little is known about kinetic parameters of E. coli growing on d- xylose or l-arabinose. One notable exception is the work of Hernandez-Montalvo and coworkers, who previously observed specific growth and sugar consumption rates that were similar for d-glucose and d-arabinose and lower for d-xylose (23).
Fig. 3.
Clusterogram of Z score profiles of physiological descriptors obtained for E. coli cells growing on various carbon sources. The 19 samples were grouped into 5 clusters based on their Z score profiles. The cluster tree is shown on the left, and the Z score profiles are shown on the right. The Manhattan function was used as distance metric, and Ward's linkage was used as clustering algorithm. The Z score profile of each individual condition in the cluster is depicted by black lines; the mean Z score profile is marked in dashed black for each cluster, and the gray shading indicates scores that fall within the noise envelope (−0.5 > Z score < 0.5). The plus or minus sign of Z scores indicates the direction of the effect.
Use of physiological descriptors to monitor response of E. coli to genetic perturbations.
The Keio collection, which encompasses 3,985 single-gene deletion mutants, provides a biological resource for testing mutational effects (4). Reliable, rapid, and high-volume processing systems are required to screen such a library. For this reason, we probed our method as a high-throughput functional genomics screening method to assess the consequence of gene deletion on E. coli glucose metabolism. Since our screen was focused on central metabolism, 94 single-gene knockout (KO) mutants with roles in metabolism were selected from the Keio collection. Three-quarters of these strains had disruptions of metabolic genes involved in carbohydrate, nucleotide, amino acid, lipid, and energy metabolism. The remaining quarter included genes involved in cellular processing and information storage. The latter genes encode transcription and sigma factors known to directly or indirectly control central metabolic enzymes (33). Next, the contributions of these genes to growth rates (μ1 and μ2); acidification and basification rates (RAc and RBa); oxygen consumption rate (RO2); and lengths of lag, growth, and transition phases (LLP, LEx1, LTP, and LEx2) were profiled. For all physiological descriptors, the fold change was calculated by dividing the mutant value by the wild-type value and the ratio was scaled to log2. In total, a matrix of 1,692 (94 single-gene knockouts × 9 physiological descriptors × 2 interplate replicates) biological measurements was generated.
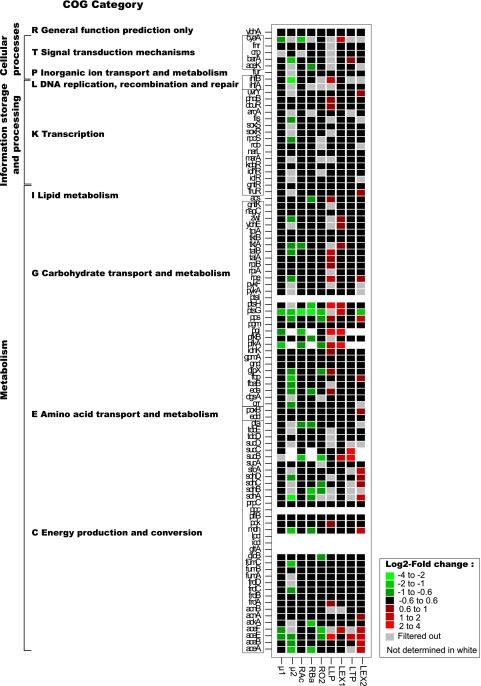
To allow rapid visual comparison of phenotypes, the effect of each gene was displayed as a function of the 9 physiological descriptors in a heat plot as presented in Fig. 4. The reported log2 fold changes correspond to the average of the two independent experiments. Of the 94 tested genes, 55 exhibited changes for at least one physiological descriptor. The greatest fraction (i.e., 41 out of 61) of genes that impacted physiological descriptors were metabolic genes. Changes were also detected in 10 out of 24 transcription and sigma factors known to have direct or indirect target genes that are involved in central carbon metabolism. For instance, mutations in the sensor kinase gene, barA, or the response regulator gene, uvrY, which is needed for switching between glycolytic and gluconeogenic carbon sources (45), affected the rate and the length of the growth on the second substrate (μ2 and LEx2) and the length of the transition phase (LTP). In contrast, gnltR, which negatively regulates the genes involved in the catabolism and uptake of d-gluconate, did not show a difference from the wild type. Except for cyaA, none of the transcription factors impacted acidification rate (RAc) or oxygen consumption rate (RO2), which is consistent with the previously published physiological data obtained with the same mutants (22). In total, only 5 KO strains (i.e., pstI, ppc, lpd, icd, and gltA mutants) did not grow at all after 24 h, which nicely agrees with previously published data (4). Among them, lpd and pstI may simply be slow growers, as their ODs after 48 h were substantially increased (4). Four KO strains (i.e., pgi, pfkA, sucC, and sucB mutants) did not show a second growth phase (i.e., no μ2) either because of both a slow growth on glucose and a long lag phase (i.e., pgi and pfkA) or because of a long transition phase (i.e., sucC and sucB). It has been shown that deletion of pgi and pfkA genes encoding the phosphoglucose isomerase and the dominant phosphofructokinase, respectively, impaired severely the specific growth and glucose consumption rates (18). For both mutants, the second growth phase escaped our screening window since cultivations were followed for 24 h. Li and coworkers showed that the deletion of sucC, one of the components of the oxoglutarate dehydrogenase complex, produced a large amount of acetate that is not utilized (39). This could also be a valuable explanation of the behavior observed for the sucB KO mutant. Finally, only 6 KO strains (i.e., cyaA, ptsG, pgi, pfkA, aceF, and aceE mutants) had altered growth rates on glucose (i.e., μ1). The reduced growth rates of cyaA and ptsG mutants, deficient in the glucose PTS system, might be explained by a lower glucose uptake rate (20, 46). This is consistent with the low acidification rate (RAc), which indicates a low acetate secretion rate. Both of the genes aceF and aceE are components of the pyruvate dehydrogenase complex, which is one of the major routes for pyruvate dissimilation in E. coli. The altered growth on glucose observed for both mutants can be explained by the inability to generate acetyl coenzyme A (acetyl-CoA) directly from glucose, although they are able to make it from acetate. A previous study has shown that an aceE and an aceF mutant of E. coli will accumulate about 50% pyruvate by mass from glucose (51a). Finally, apart from ptsG, which showed a shorter growth phase on the second substrate, no mutant exhibited physiological performance higher than that of the wild type. Such behavior can be explained by a reduced acetate production due to the limited capacity of the glucose PTS system uptake (20). Inactivation of PTS components has been successfully applied to increase production of valuable by-products at the expense of acetate production (41).
Fig. 4.
Summary of effects detected in physiological descriptors in response to genetic perturbations. The color code indicates the log2 fold change relative to the wild-type strain. Physiological descriptors showing a standard error greater than 35% are indicated in gray. Cells that did not grow are indicated in white. Data represent the averages of 2 independent cultures per condition. Genes are sorted according to their cluster of orthologous groups (COG) classification.
DISCUSSION
Although multiwell plates are undoubtedly highly suitable for high-throughput operation, they offer limited capabilities for online measurement (e.g., pH and dissolved oxygen) due to the lack of instrumentation. Here we present an assay based on oxygen- and pH-sensitive fluorescent probes that allow parallel measurement of pH, oxygen, and growth in a single well. Multiwell plates capable of parallel operation have been previously reported, but such systems require dedicated instrumentation, have limited throughput, or are not able to measure all parameters at once (7). The use of soluble fluorescent probes has numerous advantages, which are that (i) the absorbance measurement of growth is not impaired by the dual parametric fluorescence measurement of pH and oxygen, (ii) only an ordinary fluorescence reader equipped with a shaker is required, and (iii) the measurement flexibility and throughput are increased. Here the method was tested with standard plates with 48 and 96 wells, but there are no impediments to using plates with more (up to 1,536) or fewer (down to 6) wells. The low concentrations of sensors needed and the simplicity and rapidity in setting up the system provide a realistic and cost-effective alternative to existing methods (7).
The offline measurement, i.e., the time course profiles of substrate and product concentrations, represent a direct approach to assess the different cultivation phases and associated rates. Although rich in information, offline samplings are time-consuming and delicate operations in multiwell plates due to the low cultivation volumes. In contrast, with the proposed assay, online measurements are not invasive and are readily available but do not provide direct physiological information. Our data show that this physiological information is reflected and can be captured in the online measurements when the cultivation progresses from one state (phase) to another. We also observed that this information was not readily apparent from any single variable at any particular time but rather was spread across all online variables over the different time intervals. The challenge was thus to develop a tool able to extract this information in an automatic and rational manner. In this study, we present a numerical approach based on a model-free spline fitting to extract valuable information on the different phases of culture growth solely from online profiles. We demonstrate that this innovative technique was able to derive the information in the form of distinct physiological descriptors. This represents, to our knowledge, a highly valuable level of information available from online time course profiles in plates. Notably, because we used a model-free method, knowledge of the underlying mathematical model is not required. Hence, this approach can be applied to any online profiles without a priori knowledge of the relationships between what is measured online and what is happening inside the cells.
The application of this method to infer metabolic responses of E. coli was shown to work consistently for both environmental and genetic perturbations. The environmental perturbation experiments illustrated that such a method was reproducible, provided a concise summary of the patterns of carbon source utilization, and allowed examination of relationships between substrates. The genetic perturbation experiments showed that out of 60 metabolic genes, only 3 were lethal and 5 affected strongly the growth rate on glucose. Consistent with previous work (29, 37, 42), these results highlight the robustness of E. coli metabolism despite loss of a single enzyme in central carbon metabolism. Thanks to physiological descriptors, we were able to reveal not only the phenotype of silent mutations, in terms of growth, but also the phenotype of gene deletion mutants not directly involved in central metabolism (i.e., transcription and sigma factors). Using metabolomics-based approaches, it has been shown that silent mutations of yeast genes could be revealed by intracellular metabolite analysis (47). In a previous work, the extracellular medium has been used to distinguish between different physiological states of wild-type yeast and between yeast single-gene deletion mutants (1). Although metabolomics can be applied in a multiwell plate (15), measuring metabolites is time-consuming and subject to technical difficulties, especially for intracellular metabolites. Because of its speed, ease of use, and high sample processing capacity, the method proposed here could serve as a prescreening tool in metabolomics-based approaches. Further, with the exception of the metabolic regulator ptsG, no mutant exhibited physiological performance higher than that of the wild type. This agrees with the hypothesis that the wild-type E coli metabolic network represents an optimal metabolic state under the considered experimental conditions (51). In addition to E. coli, this optimal metabolic adaptation had also been observed in the yeast Saccharomyces cerevisiae, where no mutants with improved growth were detected (4, 19). Finally, noise and robustness of the method were illustrated with genes that are not induced under the tested conditions. Examples include gnltR (induced on d-gluconate) or prpc (induced on propionate), which did not affect the physiological descriptors.
In conclusion, we have here demonstrated that the proposed approach enables us to reproducibly distinguish between different environmental growth conditions and between different E. coli single-gene deletion mutants and establish the physiological basis to explain differences. This strategy is a powerful tool to generate more insights into the bioprocesses in an HT and quantitative manner and might be used for screening applications such as media, strains, or drug optimization. This method is complementary to other metabolic profiling approaches providing information on carbon source oxidation (8) or on cellular bioenergetics (17). It is reasonable also that the physiological information, brought by the methodology introduced in this work, could be very useful to constrain, check, and improve mathematical models as developed in the area of systems biology. Overall, these results provide a solid proof-of-principle for the presented workflow and suggest that there is no obstacle to applying it to other kinds of cells or processes.
ACKNOWLEDGMENTS
We thank Serguei Sokol for his help with R codes, Simon Ladeveze for his excellent experimental assistance, and Fabien Letisse for his critical reading of the manuscript.
The equipment used in this study was financed by the Région Midi-Pyrénées, the ERDF, the SICOVAL, and the French Ministry of Education & Research, which are gratefully acknowledged. This work was supported by INRA.
Footnotes
Published ahead of print on 12 August 2011.
REFERENCES
- 1. Allen J., et al. 2003. High-throughput classification of yeast mutants for functional genomics using metabolic footprinting. Nat. Biotechnol. 21:692–696 [DOI] [PubMed] [Google Scholar]
- 2. Anderlei T., Büchs J. 2001. Device for sterile online measurement of the oxygen transfer rate in shaking flasks. Biochem. Eng. J. 7:157–162 [DOI] [PubMed] [Google Scholar]
- 3. Arain S. 2006. Microrespirometry with sensor-equipped microtiter plates. Ph.D. thesis. University of Regensburg, Regensburg, Germany [Google Scholar]
- 4. Baba T., et al. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barua D., Kim J., Reed J. L. 2010. An automated phenotype-driven approach (GeneForce) for refining metabolic and regulatory models. PLoS Comput. Biol. 6:e1000970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bettenbrock K., et al. 2007. Correlation between growth rates, EIIACrr phosphorylation, and intracellular cyclic AMP levels in Escherichia coli K-12. J. Bacteriol. 189:6891–6900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Betts J. I., Baganz F. 2006. Miniature bioreactors: current practices and future opportunities. Microb. Cell Fact. 25:5–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bochner B. R. 2009. Global phenotypic characterization of bacteria. FEMS Microbiol. Rev. 33:191–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Breeuwer P., Drocourt J., Rombouts F. M., Abee T. 1996. A novel method for continuous determination of the intracellular pH in bacteria with the internally conjugated fluorescent probe 5 (and 6-)-carboxyfluorescein succinimidyl ester. Appl. Environ. Microbiol. 62:178–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Charaniya S., Hu W. S., Karypis G. 2008. Mining bioprocess data: opportunities and challenges. Trends Biotechnol. 26:690–699 [DOI] [PubMed] [Google Scholar]
- 11. Chen Y. M., Tobin J. F., Zhu Y., Schleif R. F., Lin E. C. 1987. Cross-induction of the L-fucose system by L-rhamnose in Escherichia coli. J. Bacteriol. 169:3712–3719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Desai T. A., Rao C. V. 2010. Regulation of arabinose and xylose metabolism in Escherichia coli. Appl. Environ. Microbiol. 76:1524–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Deshpande R. R., Wittmann C., Heinzle E. 2004. Microplates with integrated oxygen sensing for medium optimization in animal cell culture. Cytotechnology 46:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Doan X. T., Srinivasan R., Bapat P. M., Wangikar P. P. 2007. Detection of phase shifts in batch fermentation via statistical analysis of the online measurements: a case study with rifamycin B fermentation. J. Biotechnol. 132:156–166 [DOI] [PubMed] [Google Scholar]
- 15. Ewald J. C., Heux S., Zamboni N. 2009. High-throughput quantitative metabolomics: workflow for cultivation, quenching, and analysis of yeast in a multiwell format. Anal. Chem. 81:3623–3629 [DOI] [PubMed] [Google Scholar]
- 16. Feist A. M., et al. 2007. A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Mol. Syst. Biol. 3:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ferrick D. A., Neilson A., Beeson C. 2008. Advances in measuring cellular bioenergetics using extracellular flux. Drug Discov. Today 13:268–274 [DOI] [PubMed] [Google Scholar]
- 18. Fischer E., Sauer U. 2003. Metabolic flux profiling of Escherichia coli mutants in central carbon metabolism using GC-MS. Eur. J. Biochem. 270:880–891 [DOI] [PubMed] [Google Scholar]
- 19. Giaever G., et al. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387–391 [DOI] [PubMed] [Google Scholar]
- 20. Gosset G. 2005. Improvement of Escherichia coli production strains by modification of the phosphoenolpyruvate:sugar phosphotransferase system. Microb. Cell Fact. 4:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Guardia Alba M. J., Garcia Calvo E. 2001. Characterization of bioreaction processes: aerobic Escherichia coli cultures. J. Biotechnol. 84:107–118 [DOI] [PubMed] [Google Scholar]
- 22. Haverkorn van Rijsewijk B. R., Nanchen A., Nallet S., Kleijn R. J., Sauer U. 2011. Large-scale (13)C-flux analysis reveals distinct transcriptional control of respiratory and fermentative metabolism in Escherichia coli. Mol. Syst. Biol. 7:477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hernandez-Montalvo V., Valle F., Bolivar F., Gosset G. 2001. Characterization of sugar mixtures utilization by an Escherichia coli mutant devoid of the phosphotransferase system. Appl. Microbiol. Biotechnol. 57:186–191 [DOI] [PubMed] [Google Scholar]
- 24. Hynes J., Floyd S., Soini A. E., O'Connor R., Papkovsky D. B. 2003. Fluorescence-based cell viability screening assays using water-soluble oxygen probes. J. Biomol. Screen. 8:264–272 [DOI] [PubMed] [Google Scholar]
- 25. Hynes J., Natoli E., Jr, Will Y. 2009. Fluorescent pH and oxygen probes of the assessment of mitochondrial toxicity in isolated mitochondria and whole cells. Curr. Protoc. Toxicol. 40:2.16.1–216.22. [DOI] [PubMed] [Google Scholar]
- 26. Hynes J., et al. 2009. In vitro analysis of cell metabolism using a long-decay pH-sensitive lanthanide probe and extracellular acidification assay. Anal. Biochem. 390:21–28 [DOI] [PubMed] [Google Scholar]
- 27. Ibanez F., Grosjean P., Etienne M. 2009. Pastecs: package for analysis of space-time ecological series. R package version 1.3-11. http://CRAN.R-project.org/package=pastecs [Google Scholar]
- 28. Isett K., George H., Herber W., Amanullah A. 2007. Twenty-four-well plate miniature bioreactor high-throughput system: assessment for microbial cultivations. Biotechnol. Bioeng. 98:1017–1028 [DOI] [PubMed] [Google Scholar]
- 29. Ishii N., et al. 2007. Multiple high-throughput analyses monitor the response of E. coli to perturbations. Science 316:593–597 [DOI] [PubMed] [Google Scholar]
- 30. John G. T., Klimant I., Wittmann C., Heinzle E. 2003. Integrated optical sensing of dissolved oxygen in microtiter plates: a novel tool for microbial cultivation. Biotechnol. Bioeng. 81:829–836 [DOI] [PubMed] [Google Scholar]
- 31. Kamimura R., Konstantinov K., Stephanopoulos G. 1996. Knowledge-based systems, artificial neural networks and pattern recognition: applications to biotechnological processes. Curr. Opin. Biotechnol. 7:231. [DOI] [PubMed] [Google Scholar]
- 32. Karim M. N., Hodge D., Simon L. 2003. Data-based modeling and analysis of bioprocesses: some real experiences. Biotechnol. Prog. 19:1591–1605 [DOI] [PubMed] [Google Scholar]
- 33. Karp P. D., et al. 2002. The EcoCyc database. Nucleic Acids Res. 30:56–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kensy F. 2010. High-throughput shaken microbioreactors. Genet. Eng. Biotechnol. News 30 (6): 1 http://www.genengnews.com/gen-articles/high-throughput-shaken-microbioreactors/3224/?kwrd=m2p-labs&page=1 [Google Scholar]
- 35. Kensy F., John G. T., Hofmann B., Buchs J. 2005. Characterisation of operation conditions and online monitoring of physiological culture parameters in shaken 24-well microtiter plates. Bioprocess Biosyst. Eng. 28:75–81 [DOI] [PubMed] [Google Scholar]
- 36. Kensy F., Zang E., Faulhammer C., Tan R. K., Buchs J. 2009. Validation of a high-throughput fermentation system based on online monitoring of biomass and fluorescence in continuously shaken microtiter plates. Microb. Cell Fact. 8:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kim J., Copley S. D. 2007. Why metabolic enzymes are essential or nonessential for growth of Escherichia coli K12 on glucose. Biochemistry 46:12501–12511 [DOI] [PubMed] [Google Scholar]
- 38. Kocincova A. S., et al. 2008. Multiplex bacterial growth monitoring in 24-well microplates using a dual optical sensor for dissolved oxygen and pH. Biotechnol. Bioeng. 100:430–438 [DOI] [PubMed] [Google Scholar]
- 39. Li M., Ho P. Y., Yao S., Shimizu K. 2006. Effect of sucA or sucC gene knockout on the metabolism in Escherichia coli based on gene expressions, enzyme activities, intracellular metabolite concentrations and metabolic fluxes by 13C-labeling experiments. Biochem. Eng. J. 30:286–296 [DOI] [PubMed] [Google Scholar]
- 40. Lima S., Bonifacio R. L., Azzellini G. C., Coichev N. 2002. Ruthenium(II) tris(bipyridyl) ion as a luminescent probe for oxygen uptake on the catalyzed oxidation of HSO(3)(-). Talanta 56:547–556 [DOI] [PubMed] [Google Scholar]
- 41. Lin H., Bennett G. N., San K. Y. 2005. Metabolic engineering of aerobic succinate production systems in Escherichia coli to improve process productivity and achieve the maximum theoretical succinate yield. Metab. Eng. 7:116–127 [DOI] [PubMed] [Google Scholar]
- 42. Nicolas C., et al. 2007. Response of the central metabolism of Escherichia coli to modified expression of the gene encoding the glucose-6-phosphate dehydrogenase. FEBS Lett. 581:3771–3776 [DOI] [PubMed] [Google Scholar]
- 43. O'Mahony F. C., Papkovsky D. B. 2006. Rapid high-throughput assessment of aerobic bacteria in complex samples by fluorescence-based oxygen respirometry. Appl. Environ. Microbiol. 72:1279–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Peekhaus N., Conway T. 1998. What's for dinner? Entner-Doudoroff metabolism in Escherichia coli. J. Bacteriol. 180:3495–3502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pernestig A. K., et al. 2003. The Escherichia coli BarA-UvrY two-component system is needed for efficient switching between glycolytic and gluconeogenic carbon sources. J. Bacteriol. 185:843–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Perrenoud A., Sauer U. 2005. Impact of global transcriptional regulation by ArcA, ArcB, Cra, Crp, Cya, Fnr, and Mlc on glucose catabolism in Escherichia coli. J. Bacteriol. 187:3171–3179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Raamsdonk L. M., et al. 2001. A functional genomics strategy that uses metabolome data to reveal the phenotype of silent mutations. Nat. Biotechnol. 19:45–50 [DOI] [PubMed] [Google Scholar]
- 48. R Development Core Team 2009. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org [Google Scholar]
- 49. Riondet C., Cachon R., Waché Y., Alcaraz G., Diviès C. 1997. Measurement of the intracellular pH in Escherichia coli with the internally conjugated fluorescent probe 5- (and 6-)carboxyfluorescein succinimidyl ester. Biotechnol. Tech. 11:735–738 [Google Scholar]
- 50. Scheidle M., Klinger J., Büchs J. 2007. Combination of on-line pH and oxygen transfer rate measurement in shake flasks by fiber optical technique and respiration activity monitoring system (RAMOS). Sensors 7:3472–3480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Segre D., Vitkup D., Church G. M. 2002. Analysis of optimality in natural and perturbed metabolic networks. Proc. Natl. Acad. Sci. U. S. A. 99:15112–15117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51a. Tomar A., Eiteman M. A., Altman E. 2003. The effect of acetate pathway mutations on the production of pyravate in Escherichia Coli. Appl. Microbiol. Biotechnol. 62:76–82 [DOI] [PubMed] [Google Scholar]
- 52. Vaidyanathan S., Macaloney G., Vaughan J., McNeil B., Harvey L. M. 1999. Monitoring of submerged bioprocesses. Crit. Rev. Biotechnol. 19:277–316 [Google Scholar]
- 53. Warnes M. R., Glassey J., Montague G. A., Kara B. 1996. On data-based modelling techniques for fermentation processes. Process Biochem. 31:147 [Google Scholar]