Abstract
Acinetobacter baumannii is an important Gram-negative opportunistic pathogen causing nosocomial infections. The emergence of multiple-drug-resistant A. baumannii isolates has increased in recent years. Directed toward phage therapy, a lytic phage of A. baumannii, designated Abp53, was isolated from a sputum sample in this study. Abp53 has an isometric head and a contractile tail with tail fibers (belonging to Myoviridae), a latent period of about 10 min, and a burst size of approximately 150 PFU per infected cell. Abp53 could completely lyse 27% of the A. baumannii isolates tested, which were all multiple drug resistant, but not other bacteria. Mg2+ enhanced the adsorption and productivity of, and host lysis by, Abp53. Twenty Abp53 virion proteins were visualized in SDS-polyacrylamide gel electrophoresis, with a 47-kDa protein being the predicted major capsid protein. Abp53 has a double-stranded DNA genome of 95 kb. Sequence analyses of a 10-kb region revealed 8 open reading frames. Five of the encoded proteins, including 3 tail components and 2 hypothetical proteins, were similar to proteins encoded by A. baumannii strain ACICU. ORF1176 (one of the tail components, 1,176 amino acids [aa]), which is also similar to tail protein gp21 of Klebsiella phage phiKO2, contained repeated domains similar to those within the ACICU_02717 protein of A. baumannii ACICU and gp21. These findings suggest a common ancestry and horizontal gene transfer during evolution. As phages can expand the host range by domain duplication in tail fiber proteins, repeated domains in ORF1176 might have a similar significance in Abp53.
INTRODUCTION
Acinetobacter baumannii, which is ubiquitous in the environment, is a nonmotile, nonfermentative, aerobic Gram-negative bacterium that has emerged as an important opportunistic pathogen (41). Though it is usually harmless to healthy individuals, this organism is dangerous to hospitalized patients and especially those who are severely ill or debilitated in intensive care units (ICUs). It causes various hospital-acquired infections, including pneumonia, bloodstream infections, wound infections, septicemia, and meningitis (5, 7, 32). In addition, the role of A. baumannii in war wound infections has been identified, with rampant infections in injured soldiers returning from Iraq and Afghanistan (16, 18). A 2008 survey by the Center for Disease Control, Taiwan, indicated that this organism ranked first among bacteria causing opportunistic infections in intensive care units in Taiwan (10). A. baumannii was susceptible to most antibiotics in the 1970s, and the application of antibiotics was routinely used to treat infections caused by this organism (52). However, the treatment of A. baumannii has recently become very difficult because of its resistance to virtually all available drugs, including aminopenicillins, narrow- and expanded-spectrum cephalosporins, chloramphenicol, aminoglycosides, fluoroquinolones, and tetracyclines (34). Multidrug-resistant A. baumannii (MDRAB) isolates have increased in Taiwan since their first report in 1998 (25), similar to the situations in other countries (24, 57, 60). Thus, the treatment of MDRAB infections poses a new challenge, and finding novel treatment agents or strategies has become an absolute necessity.
The phage therapy approach was first evaluated in the late 1910s, and numerous cases of successfully treated bacterial infections from Poland, Georgia, and the former Soviet Union have been reported since (47, 51). However, phage therapy was largely abandoned in the Western world in the 1940s due to the advent of antibiotics and the inconsistency of phage-based treatments compared to antibiotics (47). Then, when bacterial resistance to antibiotics became widespread after the 1980s, interest in phage therapy was renewed, and it has been considered as an alternative treatment against antibiotic-resistant bacteria (35, 46). Studies have shown the effectiveness of using phages as therapeutic agents to treat infections in experimental mice, e.g., those infected by A. baumannii, Pseudomonas aeruginosa, and Staphylococcus aureus (43, 44). In addition, phage products have been approved for use in treating food-poisoning bacteria such as Listeria monocytogenes and Salmonella (23).
A few studies on A. baumannii phages are available, with early reports focusing on typing host A. baumannii by using specific phages (8, 21, 27). Until very recently, only two A. baumannii phages had been characterized (30, 58), AB1 and φAB2. These phages belong to the families Siphoviridae and Podoviridae, respectively (30, 58). In addition to φAB2, Lin et al. (30) isolated 7 other podophages and 2 myophages, though they remain uncharacterized. Therefore, further efforts remain to be made to develop therapeutic phage products to prevent A. baumannii infections. In this study, directed toward phage therapy, a myophage specifically infecting A. baumannii was isolated from a sputum sample collected from a hospital in Taichung, Taiwan, and designated Abp53. This paper reports the isolation, molecular properties, and phylogenetic analysis of a large predicted tail protein from Abp53, the first characterized myophage of A. baumannii.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains used in this study are listed in Table 1.A. baumannii ATCC19606 was used as the reference strain. Twenty-five strains of MDRAB were kindly donated by K. H. Shen of Taichung Veterans hospital, among which strain Ab53 was the host for propagation of phage Abp53 and the indicator host in plaque assay. Unless otherwise stated, Luria-Bertani (LB) broth and LB agar plate were the general-purpose media (31) used to cultivate A. baumannii (37°C), Escherichia coli (37°C), Xanthomonas strains (28°C), and Stenotrophomonas maltophilia (37°C). TYG contained 10 g tryptone, 6 g yeast extract, 1 mM MgSO4, 0.5% glucose, and distilled water in 1 liter. One unit of optical density at 600 nm (OD600) represented approximately 5.5 × 108 CFU/ml of A. baumannii cells.
Table 1.
Bacterial strains used in this study and their sensitivity to phage Abp53
| Species and strain(s) | Sensitivity to Abp53a | No. of strains/total no. tested for species (%) | Source or reference(s) |
|---|---|---|---|
| A. baumannii | |||
| Group I (Ab48, Ab51, Ab53, Ab55, Ab65, Ab72, ATCC 19606) | + | 7/26 (27) | Taichung Veteran Hospital |
| Group II (Ab49, Ab52, Ab61, Ab62, Ab64, Ab66, Ab67, Ab69, Ab70) | ± | 9/26 (35) | Taichung Veteran Hospital |
| Group III (Ab46, Ab47, Ab50, Ab54, Ab59, Ab63, Ab68, Ab71, Ab73, Ab74) | − | 10/26 (37) | Taichung Veteran Hospital |
| Escherichia coli DH5α, JM109 | − | 0/2 | 48, 59 |
| Stenotrophomonas maltophilia T13, T16, T17, T21, T33, T48 | − | 0/6 | 11 |
| Klebsiella pneumoniae Kpp95 | − | 0/1 | 57a |
| Pseudomonas aeruginosa 27853 | − | 0/1 | 19 |
| Xanthomonas campestris 17, P20H | − | 0/2 | 12 |
| Xanthomonas oryzae 21 | − | 0/1 | Laboratory collection |
| Xanthomonas vesicatoria 103 | − | 0/1 | Laboratory collection |
Sensitivity to Abp53 in spot test. +, formed clear clearing zones; ±, formed turbid clearing zones; −, did not form clearing zones.
Spot test and plaque assay.
For phage screening on hospital samples and to test for phage sensitivity, bacterial cells (100 μl) from overnight cultures were mixed with 3.0 ml of molten soft agar (0.7%), which was then overlaid onto the surface of the solidified regular agar (1.5%). Ten microliters of the Abp53 suspension (ca. 1.0 × 108 PFU/ml) was spotted onto the plate, which was then incubated overnight. For titer determination, 100 μl of a phage suspension after serial dilutions with sterilized, deionized water and 100 μl of cells from an overnight culture of A. baumannii were mixed with 3 ml of molten soft agar and poured onto the surface of the solidified regular agar. Plaque numbers were counted after incubating the plates overnight.
Isolation of bacteriophage.
Patient sputum samples, catheter washings of bronchopulmonary aspirate, and drainage wastewater samples were collected from Taichung Veterans General Hospital. The samples were centrifuged (10,000 × g, 10 min, 4°C), and after filtering through a membrane filter (0.45-μm pore size), the supernatants were checked for the presence of phages by spot test. The top agar within the clearing zones was picked and soaked in 100 μl of LB broth for 30 min. After appropriate dilution, the suspensions were plated for plaque formation. Two more successive single-plaque isolations were performed to purify the phage.
Purification of phage particles.
High-titer lysates of Abp53 (400 ml, ca. 1.0 × 1010 PFU/ml) were centrifuged (10,000 × g, 20 min, 4°C). The supernatants were passed through a membrane filter (0.45-μm pore size) and then centrifuged (15,000 × g, 2 h, 4°C). The phage pellets were suspended in 1.0 ml of SM buffer (50 mM Tris-HCl [pH 7.5] containing 100 mM NaCl, 10 mM MgSO4, and 0.01% gelatin) and loaded on a block gradient of CsCl (1.2, 1.35, 1.45, 1.50, and 1.70 g/ml), followed by ultracentrifugation at 28,000 rpm for 2 h at 4°C with a TH641 rotor (Sorvall OTD Combi) (28). Banded phage particles were recovered and dialyzed against SM buffer.
Separation of virion proteins by SDS-PAGE.
After dialysis, purified phage particles (ca. 5 × 107 PFU) were boiled in loading buffer for 3 min and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (10% polyacrylamide and 0.1% SDS). Protein bands were visualized by staining the gel with Coomassie brilliant blue (Bio-Rad) (39).
Assay for phage adsorption, host lysis, and one-step growth.
Phage adsorption experiments were carried out as described by Foschino et al. (20) with some modifications. Cells of A. baumannii Ab53 grown to an OD600 of 0.6 were mixed with an Abp53 suspension at a multiplicity of infection (MOI) of 0.0005 and incubated at 30°C. Aliquots (100 μl) were taken at 1-min intervals (up to 10 min) and diluted in 0.9 ml of cold LB. Following centrifugation (10,000 × g, 1 min), the supernatants containing unadsorbed phage were diluted and the titers were determined by plaque assay.
The host lysis assay was performed by mixing phage Abp53 at different MOIs (0.01 to 10) with Ab53 cells grown in LB to an OD600 of 1.0. The reaction mixtures (30 ml with and without MgSO4) were incubated at 37°C, and aliquots were taken at 1-h intervals until 8 h postinfection to measure the OD600. To test the effect of the magnesium ion, 10 mM MgSO4 was included in the reaction mixture.
One-step growth experiments were performed as described by Pajunen et al. (33) with some modifications. Cells of A. baumannii Ab53 (OD600 = 0.6) were harvested by centrifugation and then resuspended in 30 ml of fresh LB (1 × 109 CFU/ml). Phage Abp53 was added at an MOI of 0.0005 and allowed to adsorb for 5 min at room temperature. The mixture was centrifuged (10,000 × g, 1 min), and the pellets containing infected cells were suspended in 30 ml of LB, followed by incubation at 37°C. Two sets of samples were taken at intervals of 5 min. One set was immediately 10-fold diluted and centrifuged to remove host cells and determine the phage titers in the supernatant. The other set was treated with 1% (vol/vol) chloroform at 37°C to release the intracellular phage particles before titration. The LB used in these experiments was supplemented with 10 mM MgSO4.
Phage DNA isolation, restriction enzyme digestion, and pulsed-field gel electrophoresis (PFGE).
Phage particles purified by ultracentrifugation were treated with SDS (1%) and 20 U of proteinase K (P-2308; Sigma) at 58°C for 1 h, followed by addition of an equal volume of phenol-chloroform (1:1) to remove the proteinaceous materials. The extraction was repeated twice, and the DNA was precipitated as described previously (39). Restriction enzyme digestion of the phage DNA was performed following the instructions provided by the suppliers. DNA fragments were separated in 0.7% agarose gels in 1× TAE buffer (40 mM Tris acetate [pH, 8.0] containing 2 mM EDTA).
The procedures described by Tseng et al. (53) were followed for preparation of chromosomal DNA of A. baumannii, digestion of chromosomal DNA with ApaI (New England BioLabs, Beverly, MA), and PFGE of the ApaI digests in the CHEMMAPPER system from Bio-Rad (Richmond, CA). Neisseria meningitides M413 chromosomal DNA cut with NheI was used for molecular size markers (13). The same method was used to prepare Abp53 genomic DNA from the purified Abp53 phage particles after they were embedded in agarose gel plugs. PFGE was carried out in a CHEF-DR III apparatus from Bio-Rad (Richmond, CA).
DNA sequencing and bioinformatics.
The purified phage DNA was treated in a HydroShear (GeneMachines, San Carlos, CA). Fragments of 1.0 to 3.0 kb were isolated and ligated into the EcoRV site of pBluescript II SK. Inserts were subjected to nucleotide sequencing using ABI 3700 (Life Technologies, CA). The DNA sequence analysis programs used in this study were DNA Strider (17), BLAST (3), Vector NTI, and Phylip package version 3.66. Dot matrix plots were generated by a program provided by CLC bio using a 23-amino-acid scanning window. Final multiple alignment was performed with Muscle 3.8 aided by manual adjustments. Initial phylogenetic analysis was performed using the parsimony method. Bootstrap values were obtained for a consensus based on 1,000 randomly generated trees using SEQBOOT, CONSENSE, and PROTPARS in Phylip 3.69. Visualization of phylogenetic trees was carried out with Dendroscope (www.dendroscope.org). Jalview (jalview.org) was used for alignment presentation. Searches with BLASTP from NCBI using these regions of similarity as queries confirmed the internally duplicated domains. HMMER 2.3.2 was utilized to generate position-specific scoring matrices (PSSMs) for further confirmation of these domains.
The dendrogram based on ApaI pulsotypes of A. baumannii was constructed by BioNumerics software (Applied Maths, Kortrijk, Belgium), with 1% optimization and 0.8% position tolerance, using the unweighted-pair group method using average linkages (UPGMA) algorithm and the Dice similarity coefficients.
Electron microscopy.
Phage Abp53 was examined by electron microscopy of negatively stained preparations as described previously (29) using a Hitachi/H-7500 transmission electron microscope operated at 80 kV (Electron Microscopy Laboratory, Tzu Chi University).
Nucleotide sequence accession number.
The 10,290-bp sequence from phage Abp53 determined in this study was deposited in GenBank under accession number JF317274.
RESULTS AND DISCUSSION
Clinical strains of Acinetobacter baumannii isolated in Taiwan are highly diverse.
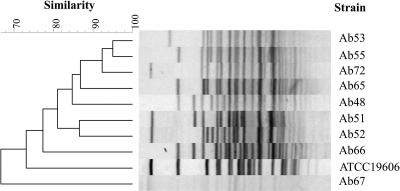
Previous reports indicate that isolates of A. baumannii are often diverse in genotype (4, 36, 55). In the host range tests for A. baumannii phage Abp53 isolated in this study, 9 Taiwan isolates from different patients and ATCC strain 19606 were found to be susceptible to Abp53 (see below). To determine how diverse these strains were, they were characterized by analyzing genomic DNA through pulsed-field gel electrophoresis (PFGE) using the restriction enzyme ApaI, as described previously (40). Figure 1 shows that each isolate exhibited a unique fingerprint pattern, and none of them had a pattern identical to that of ATCC 19606. Among these strains, which were even isolated from the same hospital, only 2 shared 95% similarity, 7 shared over 80% similarity, and 1 exhibited only 66% similarity. These results confirm that they were different strains, indicating that the Taiwan isolates are highly diverse, similar to the cases reported previously (4, 36, 55).
Fig. 1.
Dendrogram based on PFGE patterns of ApaI-digested chromosomal DNAs from 10 different A. baumannii strains. Electrophoresis was performed with 0.8% agarose at 13°C (6 V/cm; high, 35 s; low, 2.2 s; in a linear manner for 18 h). The dendrogram was constructed with BioNumerics software (Applied Maths, Kortrijk, Belgium) as described in Materials and Methods. Percent similarity is indicated on the scale.
The isolated A. baumannii phage, designated Abp53, belongs to Myoviridae type A1.
To isolate a bacteriophage specifically infecting A. baumannii, samples collected from Taichung Veterans Hospital were subjected to the spot test. One of the sputum samples was found to cause clearing zones on the lawns formed by cells of A. baumannii strain Ab53. The phage contained in the soft agar within the clearing zone was purified after three rounds of single-plaque isolation. This phage, Abp53, manifested clear plaques of 1 mm in diameter on the lawns formed by cells of strain Ab53. Ab53 cultures grown in TYG broth to the exponential phase (OD600 = 0.8) and infected with Abp53 at an MOI of 0.1 were able to produce a titer reaching 4 × 1010 PFU/ml. In contrast, the same strain grown in LB produced about 5 × 109 PFU/ml phage progeny, which was about one-eighth if that produced in TYG. Since the major differences between the two rich media lie in the supplementation of glucose and MgSO4 (10 mM) in TYG, these two components were added separately to LB to test their effects on phage yield. The results indicate that MgSO4 (10 mM) increased productivity to a level comparable to that observed in TYG, indicating that Mg+2 was responsible for the effect.
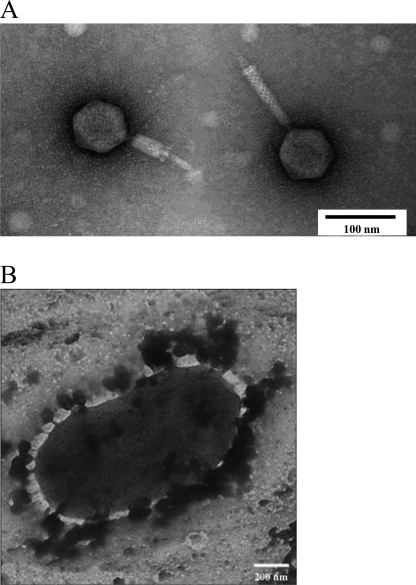
The purified phage particles were examined by electron microscopy, which revealed that Abp53 has an icosahedral head and a contractile tail (Fig. 2A) and therefore is a Myoviridae A1 type phage (http://www.mansfield.ohio-state.edu/∼sabedon/names.htm). Size estimation with 8 observed phage particles suggested that Abp53 has a head of about 59 nm in diameter and a tail of 93 to 150 nm in length and 10 to 29 nm in width. Tailed bacteriophages have a common origin and constitute an order (Caudovirales) with three families. With the very recently reported AB1 and φAB2, which belong to the families Siphoviridae and Podoviridae, respectively, each family of tailed phages now has a characterized member that specifically infects A. baumannii (30, 58).
Fig. 2.
Transmission electron microscopy of Abp53 phage particles (A) and an A. baumannii cell with Abp53 phage particles adsorbed to the surface (B).
To test for stability of Abp53 (about 4.0 × 1010 PFU/ml), titers after storage were determined for the phage lysates diluted in LB containing 10 mM MgSO4. At 4°C, 1.0 × 1010 PFU/ml of the phage was found to retain infectivity after 12 months. No significant changes in titer were found when the phage stood at room temperature for 3 months or was frozen at −85°C for 24 months.
Abp53 can rapidly adsorb to host cells and cause either cell lysis or growth inhibition.
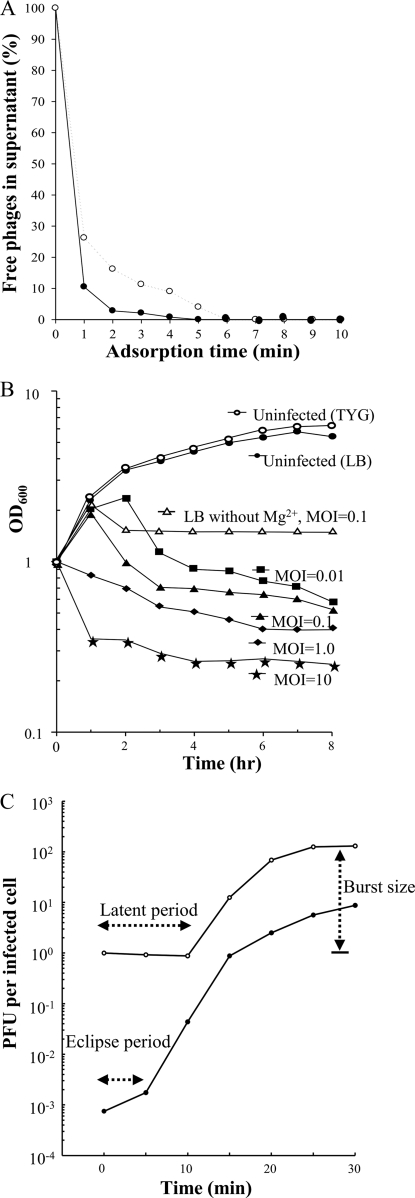
Since MgSO4 increased phage yield as described above, the adsorption of phage Abp53 to Ab53 cells was tested with and without added MgSO4 (10 mM), as described in Materials and Methods. Figure 3A shows that 90% of the phage particles adsorbed to host cells in the presence of MgSO4 at 1 min and that less than 1% of the added phage was detectable in the supernatants after 4 min. In contrast, without MgSO4, 90% of the phage particles were adsorbed at 3 min, and the adsorption rate reached 99% at 6 min (Fig. 3A). These results indicate that although Abp53 adsorption was efficient without Mg2+, this divalent cation apparently enhanced the phage adsorption. Thus, the time required for Abp53 to reach 99% adsorption is much shorter than those observed for AB1 and φAB2, which took >30 min and 6 min, respectively. Among these A. baumannii phages, AB1 requires calcium for efficient adsorption (58).
Fig. 3.
(A) Time course of phage Abp53 adsorption to the host cells of A. baumannii strain Ab53. Symbols: ○, LB broth; •, LB broth containing 10 mM MgSO4. (B) Lysis of cells of Ab53 by phage Abp53. Curves labeled with only MOI are for assays performed in LB medium with 10 mM MgSO4, while the upper three curves represent results of experiments performed for comparison. (C) One-step growth curve of Abp53 on A. baumannii strain Ab53. The medium used was LB containing 10 mM MgSO4.
To reveal the adsorption sites, Ab53 cells were infected with Abp53 at an MOI of 5.0 and the adsorption mixture was examined under a microscope. The results show that numerous phage particles attached evenly to the surfaces of Ab53 cells (Fig. 2B), similar to the case for many other bacteriophages (1, 2). This suggests that Ab53 cells have multiple receptors distributed evenly on the cell surface, which may account for their high rates of host adsorption.
The host lysis ability of Abp53 was tested by infecting Ab53 cells grown in LB broth to an OD600 of 1.0. Figure 3B shows that uninfected Ab53 cultures with and without added MgSO4 (10 mM) grew at similar rates, ca. 38 min per generation. The results of a lysis assay in the presence of MgSO4 show the following: (i) the OD600 of the cultures infected with Abp53 at an MOI of 0.01 increased to about 2.0 at 1 h, further increased at a lower rate to about 2.3 at 2 h, and then decreased sharply; (ii) at an MOI of 0.1, the OD600 increased to 1.8 at 1 h and then decreased immediately; and (iii) at higher MOIs (1.0 and 10), the OD600 decreased immediately after infection (Fig. 3B). Host lysis without supplemented MgSO4 was tested in parallel by infecting Ab53 cultures at an MOI of 0.1. Figure 3B shows that the infected cultures continued growing to an OD600 of 2.1 at 1 h and then decreased to about 1.7, showing a much lower efficiency in host lysis than those infected at the same MOI in the presence of MgSO4 (0.4 OD600 unit remained at 8 h). After initial lysis, the OD600s of these test cultures either kept decreasing gradually or remained unchanged even after prolonged incubation for 8 h (Fig. 3B). These results indicate that (i) Mg2+ is also required for the lysis of host cells by Abp53, (ii) a dose-response relationship exists in terms of host cell lysis by different MOIs of Abp53, and (iii) the nonlysed Ab53 cells cannot resume growth, causing a bactericidal effect.
Classical examples have shown that divalent ions can play a role in each step of the lytic cycle, including phage adsorption (37, 54), penetration of nucleic acid (45, 56), intracellular phage development (42, 54), and increase in phage yield (38). In addition, Ca2+ can stabilize phages and protect them from inactivation by deleterious conditions (14). In some cases, the requirement is ion specific and no effect is obtained by substituting one ion for another (14, 38). Thus, the results of phage productivity, adsorption, and host lysis suggest that the effects of Mg2+ on phage Abp53 are even broader than those observed in other phages.
Figure 3C shows the one-step growth curve of Abp53 on Ab53. The latent period was approximately 10 min, whereas the eclipse period was about 5 min. The average burst size measured was about 150 PFU per infected cell. This burst size is smaller than those of φAB2 and AB1, which were 200 and 409 PFU per infected cell, respectively.
Abp53 infects strains of A. baumannii but not other bacteria tested.
A spot test was performed to examine the host range of phage Abp53. Among the 26 A. baumannii strains tested, 7 (27%), including Ab53, supported the formation of clear clearing zones, 9 (35%) produced turbid zones, and the remaining strains (10 strains, 37%) were resistant to Abp53 (Table 1). In a plaque assay, all strains that gave clear clearing zones in the spot test supported the formation of clear plaques, but no distinct plaques were manifested by those that formed turbid or no clearing zones. Thus, the infectivity rate of Abp53 for A. baumannii isolates is low. Even lower infectivity rates were shown for the 10 A. baumannii phages (4 to 39 out of 127 strains tested), including φAB2, which were recently isolated in Taiwan (30). This might have resulted from high degrees of strain diversity, which in turn can lead to cell surface variations and phage resistance. The low infectivity rates of these individual phages suggest that the formulation of phage cocktails, as generally suggested (15, 22), is an especially important strategy for phage therapy of A. baumannii.
Bacteria other than A. baumannii were also tested for susceptibility to Abp53 by the spot test and plaque assay. The results showed that Abp53 did not infect the following bacterial species: Escherichia coli (n = 2), Pseudomonas aeruginosa 27853 (n = 1), Stenotrophomonas maltophilia (n = 6), Xanthomonas campestris (n = 2), Xanthomonas oryzae (n = 1), and Xanthomonas vesicatoria (n = 1).
The Abp53 virion consists of at least 20 proteins.
The purified phage particles were analyzed by SDS-PAGE (10% polyacrylamide) separation. More than 20 distinct protein bands, with molecular masses ranging from 20 to 94 kDa, were visualized upon staining the gel with Coomassie brilliant blue (see Fig. S1 in the supplemental material). Three bands, with molecular masses of 61, 47, and 42 kDa, were higher in abundance than the others. The 47-kDa protein, being the most abundant one, is most likely the major head protein (see Fig. S1 in the supplemental material).
Abp53 has a genomic DNA of ca. 95 kb.
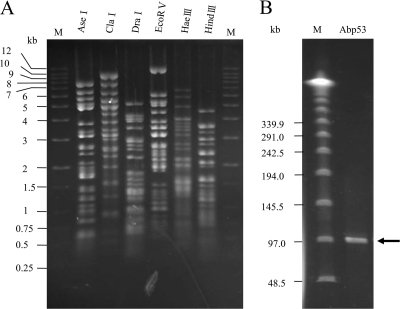
This study tested the digestibility of the Abp53 genomic DNA using 33 restriction enzymes. The results indicate that the DNA was susceptible to digestion with AseI, Ban II, ClaI, DraI, EcoRI, EcoRV, HaeIII, HincII, HindIII, NdeI, PvuII, SacII, ScaI, SpeI, SphI, XbaI, and XmnI but was resistant to ApaI, BamHI, BglII, KpnI, MluI, NaeI, NcoI, NheI, NotI, PstI, SacI, SalI, Sau3A1, SmaI, StuI, and XhoI. The enzymes AseI, ClaI, DraI, EcoRV, HaeIII, and HindIII cut the DNA into discrete fragments as revealed by agarose gel electrophoresis (Fig. 4A). The genome sizes estimated by summing the fragment lengths from the respective digests (65, 64.8, 59.6, 55.2, 58.4, and 62.6 kb from the AseI, ClaI, DraI, EcoRV, HaeIII, and HindIII digests, respectively) showed a difference of about 10 kb, which could not appropriately represent the true genome size. However, these data will be useful in the Abp53 phage genome project. For better accuracy, pulsed-field gel electrophoresis (PFGE) separation was implemented. The results show that the uncut Abp53 genomic DNA displayed a single band of about 95 kb (Fig. 4B).
Fig. 4.
(A) Agarose gel (0.7%) electrophoresis of phage Abp53 DNA digested with different restriction endonucleases (shown above the lanes). Lane M contains a 1-kb DNA ladder (from Protech Biotechnology, Taiwan) as size markers. (B) PFGE of the Abp53 genomic DNA (ca. 30 ng). Electrophoresis was performed with a 1% agarose gel at 14°C and 200 V (with an initial time of 1 s and a final time of 40 s), running for 20 h. Lane M contains a lambda DNA ladder (New England BioLabs) as molecular size markers.
Sequencing of a 10-kb region of Abp53 genome reveals several virion proteins.
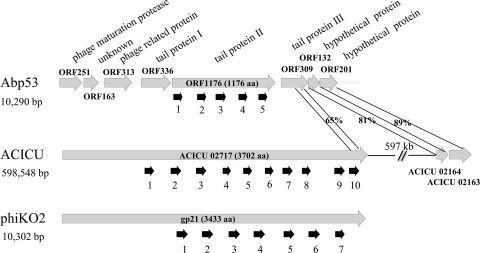
Phylogenetic trees based on virion proteins are commonly used for analysis of the relatedness of phages (49). To reveal the relatedness of Abp53 to other phages, shotgun-cloned fragments from the Abp53 genome were sequenced. One of the clones with an insert of 2.4 kb was found to encode amino acid sequences similar to those of phage tail components. Since virion protein genes are commonly clustered in phage genomes, the flanking regions might contain other virion protein genes. Therefore, PCR-based primer walking on the Abp53 genome was performed to sequence the flanking regions, with both strands being sequenced at least once on overlapping fragments, and a total of 10,290 bp was obtained. Sequence analysis indicated that this region contained 8 genes, ORF251-ORF163-ORF313-ORF336-ORF1176-ORF309-ORF132-ORF201, with the numbers designating the amino acid. Table S1 in the supplemental material lists the predicted functions of these deduced Abp53 proteins and similar proteins. ORF251 coded for a head maturation protease; the protein deduced from ORF163 had no homologue in the database; ORF313 encoded a phage-related protein; ORF336, ORF1176, and ORF309 encoded tail proteins I, II, and III, respectively; and ORF132 and ORF201 encoded hypothetical proteins (see Table S1 in the supplemental material).
The regions of ORF336 (amino acids [aa] 106 to 334), ORF1176 (the whole protein), and ORF309 (aa 162 to 308) were similar to aa 963 to 1177, 1388 to 3255, and 3549 to 3700, respectively, of ACICU_02717 from A. baumannii strain ACICU, which is a large protein of 3,702 aa assigned as a phage tail component (Fig. 5). The regions of ORF336 and ACICU_02717 shared only a low degree of identity (26%). These proteins also showed similarity to the predicted tail protein gp21 (3,433 aa in length) from Klebsiella oxytoca phage phiKO2 (Fig. 5; see Table S1 in the supplemental material). In several bacteriophage host specificity proteins of the DUF1983 superfamily, there is a functionally uncharacterized conserved domain in the C termini. This domain is conserved in aa 936 to 1017 of lambda gpJ (1,093 aa in length), a DUF1983 superfamily protein, whose C-terminal portion is responsible for phage adsorption to the host receptor, LamB (6). The C termini of ORF309 (aa 158 to 238) and ACICU_02717 (aa 3553 to 3619) also contained a similar domain, strengthening the assignment of ORF309 as a host specificity protein.
Fig. 5.
Genome organization of the 10-kb region from Abp53 (top), similar genes from A. baumannii strain ACICU, including the predicted tail protein gene ACICU02717 (middle), and predicted tail protein gene gp21 from K. oxytoca phage phiKO2 (bottom). Similar regions in the predicted proteins are linked by shading, with degrees of similarity shared being labeled. Short thick arrows indicate duplicated domains.
ORF132 and ORF201 showed high degrees of similarity (81 and 89%, respectively) to ACICU_02164 and ACICU_02163, whose genes are 597 kb apart from ACICU_02717 on the chromosome of A. baumannii strain ACICU (Fig. 5).
Abp53 ORF1176 possesses repeated domains which are homologous to those present in the predicted tail proteins of A. baumannii strain ACICU and Klebsiella phage phiKO2.
BLAST searches against the NCBI nonredundant database revealed similar regions in ORF1176, phiKO2 gp21, and ACICU_02717. Dot plot analyses identified 5, 7, and 10 similar regions (Fig. 5), respectively, for these proteins. Profile searches using HMMER confirmed that these regions are similar to one another. These repeated domains, ranging between 98 and 126 amino acid residues in length, were located at the C-terminal three-fourths of ACICU_02717 and gp21 but were dispersed in ORF1176. The N termini of ACICU_0217 and gp21 shared no similarity with ORF1176 or the genes upstream of ORF1176 (Fig. 5). The N terminus of gp21 can be viewed as a lambda gpJ-like protein, while its C terminus appears to be a large insertion made up of some repeats (9). This shows that ORF1176 lacked the region corresponding to the N terminus of the gpJ-like protein.
The similar regions detected by dot plot analyses were used to construct a multiple alignment to demonstrate sequence-level similarity among the repeats (Fig. 5; see Fig. S2 in the supplemental material). These observed similarities indicate that these domains, about 100 amino acids in length, all shared a common ancestor. The presence of multiple repetitive domains suggests the occurrence of multiple duplication events, while the presence of a nonhomologous region in the N terminus preceding the repetitive domains is indicative of a fusion event, which together resulted in the sequence composition of ACICU_02717. Multiple alignment also showed two gaps for many repeats (see Fig. S2 in the supplemental material), with one being near alignment position 15 and the other around position 100, likely due to insertion/deletion events. These repeats formed 4 distinct clusters on the phylogenetic tree (see Fig. S3 in the supplemental material). Two repeats from gp21 and one from ACICU_02717 form cluster 1, which contained the most conserved sequences according to the multiple alignment and residue conservation throughout the alignment. Cluster 2 contained 5 repeats with divergent sequences, 4 from gp21 and 1 from ORF1176. Cluster 3 contained the largest numbers of repeats, with 4 from ACICU_02717, 4 from ORF1176, and 1 from gp21. Cluster 4 included 5 repeats from ACICU_02717 exclusively. Thus, the analyses in this study establish homology between predicted tail proteins from bacterial and phage genomes, suggesting that Abp53 is a potential vehicle for horizontal gene transfer among A. baumannii and closely related species. The finding that repeats extracted from ORF1176 are clustered with bacterial domains indicates not only common ancestry but also potential exchanges during the process of evolution.
Phages can expand their host range by domain duplication in tail fiber proteins, such as in the cases of phage OP1 of Xanthomonas oryzae pv. oryzae (26) and Escherichia coli phage T4 (50). This implies that the occurrence of repeated domains in ORF1176 might have a similar significance in Abp53.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by National Science Council, Republic of China, grant NSC 96-2317-B-005-002 and by grants from Tzu Chi University (TCIRP98003-01) and Tzu Chi General Hospital (TCSP99-03-07).
We thank Chien-Shun Chiou for providing dendrogram analysis tools and Pei-Lan Lin for technical assistance.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 5 August 2011.
REFERENCES
- 1. Ackermann H. W. 2003. Bacteriophage observations and evolution. Res. Microbiol. 154:245–251 [DOI] [PubMed] [Google Scholar]
- 2. Ackermann H. W. 2001. Frequency of morphological phage descriptions in the year 2000. Arch. Virol. 146:843–857 [DOI] [PubMed] [Google Scholar]
- 3. Altschul S. F., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Anstey N. M., et al. 2002. Community-acquired bacteremic Acinetobacter pneumonia in tropical Australia is caused by diverse strains of Acinetobacter baumannii, with carriage in the throat in at-risk groups. J. Clin. Microbiol. 40:685–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Beck-Sague C. M., Jarvis W. R. 1989. Epidemic bloodstream infections associated with pressure transducers: a persistent problem. Infect. Control Hosp. Epidemiol. 10:54–59 [DOI] [PubMed] [Google Scholar]
- 6. Berkane E., et al. 2006. Interaction of bacteriophage lambda with its cell surface receptor: an in vitro study of binding of the viral tail protein gpJ to LamB (maltoporin). Biochemistry 45:2708–2720 [DOI] [PubMed] [Google Scholar]
- 7. Bou G., Cervero G., Dominguez M. A., Quereda C., Martinez-Beltran J. 2000. Characterization of a nosocomial outbreak caused by a multiresistant Acinetobacter baumannii strain with a carbapenem-hydrolyzing enzyme: high-level carbapenem resistance in A. baumannii is not due solely to the presence of beta-lactamases. J. Clin. Microbiol. 38:3299–3305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bouvet P. J., Jeanjean S., Vieu J. F., Dijkshoorn L. 1990. Species, biotype, and bacteriophage type determinations compared with cell envelope protein profiles for typing Acinetobacter strains. J. Clin. Microbiol. 28:170–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Casjens S. R., et al. 2004. The pKO2 linear plasmid prophage of Klebsiella oxytoca. J. Bacteriol. 186:1818–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Center for Disease Control 2008. Annual report of nosocomial infections surveillance system. Center for Disease Control, Taipei, Taiwan [Google Scholar]
- 11. Chang H. C., et al. 2005. Isolation and characterization of novel giant Stenotrophomonas maltophilia phage phiSMA5. Appl. Environ. Microbiol. 71:1387–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chang W. H., Lee M. C., Yang M. T., Tseng Y. H. 2005. Expression of heat-shock genes groESL in Xanthomonas campestris is upregulated by CLP in an indirect manner. FEMS Microbiol. Lett. 243:365–372 [DOI] [PubMed] [Google Scholar]
- 13. Chiou C. S., et al. 2006. Molecular epidemiology and emergence of worldwide epidemic clones of Neisseria meningitidis in Taiwan. BMC Infect. Dis. 6:25–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chow L. T., Boice L., Davidson N. 1972. Map of the partial sequence homology between DNA molecules of Bacillus subtilis bacteriophages SPO2 and phi105. J. Mol. Biol. 68:391–400 [DOI] [PubMed] [Google Scholar]
- 15. Crothers-Stomps C., Hoj L., Bourne D. G., Hall M. R., Owens L. 2010. Isolation of lytic bacteriophage against Vibrio harveyi. J. Appl. Microbiol. 108:1744–1750 [DOI] [PubMed] [Google Scholar]
- 16. Davis K. A., Moran K. A., McAllister C. K., Gray P. J. 2005. Multidrug-resistant Acinetobacter extremity infections in soldiers. Emerg. Infect. Dis. 11:1218–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Douglas S. E. 1994. DNA Strider. A Macintosh program for handling protein and nucleic acid sequences. Methods Mol. Biol. 25:181–194 [DOI] [PubMed] [Google Scholar]
- 18. Elston J. W., Bannan C. L., Chih D. T., Boutlis C. S. 2008. Acinetobacter spp. in gunshot injuries. Emerg. Infect. Dis. 14:178–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fass R. J., Barnishan J. 1979. Minimal inhibitory concentrations of 34 antimicrobial agents for control strains Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853. Antimicrob. Agents Chemother. 16:622–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Foschino R., Perrone F., Galli A. 1995. Characterization of two virulent Lactobacillus fermentum bacteriophage isolated from sour dough. J. Appl. Bacteriol. 79:677–683 [Google Scholar]
- 21. Giammanco A., Vieu J. F., Bouvet P. J., Sarzana A., Sinatra A. 1989. A comparative assay of epidemiological markers for Acinetobacter strains isolated in a hospital. Zentralbl. Bakteriol. 272:231–241 [DOI] [PubMed] [Google Scholar]
- 22. Goodridge L. D. 2010. Designing phage therapeutics. Curr. Pharm. Biotechnol. 11:15–27 [DOI] [PubMed] [Google Scholar]
- 23. Greer G. G. 2005. Bacteriophage control of foodborne bacteria. J. Food. Prot. 68:1102–1111 [DOI] [PubMed] [Google Scholar]
- 24. He C., et al. 2011. Increasing imipenem resistance and dissemination of the ISAba1 associated blaOXA-23 gene among Acinetobacter baumannii isolates in an intensive care unit. J. Med. Microbiol. 60:337–341 [DOI] [PubMed] [Google Scholar]
- 25. Hsueh P. R., et al. 2002. Pandrug-resistant Acinetobacter baumannii causing nosocomial infections in a university hospital, Taiwan. Emerg. Infect. Dis. 8:827–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Inoue Y., Matsuura T., Ohara T. 2006. Bacteriophage OP1, lytic for Xanthomonas oryzae pv. oryzae, changes its host range by duplication and deletion of the small in the deduced tail fiber gene. J. Gen. Plant Pathol. 72:111–118 [Google Scholar]
- 27. Joly-Guillou M. L., Bergogne-Berezin E., Vieu J. F. 1990. A study of the relationships between antibiotic resistance phenotypes, phage-typing and biotyping of 117 clinical isolates of Acinetobacter spp. J. Hosp. Infect. 16:49–58 [DOI] [PubMed] [Google Scholar]
- 28. Lee C. N., Lin J. W., Chow T. Y., Tseng Y. H., Weng S. F. 2006. A novel lysozyme from Xanthomonas oryzae phage phiXo411 active against Xanthomonas and Stenotrophomonas. Protein Expr. Purif. 50:229–237 [DOI] [PubMed] [Google Scholar]
- 29. Lee C. N., Lin J. W., Weng S. F., Tseng Y. H. 2009. Genomic characterization of the intron-containing T7-like phage phiL7 of Xanthomonas campestris. Appl. Environ. Microbiol. 75:7828–7837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lin N. T., Chiou P. Y., Chang K. C., Chen L. K., Lai M. J. 2010. Isolation and characterization of φAB2: a novel bacteriophage of Acinetobacter baumannii. Res. Microbiol. 161:308–314 [DOI] [PubMed] [Google Scholar]
- 31. Miller J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 32. Murray C. K., Hospenthal D. R. 2008. Acinetobacter infection in the ICU. Crit. Care Clin. 24:237–248, vii [DOI] [PubMed] [Google Scholar]
- 33. Pajunen M., Kiljunen S., Skurnik M. 2000. Bacteriophage phiYeO3-12, specific for Yersinia enterocolitica serotype O:3, is related to coliphages T3 and T7. J. Bacteriol. 182:5114–5120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Perez F., et al. 2007. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 51:3471–3484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pirisi A. 2000. Phage therapy—advantages over antibiotics? Lancet 356:1418. [DOI] [PubMed] [Google Scholar]
- 36. Qi C., Malczynski M., Parker M., Scheetz M. H. 2008. Characterization of genetic diversity of carbapenem-resistant Acinetobacter baumannii clinical strains collected from 2004 to 2007. J. Clin. Microbiol. 46:1106–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Reese J. F., Dimitracopoulos G., Bartell P. F. 1974. Factors influencing the adsorption of bacteriophage 2 to cells of Pseudomonas aeruginosa. J. Virol. 13:22–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rountree P. M. 1955. The role of divalent cations in the multiplication of Staphylococcal bacteriophages. J. Gen. Microbiol. 12:275–287 [DOI] [PubMed] [Google Scholar]
- 39. Sambrook J., Russell D. W. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 40. Seifert H., Schulze A., Baginski R., Pulverer G. 1994. Comparison of four different methods for epidemiologic typing of Acinetobacter baumannii. J. Clin. Microbiol. 32:1816–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Smith M. G., et al. 2007. New insights into Acinetobacter baumannii pathogenesis revealed by high-density pyrosequencing and transposon mutagenesis. Genes Dev. 21:601–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Snipes W., Cupp J., Sands J. A., Keith A., Davis A. 1974. Calcium requirement for assembly of the lipid-containing bacteriophage PM2. Biochim. Biophys. Acta 339:311–322 [DOI] [PubMed] [Google Scholar]
- 43. Soothill J. S. 1994. Bacteriophage prevents destruction of skin grafts by Pseudomonas aeruginosa. Burns 20:209–211 [DOI] [PubMed] [Google Scholar]
- 44. Soothill J. S. 1992. Treatment of experimental infections of mice with bacteriophages. J. Med. Microbiol. 37:258–261 [DOI] [PubMed] [Google Scholar]
- 45. Steensma H. Y., Blok J. 1979. Effect of calcium ions on the infection of Bacillus subtilis by bacteriophage SF 6. J. Gen. Virol. 42:305–314 [DOI] [PubMed] [Google Scholar]
- 46. Sulakvelidze A. 2005. Phage therapy: an attractive option for dealing with antibiotic-resistant bacterial infections. Drug Discov. Today 10:807–809 [DOI] [PubMed] [Google Scholar]
- 47. Sulakvelidze A., Alavidze Z., Morris J. G., Jr 2001. Bacteriophage therapy. Antimicrob. Agents Chemother. 45:649–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Taylor R. G., Walker D. C., McInnes R. R. 1993. E. coli host strains significantly affect the quality of small scale plasmid DNA preparations used for sequencing. Nucleic Acids Res. 21:1677–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tetart F., et al. 2001. Phylogeny of the major head and tail genes of the wide-ranging T4-type bacteriophages. J. Bacteriol. 183:358–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tetart F., Repoila F., Monod C., Krisch H. M. 1996. Bacteriophage T4 host range is expanded by duplications of a small domain of the tail fiber adhesin. J. Mol. Biol. 258:726–731 [DOI] [PubMed] [Google Scholar]
- 51. Topley W. W. C., Wilson G. S. 1929. The principles of bacteriology and immunity, p. 224-233 Willian Wood & Company, New York, NY [Google Scholar]
- 52. Towner K. J. 2009. Acinetobacter: an old friend, but a new enemy. J. Hosp. Infect. 73:355–363 [DOI] [PubMed] [Google Scholar]
- 53. Tseng Y. H., et al. 1999. Chromosome map of Xanthomonas campestris pv. campestris 17 with locations of genes involved in xanthan gum synthesis and yellow pigmentation. J. Bacteriol. 181:117–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tucker R. G. 1961. The role of magnesium ions in the growth of Salmonella phage anti-R. J. Gen. Microbiol. 26:313–323 [DOI] [PubMed] [Google Scholar]
- 55. Villari P., Iacuzio L., Vozzella E. A., Bosco U. 1999. Unusual genetic heterogeneity of Acinetobacter baumannii isolates in a university hospital in Italy. Am. J. Infect. Control 27:247–253 [DOI] [PubMed] [Google Scholar]
- 56. Watanabe K., Takesue S. 1972. The requirement for calcium in infection with Lactobacillus phage. J. Gen. Virol. 17:19–30 [DOI] [PubMed] [Google Scholar]
- 57. Wisplinghoff H., et al. 2000. Nosocomial bloodstream infections caused by Acinetobacter species in United States hospitals: clinical features, molecular epidemiology, and antimicrobial susceptibility. Clin. Infect. Dis. 31:690–697 [DOI] [PubMed] [Google Scholar]
- 57a. Wu L. T., Chang S. Y., Yen M. R., Yang T. C., Tseng Y. H. 2007. Characterization of extended-host-range pseudo-T-even bacteriophage Kpp95 isolated on Klebsiella pneumoniae. Appl. Environ. Microbiol. 73:2532–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yang H., Liang L., Lin S., Jia S. 2010. Isolation and characterization of a virulent bacteriophage AB1 of Acinetobacter baumannii. BMC Microbiol. 10:131–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yanisch-Perron C., Vieira J., Messing J. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103–119 [DOI] [PubMed] [Google Scholar]
- 60. Yuji K., Oiso G., Matsumura T., Murashige N., Kami M. Police investigation into multidrug-resistant Acinetobacter baumannii outbreak in Japan. Clin. Infect. Dis. 52:422. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.